Abstract
INTRODUCTION:
To faithfully distribute genetic material to daughter cells during cell division, spindle fibers must couple to DNA by means of a structure called the kinetochore, which assembles at each chromosome’s centromere. Human centromeres are located within large arrays of tandemly repeated DNA sequences known as alpha satellite (αSat), which often span millions of base pairs on each chromosome. Arrays of αSat are frequently surrounded by other types of tandem satellite repeats, which have poorly understood functions, along with nonrepetitive sequences, including transcribed genes. Previous genome sequencing efforts have been unable to generate complete assemblies of satellite-rich regions because of their scale and repetitive nature, limiting the ability to study their organization, variation, and function.
RATIONALE:
Pericentromeric and centromeric (peri/centromeric) satellite DNA sequences have remained almost entirely missing from the assembled human reference genome for the past 20 years. Using a complete, telomere-to-telomere (T2T) assembly of a human genome, we developed and deployed tailored computational approaches to reveal the organization and evolutionary patterns of these satellite arrays at both large and small length scales. We also performed experiments to map precisely which αSat repeats interact with kinetochore proteins. Last, we compared peri/centromeric regions among multiple individuals to understand how these sequences vary across diverse genetic backgrounds.
RESULTS:
Satellite repeats constitute 6.2% of the T2T-CHM13 genome assembly, with αSat representing the single largest component (2.8% of the genome). By studying the sequence relationships of αSat repeats in detail across each centromere, we found genome-wide evidence that human centromeres evolve through “layered expansions.” Specifically, distinct repetitive variants arise within each centromeric region and expand through mechanisms that resemble successive tandem duplications, whereas older flanking sequences shrink and diverge over time. We also revealed that the most recently expanded repeats within each αSat array are more likely to interact with the inner kinetochore protein Centromere Protein A (CENP-A), which coincides with regions of reduced CpG methylation. This suggests a strong relationship between local satellite repeat expansion, kinetochore positioning, and DNA hypomethylation. Furthermore, we uncovered large and unexpected structural rearrangements that affect multiple satellite repeat types, including active centromeric αSat arrays. Last, by comparing sequence information from nearly 1600 individuals’ Xchromosomes, we observed that individuals with recent African ancestry possess the greatest genetic diversity in the region surrounding the centromere, which sometimes contains a predominantly African αSat sequence variant.
CONCLUSION:
The genetic and epigenetic properties of centromeres are closely interwoven through evolution. These findings raise important questions about the specific molecular mechanisms responsible for the relationship between inner kinetochore proteins, DNA hypomethylation, and layered αSat expansions. Even more questions remain about the function and evolution of non-αSat repeats. To begin answering these questions, we have produced a comprehensive encyclopedia of peri/centromeric sequences in a human genome, and we demonstrated how these regions can be studied with modern genomic tools. Our work also illuminates the rich genetic variation hidden within these formerly missing regions of the genome, which may contribute to health and disease. This unexplored variation underlines the need for more T2T human genome assemblies from genetically diverse individuals.
Existing human genome assemblies have almost entirely excluded repetitive sequences within and near centromeres, limiting our understanding of their organization, evolution, and functions, which include facilitating proper chromosome segregation. Now, a complete, telomere-to-telomere human genome assembly (T2T-CHM13) has enabled us to comprehensively characterize pericentromeric and centromeric repeats, which constitute 6.2% of the genome (189.9 megabases). Detailed maps of these regions revealed multimegabase structural rearrangements, including in active centromeric repeat arrays. Analysis of centromere-associated sequences uncovered a strong relationship between the position of the centromere and the evolution of the surrounding DNA through layered repeat expansions. Furthermore, comparisons of chromosome X centromeres across a diverse panel of individuals illuminated high degrees of structural, epigenetic, and sequence variation in these complex and rapidly evolving regions.
Graphical Abstract

Gapless assemblies illuminate centromere evolution. (Top) The organization of peri/centromeric satellite repeats. (Bottom left) A schematic portraying (i) evidence for centromere evolution through layered expansions and (ii) the localization of inner-kinetochore proteins in the youngest, most recently expanded repeats, which coincide with a region of DNA hypomethylation. (Bottom right) An illustration of the global distribution of chrX centromere haplotypes, showing increased diversity in populations with recent African ancestry.
For two decades, genome sequencing and assembly efforts have excluded an estimated 5 to 10% of the human genome, most of which is found in and around each chromosome’s centromere (1, 2). These large regions contain highly repetitive DNA sequences, which impede assembly from short DNA sequencing reads (1, 3). Centromeres function to ensure proper distribution of genetic material to daughter cells during cell division, making them critical for genome stability, fertility, and healthy development (4). Nearly everything known about the sequence composition of human centromeres and their surrounding regions, called pericentromeres, stems from individual experimental observations (5–8), low-resolution classical mapping techniques (9, 10), analyses of unassembled sequencing reads (11–14), or recent studies of centromeric sequences on individual chromosomes (15–17). As a result, millions of bases in the pericentromeric and centromeric regions (hereafter peri/centromeres) remain largely uncharacterized and omitted from contemporary genetic and epigenetic studies. Recently, long-read sequencing and assembly methods enabled the Telomere-to-Telomere Consortium to produce a complete assembly of an entire human genome (T2T-CHM13) (2). This effort relied on careful measures to correctly assemble, polish, and validate entire peri/centromeric repeat arrays (2, 18). By deeply characterizing these recently assembled sequences, we present a high-resolution, genome-wide atlas of the sequence content and organization of human peri/centromeric regions.
Centromeres provide a robust assembly point for kinetochore proteins, which physically couple each chromosome to spindle fibers during cell division (4). Compromised centromere function can lead to nondisjunction, a major cause of somatic and germline disease (19). In many eukaryotes, the centromere is composed of tandemly repeated DNA sequences, called satellite DNA, but these sequences differ widely among species (20, 21). In humans, centromeres are defined by alpha satellite DNA (αSat), an AT-rich repeat family composed of ~171 base pair (bp) monomers, which can occur as different subtypes repeated in a head-to-tail orientation for millions of bases (22, 23). In the largest αSat arrays, different monomer subtypes belong to higher-order repeats (HORs); for example, monomer subtypes a, b, and c can repeat as abc-abc-abc (24, 25). HOR arrays tend to be large and highly homogeneous, often containing thousands of nearly identical HOR units. However, kinetochore proteins associate with only a subset of these HOR units, usually within the largest HOR array on each chromosome, which is called the active array (25, 26). Distinct αSat HOR arrays tend to differ in sequence and structure (27, 28), and like other satellite repeats, they evolve rapidly through mechanisms such as unequal crossover and gene conversion (29, 30). Consequently, satellite arrays frequently expand and contract in size and generate a high degree of interindividual polymorphism (29–31). Active αSat HOR arrays are flanked by inactive pericentromeric regions, which often include (i) smaller arrays of diverged αSat monomers that lack HORs (27, 32); (ii) transposable elements (TEs); (iii) segmental duplications, which sometimes include expressed genes (33, 34); and (iv) non-αSat satellite repeat families (35), which have poorly understood functions. Given the opportunity to explore these regions in a complete human genome assembly, we investigated the precise localization of inner kinetochore proteins within large αSat arrays and surveyed sequence-based trends in the structure, function, variation, and evolution of peri/centromeric DNA.
A comprehensive map of peri/centromeric satellite DNA
Human peri/centromeric satellite DNAs represent 6.2% of the T2T-CHM13v1.1 genome (~189.9 Mb) (tables S1 and S2 and figs. S1 and S2). Nearly all of this sequence remains unassembled or belongs to simulated arrays called reference models (12) in the current GRCh38/hg38 reference sequence (hereafter, hg38), including pericentromeric satellite DNA families that extend into each of the five acrocentric short arms. From decades of individual observations, a framework for the overall organization of a typical human peri/centromeric region has been proposed (Fig. 1A). By annotating and examining the repeat content of these regions in the CHM13 assembly (Fig. 1, B and C; figs. S1 and S2; table S1; and database S1), we tested and largely confirmed this broad framework genome-wide at base-pair resolution. However, we uncovered unexpected large-scale structural rearrangements and previously unresolved satellite variants (fig. S1).
Fig. 1. Overview of all peri/centromeric regions in CHM13.
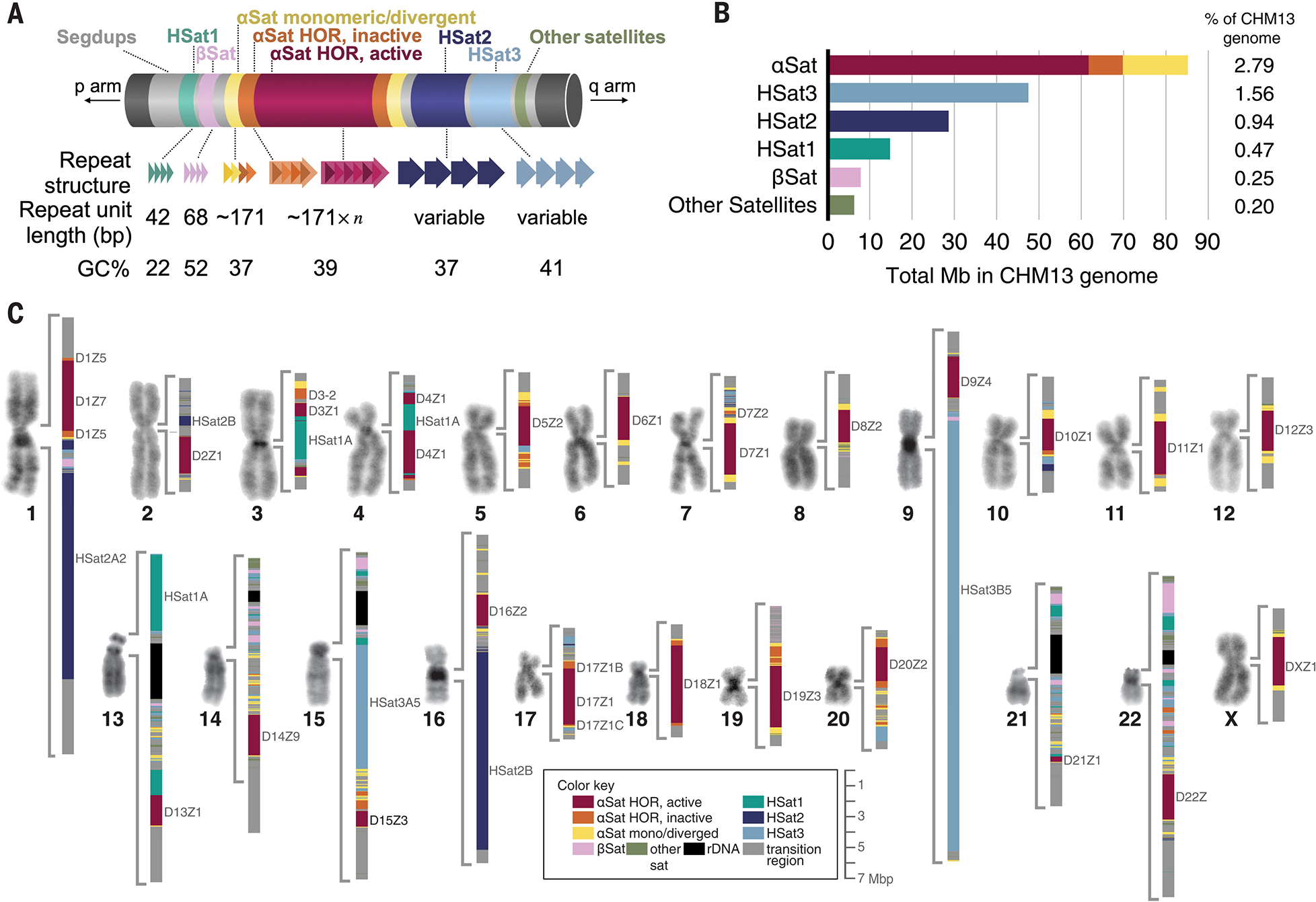
(A) Schematic of a generalized human peri/centromeric region, identifying major sequence components and their properties (not to scale). HSat2,3 repeat unit lengths vary by genomic region. (B) Barplots of the total lengths of each major satellite family genome-wide. (C) Micrographs of representative 4′,6-diamidino-2-phenylindole (DAPI)–stained chromosomes from CHM13 metaphase spreads, next to a color-coded map of peri/centromeric satellite DNA arrays [available as a browser track (database S1)]. Large satellite arrays are labeled.
All centromeric regions contain long tracts, or arrays, of tandemly repeated αSat monomers (85.2 Mb total genome-wide) (Fig. 1, B and C) (36). Most chromosomes also contain classical human satellites 2 and/or 3 (HSat2 and HSat3, totaling 28.7 and 47.6 Mb, respectively). HSat2 and HSat3 are derived from the simple repeat (CATTC)n and constitute the largest contiguous satellite arrays found in the human genome, including a 27.6-Mb array on chromosome 9 (chr9) (Fig. 1, B and C) (11, 37, 38). Furthermore, two distinct satellite DNA families constitute the most AT-rich regions of the genome (37, 39), which we refer to as HSat1A (13.4 Mb total, found mostly on chr3, chr4, and chr13) and HSat1B (found mostly on chrY, with 1.2 Mb on the acrocentrics) (table S1). Two additional large families, beta satellite (bSat; 7.7 Mb total) and gamma satellite (γSat; 630 kb total), are more GC-rich than αSat and contain dense CpG methylation (fig. S3). All remaining annotated pericentromeric satellite DNAs total 5.6 Mb, with 1.2 Mb representing previously unresolved types of satellite DNA (table S1 and fig. S2) (40). Nonsatellite bases between satellite arrays and extending into the p-arms and q-arms are considered “centric transition” regions, which largely represent long tracts of segmental duplications, including expressed genes (Fig. 1C and fig. S1) (2, 41, 42). These annotations provide a complete and detailed map of all the peri/centromeric sequences in a human genome.
Complete assessment of αSat substructure and genomic organization
To better understand the organization and evolution of αSat arrays, we generated a genome-wide database of αSat monomers (42). We grouped these monomers into distinct classes belonging to 20 suprachromosomal families (SFs) (tables S2 and S3 and database S2) (32, 43, 44) and identified 80 different HOR arrays and more than 1000 different monomers in HORs across the genome (70 Mb total) (table S4 and database S3) (38). Although 18 out of 23 chromosomes contain multiple, distinct HOR arrays (up to nine), only one HOR array per chromosome is active, meaning that it consistently associates with the kinetochore across individuals (Fig. 1, B and C, and table S4) (25). The active array on each chromosome ranges in size from 4.8 Mb on chr18 down to 340 kb on chr21, which is near the low end of the estimated αSat size range for chr21 among healthy individuals (45). Inactive HOR arrays tend to be much smaller (8 Mb total genome-wide) (Fig. 1, B and C, and table S4). Adjacent to many homogeneous HOR arrays are regions of divergent αSat HORs, in which HOR periodicity is somewhat or even completely eroded (44), as well as highly divergent αSat monomeric layers that lack HOR structure (32), totaling 15.2 Mb in CHM13.
The completeness and quality of the T2T-CHM13 assembly also allowed us to resolve HOR arrays that are highly similar between chromosomes, such as those found on chromosomes 13/21, 14/22, and 1/5/19, which have confounded studies in the past (7, 27, 36). Within these arrays, we identified chromosome-specific sequence variants and patterns of structural variants, which we validated using flow-sorted chromosome libraries for the chromosome 1/5/19 arrays (fig. S4) (42). This enabled us to infer their evolutionary history, providing evidence that the 1/5/19 HOR first originated on chr19 (42).
Large structural rearrangements in peri/centromeric regions
Producing complete maps of peri/centromeric regions revealed the large-scale organization of satellite DNAs and their embedded nonsatellite sequences, including TEs and genes (Fig. 2, A to E). Although divergent αSats contain many inversions (46) and TE insertions (47), such events within active HOR arrays are unexpected because they were considered to be homogeneous (48, 49). Quantifying strand inversions across entire satellite arrays revealed unexpected anomalies (Fig. 2, A, B, and E; fig. S1; table S5; and databases S4 and S5). For example, we uncovered a 1.7-Mb inversion inside the active αSat HOR array on chr1 (Fig. 2A), along with inversions in inactive HOR arrays on chr3, chr16, and chr20 (figs. S1 and S5). Unexpectedly, the large pericentromeric HSat3 array on chr9 and the βSat arrays on chr1 and the acrocentrics contain more than 200 inversion breakpoints (Fig. 2A and fig. S5), whereas in other arrays inversions are rare.
Fig. 2. Structural rearrangements, genes, and TEs in peri/centromeric regions.

(A) The peri/centromeric region of chr1 (cylindrical schematic at top), zooming into the transition region between the large αSat and HSat2 arrays (tracks 1 to 4). Track 1, satellite families (color key at bottom left), with vertical placement indicating the strand with canonical satellite repeat polarity. Track 2, positions of TEs overlapping αSat or HSat1,2,3, colored by TE type. Track 3, annotated transcription start sites, colored by gene type. Track 4, HSat2,3 subfamily assignments [as in (11)] and αSat SF assignments, with large arrays labeled. (B) As in (A) but for chr17, with the previously unresolved HSat3B1 array indicated with an asterisk. (C) Gene annotations between the αSat and HSat3 arrays on chr17. (D) Heatmap showing the major and minor localizations of each αSat HOR SF (top; red) and each HSat2,3 subfamily (bottom; blue). “N” indicates localizations not described in (11). Dash “–” indicates the chr1 HSat3B2 array deleted in CHM13. HSat3A3 and 3A6 are predominantly found on chrY (not in CHM13). (E) Barplots illustrate the number of inversion breakpoints (strand switches) or the number and type of TEs detected per megabase within different satellite families. div, divergent αSat (dHORs + monomeric).
Apart from inversions, two multimegabase HSat1A arrays appear to have inserted in and split the active HOR arrays on chr3 and chr4 (fig. S1 and table S6). We also found evidence for an ancient duplication event that predated African ape divergence and involved a large segment of the ancient chr6 centromere plus about 1 Mb of adjacent p-arm sequence (database S6) (42). This duplication created a different centromere locus that hosts the current active chr6 HOR array.
We also assigned HSat2 and HSat3 arrays to their respective sequence subfamilies from (11) and found previously unresolved chromosomal localizations of several HSat3 subfamilies (such as HSat3B1 on chr17) (Fig. 2, B and D). However, we also noticed a lack of HSat3B2 on chr1, contrary to expectations based on different cell lines (11), implying a large deletion of this subfamily on chr1 in CHM13.
To better understand whether these satellite inversions, insertions, and deletions are common outside of the CHM13 genome, we searched for them across 16 haplotype-resolved draft diploid assemblies of genetically diverse individuals from the Human Pangenome Reference Consortium (HPRC) (50). This revealed that the inversion in the active αSat HOR array on chr1 is polymorphic across individuals and evident in about half of ascertainable haplotypes (11 of 24) (fig. S6). However, the HSat1A insertions on chr3 and chr4 are present in all ascertainable haplotypes (32 of 32 and 33 of 33, respectively) (fig. S7). Furthermore, CHM13’s missing chr1 HSat3B2 array is contained within a 400-kb polymorphic deletion, which we detected in 29% (8 of 28) of haplotypes examined (Fig. 2A and fig. S7). Thus, these peri/centromeric structural rearrangements are not specific to the CHM13 genome but are present either variably or fixed across humans.
TE and gene interspersion in peri/centromeric regions
Like inversions and insertions, TEs are virtually absent from homogeneous HOR arrays but are enriched in divergent αSat in CHM13 (Fig. 2E and database S7) (47, 51). The CHM13 assembly also revealed regions where combinations of TE sequences have been tandemly duplicated, forming “composite satellites” [described in (40)]. We also found that other satellites–such as HSat1A/B, HSat3, and βSat–often include fragments of ancient TEs as part of their repeating units, a phenomenon rarely observed in αSat HOR arrays (Fig. 2, A, B, and E, and fig. S8) (39, 52, 53).
We also compared our pericentromeric maps with gene annotations reported for T2T-CHM13, revealing 676 gene and pseudogene annotations embedded between large satellite arrays, including 23 protein coding genes and 141 long noncoding RNAs (lncRNAs) (excluding the acrocentric short arms) (table S7 and database S8) (2). One region on chr17, located between the large HSat3 and αSat arrays (Fig. 2C), contains two protein-coding genes: KCNJ17, which encodes a disease-associated potassium channel in muscle cells (54), and UBBP4, which encodes a functional ubiquitin variant that may play a role in regulation of nuclear lamins (55). KCNJ17 is missing from GRCh38, which likely has caused inaccurate and missed variant calls in paralogous genes KCNJ12 and KCNJ18 (56). This region also contains a lncRNA annotation (LINC02002), which starts inside an SST1 element and continues into an adjacent 33-kb array of divergent αSat. Furthermore, we identified a processed paralog of an apoptosis-related protein-coding gene, BCLAF1 (BCL2 Associated Transcription Factor 1), as part of a segmental duplication embedded within an inactive αSat HOR array on chr16 (fig. S9).
The fine repeat structure of satellite DNA arrays
To further chart the structure of peri/centromeric regions at high resolution, we compared individual repeat units within and between different satellite arrays. We decomposed each αSat HOR array first into individual monomers and then into entire HORs, revealing the positions of full-size canonical HORs and structural variant HORs resulting from insertions or deletions (databases S9 and S10) (42). Whereas some chromosomes, such as chr7, are composed almost entirely of canonical HOR units, others, such as chr10, contain many structural variant HOR types, with high variation in the relative frequency of these structural variants across individuals (Fig. 3A and fig. S10).
Fig. 3. Genome-wide evidence of layered expansions in centromeric αSat arrays.

(A) (Top) HOR structural variant positions across the active αSat arrays on chr7 and chr10 (gray, canonical HORs; other colors, structural variants). (Bottom) Percentages of HOR structural variant types on HiFi sequencing reads from 16 HPRC cell lines. Variant nomenclature is described in (42); canonical HOR percentages are listed on the plot. (B) Repeat periodicities identified with NTRprism for the HSat3B1 array on chr17. (C) Comparison of the age and divergence of LINE TEs embedded in different αSat SF layers. (D) (i) Four centromeres in which an active HOR array of distinct origin appears to have expanded within a now-inactive HOR array. (ii) and (iii) Monomeric SFs (rainbow colors) surrounding active HOR arrays on eight chromosomes, with major HOR-haps shown (k = 2 to 3). Red, younger, emphasized below with red rectangles; gray, older, emphasized below with asterisks. (E) Zoomed-in view of chr3 αSat HOR arrays, divided into finer symmetrical HOR-haps (k = 7). (F) (Left) Minimum evolution tree showing the phylogenetic relationships between all HORs, colored by fine (k = 7) HOR-hap assignments. Red and gray ellipses group major HOR-hap divisions into younger and older variants, respectively (42). (Right) Phylogenetic tree built from HOR-hap consensus sequences derived from branches in the left tree, rooted with a reconstructed ancestral cen3 active HOR sequence (ANC) (42). Branch lengths indicate base substitutions per position.
Unlike αSat, some families such as HSat2 and HSat3 have inconsistent or unknown repeat unit lengths and often contain an irregular hierarchy of smaller repeating units. We refer to these repeat units as nested tandem repeats (NTRs), a more general term than HORs, which are composed of discrete numbers of monomers of similar lengths. To expand our ability to annotate repeat structure within assembled satellite DNA arrays, we developed NTRprism, an algorithm to discover and visualize satellite repeat periodicity [(42), similar to (57)]. Using this tool, we discovered HORs in HSat1 and βSat arrays, as well as NTRs in multiple HSat2,3 arrays, such as a 2235-bp repeating unit in the HSat3B1 array on chr17 (Fig. 3B and fig. S11). We also applied this tool in smaller windows across individual arrays, showing that repeat periodicity can vary across an array, which is consistent with NTRs evolving and expanding hyper-locally in some cases (fig. S11).
Genome-wide evidence of layered expansions in centromeric arrays
The T2T-CHM13 assembly also provides an opportunity to examine how peri/centromeric sequences evolve. A “layered expansion” model for centromeric αSat evolution has been hypothesized on the basis of limited observations of the most diverged αSat sequences in the human genome [reviewed in (36)]. This model postulates that distinct αSat repeats periodically emerge and expand within an active array, displacing the older repeats sideways and becoming the site of kinetochore assembly. The newer, expanding αSat sequences can originate from within the same array (32) or from a different array (intra-versus interarray seeding) (58, 59). As this process iterates over time, the displaced sequences form distinct layers that flank the active centromere with mirror symmetry (Fig. 3C), and these flanking layers rapidly shrink and decay (17, 32, 44). We used the T2T-CHM13 assembly to infer the evolutionary dynamics of αSat repeat arrays genome-wide to test the layered expansion model and understand how it may relate to centromere function. In doing so, we detected evidence of layered expansions across all αSat sequences, from the most diverged fringes of monomeric αSat to the cores of active HOR arrays.
First, we confirmed that two types of divergent αSat symmetrically flank HOR arrays across the genome: divergent HORs (dHORs) (database S11) and monomeric αSats (table S8), which represent ancient, decayed centromeres of primate ancestors (32). We classified divergent αSat into distinct SFs and dHOR families and demonstrated how these sequences accumulate mutations, inversions, TE insertions, and non-αSat satellite expansions over time (Fig. 3C; tables S5, S6, and S9; and databases S4, S5, and S7). We also found gradients of size and intra-array divergence (17 to 26%) in monomeric αSat layers, a steep (~10%) divergence increase between HORs and dHORs, and a gradient of embedded TE quantity and age that parallels the age of monomeric layers (Figs. 2E and 3C, table S9, and database S7) (17, 32, 44).
We next asked whether the layered expansion pattern extends into the active αSat HOR arrays. On four chromosomes, the active HOR array is surrounded symmetrically by inactive HORs of a distinct type, which is consistent with interarray seeding [chr1 (60), chr2, chr16, and chr18] (Fig. 3D). In the assembled centromeres from chrX (16, 61, 62) and chr8 (17), the central part of the active array was found to contain HOR variants slightly different from those on the flanks. To test whether this array structure is typical, we aligned individual HOR units within the same array and clustered them on the basis of their shared sequence variants (49, 63, 64) into “HOR-haplotypes” or “HOR-haps” (42). Initial broad classifications of HOR units into two to four distinct HOR-haps per array revealed symmetrical layering, which typically expands from the middle of the array and is consistent with intra-array seeding and expansion (Fig. 3D, dark red versus gray). Further classification into a larger number of HOR-haps (5 to 10) found additional evidence for symmetric patterns (Fig. 3E) (42).
By building rooted phylogenetic trees of consensus HOR-haps, we confirmed that the middle HOR-haps are the most recently evolved (Fig. 3F) (42). We also verified this using complete phylogenetic analysis of all HOR units on chr3, chr8, and chrX (shown for chr3 in Fig. 3F) (42). In addition, the intra-array divergence in central HOR-haps is often slightly lower than in the flanking arrays, indicating that the central HOR-haps have expanded more recently (Fig. 3F) (42). Together, these findings present genome-wide evidence that active αSat HOR arrays evolve rapidly through layered expansions, raising the question of how this dynamic evolutionary process relates to the positioning of the centromere.
Precise mapping of sites of kinetochore assembly
Human centromeres are defined epigenetically as the specific subregion bound by inner kinetochore proteins within each active αSat HOR array (21, 65). Centromeres contain a combination of epigenetic marks that distinguish them from the surrounding pericentromeric heterochromatin. For example, the histone variant Centromere Protein A (CENP-A) is constitutively present at centromeres (66) and is often accompanied by “centrochromatin”-associated modifications to canonical histones (67). Active αSat arrays also have generally high CpG methylation compared with that of neighboring inactive arrays (26) and contain local regions of reduced CpG methylation called centromere dip regions (CDRs) (16, 17, 26). To study HOR organization at sites of kinetochore assembly, we identified discrete regions of CENP-A enrichment within each active array using sequencing data from native chromatin immunoprecipitation (NChIP-seq) [data from (17)] and from CUT&RUN [data from this study (42)] (table S10) (68). To map these short sequencing reads within αSat arrays, we developed specialized, repeat-sensitive alignment approaches (42).
We confirmed that CENP-A binding is almost exclusively localized within αSat HOR arrays, with one active array per chromosome (tables S4 and S11) (25). We also found the strongest CENP-A enrichment near and within CDRs on all chromosomes (17, 26). We found that the complete span of each centromere position, defined as a window with high CENP-A enrichment, extends outside of the CDR and totals 190 to 570 kb on each chromosome (Fig. 4, A and B, and table S11). Each CENP-A span occupies 7 to 24% of the total length of the active HOR array in which it is embedded (table S11), which is contrary to predictions from previous work on chrX and chrY in different cell lines (69). However, we cannot exclude the possibility that lower levels of CENP-A extend beyond these windows of strong enrichment, or that the sizes of these windows vary among cells or cell types. We detected smaller regions of CENP-A enrichment outside of the primary CDR, with some overlapping a minor, secondary CDR (chr 4, chr16, and chr22) or no CDR at all (chr18) (Fig. 4B, fig. S12, and table S11). Furthermore, similar dips in CpG methylation, although infrequent, do occur outside CENP-A–associated regions, as observed in a 5S RNA composite satellite array (40) and within a 10-kb region in the active αSat HOR array on chr5 (fig. S12).
Fig. 4. Inner kinetochore associates with recently expanded αSat HORs.

(A) Active αSat HOR array on chr12 (coordinates at top). Track 1, CENP-A NChIP-seq marker-assisted mapping coverage. Track 2, reference-free region-specific marker enrichment (black indicates no markers in bin) (42). Track 3, percent of CpG sites methylated. Tracks 4 and 5, HOR-haps (k = 5 or 2 clusters, respectively). Track 6, number of HOR units (out of 10 per bin) that have at least one identical copy in the array. (Bottom) Self-alignment dotplot (exact-match word size 2000), with arrows pointing to a zone of recent duplication. (Inset) Smaller dotplot of the entire array (word size 500, allowing for detection of older duplications), with positions of two large macro-repeats indicated with blue lines. (B) As in (A) but for chr4. (Inset) Highlighting of a secondary CENP-A enrichment site and minor CDR on the other side of the interrupting HSat1A array. (C) As in (A) but for chr6, with CENP-A enrichment over an older HOR-hap region. (D) Rooted HOR-hap consensus phylogenetic trees as in Fig. 3F, with CENP-A–enriched region(s) indicated with arrows.
We also found that CENP-A is typically enriched in young, recently expanded HOR-haps (Fig. 4, A to D, and table S11). For example, in the active array on chr12, CENP-A is enriched on only one of two large macro-repeat structures, both of which contain similar young HOR-haps (Fig. 4A and fig. S13). Further investigation revealed that CENP-A and the CDR coincide with a zone of very recent HOR expansions (eight sites of nearly identical duplications within a ~365-kb region) (fig. S13) (42) that distinguish one macro-repeat region from the other (Fig. 4, A and D). On most other chromosomes, we similarly observed a predominant zone of recently expanded young HOR-haps (42), which tends to associate with CENP-A (eight more examples are shown in fig. S14 and table S11).
However, we identified a few notable exceptions to this general trend. On chr4, which has two CENP-A regions occurring on either side of a 1.7-Mb HSat1A array, we found that the larger CENP-A region spans a slightly younger HOR-hap, and the minor CENP-A region spans an older HOR-hap (Fig. 4, B and D). On chr5, chr7, and chr13, CENP-A overlaps young HOR-haps but not near the predominant zone of recent expansions on that chromosome (fig. S15 and table S11) (42). Inversely, CENP-A overlaps the zone of recent expansion on chr2, but this zone is composed of older HOR-haps (fig. S15). On chr6, we observed CENP-A enrichment within an older HOR-hap layer, more than a megabase away from the major zone of recent duplications and expansions in this centromere (Fig. 4, C and D). Last, chr21 shows enrichment across the entire active HOR array (the smallest in CHM13) (table S11). We observed that human centromeres and CDRs are typically, although not universally, positioned over young and/or recently expanded layers within active HOR arrays in CHM13, indicating that centromere function is closely related to the rapid evolution of αSat sequences.
Genetic variation across human X centromeres
Satellite DNA arrays are highly variable in size across individuals. The extremes of satellite size variation are often plainly visible under the microscope in chromosomal karyotypes (30), yet the clinical relevance of these variants remains unknown and largely unexplored. Studies have provided low-resolution sequencing-based evidence for variability in both satellite array lengths and in the frequency of certain sequence and structural variants within human populations (11–13, 29). However, satellite array variation and evolution have remained poorly understood at base-level resolution owing to a lack of complete centromere assemblies.
Therefore, we characterized and compared centromere array assemblies from chrX across seven XY individuals with diverse genetic ancestry [lymphoblastoid cell lines from (70)] (Fig. 5A, fig. S16, and table S12). We assigned repeats in the cenX active array to seven HOR-haps, revealing both localized and broad variation within each array (42). For example, we identified duplications spanning hundreds of kilobases in two assemblies relative to CHM13 (HG01109 and HG03492) (Fig. 5A and fig. S17). Four of the seven arrays contain zones of recent HOR expansion in the younger HOR-hap (CHM13, HG01109, HG02145, and HG03098). The remaining three assemblies show a trend of recent expansion within older HOR-haps closer to the p-arm (HG03492, HG01243, and HG02055). We also found evidence for a recently expanded HOR-hap type (HOR-hap 6) present in three individuals with recent African ancestry but absent in the other individuals, including CHM13 (Fig. 5A, dark red).
Fig. 5. Substantial genetic and epigenetic variation in and around the chrX centromere.
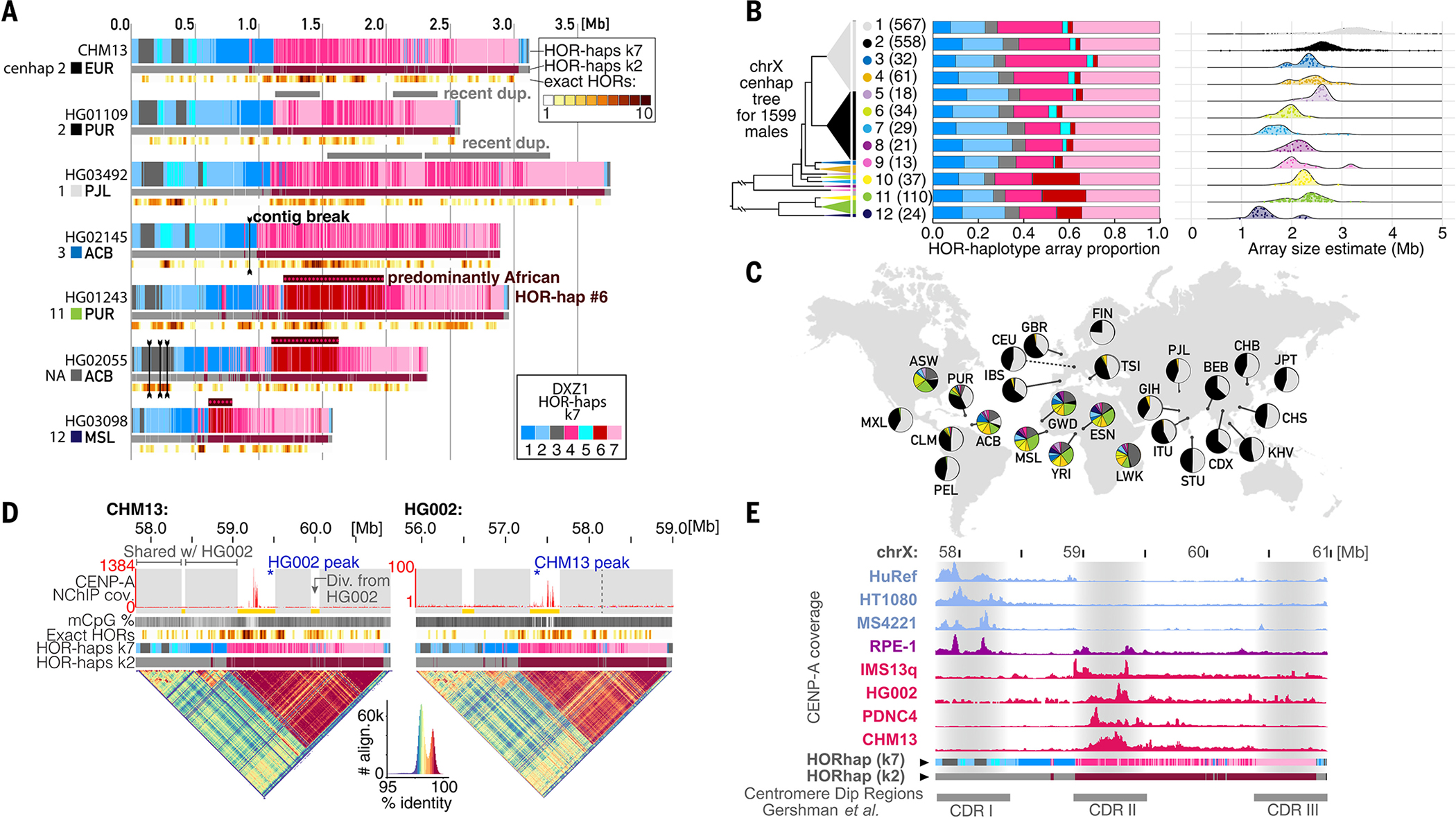
(A) Comparing the active αSat HOR array on chrX (DXZ1) between (top) CHM13 and six HPRC cell line HiFi read assemblies. Tracks indicate HOR-haps (top, k = 7; bottom, k = 2) and recent HOR duplication events (bottom, as in Fig. 4A). (B) (Left) Phylogenetic tree illustrating the relationships of 12 cenhaps defined by using short-read data from 1599 XY genomes from (70, 73) plus HG002, CHM13, and HuRef. Triangle vertical length is proportional to the number of individuals in that cenhap (98 individuals, labeled NA and colored dark gray, belong to small clades not among the 12 major cenhaps). (Middle) Barplots illustrating the average HOR-hap compositions for all individuals within each cenhap, colored as in (A). (Right) Ridgeline plots indicating the distribution of estimated total array sizes for all individuals within each cenhap, with individual values represented as jittered points. (C) Populations represented among the 1599 XY genomes, with pie charts indicating the proportion of cenhap assignments within each population, with the same colors used as in the tree in (B). Population descriptions are in (42). (D) Comparison of the DXZ1 assembly for CHM13 and HG002, which are both in cenhap 2. Tracks are as in (A), with the addition of a top track to indicate regions that align closely (gray) or are diverged (yellow) between the two individuals. Vertical dotted line indicates the homologous site of a CHM13 expansion on the HG002 array. (Bottom) StainedGlass dotplots representing the percent identity of self-alignments within the array, with a color-key and histogram below (88). (E) A comparison of CENP-A coverage (NChIP-seq or CUT&RUN) in eight cell lines relative to the CHM13 chrX centromere assembly. Each track is normalized to its maximum peak height in the array. Below are CDR positions from (26).
Next, we studied how this variation within αSats relates to variation across single-nucleotide variants (SNVs) that tend to be co-inherited with the centromere. Because meiotic crossover rates are low in peri/centromeric regions (71), centromeres are embedded in long haplotypes, called cenhaps (72). Cenhaps are identified by clustering pericentromeric SNVs into phylogenetic trees and then splitting them into large clades of shared descent. We divided a group of 1599 XY individuals genotyped using published short-read sequencing data (73) into 12 cenhaps (with 98 individuals remaining unclassified) (Fig. 5B, fig. S18, and database S12). We also used these short-read sequencing data to estimate the absolute size of each individual’s chrX active HOR array (fig. S19 and database S12) (12, 72), along with the relative proportion of that individual’s array belonging to each HOR-hap (42). This analysis revealed that distinct cenhaps have different αSat array size distributions and different average HOR-hap compositions (Fig. 5B and fig. S18). For example, HOR arrays belonging to cenhaps 1 and 2 tend to be larger overall than those belonging to cenhaps 3 to 12. We found a recent duplication in the chrX HOR array, representing hundreds of kilobases, that is common in cenhap 1 and can explain the relatively larger average array sizes in this cenhap (Fig. 5B).
Two of the 12 cenhaps (1 and 2) are very common in non-African populations (49 and 47% of individuals, respectively) and rare in African populations (1.7 and 3.5%, respectively) (Fig. 5C). The remaining 10 cenhaps are almost exclusive to African populations as well as those with recent African admixture (ASW, PUR, CLM, and ACB). The relatively low cenhap diversity in non-African populations is consistent with their lower overall genetic diversity, which is attributable to demographic bottlenecks during early human migrations out of Africa (70). This analysis also revealed that HOR-hap 6 appears to be almost exclusively found in cenhaps 10 to 12, which form an anciently diverged clade within African populations (Fig. 5B). These findings demonstrate that centromere-linked SNVs can be used to tag and track the evolution of αSat, and they underline the need for greater representation of African genomes in pan-genome assembly efforts.
Last, to dissect the sequence differences between two arrays from the same cenhap, we compared two finished centromere assemblies from CHM13 and HG002, a cell line whose chrX array had been constructed by use of T2T assembly methods and whose array structure had been experimentally validated (2). We found both genomes to be highly concordant across the array, apart from three regions, where we observed recent amplifications and/or deletions of repeats (Fig. 5D and fig. S20). These comparisons of completely assembled centromeres demonstrate that satellite DNA variation is common at both coarse and fine scales, raising the question of how this genetic variation relates to possible epigenetic variation in centromere positioning.
Epigenetic variation across human X centromeres
To examine how centromere positioning varies among individuals, we compared patterns of CENP-A CUT&RUN enrichment on the fully assembled chrX centromeres from HG002 and CHM13 (26). The region with the strongest CENP-A enrichment in both arrays coincides with the most pronounced sequence differences between CHM13 and HG002, mostly because of structural rearrangements (Fig. 5D, yellow, and fig. S20). Despite these local structural differences, CENP-A remains positioned over CDRs and young HOR-haps in both individuals.
Last, we asked whether CENP-A enrichment patterns were consistently found in younger HOR-haps, as observed in CHM13 and HG002, across seven additional cell lines with publicly available CENP-A NChIP-seq and CUT&RUN datasets (Fig. 5E and fig. S21). Unlike CHM13, in three XY individuals we observed CENP-A enrichment within the older HOR-hap subregion, proximal to the p-arm, indicating the presence of an epiallele [HuRef (74), HT1080b (75), and MS4221 (76)]. This coincides with an alternative CDR observed in the HG03098 cell line [CDR I from (26)] (Fig. 5E). Further, we examined two independent CUT&RUN experiments from the RPE-1 cell line (XX) (77) and found enrichment on both older and younger HOR-haps, which could be explained if the two chrX homologs carry different functional epialleles. Three additional XX cell lines were consistent with CHM13, providing evidence that the same CENP-A–enriched HOR-hap is shared across both chrX homologs in each line (IMS13q, PDNC4, and K562) (Fig. 5E and fig. S21) (78). These overlap a CDR also seen in the HG01109 cell line [CDR II from (26)] (Fig. 5E). A third CDR proximal to the q-arm was observed in the HG01243 and HG03492 cell lines (26), which is indicative of a third possible CENP-A epiallele. These findings uncover frequent variation in the position of the chrX centromere, with some XX individuals potentially harboring heterozygous epialleles.
Discussion
This study provides comprehensive maps of recently assembled human peri/centromeric regions to facilitate exploration of their function, variation, and evolution. Using this resource, we uncovered strong evidence that the genetic and epigenetic fates of centromeres are intertwined through evolution: αSat arrays evolve through layered expansions, and the inner-kinetochore protein CENP-A tends to associate with the most recently expanded sequences. The kinetochore frequently shifts to new loci, and the old loci rapidly shrink and decay.
One possible explanation for this relationship is that αSat expansions occur independently of the kinetochore, but the kinetochore maintains an affinity for some property of recently expanded sequences, such as their homogeneity (the “independent expansion hypothesis”). Kinetochore-independent expansion is feasible in light of our observation of large duplications and localized repeat expansions in noncentromeric satellites such as HSat3 arrays, which are not associated with kinetochores (fig. S11). Another possibility is that kinetochore proteins–or other proteins that may associate with the centromere such as loading, replication, recombination, or repair factors—play a causal role in the expansion of particular HOR variants [the “kinetochore selection hypothesis” (36)]. This aligns with the proposed recombination-based homogenization process in Arabidopsis (79). Further, experiments in model organisms have demonstrated that extreme array sequence variants increase meiotic and mitotic nondisjunction rates and can promote both mutational drive and/or female meiotic drive (20, 80–82). Similar drive mechanisms (83), along with selection for variants that promote high-fidelity chromosome transmission, may also play a role in shaping centromeric sequence evolution in humans. Exploring these evolutionary models, as well as studying why CENP-A colocalizes with CDRs, will require precise experimental methods for measuring interactions between kinetochore proteins and repetitive DNA [such as DiMeLo-seq (84)].
Fully assembled peri/centromeric regions also provide a reference against which sequencing information from multiple individuals can be aligned and compared. By doing so, we uncovered a 400-kb polymorphic deletion of an entire HSat3 array and a 1.7-Mb polymorphic inversion in an active αSat HOR array, both on chr1. We also detected an expansion of a particular αSat sequence variant on chrX in individuals with recent African ancestry. This high degree of satellite DNA polymorphism underlines the need to produce T2T assemblies from genetically diverse individuals, to fully capture the extent of human variation in these regions, and to shed light on their recent evolution. Measuring this variation will also be essential to understand the functional consequences of satellite variation on centromere function or, in the case of HSat3, on phenomena such as satellite transcription in response to stress [reviewed in (38)].
Along with genetic variation, we identified epigenetic variation in the location of CENP-A within the αSat array on chrX, similar to a rare but well-studied epiallele on chr17 (85–87). CENP-A is typically positioned on young HOR-haps on chrX, as seen for most chromosomes in CHM13. However, in some cell lines, CENP-A appears to be positioned over older chrX HOR-haps more than a megabase away (Fig. 5E), which is similar to the positioning of the chr6 CENP-A locus in CHM13. Thus, although CENP-A tends to localize to the most recently expanded HORs, there are exceptions on at least some chromosomes in some individuals. Studying centromere positioning across many samples, across families, and across different tissues from the same individuals will reveal the extent of this epigenetic plasticity in centromere localization and how this epigenetic variation relates to genetic variation and evolution. This will potentially illuminate how human cells maintain essential centromere functions despite the rapid evolution of centromeric DNA and inner-kinetochore proteins, an anomaly referred to as the “centromere paradox” (20).
Materials and methods
A very brief methods overview is provided here. Detailed methods are provided in (42). Repeats in the T2T-CHM13 assembly were annotated by parsing and combining output from RepeatMasker [provided in (40)] along with custom-built pipelines for annotating αSat and HSat2,3 (42). Regions identified as “SAR” by RepeatMasker were annotated as HSat1A, and regions annotated as “HSATI” by RepeatMasker were annotated as HSat1B. αSat HOR-haps were identified by (i) generating multiple alignments of all HOR units (or subregions of HOR units) from an array, (ii) deriving a consensus sequence, (iii) recoding the individual sequences into binary vectors based on matches to the consensus, and (iv) clustering these binary vectors by use of k-means clustering. Phylogenetic analyses of αSat sequences were performed with MEGA5. Dotplots colored by percent identity were produced with StainedGlass (88).
To analyze short-read NChIP-seq and CUT&RUN data, two parallel methods were developed: (i) marker-assisted mapping to the T2T-CHM13 reference and (ii) reference-free region-specific marker enrichment. For marker-assisted mapping, reads were aligned to the reference then filtered to include only alignments that overlap precomputed nucleotide oligomers of length k (k-mers) that occur in only one distinct position in the reference. For reference-free enrichment analysis, a set of k-mers that are enriched in CENP-A–targeted sequencing reads (relative to reads from input or immunoglobulin G controls) were first identified. Next, these enriched k-mers were compared with precomputed k-mers in the reference that occur exclusively within a single window of a given size (“region-specific markers”). Windows with multiple matches to enriched k-mers were reported as enriched for CENP-A. We performed a similar analysis using HOR-hap–specific markers on chrX, to reveal the broad enrichment of CENP-A on each HOR-hap across multiple individuals (fig. S21).
Supplementary Material
ACKNOWLEDGMENTS
This work used the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov). Certain commercial equipment, instruments, or materials are identified to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Funding:
This work was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (S.N., S.K., A.R., A.M.M., S.B., A.Y., and A.M.P.), the Intramural funding at the National Institute of Standards and Technology (J.M.Z.), HHMI Hanna H. Gray Fellowship (N.A.), Damon Runyon Postdoctoral Fellowship, and Pew Latin American Fellowship (G.V.C.). Grants supporting this work are from the US National Institutes of Health (NIH/NIGMS F32 GM134558 to G.A.L.; NIH/NHGRI R01 GM074728 to P.S. and A.F.S.; NIH R01GM123312-02 to S.J.H., G.A.H., and R.J.O.; NIH R21CA240199 to R.J.O.; NIH/NHGRI F31HG011205 to C.J.S.; NIH/NHGRI R01HG009190 to A.G. and W.T.; NIH/NHGRI R01HG010485, U41HG010972, and U01HG010961 to K.S.; NIH/NIGMS R01GM132600, P20GM103546, and NIH/NHGRI U24HG010136 to D.O. and T.J.W.; NIH/NHGRI R01HG010329 to S.R.S.; NIH/NHGRI R01HG010485, U41HG010972, U01HG010961, U24HG011853, and OT2OD026682 to B.P.; NIH R01AG054712 to E.I.R.; NIH/GM R35GM139653 and R01GM117420 to G.H.K.; NIH R01GM124041, R01GM129263, and R21CA238758 to B.A.S.; NIH/NHGRI R01HG010169 and U01HG010971 to E.E.E.; NIH/OD/NIMH DP2MH119424 to M.Y.D.; NIH/NHGRI U24HG010263; NHGRI U24HG006620; NCI U01CA253481; NIDDK R24DK106766-01A1 to M.C.S.; NIH/NHGRI U41HG007234 to M.D.; NIH/NHGRI R01HG011274-01 and NIH/NHGRI R21HG010548-01 to K.H.M.), National Science Foundation (NSF 1613806 to S.J.H., G.A.H., and R.J.O.; NSF 1643825 to R.J.O.; NSF DBI-1627442, NSF IOS-1732253, and NSF IOS-1758800 to M.C.S.); Mark Foundation for Cancer Research to SA and MCS (19-033-ASP); Russian Science Foundation RSF 19-75-30039 (analysis of genomic repeats) (I.A.A.); St. Petersburg State University (grant ID PURE 73023573 to A.M., T.D., and I.A.A. and grant ID PURE 51555639 to O.K.); Supported by the Sirius University (L.U.); Ministry of Science and Higher Education of the Russian Federation [075-10-2020-116 (13.1902.21.0023)] (F.G.); Connecticut Innovations to R.J.O.; and Stowers Institute for Medical Research to J.L.G.; A.S. is a Chan Zuckerberg Biohub Investigator. E.E.E. and A.F.D. are investigators of the Howard Hughes Medical Institute.
Footnotes
Competing interests: S.K. and K.H.M. have received travel funds to speak at symposia organized by Oxford Nanopore Technologies. W.T. has two patents (8,748,091 and 8,394,584) licensed to Oxford Nanopore Technologies. S.A. is an employee of Oxford Nanopore Technologies. P.K. owns and receives income from Reservoir Genomics. E.E.E. is a scientific advisory board (SAB) member of Variant Bio. K.H.M. is a SAB member of Centaura. No other competing interests are declared from other authors.
Data and materials availability: CHM13hTERT cells were obtained for research use through a materials transfer agreement with U. Surti and the University of Pittsburgh. Genomic and CUT&RUN sequencing data are available through BioProjects PRJNA559484 (CHM13) and PRJNA752795 (HG002). Additional data described in the paper are presented, curated, or archived in the supplementary materials. The Human Pangenome Reference Consortium (HPRC) data for 16 human samples (HG002, HG003, HG004, HG005, HG006, HG007, HG01243, HG02055, HG02109, HG02723, HG03492, HG01109, HG01442, HG02080, HG02145, and HG03098) were obtained from BioProject PRJNA730822. Nonhuman primate reference sequences cited in database S6 were obtained from Genbank (accessions NW_019932813, NW_019932814, NW_019933443, NW_019932985, NW_019932799, NW_022149050, NW_022149051, SRLZ01006095, NW_022149053, NW_022149054, NW_019937219, and NC_041757). Data tracks and satellite annotations can be visualized on the UCSC Genome Browser (89, 90), linked from https://github.com/marbl/chm13. Code used for analyzing the data are available at (91, 92). Phylogenetic data are available at (93).
SUPPLEMENTARY MATERIALS
science.org/doi/10.1126/science.abl4178 Materials and Methods Supplementary Text Figs. S1 to S43 Tables S1 to S16 References (94–155) MDAR Reproducibility Checklist Databases S1 to S21
View/request a protocol for this paper from Bio-protocol.
REFERENCES AND NOTES
- 1.Eichler EE, Clark RA, She X, An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 5, 345–354 (2004). doi: 10.1038/nrg1322 [DOI] [PubMed] [Google Scholar]
- 2.Nurk S et al. , The complete sequence of a human genome. Science 376, 44 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miga KH, Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosome Res. 23, 421–426 (2015). doi: 10.1007/s10577-015-9488-2 [DOI] [PubMed] [Google Scholar]
- 4.McKinley KL, Cheeseman IM, The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 (2016). doi: 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wevrick R, Willard HF, Physical map of the centromeric region of human chromosome 7: Relationship between two distinct alpha satellite arrays. Nucleic Acids Res. 19, 2295–2301 (1991). doi: 10.1093/nar/19.9.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson MS, Slijepcevic P, Ponder BA, The organisation of repetitive sequences in the pericentromeric region of human chromosome 10. Nucleic Acids Res. 21, 5865–5874 (1993). doi: 10.1093/nar/21.25.5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trowell HE, Nagy A, Vissel B, Choo KH, Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: Identification of a narrow domain containing two key centromeric DNA elements. Hum. Mol. Genet. 2, 1639–1649 (1993). doi: 10.1093/hmg/2.10.1639 [DOI] [PubMed] [Google Scholar]
- 8.Tyler-Smith C, Structure of repeated sequences in the centromeric region of the human Y chromosome. Development 101 (suppl.), 93–100 (1987). doi: 10.1242/dev.101.Supplement.93 [DOI] [PubMed] [Google Scholar]
- 9.Tagarro I, Fernández-Peralta AM, González-Aguilera JJ, Chromosomal localization of human satellites 2 and 3 by a FISH method using oligonucleotides as probes. Hum. Genet. 93, 383–388 (1994). doi: 10.1007/BF00201662 [DOI] [PubMed] [Google Scholar]
- 10.Archidiacono N et al. , Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics 25, 477–484 (1995). doi: 10.1016/0888-7543(95)80048-Q [DOI] [PubMed] [Google Scholar]
- 11.Altemose N, Miga KH, Maggioni M, Willard HF, Genomic characterization of large heterochromatic gaps in the human genome assembly. PLOS Comput. Biol. 10, e1003628 (2014). doi: 10.1371/journal.pcbi.1003628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miga KH et al. , Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24, 697–707 (2014). doi: 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y, Myers EW, Morishita S, Rapid and ongoing evolution of repetitive sequence structures in human centromeres. Sci. Adv. 6, eabd9230 (2020). doi: 10.1126/sciadv.abd9230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkan C et al. , Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLOS Comput. Biol. 3, 1807–1818 (2007). doi: 10.1371/journal.pcbi.0030181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M et al. , Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 36, 321–323 (2018). doi: 10.1038/nbt.4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miga KH et al. , Telomere-to-telomere assembly of a complete human X chromosome. Nature 585,79–84 (2020). doi: 10.1038/s41586-020-2547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logsdon GA et al. , The structure, function and evolution of a complete human chromosome 8. Nature 593, 101–107 (2021). doi: 10.1038/s41586-021-03420-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cartney AMM et al. , Chasing perfection: validation and polishing strategies for telomere-to-telomere genome assemblies. bioRxiv 450803 [Preprint] (2021); doi: 10.1101/2021.07.02.450803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaoka SI, Hassold TJ, Hunt PA, Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012). doi: 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff S, Ahmad K, Malik HS, The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001). doi: 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- 21.Karpen GH, Allshire RC, The case for epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496 (1997). doi: 10.1016/S0168-9525(97)01298-5 [DOI] [PubMed] [Google Scholar]
- 22.Wu JC, Manuelidis L, Sequence definition and organization of a human repeated DNA. J. Mol. Biol. 142, 363–386 (1980). doi: 10.1016/0022-2836(80)90277-6 [DOI] [PubMed] [Google Scholar]
- 23.Willard HF, The genomics of long tandem arrays of satellite DNA in the human genome. Genome 31, 737–744 (1989). doi: 10.1139/g89-132 [DOI] [PubMed] [Google Scholar]
- 24.Willard HF, Waye JS, Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 3, 192–198 (1987). doi: 10.1016/0168-9525(87)90232-0 [DOI] [Google Scholar]
- 25.McNulty SM, Sullivan BA, Alpha satellite DNA biology: Finding function in the recesses of the genome. Chromosome Res. 26, 115–138 (2018). doi: 10.1007/s10577-018-9582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gershman A et al. , Epigenetic patterns in a complete human genome. Science 376, eabj5089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y, Alpha-satellite DNA of primates: Old and new families. Chromosoma 110, 253–266 (2001). doi: 10.1007/s004120100146 [DOI] [PubMed] [Google Scholar]
- 28.Willard HF, Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 37, 524–532 (1985). [PMC free article] [PubMed] [Google Scholar]
- 29.Miga KH, Centromeric satellite DNAs: Hidden sequence variation in the human population. Genes 10, 352 (2019). doi: 10.3390/genes10050352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig-Holmes AP, Shaw MW, Polymorphism of human constitutive heterochromatin. Science 174, 702–704 (1971). doi: 10.1126/science.174.4010.702 [DOI] [PubMed] [Google Scholar]
- 31.Mahtani MM, Willard HF, Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: High-frequency polymorphisms and array size estimate. Genomics 7, 607–613 (1990). doi: 10.1016/0888-7543(90)90206-A [DOI] [PubMed] [Google Scholar]
- 32.Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA, The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLOS Genet. 5, e1000641 (2009). doi: 10.1371/journal.pgen.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.She X et al. , The structure and evolution of centromeric transition regions within the human genome. Nature 430, 857–864 (2004). doi: 10.1038/nature02806 [DOI] [PubMed] [Google Scholar]
- 34.Genovese G et al. , Using population admixture to help complete maps of the human genome. Nat. Genet. 45, 406–414, 414e1–2 (2013). doi: 10.1038/ng.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC, Human centromeric DNAs. Hum. Genet. 100, 291–304 (1997). doi: 10.1007/s004390050508 [DOI] [PubMed] [Google Scholar]
- 36.Miga KH, Alexandrov IA, Variation and evolution of human centromeres: A field guide and perspective. Annu. Rev. Genet. 55, 583–602 (2021). doi: 10.1146/annurev-genet-071719-020519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosser J, Frommer M, Paul C, Vincent PC, Sequence relationships of three human satellite DNAs. J. Mol. Biol. 187, 145–155 (1986). doi: 10.1016/0022-2836(86)90224-X [DOI] [PubMed] [Google Scholar]
- 38.Altemose N, A classical revival: Human satellite DNAs enter the genomics era. Preprints (2022), doi: 10.20944/preprints202202.0009.v1 [DOI] [PubMed]
- 39.Frommer M, Prosser J, Vincent PC, Human satellite I sequences include a male specific 2.47 kb tandemly repeated unit containing one Alu family member per repeat. Nucleic Acids Res. 12, 2887–2900 (1984). doi: 10.1093/nar/12.6.2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoyt SJ et al. , From telomere to telomere: The transcriptional and epigenetic state of human repeat elements. Science 376, eabk3112 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollger MR et al. , Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Materials and methods are available as supplementary materials.
- 43.Shepelev VA et al. , Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom. Data 5, 139–146 (2015). doi: 10.1016/j.gdata.2015.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uralsky LI et al. , Classification and monomer-by-monomer annotation dataset of suprachromosomal family 1 alpha satellite higher-order repeats in hg38 human genome assembly. Data Brief 24, 103708 (2019). doi: 10.1016/j.dib.2019.103708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo AW, Liao GC, Rocchi M, Choo KH, Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res. 9, 895–908 (1999). doi: 10.1101/gr.9.10.895 [DOI] [PubMed] [Google Scholar]
- 46.Rudd MK, Willard HF, Analysis of the centromeric regions of the human genome assembly. Trends Genet. 20, 529–533 (2004). doi: 10.1016/j.tig.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 47.Kazakov AE et al. , Interspersed repeats are found predominantly in the “old” α satellite families. Genomics 82, 619–627 (2003). doi: 10.1016/S0888-7543(03)00182-4 [DOI] [PubMed] [Google Scholar]
- 48.Warburton PE, Willard HF, Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J. Mol. Biol. 216, 3–16 (1990). doi: 10.1016/S0022-2836(05)80056-7 [DOI] [PubMed] [Google Scholar]
- 49.Warburton PE, Willard HF, Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: Evidence for concerted evolution along haplotypic lineages. J. Mol. Evol. 41, 1006–1015 (1995). doi: 10.1007/BF00173182 [DOI] [PubMed] [Google Scholar]
- 50.Miga KH, Wang T, The need for a human pangenome reference sequence. Annu. Rev. Genomics Hum. Genet. 22, 81–102 (2021). doi: 10.1146/annurev-genom-120120-081921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF, Genomic and genetic definition of a functional human centromere. Science 294, 109–115 (2001). doi: 10.1126/science.1065042 [DOI] [PubMed] [Google Scholar]
- 52.Bandyopadhyay R, McQuillan C, Page SL, Choo KH, Shaffer LG, Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res. 9, 223–233 (2001). doi: 10.1023/A:1016648404388 [DOI] [PubMed] [Google Scholar]
- 53.Agresti A et al. , Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family). J. Mol. Biol. 205, 625–631 (1989). doi: 10.1016/0022-2836(89)90308-2 [DOI] [PubMed] [Google Scholar]
- 54.Ryan DP et al. , Mutations in potassium channel Kir2.6 cause susceptibility to thyrotoxic hypokalemic periodic paralysis. Cell 140, 88–98 (2010). doi: 10.1016/j.cell.2009.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubois M-L et al. , UBB pseudogene 4 encodes functional ubiquitin variants. Nat. Commun. 11, 1306 (2020). doi: 10.1038/s41467-020-15090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aganezov S et al. , A complete reference genome improves analysis of human genetic variation. Science 376, eabl3533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paar V, Basar I, Rosandić M, Gluncić M, Consensus higher order repeats and frequency of string distributions in human genome. Curr. Genomics 8, 93–111 (2007). doi: 10.2174/138920207780368169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandrov IA, Mitkevich SP, Yurov YB, The phylogeny of human chromosome specific alpha satellites. Chromosoma 96, 443–453 (1988). doi: 10.1007/BF00303039 [DOI] [PubMed] [Google Scholar]
- 59.Willard HF, Waye JS, Chromosome-specific subsets of human alpha satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 25, 207–214 (1987). doi: 10.1007/BF02100014 [DOI] [PubMed] [Google Scholar]
- 60.Finelli P et al. , Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics 38, 325–330 (1996). doi: 10.1006/geno.1996.0635 [DOI] [PubMed] [Google Scholar]
- 61.Bzikadze AV, Pevzner PA, Automated assembly of centromeres from ultra-long error-prone reads. Nat. Biotechnol. 38, 1309–1316 (2020). doi: 10.1038/s41587-020-0582-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miga KH, Centromere studies in the era of ‘telomere-to-telomere’ genomics. Exp. Cell Res. 394, 112127 (2020). doi: 10.1016/j.yexcr.2020.112127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warburton PE, Wevrick R, Mahtani MM, Willard HF, Pulsed-field and two-dimensional gel electrophoresis of long arrays of tandemly repeated DNA: Analysis of human centromeric alpha satellite. Methods Mol. Biol. 12, 299–317 (1992). doi: 10.1385/0-89603-229-9:299 [DOI] [PubMed] [Google Scholar]
- 64.Durfy SJ, Willard HF, Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: Evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics 5, 810–821 (1989). doi: 10.1016/0888-7543(89)90123-7 [DOI] [PubMed] [Google Scholar]
- 65.Blower MD, Sullivan BA, Karpen GH, Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 (2002). doi: 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Hooser AA et al. , Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542 (2001). doi: 10.1242/jcs.114.19.3529 [DOI] [PubMed] [Google Scholar]
- 67.Sullivan BA, Karpen GH, Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076–1083 (2004). doi: 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skene PJ, Henikoff S, An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). doi: 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA, Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 19, 457–470 (2011). doi: 10.1007/s10577-011-9208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.1000 Genomes Project Consortium, A global reference for human genetic variation. Nature 526, 68–74 (2015). doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nambiar M, Smith GR, Repression of harmful meiotic recombination in centromeric regions. Semin. Cell Dev. Biol. 54, 188–197 (2016). doi: 10.1016/j.semcdb.2016.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langley SA, Miga KH, Karpen GH, Langley CH, Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA. eLife 8, e42989 (2019). doi: 10.7554/eLife.42989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byrska-Bishop M et al. , High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. bioRxiv 430068 (2021); doi: 10.1101/2021.02.06.430068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henikoff JG, Thakur J, Kasinathan S, Henikoff S, A unique chromatin complex occupies young a-satellite arrays of human centromeres. Sci. Adv. 1, e1400234 (2015). doi: 10.1126/sciadv.1400234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thakur J, Henikoff S, CENPT bridges adjacent CENPA nucleosomes on young human a-satellite dimers. Genome Res. 26, 1178–1187 (2016). doi: 10.1101/gr.204784.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hasson D et al. , The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20, 687–695 (2013). doi: 10.1038/nsmb.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dumont M et al. , Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 39, e102924 (2020). doi: 10.15252/embj.2019102924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Falk SJ et al. , Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703 (2015). doi: 10.1126/science.1259308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Naish M et al. , The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374, eabi7489 (2021). doi: 10.1126/science.abi7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fishman L, Kelly JK, Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution 69, 1208–1218 (2015). doi: 10.1111/evo.12661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kursel LE, Malik HS, The cellular mechanisms and consequences of centromere drive. Curr. Opin. Cell Biol. 52, 58–65 (2018). doi: 10.1016/j.ceb.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chmátal L et al. , Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24, 2295–2300 (2014). doi: 10.1016/j.cub.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rice WR, A game of thrones at human centromeres II. A new molecular/evolutionary model. bioRxiv 731471 [Preprint] (2019); doi: 10.1101/731471 [DOI] [Google Scholar]
- 84.Altemose N et al. , DiMeLo-seq: a long-read, single-molecule method for mapping protein-DNA interactions genome-wide. bioRxiv 451383 [Preprint] (2021); doi: 10.1101/2021.07.06.451383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maloney KA et al. , Functional epialleles at an endogenous human centromere. Proc. Natl. Acad. Sci. U.S.A. 109, 13704–13709 (2012). doi: 10.1073/pnas.1203126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA, Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26, 1301–1311 (2016). doi: 10.1101/gr.206706.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayden KE et al. , Sequences associated with centromere competency in the human genome. Mol. Cell. Biol. 33, 763–772 (2013). doi: 10.1128/MCB.01198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vollger MR, Kerpedjiev P, Phillippy AM, Eichler EE, StainedGlass: Interactive visualization of massive tandem repeat structures with identity heatmaps. Bioinformatics, btac018 (2022). doi: 10.1093/bioinformatics/btac018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raney BJ et al. , Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30, 1003–1005 (2014) doi: 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kent WJ et al. , The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). doi: 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lucas J, kmiga/alphaAnnotation: HumAS-HMMER_for_AnVIL. Zenodo (2021); doi: 10.5281/zenodo.5715444 [DOI] [Google Scholar]
- 92.Altemose N, altemose/NTRprism: NTRprism-v1.0.0. Zenodo (2021); doi: 10.5281/zenodo.5715473 [DOI] [Google Scholar]
- 93.Uralsky L, Phylogenetic trees and supp. alignments for Altemose et al. 2021, T2T Consortium. FigShare (2022); doi: 10.6084/m9.figshare.19299857.v1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


