Abstract
Ubiquitin-like proteins (Ubls) share some features with ubiquitin (Ub) such as their globular 3D structure and the ability to attach covalently to other proteins. Interferon Stimulated Gene 15 (ISG15) is an abundant Ubl that similar to Ub, marks many hundreds of cellular proteins, altering their fate. In contrast to Ub, , ISG15 requires interferon (IFN) induction to conjugate efficiently to other proteins. Moreover, despite the multitude of E3 ligases for Ub-modified targets, a single E3 ligase termed HERC5 (in humans) is responsible for the bulk of ISG15 conjugation. Targets include both viral and cellular proteins spanning an array of cellular compartments and metabolic pathways. So far, no common structural or biochemical feature has been attributed to these diverse substrates, raising questions about how and why they are selected. Conjugation of ISG15 mitigates some viral and bacterial infections and is linked to a lower viral load pointing to the role of ISG15 in the cellular immune response. In an apparent attempt to evade the immune response, some viruses try to interfere with the ISG15 pathway. For example, deconjugation of ISG15 appears to be an approach taken by coronaviruses to interfere with ISG15 conjugates. Specifically, coronaviruses such as SARS-CoV, MERS-CoV, and SARS-CoV-2, encode papain-like proteases (PL1pro) that bear striking structural and catalytic similarities to the catalytic core domain of eukaryotic deubiquitinating enzymes of the Ubiquitin-Specific Protease (USP) sub-family. The cleavage specificity of these PLpro enzymes is for flexible polypeptides containing a consensus sequence (R/K)LXGG, enabling them to function on two seemingly unrelated categories of substrates: (i) the viral polyprotein 1 (PP1a, PP1ab) and (ii) Ub- or ISG15-conjugates. As a result, PLpro enzymes process the viral polyprotein 1 into an array of functional proteins for viral replication (termed non-structural proteins; NSPs), and it can remove Ub or ISG15 units from conjugates. However, by de-conjugating ISG15, the virus also creates free ISG15, which in turn may affect the immune response in two opposite pathways: free ISG15 negatively regulates IFN signaling in humans by binding non-catalytically to USP18, yet at the same time free ISG15 can be secreted from the cell and induce the IFN pathway of the neighboring cells. A deeper understanding of this protein-modification pathway and the mechanisms of the enzymes that counteract it will bring about effective clinical strategies related to viral and bacterial infections.
Abbreviations: CoV, Coronavirus; Da, Dalton; DIG, deISGylation enzymes; DUB, deubiquitinase enzyme; IP, Immunoprecipitation; IAV, Influenza A viruses; IRF3, IFN regulatory factor 3; IFN, Interferon; IFNAR, Interferon receptor; ISG15, Interferon Stimulating Gene 15; JAK, Janus Kinase; LPS, Liposaccharide; MS, Mass Spectrometry; MERS, Middle eastern respiratory syndrome; MW, Molecular weight; NP, Nucleoprotein; PLpro, Papain-Like proteases; PCLγ, Phospholipase Cgamma1; interacting 1 Pin1, Peptidyl-prolyl isomerase NIMA; PolyUb, Poly ubiquitin; RIG1, Retinoic Acid-Inducible Gene 1; SARS, Severe Acute Respiratory Syndrome; SDS PAGE, Sodium dodecyl sulfate Polyacrylamide gel; Ub, Ubiquitin; UCRP, Ubiquitin Cross Reactive Protein; Ubls, Ubiquitin Like proteins; UPS, Ubiquitin Proteasome System; USP, Ubiquitin Specific Peptidase; WT, Wild Type
Keywords: ISG15, Interferon, Ubiquitin, Ubiquitin-Like proteins, Ubiquitin-specific proteases, USP, Deubiquitinating enzymes, DUB, Ubiquitin E3 ligases, HERC5, Papain-like proteases, PLpro, Coronavirus, MERS-CoV, SARS-CoV, SARS-CoV2
1. Overview of ISG15 - a unique member of the Ubl Family
Ubiquitin (Ub) is the most conserved protein in eukaryotic organisms. It is a small, globular protein that is conjugated to a variety of other proteins, altering their fate or function. This post-translational modification functions in many cellular processes and affects virtually every aspect of cell biology. Protein degradation by proteasome or autophagy, transcriptional regulation, cell cycle, endocytosis, DNA repair, mitochondria morphology or clearance, and regulation of vesicular trafficking, among others, are all tightly regulated by ubiquitination of key factors in these pathways [1]. The broad outcome of this post-translation modification is due, in part, to the ability of ubiquitin to form different structures (mono-ubiquitin, polyubiquitin chains of different lengths or linkages, mixed chains, or branched chains), each presenting a different surface area and hence a different signal to target the modified protein to a different pathway.
Following its discovery, it became apparent that Ub is a member of a larger group of Ubiquitin-like (Ubl) proteins that in several cases retain the ability to conjugate to other proteins [2], [3]. Members of the Ubl family share a similar globular structure: a characteristic β-grasp fold of a 5-stranded beta-sheet that curves around a central alpha-helix [3]. Each Ubl modifier has a dedicated E1 activating enzyme that uses energy from ATP hydrolysis to generate a thiol-ester intermediate at the free carboxy-terminus of the modifier and transfers it to an E2 conjugating enzyme. To complete the conjugation of small-protein modifiers to their targets, a third enzyme termed an E3 ligase usually bridges the Ub(l)-bound E2 and the target, and catalyzes the transfer reaction [4]. Just like Ub, conjugation of Ubls is involved in different processes including, transcription, DNA repair, signal transduction, autophagy, immune response and inflammation, and cell-cycle control.
Interferon Stimulated Gene 15 (ISG15), stands apart from other Ubl family members by being found only in vertebrates, requiring stimulation for its expression, and is composed of tandem Ubl domains ( Fig. 1). First identified in 1979 by Peter Lengyel and colleagues [5], ISG15 is induced in an interferon (IFN)-dependent pathway in response to infection (viral, bacterial, parasite), or upon exposure to poly I: C or liposaccharide [6], [7]. ISG15 expression is also associated with certain types of cancer [8] and even with several genotoxic stresses [9]. ISG15 was first referred to as Ubiquitin Cross Reactive Protein (UCRP) because some Ub antibodies recognize ISG15 as well [10]. As mentioned, the structure of ISG15 is composed of two Ubl domains (the N-terminal and C-terminal domains, or the distal and proximal Ubls) [11], [12], [13] connected by a flexible linker of highly charged and hydrophilic residues (Asp76-Lys77-Cys78/Ser78-Asp79-Glu80) that enables different orientations to be formed in the event of interactions with different molecules [14]. The structure-function relationship of this tandem architecture is still unclear – does it simply deliver double the Ubl dose per conjugation event, or do each of the domains provide distinct binding surfaces to downstream receptors or binding partners? Supporting the notion that each domain contributes a different set of interactions, the C-terminal Ubl domain (independently from the N-terminal domain) was found to be sufficient for recognition by the E1 activating enzyme of ISG15, UBE1L (alias: UBA7), but conjugation to targets was dependent on both Ubl domains of ISG15 [15]. A dedicated protease for disassembling ISG15-conjugates, USP18, also recognizes the C-terminal Ubl domain of ISG15 leaving the distal N-terminal domain free for other potential interactions [16], [17]. By contrast, a viral protease, PLpro, interacts with both domains of ISG15 enabling PLpro from different coronaviruses to distinguish between ISG15 and monoUb or diUb modifications [18], [19], [20], [21], [22], [23].
Fig. 1.
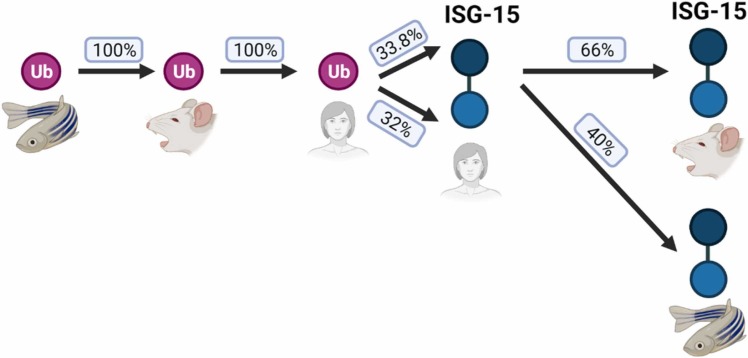
Conservation of ubiquitin and ISG-15 across vertebrates. Sequence alignment of ubiquitin from zebrafish, mice, and humans indicates perfect conservation of ubiquitin. The ISG15 protein contains two ubiquitin-like domains (UBLs). Although the 3D structure of each of the ISG15 domains resembles the 3D fold of Ub, a comparison of each domain of human ISG15 to the Ub sequence results in roughly 33 % sequence identity by BLAST [129]. In comparison to ubiquitin, ISG15 is less conserved across vertebrates, dropping to 66 % and 40 % identity between humans and mice or zebrafish, respectively.
The ISG15-dedicated E1activatingenzyme, UBE1L is the bottleneck of the conjugation pathway. Although it shares a 45 % sequence identity with the ubiquitin E1 activating enzyme in humans, UBE1 (alias: UBA1), UBE1L is specific to ISG15 and is unable to form thioester bonds with other Ubls. Congruently, deletion of UBE1L in mice eliminates ISGylation with no effect on the ubiquitin landscape [24], [25]. The second enzyme in the ISG15 conjugation cascade is the E2 conjugating enzyme, UBE2L6 (aka UbcH8 in humans). This enzyme is not unique to ISG15 as it can also conjugate Ub to targets [26], [27]. Several E3 ligases have been documented to catalyze ISG15 conjugation: HERC5, HERC6, HHARI, and EFP (aka TRIM25) [28], [29], [30]. By far the dominant E3 ligase for ISG15 in humans is HERC5 (and HERC6 in mouse), since in their absence,little, if any, conjugates are detectable [31], [32], [33], [34]. Interestingly, both EFP and HERC5 can undergo ISGylation themselves, though further research is necessary to understand the reason for ISGylation of ISGylation enzymes. In addition to ISG15 itself, the entire conjugation machinery (comprising the dedicated E1, E2, and the few known E3s) is induced by IFN type I signaling underlying the massive increase in ISG15-conjugates in response to infection [12].
As menthioned above, a single E3 ligase in human cells, HERC5, appears to be responsible for the bulk of ISG15-conjugation following IFN induction [35]. The heterogeneous “smear” of high MW ISG15-conjugates resolved by SDS-PAGE, characteristic of a wide range of ISGylated proteins, disappears almost completely in extracts of HERC5-/- U2OS cells. The fact that only one enzyme can account for more than 500 identified substrates [36], implies that the enzyme recognizes a "general feature" of the target. Several models have been raised to explain how HERC5 selects its targets. Thus, the team of Dong-Er Zhang found Ub to be one of the substrates of ISG15 conjugation implying that ISG15 and Ub can form mixed chains (through K29/K48 residues of Ub) on targets [37]. Given that Ub is an extremely prevalent post-translational modification [38], [39], recognition of Ub as a common feature of its substrates might explain ISGylation of otherwise seemingly unrelated substrates by a single E3 ligase. In this manner, ISGylation in response to IFN (or other stimuli) would change the fate of proteins by converting conjugated Ub into mixed ISG15-Ub modifications. Another explanation for the plethora of ISG15 targets could be the co-localization of the dedicated E3 ligase with its targets and the timing of the conjugation step. Thus, the Huibregtse lab failed to achieve ISGylation of previously reported substrates unless co-transfection of the relevant substrate was done simultaneously with the expression of ISGylation enzymes [34], [40]. This led the authors to consider the possibility of a "timeline" boundary for the ISGylation process. In addition, the authors found co-localization between HERC5 and polysomes. Thus, they concluded that the bulk of ISGylation occurs on proteins that are concurrently synthesized ( Fig. 2). During infection, a significant portion of actively synthesized proteins would be viral proteins, guaranteeing their modification by ISG15. However, the purpose of this "time frame" as well as the fate of the ISGylated proteins (host and viral) is not understood. Further investigation on the ISGylation process needs to be done to formulate solid conclusions.
Fig. 2.
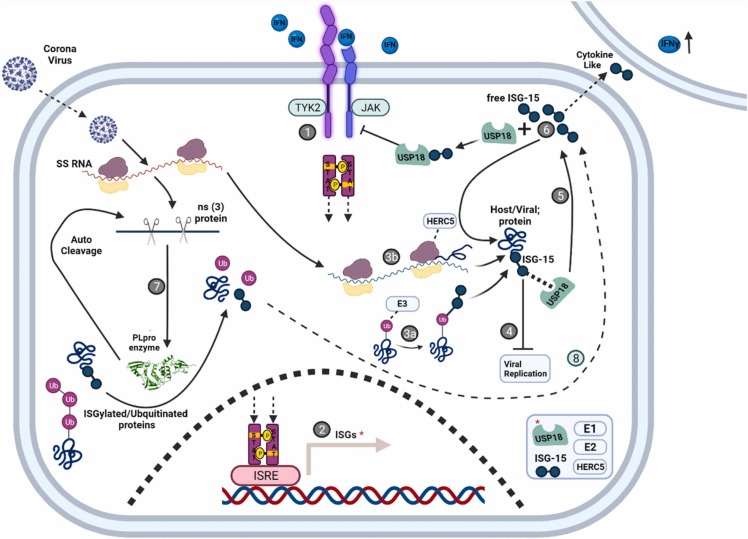
Factors that balance free and conjugated ISG15 during the immune response. 1. Binding of interferon (IFN) to the interferon receptor (IFNR) results in a conformational change that enables the transphosphorylation of associated Janus Kinase (JAK). The activated JAK then phosphorylates adjacent signal transducer and activator of transcription (STAT) proteins. 2. Phosphorylated STAT dimers migrate to the nucleus where they bind to interferon-sensitive response elements (ISRE) inducing hundreds of interferon-stimulated genes (ISGs), among them are ISG15, ISG15 conjugating enzymes (e.g. UBE1L, UBE2L6, HERC5), and its specific deISGylase (DIG), USP18). 3. HERC5, the major ISG15 E3 ligase may interact with the ribosome where it massively labels synthesized proteins (from both host or viral transcripts) as they emerge from the ribosome (3a). An as-yet-unidentified E3 may interact with a ubiquitous protein signal such as ubiquitin to generate mixed Ub-ISG15 modifications on target proteins (3b). 4. ISGylation contributes to innate immune response and inhibits viral replication, though the precise mechanism remains elusive. 5. ISGylation is overturned by USP18, a DIG that can remove ISG15 from a substrate releasing free ISG15. 6. The free form of ISG15 can be conjugated to new targets (by HERC5 and other E3 ligases), or stabilize USP18, a known inhibitor of the STAT-JAK pathway (unrelated to its catalytic DIG activity) thereby inhibiting the innate immune response. Free ISG15 can also be secreted from the cell and act as a cytokine-like protein thereby enhancing the host immune response. 7. Some viruses evolved the ability to bypass the ISG15 pathway. Coronaviruses encode a PLpro enzyme that can cleave Ub or ISG15 from conjugates (in addition to cutting the viral polypeptide precursors, PP1a, PP1ab, into an array of functional proteins (NSPs) initiating the replication of the virus). 8. By counteracting the conjugating pathway, the virus generates free ISG15. |Deconjugating ISG15 may benefit the virus to bypass the host's innate immune response, but the formation of free ISG15 could also alert neighboring cells (6).
In addition to mixed modifications on targets, a crosstalk between the Ubiquitin Proteasome System (UPS) and ISG15 modification pathways has been deduced from an observation that levels of ubiquitinated proteins decreased in ISG15-overexpressing breast cancer cells [10]. Since ISG15 and Ub each conjugate to lysine residues on their targets, ISG15 conjugation may block the formation of Ub chains and thereby interfere with proteasome-mediated protein degradation. Indeed, ISGylation appears to correlate with the stabilization of its targets [41]. However, inhibition of the proteasome and not the lysosome led to the accumulation of ISG15 chains [42], suggesting that some ISG15 conjugates could be targeted to the proteasome. It is unclear if ISG15 alone or rather mixed ISG15-Ub chains are sufficient for proteasome targeting [37].
ISGylation is a reversible process. Deconjugating enzymes (referred to as deISGylases or DIGs) can remove ISG15 from a chain or a substrate. Not all deconjugating enzymes are specific: a few proteases can hydrolyze isopeptide bonds linking either Ub or ISG15 to their targets [43], however, Ubiquitin-Specific Peptidase 18 (USP18; aka UBP43) is exclusive for ISG15 [16], [17]. Accordingly, a study on USP18−/− Chronic myeloid leukemia (CML)-derived HAP1 cells identified hundreds of ISGylation targets, emphasizing the potential of USP18 to counteract ISG15-conjugation [41]. In addition to its catalytic effect, human USP18 has a regulatory effect on IFN signaling, unrelated to its catalytic effect [44], [45], [46] (see the anti-viral section below).
In contrast to the detailed knowledge of Ub as a post-translational modification, the role of ISG15 is still unraveling. Difficulties arise from comparing experiments across species since ISG15 is only partly conserved (in contrast to ubiquitin, which is almost 100 % conserved among species; Fig. 1). Compounding the challenges to ISG15 research, ISG15 can be found both inside and outside the cell, and as part of a chain or as a free unconjugated molecule [47]. In the next paragraphs, we will summarize the results of different groups illuminating the current understanding of ISG15 as a molecular signal and try to rationalize how this contributes to cellular anti-viral defense.
2. Protein ISGylation
ISG15 was identified over four decades ago, but despite the understanding that there are numerous substrates, comprehensive lists of targets have not been compiled until recently. Challenges included the difficulty to distinguish between Ub and ISG15 conjugation due to cross-reactivity of early antibodies [48] and the identical Gly-Gly signature remnant on targets following the typical use of trypsin during sample preparation for Mass Spectrometry (MS) [49], [50]. Actually, the first bona fide ISGylated substrate was reported only in 2002 [51]. The authors had noticed an elevation in serine protease inhibitor 2 (Spi2a) in response to bacterial infection in macrophages. Then, by SDS-PAGE resolution they had noticed a form of Spi2a migrating at a higher MW than expected. Immunoprecipitation (IP) combined with MS analysis revealed the specie to be Spi2a conjugated to ISG15. Improvement in high throughput screening methodologies enabled the discovery of hundreds of ISGylated proteins over the following two decades. An early study used high throughput immunoblotting to identify ISGylated targets and found that key regulators of signal transduction such as phospholipase Cgamma1 (PLCγ1), the kinases ERK1 and JAK1, and the transcription factor STAT1, an immediate substrate of JAK1, are all modified by ISG15 [52]. It is interesting to note that activated STAT1 binds to elements in the promotor of ISG15 (among a few hundred other IFN-stimulated genes) and induces its transcription [53] (see also Fig. 2).
Proteomic studies have provided valuable information on the identity of specific ISG15 targets, but the precise makeup of the ISGylome is still unclear due to the current limitations of the method [36]. A comprehensive review on proteomics mapping of the ISG15-conjugation landscape has been published [36]; here, we will briefly summarize some of the milestones ending with the most recent findings. One of the first proteomic studies to identify ISG15 targets employed immunoprecipitation to enrich ISG15 from IFN-treated humans or from mouse cell lines lacking USP18, yielded 76 proteins [54]. Notable candidates included STAT1, and VCP/p97, a central component of the ubiquitin-proteasome system for protein turnover. In another example, a proteomic study of liposaccharide (LPS)-stimulated microglia identified 692 putative ISGylation candidates spanning proteins that are usually associated with translation, peptide metabolic, organonitrogen compound metabolic, amide biosynthetic, or cellular amide metabolic processes (and including once again, STAT1) [55]. Interestingly, in addition to host proteins from a variety of cellular pathways, also viral proteins were modified by ISG15, dependent on the E1 activating enzyme UBE1L [55], [56].
Recent advances in MS have yielded hundreds of new ISGylation targets, many of which have been confirmed by other methods. Enrichment of ISGylated proteins is usually performed for MS/MS analysis. Enrichment can be achieved by overexpression of tagged ISG15 to pullout ISG15 conjugates for analysis, though caution is warranted as abnormal ISG15 levels can perturb the conjugation system. Overexpressed ISG15 can lead to nonspecific conjugation, while in parallel, excess unconjugated ISG15 can suppress IFN-signaling by stabilizing USP18, a known inhibitor of the IFN pathway [57]. ISG15 enrichment can also be obtained by using ISG15 antibodies to trap the conjugates for analysis. Immunoprecipitation is typically less efficient than epitope isolation and may pose hurdles in the identification of low abundance hits. Both approaches do not distinguish conjugated ISG15 from free ISG15, limiting their efficiency in identifying targets. Using a USP18 -/- cell line is yet another approach to stabilize ISG15-modified conjugates and increase their abundance [58], yet one needs to take into account that, as mentioned above, USP18 itself plays an independent regulatory role by interacting with the IFN receptor and inhibiting the JAK-STAT signaling pathway [59]. Regardless of the enrichment method, robust identification by MS/MS requires appropriate reference data sets (such as cells null for ISG15, expressing non-tagged ISG15, or mock treatment/ mock beads for isolation).
In an attempt to obtain direct evidence for ISG15 modifications on targets, an anti-GlyGly antibody was used to enrich branched peptides from bacteria-infected mouse liver [60]. As a reminder, since the ISG15 primary sequence ends with the amino acids Arg-Gly-Gly, trypsinization of ISG15-conjugates generates a di-glycine modification on lysine-containing peptides of the target. Monitoring the changes to liver proteome expressing normal or catalytically inactive USP18 (to stabilize ISG15 conjugates), revealed 434 different proteins with GlyGly remnants on lysine residues that could be attributed to ISG15 [60]. In this study too, ISG15 modified a broad range of targets including extracellular proteins, mitochondrial proteins, metabolic enzymes, key regulators of autophagy, and even ubiquitin. Another study that used anti-GlyGly antibodies to enrich for ISGylation signature peptides following trypsinization compared putative ISGylated targets in IFN-treated wild-type (WT) and USP18−/− (to stabilize ISGylated targets) chronic myeloid leukemia (CML)-derived HAP1 cell lines [41]. In this manner, 312 proteins were identified as undergoing ISGylation, including HERC5, ADAR, PKR, and IFIT3 (110 of which were also verified by an ISG15 immunoprecipitation). Immobilized anti-ISG15 to enrich for ISGylated proteins was similarly used in another proteomic study [61] that identified 98 targets involved in metabolism, nucleotide pathways, and antiviral defenses including several interferon-stimulated genes (such as HERC6). Possibly explaining the lower number of targets identified by immunoprecipitation of ISG15 relative to other enrichment approaches, another recent study found anti-ISG15 antibodies ineffective to isolate ISG15 conjugates and therefore opted to overexpress FLAG-ISG15 in ISG15-/- beige adipocyte cells treated with LPS. Following pulldown with anti-FLAG (vs. IgG control), the researchers performed isobaric labeling with tandem mass tags (TMT) and mass spectrometry [62]. In this manner, 529 ISGylated proteins were identified, spanning carbohydrate metabolism, stress response, cell structure, and in particular, most glycolytic enzymes. Focusing on LDHA, the enzyme that converts pyruvate to lactate, the authors found that ISGylation suppressed its activity and decreased lactate production, involving IRF3, ISG15, and USP18 in adipose thermogenesis [62]. Another study used TMT to compare the total proteomic shift between viral-infected and naïve ISG15 null cell lines [63]. Entire proteomes of pseudorabies virus (PRV)-infected and mock ISG15−/−-porcine kidney epithelial (PK15) were comparted by trypsinization of whole-cell extracts, TMT-labeling coupled with LC-MS/MS analysis. 162 proteins were markedly upregulated and 79 were significantly downregulated in PRV-infected ISG15−/−-PK15 cells, once again spanning a broad range of metabolic pathways and cellular compartments [63]. Downregulation of proteins such as IL18, Strap, Hsp40, or Mccc1 may aid PRV propagation. An innovative attempt at deciphering the outcome of host-protein ISGylation benefitted from the deISGylation enzyme, PLpro, encoded by the SARS-CoV-2 virus [64]. Shotgun proteomics and captured GG-modified peptides of SARS-CoV-2-infected macrophages identified 181 bona fide ISGylation targets 95 of which were substrates for deISGylation by the SARS-CoV-2 PLpro. Interestingly, many were enzymes of the glycolytic pathway, indicating that PLpro counteracts the anti-inflammatory effects of ISGylation in infected cells [64]. The combined effect of substrate deISGylation with increased free ISG15 led to hyper-inflammation and reduced antigen presentation, both of which are key features of severe COVID-19 pathology.
One of the remaining challenges in identifying unique targets of ISG15-conjugation by mass spectrometry is to positively assign the Gly-Gly signature modification on substrate lysins to ISG15 following the use of trypsin for standard MS sample preparation [49], [50], [65], [66]. MS analysis has yet to develop a methodology to discriminate between ISGylation and ubiquitination at extremely high confidence. One approach would be to develop an immuno-enrichment strategy of unique ISG15-sites by innovatively employing proteases to generate a unique ISG15-site that could be isolated with a specific antibody (as has been successfully achieved for ubiquitin) [67]). Another approach to distinguish between ISG15 and Ub is using Lys C instead of Trypsin. As LysC cleave only after lysine residues, a different “signature peptide” is created if one is using mouse ISG15. In the interim, MS/MS results should be verified by other methods and analyzed in light of the manipulation that was done on the cells and the controls.
While it is clear that modification of cellular proteins by ISG15 is prevalent following exposure to IFN, how are the targets identified as substrates is still an enigma. So far, no discernable consensus motif or common feature of targets has been identified; on the contrary, the sheer diversity of the substrates points to indiscriminate conjugation. This observation suggests that answer does not lie with the targets but rather with the properties of the E3 ligases that direct the ligation, and the general molecular signal posed by ISG15 conjugation. However, regardless of how they are selected, to what extent each target is modified and how does ISGylation alters its fate remain inconclusive for many targets. For instance, some ISGylated proteins enhance the immune response, whereas modification of others interferes with innate immunity.
Evidence of the positive effect of ISGylation on the immune response can be found in the specific modification of the IFN regulatory factor 3 (IRF3) [33]. HERC5 binds and ISGylates IRF3, preventing the interaction between peptidyl-prolyl isomerase NIMA-interacting 1 (Pin1) and IRF3, thus antagonizing IRF3 ubiquitination and its subsequence degradation. By stabilizing IRF3, ISGylation directly prolongs the IFN response. By contrast, ISGylation of other proteins can suppress the immune response. For example, Retinoic Acid-Inducible Gene 1 (RIG1) is a cytosolic protein [68], [69] that contains an RNA binding site, a region that can sense invasion of an RNA virus, and initiate IFN production [27], [70], [71], [72]. Enhanced RIG1 mediated antiviral response was observed in UBE1L-/- cells. In addition, proteasome inhibition resulted in the accumulation of the ISG15-conjugated RIG1 at the expense of the unconjugated form [71]. Hence, it was suggested that ISGylated RIG1 is probably not active. Viral proteins are also found among the numerous ISGylated proteins. One example, is the NS1 protein, one of 11 proteins encoded by Influenza A viruses (IAVs) [73]. Expression of NS1 has various destructive effects on the host cell, including inhibition of IFN1 induction. As part of the innate immune response, the host cell ISGylates NS1, impeding its interactions and limiting its actions.
To summarize, following infection or induction of IFN signaling, ISGylation occurs on hundreds of proteins, both viral and host. Targets originate from different cellular compartments and partake in a whole slew of cellular pathways [72] suggesting that deciphering the molecular role of ISG15 may not be tied to the precise identity of its targets. Although the mechanism of the ISG15 protein as an antiviral agent is somewhat illusive, genetic studies have taught us that expression of ISG15 and its counterparts (USP18, UBEL1, etc.) do have an importance in stimulating the immune system.
3. The anti-viral role of ISG15
Elevated levels of ISG15 are correlated with a number of human ailments such as cancer, neurodegeneration, inflammation, and assaults from pathogens [74], [75]. Indeed, IFN has been used to treat certain types of cancer and specific viruses [76]. Since ISG15 is most abundant in the cell following IFN induction, efforts have been made to try to understand the anti-viral properties of ISG15 [77], [78], [79]. Data derived from ISG15-deficient patients, from ISG15 -/-, UBE1L -/-, and USP18 -/- animal models, and from cellular models ( Table 1) point to a role of ISG15 in the immune response that depends on many factors such as the source of infection (virus, mycobacteria, chronic inflammation, etc.), the host (mouse or human), the ratio of conjugated to free ISG15, or the cellular location of ISG15 (cells cytosol or secreted). A number of recent reviews have detailed the antiviral properties of ISG15 – for example [77], [78], [79], [80] – here we will provide a brief overview and highlight the complexity of recent results.
Table 1.
Manipulations of ISG15 and their effects on virus infections in different models.
| Alteration of ISG15 | Virus/Pathogen | Model system | Effect | Ref. |
|---|---|---|---|---|
| overexpression of ISG15 | Pseudorabies Virus (PRV) | inducible cell line stably expressing the pISG15 gene | Inhibition of PRV replication reducing the viral titers and mRNA levels of PRV | [120] |
| Stable overexpression or knockdown of ISG15 | Classical Swine Fever Virus (CSFV) | Porcine Alveolar Macrophages (PAMs) | Loss of ISG15 led to abnormal proliferation of CSFV. Mechanism: the loss of ISG15 lead to loss of beclin-1 (BECN1) ISGylation that inhibits the autophagy process, which is necessary for CSFV replication | [121] |
| siRNA against ISG15 | Zika Virus (ZIKV) | Human Corneal Epithelial Cells (Pr. HCEC) | Silencing ISG15 results in increase ZIKV | [122] |
| ISG15 KO | SARS-CoV-2flaviviruses and picornaviruses | Mainly KO cell line: MEF / HeLa | ISGylation of MDA5 protein is crucial for sensing the viral nucleic acids recognition and triggers the immune response | [123] |
| ISG15 KO | Paramyxoviruses | A549-ISG15−/−cells | Rapid induction of ISGs synthesis | [124] |
| ISG15 KO | HIV | Human B cell precursor leukemia cell line, BlaER1 and THP-1 cells | Accumulation of misfolded dominant negative p53 resulting in enhance of HIV replication (functional p53 is required for p21 function: p21 acts as an inactivator of dNTP synthesis and activator of the restriction factor SAMHD1) |
[125] |
| ISG15 KO | Toxoplasma gondii | HeLa (cervical) and A549 (lung) cells. | ISG15 KO resulted in a decrease of the IFNγ response to the pathogen, impaired recruitment of the following adaptors: p62, NDP52, and LC3 to the parasitophorous vacuole. These effects prevent the restriction of the parasite growth | [126] |
| Cell lines expressing sub genomic replicons and replicon virus-like particles+ siRNA against ISG15+ O.E of ISG15/HERC | Flaviviruses | A549 cells | O.E of ISG15 (and HERC5) leads to reduced replication by suppressing the ALIX and CHMP4A pathway, two of the host’s proteins that are necessary for viral replication. The depletion of ISG15 results in enhancement of the effect observed in ALIX KO cell line. | [127] |
| ISG15 KO cells | Viral Hemorrhagic Septicemia Virus (VHSV) | EPC cells | Impaired viral replication, as compared to MX1 KO cell, even if the cells are pretreated with poly I:C | [128] |
A thorough summary of the impact of ISG15 on viral infection in human and mouse models was published by Lenchow D. in 2013 [78]. Here we expand the list with data published after 2013.
ISG15-/- mice were fertile and viable [81]. Somewhat unexpectedly, cells derived from ISG15 -/- mice were not sensitive to vesicular stomatitis virus or lymphocytic choriomeningitis virus (VSV virus and LCMV virus). In addition, STAT1 signaling pathway in ISG15 -/- mice seemed unaffected relative to wild-type (WT) mice. In contrast to these observations, another study revealed that ISG15 -/- mice were susceptible to both influenza A/WSN/33 and influenza B infections, in addition to herpes simplex virus type 1, murine gamma-herpesvirus infections, and Sindbis virus infection [82]. Moreover, the host immune response to the Sindbis virus was restored upon overexpression of ISG15 (but not of the non-conjugatable mutant ISG15), indicating that a functional ISG15 conjugation system is required for the antiviral properties of ISG15 [83].
USP18 knock out mice were more resistant than WT to complications caused by lymphocytic choriomeningitis virus (LCMV) or vesicular stomatitis virus (VSV), and displayed increased resistance to Hepatitis B Virus [84], [85]. These mice were also more resistant to Salmonella typhimurium infection, a bacterial infection [86]. USP18 can specifically remove ISG15 from a chain or a substrate. Thus, ISG15 conjugates accumulated in USP18 -/- mice [87], [88], in agreement with a role for ISG15 conjugates in combating infections. Interestingly, USP18 in humans (but not in mice) can also interfere with the anti-viral outcome of IFN signaling by an additional mechanism: USP18 can bind the interferon receptor 2 (IFNAR2), block the association of JAK, and possibly limit IFNAR2-IFNAR1 dimerization, thereby interfering with the JAK-STAT signaling pathway [89]. This inhibitory effect does not depend on its catalytic activity but does seems to be dependent on ISG15, specifically on intact ISG15 containing its GG tail tough not necessarily conjugated [58], [59], [85], [89], [90]. The proposal was that ISG15 stabilizes USP18 by protecting it from ubiquitination and degradation, thereby increasing its availability for IFNAR binding [59], [87]. By contrast, ISG15-conjugates engage USP18, apparently interfering with its affinity for IFNAR. Consequently, the balance of free-to-conjugated ISG15 is crucial for the proper function of the immune response. (Fig. 2). Mice lacking USP18 displayed upregulated IFN-inducible ISGylation, and showed resistance to certain viral infections and cells derived from these mice showed lower viral replication [85]. These findings in mice point to an immunosuppressive effect by USP18-driven deISGylation. Loss of USP18 activity in human cells also led to enhanced ISGylation. A study on USP18 −/− Chronic myeloid leukemia (CML)-derived HAP1 cells identified hundreds of ISGylation targets including many involved in translation, dsRNA binding, innate immune response, and viral defense [41]. This observation supports the role of human USP18 in partially counteracting the innate immune response conferenced by ISG15-conjugation.
UBE1L-/- mice were also fertile and showed no significant phenotypes. As expected, without the E1 component, the remainder of the ISGylation machinery failed to achieve ISG15 conjugation without any noticeable effect on free ISG15 [24], [25]. In addition, cells derived from UBE1L -/- knockout mice showed typical levels of phosphorylated STAT, indicating a normal activation of the IFN pathway. Greater susceptibility to influenza B virus infection was detected in these mice, but the lack of ISG15-conjugation did not affect the resistance to two other viruses: VSV and LCMV [91]. Once again, one may conclude that at least part of the defense against influenza B is dependent on ISG15 conjugation. Supporting this claim, one of the major ISGylated targets upon influenza B infection was found to be the viral nucleoprotein (NP) necessary for viral RNA synthesis. ISGylation of NP blocks NP oligomerization, effectively acting as an inhibitor of the viral replication process [92]. In UBE1L -/- mice, the NP would not be ISGylated enabling uninterrupted viral replication. In counter attempt, influenza B interfere with ISG15 conjugation through a specific region in the NS1B protein that was found to inhibit ISGylation [93]. Interestingly, these observations were not made for the influenza A virus. These findings suggest that every virus evolved a different approach to evade the immune response and indicate that some viruses try to interfere with the ISG15-conjugation machinery to do so [94]. This may explain the contradictory results emanating from different model systems or experimental conditions.
The clinical picture is even more complex. Human patients were identified with inherited (recessive inheritance) ISG15 deficiency (ISG15 -/-) [95]. Based on the mice data, we would have expected that they would be partially susceptible to viral attacks. However, cells derived from these patients were not susceptible to any virus that was tested [95]. In addition, humans lacking functional ISG15 displayed no enhanced susceptibility to viruses [80] and did not report suffering from repeated infections. Actually, the patients were only identified after they developed a serious illness in response to a live tuberculosis vaccine (Bacillus-Calmette-Guerin vaccine), a bacterial infection. These observations can be partially explained by another feature of the ISG15 protein: the free form can be secreted from cells. Early in 1991 an "IFN-induced 15-kDa protein" was found in the media of lymphocytes or monocytes cells [96]. In parallel, the presence of this "15,000 molecular weight protein" in the media, resulted in neopterin secretion (a marker for the activation of the immune response) [97]. In addition, five years later, secreted ISG15 was found to induce the proliferation of natural killer cells and induced the production of IFNγ [98], [99]. The absence of conjugated ISG15 in the media indicated that the ISG15 presence is not due to cell lysis [100], [101]. A further hint at the function of secreted ISG15 came from a recent study that demonstrated that merely adding purified ISG15 to cell media resulted in IFNγ induction [100], [102]. As the targeting sequence to the endoplasmic reticulum has not been identified, the secretion of ISG15 is probably not through the "traditional" pathway. These findings imply that secreted ISG15 activates the immune response and acts as a cytokine-like protein. As IFNγ is related to mycobacterial infections, it could explain the observed illness of ISG15 -/- patients that would lack this stimulatory mechanism, but it does not satisfactorily explain the lack of susceptibility to viral infection in those patients. Interestingly, some human ISG15 KO cell lines can be more susceptible to viruses (Table 1), meaning that there is a gap between in vivo and in vitro results and the full picture is not fully understood. It is worth mentioning that some ISG deficient patients do present additional phenotypes, for example, in China, a family with inherited ISG15 deficiency, presented with intermittent seizures stemming caused by intracranial calcifications [57]. In addition, a dermatological presentation can be found in some patients [103]. These observations are probably related to the inhibitory role of USP18 and not to the inflammatory pathway.
In summary, the ISG15 pathway is one of the mechanisms by which the host responds to infection. Different viruses try to evade this pathway by taking different approaches, with some altering the conjugated ISG15 landscape and others influencing the levels of free ISG15 [75]. Different findings were found by different groups, depending on the virus tested and the manipulation that was done to the system (Table 1). Understanding the mechanism by which a specific virus interferes with the immune response by manipulating the ISG15 pathway would provide clinical benefits. In the following section we overview a pertinent and fascinating example from coronaviruses that encode a deISGylase that is related to ubiquitin-specific peptidases (USPs) in order to directly interfere with ISGylation by the host cell.
4. The Coronavirus papain-like proteases (PLPs or PLpro) as USP-related DUBs/DIGs
Coronavirus Papain-Like proteases (PLpro) are unusual enzymes that are worth mentioning in the context of ISG15 and its anti-viral function. Coronaviruses can cause serious respiratory diseases, the common symptoms of which include fever, dry cough, and shortness of breath. In more serious cases, renal damage, acute hepatic, blood clots in multiple organs, and meningitis have been reported [104]. Some coronaviruses alter the patient’s immune system response, which delays the patient's recovery and is the number one cause of prolonged hospitalization. For this reason, many efforts have been made to try to understand the mechanism by which coronaviruses interfere with the proper function of the host immune system. One potential candidate is the viral PLpro enzyme [19], [22], [105]. By cleaving the virus polyprotein into functional subunits of the replicase complex (Fig. 2), this enzyme enables the initiation of the replication and transcription of the viral genome. However, this enzyme also deconjugates ubiquitin or ISG15 modifications from target proteins in the host cell [19], [106], [107]. A narrow cleft restricts access to the active site of PLpro to glycine at positions P1 and P2, limiting substrates to specific sequences found in the viral polypeptide precursors (PP1a, PP1ab) or in the flexible C-termini of Ub and ISG15 [18], [108], [109]. As a consequence, PLpro enzymes from MERS, SARS, and SARS-Cov2 are able to hydrolyze the isopeptide bond at either the C-terminus of Ub or ISG15 [18], [19], [21], [22], [109], [110].
Other than PLpro, the only documented deISGylation enzymes (DIGs) are certain members of the eukaryotic ubiquitin-specific protease family (USP): USP2, USP5, USP13, USP14, and USP18. USP18 is unique as it is specific for ISG15, whereas the other USPs function primarily as deubiquitination enzymes (DUBs) with only marginal efficiency at processing ISG15 conjugates [43]. PLpro enzymes also double up as both DUBs and DIGs with different efficiencies (in each Coronavirus). Since they act on similar substrates it has been suggested that PLpros and DUBs share a common mode of action [43], [106], [109], [111], [112]. With this in mind, we superimposed the 3D structures of three Coronavirus PLpro enzymes with human USP18 and USP14 ( Fig. 3A). Superimposition of these structures highlights their similar architectures and common catalytic core. The active site of USP family members, as well as of PLpro enzymes, is buried in the palm of a hand-like structure, from which beta-strand fingers extend opposite an alpha-helical thumb (Fig. 3A). In all PLpros and in USP14 (Fig. 3A i-iii, v), this thumb extends into a ubiquitin-like domain. Most other USP family members lack this N-terminal UBL domain (exemplified here by USP18; Fig. 3A iv), indicating that the UBL domain of PLpro is not integral to the catalytic mechanism. Comparing substrate-bound enzyme structures emphasizes their similar catalytic mechanism (Fig. 3B). The substrate (Ub/ISG15) “rests” on the palm surface with its flexible C-terminal extension entering through a cleft reaching the active site. The narrow access is limited to the flexible consensus sequence (R/K)LXGG, found in the viral polyprotein precursor (viral polypeptide precursors (PP1a, PP1ab; [113])), as well as at the C-terminus of ISG15 and Ub [18], [19], [22], [23], explaining the similar cleavage specificities of PLpros and USPs. The catalytic mechanism of both PLpros and USPs depends on a catalytic triad typical of cysteine proteases (Cis-His-Asp/Asn). Since these three active sites residues precisely overlay and originate from the same secondary structures (Fig. 3C), PLpro and USPs could be considered part of the same greater protease family.
Fig. 3.
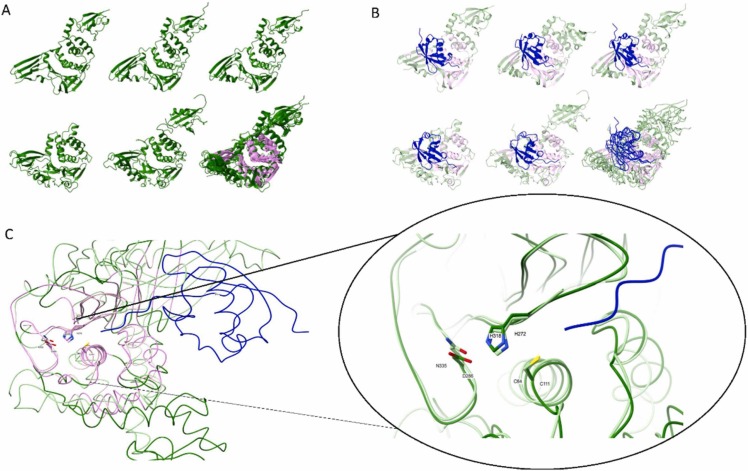
Structural alignment of viral PLpro and select USP enzymes. A. Ribbon illustration of apoenzyme 3D structures: i) MERS-PLpro (pdb:4rna), ii) SARS-CoV-PLpro (pdb:2fe8), iii) SARS-CoV2-PLpro (pdb:6wrh), iv) hUSP18 (AF-Q9UMW8-F1; [130]), and v) hUSP14 (pdb:6iik+1wgg). PDB entries were taken from https://www.rcsb.org/ and AlphaFold models from https://alphafold.ebi.ac.uk/. A full 3D structure of hUSP14 was generated by superimposing the USP domain (6iik) and N-terminal UBL domain (1wgg) onto the PLpro structure. A model of hUSP18 was generated by Alphafold [130]. vi) Superimposition of all five enzymes highlights structurally equivalent residues shared by all five enzymes (i.e. the common core shown in pink) calculated by mTM-align [131]. B. Ribbon illustration of PLpro and USP 3D structures in complex with substrate, either Ub (hUSP14) or proximal domain of ISG15: i) MERS-PLpro (pdb:5w8u), ii) SARS-CoV-PLpro (pdb:5tl7), iii) SARS-CoV2-PLpro (pdb:6xa9), iv) hUSP18 (AlphaFold model:AF-Q9UMW8-F1 +PDB:2hj8), and v) hUSP14 (PDB:2ayo+1wgg). Full 3D structure of Ub-bound hUSP14 was generated by the superimposition of 2ayo for the Ub-aldehyde domain bound to the USP domain and 1wgg for the N-terminal UBL domain of hUSP14, onto the structure of PLpro. A model of ISG15-bound hUSP18 was generated by Alphafold [130] for hUSP18 and the solution structure of the proximal domain of hISG15 (PDB:2hj8) superimposed on complexed mUSP18-mISG15 (PDB:5CHV). vi) Superimposition of substrate-bound enzymes defines the common core. Structurally equivalent residues of all five enzymes are shown in pink as computed by mTM-align [131]. C. Superimposition of the catalytic core of hUSP18 and ISG15-bound SARS-CoV2-PLpro (as determined in panel A). The shared residues of both these proteases calculated by mTM-align[131] are shown in pink; left. The catalytic triad residues overlay precisely in the active site; right. The narrow cleft leading to this active site restricts access to substrates with flexible stretches (shown in blue). Cleavage is limited to the presence of Glycine at P1 and P2 sites. Images were generated using ChimeraX [132].
Regions beyond the catalytic core are responsible for substrate selectivity. It is these interactions with substrate, which are distal to the active site involving the beta-strand fingers and the thumb, that determine the substrate preference of each enzyme (Fig. 3B). For instance, MERS-CoV, SARS-CoV, and SARS-CoV2 PLpros can all cleave conjugated Ub or ISG15, though only the latter prefers ISG15 as a substrate [105], [110], [113]. Most USPs also prefer Ub, except for USP18 which is specific for ISG15. For this reason, we compared SARS-CoV2 PLpro and USP18 to emphasize the primary interactions of residues on the palm and beta fingers with the proximal Ubl domain of ISG15 or the ubiquitin chain (Fig. 3B). Indeed, point mutations in the MERS-CoV PLpro enzyme were designed to create enzymes with alternate substrate specificities – a DUB deficient enzyme, a DIG deficient enzyme, and even a protease hyperactive enzyme [21]. However, substrate selectivity is driven by secondary interactions of the distal Ubl of ISG15 with the thumb motif of the enzyme. Thus SARS-CoV2-PLpro can differentiate between ISG15 and K48-linked diUb through such distal interactions [110]. These properties raise the potential to engineer differential inhibitors of PLpro for each of its substrates. For instance, interfering with the ability to associate with ISG15, may provide a milder approach than inhibiting its catalytic activity that could interfere with other substrates or cross-target other cellular USPs.
Among Corona and other Noroviruses, SARS-CoV2 stands out as particularly potent at interfering with the host immune response [64], [110]. Part of this effect can be attributed to the preference of the SARS-CoV2 PLpro for deconjugating ISG15 over Ub [19], [22], [69], [105], [106], [107], [110], [113], [114]. For instance, a non-covalent inhibitor GRL-0617 that inhibits the catalytic activity of SARS-CoV2-PLpro could reinstate ISGylation of host proteins, and was sufficient for SARS-CoV2 infected cells to recover their IFN-signaling and decrease the number of viral particles observed in the supernatant [110]. Inhibiting a single viral enzyme, PLpro, demonstrates the broad effects of the ISG15 pathway and the consequence of virus interference with the balance of conjugated to free ISG15.
5. Summary
ISG15 is one of the strongest IFN-stimulated genes resulting in conjugation to hundreds of targets both viral and host proteins, but the full picture of the ISG15 landscape is complicated and elusive at this stage. Overall, modification of proteins by ISG15 appears to interfere with cellular processes that are conductive to viral replications. These include both cellular targets and synthesized viral proteins. It is not surprising that some viruses have adopted strategies to counteract the host ISGylation pathway by suppressing ISG15 expression, blocking ISG15 conjugation, sequestering ISG15 or ISG15-conjugates, or enhancing deconjugation by encoding for exogenous DIGs. Many viruses encode proteases that double up as both a processing enzyme of viral pre-proteins and as DUBs that counteract host ubiquitination of host or viral proteins. Some viral DUBs are members of the OTU sub-family [115], [116], [117], for instance, arterivirus PLP2 or tymovirus PRO that are required for viral replication due to their primary role in polyprotein maturation but also harbor inherent DUB activity. Initially, these enzymes were classified as papain-like proteases, however now with their 3-D structures resolved, it is “crystal clear” that they are members of the OTU sub class of DUBs [115]. The list of viruses that encode OTU-like proteases has increased significantly covering both DNA and RNA viruses [116], [117]. Coronaviral proteases are still generally referred to as papain-like proteases. In light of the structural alignment (Fig. 3), it may be prudent to consider PPpro as USP-like enzymes.
After controlling the pathogen assault, cells need to decelerate the ISGylation pathway or risk chronic inflammation or even cell death [118], [119]. For this purpose, cells encode an endogenous DIG, USP18, that removes ISG15 from conjugates while releasing unconjugated ISG15. Since catalytically inactive USP18 does not lead to any obvious abnormalities (at least in mice), designing a specific active site inhibitor for USP18 may enhance the anti-viral response with only limited side effects [58]. The catalytic activity of USP18 interfere with the host ISG15 landscape, though it is still unclear how the balance between free and conjugated ISG15 contributes to the anti-viral response. Liberating free ISG15 from conjugates may trigger ISG15 secretion and thus contribute to inflammation associated with infection [64]. As free ISG15 may be secreted from infected cells, the balance of free to conjugated ISG15 not only affects IFN-induced signaling in a given cell but could potentially affect neighboring cells as well [57], [64], [101]. In contrast, binding this free ISG15, USP18 suppresses JAK-STAT signaling further counteracting IFN signaling.By better understanding the ISG15 pathway, it may be possible to target certain illnesses on a case-by-case basis without the need for general activation of IFN signaling with its hundreds of downstream targets.
Declaration of Competing Interest
The authors state that we do not have any financial and personal relationships with other people or organizations that could inappropriately influence our work or this manuscript that we have written.
Acknowledgments
We dedicate this review to the memory of our dear colleague Prof Huib Ovaa (NKI/LUMC), a pioneer of ubiquitin and ubiquitin-like molecules (including ISG15) research using innovative chemical tools. Huib, your leadership, and insight will be sorely missed. We thank Dr. Klaus-Peter Knobeloch for introducing us to the field of ISG15 and providing endless information and Dr. Oded Kleifeld for critical discussions on ISG15 proteomic approaches. Research in the lab of MHG is supported in part by grants from the Israel Science Foundation (ISF 755/19) for studying cellular stress conditions, Israel Science Foundation - National Natural Science Foundation of China (ISF-NFSC 2512/18) for research on proteasome mechanisms, and from National Science Foundation - USA-Israel Binational Science Foundation (NSF-BSF 1818280) for dissecting properties of Ubl.
References
- 1.Komander D., Rape M. The ubiquitin code. Annu Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu Rev. Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 3.Cappadocia L., Lima C.D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2018;118(3):889–918. doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell P.J., Broeze R.J., Lengyel P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279(5713):523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- 6.Vargas-Inchaustegui D.A., Xin L., Soong L. Leishmania braziliensis infection induces dendritic cell activation, ISG15 transcription, and the generation of protective immune responses. J. Immunol. 2008;180(11):7537–7545. doi: 10.4049/jimmunol.180.11.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Peet G.W., Balzarano D., Li X., Massa P., Barton R.W., Marcu K.B. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J. Biol. Chem. 2001;276(21):18579–18590. doi: 10.1074/jbc.M100846200. [DOI] [PubMed] [Google Scholar]
- 8.Han H.G., Moon H.W., Jeon Y.J. ISG15 in cancer: beyond ubiquitin-like protein. Cancer Lett. 2018;438:52–62. doi: 10.1016/j.canlet.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Liu M., Hummer B.T., Li X., Hassel B.A. Camptothecin induces the ubiquitin-like protein, ISG15, and enhances ISG15 conjugation in response to interferon. J. Interferon Cytokine Res. 2004;24(11):647–654. doi: 10.1089/jir.2004.24.647. [DOI] [PubMed] [Google Scholar]
- 10.Desai S.D., Haas A.L., Wood L.M., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur E.K.A., Liu L.F. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66(2):921–928. doi: 10.1158/0008-5472.CAN-05-1123. [DOI] [PubMed] [Google Scholar]
- 11.Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262(23):11315–11323. [PubMed] [Google Scholar]
- 12.Loeb K.R., Haas A.L. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 1992;267(11):7806–7813. [PubMed] [Google Scholar]
- 13.Narasimhan J., Potter J.L., Haas A.L. Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 1996;271(1):324–330. doi: 10.1074/jbc.271.1.324. [DOI] [PubMed] [Google Scholar]
- 14.Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005;280(29):27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y.G., Yan X.Z., Xie Y.Y., Gao X.C., Song A.X., Zhang D.E., Hu H.Y. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J. Biol. Chem. 2008;283(19):13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- 16.Basters A., Geurink P.P., Rocker A., Witting K.F., Tadayon R., Hess S., Semrau M.S., Storici P., Ovaa H., Knobeloch K.P., Fritz G. Structural basis of the specificity of USP18 toward ISG15. Nat. Struct. Mol. Biol. 2017;24(3):270–278. doi: 10.1038/nsmb.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basters A., Knobeloch K.P., Fritz G. How USP18 deals with ISG15-modified proteins: structural basis for the specificity of the protease. FEBS J. 2018;285(6):1024–1029. doi: 10.1111/febs.14260. [DOI] [PubMed] [Google Scholar]
- 18.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem Biophys. 2007;466(1):8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39(18) doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bekes M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-Linked Di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol. Cell. 2016;62(4):572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clasman J.R., Everett R.K., Srinivasan K., Mesecar A.D. Decoupling deISGylating and deubiquitinating activities of the MERS virus papain-like protease. Antivir. Res. 2020;174 doi: 10.1016/j.antiviral.2019.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K.I., Yan M., Malakhova O., Luo J.K., Shen M.F., Zou W., de la Torre J.C., Zhang D.E. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol. Cell Biol. 2006;26(2):472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krug R.M., Zhao C., Beaudenon S. Properties of the ISG15 E1 enzyme UbE1L. Methods Enzym. 2005;398:32–40. doi: 10.1016/S0076-6879(05)98004-X. [DOI] [PubMed] [Google Scholar]
- 26.Serniwka S.A., Shaw G.S. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry. 2009;48(51):12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- 27.Arimoto K., Konishi H., Shimotohno K. UbcH8 regulates ubiquitin and ISG15 conjugation to RIG-I. Mol. Immunol. 2008;45(4):1078–1084. doi: 10.1016/j.molimm.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Okumura F., Zou W., Zhang D.E. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21(3):255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimoto K., Hishiki T., Kiyonari H., Abe T., Cheng C., Yan M., Fan J.B., Futakuchi M., Tsuda H., Murakami Y., Suzuki H., Zhang D.E., Shimotohno K. Murine Herc6 plays a critical role in protein ISGylation in vivo and has an ISGylation-independent function in seminal vesicles. J. Interferon Cytokine Res. 2015;35(5):351–358. doi: 10.1089/jir.2014.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oudshoorn D., van Boheemen S., Sanchez-Aparicio M.T., Rajsbaum R., Garcia-Sastre A., Versteeg G.A. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou W., Zhang D.E. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 2006;281(7):3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 32.Dastur A., Beaudenon S., Kelley M., Krug R.M., Huibregtse J.M. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 2006;281(7):4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 33.Shi H.X., Yang K., Liu X., Liu X.Y., Wei B., Shan Y.F., Zhu L.H., Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell Biol. 2010;30(10):2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durfee L.A., Lyon N., Seo K., Huibregtse J.M. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell. 2010;38(5):722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domingues P., Bamford C.G.G., Boutell C., McLauchlan J. Inhibition of hepatitis C virus RNA replication by ISG15 does not require its conjugation to protein substrates by the HERC5 E3 ligase. J. Gen. Virol. 2015;96(11):3236–3242. doi: 10.1099/jgv.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thery F., Eggermont D., Impens F. Proteomics mapping of the ISGylation landscape in innate immunity. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.720765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J.B., Arimoto K., Motamedchaboki K., Yan M., Wolf D.A., Zhang D.E. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 2015;5:12704. doi: 10.1038/srep12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haakonsen D.L., Rape M. Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol. 2019;29(9):704–716. doi: 10.1016/j.tcb.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Rusilowicz-Jones E.V., Urbe S., Clague M.J. Protein degradation on the global scale. Mol. Cell. 2022;82(8):1414–1423. doi: 10.1016/j.molcel.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Durfee L.A., Huibregtse J.M. Identification and validation of ISG15 target proteins. Subcell. Biochem. 2010;54:228–237. doi: 10.1007/978-1-4419-6676-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto-Fernandez A., Salio M., Partridge T., Chen J., Vere G., Greenwood H., Olie C.S., Damianou A., Scott H.C., Pegg H.J., Chiarenza A., Diaz-Saez L., Smith P., Gonzalez-Lopez C., Patel B., Anderton E., Jones N., Hammonds T.R., Huber K., Muschel R., Borrow P., Cerundolo V., Kessler B.M. Deletion of the deISGylating enzyme USP18 enhances tumour cell antigenicity and radiosensitivity. Br. J. Cancer. 2021;124(4):817–830. doi: 10.1038/s41416-020-01167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu M., Li X.L., Hassel B.A. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 2003;278(3):1594–1602. doi: 10.1074/jbc.M208123200. [DOI] [PubMed] [Google Scholar]
- 43.Catic A., Fiebiger E., Korbel G.A., Blom D., Galardy P.J., Ploegh H.L. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2(7) doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber C., Martin-Fernandez M., Ailal F., Qiu X., Taft J., Altman J., Rosain J., Buta S., Bousfiha A., Casanova J.L., Bustamante J., Bogunovic D. Homozygous STAT2 gain-of-function mutation by loss of USP18 activity in a patient with type I interferonopathy. J. Exp. Med. 2020;217(5) doi: 10.1084/jem.20192319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ketscher L., Knobeloch K.P. ISG15 uncut: Dissecting enzymatic and non-enzymatic functions of USP18 in vivo. Cytokine. 2015;76(2):569–571. doi: 10.1016/j.cyto.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Vasou A., Nightingale K., Cetkovská V., Bamford C.G.G., Andrejeva J., R. E., J. McLauchlan M.P., Weekes D.J., Hughes A co-opted ISG15-USP18 binding mechanism normally reserved for deISGylation controls type I IFN signalling. BioRxiv. 2021 doi: 10.1101/2021.06.01.446527. [DOI] [Google Scholar]
- 47.Bogunovic D., Boisson-Dupuis S., Casanova J.L. ISG15: leading a double life as a secreted molecule. Exp. Mol. Med. 2013;45 doi: 10.1038/emm.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potter J.L., Narasimhan J., Mende-Mueller L., Haas A.L. Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J. Biol. Chem. 1999;274(35):25061–25068. doi: 10.1074/jbc.274.35.25061. [DOI] [PubMed] [Google Scholar]
- 49.Mendes M.L., Fougeras M.R., Dittmar G. Analysis of ubiquitin signaling and chain topology cross-talk. J. Proteom. 2020;215 doi: 10.1016/j.jprot.2020.103634. [DOI] [PubMed] [Google Scholar]
- 50.Denison C., Kirkpatrick D.S., Gygi S.P. Proteomic insights into ubiquitin and ubiquitin-like proteins. Curr. Opin. Chem. Biol. 2005;9(1):69–75. doi: 10.1016/j.cbpa.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Hamerman J.A., Hayashi F., Schroeder L.A., Gygi S.P., Haas A.L., Hampson L., Coughlin P., Aebersold R., Aderem A. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J. Immunol. 2002;168(5):2415–2423. doi: 10.4049/jimmunol.168.5.2415. [DOI] [PubMed] [Google Scholar]
- 52.Malakhov M.P., Kim K.I., Malakhova O.A., Jacobs B.S., Borden E.C., Zhang D.E. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2003;278(19):16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 53.Tolomeo M., Cavalli A., Cascio A. STAT1 and its crucial role in the control of viral infections. Int J. Mol. Sci. 2022;23(8) doi: 10.3390/ijms23084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giannakopoulos N.V., Luo J.K., Papov V., Zou W., Lenschow D.J., Jacobs B.S., Borden E.C., Li J., Virgin H.W., Zhang D.E. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys. Res. Commun. 2005;336(2):496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 55.Przanowski P., Loska S., Cysewski D., Dabrowski M., Kaminska B. ISG’ylation increases stability of numerous proteins including Stat1, which prevents premature termination of immune response in LPS-stimulated microglia. Neurochem. Int. 2018;112:227–233. doi: 10.1016/j.neuint.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Jeon Y.J., Choi J.S., Lee J.Y., Yu K.R., Kim S.M., Ka S.H., Oh K.H., Kim K.I., Zhang D.E., Bang O.S., Chung C.H. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009;10(4):374–380. doi: 10.1038/embor.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., Tezcan I., Rice G.I., Chen C., Mansouri N., Mahdaviani S.A., Itan Y., Boisson B., Okada S., Zeng L., Wang X., Jiang H., Liu W., Han T., Liu D., Ma T., Wang B., Liu M., Liu J.Y., Wang Q.K., Yalnizoglu D., Radoshevich L., Uze G., Gros P., Rozenberg F., Zhang S.Y., Jouanguy E., Bustamante J., Garcia-Sastre A., Abel L., Lebon P., Notarangelo L.D., Crow Y.J., Boisson-Dupuis S., Casanova J.L., Pellegrini S. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517(7532):89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ketscher L., Hannss R., Morales D.J., Basters A., Guerra S., Goldmann T., Hausmann A., Prinz M., Naumann R., Pekosz A., Utermohlen O., Lenschow D.J., Knobeloch K.P. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proc. Natl. Acad. Sci. U.S.A. 2015;112(5):1577–1582. doi: 10.1073/pnas.1412881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francois-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J., Pellegrini S., Uze G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Thery F., Wu N.C., Luhmann E.K., Dussurget O., Foecke M., Bredow C., Jimenez-Fernandez D., Leandro K., Beling A., Knobeloch K.P., Impens F., Cossart P., Radoshevich L. The in vivo ISGylome links ISG15 to metabolic pathways and autophagy upon Listeria monocytogenes infection. Nat. Commun. 2019;10(1):5383. doi: 10.1038/s41467-019-13393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu C., Li J., Tian C., Qin M., Wang Z., Shi B., Qu G., Wu C., Nan Y. Proteomic analysis of ISGylation in immortalized porcine alveolar macrophage cell lines induced by type I interferon. Vaccines. 2021;9(2) doi: 10.3390/vaccines9020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan S., Kumari M., Xiao H., Jacobs C., Kochumon S., Jedrychowski M., Chouchani E., Ahmad R., Rosen E.D. IRF3 reduces adipose thermogenesis via ISG15-mediated reprogramming of glycolysis. J. Clin. Investig. 2021;131(7) doi: 10.1172/JCI144888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He W., Li C., Dong L., Yang G., Liu H. Vol. 12. Basel; 2021. Tandem mass tag-based quantitative proteomic analysis of ISG15 knockout PK15 cells in pseudorabies virus infection. (Genes). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munnur D., Teo Q., Eggermont D., Lee H.H.Y., Thery F., Ho J., van Leur S.W., Ng W.W.S., Siu L.Y.L., Beling A., Ploegh H., Pinto-Fernandez A., Damianou A., Kessler B., Impens F., Mok C.K.P., Sanyal S. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021;22(11):1416–1427. doi: 10.1038/s41590-021-01035-8. [DOI] [PubMed] [Google Scholar]
- 65.Sulkshane P., Duek I., Ram J., Thakur A., Reis N., Ziv T., Glickman M.H. Inhibition of proteasome reveals basal mitochondrial ubiquitination. J. Proteom. 2020;229 doi: 10.1016/j.jprot.2020.103949. [DOI] [PubMed] [Google Scholar]
- 66.Ziv I., Matiuhin Y., Kirkpatrick D.S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S.P., Haguenauer-Tsapis R., Reis N., Glickman M.H., Kleifeld O. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell. Proteom.: MCP. 2011;10(5) doi: 10.1074/mcp.M111.009753. M111 009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akimov V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M., Kratchmarova I., Kelstrup C.D., Olsen J.V., Blagoev B. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018;25(7):631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 68.Dzimianski J.V., Scholte F.E.M., Bergeron E., Pegan S.D. ISG15: it’s complicated. J. Mol. Biol. 2019;431(21):4203–4216. doi: 10.1016/j.jmb.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiang C., Liu G., Gack M.U. Viral evasion of RIG-I-Like receptor-mediated immunity through dysregulation of ubiquitination and ISGylation. Viruses. 2021;13(2) doi: 10.3390/v13020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 71.Kim M.J., Hwang S.Y., Imaizumi T., Yoo J.Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 2008;82(3):1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 2005;102(29):10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang Y., Zhong G., Zhu L., Liu X., Shan Y., Feng H., Bu Z., Chen H., Wang C. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 2010;184(10):5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- 74.Mirzalieva O., Juncker M., Schwartzenburg J., Desai S. ISG15 and ISGylation in human diseases. Cells. 2022;11(3) doi: 10.3390/cells11030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Villarroya-Beltri C., Guerra S., Sanchez-Madrid F. ISGylation - a key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017;130(18):2961–2969. doi: 10.1242/jcs.205468. [DOI] [PubMed] [Google Scholar]
- 76.Friedman R.M. Clinical uses of interferons. Br. J. Clin. Pharmacol. 2008;65(2):158–162. doi: 10.1111/j.1365-2125.2007.03055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campbell J.A., Lenschow D.J. Emerging roles for immunomodulatory functions of free ISG15. J. Interferon Cytokine Res. 2013;33(12):728–738. doi: 10.1089/jir.2013.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morales D.J., Lenschow D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013;425(24):4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perng Y.C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16(7):423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Speer S.D., Li Z., Buta S., Payelle-Brogard B., Qian L., Vigant F., Rubino E., Gardner T.J., Wedeking T., Hermann M., Duehr J., Sanal O., Tezcan I., Mansouri N., Tabarsi P., Mansouri D., Francois-Newton V., Daussy C.F., Rodriguez M.R., Lenschow D.J., Freiberg A.N., Tortorella D., Piehler J., Lee B., Garcia-Sastre A., Pellegrini S., Bogunovic D. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 2016;7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Osiak A., Utermohlen O., Niendorf S., Horak I., Knobeloch K.P. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell Biol. 2005;25(15):6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T., Osiak A., Levine B., Schmidt R.E., Garcia-Sastre A., Leib D.A., Pekosz A., Knobeloch K.P., Horak I., Virgin H.Wt. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U.S.A. 2007;104(4):1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lenschow D.J., Giannakopoulos N.V., Gunn L.J., Johnston C., O’Guin A.K., Schmidt R.E., Levine B., Virgin H.Wt. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 2005;79(22):13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J.H., Luo J.K., Zhang D.E. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J. Immunol. 2008;181(9):6467–6472. doi: 10.4049/jimmunol.181.9.6467. [DOI] [PubMed] [Google Scholar]
- 85.Ritchie K.J., Hahn C.S., Kim K.I., Yan M., Rosario D., Li L., de la Torre J.C., Zhang D.E. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 2004;10(12):1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 86.Kim K.I., Malakhova O.A., Hoebe K., Yan M., Beutler B., Zhang D.E. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J. Immunol. 2005;175(2):847–854. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- 87.Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y., Shuai K., Zhang D.E. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25(11):2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malakhova O.A., Yan M., Malakhov M.P., Yuan Y., Ritchie K.J., Kim K.I., Peterson L.F., Shuai K., Zhang D.E. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17(4):455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Honke N., Shaabani N., Zhang D.E., Hardt C., Lang K.S. Multiple functions of USP18. Cell Death Dis. 2016;7(11) doi: 10.1038/cddis.2016.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Basters A., Knobeloch K.P., Fritz G. USP18 - a multifunctional component in the interferon response. Biosci. Rep. 2018;38(6) doi: 10.1042/BSR20180250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lai C., Struckhoff J.J., Schneider J., Martinez-Sobrido L., Wolff T., Garcia-Sastre A., Zhang D.E., Lenschow D.J. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 2009;83(2):1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao C., Sridharan H., Chen R., Baker D.P., Wang S., Krug R.M. Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat. Commun. 2016;7:12754. doi: 10.1038/ncomms12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan W., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20(3):362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giannakopoulos N.V., Arutyunova E., Lai C., Lenschow D.J., Haas A.L., Virgin H.W. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J. Virol. 2009;83(4):1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A.V., Adimi P., Mansouri N., Okada S., Bryant V.L., Kong X.F., Kreins A., Velez M.M., Boisson B., Khalilzadeh S., Ozcelik U., Darazam I.A., Schoggins J.W., Rice C.M., Al-Muhsen S., Behr M., Vogt G., Puel A., Bustamante J., Gros P., Huibregtse J.M., Abel L., Boisson-Dupuis S., Casanova J.L. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337(6102):1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knight E., Jr., Cordova B. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J. Immunol. 1991;146(7):2280–2284. [PubMed] [Google Scholar]
- 97.Recht M., Borden E.C., Knight E., Jr. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 1991;147(8):2617–2623. [PubMed] [Google Scholar]
- 98.D’Cunha J., Knight E., Jr., Haas A.L., Truitt R.L., Borden E.C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. U.S.A. 1996;93(1):211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.D’Cunha J., Ramanujam S., Wagner R.J., Witt P.L., Knight E., Jr., Borden E.C. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 1996;157(9):4100–4108. [PubMed] [Google Scholar]
- 100.Swaim C.D., Canadeo L.A., Huibregtse J.M. Approaches for investigating the extracellular signaling function of ISG15. Methods Enzym. 2019;618:211–227. doi: 10.1016/bs.mie.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Swaim C.D., Canadeo L.A., Monte K.J., Khanna S., Lenschow D.J., Huibregtse J.M. Modulation of extracellular ISG15 signaling by pathogens and viral effector proteins. Cell Rep. 2020;31(11) doi: 10.1016/j.celrep.2020.107772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swaim C.D., Scott A.F., Canadeo L.A., Huibregtse J.M. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol. Cell. 2017;68(3):581–590 e5. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martin-Fernandez M., Bravo Garcia-Morato M., Gruber C., Murias Loza S., Malik M.N.H., Alsohime F., Alakeel A., Valdez R., Buta S., Buda G., Marti M.A., Larralde M., Boisson B., Feito Rodriguez M., Qiu X., Chrabieh M., Al Ayed M., Al Muhsen S., Desai J.V., Ferre E.M.N., Rosenzweig S.D., Amador-Borrero B., Bravo-Gallego L.Y., Olmer R., Merkert S., Bret M., Sood A.K., Al-Rabiaah A., Temsah M.H., Halwani R., Hernandez M., Pessler F., Casanova J.L., Bustamante J., Lionakis M.S., Bogunovic D. Systemic type I IFN inflammation in human ISG15 deficiency leads to necrotizing skin lesions. Cell Rep. 2020;31(6) doi: 10.1016/j.celrep.2020.107633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Biswas S., Thakur V., Kaur P., Khan A., Kulshrestha S., Kumar P. Blood clots in COVID-19 patients: simplifying the curious mystery. Med. Hypotheses. 2021;146 doi: 10.1016/j.mehy.2020.110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yan S., Wu G. Spatial and temporal roles of SARS-CoV PL(pro) -A snapshot. FASEB J. 2021;35(1) doi: 10.1096/fj.202002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U.S.A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79(7):4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Muller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587(7835):657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen X., Chou C.Y., Chang G.G. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir. Chem. Chemother. 2009;19(4):151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 112.Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q., Luo R., Liu X., Li K., Chen H., Chen Z., Xiao S. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 2011;85(8):3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Freitas B.T., Scholte F.E.M., Bergeron E., Pegan S.D. How ISG15 combats viral infection. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bailey-Elkin B.A., van Kasteren P.B., Snijder E.J., Kikkert M., Mark B.L. Viral OTU deubiquitinases: a structural and functional comparison. PLoS Pathog. 2014;10(3) doi: 10.1371/journal.ppat.1003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Proulx J., Borgmann K., Park I.W. Role of virally-encoded deubiquitinating enzymes in regulation of the virus life cycle. Int J. Mol. Sci. 2021;22(9) doi: 10.3390/ijms22094438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bailey-Elkin B.A., Knaap R.C.M., Kikkert M., Mark B.L. Structure and function of viral deubiquitinating enzymes. J. Mol. Biol. 2017;429(22):3441–3470. doi: 10.1016/j.jmb.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murira A., Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crow M.K., Ronnblom L. Type I interferons in host defence and inflammatory diseases. Lupus Sci. Med. 2019;6(1) doi: 10.1136/lupus-2019-000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu H., Li S., Yang X., Wang X., Li Y., Wang C., Chen L., Chang H. Porcine ISG15 modulates the antiviral response during pseudorabies virus replication. Gene. 2018;679:212–218. doi: 10.1016/j.gene.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 121.Li C., Wang Y., Zheng H., Dong W., Lv H., Lin J., Guo K., Zhang Y. Antiviral activity of ISG15 against classical swine fever virus replication in porcine alveolar macrophages via inhibition of autophagy by ISGylating BECN1. Vet. Res. 2020;51(1):22. doi: 10.1186/s13567-020-00753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singh P.K., Singh S., Farr D., Kumar A. Interferon-stimulated gene 15 (ISG15) restricts Zika virus replication in primary human corneal epithelial cells. Ocul. Surf. 2019;17(3):551–559. doi: 10.1016/j.jtos.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu G., Lee J.H., Parker Z.M., Acharya D., Chiang J.J., van Gent M., Riedl W., Davis-Gardner M.E., Wies E., Chiang C., Gack M.U. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021;6(4):467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holthaus D., Vasou A., Bamford C.G.G., Andrejeva J., Paulus C., Randall R.E., McLauchlan J., Hughes D.J. Direct antiviral activity of IFN-stimulated genes is responsible for resistance to paramyxoviruses in ISG15-deficient cells. J. Immunol. 2020;205(1):261–271. doi: 10.4049/jimmunol.1901472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Osei Kuffour E., Konig R., Haussinger D., Schulz W.A., Munk C. ISG15 deficiency enhances HIV-1 infection by accumulating misfolded p53. mBio. 2019;10(4) doi: 10.1128/mBio.01342-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhushan J., Radke J.B., Perng Y.C., McAllaster M., Lenschow D.J., Virgin H.W., Sibley L.D. ISG15 connects autophagy and IFN-gamma-dependent control of toxoplasma gondii Infection in human cells. mBio. 2020;11(5) doi: 10.1128/mBio.00852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tran P.T., Chiramel A.I., Johansson M., Melik W. Roles of ESCRT proteins ALIX and CHMP4A and their interplay with interferon-stimulated gene 15 during tick-borne flavivirus infection. J. Virol. 2022;96(3) doi: 10.1128/JVI.01624-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim M.S., Kim K.H. Effect of CRISPR/Cas9-mediated knockout of either Mx1 or ISG15 gene in EPC cells on resistance against VHSV infection. Fish. Shellfish Immunol. 2019;93:1041–1046. doi: 10.1016/j.fsi.2019.08.058. [DOI] [PubMed] [Google Scholar]
- 129.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Vol. 596. 2021. Highly accurate protein structure prediction with AlphaFold; pp. 583–589. (Nature). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dong R., Peng Z., Zhang Y., Yang J. mTM-align: an algorithm for fast and accurate multiple protein structure alignment. Bioinformatics. 2018;34(10):1719–1725. doi: 10.1093/bioinformatics/btx828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]