Abstract
Cells of the gram-negative bacterium Ralstonia sp. strain SBUG 290 grown in the presence of biphenyl are able to cooxidize dibenzofuran which has been 1,2-hydroxylated. Meta cleavage of the 1,2-dihydroxydibenzofuran between carbon atoms 1 and 9b produced 2-hydroxy-4-(3′-oxo-3′H-benzofuran-2′-yliden)but-2-enoic acid, which was degraded completely via salicylic acid. The presence of these intermediates indicates a degradation mechanism for dibenzofuran via lateral dioxygenation by Ralstonia sp. strain SBUG 290.
The search for microorganisms able to grow with dibenzofuran, a model compound for highly toxic environmental pollutants like polychlorinated dibenzofurans, as a sole source of carbon and energy has led to the isolation of several bacterial strains belonging to the genera Sphingomonas, Brevibacterium (Terrabacter), and Staphylococcus (4, 6, 7, 12, 16, 20). Degradation of dibenzofuran starts with an oxygenolytic attack at the angular position 4 and 4a adjacent to the ether bridge, resulting in the formation of 2,2′,3-trihydroxybiphenyl. This intermediate, in turn, is transformed by meta cleavage to a 2-hydroxyphenyl hexadienoic acid derivative and salicylic acid (18). There are also reports of cometabolic activities and unspecific cooxidation of dibenzofuran derivatives by biphenyl- or naphthalene-degrading bacteria. Aromatic ring fission of such hydroxylated compounds was assumed (2, 5) but was demonstrated in only one case (17).
This paper describes the formation and identification of a ring fission product and further metabolites during cometabolic degradation of dibenzofuran by the biphenyl-utilizing bacterium Ralstonia sp. strain SBUG 290, which indicate an alternative degradation pathway.
The bacterial strain SBUG 290 was isolated from compost soil by enrichment cultures with 4-chlorobiphenyl as a growth substrate. It was assigned to the genus Ralstonia on the basis of physiological data (9). Additionally, strain SBUG 290 was analyzed for 16S rRNA sequences by E. Stackebrandt and coworkers (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) and was suggested to be closely related to Ralstonia eutropha. However, DNA-DNA hybridization experiments demonstrated a similarity of only 34% with Ralstonia eutropha DSM 531T. Therefore, this strain represents a new species of the genus Ralstonia. For incubations with dibenzofuran, cells were grown with biphenyl (10) up to an optical density at 600 nm (OD600) of about 2, harvested by centrifugation (30 min, 20,000 × g), washed with sodium phosphate buffer (0.067 M, pH 7.0), and resuspended in mineral salts medium to obtain an OD600 of 5.0. The formation of metabolites from dibenzofuran, added to the medium as solid matter (1.49 mM), was determined by analyzing the aqueous culture supernatant with high-performance liquid chromatography (HPLC) according to the method of Hammer et al. (8). For the determination of dibenzofuran consumption, whole cultures were removed and frozen at −20°C. At the end of the incubation, all flasks were thawed and shaken for 5 min with 2 ml of trichloromethane. One-microliter samples of the trichloromethane phase were directly analyzed by gas chromatography (GC) (8). Controls were carried out with heat-killed cells and with cells not exposed to dibenzofuran. The data are reported as mean values for two separate experiments with replicate batch cultures.
Metabolites were isolated from 500-ml flasks containing 100 ml of the biphenyl-grown cell suspension and 0.02% (1.2 mM) dibenzofuran by extraction of the centrifuged supernatant with ethyl acetate at pHs of 7 and 2 (8). The purification of metabolite I was carried out by washing the residue of the acidic extract several times with small amounts of methanol because the metabolite showed only limited solubility in solvents less polar than water. Before GC-mass spectrometry (GC-MS) analyses (8), the acid-extractable compounds were derivatized by methylation (3) in a microapparatus (Aldrich, Steinheim, Germany) or by silylation with N,O-bis(trimethylsilyl)-trifluoroacetamide and -trimethylchlorosilane (Silyl 991; Macherey-Nagel, Düren, Germany). Direct-insertion mass spectra were recorded on an HP5989A MS engine (Hewlett-Packard, Bad Homburg, Germany). The 1H and 13C nuclear magnetic resonance (NMR) experiments were carried out on a DRX 500 instrument (Bruker, Karlsruhe, Germany) in deuterated methanol or benzene solution. Dibenzofuran was purchased from Aldrich. 1,2-Dihydroxydibenzofuran was prepared according to the method of Tashiro et al. (18).
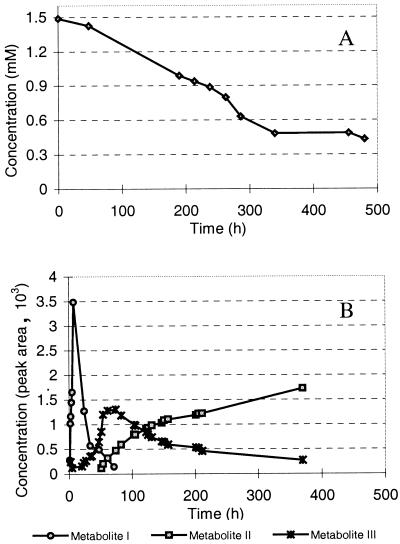
Ralstonia sp. strain SBUG 290 was able to use the aromatic compounds biphenyl and 4-chlorobiphenyl as well as salicylic acid, benzoic acid, and phenol for growth, whereas some conventional substrates, like glucose, were not metabolized. Ralstonia sp. strain SBUG 290 growing in mineral salts medium consumed about 0.122 mmol of biphenyl liter−1 h−1, reaching an OD600 of 3 within 48 h. Apart from small amounts of benzoic acid, no other metabolites accumulated. Dibenzofuran did not support growth but was metabolized rapidly by biphenyl-grown cells at a rate of up to 0.0029 mmol liter−1 h−1 (Fig. 1). No induction of dibenzofuran-oxidizing enzymes was observed with nonaromatic growth substrates, like nutrient broth. The consumption of dibenzofuran was accompanied by a change of color in the medium to bright orange, a change in the absorption maximum of the culture fluid at 462 nm, and the formation of at least 11 metabolites detected by HPLC, with retention times (RT) of 4 to 12 min (dibenzofuran RT = 13.9 min). Metabolite I, with an RT of 8.7 min, accumulated first and at high concentrations (Fig. 1). At least 10% of the dibenzofuran added was excreted into the culture medium as metabolite I for a short period before being transformed completely. Within that time, two other major metabolites, named II and III (RT = 4.3 and 6.5 min, respectively), accumulated over an incubation period of more than 10 days. Furthermore, all four monohydroxylated dibenzofurans were found in the incubation medium. These other dibenzofurans were identified by HPLC and GC-MS and may possibly be formed from the initially produced cis-1,2- and cis-3,4-dihydrodiols, as described for the formation of 9-fluorenol from fluorene by Resnick and Gibson (15). Additionally, salicylic acid was identified by HPLC and GC-MS analyses of the ethyl acetate extract. After more than 30 days, none of these metabolites was detectable.
FIG. 1.
Decrease of dibenzofuran concentration (A) and formation of the major metabolites, I, II, and III, (B) during incubation with biphenyl-grown cells of Ralstonia sp. strain SBUG 290 (OD600 = 5.0). In control experiments with heat-killed cells, no decrease of substrate concentration in the culture medium occurred (data not shown).
The kinetics of formation and the concentration of metabolite I correlated with the orange coloration of the medium, although no absorption maximum at 462 nm in the UV spectrum was observed under the HPLC conditions (pH 3) used. However, purified metabolite I dissolved in phosphate buffer showed a pH-dependent shift (pH 3→pH 7) in the absorption maximum from 400 to 462 nm. This shift indicates a keto-enol tautomerism of a meta-cleavage product, similar to that which is well known from the biphenyl meta-cleavage degradation pathway (1, 12).
The formation of yellow degradation products during cometabolic degradation of dibenzofuran was described previously by Cerniglia et al. (2) for the naphthalene- and biphenyl-degrading Beijerinckia sp. strain B8/36. This strain oxidizes dibenzofuran to 2,3-dihydroxy-2,3-dihydrodibenzofuran and 1,2-dihydroxy-1,2-dihydrodibenzofuran, which are converted to 2,3-dihydroxydibenzofuran and 1,2-dihydroxydibenzofuran. Cometabolic ring fission was also assumed, based on the yellow coloration of the medium during dibenzofuran degradation by glucose-grown cells of Pseudomonas sp. strain HL7b, but neither a description of the mechanism nor the structure of the product was given (5).
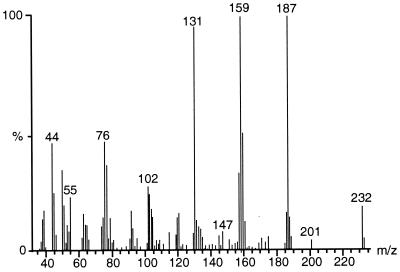
For structural identification of metabolite I, a mass spectrum (Fig. 2) was recorded by direct insertion, because no signal was obtainable by GC-MS analysis. The mass spectrum revealed a molecular weight of 232, which is in accordance with the molecular formula of C12H8O5. The fragmentation to m/z 187 (M+ −45) indicates the loss of a carboxyl group, and m/z 76 indicates the presence of an aromatic ring. After methylation of this compound, two derivatives were obtained with nearly identical mass spectra but different retention times (25.2 and 25.5 min) in GC-MS analysis. The investigation of the derivatives by high-resolution MS revealed a molecular weight of 260.06808 (260.06847 was calculated for the molecular formula of C14H12O5). This finding indicates that in addition to the carboxyl group a hydroxyl group is present in the molecule. Preparative separation of these two derivatives was not possible.
FIG. 2.
Mass spectrum of purified dibenzofuran metabolite I. Diagnostic signals are present at m/z 232 (M+), m/z 187 (M+ − COOH), m/z 159 (M+ − COOH − CO), m/z 131 (M+ − COOH − 2 CO), and m/z 76 (C6H4+).
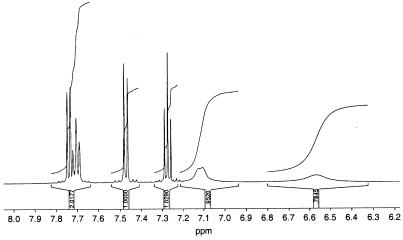
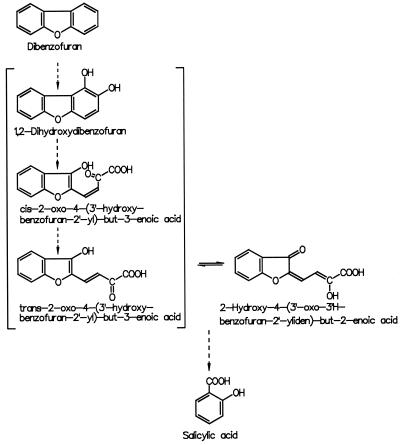
The 1H NMR spectrum of the unmethylated metabolite I (Fig. 3) showed a very diffuse resonance of the methylene protons, indicating a labile molecule forming isomers and most likely showing keto-enol tautomerism. To suppress isomerization and to obtain interpretable NMR spectra, derivatization of the enol structure was performed. Methylation yielded a mixture not suitable for investigation but silylation was achieved without by-products. For exact structure elucidation, two-dimensional NMR experiments were carried out for the determination of 1H-1H connectivity and direct as well as long-range 1H-13C connectivity of the silylated metabolite. On the basis of these experiments, all C and H atoms were unambiguously assigned due to chemical shifts, long-range couplings, and coupling constants (Table 1). Thus, the structure of metabolite I was determined to be 2-hydroxy-4-(3′-oxo-3′H-benzofuran-2′-yliden)but-2-enoic acid (HOBB). To explore the origin of HOBB, the possible ring cleavage substrate 1,2-dihydroxydibenzofuran was added to biphenyl-grown cells. The typical coloration of the culture supernatant was observed within 2 min and was accompanied by the formation of HOBB. In these incubation assays metabolites II and III were also observed. HOBB was degraded completely within 24 h, but no metabolite accumulation was detectable in the aqueous supernatant by HPLC. Because metabolites II and III (which were unstable during the purification procedure and therefore could not be further characterized) were not observed during degradation of HOBB, a second transformation pathway for dibenzofuran can be assumed. The formation of salicylic acid from both substrates was proven by GC-MS analysis of the organic extracts. Further investigations showed that Ralstonia sp. strain SBUG 290 is able to cleave 1,2-dihydroxydibenzofuran to produce HOBB after growth on nutrient broth alone and does not require induction by biphenyl.
FIG. 3.
1H NMR spectrum of metabolite I in CD3OD with dimethyl sulfoxide as the internal standard. The two doublets (δ = 7.74 and 7.47 ppm) and two triplets (δ = 7.27 and 7.70 ppm) show the proton signals of the aromatic ring. The diffuse resonances at δ of 7.12 and 6.56 ppm belong to the methylene protons of the cleaved ring.
TABLE 1.
1H NMR (500 MHz) and 13C NMR (126 MHz) results with the silylated metabolite C6D6
| δ (ppm) | Proton assignment | J (Hz) | δ (ppm) | Atom assignment |
|---|---|---|---|---|
| 6.56 | 5′-H, dd | 3J4′,5′ = 7.3; 4J4′,6′ = 8.5 | 106.9 | C-4 |
| 6.68 | 7′-H, d | 3J6′,7′ = 8.5 | 112.6 | C-3 |
| 6.88 | 6′-H, dd | 3J6′,7′ = 8.5; 4J4′,6′ = 8.5 | 112.7 | C-7′ |
| 7.38 | 4-H, d | 3J3,4 = 12.2 | 122.9 | C-3′a |
| 7.44 | 3-H, d | 3J3,4 = 12.2 | 123.4 | C-5′ |
| 7.53 | 4′-H, d | 3J4′,5′ = 7.3 | 124.7 | C-4′ |
| 136.4 | C-6′ | |||
| 147.7 | C-2 | |||
| 148.9 | C-2′ | |||
| 164.1 | C-1 | |||
| 165.7 | C-7′a | |||
| 183.1 | C-3′ |
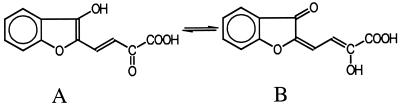
A similar attempt to characterize a yellow ring fission product formed from dibenzofuran during cooxidation was carried out by Selifonov et al. for naphthalene-grown Pseudomonas strains (17). Because of the instability of the pure ring cleavage product, the authors used the methylated compound for separation and structural characterization by GC-MS and NMR. The substance was shown to be the methyl ester of one of the two possible primary products formed upon meta cleavage of 1,2-dihydroxydibenzofuran between carbon atoms 1 and 9b (Fig. 4). By comparing the mass spectra of the methylated compounds produced, we deduced that the same ring cleavage mechanism is utilized by Ralstonia sp. strain SBUG 290 and the Pseudomonas strains. In Pseudomonas, further degradation of the ring fission product was not observed because the hydrolase encoded by the nahE gene of the nah operon was not able to cleave pyruvate from this molecule (13). In contrast, Ralstonia sp. strain SBUG 290 was able to degrade HOBB to salicylic acid. Considering that salicylic acid can be used as a growth substrate, it remains unclear why Ralstonia sp. strain SBUG 290 is not able to use dibenzofuran as a growth substrate. Cometabolic degradation of chlorinated dibenzofuran by biphenyl-utilizing Burkholderia (previously Alcaligenes) sp. strain JB1 was reported by Parsons et al. (14). The identification of 3-chloro-2′,3,3′-trihydroxybiphenyl and 5-chlorosalicylic acid (13) shows that Burkholderia sp. strain JB1 degrades 2-chlorodibenzofuran by the same pathway as described for bacteria able to use dibenzofuran as the sole source of carbon and energy. Until now it was assumed that only bacteria using dibenzofuran as the sole source for carbon and energy are able to completely degrade dibenzofuran (21). Our work shows that a complete degradation of dibenzofuran via lateral dioxygenation and meta cleavage of the aromatic structure (Fig. 5) is also possible during cometabolic processes.
FIG. 4.
Structures of the two possible primary products formed upon meta cleavage of 1,2-dihydroxydibenzofuran between carbon atoms 1 and 9b. (A) Structure of the methyl ester, proven by Selifonov et al. (17). (B) Metabolite I, HOBB.
FIG. 5.
Proposed pathway of dibenzofuran degradation by Ralstonia sp. strain SBUG 290.
Acknowledgments
This study was supported by grant no. 1450638 R9 from the Bundesministerium für Forschung und Technologie and by the Grube-Land-und Umwelttechnik GmbH, Brake, Germany.
We thank R. Thede, Institute of Physical Chemistry, University of Greifswald, for analysis of mass spectra after direct insertion; J. Deck, National Center for Toxicological Research, Food and Drug Administration, Jefferson, Ark., for recording of 1H NMR spectra; and E. Stackebrandt and J. Burghard (DSMZ) for performing rRNA analyses and DNA reassociation experiments. For editorial help, we thank R. Jack.
Footnotes
Dedicated to Günter Grube and his contribution to science and technology.
REFERENCES
- 1.Catelani D, Colombi A. Metabolism of biphenyl. Biochem J. 1974;143:431–434. doi: 10.1042/bj1430431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerniglia C E, Morgan J C, Gibson D T. Bacterial and fungal oxidation of dibenzofuran. Biochem J. 1979;180:175–185. doi: 10.1042/bj1800175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Boer T D, Backer H J. Diazomethane. In: Leonard N J, editor. Organic synthesis. Vol. 36. New York, N.Y: John Wiley & Sons; 1956. pp. 14–16. [Google Scholar]
- 4.Engesser K H, Strubel V, Christoglou K, Fischer P, Rast H G. Dioxygenolytic cleavage of aryl ether bonds: 1,10-dihydrodiol-1,10-dihydroxyfluoren-9-one, a novel arene dihydrodiol, as evidence for angular dioxygenation of dibenzofuran. FEMS Microbiol Lett. 1989;65:205–210. doi: 10.1016/0378-1097(89)90392-3. [DOI] [PubMed] [Google Scholar]
- 5.Foght J M, Westlake D W S. Degradation of polycyclic aromatic hydrocarbons and aromatic heterocycles by a Pseudomonas species. Can J Microbiol. 1988;34:1135–1141. doi: 10.1139/m88-200. [DOI] [PubMed] [Google Scholar]
- 6.Fortnagel P, Harms H, Wittich R-M, Francke W, Krohn S, Meyer H. Cleavage of dibenzofuran and dibenzodioxin ring systems by a Pseudomonas bacterium. Naturwissenschaften. 1989;76:222–223. doi: 10.1007/BF00627694. [DOI] [PubMed] [Google Scholar]
- 7.Fortnagel P, Harms H, Wittich R-M, Krohn S, Meyer H, Sinnwell V, Wilkes H, Francke W. Metabolism of dibenzofuran by Pseudomonas sp. strain HH69 and the mixed culture HH27. Appl Environ Microbiol. 1990;56:1148–1156. doi: 10.1128/aem.56.4.1148-1156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer E, Krowas D, Schäfer A, Specht M, Francke W, Schauer F. Isolation and characterization of a dibenzofuran-degrading yeast: identification of oxidation and ring cleavage products. Appl Environ Microbiol. 1998;64:2215–2219. doi: 10.1128/aem.64.6.2215-2219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. [Google Scholar]
- 10.Hundt K, Wagner M, Becher D, Hammer E, Schauer F. Effect of selected environmental factors on degradation and mineralization of biaryl compounds by the bacterium Ralstonia pickettii in soil and compost. Chemosphere. 1998;36:2321–2335. doi: 10.1016/s0045-6535(97)10201-6. [DOI] [PubMed] [Google Scholar]
- 11.Lunt D, Evans C. The microbial metabolism of biphenyl. Biochem J. 1970;118:54–55. doi: 10.1042/bj1180054pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monna L, Omori T, Kodama T. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl Environ Microbiol. 1993;59:285–289. doi: 10.1128/aem.59.1.285-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons J R, de Bruijne J A, Weiland A R. Biodegradation pathway of 2-chlorodibenzo-p-dioxin and 2-chlorodibenzofuran in the biphenyl-utilizing strain JB1. Chemosphere. 1998;37:1915–1922. doi: 10.1016/s0045-6535(98)00258-6. [DOI] [PubMed] [Google Scholar]
- 14.Parsons J R, Ratsak C, Siekerman C. Biodegradation of chlorinated dibenzofurans by an Alcaligenes strain. In: Hutzinger O, Fiedler H, editors. Dioxin '90, organohalogen compounds 1, EPRI seminar. Bayreuth, Germany: ECO-Informa Press; 1990. pp. 377–380. [Google Scholar]
- 15.Resnick S M, Gibson D T. Regio- and stereospecific oxidation of fluorene, dibenzofuran, and dibenzothiophene by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl Environ Microbiol. 1996;62:4073–4080. doi: 10.1128/aem.62.11.4073-4080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid A, Rothe B, Altenbuchner J, Ludwig W, Engesser K-H. Characterization of three distinct extradiol dioxygenases involved in mineralization of dibenzofuran by Terrabacter sp. strain DPO360. J Bacteriol. 1997;179:53–62. doi: 10.1128/jb.179.1.53-62.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selifonov S A, Slepenkin A V, Adanin V M, Nefedova M Y, Starovoitov I I. Oxidation of dibenzofuran by Pseudomonas strains harbouring plasmids of naphthalene degradation. Mikrobiologiya. 1991;60:67–71. [PubMed] [Google Scholar]
- 18.Tashiro M, Yoshiya H, Fukata G. Bromination of 2,2′-dihydroxy-3,3′,5,5′-tetra-tert-butylbiphenyl and preparation of hydroxydibenzofurans. J Org Chem. 1982;47:4425–4429. [Google Scholar]
- 19.Wilkes H, Francke W, Wittich R-M, Harms H, Schmidt S, Fortnagel P. Mechanistic investigations on microbial degradation of diaryl ethers. Naturwissenschaften. 1992;79:269–271. [Google Scholar]
- 20.Wilkes H, Wittich R-M, Timmis K M, Fortnagel P, Francke W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1996;62:367–371. doi: 10.1128/aem.62.2.367-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittich R-M. Degradation of dioxin-like compounds by microorganisms. Appl Microbiol Biotechnol. 1998;49:489–499. doi: 10.1007/s002530051203. [DOI] [PubMed] [Google Scholar]