Abstract
The diversity of ammonia-oxidizing bacteria in aquatic sediments was studied by retrieving ammonia monooxygenase and methane monooxygenase gene sequences. Methanotrophs dominated freshwater sediments, while β-proteobacterial ammonia oxidizers dominated marine sediments. These results suggest that γ-proteobacteria such as Nitrosococcus oceani are minor members of marine sediment ammonia-oxidizing communities.
Recent studies of ammonia-oxidizing bacteria have used PCR primers that target β-proteobacteria such as Nitrosomonas and Nitrosospira (17). This approach ignores the ammonia oxidizing bacteria in the γ subdivision of the Proteobacteria. This is unfortunate since γ-proteobacteria such as Nitrosococcus oceani may be important ammonia oxidizers, especially in marine habitats (18, 19), where β-proteobacteria are rarely found (3, 13).
To test the hypothesis that γ-proteobacteria dominate marine sediment ammonia-oxidizing communities, we used a conserved primer set to amplify the ammonia monooxygenase (amoA) and methane monooxygenase (pmoA) genes from all known ammonia oxidizers and methanotrophs. These primers anneal to conserved regions of these genes (9) and have been applied to soil and freshwater habitats (e.g., see references 2, 6, and 10). Since the topology of the amoA-pmoA phylogenetic tree is in good agreement with the corresponding 16S ribosomal DNA (rDNA) tree (14), we can tentatively identify the dominant ammonia oxidizers using this approach. We constructed clone libraries of amoA and pmoA genes from freshwater and marine sediments. Our marine clone libraries were dominated by β-proteobacterial amoA genes, leading us to reject the hypothesis that γ-proteobacteria dominate ammonia-oxidizing communities in marine sediments.
Sites and sampling.
Marine sediments were collected from three well-characterized Pacific Northwest sampling sites. Two were located along the Washington continental margin (Pacific Ocean) and one was located in Puget Sound, Wash. As a control, freshwater sediment was collected from Wintergreen Lake in Michigan. Sample designations (in parentheses) and specific locations are as follows: Washington continental margin site 301 (WC301), 46°48.60′N, 124°37.20′W, 119-m water depth; Washington continental margin site 306 (WC306), 48°29.60′N, 126°43.22′W, 630-m water depth; Puget Sound (PS), 47°43.50′N, 122°23.90′W, 182-m water depth; Wintergreen Lake (FW), 42°23′58"N, 85°23′00"W, 3-m water depth. The report of Braker et al. (1) contains more-detailed information about the sampling sites. Surface (depth, 0 to 2 cm) sediment samples were collected and stored frozen until DNA extraction. Oxygen was available for ammonia oxidation at the freshwater sampling site (by direct measurement), and ammonia oxidation rates at sites WC301, WC306, and PS were 1.78, 0.29, and 2.24 mmol NH4+ · m2 · day−1, respectively (5).
Community analysis.
DNA was extracted from sediments using the procedure of Gray and Herwig (4). We amplified amoA and pmoA functional genes using PCR primers that were slightly modified from those described by Holmes et al. (9) to favor γ-proteobacteria (Table 1). PCR reaction conditions were similar to those used by Holmes et al. (9), with the exception of adding 200 ng of bovine serum albumin (Roche Molecular Biochemicals, Indianapolis, Ind.) per μl to the reaction mixture. After amplification, the 530-bp product was excised from a 2% agarose gel and purified using the Qiaquick gel extraction kit (Qiagen, Valencia, Calif.) to ensure a properly sized insert. Clone libraries of the PCR products were constructed for each sediment using the TA cloning kit (Invitrogen, Carlsbad, Calif.). Clones containing a 530-bp insert were screened by restriction fragment length polymorphism (RFLP) analysis as previously described (20) using the restriction enzymes HhaI (GCG↓G; Gibco BRL, Rockville, Md.) and MspI (C↓CGG; Gibco BRL). Unique RFLP patterns were enumerated, and nucleotide sequence data were obtained from the 10 most abundant restriction fragment types in each sediment sample. Sequences were initially compared to all GenBank database sequences using the BLAST algorithm found at the GenBank web site (http://www.ncbi.nlm.nih.gov/BLAST/). Those sequences similar to amoA or pmoA were aligned and compiled in the Genetic Data Environment software package. A neighbor-joining phylogenetic tree was constructed from derived amino acid sequences using the programs SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE of Phylip (version 3.5) found at the Phylip web site (http: //evolution.genetics.washington.edu/phylip.html).
TABLE 1.
Primer and target amoA and pmoA gene sequences
| Primer pair or target | No. of repsa | Primer or target sequence
|
|
|---|---|---|---|
| Forward | Reverse | ||
| Primer pairs | |||
| Original primer pairb | 5′ GGN GAC TGG GAC TTC TGG 3′ | 5′ GAA SGC NGA GAA GAA SGC 3′ | |
| Modified primer pairc | 5′ GGN GAC TGG GAC TTC TGG 3′ | 5′ –AA VGC VGA GAA GAA WGC 3′ | |
| Targetsd | |||
| Consensus γ-amoA | 2 | 5′ GGG GAC TGG GAY TTC TGG 3′ | 5′ AAA ACC CGM RAA RAA RGC 3′ |
| Consensus β-amoA | 32 F, 68 R | 5′ GGH GAC TGG GAY TTC TGG 3′ | 5′ RAA NCC GGM GAA GAA BGC 3′ |
| Consensus γ-pmoA | 5 | 5′ GGG GAC TGG GAC TTC TGG 3′ | 5′ GAA GGC GGA GAA GAA BGC 3′ |
Number of available representatives (reps) from each major subdivision that were compared to create the primer sequence. F, forward (corresponding to Nitrosomonas europaea amoA positions 172 to 189); R, reverse (corresponding to N. europaea amoA positions 665 to 681).
For a description, see reference 9.
Primer pair used in this study.
Consensus sequences were constructed by comparing all available representatives from each proteobacterial subdivision. If 80% or more of the sequences had an individual nucleotide at a position, that base is displayed. Otherwise, the ambiguous base is displayed. No α-proteobacterial pmoA sequences are available for these regions. B, cytosine, thymine, or guanine; H, adenine, cytosine, or thymine; M, adenine or cytosine; N, any nucleotide; R, adenine or guanine; S, cytosine or guanine; V, adenine, cytosine, or guanine; W, adenine or thymine; Y, cytosine or thymine.
Community profiles of ammonia oxidizing bacteria.
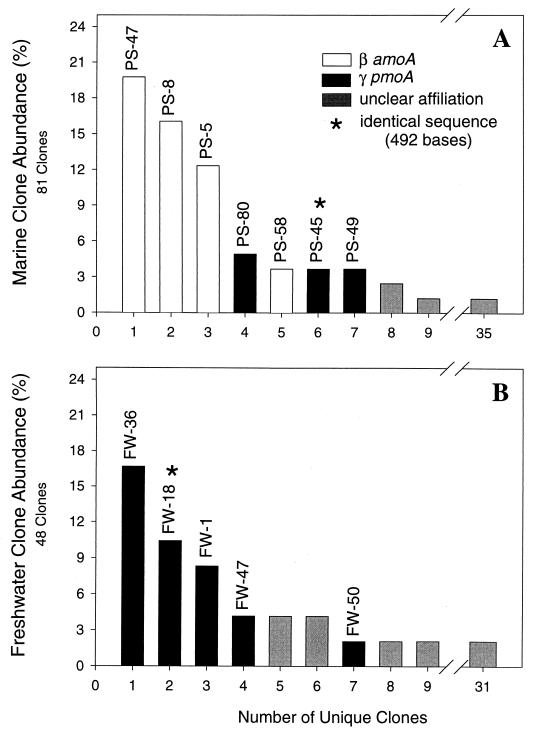
The Puget Sound clone library was representative of the other marine libraries. This library was dominated by three restriction fragment types but also contained diverse, rare fragments (Fig. 1A). Compared to the sequences in the GenBank database, most of these clones (75%) were amoA sequences most similar to β-proteobacterial ammonia oxidizers (Fig. 1A). Three of these sequences, PS-5, PS-8, and PS-47, accounted for 70% of the identified clones. Nine additional clones (16%) were associated with pmoA sequences similar to γ-proteobacterial methanotrophs. The five remaining marine clones (9%) displayed no similarity to known amoA or pmoA sequences. In contrast, the freshwater clone library (48 total clones) was composed entirely of γ-proteobacterial pmoA sequences from methanotrophs (Fig. 1B). Interestingly, one pmoA sequence type was found in both freshwater and marine habitats (FW-18 and PS-45, respectively). The sequences of FW-18 and PS-45 were identical over the 490 available nucleotides. We failed to detect γ-proteobacterial amoA sequences similar to the Nitrosococcus oceani sequence in either habitat.
FIG. 1.
Abundance of RFLP types in Puget Sound (A) and Wintergreen Lake (B) sediment clone libraries. Representative sequence types appear above each bar, and the phylogenetic affiliations and functional gene types are shown.
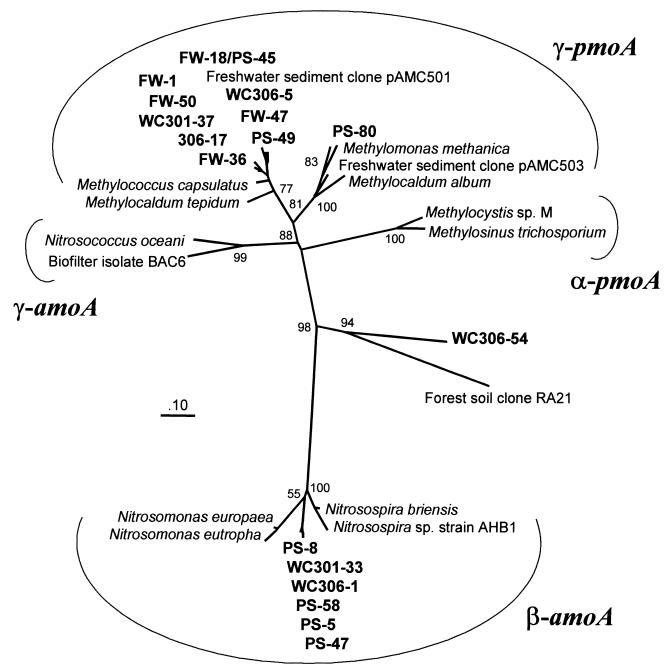
The evolutionary relationships among our retrieved sequences are explored in Fig. 2. This phylogenetic tree contains all marine sequences, including those we retrieved from the Washington continental margin sediments. Marine amoA sequences clustered together, but distinct from Nitrosomonas and Nitrosospira sequences. Most marine pmoA sequences were similar to Methylococcus capsulatus, a type I methanotroph. This cluster includes freshwater sediment sequences retrieved from Lake Washington in Seattle, Wash. (represented by clone pAMC501 [2]). One clone (WC306-54) showed no clear affiliation to amoA or pmoA sequences, indicating a possibly novel lineage. Although this clone appears to cluster with a sequence retrieved from a beech forest soil in Denmark (clone RA21) (10), the nucleotide and amino acid sequence similarities are very low (58.0 and 55.7%, respectively). Freshwater clones clustered with type I methanotrophs. Again, no γ-proteobacterial amoA sequences were retrieved.
FIG. 2.
Neighbor-joining phylogenetic tree of derived amoA and pmoA amino acid sequences. Major proteobacterial lineages are denoted. Trees are based on comparison of 163 amino acids. This figure also contains sequences from clone libraries constructed from sediment samples collected from the Washington continental margin. Bootstrap values from 100 resamplings are shown, except for those nodes with values below 50 or where branching orders between the consensus tree and this tree were inconsistent.
Marine sediments contain β-proteobacterial ammonia oxidizers.
We failed to detect any γ-proteobacterial amoA sequences, leading us to reject the hypothesis that γ-proteobacteria dominate ammonia-oxidizing communities in marine sediments. Our clone libraries were instead populated by β-proteobacterial amoA sequences. These sequences form a distinct branch, evolutionarily separate from the Nitrosomonas and Nitrosospira lineages (Fig. 2). Stephen et al. (16) found a similar pattern of ammonia oxidizer diversity in polluted marine sediments. Using PCR primers specific to the β-proteobacterial ammonia oxidizers, these authors discovered two clusters of environmental 16S rDNA sequences (clusters 1 and 5) distinct from the Nitrosomonas and Nitrosospira lineages. These novel clusters contain no cultivated representatives. The functional amoA genes we retrieved from marine sediments may come from the same organisms as the 16S rDNA sequences found in cluster 1 or cluster 5 ammonia oxidizers.
Aquatic sediments contain type I (12) or type II (11) methanotrophs, or both (2, 6). Our pmoA sequences were exclusively type I (γ-proteobacterial). We were surprised to discover remarkable similarity between freshwater and marine pmoA sequences, including an identical sequence in both libraries (clones PS-45 and FW-18). Salinity is a strong adaptive barrier. Microorganisms in equivalent marine and freshwater habitats are generally very different (3, 13), even though they perform similar functions (8). We expected to find phylogenetically distinct clusters of freshwater and marine sequences, similar to those observed in environmental 16S rDNA surveys (7, 15, 21). Instead, pmoA sequences of freshwater and marine habitats display no evolutionary divergence.
This study should not be used to prove the importance of the β-proteobacterial ammonia oxidizers. It is likely that we are missing important community members by using selective techniques such as probing, PCR amplification, and culturing. By limiting studies to the β-proteobacterial ammonia oxidizers, we may still be ignoring important components of native bacterial communities.
Nucleotide sequence accession numbers.
The amoA and pmoA gene sequences presented in this paper were submitted to GenBank under the following accession numbers: PS-5, AF211883; PS-8, AF211884; PS-45, AF211873; PS-47, AF211885; PS-49, AF211874; PS-58, AF211886; PS-80, AF211872; WC301-33, AF211887; WC301-37, AF211877; WC306-1, AF211888; WC306-5, AF211875; WC306-17, AF211876; WC306-54, AF211889; FW-1, AF211878; FW-18, AF211879; FW-36, AF211881; FW-47, AF211880; FW-50, AF211882.
Acknowledgments
This work was supported by Department of Energy grant DE-FG02-98ER62535, with contributions from the Center for Microbial Ecology NSF Grant DEB91200006.
REFERENCES
- 1.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirS and nirK) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello A, Lidstrom M E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol. 1999;65:5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glockner F O, Fuchs B M, Amann R. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartnett H E. Organic carbon input, degradation and preservation in continental margin sediments: an assessment of the role of a strong oxygen deficient zone. Ph.D. thesis. Seattle: University of Washington; 1998. [Google Scholar]
- 6.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 8.Hobbie J E. A comparison of the ecology of planktonic bacteria in fresh and salt water. Limnol Oceanogr. 1988;33:750–764. [Google Scholar]
- 9.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microb Ecol. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 10.Holmes A J, Roslev P, McDonald I R, Iversen N, Henriksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microb Ecol. 1996;21:197–211. [Google Scholar]
- 12.Nold S C, Boschker H T S, Pel R, Laanbroek H J. Ammonium addition inhibits 13C-methane incorporation into methanotroph membrane lipids in a freshwater sediment. FEMS Microb Ecol. 1999;29:81–89. [Google Scholar]
- 13.Nold S C, Zwart G. Patterns and governing forces in aquatic microbial communities. Aquat Ecol. 1998;32:17–35. [Google Scholar]
- 14.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stahl D A, Key R, Flesher B, Smit J. The phylogeny of marine and freshwater caulobacters reflects their habitat. J Bacteriol. 1992;174:2193–2198. doi: 10.1128/jb.174.7.2193-2198.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utaker J B, Nes I F. A qualitative evaluation of the published oligonucleotides specific for the 16S rRNA gene sequences of the ammonia-oxidizing bacteria. Syst Appl Microbiol. 1998;21:72–88. doi: 10.1016/S0723-2020(98)80010-6. [DOI] [PubMed] [Google Scholar]
- 18.Ward B B. Nitrogen transformations in the Southern California Bight. Deep Sea Res. 1987;34:785–805. [Google Scholar]
- 19.Watson S W. Characteristics of a marine nitrifying bacterium, Nitrosocystis oceanus sp. n. Limnol Oceanogr. 1965;10:R274–R289. [Google Scholar]
- 20.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]
- 21.Zwart G, Hiorns W D, van Agterveld M P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]