Abstract
The Streptococcus pneumoniae Rgg144/SHP144 regulator‐peptide quorum sensing (QS) system is critical for nutrient utilization, oxidative stress response, and virulence. Here, we characterized this system by assessing the importance of each residue within the active short hydrophobic peptide (SHP) by alanine‐scanning mutagenesis and testing the resulting peptides for receptor binding and activation of the receptor. Interestingly, several of the mutations had little effect on binding to Rgg144 but reduced transcriptional activation appreciably. In particular, a proline substitution (P21A) reduced transcriptional activation by 29‐fold but bound with a 3‐fold higher affinity than the wild‐type SHP. Consistent with the function of Rgg144, the mutant peptide led to decreased utilization of mannose and increased susceptibility to superoxide generator paraquat. Pangenome comparison showed full conservation of P21 across SHP144 allelic variants. Crystallization of Rgg144 in the absence of peptide revealed a comparable structure to the DNA bound and free forms of its homologs suggesting similar mechanisms of activation. Together, these analyses identify key interactions in a critical pneumococcal QS system. Further manipulation of the SHP has the potential to facilitate the development of inhibitors that are functional across strains. The approach described here is likely to be effective across QS systems in multiple species.
Keywords: inhibitor design, quorum sensing, Rgg transcriptional regulators, Streptococcus pneumoniae, structure‐function
Streptococcus pneumoniae utilizes a network of cell‐cell communication molecules to respond to extracellular cues. The regulator‐peptide system, Rgg144/SHP144, plays a central role in this network, making it a major virulence determinant. Here we employ structure‐function studies to identify residues that contribute to the affinity and activities of Rgg144 signalling that in turn modulate pneumococcal pathogenesis.
![]()
1. INTRODUCTION
Bacterial populations can synchronize their responses to different stimuli, including nutrient sources, temperature, or acidity, through cell density‐linked signaling circuits, known as quorum‐sensing (QS) systems (Aggarwal et al., 2020; Rutherford & Bassler, 2012). QS systems coordinate the detection and processing of chemical signals and activation of cognate receptors for downstream gene expression.
In general, bacterial QS circuits rely on three basic elements: (i) production of a signaling molecule or auto‐inducing peptide, (ii) processing and/or accumulation of this molecule in the extracellular milieu, and (iii) detection of signals by cognate receptors (Aggarwal et al., 2020). The composition and structure of the signaling molecules, as well as the mechanisms of recognition and sense of the signals, are different in Gram‐positive and Gram‐negative bacteria. N‐acylhomoserine lactones (AHLs) are the most dominant signaling molecules utilized by Gram‐negative bacteria (Fetzner, 2015). On the contrary, Gram‐positive bacteria rely on small modified oligopeptides to modulate the expression of their target genes. These peptide pheromones are ribosomally synthesized, and actively transported out of producing cells by the action of specific transporters. They are often modified by proteolytic processing during the export by membrane‐bound peptidases and/or cyclization (LaSarre & Federle, 2013). Following secretion, the peptides are detected either by membrane‐bound sensor kinases on the cell surface or imported into the cell by an oligopeptide permease (Opp or Ami) after which they interact with their cognate receptors.
The Rgg (regulator gene of glucosyltransferase) family of regulators are members of the RRNPP (Rap, Rgg, NprR, PlcR, PrgX) QS systems, and are common in AT‐rich Gram‐positive bacteria, including in Streptococcaceae, Lactobacillales, and Listeriaceae (Cook & Federle, 2014; Monnet & Gardan, 2015). They are characterized by an N‐terminal helix‐turn‐helix motif (HTH), that binds to the promoter of Rgg‐regulated genes, and by a conserved C‐terminal regulatory domain (Cook & Federle, 2014). The rgg genes are located close to the ORF for short hydrophobic peptides (SHP) required for signaling. There are three distinct SHP groups that are categorized, based on their amino acid compositions and their genomic location relative to their cognate Rggs. Group I and II SHPs include an N‐terminal aspartate or glutamate, respectively, and the SHP genes are divergently transcribed in relation to their cognate Rgg‐encoding genes (Fleuchot et al., 2011). Group III SHPs also contain an N‐terminal aspartate or glutamate, but the genes coding these SHPs typically overlap with the ends of the rgg genes from which they are convergently transcribed. Recently, the structure of Streptococcus thermophilus Rgg3 (Group 1) alone and in complex with its cognate pheromone SHP3 was determined (Capodagli et al., 2020; Fleuchot et al., 2011). Furthermore, structures of two Rgg transcriptional activators, Streptococcus dysgalactiae Rgg2 and S. thermophilus Rgg3, solved in complex with their target promoters have revealed how these Rgg proteins specifically recognize their target DNA sequences (Capodagli et al., 2020; Parashar et al., 2015).
The type 2 D39 strain, used in this study, contains six rgg homologs but pneumococcal genomes can have up to 8 rgg homologs (Wang et al., 2020; Zhi et al., 2018). Of these, rgg144, rgg939, and rgg1518 are associated with shp144, shp939, and shp1518, respectively. Our previous work has shown that Rgg144, Rgg939, and Rgg1518 QS systems regulate host glycan metabolism, capsule synthesis, and virulence (Shlla et al., 2021; Zhi et al., 2018). However, the mechanisms by which these Rggs interact with their cognate pheromones and the phenotypic impact of these interactions are not known. Rgg144 is of particular interest because unlike the Rgg939 and Rgg1518 systems, it is part of the core pneumococcal genome. The regulon of this QS system is sugar‐specific, with a broad response in mannose and a limited response in galactose (Zhi et al., 2018). Unlike the previously characterized Rgg2 and Rgg3 of S. thermophilus and S. pyogenes, which are activated by Group 1 pheromones, Rgg144 is activated by a Group 3 SHP for which relatively little is known (Fleuchot et al., 2011). In this work, we have determined the structure of Rgg144 and investigated its interaction with SHP144. Using site‐directed mutagenesis we identified a peptide inhibitor of Rgg144 that binds to the Rgg but fails to induce transcription of Rgg‐controlled genes. This inhibitor prevents Rgg‐dependent transcription and its associated phenotypes.
2. RESULTS
2.1. Structure of Rgg144
Recombinant Rgg144 was produced by expression in E. coli and refolded from inclusion bodies. Gel filtration indicated that Rgg144 is a homodimer in a solution with an apparent molecular mass of ~60 kDa (monomer molecular mass = 33.9 kDa). Crystals were grown at pH 7.5 using both native Rgg144 and selenomethionine‐substituted protein, and data collection and refinement statistics for the native structure are provided in Table S1 (PDB ID: 7ZCV). The native crystals diffracted to 1.9 Å and selenomethionine crystals to ~2.5 Å. Initial phases were determined by selenium single‐wavelength anomalous diffraction and the preliminary model was used to determine the structure using the native dataset. The asymmetric unit of crystals contained a single Rgg dimer (Figure 1). Rgg144 has a tetratricopeptide‐like fold, typical of RRNPP family members (Parashar et al., 2015; Perez‐Pascual et al., 2016). The stereochemistry of the model is of high quality with over 99% of residues in Ramachandran‐favored regions. Each polypeptide is aligned parallel to its partner and comprises an N‐terminal helix‐turn‐helix domain and a C‐terminal regulatory domain featuring a pronounced groove that forms the binding site for the SHP (Fleuchot et al., 2011). The interface is extensive and spans both domains with a total buried surface area of 6648 Å2. A long loop connects the two domains enabling the helix‐turn‐helix domain of one protomer to pack against the regulatory domain of its partner as well as to the adjacent helix‐turn‐helix domain (Figure 1).
FIGURE 1.
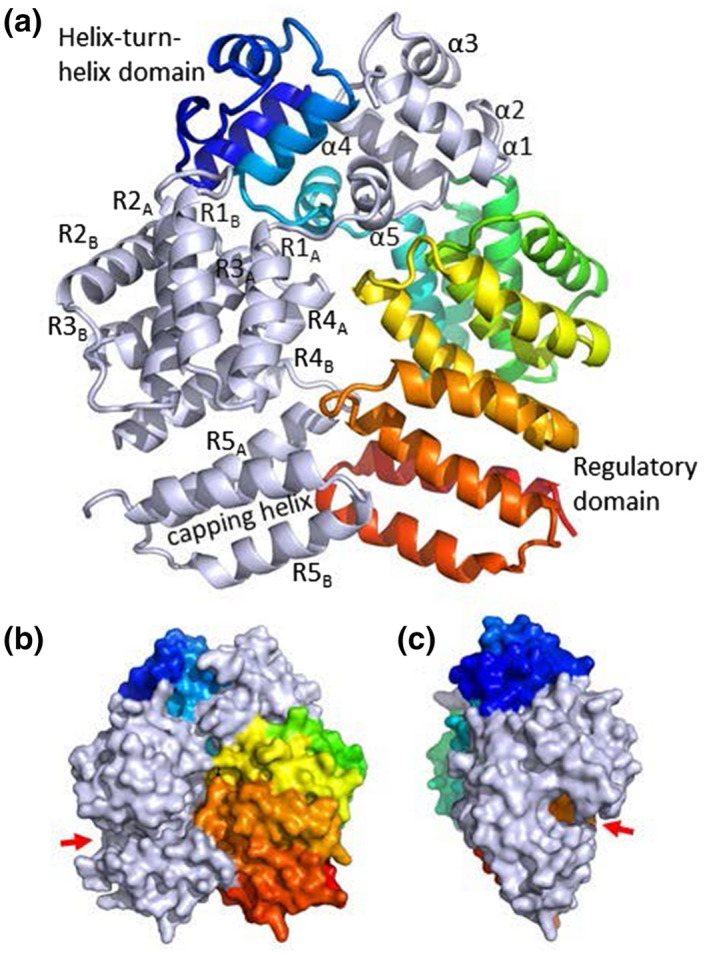
Crystal structure of Rgg144. (a) The Rgg homodimer. Polypeptides are light gray or rainbow colored, with the N‐terminus in blue and the C‐terminus in red. Helices have been numbered according to Parashar et al. (2015). The DNA‐binding helix is α3. (b) and (c) surface representation of the front and side views of the Rgg homodimer. The position of the putative peptide‐binding groove is shown by the red arrow.
Of the two other Rgg proteins for which structures are available (Capodagli et al., 2020; Parashar et al., 2015), Rgg144 shares 23.3% sequence identity with Rgg3 from S. thermophilus and 21.8% identity with Rgg2 from S. dysgalactiae (Parashar et al., 2015). Rgg144 has a similar overall structure to the unbound form of Rgg2 with a RMS deviation of 2.85 Å over 242 residues for each polypeptide. The helix‐turn‐helix domain consists of 5 α‐helices (designated α1 – α5; Figure 1a) in which helix α3 is the DNA‐binding helix. Adjacent DNA‐binding helices are separated by ~26 Å, similar to the separation observed in Rgg2 and Rgg3 bound to DNA (PDB: 6W1F and 6WIA). The C‐terminal domain comprises 11 α‐helices (designated R1A and R1B, R2A and R2B, R3A and R3B, R4A and R4B, R5A and R5B, and the capping helix), with adjacent helices running antiparallel to form a right‐handed superhelical structure with a C‐shaped conformation and with the putative SHP‐binding groove located at the center of the C (Figure 1b,c). Thus, our structure suggests conservation in the structure‐function of Rggs across streptococcal species, despite relatively low sequence identity, where the N‐termini bind DNA and control transcription of Rgg targets, and the C‐termini recognize the cognate peptides and dictate peptide specificity. The allosteric nature of the Rgg/SHP interaction involving an extensive region of contacts between components (Fleuchot et al., 2011) suggests that the SHP can be manipulated to regulate Rgg activity.
2.2. Quantifying the functional importance of SHP144 amino acid residues for Rgg144‐dependent transcription
Our previous work (Zhi et al., 2018) showed that Rgg144 drives shp144 transcription and that a 13 amino acid synthetic SHP is sufficient to stimulate this Rgg‐dependent transcription: S14EWVIVIPFLTNL26, designated SHP144, where the numbering refers to the sequence of the full‐length SHP before processing. SHP144 is considerably longer than most other peptide pheromones in the RRNPP family, which are generally between 5 and 8 residues long (Aggarwal et al., 2014; Bouillaut et al., 2008). Despite considerable efforts, we were unable to crystallize Rgg144 in the presence of its SHP. We, therefore, adopted a different strategy to probe the Rgg144‐SHP144 interaction, in which we mutated, in turn, each amino acid residue in the mature SHP144 to alanine and then measured the effect of each change on Rgg‐dependent transcription. Transcription was measured using a reporter strain containing the lacZ gene under the transcriptional control of the shp144 promoter (Pshp144), in a shp144 mutant. Complementation of this strain was tested by expressing either wild‐type or mutant shp144. Of the 13 strains expressing a modified shp144, five were completely unable to induce P shp144 ‐driven β‐galactosidase activity: those encoding the mutations W16A, V17A, I18A, I20A, and P21A (p < .0001), whereas seven mutations S14A, E15A, F22A, L23A, T24A, N25A, and L26A resulted in a significant reduction in the activity, compared to the strain containing an intact copy of shp144 (Figure 2, Table 1). The reduction in transcription observed with S14A, E15A, F22A, and L23A mutations was higher than those with T24A, N25A, and L26A, reflecting the importance of the N‐terminal end of the peptide for the activity. Interestingly, the mutation V19A had no impact on transcription, indicating that V19 of SHP144 is not essential for Rgg‐dependent transcription.
FIGURE 2.
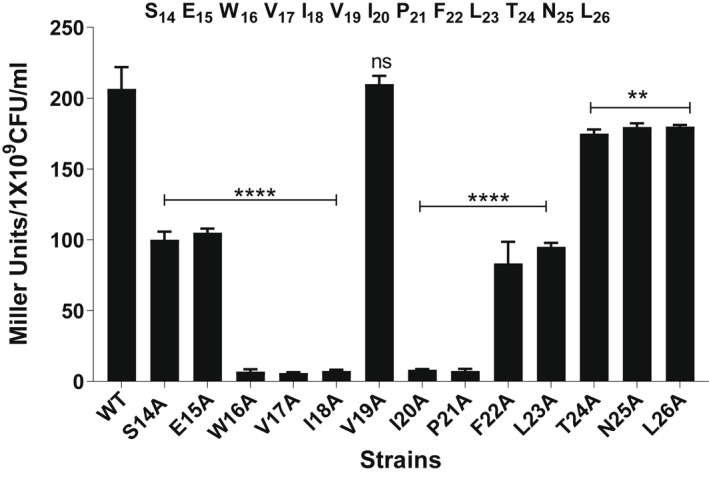
The effect of mutations to SHP144 on transcriptional activation by Rgg144 using a LacZ reporter assay. The reporter strain carrying a lacZ under the putative promoter of shp144 was genetically complemented with either wild‐type (WT) or mutant shp144 in a Δshp144 background. Strains were grown in a chemically defined medium supplemented with 55 mM mannose to late exponential phase. Rgg‐dependent transcription from the Pshp144 (where P is a promoter) was measured by β‐galactosidase activity relative to Pshp144::lacZ‐Δshp144com, which expresses wild type shp144 (WT). The sequence of the 13‐residue long active SHP144 representing the C‐terminal end is shown on top of the bar chart, and the numbers are relative to the full‐length SHP144. **p < .01, ****p < .0001, “ns” non‐significant compared to the WT. The error bars represent the standard error of the mean for at least three independent replicates.
TABLE 1.
Transcription activation and binding capabilities of native and modified SHPs
| Peptide a | Transcription activation b (Miller units/109 cells) | Binding affinities (KD c in μM) |
|---|---|---|
| C13 (WT) | 215 ± 5 | 6.6 ± 0.3 |
| S14A | 105 ± 5 | 8.1 ± 0.9 |
| E15A | 107 ± 2.5 | 3.3 ± 0.1 |
| W16A | 7.5 ± 2.5 | 50.0 ± 3.6 |
| V17A | 6 ± 1 | 9.7 ± 0.6 |
| I18A | 7.5 ± 1.5 | >100 |
| V19A | 215 ± 5 | 4.0 ± 0.8 |
| I20A | 8.5 ± 0.5 | 23.54 |
| P21A | 7.5 ± 2.5 | 1.7 ± 0.1 |
| F22A | 90 ± 10 | 7.1 ± 0.4 |
| L23A | 95 ± 5 | 4.3 ± 0.2 |
| T24A | 175 ± 5 | 3.3 ± 0.1 |
| N25A | 185 ± 5 | 3.3 ± 0.2 |
| L26A | 182 ± 2.5 | 3.4 ± 0.3 |
WT C13 or modified peptides were used. The numbers show the position of alanine replacement within the full‐length Shp144.
Mean ± SEM of three independent experiments.
Dissociation constant between the Rgg144 and its ligand SHP144.
2.3. The role of SHP144 residues in binding to Rgg144 assessed by fluorescence polarization
Upon binding, SHPs induce a conformational change in their cognate Rggs that initiates transcription from Rgg‐dependent genes. Mutations to SHP144 could reduce transcription by disrupting binding to Rgg144 and/or by failing to induce the conformational change in the Rgg. To further investigate the roles of individual residues in SHP144, we established a specific binding assay using synthetic FITC‐labeled SHP144 and purified recombinant Rgg144. Binding was detected using fluorescence polarization. The FITC‐labeled SHP (SHP144‐C13) is bound to Rgg144 with a K D of 6.6 μM (Figure 3a, Table 1). No binding was detected using a non‐specific FITC‐labeled peptide of the same size, or when Rgg144 was replaced by BSA, indicating that binding is specific (Figure 4).
FIGURE 3.
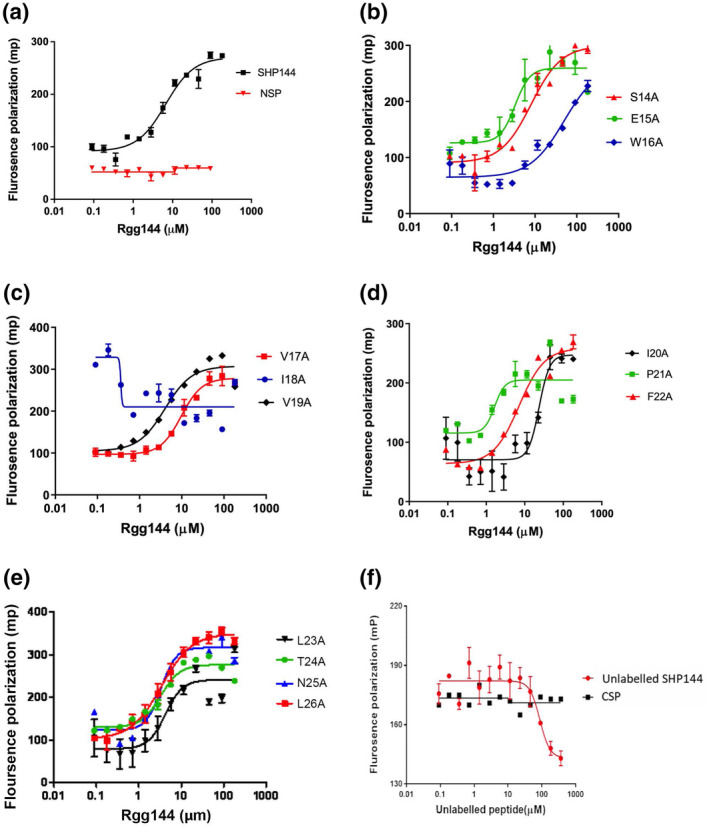
Rgg/SHP144 binding measured by fluorescence polarization using FITC‐SHP144 peptides. (a) Binding by the 13 residue WT peptide FITC‐SHP144 and by FITC‐NEC‐13 non‐specific peptide. (b–e) Rgg binding by FITC‐labeled mutant peptides. The position of substitution is indicated. In each case, peptide (10 μM) was incubated with serially diluted recombinant Rgg144 (0.09 to 182 μM) for 20 min at 20°C. (f) Assessment of the capability of unlabeled peptide SHP144 to competitively displace Rgg/FITC‐SHP144 complex. The 10 nm FITC‐SHP144 was initially mixed with 6.6 μm Rgg144 (KD value was taken from previous direct binding assay) and then incubated with serially diluted unlabeled SHP144 (0.09–364 μm) peptide (red circles) for 30 min at 20°C. The millipolarization values (mp) values fell with increasing amounts of unlabeled peptide SHP144 whereas those values remained constant after the addition of the control peptide, CSP (black squares). Data are the mean ± SEM from three independent experiments. Mp values were measured at wavelength 485 nm excitation and 520 nm emission using a Hidex Sense Microplate Reader.
FIGURE 4.
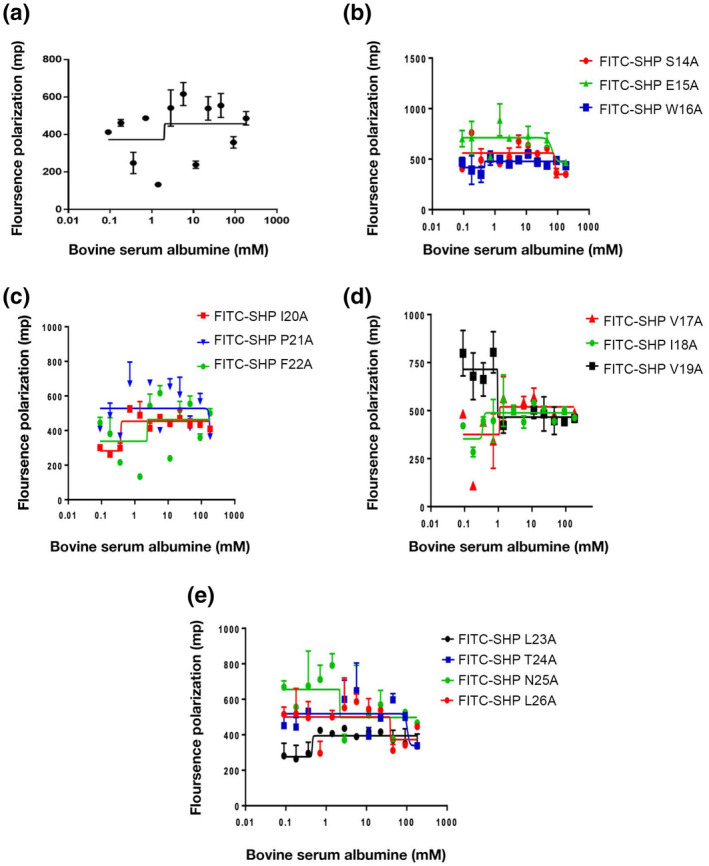
Diagram showing intermolecular interaction between bovine serum albumin and modified or unmodified fluorescent SHP144 peptide using fluorescence polarization technique. (a) indicates the binding of native FITC‐SHP144 to BSA whereas (b–e) represent BSA interaction with modified FITC‐SHP144 peptides. Polarization values (mp) were measured using Hidex Sense Microplate Reader at excitation 485 nm and emission 520 nm, and each value was plotted against BSA concentration. The values represent the average of three independent experiments.
To further investigate the interaction between Rgg144 and the SHP144, a competition assay was developed in which serial dilutions of unlabeled SHP144 were mixed with the labeled SHP‐Rgg complex (Figure 3f). The unlabeled peptide competed for binding with an IC50 (the concentration of competitor required to reduce FITC‐SHP144 binding to 50%) of 86.8 μM. By contrast, competence stimulating peptide (CSP), which does not bind to Rgg144, did not inhibit SHP binding at the highest concentration tested (Figure 3f).
We next evaluated the contribution of each residue toward Rgg binding using FITC‐labeled peptides each containing a single alanine substitution (Figure 3). Most of the amino acid substitutions had little effect on binding: S14A, E15A, V17A, V19A, F22A, L23A, T24A, N25A, and L26A, with K D s similar to those of the WT SHP (Figure 3b–e). In contrast, W16A and I20A reduced binding by ~8‐fold and ~4‐fold (Figure 3b,d), respectively, while I18A abolished binding completely (KD > 100 μM; >15‐fold decrease in affinity) (Figure 3c).
Of the five mutations that reduced Rgg‐dependent transcription to background levels (W16A, V17A, I18A, I20A, and P21A), three (W16A, I18A, and I20A) can be explained, at least in part, by exhibiting a reduced binding to the Rgg. However, the P21A peptide is bound threefold more tightly than the WT peptide, while the V17A peptide is bound with comparable affinity, suggesting that, upon binding, these peptides fail to induce the conformational change that is necessary to initiate Rgg‐dependent transcription. Other mutations have more subtle but nonetheless notable effects. S14A, F22A, and L23A each reduced transcription by twofold but did not reduce binding. Thus, peptides containing V17A and P21A (and to a lesser extent S14A, F22A, and L23A) are candidates for competitive inhibitors of Rgg‐dependent transcription.
2.4. SHP144 in the context of the pangenome
Our data were generated in an important model strain, yet, because there is extensive genetic diversity across the pneumococcal pangenome, we employed comparative genomics to interpret these data across a multitude of pneumococcal strains. We used a database of 7548 genomes downloaded from PubMLST (genome IDs listed in SFile1‐Listof7548GenomeID). To extract all the SHP144 sequences, the analysis was anchored on Rgg144. Using tblastN, we identified the Rgg144 gene and the 4Kb sequence downstream. We searched (using tblastN) this genomic region for sequences similar to SHP144. Analysis of the sequences revealed 7457 SHP144 sequences, that could be organized into 12 SHP144 alleles (Figure 5). Alleles that were present in less than 1% of the set were excluded. An alignment of the SHP144 sequences highlights three prominent features for SHP144: (i) a positively charged N‐terminus, (ii) a hydrophobic central region containing a negatively charged residue, and (iii) a highly conserved C‐terminus ending in “PFL(T/I)N(R/L)” (Figure 5).
FIGURE 5.
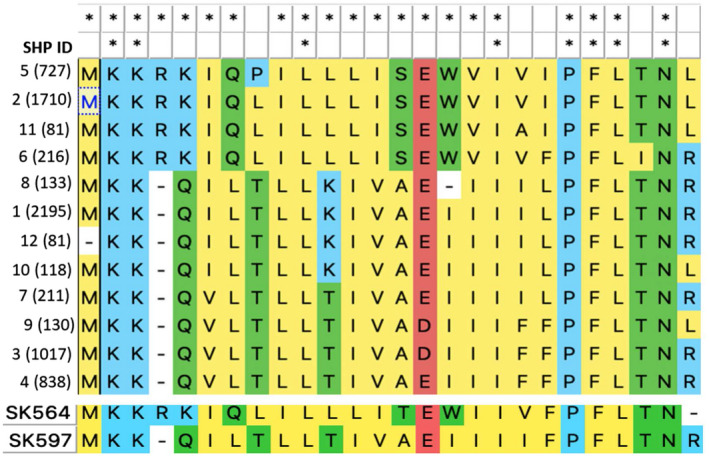
Alignment of SHP144 alleles. Upper panel: Alignment of 12 SHP144 alleles identified in 7548 pneumococcal genomes. SHP alleles and the number of strains that encode each allele (in parenthesis) are shown in the left‐hand side column. “*” highlights positions with 100% identity across the set of sequences. The first row of “*”, indicates the positions with 100% identity across 4 alleles from set 1 that includes sequences 2, 5, 6, and 11, and the second row indicates the identity among all pneumococcal SHP alleles. Alleles are named based on their frequency across the genomic set. Alleles that were encoded by less than 1% of genomes are excluded. Lower panel: Two representative alleles from Streptococcus mitis, the top one from strain SK564 resembles alleles from pneumococcal set 1 and the lower one from strain SK597 resembles alleles from pneumococcal set 2.
Across SHP144 alleles we observed variation in the central region, such that only 30% of residues are fully conserved across all alleles. The SHP144 sequence can be divided into two sets, such that over 65% of the residues are identical within each set (Figure 5, alignment of set 1). The peptide from D39 is part of Set 1, which includes 37% of the SHP144 sequence in our data set. Set 1 is distinguished by the presence of “SEW” at positions 14–16, where the functional peptide begins (Zhi et al., 2018). In contrast, set 2, which contains the remaining 63% of sequences, has “AEI” at that position. The central hydrophobic stretch from position 17–20 varies between the groups, yet has highly conserved isoleucine at position 18. Of note, when this residue is mutated to an alanine residue, the resulting peptide has dramatically reduced binding to Rgg144 (and reduced Rgg activation), consistent with I18 having a key binding role in SHPs (Figure 5, upper panel). Finally, the six residues at the C‐terminus are highly conserved across both sets, despite relatively modest effects on receptor binding and/or transcription (Figures 2 and 3). The exception is P21 that is essential for Rgg‐dependent activation but not binding in our model strain. Interestingly, this residue was completely conserved in all pneumococcal genomes, highlighting its functional importance. Finally, we expanded our search beyond pneumococcus and investigated whether SHP144 orthologues are conserved in Streptococcus mitis, a commensal that often inhabits the same niche as pneumococcus. By searching for short peptides neighboring the Rgg144 orthologue, we identified alleles that resemble SHP144 from both set1 and set 2 (Figure 5, lower panel). While functional testing beyond set 1 is not in the scope of this study, we hypothesize that inhibitors generated by replacing P21A may be effective against all pneumococcal SHP144 peptides, while those focused on other regions of the SHP are more likely to be strain specific.
2.5. Inhibition of Rgg‐dependent transcription by mutant SHP peptides
To assess the inhibitory effects of the V17A and P21A mutations, the spent culture supernatants of strains producing these mutant SHPs were collected at the late exponential phase, when the activity of the QS system is high, and was added to cultures containing the reporter strain that produces native SHP144: Pshp144::lacZ‐wt strain. Inhibition of Rgg‐mediated transcription was predicted to diminish activation of Pshp144 by the endogenously produced wild type SHP144 peptide. The results showed, indeed, that culture supernatant containing SHP P21A significantly decreased transcriptional activity (Figure 6a), and that inhibition was concentration‐dependent (Figure 6b). A reduction in transcriptional activity was also seen by culture supernatant containing SHP V17A (Figure 6a) but to a lesser extent.
FIGURE 6.
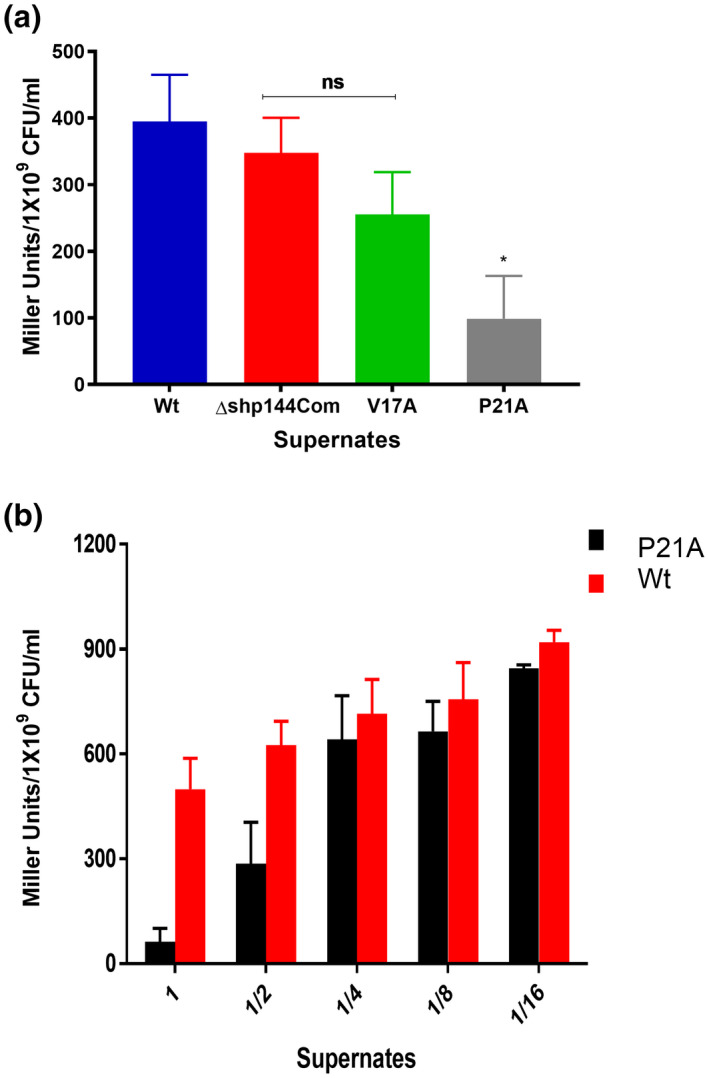
Inhibition of Pshp144 transcription using supernatants containing mutant SHPs. (a) Cells from the reporter strain (Pshp144:: lacZ‐wt) were incubated with the supernatants of wild type, or mutant Shp144 producing strains. The supernatants were obtained at the late exponential phase. (b) Dose‐dependent inhibition of Pshp144 transcription by using supernate containing SHP P21A. Wild type and modified supernates were serially diluted, and incubated with Pshp144:: lacZ‐wt to an OD600 (~0.6), and the Pshp144 activity was measured by β‐galactosidase assay (n = 3). The error bars represent the standard error of the mean. *p < .05, “ns” not significant compared to the wild‐type.
To confirm that SHP P21A inhibits Rgg‐dependent transcription, we mixed supernatants from the wild type and strains producing mutated peptides collected at the late exponential phase. Different mixtures (1:1, 1:4, and 1:10 of WT:mutant) were tested using the β‐galactosidase reporter strain in which shp144 has been deleted (P shp144: : lacZ‐Δshp144). A 1:10 ratio of wt:mutant supernatant significantly decreased transcriptional activation of the system (Figure 7). Given that P21 is completely conserved across SHP144 alleles, we speculate that this effect will be observed across strains.
FIGURE 7.
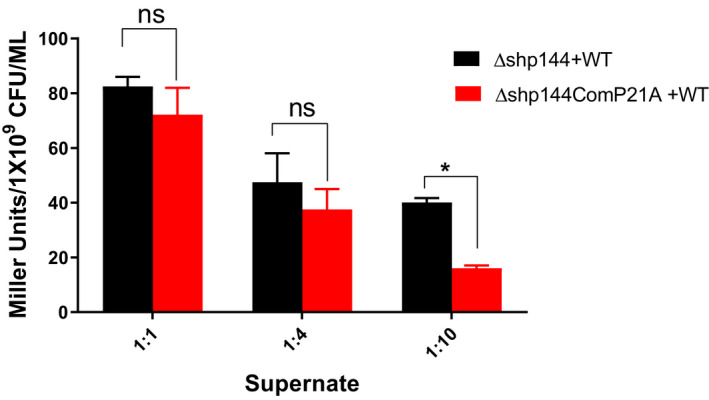
The P21A peptide inhibits Rgg‐dependent transcription by SHP144. Decreasing ratios of Δshp144comP21A and wild type (WT) supernatants were incubated with a pellet of reporter strain Pshp144:: lacZ‐Δshp144, and the Pshp144 driven β‐galactosidase activity was measured (red columns). As a control dilutions of wild type supernatant in Δshp144 supernate were used (black columns). Comparisons are made relative to shp144 transcription levels in culture containing a mixture of WT and Δshp144 supernates. *p < .05 and “n” non‐significant (n = 3).
2.6. Phenotypic impact of inhibitor peptide on pneumococcal growth and oxidative stress resistance
Having established that the P21A mutation inhibits Rgg‐dependent transcription, we went on to test whether it also inhibited the Rgg/SHP144 conferred phenotypes, namely utilization of mannose and oxidative stress resistance (Zhi et al., 2018). As demonstrated in Figure 8a, pneumococcal growth in the absence of SHP144 is attenuated in chemically defined medium supplemented with 55 mM mannose relative to the wild type strain. The addition of wild type SHP reconstituted the growth of the Δshp144strain in a dose‐dependent manner (Figure 8b). This phenotypic complementation, however, was not observed upon the addition of the same concentration of modified S14A or P21A peptides, shown previously to reduce transcription, highlighting the specificity of the peptide for functional complementation. As expected, no growth complementation was seen in glucose, N‐acetyl glucosamine, or galactose as the main source of carbon (Figure S1), emphasizing the importance of mannose for stimulation of Rgg/SHP144 QS.
FIGURE 8.
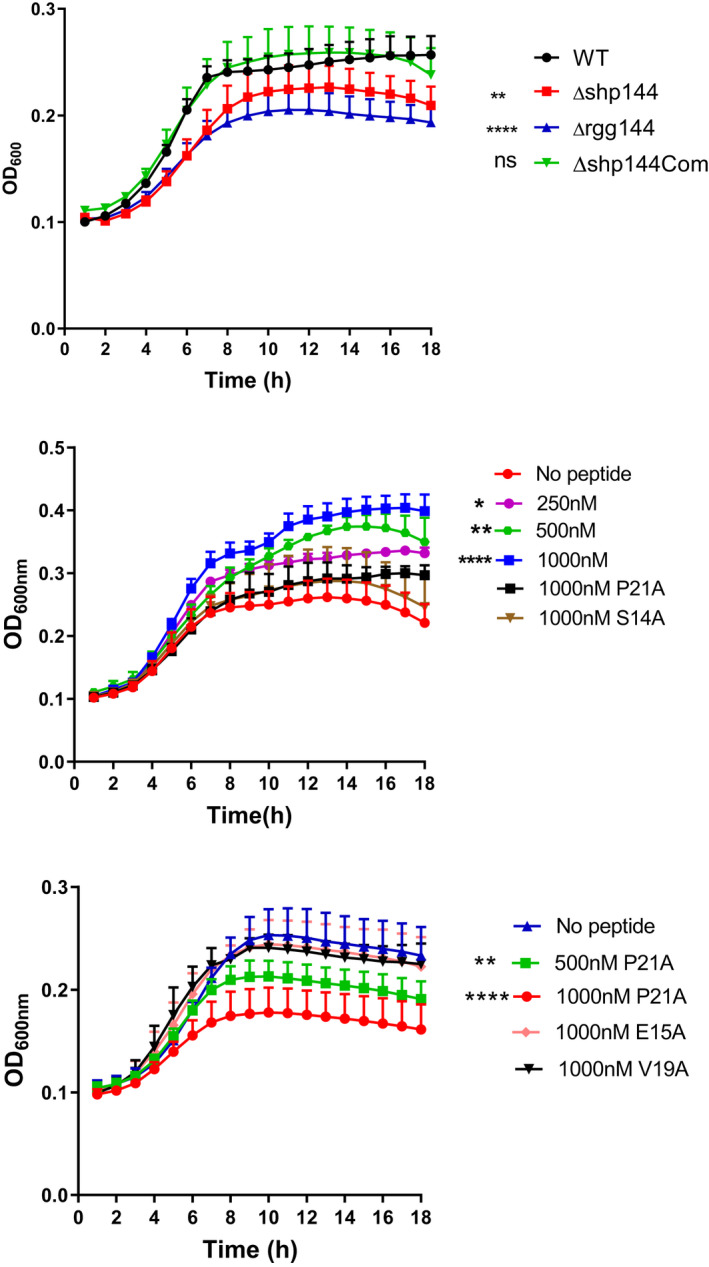
The impact of peptides on pneumococcal growth in CDM supplemented with mannose. (a) Growth profiles of pneumococcal strains in CDM containing 55 mM mannose. (b) Exogenous addition of wild type C13 peptide (but not mutant peptides) restores mutant shp144 growth on mannose. Comparisons are made relative to Δshp144 culture without peptide. (c) Inhibition of growth of wild type Streptococcus pneumoniae on mannose by SHP P21A. Error bars show the standard error of the mean from at least three independent experiments. *p < .05, **p < .01, ****p < .0001.
We next tested the P21A peptide as a potential inhibitor of wild type Streptococcus pneumoniae growth on mannose. SHP P21A reduced pneumococcal growth in a concentration‐dependent manner, despite the presence of a functional shp allele in the wild type strain, confirming its efficacy as a Rgg inhibitor. In contrast, SHP E15A or V19A, which had little or no effect on binding and did not influence transcriptional activity, did not show any significant inhibitory activity (Figure 8c).
Next, we determined the phenotypic impact of SHP P21A on pneumococcal oxidative stress resistance. When pneumococci were treated with 1 mM of the superoxide radical generator paraquat, a significant adverse impact on Δshp144 growth was seen relative to the wild type strain (p < .05) (Figure 9a). For example, after 1‐h incubation 91% of wild type pneumococci survived, while only 65% of the Δshp144 mutant could be recovered (Figure 9b). As expected, the addition of the unmodified SHP to Δshp144 reconstituted the pneumococcal resistance to paraquat. Strikingly, the addition of 10 μM SHP P21A, but not SHP S14A rendered the wild type S. pneumoniae susceptible to killing (Figure 9c).In conclusion, systematic mutation of SHP144 has identified a Rgg inhibitor SHP P21A that binds with a higher affinity than SHP144. SHP P21A blocks Rgg‐dependent transcription by the wild type SHP144 and the downstream Rgg‐dependent phenotypes. Other residues in the Rgg, notably V17 and to lesser extents S14, F22, and L23 can be replaced with little impact on binding but with reduced transcription by the Rgg, highlighting their potential as targets in developing future inhibitors.
FIGURE 9.
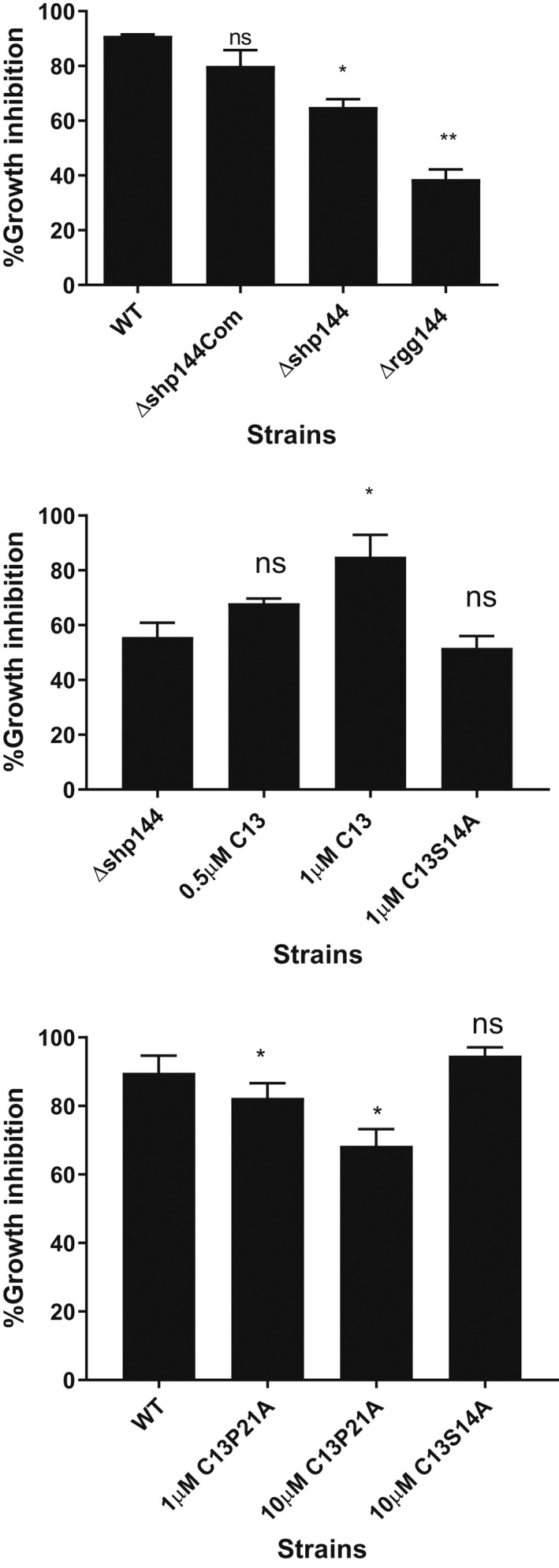
Evaluation of selected peptides against pneumococcal oxidative stress resistance. (a) Survival of pneumococcal strains after treatment with 1 mM paraquat. (b) The addition of 1 μM SHP144 reconstitutes Δshp144 oxidative stress resistance but not SHP S14A. (c) Inhibition of pneumococcal oxidative stress resistance using 10 μM SHP P21A. This inhibition was not seen with SHP S14A. The survival percentages were calculated for each strain grown in THY in the presence or absence of the selected peptide. Comparisons were made relative to wild type or Δshp144. Data represent the average of at least three independent experiments, each with triplicates. *p < .05, **p < .01, “ns” p > .05.
2.7. In vivo studies
Previously, we demonstrated a requirement for Rgg/SHP systems for efficient nasopharyngeal colonization. Therefore, we tested the hypothesis that modifications of selected amino acid residues shown to be important for binding and transcriptional activation in SHP144 will lead to a decreased pneumococcal colonization. This hypothesis was tested using strains where the wild type allele was replaced with the mutant form of shp144 encoding E15A that has little effect on binding or transcription, I20A that reduces binding and transcription, or P21A that impairs transcription alone. One hour after intranasal administration, the bacterial load in the nasopharyngeal wash for all strains was similar (p > .05) (Figure 10a). At 7 days post‐infection, however, colony counts for Δshp144 (log10 1.02 ± 0.63 CFU/ml, n = 5) were significantly lower than counts of the wild type strain (log10 4.18 ± 0.21 CFU/ml, n = 5) (p < .01) or of the complemented strain (log10 4.33 ± 0.13 CFU/ml, n = 5). Significantly, the colony counts for Δshp144ComI20A and Δshp144ComP21A (log10 1.48 ± 0.88 and 1.43 ± 0.84 CFU/ml, respectively, n = 5), were also less than the wild type complemented strain (log10 4.325 ± 0.13 CFU/ml, n = 5) (p < .05) (Figure 10) implying that SHP I20A and SHP P21A have reduced activity in vivo as well as in vitro. As expected, no significant difference in the bacterial load of Δshp144ComE15A was seen, consistent with its near wild‐type activity (p > .05). These results support our in vitro findings that residues important for transcriptional activation and binding play a vital role in the proper functioning of Rgg144/Shp144 function in vivo.
FIGURE 10.
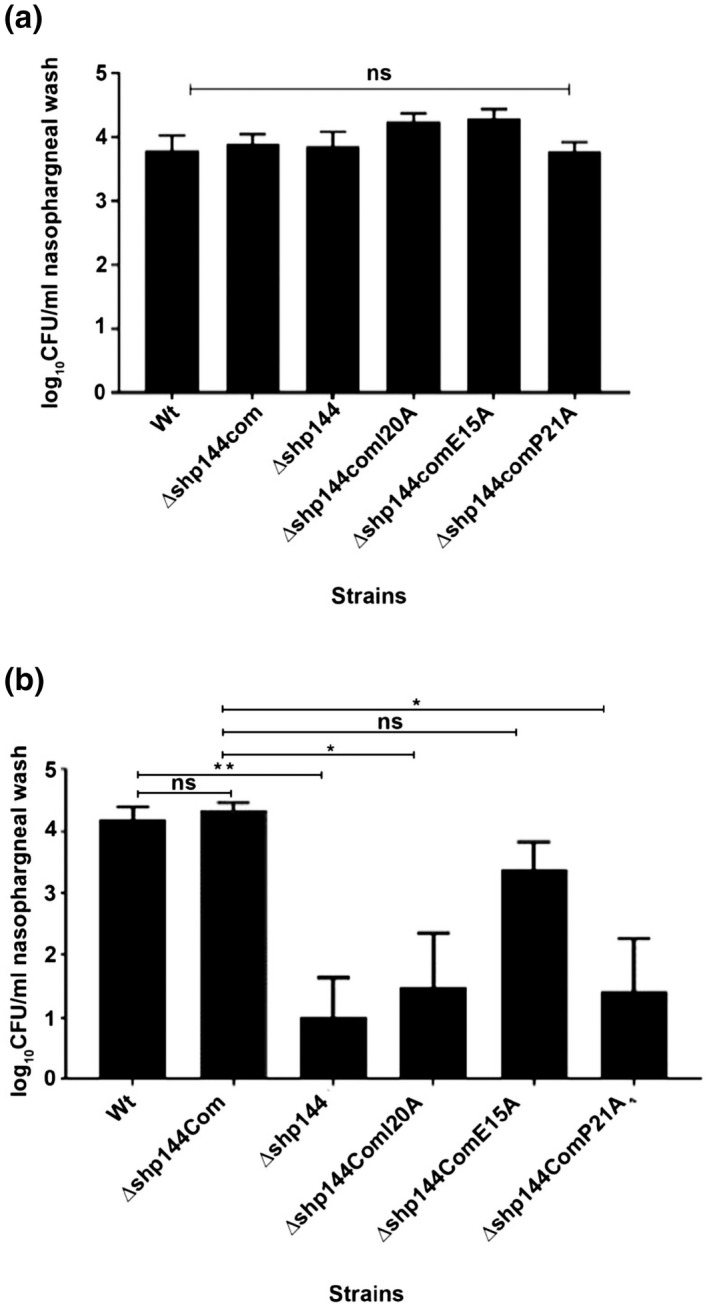
Pneumococcal strains lacking shp144 or having modified shp144 are less able to colonize nasopharynx. Mice were challenged with approximately 2.5 × 105 CFU pneumococci. Infected mice were culled on day 0 (a) and day 7 (b), and CFU/ml of bacteria were calculated by serial dilutions of nasopharyngeal wash. Each bar represents the mean of data collected from five mice. Error bars show the standard error of the mean. Significance changes in bacterial counts are compared with wild type and complemented strains using one‐way ANOVA and Tukey's multiple comparisons test. (*p < .05, **p < .01, “ns” not significant).
Perhaps unsurprisingly, it was not possible to complement a Δshp144 strain with exogenously added SHP by intranasal administration, so it was not possible to test the efficacy of peptide inhibitors in vivo (Figures S3a,b). This is likely to be due to rapid degradation of the SHP or clearance by the lung. Thus, the presentation of inhibitors in a stable, accessible form needs to be optimized. Nevertheless, the approach adopted here has the potential to develop inhibitors that can modify bacterial behavior given the correct presentation. Moreover, it is potentially applicable to a wild variety of QS systems.
3. DISCUSSION
Quorum sensing systems have been suggested as an effective anti‐infective target because inhibitors change the collective behavior of a population of bacteria and prevent efficient microbial adaptation to environmental conditions (Kalia, 2013). The resulting failure to thrive is likely to render strains more susceptible to clearance by the host's immune system (Kalia, 2013; Rutherford & Bassler, 2012). As QS inhibitors do not kill bacteria but switch off the adaptive and virulence capabilities of the target microbe, their impact on microbiota would be less than the broad‐spectrum traditional antibiotics. Consequently, the development of resistance to QS inhibitors may be lower than traditional antibiotics because QS systems are only required for specific adaptations rather than being essential for microbial survival.
In this work, we targeted the Rgg144 system in S. pneumoniae D39 because of its importance in pneumococcal colonization and virulence and our previous knowledge regarding the Rgg144 phenotype (Zhi et al., 2018). The Rgg144 QS system plays a crucial role in colonization, which is exemplified by the significant reduction in colony counts at 7 days post‐infection observed for the deletion strain when compared to the isogenic wild type. Similarly, the median survival time of mice‐infected intranasally with ∆rgg144 is significantly higher than the wild type‐infected group (Zhi et al., 2018). The observed impact of this QS system on colonization is likely due to its role in the control of mannose utilization and capsule expression.
Despite relatively low sequence identity (<25%), the three‐dimensional‐fold of Rgg144 resembles that of Rgg2 of S. dysgalactiae and Rgg3 of S. thermophilus (Capodagli et al., 2020). Adjacent DNA‐binding helices are separated by ~26 Å, similar to the distance between the corresponding helices of Rgg2 and Rgg3 bound to DNA (PDB: 6W1A and 6WIF; Figure S3). The DNA is significantly bent such that the DNA‐binding helices of the Rgg interact with adjacent major grooves in the target DNA sequence. Upon SHP binding, there are relatively large changes to the C‐terminal domain, and the helix‐turn‐helix domains become poorly defined in the structure suggesting considerable conformational flexibility and potentially disrupting the interaction with DNA. The structure of Rgg144 is also compatible with the DNA‐binding conformation of Rgg2 suggesting a similar mechanism of action. Consistent with this hypothesis, Rgg144 binds to DNA in the absence of its SHP. However, additional structural and biochemical data in both the presence of DNA and the SHP are required to determine the precise mechanism of transcription regulation.
Alanine‐scanning mutagenesis of the SHP144 revealed that only I18 is essential for binding SHP144 to Rgg144. Mutations W16A and I20A also reduced Rgg binding (by ~8‐ and 4‐fold) and displayed a corresponding reduction in the ability to induce transcription from a Rgg‐control promoter in each case. Many of the other mutations had little impact on Rgg binding and modest effects on transcription. The exceptions were SHP P21A and SHP V17A, both of which abolished transcription to basal levels despite still binding to the Rgg. SHP F22A and SHP L23A showed similar phenotypes but had lesser effects on transcription. The most likely explanation for these observations is that these mutant SHPs fail to induce the change in the Rgg that initiates transcription. SHP P21A, in particular, was an effective Rgg inhibitor, preventing Rgg‐dependent transcription and the associated phenotypes of mannose‐dependent growth and oxidative stress resistance. Interestingly, the binding affinity of SHP P21A was 3‐fold higher (1.66 μM) than the wild type SHP (6.6 μM) (Table 1), raising the possibility that Rgg inhibition could be improved by rational approaches aimed at increasing the affinity of binding further using phage display or through the use of peptide libraries. Residues V19, P21, F22, and L23 of SHP144 are attractive targets for mutagenesis because they are important for transcription activation but not for Rgg144 binding. Such strategies have been used effectively for a wide variety of inhibitors (Fosgerau & Hoffmann, 2015) including cytotoxic T‐cell inhibitors (Tretiakova et al., 2000), complement inhibitors (Qu et al., 2013), HIV fusion inhibitors (Eggink et al., 2010), and protease inhibitors (Hong et al., 2000). The extensive SHP‐binding groove represents an attractive target for such approaches.
Our previous work has shown that Rgg144 is conserved across viridian species (Zhi et al., 2018). Here, we perform a detailed analysis of 7548 pneumococcal genomes: It revealed 12 distinct SHP144 pneumococcal alleles. This level of diversity may be due to niche adaptation linked to Rgg144/SHP144 function or intraspecies competition and is consistent with selective pressure on the Rgg144 system. In this study, we limited experimentation to strain D39. The extent to which our data is relevant to a broad range of strains is likely to depend on the variability in the Rgg144/SHP144 system, as well as the extent to which Rgg144/SP144 interacts with components of the accessory genome. Based on the conservation of P21 in strains with the set 1 allele, we speculate that the role of P21 is conserved for most strains with this allele, provided the function of Rgg144/SHP144 is not dependent on strain background. Additional studies are planned to understand conserved and strain‐specific features, as well as relevance to other species.
Antimicrobial peptides have been indicated as a potential solution to the rising trend of antibiotic resistance. Some bacteria, such as Enterobacteria, produces ribosomally synthesized antimicrobial peptides or bacteriocins, which kill closely related bacteria by creating membranous pores, interfering with the cell wall or protein synthesis, or via degradation of nucleic acids (Almeida‐Santos et al., 2021). Therefore, conceptually, the use of peptides is recognized as a strategy to treat infectious diseases. In this study, we created an antagonistic peptide targeting a pneumococcal virulence determinant, the Rgg144 QS system. One of the antagonistic peptides, with a proline replacement, was effective in interfering with Rgg144‐conferred phenotypes. A similar approach has been used to develop inhibitory peptidic analogs to the PlcR/PapR QS system of Bacillus cereus (Yehuda et al., 2018; 2019). Using seven residues long‐modified PapR peptides (PapR7), the hemolytic activity regulated by PlcR/PapR QS system was abolished, without affecting bacterial growth. By creating and testing peptides with multiple mutations, the importance of proline and glutamic acid residues was shown in PapR/PlcR interactions and used in designing strong inhibitors (Yehuda et al., 2019). While all the PapR sequences from different strains of the B. cereus group showed divergences in their three N‐terminal residues, the glutamic acid residue in position six was found to be conserved (Slamti & Lereclus, 2005).
Similarly, we also found a proline amino acid conserved in a large subset of SHP144 sequences, which, when modified, generated an inhibitory peptide consistent with the importance of this proline residue in the conformational arrangement of SHP144 and the interaction with its cognate receptor.
When tested in vivo, the S. pneumoniae Δshp144 strain could be complemented with an intact copy of shp144, however, complementation with the sequence carrying the P21A alteration failed to restore function, consistent with the in vitro data. Moreover, in vivo, it was not possible to complement a shp mutant strain with externally added synthetic wild type SHP indicating that stability and delivery of the peptides need to be improved to test the therapeutic potential of inhibitors. One reason why the inhibitory synthetic peptide was not efficient in vivo could be due to its “suboptimal” hydrophobicity. A study investigating the role of peptide hydrophobicity in the action mechanism of α‐helical antimicrobial peptides found that decreasing hydrophobicity diminishes antimicrobial activity, while its enhancement to a certain level will improve antimicrobial activity and an additional increase would lead to a decrease in the activity, possibly due to increased dimerization, which inhibits access to the prokaryotic cell membrane (Chen et al., 2007). Peptide stability has been shown to be improved by amino acid residue replacement to prevent proteolytic degradation. This can be achieved by L‐, D‐, or nonnatural amino acid residues, or by a targetted chemical modification by acetylation and/or amidation (De Waard & Sabatier, 2006). Once an optimized peptide is obtained, we expect that these peptides should act as narrow‐spectrum antimicrobials targeting S. pneumoniae fitness without any effect on its survival potentially through its impact on oxidative stress resistance and utilization of host‐derived sugars, such as mannose.
Several approaches have been taken to inhibit Gram‐positive QS systems. These include screening combinatorial libraries to identify competitive peptidomimetic inhibitors (Karathanasi et al., 2018), the use of natural microbial products that target virulence gene expression (Mansson et al., 2011), the use of molecularly imprinted polymers (Motib et al., 2017), or immunopharmacotherapeutic approaches that use monoclonal antibodies to neutralize the signal peptide via sequestration (Park et al., 2007). Recently, cyclosporine was identified as a Rgg inhibitor in S. dysgalactiae by screening a large inhibitor library (Parashar et al., 2015). While cyclosporine could not be used as an anti‐infective, due to its immunosuppressive properties, it highlights the tractability of the Rgg QS system as a target for inhibitors. The approach described in this manuscript represents another potential strategy to develop QS inhibitors. A particular advantage is that it utilizes the naturally occurring peptide ligand as a starting point for inhibitor design. Our approach could be easily adapted to target other systems involving transcription regulators that are controlled by peptide ligands.
4. MATERIALS AND METHODS
4.1. Bacterial strains and growth conditions
S. pneumoniae D39 strains and plasmids constructed and used in this study are listed in Table S2. Pneumococci were grown either in brain heart infusion or Todd Hewitt broth supplemented with 0.5% (w/v) yeast extract. Blood agar plates supplemented with 5% (v/v) defibrinated horse blood was used for the cultivation of pneumococci at 37°C in 5% CO2. In addition, a chemically defined medium (CDM) supplemented with 55 mM of selected sugar was used for phenotypic characterization studies. All bacterial stocks were kept in 15% (v/v) glycerol at −80°C. For Escherichia coli, Luria Bertani (LB) broth was used for growth at 37°C in a shaking incubator or on LB agar plates (Oxoid, UK). When necessary, 100 μg/ml spectinomycin, 250 μg/ml kanamycin, and 3 μg/ml tetracycline were added to the pneumococcal growth medium, whereas 100 μg/ml ampicillin and 50 μg/ml kanamycin were used for E. coli.
4.2. Construction of genetically modified strains
An intact copy of the shp144 coding sequence (located between 150,352 and 150,617 in S. pneumoniae type 2 D39 strain according to the KEGG database) was introduced into the pneumococcal genome at a transcriptionally silent site using non‐replicative pneumococci plasmid pCEP as described previously (Guiral et al., 2006). The gene of interest with its native promoter was amplified using pneumococcal DNA and the primers listed in Table S3. After amplification, the PCR product was digested and ligated into the BamH1 and Nco1 restriction sites of pCEP. The recombinant plasmid was transformed into Δshp144. The successful integration of shp144 was confirmed by PCR and sequence analysis. The construct was designated as ∆shp144com.
For alanine‐scanning mutagenesis, mutations were introduced into the gene encoding the SHP144 polypeptide, replacing, individually, each residue in the mature polypeptide with alanine, using overlap extension PCR (SOEing PCR) and the primers listed in Table S3 (Al‐Bayati et al., 2017; Kahya et al., 2017). The purified product was then digested and ligated into pCEP, as for the wild type gene. The resulting recombinant plasmids were sequenced and transformed into Δshp144 (Table S2).
4.3. Construction of pneumococcal transcriptional reporter strains
The list of transcriptional reporter strains used in this study is provided in Table S2. Pneumococcal transcriptional reporter strains were generated following the protocol described previously (Al‐Bayati et al., 2017). After in silico identification using BPROM (Softberry), the putative promoter region of shp144 (Pshp144) was amplified and fused on to the promoterless lacZ gene in an integrative reporter plasmid pPP2 (Halfmann et al., 2007). The recombinant pPP2 was transformed into different pneumococcal backgrounds via double crossover in the bgaA gene. The successful integration of fusion into wild type and mutant constructs was confirmed by colony PCR using primer pairs Fusion seq/UF and Fusion seq/DR (Table S3).
4.4. Expression and purification of Rgg144
The rgg144 (SPD_0144) was amplified and cloned, into the ampicillin‐resistant plasmid pLEICS‐01. The recombinant plasmid was verified by DNA sequencing using T7 Promoter‐F and pLEICS‐01‐Seq‐R primers shown in Table S3. The sequenced plasmid was transformed into E. coli BL21 (DE3) competent cells using heat shock. A single colony containing the desired construct was inoculated into power prime broth containing 100 μg/ml ampicillin and incubated overnight at 37°C to an OD600 ~ 1.4–1.5 in a shaking incubator. The culture was induced with 1 mM IPTG and left overnight at 37°C. The cell pellet was resuspended with lysis buffer containing 1 mM EDTA and 0.5 mg/ml lysozyme. A complete EDTA‐free protease inhibitor cocktail tablet (Roche, Basel, Switzerland), 5 μg/ml DNase, and 5 mM MgCl2 was added to the mixture and incubated at room temperature for 15 min. The sample was then sonicated and cleared by centrifugation at 10,000g for 20 min at 4° C. The pellet was washed several times with buffer containing 1% (v/v) Triton X‐100 and 1 mg/ml sodium deoxycholate to ensure complete disruption of cell membranes, and release of intracellular protein. Finally, the washed inclusion bodies were resuspended in 50 mM Tris–HCl buffer, pH 8.5, and kept at −80°C. Purified inclusion bodies were solubilized using 6 M Guanidine HCl and 5 mM DTT at 37°C and refolded by diluting the sample to about 2 mg/ml and dripping slowly into a 10‐fold excess of refolding buffer at 4°C. The refolded protein was then dialyzed into 50 mM Tris–HCL pH 7.5 containing 500 mM NaCl and 5 mM imidazole and passed through a Ni‐NTA affinity column. After washing with loading buffer, bound protein was eluted with 500 mM imidazole. Fractions containing Rgg144 were passed through a Superdex 200 16/60 HiLoad column in 20 mM Tris–HCl containing 50 mM NaCl, and fractions were collected across the elution peak and analyzed on SDS‐PAGE. Finally, the selected fractions were concentrated using an Amicon Ultra‐15 centrifugal filter unit to ~5 mg/ml (Millipore, UK), and stored at −80°C for further use. The identity of the purified recombinant protein was verified by matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) by PNACL (University of Leicester). For the production of selenomethionine‐substituted Rgg144, the expression plasmid was introduced into B834(DE3) cells, and cells were grown in SelenoMet™ Medium (Molecular dimensions, UK) following the protocol provided. Protein was isolated, refolded, and purified as described above for the native Rgg144 protein.
4.5. Crystallization and structure determination
All crystals were grown using the sitting‐drop vapor diffusion method by mixing equal volumes (1.0 + 1.0 μl) of protein and reservoir solution. Protein at 4–5 mg/ml was mixed with reservoir solution containing 100 mM Bis‐Tris propane pH 7.5, containing 200 mM potassium sodium tartrate tetrahydrate and PEG4K (18%–26%). All crystals were maintained at 100°K during data collection. Diffraction data were collected at beamline I02 at the Diamond Light Source and were processed with iMosflm. Phases were determined by selenium single‐wavelength anomalous diffraction using selenomethionine‐substituted Rgg144 crystals using the Auto‐Rickshaw platform (Panjikar et al., 2005). Initial models were refined using higher resolution data collected from native crystals. Models were optimized using cycles of manual refinement with Coot and refinement in Refmac5, part of the CCP4 software suite, and in Phenix.
4.6. Pangenome analysis
We employed a database of 7548 S. pneumoniae genomes downloaded from PubMLST and selected for quality by Melissa Jansen van Rensburg and Angela Brueggemann (personal communication). The genome IDs are listed in SFile1‐Listof7548GenomeID. The sequences were downloaded from the PubMLST website (PubMLST Pneumococcal Genome Library, Organisms tab, and Streptococcus penumoniae).
All known SHP144 sequences are adjacent to their cognate regulator Rgg144. Thus, we made neighboring to Rgg144 a criteria for SHP144 identification. First, we identified the genomic region encoding Rgg144. To this end, we ran tblastn using the sequence for Rgg144 from D39 as a query. We parsed the top hit in every genome to select sequences that were at least 80% identical over a minimum length of 200 amino acids. Next, we used a python code to extract the 4Kb downstream of Rgg144, creating a database of 4Kb sequences/genome to search for SHP144 sequences.
To capture the SHP144 alleles, we employed tblastn. For queries, we used the SHP144 alleles from D39, PN4595, TIGR4, SP23BS72, and CDC108700, as these represent diverse sequences of SHP144. As a database, we used the 4Kb regions described above. We selected the top hit in each genome, with a minimum of 80% identity over 13 amino acids to any of the queries.
To determine the number of unique SHP144 alleles we consolidated all the hits from every genome and used Python to generate a list of unique alleles. A representative of each allele was used to generate an alignment using the MEGA clustalW alignment tool and visualized using MEGA. The number of alleles per genome was enumerated using a Python script and is represented, within parenthesis, by the allele name in Figure 5.
We used a parallel approach to identify the SHP144 sequence on a select set of S. mitis genomes (set from [Zhi et al., 2018]). We identified the Rgg144 using blastp, then searched the neighboring region for short peptides with sequence similarity to the pneumococcal Rggs. The figure shows two representative sequences, one that resembled pneumococcal alleles from set 1, and the other pneumococcal alleles from set 2.
4.7. β‐galactosidase activity assay
The β‐galactosidase assay was used to assess Pshp144 expression levels in different backgrounds, following the procedure described previously (Zhang & Bremer, 1995). Reporter cells were grown in a chemically defined medium supplemented with 55 mM of the selected sugar, with or without synthetic peptide, to the late exponential phase (Table S4). The enzymatic activity was calculated and expressed in Miller Units (nmol p‐nitrophenol/min per 109 pneumococcal cells.
4.8. Peptides
Synthetic SHP144 peptides representing the mature SHP144, with or without FITC‐tags were purchased from COVLAB, UK (>98% pure) as shown in Table S4. Peptides were dissolved in DMSO at 6 mM and stored at −20°C for further use.
4.9. Growth assays
Chemically defined medium supplemented with 55 mM of desired sugar, and in the presence of varying concentrations of native and modified SHP144 synthetic peptide (Table S4), was used to characterize the growth properties of wild type D39 and its mutants, using a Multiskan TM GO Microplate Spectrophotometer (Thermo Scientific, UK). Growth was monitored by measuring the oOD600nm every hour at 37°C. This experiment was done in triplicate and repeated at least three times.
4.10. Pneumococcal survival assay
Oxidative stress resistance of pneumococcal cells, with or without synthetic SHP144 peptides, was tested using the superoxide generator paraquat. The pneumococcal strains were grown in THY supplemented with 0.5% (w/v) yeast extract to the early exponential phase. The bacterial sample was then exposed to 1 mM paraquat for 1 h at 37°C. The bacterial culture without paraquat was used as a control. The number of viable cells was determined by serial dilution and plating onto blood agar plates and survival percentages were calculated by comparing the CFU/ml of exposed culture to the CFU/ml of the control.
4.11. Inhibition of Rgg‐dependent transcription using spent culture supernatants
To testif‐modified SHP144 peptides inhibited endogenous SHP144 produced by wild type D39, the cell‐free culture supernatants were collected from wild type D39 and modified shp144 strains grown on mannose to late exponential phase. Supernatants were added to the wild type reporter strain Wt‐Pshp144::lacZ cells, and the impact on Pshp144 transcription was examined using a β‐galactosidase assay, as described above. The inhibitory effect of the modified peptide was also confirmed by the addition of a mixture of wild type and mutant supernatants to cells of Δshp144‐Pshp144::lacZ which is deficient SHP144, and assessing the promotor activity as described above.
4.12. Measurement of Rgg‐SHP144 binding using fluorescence polarization
A fluorescence polarization assay was used to measure the binding affinities of native and mutant SHPs to purified Rgg144. To do this, the purified protein was serially diluted in 50 mM Tris–HCl pH 7.4, containing 150 mM NaCl, into a 96‐well, black polystyrene plate. Samples were incubated with 10 nM wild‐type or mutant SHP144 for 20 min at 20°C. Millipolarization values (mp) were measured at wavelength 485 nm excitation and 520 nm emission using a Hidex Sense Microplate Reader. The values were plotted against protein concentration and the KD was calculated using a non‐linear regression dose‐response curve (Graph Pad Prism version7.02). Control experiments were performed using bovine serum albumin (BSA) and FITC‐NEC‐13 peptide. For competition FP binding assays, 10 nm of FITC‐SHP was mixed with Rgg144 protein to a final concentration of 6.6 μM. This mixture was incubated with diluted unlabeled SHP for 30 min, and polarization values were detected as described above.
4.13. Mouse studies
The significance of selected residues and the impact of the unmodified synthetic peptides on pneumococcal colonization was tested in the mouse nasopharyngeal colonization model (Kahya et al., 2017). For infection studies, female CD1 outbred mice, 8–10 weeks old, were used. Each individual mouse received approximately 2.5 × 105 CFU in 20 μl PBS intranasally under light anesthesia using 2.5% (v/v) isoflurane (Isocare) over oxygen (1.4–1.6 liters/min). When required synthetic peptides were also administered in 20 μl PBS. After administration of the dose, the inoculum size was further confirmed by platting. During the course of infection, the animals were observed daily for disease signs such as lethargy, hunched, or piloerect postures. To determine the bacterial counts, a nasopharyngeal wash was obtained after culling the mice. The bacterial counts in these samples were determined by plating the serially diluted samples on blood agar plates containing 5 μg/ml of gentamicin to prevent contamination.
4.14. Statistical analysis
Graph Pad Prism version 7.02 (Graphpad, California, USA) was used to analyze all data presented in this study. Data were expressed as means ± standard error of the mean (SEM). One‐way analysis of variance (ANOVA) followed by Dunnett's multiple comparison test was used to compare the differences between the groups for growth analysis and enzymatic activity. A nonlinear regression dose‐response curve (stimulation and inhibition) was used to determine the K D and IC50 of direct and indirect fluorescence polarization data, respectively.
AUTHOR CONTRIBUTIONS
All authors contributed to the conception and design of the study; the acquisition, analysis, and interpretation of the data, and the writing of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Mouse studies were performed under the project (permit no. 60/4327) and personal (permit no. 80/10279) licenses according to the United Kingdom Home Office guidelines under the Animals Scientific Procedures Act 1986, and the University of Leicester ethics committee approval. The protocol used was approved by both the U.K. Home Office and the University of Leicester ethics committee. Where specified, the procedures were done under anesthetic with isoflurane. Animals were kept in individually ventilated cages in a controlled environment and were regularly monitored after infection to reduce suffering.
Supporting information
Supinfo
ACKNOWLEDGMENTS
We would like to thank Melissa Jansen van Renburgh and Angela Brueggemann for their guidance with the selection of genomes included in the dataset. We are grateful for the support from the NIH (R01 AI139077‐01A1 to NLH and R01 AI135060‐01A1 to AU). We also gratefully acknowledge the valuable support of the Preclinical Facility staff for the in vivo experiments in Leicester.
Abdullah, I. T. , Ulijasz, A. T. , Girija, U. V. , Tam, S. , Andrew, P. , Hiller, N. L. , Wallis, R. & Yesilkaya, H. (2022). Structure‐function analysis for the development of peptide inhibitors for a Gram‐positive quorum sensing system. Molecular Microbiology, 117, 1464–1478. 10.1111/mmi.14921
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aggarwal, C. , Jimenez, J.C. , Nanavati, D. & Federle, M.J. (2014) Multiple length peptide‐pheromone variants produced by Streptococcus pyogenes directly bind Rgg proteins to confer transcriptional regulation. The Journal of Biological Chemistry, 289(32), 22427–22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal, S.D. , Yesilkaya, H. , Dawid, S. & Hiller, N.L. (2020) The pneumococcal social network. PLoS Pathogens, 16(10), e1008931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Bayati, F.A. , Kahya, H.F. , Damianou, A. , Shafeeq, S. , Kuipers, O.P. , Andrew, P.W. et al. (2017) Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase. Scientific Reports, 7, 43587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida‐Santos, A.C. , Novais, C. , Peixe, L. & Freitas, A.R. (2021) Enterococcus spp. as a producer and target of bacteriocins: a double‐edged sword in the antimicrobial resistance crisis context. Antibiotics (Basel), 10(10), 1215–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaut, L. , Perchat, S. , Arold, S. , Zorrilla, S. , Slamti, L. , Henry, C. et al. (2008) Molecular basis for group‐specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Research, 36(11), 3791–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodagli, G.C. , Tylor, K.M. , Kaelber, J.T. , Petrou, V.I. , Federle, M.J. & Neiditch, M.B. (2020) Structure‐function studies of Rgg binding to pheromones and target promoters reveal a model of transcription factor interplay. Proceedings of the National Academy of Sciences of the United States of America, 117(39), 24494–24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Guarnieri, M.T. , Vasil, A.I. , Vasil, M.L. , Mant, C.T. & Hodges, R.S. (2007) Role of peptide hydrophobicity in the mechanism of action of alpha‐helical antimicrobial peptides. Antimicrobial Agents and Chemotherapy, 51(4), 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, L.C. & Federle, M.J. (2014) Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiology Reviews, 38(3), 473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Waard, M. & Sabatier, J.‐M. (2006) Structure‐function strategies to improve the pharmacological value of animal toxins. In: Kastin, A.J. (Ed.) Handbook of biologically active peptides. Cambridge, MA: Academic Press, pp. 415–419. [Google Scholar]
- Eggink, D. , Berkhout, B. & Sanders, R.W. (2010) Inhibition of HIV‐1 by fusion inhibitors. Current Pharmaceutical Design, 16(33), 3716–3728. [DOI] [PubMed] [Google Scholar]
- Fetzner, S. (2015) Quorum quenching enzymes. Journal of Biotechnology, 201, 2–14. [DOI] [PubMed] [Google Scholar]
- Fleuchot, B. , Gitton, C. , Guillot, A. , Vidic, J. , Nicolas, P. , Besset, C. et al. (2011) Rgg proteins associated with internalized small hydrophobic peptides: a new quorum‐sensing mechanism in streptococci. Molecular Microbiology, 80(4), 1102–1119. [DOI] [PubMed] [Google Scholar]
- Fosgerau, K. & Hoffmann, T. (2015) Peptide therapeutics: current status and future directions. Drug Discovery Today, 20(1), 122–128. [DOI] [PubMed] [Google Scholar]
- Guiral, S. , Henard, V. , Laaberki, M.H. , Granadel, C. , Prudhomme, M. , Martin, B. et al. (2006) Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose‐driven gene expression in Streptococcus pneumoniae . Microbiology (Reading), 152(Pt 2), 343–349. [DOI] [PubMed] [Google Scholar]
- Halfmann, A. , Hakenbeck, R. & Bruckner, R. (2007) A new integrative reporter plasmid for Streptococcus pneumoniae . FEMS Microbiology Letters, 268(2), 217–224. [DOI] [PubMed] [Google Scholar]
- Hong, L. , Zhang, X.C. , Hartsuck, J.A. & Tang, J. (2000) Crystal structure of an in vivo HIV‐1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Science, 9(10), 1898–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahya, H.F. , Andrew, P.W. & Yesilkaya, H. (2017) Deacetylation of sialic acid by esterases potentiates pneumococcal neuraminidase activity for mucin utilization, colonization and virulence. PLoS Pathogens, 13(3), e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia, V.C. (2013) Quorum sensing inhibitors: an overview. Biotechnology Advances, 31(2), 224–245. [DOI] [PubMed] [Google Scholar]
- Karathanasi, G. , Bojer, M.S. , Baldry, M. , Johannessen, B.A. , Wolff, S. , Greco, I. et al. (2018) Linear peptidomimetics as potent antagonists of Staphylococcus aureus agr quorum sensing. Scientific Reports, 8(1), 3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre, B. & Federle, M.J. (2013) Exploiting quorum sensing to confuse bacterial pathogens. Microbiology and Molecular Biology Reviews, 77(1), 73–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson, M. , Nielsen, A. , Kjaerulff, L. , Gotfredsen, C.H. , Wietz, M. , Ingmer, H. et al. (2011) Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine photobacterium. Marine Drugs, 9(12), 2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnet, V. & Gardan, R. (2015) Quorum‐sensing regulators in Gram‐positive bacteria: “cherchez le peptide”. Molecular Microbiology, 97(2), 181–184. [DOI] [PubMed] [Google Scholar]
- Motib, A. , Guerreiro, A. , Al‐Bayati, F. , Piletska, E. , Manzoor, I. , Shafeeq, S. et al. (2017) Modulation of quorum sensing in a gram‐positive pathogen by linear molecularly imprinted polymers with anti‐infective properties. Angewandte Chemie (International Ed. in English), 56(52), 16555–16558. [DOI] [PubMed] [Google Scholar]
- Panjikar, S. , Parthasarathy, V. , Lamzin, V.S. , Weiss, M.S. & Tucker, P.A. (2005) Auto‐rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X‐ray diffraction experiment. Acta Crystallographica Section D, Biological Crystallography, 61(Pt 4), 449–457. [DOI] [PubMed] [Google Scholar]
- Parashar, V. , Aggarwal, C. , Federle, M.J. & Neiditch, M.B. (2015) Rgg protein structure‐function and inhibition by cyclic peptide compounds. Proceedings of the National Academy of Sciences of the United States of America, 112(16), 5177–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Jagasia, R. , Kaufmann, G.F. , Mathison, J.C. , Ruiz, D.I. , Moss, J.A. et al. (2007) Infection control by antibody disruption of bacterial quorum sensing signaling. Chemistry & Biology, 14(10), 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Pascual, D. , Monnet, V. & Gardan, R. (2016) Bacterial cell‐cell communication in the host via RRNPP peptide‐binding regulators. Frontiers in Microbiology, 7, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, H. , Ricklin, D. , Bai, H. , Chen, H. , Reis, E.S. , Maciejewski, M. et al. (2013) New analogs of the clinical complement inhibitor compstatin with subnanomolar affinity and enhanced pharmacokinetic properties. Immunobiology, 218(4), 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S.T. & Bassler, B.L. (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspectives in Medicine, 2, a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlla, B. , Gazioglu, O. , Shafeeq, S. , Manzoor, I. , Kuipers, O.P. , Ulijasz, A. et al. (2021) The Rgg1518 transcriptional regulator is a necessary facet of sugar metabolism and virulence in Streptococcus pneumoniae . Molecular Microbiology, 116, 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti, L. & Lereclus, D. (2005) Specificity and polymorphism of the PlcR‐PapR quorum‐sensing system in the Bacillus cereus group. Journal of Bacteriology, 187(3), 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretiakova, A.P. , Little, C.S. , Blank, K.J. & Jameson, B.A. (2000) Rational design of cytotoxic T‐cell inhibitors. Nature Biotechnology, 18(9), 984–988. [DOI] [PubMed] [Google Scholar]
- Wang, C.Y. , Medlin, J.S. , Nguyen, D.R. , Disbennett, W.M. & Dawid, S. (2020) Molecular determinants of substrate selectivity of a pneumococcal Rgg‐regulated peptidase‐containing ABC transporter. mBio, 11(1),e02502–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda, A. , Slamti, L. , Bochnik‐Tamir, R. , Malach, E. , Lereclus, D. & Hayouka, Z. (2018) Turning off Bacillus cereus quorum sensing system with peptidic analogs. Chemical Communications (Camb), 54(70), 9777–9780. [DOI] [PubMed] [Google Scholar]
- Yehuda, A. , Slamti, L. , Malach, E. , Lereclus, D. & Hayouka, Z. (2019) Elucidating the hot spot residues of quorum sensing peptidic autoinducer PapR by multiple amino acid replacements. Frontiers in Microbiology, 10, 1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. & Bremer, H. (1995) Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. The Journal of Biological Chemistry, 270(19), 11181–11189. [DOI] [PubMed] [Google Scholar]
- Zhi, X. , Abdullah, I.T. , Gazioglu, O. , Manzoor, I. , Shafeeq, S. , Kuipers, O.P. et al. (2018) Rgg‐Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Scientific Reports, 8(1), 6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supinfo
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


