Abstract
Penicillium chrysogenum uses sulfate as a source of sulfur for the biosynthesis of penicillin. Sulfate uptake and the mRNA levels of the sulfate transporter-encoding sutB and sutA genes are all reduced by high sulfate concentrations and are elevated by sulfate starvation. In a high-penicillin-yielding strain, sutB is effectively transcribed even in the presence of excess sulfate. This deregulation may facilitate the efficient incorporation of sulfur into cysteine and penicillin.
Penicillin biosynthesis by the filamentous fungus Penicillium chrysogenum starts with the condensation of three amino acids, l-α-aminoadipic acid, l-cysteine, and l-valine, by δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (20). Inorganic sulfate serves as a precursor for the formation of cysteine (9) during industrial penicillin production (16, 17). Sulfate is actively taken up (3, 10) and enters the sulfate assimilation pathway.
Sulfate uptake is generally an important point of regulation of sulfur metabolism in filamentous fungi. In Neurospora crassa (11, 17) and Aspergillus nidulans (1, 12, 13), sulfate uptake is subject to sulfur (metabolite) repression, which effects the expression of sulfate permease encoding genes. Recently, we isolated two genes of P. chrysogenum that encode sulfate transporters: SutB, the major sulfate transporter, and SutA, which is similar to SutB but whose function has not been elucidated (21). In P. chrysogenum Wisconsin 54-1255, the expression of both genes is elevated under sulfur starvation conditions (21). sutA is much more weakly expressed than sutB.
Penicillin fermentation normally occurs in the presence of excess sulfate (9), which represses the transcription of sutB and sutA (21). Our working hypothesis is that sulfate uptake is a factor that limits penicillin production. The objective of this study was to determine the relationship between penicillin biosynthesis and sulfate metabolism. We measured mRNA levels of sutB, sutA, and parA, a gene with a high degree of homology to 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductases. We concluded that the regulation of sulfate assimilation occurs by modulation of the enzyme activities and at the level of transcription and that sutB expression is deregulated in high-penicillin-producing strains.
Cloning of parA.
The sulfate assimilation pathway catalyzes the uptake and reduction of sulfate, yielding sulfide that is incorporated into Cys. PAPS reductase reduces the intermediate PAPS to sulfite. We obtained a partial cDNA clone with sequence similarity to the PAPS reductase-encoding genes from A. nidulans (2) and Saccharomyces cerevisiae (18) during a PCR-based strategy aimed at cloning genes with an ATP-binding motif. We isolated a full-length parA cDNA clone by inverse PCR with the primers parA-rev (5′-CTC GTT GTA GGG CAC ATC GTT CTC CTT GAC-3′) and parA-forw (5′-CTC CTC GAC CGT GGT TAT AAG AGC ATT GG-3′) on a cDNA library of a high-penicillin-yielding P. chrysogenum strain using the Expand Long Template PCR system (Boehringer-Mannheim, Mannheim, Germany). The blunt-ended (5.7-kb) PCR product (including the vector sequences) was phosphorylated, self-ligated, and transformed into Escherichia coli. The encoded gene product (GenBank accession no. AF227433) had high amino acid sequence similarity with PAPS reductases encoded by the A. nidulans sA gene (79% identity) (GenBank no. X82555) and the MET16 genes of S. cerevisiae (59% identity) (GenBank no. AAA34774), and Schizosaccharomyces pombe (56% identity) (GenBank no. Z69729). Because of the high sequence homology with known PAPS reductases, the cloned gene product is tentatively assigned as the P. chrysogenum ParA protein.
Sulfate uptake studies.
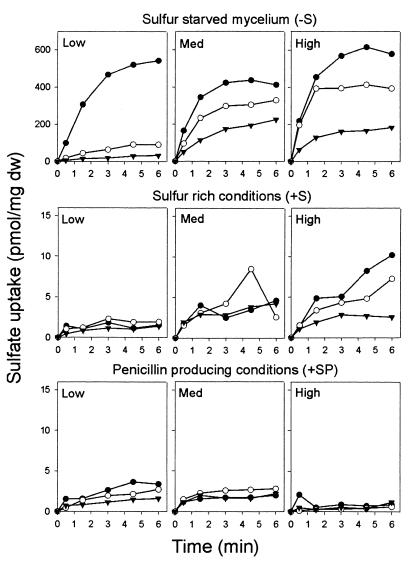
We grew three P. chrysogenum strains under different conditions and measured the uptake of radiolabeled sulfate ([35S]Na2SO4) (21). These strains mainly differ in the amount of penicillin produced (9): P. chrysogenum Wisconsin 54-1255 (5) produces relatively low amounts of penicillin; Panlabs P2 (14) is an intermediate penicillin-producing strain; and GB8 (8) produces relatively high amounts of penicillin. These strains were grown under various conditions. First, standard main-culture medium facilitating penicillin production (with lactose and sulfate as C and S sources, respectively) was used, and growth was allowed to proceed for 40, 64, and 88 h without medium exchange (designated +SP, for high sulfate and penicillin production). Second, growth occurred on the same medium for 24, 48, and 72 h, after which the medium was exchanged for fresh medium, and growth continued for another 16 h (sulfur sufficient [+S]) (final growth times, 40, 64, and 88 h). Third, growth proceeded as described above except that the sulfate salts were replaced by chloride salts in the exchange medium (sulfur starvation [−S]). Finally, strain GB8 was grown for 40, 64, and 88 h on standard medium but with glucose instead of lactose as the C source. Under these conditions, penicillin biosynthesis is repressed (4) (+SnP). Sulfate uptake is almost completely repressed in all three strains during growth on sulfur-sufficient medium (Fig. 1, +S). Only for strain GB8 was sulfate uptake significantly higher than the detection limit of 1 to 2 pmol/mg (dry weight). Growth under sulfur starvation conditions resulted in a strong induction of sulfate uptake (Fig. 1, −S). Initial sulfate uptake rates were highest for GB8 and lowest for Wisconsin 54-1255 but decreased with time to a rate that related inversely to the penicillin production capacity (Fig. 1, −S).
FIG. 1.
Sulfate uptake by mycelium of P. chrysogenum strains Wisconsin 54-1255 (Low, low-penicillin-yielding strain), Panlabs P2 (Med, moderately yielding strain), and GB8 (High, relatively high yielding strain). Prior to the sulfate uptake experiments, the mycelia were grown under different growth conditions. Sulfate uptake was monitored after 40 h (▾), 64 h (○), and 88 h (●) of growth. The data shown are for a single experiment, but similar results were obtained in two other independent experiments. The detection limit of the assay is 2 pmol/mg (dry weight [dw]).
Northern analyses.
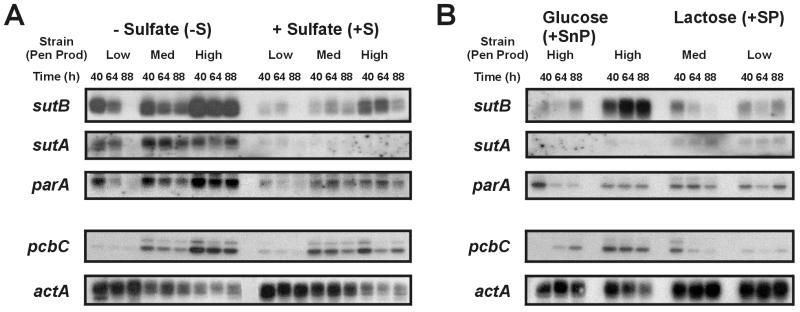
From the same mycelium used in the sulfate uptake experiments we also isolated total RNA and used Northern analyses to monitor the expression of sutA, sutB, parA, and pcbC, which encodes the second enzyme in the penicillin biosynthetic pathway (isopenicillin N synthase). Actin was used as an internal control. Sulfur starvation (−S) led to elevated levels of sutA, sutB, and parA transcripts compared to growth on sulfur-sufficient medium (+S) (Fig. 2A). In the high-yielding GB8 strain, the sutB mRNA level was strongly elevated and was relatively stably with time (Fig. 2A, −S), along with the sulfate uptake capacity (Fig. 1, −S). The extent of repression for parA is less than that for sutA and sutB.
FIG. 2.
Northern analysis of expression of sutA, sutB, parA, pcbC, and actA in mycelium of the P. chrysogenum strains Wisconsin 54-1255 (Low), Panlabs P2 (Med), and GB8 (High) after growth under (A) sulfur starvation (−S) and sulfur-sufficient (+S) conditions and (B) under penicillin-producing (+SP) and glucose-repressed (+SnP) conditions. Total RNAs (A, 10 μg for Wisconsin 54-1255 and Panlabs P2 and 3 μg for GB8; B, 10 μg for all strains) were separated by agarose-formaldehyde gel electrophoresis, transferred to nylon membranes (Bio-Rad Laboratories, Hercules, Calif.), and hybridized with 32P-labeled probes, as previously described (21). The data shown are for a single experiment, but similar results were obtained in two other independent experiments.
With time, the sutB and parA mRNA levels clearly decreased (Fig. 2A). For sulfur-starved mycelium, this decrease appears to follow the observed decrease in sulfate uptake activity (Fig. 1, −S). Both sulfate uptake and sutB mRNA levels dropped more quickly in Wisconsin 54-1255 than in GB8 (Fig. 2A). These observations cannot be attributed to differences in the sutB gene copy number, since all three strains carry only one copy of the sutB gene (results not shown). Although less evident due to the low mRNA levels, the decrease in the sutB transcript level with time also was observed for mycelium grown under sulfur-sufficient conditions (+S, +SP) (Fig. 2), for which sulfate uptake activity was at most barely detectable (Fig. 1). SutB is needed to acquire the sulfate under the +SP conditions (24). Therefore, it appears that in addition to the regulation of sutB transcript levels, SutB activity may be regulated also at the enzymatic level.
The mRNA levels of sutB and pcbC, especially in sulfur-starved mycelium (−S), increased with the penicillin-yielding capability of the strains (Fig. 2). Furthermore, the sutB mRNA level in GB8 was much higher under penicillin biosynthesis-inducing conditions (lactose as the C source, +SP) than under penicillin biosynthesis-repressing conditions (glucose as the C source, +SnP) (Fig. 2B), while the sutA mRNA levels were low under both conditions (Fig. 2B). The parA mRNA levels seemed not to relate to the penicillin-producing capacity of the strains used (Fig. 2B). We conclude that penicillin biosynthesis is correlated with sutB mRNA levels but not with the sutA and parA transcripts.
In S. cerevisiae, expression of the PAPS reductase-encoding MET16 gene is under both sulfur and amino acid control (18). However, for filamentous fungi, no information on the regulation of genes involved in sulfate assimilation is available (2, 6, 11). Since ATP sulfurylase is feedback inhibited by PAPS (2, 7, 15), it may seem unnecessary to regulate the transcription of genes in the pathway. Yet we found that the mRNA levels of the putative P. chrysogenum parA gene are controlled by the sulfur content of the medium. Consistent with earlier observations that SutB is the major sulfate permease of P. chrysogenum (21) and the observed correlation between the sulfate uptake activity and sutB mRNA levels (Fig. 1 and 2A), it seems likely that most sulfate enters the cell via SutB. The molecular basis of the relationship between penicillin biosynthesis and the sutB mRNA level has not been resolved. However, we hypothesize that under conditions of high penicillin biosynthesis, the elevated demand for sulfur and associated rapid consumption of Cys results in a depletion of an internal pool of sulfur-containing metabolites. The levels in this pool may be sensed by the regulatory system that monitors the sulfur requirement of the organism (19). Alternatively, the extensive mutagenesis and selection for high-penicillin-yielding P. chrysogenum strains may have resulted in mutations in the scon genes (11, 12) that have been implicated in the regulation of sulfur metabolism in other fungi. Taken together, these data suggest that in strains producing relatively high amounts of penicillin, sulfate metabolism and transport is effectively de-regulated.
Acknowledgments
This project was supported by the EC Eurofung Cell Factory RTD4CY96-0535.
We thank DSM-Gist (Delft, The Netherlands) for providing the cDNA library and the pcbC probe.
REFERENCES
- 1.Arst H N., Jr Genetic analysis of the first steps of sulfate metabolism in Aspergillus nidulans. Nature. 1968;219:268–270. doi: 10.1038/219268a0. [DOI] [PubMed] [Google Scholar]
- 2.Borges-Walmsley M I, Turner G, Bailey A M, Brown J, Lehmbeck J, Clausen I G. Isolation and characterisation of genes for sulfate activation and reduction in Aspergillus nidulans: implications for evolution of an allosteric control region by gene duplication. Mol Gen Genet. 1995;247:423–429. doi: 10.1007/BF00293143. [DOI] [PubMed] [Google Scholar]
- 3.Bradfield G, Somerfield P, Meyn T, Holby M, Babcock D, Bradley D, Segel I H. Regulation of sulfate transport in filamentous fungi. Plant Physiol. 1970;46:720–727. doi: 10.1104/pp.46.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakhage A A. Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–585. doi: 10.1128/mmbr.62.3.547-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantoral J M, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H, Martín J F. Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenumimpaired in penicillin biosynthesis. J Biol Chem. 1993;268:737–744. [PubMed] [Google Scholar]
- 6.Clarke D L, Newbert R W, Turner G. Cloning and characterisation of the adenosyl phophosulfate kinase gene from Aspergillus nidulans. Curr Genet. 1997;32:408–412. doi: 10.1007/s002940050295. [DOI] [PubMed] [Google Scholar]
- 7.Foster B A, Thomas S M, Mahr J A, Renosto F, Patel H C, Segel I H. Cloning and sequencing of ATP sulfurylase from Penicillium chrysogenum. Identification of a likely allosteric domain. J Biol Chem. 1994;269:19777–19786. [PubMed] [Google Scholar]
- 8.Gouka R J, van Hartingsveldt W, Bovenberg R A L, van Zeijl C M J, van den Hondel C A M J J, van Gorcom R F M. Development of a new transformant selection system for Penicillium chrysogenum: isolation and characterization of the P. chrysogenum acetyl-coezyme A synthetase gene (facA) and its use as a homologous selection marker. Appl Microbiol Biotechnol. 1993;38:514–519. doi: 10.1007/BF00242947. [DOI] [PubMed] [Google Scholar]
- 9.Herschbach G J M, van der Beek C P, van Dijck P W M. The penicillins: properties, biosynthesis, and fermentation. In: Van Damme E, editor. Biotechnology of industrial antibiotics. New York, N.Y: Marcel Dekker; 1984. pp. 45–140. [Google Scholar]
- 10.Hillenga D J, Versantvoort H J M, Driessen A J M, Konings W N. Sulfate transport in Penicillium chrysogenumplasma membranes. J Bacteriol. 1996;178:3953–3956. doi: 10.1128/jb.178.13.3953-3956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzluf G A. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- 12.Natorff R, Balinska M, Paszewski A. At least four regulatory genes control sulfur metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1993;238:185–192. doi: 10.1007/BF00279546. [DOI] [PubMed] [Google Scholar]
- 13.Natorff R, Piotrowska M, Paszewski A. The Aspergillus nidulans sulfur regulatory gene sconBencodes a protein with WD40 repeats and an F-box. Mol Gen Genet. 1998;257:255–263. doi: 10.1007/s004380050646. [DOI] [PubMed] [Google Scholar]
- 14.Newbert R W, Barton B, Greaves P, Harper J, Turner G. Analysis of a commercially improved Penicillium chrysogenumstrain series: involvement of recombigenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J Ind Microbiol Biotechnol. 1997;19:18–27. doi: 10.1038/sj.jim.2900411. [DOI] [PubMed] [Google Scholar]
- 15.Renosto F, Martin R L, Wailes L M, Daley L A, Segel I H. Regulation of inorganic sulfate activation in filamentous fungi. Allosteric inhibition of ATP sulfurylase by 3′-phosphoadenosine-5′-phosphosulfate. J Biol Chem. 1990;265:10300–10308. [PubMed] [Google Scholar]
- 16.Segel I H, Johnson M J. Accumulation of intracellular inorganic sulfate by Penicillium chrysogenum. J Bacteriol. 1961;81:91–98. doi: 10.1128/jb.81.1.91-98.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao Y, Marzluf G A. Synthesis and differential turnover of the CYS3 regulatory protein of Neurospora crassaare subject to sulfur control. J Bacteriol. 1998;180:478–482. doi: 10.1128/jb.180.3.478-482.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas D, Barbey R, Surdin-Kerjan Y. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3′-phosphoadenylylsulfate reductase structural gene. J Biol Chem. 1990;265:15518–15524. [PubMed] [Google Scholar]
- 19.Tweedie J W, Segel I H. Specificity of transport processes for sulfur, selenium, and molybdenum anions by filamentous fungi. Biochim Biophys Acta. 1970;196:95–106. doi: 10.1016/0005-2736(70)90170-7. [DOI] [PubMed] [Google Scholar]
- 20.van de Kamp M, Driessen A J M, Konings W N. Compartmentalization and transport in β-lactam antibiotic biosynthesis by filamentous fungi. Antonie Leeuwenhoek. 1999;75:41–78. doi: 10.1023/a:1001775932202. [DOI] [PubMed] [Google Scholar]
- 21.van de Kamp M, Pizzinini E, Vos A, van der Lende T R, Schuurs Theo A, Newbert R W, Turner G, Konings W N, Driessen A J M. Sulfate transport in Penicillium chrysogenum: cloning and characterization of the sutA and sutBgenes. J Bacteriol. 1999;181:7228–7234. doi: 10.1128/jb.181.23.7228-7234.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]