Abstract
Background
The role of Helicobacter pylori (H. pylori) virulence factors of such as vacA s1m1 and cagA in designating clinical outcomes and eradication rate has been deeply challenged in the last decade. The goal of this analysis was to identify the potential relevance between cagA and vacA genotypes with reported antibiotic resistance observed in clinical H. pylori isolates.
Methods
This literature search was conducted in databases such as Clarivate analytics, PubMed, Scopus, EMBASE, DOAJ, and Google Scholar by April 2022, regardless of language restrictions and publication date. Quality of the included studies was assessed by the Newcastle–Ottawa scale. Statistical analysis of retrieved studies was fulfilled using Comprehensive Meta-Analysis software version 2.2. Following quality appraisal of eligible studies, potential association between the status of cagA and vacA genes with resistance to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin was measured using odds ratio with 95% confidence interval. We also used sensitivity analyses and meta-regression to eliminate the source of heterogeneity from the overall estimates. Publication bias was assessed using funnel plot, Egger’s test, Begg’s test with the trim and fill procedure to assess the presence and magnitude of publication bias in the included studies.
Results
Our findings suggested that a significant relationship between cagA status and increase resistance to metronidazole (OR: 2.69; 95% CI: 1.24–5.83). In subgroup analysis, we found that in the Western population, infection with cagA-positive strains could be led to increase in the resistance to metronidazole (OR: 1.59; 95% CI: 0.78–3.21), amoxicillin (OR: 19.68; 95% CI: 2.74–141.18), and levofloxacin (OR: 11.33; 95% CI: 1.39–91.85). After implementation of trim and fill method, the adjusted OR was not significantly differed from original estimates which in turn represented our subgroup analysis was statistically robust. On the other hand, vacA genotypes usually reduce the antibiotic resistance of this bacterium, so that vacA s1m1 significantly reduces the resistance to metronidazole (OR: 0.41; 95% CI: 0.20–0.86). Surprisingly, resistance of vacA s2m2 strains to antibiotics was low, the reason may be due to the non-inflammatory properties of strains containing vacA s2m2. The meta-regression and sensitivity analyses successfully reduced the effect of heterogeneity from the overall estimates. In addition, although the pooled OR is reduced after trim and fill adjustment but results do not change the conclusion regarding vacA genotypes and antibiotic resistance.
Conclusions
According to our findings, it was clearly demonstrated that cagA-positive strains are resistance to metronidazole, especially in Western countries. In Western countries, vacA s1m1 increases resistance to amoxicillin and levofloxacin. Based on the present findings, the vacA s1m1 genotype significantly increases resistance to metronidazole, while the vacA s1m2 decreases resistance to clarithromycin and metronidazole. Resistance to antibiotics in less virulent (vacA s2m2) strains is statistically significant lower than others.
Keywords: Antibiotic resistance, cagA, H. pylori, Treatment, vacA
Background
Helicobacter pylori (H. pylori) is a S-shaped microorganism that colonize in the surface of gastric mucosa of half the world’s population, maybe even more [1]. Long last colonization with this bacterium leads to a chronic progressive gastric inflammation associated with severe gastrointestinal effects [2]. Nowadays, eradication of H. pylori is the main therapeutic strategy in management of patients who suffering from different complications including peptic ulcer disease (PUD), gastric cancer (GC), mucosa associated-lymphoid tissue (MALT) lymphoma, and atrophic gastritis [3]. According to the Kyoto Global Consensus Conference, eradication of H. pylori infection among the asymptomatic subjects seems an necessity [4]. Nevertheless, the rate of the treatment for H. pylori infection is declining annually; the emergence of clarithromycin-resistant strains has been declared a global threat by the World Health Organization (WHO) [5, 6].
The cure rate of H. pylori infection could be affected by both microbial (high bacterial load, point mutations, biofilm formation, efflux pumps, and virulence factors), and non-microbial (cytochrome P450 2C19 polymorphism, multidrug resistance transporter-1, pro-inflammatory cytokines polymorphism, smoking, life style, duration of treatment, high gastric acidity, poor patient compliance) factors; all of these factors play a role in the severity of the infection [3, 7, 8]. Vacuolating cytotoxin A (vacA) and cytotoxin associated gene A (cagA) are considered as the main virulence factors of H. pylori [9]. The toxin encoded by the vacA gene causes apoptosis, T-cell activation, and persistent infection (through inhibition of immune system), which these changes are lead to severe gastrointestinal outcomes [10]. Full-length sequence analysis of the vacA gene showed that this gene has a mosaic structure and is encoded by different subfamilies s1, m1 and m2 alleles, with its own biological activities [11]. The vacA s1/m1 genotype possess the highest toxicity property for host cells, while the vacA s2/m2 genotype biologically is inactive [12, 13]. CagA is encoded by cagA gene; this toxin is highly immunogenic, and upon entering the host cell, it activates kinases through EPIYA motifs in its C-terminal, which in turn disrupt signaling pathways [14].
Studies have shown that this protein induces IL-8 expression, which contributes to the formation of cytokine storms and eventually susceptibility to PUD as well as GC [15]. Both CagA and VacA antigens significantly affect the colonization and pathogenesis of this bacterium, and play a determining role in cure rate of disease [16, 17]. Although chromosomal mutations are considered to be the main mechanism of antibiotic resistance, but, the location of these single nucleotide polymorphisms (SNPs) is not the same in all populations, and therefore, understanding the mechanisms of antibiotic resistance of H. pylori is essential for the introduction of rational antibiotic combinations [18]. In recent studies, the eradication results associated with CagA and VacA status are highly inconsistent [19–22]. Interestingly, in meta-analysis by Wang et al. (collecting the data from 26 papers), it was represented that the eradication rate of infection in patients infected with vacA s1/cagA positive strains was more conducive compared to less virulent strains [8].
In this study, we performed a comprehensive literature search to demonstrate the relationship between cagA or vacA status and antibiotic resistance in H. pylori.
Methods
Eligibility of relevant studies
Using international databases such as the Clarivate analytics, PubMed, Scopus, EMBASE, DOAJ, and Google Scholar, related articles to the effect of cagA and vacA on the antibiotic resistance of H. pylori were reviewed, regardless of publication and language restrictions until April 2022. In this regard, we used keywords based on MeSH terms such as “Genotype”, “Antibiotic resistance”, “Helicobacter pylori”, “H. pylori”, “VacA”, “CagA”, and “Antimicrobial resistance”. The bibliography of articles was reviewed manually to retrieve missing related studies.
Inclusion and exclusion criteria
Our inclusion criteria were the following: (1) studies on the association between cagA/vacA status and antibiotic resistance; (2) studies on human subjects; (3) studies based on standard methodology (CLSI); (4) studies without repetitive samples. On the other hand, studies such as case reports, reviews, congress abstracts, duplicates, studies on non cagA/vacA genes, in vitro studies, as well as studies without clear results were excluded from this study.
Data extraction
Eligibility of studies was evaluated by the two authors separately, and conflicting of interest was resolved by discussion. The main items were including: first author, country, year of publication, number of H. pylori isolates, number of cagA + isolates, number of vacA s1m1 + isolates, antimicrobial susceptibility tests, and frequency of each genotype (cagA and vacA s1m1) resistant to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin (Table 1) [23–63].
Table 1.
Characteristics of included studies
| First author | Country | Year | Number of H. pylori isolates | Number of cagA + H. pylori isolates | Number of vacA s1m1 + H. pylori isolates | Methods | Number of H. pylori resistant to clarithromycin | Number of H. pylori resistant to metronidazole | Number of H. pylori resistant to amoxicillin | Number of H. pylori resistant to tetracycline | Number of H. pylori resistant to levofloxacin | Refs. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cagA+ | vacA s1m1+ | cagA+ | vacA s1m1+ | cagA+ | vacA s1m1+ | cagA+ | vacA s1m1+ | cagA+ | vacA s1m1+ | ||||||||
| Broutet | France | 2001 | 156 | 84 | NR | E-test | 8/16 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [23] |
| Solca | Switzerland | 2001 | 71 | 38 | 24 | NR | 6/12 | 4/12 | 12/28 | 9/28 | NR | NR | NR | NR | NR | NR | [24] |
| Toro | Spain | 2003 | 98 | 63 | NR | E-test | 1/10 | NR | 22/38 | NR | NR | NR | NR | NR | NR | NR | [25] |
| Elviss | UK | 2004 | 363 | 287 | 149 | E-test | 1/3 | 0/3 | 19/31 | 6/31 | NR | NR | NR | NR | NR | NR | [26] |
| Elviss | UK | 2005 | 101 | 81 | 48 | E-test | 6/12 | 5/12 | 38/53 | 27/53 | NR | NR | NR | NR | NR | NR | [27] |
| Chihu | Mexico | 2005 | 49 | 38 | NR | E-test | NR | NR | 9/11 | NR | NR | NR | NR | NR | NR | NR | [28] |
| Francesco | Italy | 2006 | 62 | 40 | 23 | E-test | 10/15 | 4/15 | NR | NR | NR | NR | NR | NR | NR | NR | [29] |
| Lai | Taiwan | 2006 | 31 | 31 | NR | E-test | 13/53 | NR | 20/53 | NR | NR | NR | NR | NR | NR | NR | [30] |
| Boyanova | Bulgaria | 2009 | 108 | 88 | NR | Agar dilution method | 22/31 | NR | 35/45 | NR | NR | NR | NR | NR | NR | NR | [31] |
| Taneike | Ireland | 2009 | 103 | 70 | 19 | E-test | 3/17 | 2/17 | 10/39 | 12/39 | NR | NR | NR | NR | NR | NR | [32] |
| Hu | Taiwan | 2009 | 133 | 127 | 59 | PCR-RFLP | 18/18 | 8/18 | NR | NR | NR | NR | NR | NR | NR | NR | [33] |
| Trespalacios | Columbia | 2010 | 79 | NR | NR | E-test | 7/14 | 5/14 | 21/64 | 17/64 | 3/3 | 2/3 | NR | NR | NR | NR | [34] |
| Agudo | Spain | 2010 | 117 | 44 | NR | E-test | 5/34 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [35] |
| Vega | Argentina | 2010 | 299 | 122 | 200 | Agar dilution | 44/83 | 73/83 | 73/113 | 84/113 | NR | NR | NR | NR | NR | NR | [36] |
| Ayala | Mexico | 2011 | 90 | NR | NR | E-test | OR: 0.79; 95% CI: 0.11–5.33 | OR: 4.76; 95% CI: 0.2–109.7 | OR: 0.69; 95% CI: 0.21–2.28 | OR: 2.58; 95% CI: 0.59–11.3 | NR | NR | NR | NR | NR | NR | [37] |
| Babab | Japan | 2011 | 35 | 35 | NR | PCR | 12/35 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [38] |
| Khan | Pakistan | 2012 | 178 | 83 | NR | Agar dilution | 35/64 | NR | 67/149 | NR | 20/66 | NR | NR | NR | NR | NR | [39] |
| Yula | Turkey | 2013 | 91 | 68 | NR | Agar dilution | 6/7 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [40] |
| Ghotaslou | Iran | 2013 | 99 | 84 | NR | Modified disk diffusion test | 16/21 | NR | 67/97 | NR | 24/34 | NR | NR | NR | NR | NR | [41] |
| Alfizah | Malaysia | 2013 | 95 | NR | 49 | E-test | NR | NR | NR | 15/28 | NR | NR | NR | NR | NR | NR | [42] |
| Rengifo | Colombia | 2013 | 149 | NR | NR | Agar dilution | 6/7 | 6/7 | NR | NR | 7/8 | 7/8 | NR | NR | NR | NR | [43] |
| Karabiber | Turkey | 2014 | 98 | 50 | NR | Disk-diffusion | 4/12 | NR | 3/6 | NR | NR | NR | NR | NR | NR | NR | [44] |
| Rasheed | Pakistan | 2014 | 46 | 37 | 27 | E-test | 17/22 | 13/22 | 28/34 | 18/34 | 19/25 | 14/25 | 0/2 | 2/2 | NR | NR | [45] |
| Hussein | Iraq | 2015 | 74 | 35 | 42 | GenoType HelicoDR kit | 3/12 | 2/12 | NR | NR | NR | NR | NR | NR | 1/3 | 2/3 | [46] |
| Boyanova | Bulgaria | 2015 | 84 | 64 | 21 | E-test | 26/26 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [47] |
| Fasciana | Italy | 2015 | 100 | 48 | 35 | E-test | 9/25 | 12/25 | NR | NR | NR | NR | NR | NR | NR | NR | [48] |
| Liou | Taiwan | 2015 | 1395 | 597 | 300 | Agar dilution | 135/1175 | 63/578 | 294/1176 | 155/577 | 29/1177 | 14/579 | 36/1159 | 24/564 | 103/1180 | 44/581 | [49] |
| Mill´an | Mexico | 2016 | 45 | 35 | 36 | Disk-diffusion | 3/8 | 3/8 | NR | NR | NR | NR | NR | NR | NR | NR | [50] |
| Miftahussurur | Indonesia | 2016 | 77 | 73 | 52 | E-test | 7/7 | 6/7 | 34/36 | 21/36 | NR | NR | NR | NR | 22/24 | 16/24 | [51] |
| Schwetz | Austria | 2016 | 178 | 100 | 72 | E-test | 27/54 | 21/54 | 21/35 | 16/35 | NR | NR | NR | NR | 17/21 | 15/21 | [52] |
| Bachir | Algeria | 2018 | 163 | 97 | 100 | E-test | 18/151 | 18/151 | 65/151 | 66/151 | NR | NR | NR | NR | NR | NR | [53] |
| Farzi | Iran | 2019 | 68 | 57 | 26 | Agar dilution | 20/23 | 10/23 | 52/56 | 23/56 | 18/21 | 8/21 | 3/3 | 1/3 | 17/19 | 7/19 | [54] |
| Imkamp | Switzerland | 2019 | 41 | 19 | NR | E-test | 14/35 | NR | 15/30 | NR | NR | NR | NR | NR | 7/12 | NR | [55] |
| Khani | Iran | 2019 | 61 | 40 | 25 | E-test | 13/48 | 13/48 | NR | NR | NR | NR | NR | NR | NR | NR | [56] |
| Abdollahi | Iran | 2019 | 63 | 37 | NR | Modified disk diffusion | 15/20 | NR | 22/35 | NR | 14/17 | NR | 1/2 | NR | NR | NR | [57] |
| Farzi | Iran | 2019 | 33 | 29 | 12 | Agar dilution | 11/12 | 4/12 | 25/33 | 9/33 | 9/10 | 3/10 | 2/2 | 1/2 | 9/9 | 2/9 | [58] |
| Wang | China | 2019 | 100 | 87 | 42 | E-test | OR: 2.192; 95% CI: 0.427–11.235 | OR: 0.763; 95% CI: 0.287–2.027 | OR: 1.509; 95% CI: 0.409–5.561 | OR: 0.287; 95% CI: 0.096–0.863 | 0 | OR: 0.434; 95% CI: 0.078–2.420 | 0 | OR: 0.758; 95% CI: 0.215–2.667 | OR: 5.133; 95% CI: 1.297–20.319 | OR: 0.749; 95% CI: 0.311–1.804 | [59] |
| Glowniak | Poland | 2019 | 62 | 35 | 12 | E-test | 3/4 | 2/4 | 6/8 | 2/8 | 0 | 0 | 0 | 0 | 3/4 | 1/4 | [60] |
| Hamidi | Iran | 2020 | 50 | 27 | 8 | Agar dilution | 7/11 | 3/11 | 17/34 | 3/34 | 11/16 | 3/16 | 5/8 | 1/8 | 11/14 | 3/14 | [61] |
| Haddadi | Iran | 2020 | 128 | 72 | NR | Disk diffusion | 4/4 | NR | 47/52 | NR | 20/23 | NR | 5/5 | NR | NR | NR | [62] |
| Okullu | Turkey | 2020 | 33 | 11 | NR | GenoType HelicoDR kit | 4/13 | NR | NR | NR | NR | NR | NR | NR | NR | NR | [63] |
| NR not reported | |||||||||||||||||
According to the literature, vacA s1m1 is the most virulent genotype of H. pylori, nevertheless, in the present meta-analysis, we evaluated the frequency of other vacA genotypes in all eligible studies. The distribution of antibiotic resistance of three genotypes vacA s1m2, vacA s2m1, and vacA s2m2 was assessed and their results are shown in Table 2.
Table 2.
Distribution of antibiotic resistance in vacA genotypes
| First author | vacA genotypes | Clarithromycin | Metronidazole | Amoxicillin | Tetracycline | Levofloxacin | Refs. |
|---|---|---|---|---|---|---|---|
| Solca | vacA s1/m2 | 4/12 | 8/28 | NR | NR | NR | [24] |
| vacA s2/m1 | 1/12 | 1/28 | NR | NR | NR | ||
| vacA s2/m2 | 3/12 | 10/28 | NR | NR | NR | ||
| Elviss | vacA s1/m2 | 1/3 | NR | NR | NR | NR | [26] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 0/3 | 2/8 | NR | NR | NR | ||
| Elviss | vacA s1/m2 | 2/3 | 22/31 | NR | NR | NR | [27] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 0/3 | 1/31 | NR | NR | NR | ||
| Francesco | vacA s1/m2 | 6/15 | NR | NR | NR | NR | [29] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 4/15 | NR | NR | NR | NR | ||
| Trespalacios | vacA s1/m2 | NR | NR | NR | NR | NR | [34] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 2/15 | 9/15 | 2/15 | NR | NR | ||
| Vega | vacA s1/m2 | NR | NR | NR | NR | NR | [36] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 10/83 | 29/113 | NR | NR | NR | ||
| Alfizah | vacA s1/m2 | NR | 12/28 | NR | NR | NR | [42] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | NR | NR | NR | NR | NR | ||
| Rasheed | vacA s1/m2 | 7/22 | 13/34 | 9/25 | 0/2 | NR | [45] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 2/22 | 3/34 | 2/25 | 0/2 | NR | ||
| Hussein | vacA s1/m2 | 2/12 | NR | NR | NR | 1/3 | [46] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 3/12 | NR | NR | NR | 0/3 | ||
| Fasciana | vacA s1/m2 | 4/25 | NR | NR | NR | NR | [48] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 9/25 | NR | NR | NR | NR | ||
| Liou | vacA s1/m2 | 76/643 | 162/646 | 13/645 | 11/634 | 62/646 | [49] |
| vacA s2/m1 | 0/3 | 2/3 | 0/3 | 0/3 | 0/3 | ||
| vacA s2/m2 | 0/5 | 0/5 | 0/5 | 0/5 | 1/5 | ||
| Mill´an | vacA s1/m2 | 0/8 | NR | NR | NR | NR | [50] |
| vacA s2/m1 | 0/8 | NR | NR | NR | NR | ||
| vacA s2/m2 | 2/8 | NR | NR | NR | NR | ||
| Schwetz | vacA s1/m2 | 14/54 | 6/35 | NR | NR | 3/21 | [52] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 19/54 | 13/35 | NR | NR | 3/21 | ||
| Bachir | vacA s1/m2 | 6/38 | 13/102 | NR | NR | NR | [53] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 9/38 | 19/102 | NR | NR | NR | ||
| Farzi | vacA s1/m2 | 11/23 | 29/56 | 9/21 | 2/3 | 9/19 | [54] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 2/23 | 4/56 | 4/21 | 0/3 | 3/19 | ||
| Khani | vacA s1/m2 | 12/48 | NR | NR | NR | NR | [56] |
| vacA s2/m1 | 9/48 | NR | NR | NR | NR | ||
| vacA s2/m2 | 14/48 | NR | NR | NR | NR | ||
| Farzi | vacA s1/m2 | 7/12 | 14/27 | 4/10 | 1/9 | 5/9 | [58] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 1/12 | 4/27 | 3/10 | 2/9 | 0/2 | ||
| Hamidi | vacA s1/m2 | 4/11 | 14/34 | 7/16 | 4/8 | 5/14 | [61] |
| vacA s2/m1 | NR | NR | NR | NR | NR | ||
| vacA s2/m2 | 2/11 | 4/34 | 3/16 | 1/8 | 1/14 | ||
| Glowniak | vacA s1/m2 | 1/4 | 3/8 | NR | NR | 1/4 | [60] |
| vacA s2/m2 | 1/4 | 3/8 | NR | NR | 2/4 |
Quality assessment
The Newcastle–Ottawa scale (NOS) was used to assess the quality of the included studies. The quality of studies was evaluated based on the items such as selection, comparability, and outcome, so that NOS scores in the range of 1–3, 4–6, and 7–9 were considered low, medium, and high respectively. The quality appraisal process was performed separately by the two authors, and the disagreement was resolved through discussion.
Statistical analysis
Retrieved studies was analyzed using Comprehensive Meta-Analysis (CMA) software version 2.2 (Biostat, Englewood, NJ, USA). Frequency of cagA- and vacA-positive strains was measured based on the event rate with 95% confidence interval (95%CI). Finally, the association between the genotypes of these virulence factors and resistance to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin was calculated using the odds ratio (OR) and corresponding 95% CI. For measuring heterogeneity, we used from two parameters Cochran’s Q statistic and I2 statistic. The fixed-effects model was used when there was no significant heterogeneity (p value ≥ 0.10 and I2 ≤ 50%) between the studies [64]; a random-effect model based on the Dersimonian and Laird method was used if significant heterogeneity was identified [65]. Eventually, publication bias was assessed by Egger’s p value test, Begg’s p value test, and asymmetry of funnel plot [66]. We also used the “trim-fill” method to prove the correction effect on publication bias according to Duval and Tweedie [67, 68]. We performed subgroup analysis based on several items such as ethnicity, study sample size, diagnostic test, and developing/developed status of country. Moreover, the leave-one-out method as sensitivity analyses were performed to estimate the effect of each included study on overall effect [69]. A random effects meta-regression analysis was performed to assess the potential sources of heterogeneity to explore factors that may be associated with between-study variations in H. pylori antibiotic resistance.
Results
Characteristics of the included studies
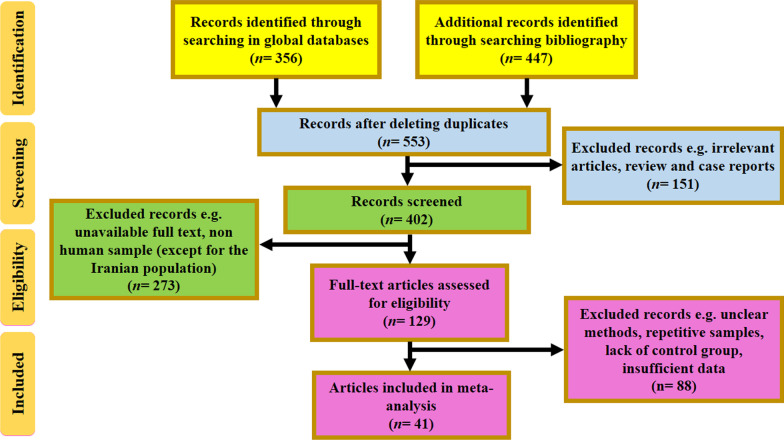
A systematic literature search was conducted based on PRISMA guideline. In the first stage, 509 articles were selected as potential documents. According to the inclusion criteria 471 articles were deleted and finally 38 eligible articles were entered in the present research (Fig. 1). Of all eligible studies, 38 articles had evaluated the relationship of cagA and antibiotic resistance, while 23 articles had assessed the effect of vacA genotypes on antibiotic resistance. The NOS results showed that the quality of eligible studies was ranged between 6 and 8. All studies in had been performed in regions such as Asia, Europe, and Latin America during 2001–2020. Standard methods for detecting antibiotic resistance included agar dilution, modified disk-diffusion agar, E-test, PCR-RFLP, GenoType HelicoDR kit. In the present study, 5156 of clinical positive samples were evaluated, and consequently the frequency of infection with cagA and vacA s1m1 was computed 64.6% (95% CI: 58.4–70.4) and 41.9% (95% CI: 34.3–50.0), respectively.
Fig. 1.
The flowchart of included studies
The vacA status and antibiotic resistance
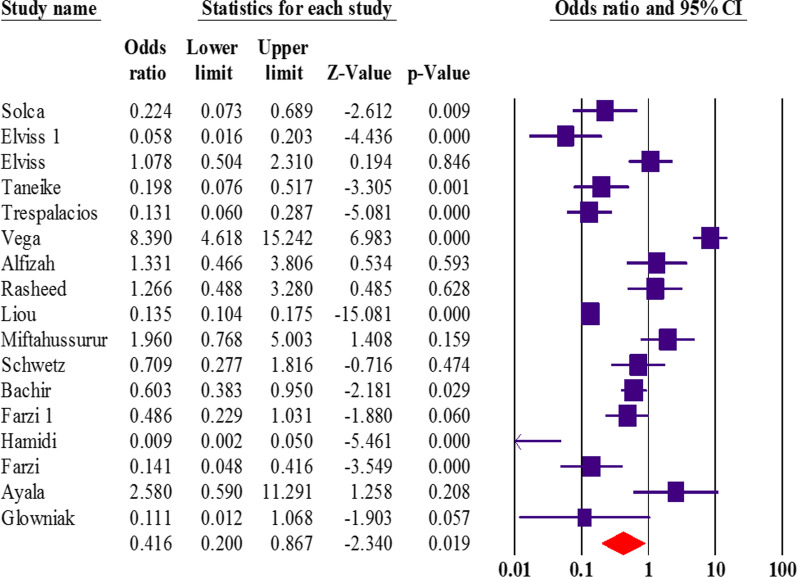
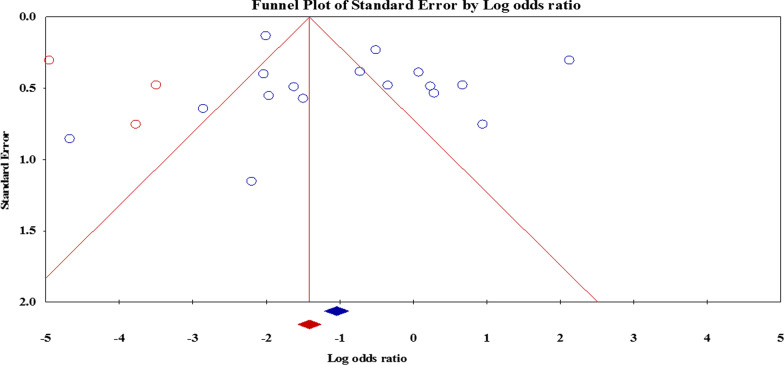
Overall, 23 articles had appraised the vacA genotypes status and resistance to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin. Interestingly, we found that the vacA s1m1 significantly reduced the risk of resistance to metronidazole (OR: 0.41; 95% CI: 0.20–0.86) (Fig. 2). After exclusion 4 studies, the sensitivity analysis was similar (OR: 0.34; 95% CI: 0.29–0.40) without significant heterogeneity rate. Moreover, the results were not significant for other antibiotics (Table 3). Due to the presence of a significant asymmetry in funnel plots, we performed trim and fill method to exclude potential publication bias. Adjusted OR according to the trim-and-fill method was lower than the original estimates but results were similar to the original findings (OR: 0.25; 95% CI: 0.11–0.57); however, a significant difference was not noted between before and after filling the potential missing studies (Fig. 3). Thus, trim and fill method did not change conclusion, indicating that our results were statistically robust regarding potential association between vacA s1m1 and resistance to metronidazole.
Fig. 2.
The forest plot associated with vacA s1m1 and resistance to metronidazole
Table 3.
Odds ratio (OR) with 95% CI for vacA s1m1 genotype and antibiotic resistance in H. pylori
| Antibiotic resistance | Random effects model | Heterogeneity | Publication bias | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p value | p value | I-squared | Egger’s p value | Begg’s p value | |
| Clarithromycin | 0.40 (0.13–1.22) | 0.1 | 0.01 | 94.69 | 0.01 | 0.79 |
| Metronidazole | 0.41 (0.20–0.86) | 0.01 | 0.01 | 93.54 | 0.37 | 0.23 |
| Amoxicillin | 0.32 (0.01–5.78) | 0.4 | 0.01 | 96.70 | 0.05 | 0.5 |
| Tetracycline | 0.19 (0.007–5.49) | 0.3 | 0.01 | 94.80 | 0.1 | 0.2 |
| Levofloxacin | 0.40 (0.03–4.18) | 0.4 | 0.01 | 97.0 | 0.04 | 0.9 |
Fig. 3.
The funnel plot adjusted using a trim and fill method for evaluation of possible link between vacA s1m1 and resistance to metronidazole
The details of overall estimates related to vacA s1m1 based on the sample size of the study, diagnostic test, and developing/developed status of country are given in the Table 4.
Table 4.
The vacA s1m1-positive status and metronidazole resistance
| Factors | Random-effects model | Heterogeneity | ||||
|---|---|---|---|---|---|---|
| OR | 95%CI | p value | p value | I-squared | ||
| Level of country | Developing country | 0.30 | 0.13–0.68 | 0.01 | 0.01 | 86.26 |
| Developed country | 0.55 | 0.18–1.65 | 0.01 | 0.01 | 93.33 | |
| Sample size | ≥ 100 | 1.13 | 0.84–1.52 | 0.01 | 0.31 | 24.65 |
| ≤ 100 | 0.28 | 0.13–0.60 | 0.01 | 0.05 | 64.32 | |
| Diagnostic test | E-test | 0.64 | 0.26–1.57 | 0.3 | 0.02 | 58.32 |
| Agar dilution based | 0.25 | 0.03–1.79 | 0.17 | 0.5 | 32.81 | |
| Disk diffusion based | 2.12 | 0.96–4.67 | 0.05 | 0.9 | 0.00 | |
| Molecular based | 1.33 | 0.46–3.80 | 0.03 | 0.9 | 0.00 | |
In subgroup analysis, the results showed that in an Asian population vacA s1m1 significantly increases the resistance of H. pylori to metronidazole (OR: 0.37; 95% CI: 0.15–0.90), while in Western countries, vacA s1m1 increases resistance to amoxicillin and levofloxacin. (OR: 16.58; 95% CI: 1.77–154.58, and OR: 6.25; 95% CI: 1.63–23.84, respectively). We showed that vacA s2m2 decreases resistance to all five antibiotics (clarithromycin, metronidazole, amoxicillin, tetracycline and levofloxacin). On the other hand, vacA s1m2 decreases resistance to clarithromycin and metronidazole, while vacA s2m1 only decreases resistance to clarithromycin. Details on the relationship between non-vacA s1m1 genotypes and antibiotic resistance are summarized in Table 5.
Table 5.
Odds ratio (OR) with 95% CI for Non-vacA s1m1 genotypes and antibiotic resistance in H. pylori
| Non-vacA s1m1 genotypes | Antibiotic resistance | Random-effects model | Heterogeneity | Publication bias | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI ) | p value | p value | I-squared | Egger’s p value | Begg’s p value | ||
| vacA s1m2 | Clarithromycin | 0.13 (0.05–0.16) | 0.01 | 0.01 | 81.34 | 0.78 | 0.88 |
| Metronidazole | 0.32 (0.12–0.81) | 0.01 | 0.01 | 92.01 | 0.50 | 0.20 | |
| Amoxicillin | 0.11 (0.003-3.9) | 0.2 | 0.01 | 97.76 | 0.02 | 0.5 | |
| Tetracycline | 0.05 (0.001-4.6) | 0.2 | 0.01 | 95.01 | 0.05 | 0.5 | |
| Levofloxacin | 0.16 (0.02–1.36) | 0.09 | 0.01 | 93.48 | 0.04 | 0.5 | |
| vacA s2m1 | Clarithromycin | 0.03 (0.01–0.09) | 0.01 | 0.01 | 0.00 | 0.05 | 0.7 |
| Metronidazole | 0.07 (0.00-173.5) | 0.5 | 0.01 | 92.02 | NA | NA | |
| Amoxicillin | 0.02 (0.00-1.34) | 0.06 | 0.9 | 0.00 | NA | NA | |
| Tetracycline | 0.02 (0.00-1.34) | 0.06 | 0.9 | 0.00 | NA | NA | |
| Levofloxacin | 0.02 (0.00-1.34) | 0.06 | 0.9 | 0.00 | NA | NA | |
| vacA s2m2 | Clarithromycin | 0.07 (0.03–0.13) | 0.01 | 0.01 | 55.47 | 0.02 | 0.04 |
| Metronidazole | 0.06 (0.02–0.15) | 0.01 | 0.01 | 84.67 | 0.52 | 0.50 | |
| Amoxicillin | 0.04 (0.01–0.09) | 0.01 | 0.01 | 17.61 | 0.18 | 0.1 | |
| Tetracycline | 0.03 (0.00-0.14) | 0.01 | 0.01 | 0.00 | 0.07 | 0.5 | |
| Levofloxacin | 0.03 (0.01–0.12) | 0.01 | 0.01 | 21.26 | 0.78 | 0.5 | |
| NA not available | |||||||
A meta-regression was performed to examine the sources of heterogeneity according to the publication year or NOS score; the results of meta-regression showed that H. pylori antibiotic resistance was significantly influenced by publication year (Slope intercept: -0.18; 95% CI: -0.24 to -0.12; SE: 0.029; p value: 0.01) or NOS score scale (Slope intercept: -7.30; 95% CI: -8.98 to -5.63; SE: 0.85; p value: 0.01). In subgroup analysis, we found no association between the high virulent strains containing cagA-vacA s1m1 and antibiotic resistance (Fig. 4). In general, it seems that the degree of antibiotic resistance in strains with high pathogenicity is not different from the strains with low virulence. Due to heterogeneity and publication bias, we need further studies with larger sample sizes.
Fig. 4.
The forest plot associated with cagA-vacA s1m1 and resistance to clarithromycin and metronidazole
The cagA status and antibiotic resistance
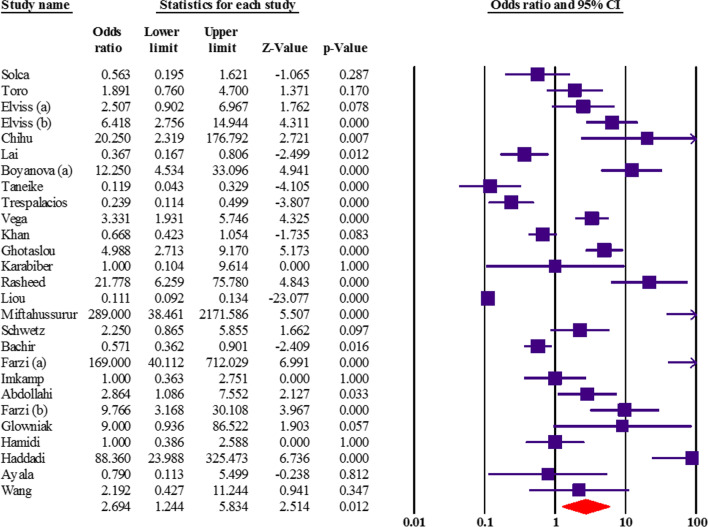
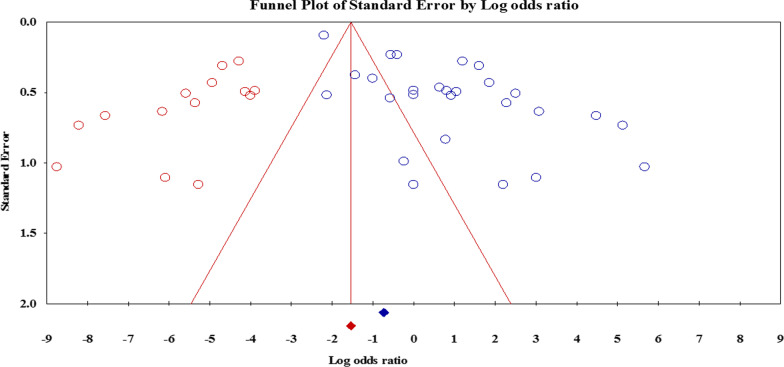
Association between cagA status and resistance to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin had been measured in 40 articles. Based on the current results, it seems that cagA significantly increases metronidazole resistance (OR: 2.69; 95% CI: 1.24–5.83; p value: 0.01), especially in Western countries (Fig. 5). By discovering the potential sources of heterogeneity, we excluded 3 studies. Sensitivity analysis showed a similar OR: 2.67 (95% CI: 1.20–5.94; p value: 0.01). The details of overall estimates related to cagA based on the sample size of the study, diagnostic test, and developing/developed status of country are addressed in the Table 6. However, the results of Egger’s regression test and asymmetry of funnel plot showed evidence of publication bias in overall estimates. Thus, we have performed the trim and fill method to adjust for publication bias. The pooled OR did not show the correlation between cagA status and antibiotic resistance (OR: 0.29; 95% CI: 0.13–0.64; p value: 0.001). Hence, after imputed missing studies by the trim and fill method, the adjusted estimate significantly dropped from OR: 2.69 (95% CI: 1.24–5.83) to OR: 0.29 (95% CI: 0.13–0.64) that revealed there is no relationship between cagA status and resistance to metronidazole. The population sample size was low in some included studies that may cause to this significant difference between adjusted OR and original estimates. More extensive research is needed to confirm the present findings.
Fig. 5.
The forest plot associated with cagA and resistance to metronidazole
Table 6.
The cagA-positive status and metronidazole resistance
| Factors | Random-effects model | Heterogeneity | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p value | p value | I-squared | ||
| Level of country | Developing country | 2.02 | 0.84–4.81 | 0.01 | 0.01 | 55.28 |
| Developed country | 3.36 | 1.14 | 0.02 | 0.03 | 67.45 | |
| Sample size | ≥ 100 | 2.02 | 0.53–7.60 | 0.01 | 0.01 | 98.05 |
| ≤ 100 | 3.02 | 1.29–7.043 | 0.01 | 0.01 | 90.97 | |
| Diagnostic test | E-test | 2.50 | 1.07–5.83 | 0.03 | 0.06 | 51.69 |
| Agar dilution based | 3.67 | 0.69–19.47 | 0.12 | 0.04 | 63.97 | |
| Disk diffusion based | 1.17 | 0.41–3.33 | 0.01 | 0.93 | 0.00 | |
| Molecular based | 0.23 | 0.11–0.49 | 0.01 | 0.9 | 0.00 | |
In addition, our findings showed a non-significant association between cagA status and resistance to clarithromycin, amoxicillin, tetracycline, and levofloxacin. The results of cagA status and resistance to these antibiotics are listed in Table 7. Sensitivity analysis also confirmed the stability of the overall estimates after excluding studies that may cause significant heterogeneity.
Table 7.
Odds ratio (OR) with 95% CI for cagA genotype and antibiotic resistance in H. pylori
| Resistance to | Random-effects model | Heterogeneity | Publication bias | |||
|---|---|---|---|---|---|---|
| OR (95%CI ) | p value | p value | I-squared | Egger’s p value | Begg’s p value | |
| Clarithromycin | 1.61 (0.63–4.11) | 0.31 | 0.01 | 95.90 | 0.01 | 0.62 |
| Metronidazole | 2.69 (1.24–5.83) | 0.01 | 0.01 | 96.42 | 0.01 | 0.27 |
| Amoxicillin | 5.14 (0.23–114.5) | 0.33 | 0.01 | 98.46 | 0.02 | 0.21 |
| Tetracycline | 1.32 (0.01–122.0) | 0.95 | 0.01 | 95.59 | 0.01 | 0.50 |
| Levofloxacin | 8.77 (0.24–310.8) | 0.21 | 0.01 | 98.21 | 0.01 | 0.50 |
A meta-regression was performed to examine the sources of heterogeneity according to the publication year or NOS score; the results of meta-regression showed that publication year (Slope intercept: − 0.150; 95% CI: − 0.20 to − 0.10; SE: 0.025; p value: 0.01) or NOS score scale (Slope intercept: − 5.26; 95% CI: − 6.82 to − 3.69; SE: 0.79; p value: 0.01) was disrupted the association between cagA status and H. pylori antibiotic resistance. In the subgroup analysis, our results showed that cagA increases resistance to metronidazole, amoxicillin, and levofloxacin only in the Western population (OR: 1.59; 95% CI: 0.78–3.21, OR: 19.68; 95% CI: 2.74–141.18, and OR: 11.33; 95% CI: 1.39–91.85, respectively), nonetheless, the results associated with the Asian countries were not significant (Table 8). After the trim and fill method, the adjusted OR was slightly lower than original estimates (but not significant difference) which indicates the reliability of the overall estimates.
Table 8.
Results of subgroup analysis for both Asian and Europe/America (West) populations
| Virulence factor/region | Clarithromycin | Metronidazole | Amoxicillin | Tetracycline | Levofloxacin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virulence factor | Region | OR | 95% CI | p value | OR | 95% CI | p value | OR | 95% CI | p value | OR | 95% CI | p value | OR | 95% CI | p value |
| cagA | Asia | 3.12 | 0.64–15.17 | 0.1 | 5.06 | 1.24–20.12 | 0.02 | 3.26 | 0.10-97.37 | 0.49 | 0.73 | 0.007–83.60 | 0.9 | 5.34 | 0.04–600.0 | 0.48 |
| West | 0.87 | 0.31–2.43 | 0.7 | 1.59 | 0.78–3.21 | 0.1 | 19.68 | 2.74-141.18 | 0.03 | NA | NA | NA | 11.33 | 1.39–91.85 | 0.02 | |
| vacA s1m1 | Asia | 0.22 | 0.06–0.81 | 0.02 | 0.37 | 0.15–0.90 | 0.03 | 0.08 | 0.002–2.91 | 0.16 | 0.13 | 0.004–4.76 | 0.27 | 0.22 | 0.01–0.03 | 0.27 |
| West | 0.65 | 0.16–2.52 | 0.5 | 0.46 | 0.13–1.58 | 0.21 | 16.58 | 1.77-154.58 | 0.01 | NA | NA | NA | 6.25 | 1.63–23.84 | 0.01 | |
| vacA s1m2 | Asia | 0.17 | 0.04–0.71 | 0.01 | 0.47 | 0.14–1.51 | 0.2 | 0.11 | 0.003–3.94 | 0.22 | 0.05 | 0.001–4.66 | 0.20 | 0.23 | 0.01–3.05 | 0.26 |
| West | 0.10 | 0.03–0.29 | 0.01 | 0.23 | 0.03–1.41 | 0.1 | NA | NA | NA | NA | NA | NA | 0.033 | 0.006–0.17 | 0.01 | |
| vacA s2m1 | Asia | 0.04 | 0.01–0.12 | 0.07 | 4.00 | 0.13-119.23 | 0.40 | 0.02 | 0.01–1.34 | 0.06 | 0.02 | 0.01–1.34 | 0.06 | 0.02 | 0.01–1.34 | 0.06 |
| West | 0.06 | 0.01–0.06 | 0.09 | 0.01 | 0.00-0.02 | 0.5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| vacA s2m2 | Asia | 0.06 | 0.02–0.19 | 0.01 | 0.012 | 0.006–0.02 | 0.01 | 0.044 | 0.019–0.12 | 0.01 | 0.035 | 0.008–0.14 | 0.001 | 0.02 | 0.007–0.08 | 0.01 |
| West | 0.07 | 0.03–0.15 | 0.01 | 0.15 | 0.05–0.42 | 0.01 | 0.024 | 0.003–0.19 | 0.001 | NA | NA | NA | 0.12 | 0.003–5.75 | 0.28 | |
| cagA-vacA s1m1 | Asia | 0.53 | 0.38–0.75 | 0.07 | 1.31 | 0.88–1.94 | 0.17 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| West | 1.87 | 0.67–4.86 | 0.23 | 0.42 | 0.07–2.45 | 0.33 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
NA not available
Publication bias
The results of Egger’s and Begg’s tests, as well as funnel plot asymmetry showed a significant publication bias; however, when the trim-and-fill method was performed to correct the results, the adjusted OR for vacA genotypes was decreased but no significant difference was observed compared to original estimates (Fig. 6). However, the adjusted OR for cagA status and resistance to metronidazole was dropped significantly that represents there is no association between cagA status and antibiotic resistance.
Fig. 6.
The trimmed and filled funnel plot represent the correlation between the standardized OR and its standard error with pseudo 95% confidence limits regarding possible association between cagA status and resistance to metronidazole
Discussion
The cagA and vacA genes are the most well-known virulence factors of H. pylori, and previous studies have demonstrated that infection with cagA-vacA s1m1 positive strains can increase the risk of severe gastrointestinal disorders [70, 71]. Wang et al. understood that infection with strains carrying both cagA and vacA products could increase the chance of eradicating H. pylori infection, however, the reported heterogeneity was significant [8]. Infection with cagA-positive strains can be led to gastric mucosal inflammation, which in turn increases the diffusion of antibiotic (following an increase in blood flow, disruption of mucosal barrier, and inhibition of IL-1β-induced gastric acid secretion) and ultimately high cure rate [72, 73]. Interestingly, vacA s1-positive strains reduce the risk of treatment failure due to induce sever gastric inflammation and lower expression of somatostatin [74, 75].
To the best of our knowledge, this is the first meta-analysis study that investigated the potential association between H. pylori virulence factor and antibiotic resistance. Based on this analysis, a considerable association exists between the status of vacA-cagA genes and resistance of H. pylori to commonly used antibiotic agents. The results of the present study indicated that cagA-positive strains can significantly increase resistance to metronidazole (OR: 2.69; 95% CI: 1.24–5.83; p value: 0.01). Although, s1m1 genotype of vacA significantly reduces resistance to metronidazole, vacA s1m2 reduces resistance to both clarithromycin and metronidazole. Moreover, vacA s2m1 decreased resistance to clarithromycin, as well as vacA s2m2 decreased resistance to metronidazole, clarithromycin, amoxicillin, tetracycline, and levofloxacin. We showed that cagA-positive strains in particular in Western countries increase the risk of resistance to metronidazole, amoxicillin, and ciprofloxacin.
In their study, Chisholm et al. asserted that resistance against metronidazole was not merely due to mutation in the rdxA gene, but was influenced by a variety of mechanisms [76]. In a study by Kim et al., they showed that resistance to metronidazole could occur even in the lack of rdxA expression or truncated RdxA [77]. Correlation between cagA pathogenicity islands (PIA) and resistance to metronidazole first was investigated by Alfizah et al.; they found that strains containing an intact cagPAI region were sensitive to metronidazole, while strains possessing partially deleted cagPAI regions were resistant to metronidazole [42]. Variations in the 3’ terminal of cagA lead to the differentiation of new subclones with unique genetic characteristics, and due to this fact, Rengifo et al. in their study demonstrated that genetic changes in this region cause the formation of antibiotic-resistant subclones [43, 78]. Recent studies show that in patients treated with antibiotics, new subclones of cagA are formed due to recombination and quorum sensing, which differ in some features and this phenomenon is effective in antibiotic resistance [79, 80]. We showed that gastric colonization with cagA-positive strains, especially in Western countries, can potentially increase the risk of resistance to common antibiotics. In a study conducted by Yue et al., they realized that the prevalence of resistance to metronidazole in strains with Western-type cagA 3’ variable region was significantly higher than East Asian-type strains [81, 82]. Today, evidence suggests that CagA protein is involved in processes such as integron acquisition, biofilm formation, and efflux pump function [83–85]. In general, cagA-positive strains, especially in the Western population, seem to be considered as diagnostic biomarkers in the phenomenon of antibiotic resistance. Recently, Ayibatari et al. revealed that patients carrying Western-type cagA had higher rates of gastritis than East Asian-type cagA [86].
Our results showed that vacA s2m2 genotype was associated with a significant decrease in resistance to antibiotics. Strains containing vacA s2m2 genotype are not able to produce VacA cytotoxic antigen [87]. Krzyżek et al. observed that the change to coccoid form in vacA s1m1 strains was significantly higher than vacA s2m2 strains [88]. Studies show that vacA s2m2 strains have higher nutritional requirements and are also less compatible with antibiotics, so they are more sensitive to antibiotics [89–91]. Though, our results suggested that there is no meaningful association between cagA/vacA s1m1 double positive H. pylori infection and antibiotic resistance. The biofilm formation capacity of vacA s1m1 genotype is higher than other genotypes, which in turn is an effective strategy in antibiotic resistance [92, 93]. Our results (as several cross-sectional studies) showed that the s1m1 and s1m2 genotypes reduce the risk of resistance to metronidazole and clarithromycin [59, 94–96]. Strains containing s1 or m1 are strong immunogens to stimulate the immune system and gastritis, so antibiotic delivery in the stomach lumen increases due to increased blood flow [39]. Nevertheless, the effect of other virulence factors may be ignored, for example Brennan et al. showed that the incidence of infection with s1m1/s1m2 strains was higher in treatment-naïve patients than in those previously treated [91].
Overall, our statistical analysis showed that metronidazole resistance was significantly high in cagA-positive H. pylori strains. As well as, less virulent vacA s2m2 genotype was sensitive to all antibiotics. Our study had several limitation including: (1) small ample size; (2) study only on adult population; (3) high heterogeneity among the included studies; (4) imbalanced geographical distribution; (5) inaccessibility to raw data to assess bacterial density and other factors in cag PAI; (6) publication bias. However, we performed meta-regression and sensitivity analyses to diminish the effects of heterogeneity on the reliability of the pooled estimates. Meta-regression and sensitivity analyses assisted us exclude the impact of some positive data on the overall estimates. Moreover, we used random-effects models to establish associations among the moderate variables with high heterogeneity. Therefore, it is appropriate to present evidence, but the findings should be interpreted with more caution. In the current meta-analysis, publication bias considerably changed the association between cagA status and resistance to metronidazole according to the trim-and-fill method. Meanwhile, adjusted OR for vacA genotype and antibiotic resistance after implementation of the trim and fill producer revealed that results were slightly lower without significant difference with overall estimates.
Conclusions
In the current meta-analysis, our findings showed that infection with cagA-positive strains of H. pylori significantly increases the risk of metronidazole resistance in Western countries. In addition, vacA s1m1 increases resistance to amoxicillin and levofloxacin in Western countries. According to our findings, the vacA s1m1 significantly increases resistance to metronidazole, while the vacA s1m2 decreases resistance to clarithromycin and metronidazole. Additionally, antibiotic resistance to clarithromycin, metronidazole, amoxicillin, tetracycline, and levofloxacin in less virulent H. pylori strains (carrying vacAs2m2 genotype) is significantly lower than others. We also performed the trim and fill method to exclude the potential bias from the overall estimates. Although, the adjusted OR was slightly lower than original estimates but this difference was not significant.
Acknowledgements
We appreciate from both Mashhad University of Medical Sciences and Jiroft University of Medical Sciences.
Abbreviations
- H. pylori
Helicobacter pylori
- PU
Peptic ulcer
- MALT
Gastric mucosa associated-lymphoid tissue
- GC
Gastric cancer
- WHO
World Health Organization
- vacA
Vacuolating cytotoxin A
- cagA
Cytotoxin associated gene A
- PUD
Peptic ulcer disease
- SNPs
Single nucleotide polymorphisms
- MOS
Newcastle–Ottawa scale
- CMA
Comprehensive Meta-Analysis
- CI
Confidence interval
- OR
Odds ratio
Author contributions
ATB and MK2 have contributed to design of the work and analysis of data. MK1 and MK2 have drafted the work and substantively revised it. ATB and MK2 have reviewed and revised the draft manuscript. All authors read and approved the final manuscript.
Funding
We have not received any funding for this research.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable (this paper was provided based on researching in global databases).
Consent for publication
Not applicable.
Competing interests
There is no any conflict of interest among the all authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amin Talebi Bezmin Abadi, Email: amin.talebi@modares.ac.ir.
Masoud Keikha, Email: Masoud.keykha90@gmail.com.
References
- 1.Hooi JK, Lai WY, Ng WK, Suen MM, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VW, Wu JC. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology. 2018;155(5):1372–1382e1317. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugimoto M, Yamaoka Y. Virulence factor genotypes of Helicobacter pylori affect cure rates of eradication therapy. Arch Immunol Ther Exp. 2009;57(1):45–56. doi: 10.1007/s00005-009-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64(9):1353–1367. doi: 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keikha M, Askari P, Ghazvini K, Karbalaei M. Levofloxacin-based therapy as an efficient alternative for eradicating Helicobacter pylori infection in Iran: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2021;29:420–429. doi: 10.1016/j.jgar.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Organization WH. WHO publishes list of bacteria for which new antibiotics are urgently needed. 2017.
- 7.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment of Helicobacter pylori infection: meeting the challenge of antimicrobial resistance. World J Gastroenterol WJG. 2014;20(29):9898. doi: 10.3748/wjg.v20.i29.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Li Q, Gong Y, Yuan Y. The association between vacA or cagA status and eradication outcome of Helicobacter pylori infection: a meta-analysis. PLoS ONE. 2017;12(5):e0177455. doi: 10.1371/journal.pone.0177455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim I-J, Blanke SR. Remodeling the host environment: modulation of the gastric epithelium by the Helicobacter pylori vacuolating toxin (VacA) Front Cell Infect Microbiol. 2012;2:37. doi: 10.3389/fcimb.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugimoto M, Zali M, Yamaoka Y. The association of vacA genotypes and Helicobacter pylori-related gastroduodenal diseases in the Middle East. Eur J Clin Microbiol Infect Dis. 2009;28(10):1227–1236. doi: 10.1007/s10096-009-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Hosseini ME, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133(3):926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 12.Letley DP, Rhead JL, Twells RJ, Dove B, Atherton JC. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. J Biol Chem. 2003;278(29):26734–26741. doi: 10.1074/jbc.M304071200. [DOI] [PubMed] [Google Scholar]
- 13.Keikha M, Ali-Hassanzadeh M, Karbalaei M. Association of Helicobacter pylori vacA genotypes and peptic ulcer in Iranian population: a systematic review and meta-analysis. BMC Gastroenterol. 2020;20(1):1–11. doi: 10.1186/s12876-020-01406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keikha M, Askari P, Ghazvini K, Karbalaei M. Levofloxacin-based therapy as an efficient alternative for eradicating Helicobacter pylori infection in Iran: a systematic review and meta-analysis. J Global Antimicrob Resist. 2021;29:420–429. doi: 10.1016/j.jgar.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Yamaoka Y, Kita M, Kodama T, Sawai N, Imanishi J. Helicobacter pylori cagA gene and expression of cytokine messenger RNA in gastric mucosa. Gastroenterology. 1996;110(6):1744–1752. doi: 10.1053/gast.1996.v110.pm8964399. [DOI] [PubMed] [Google Scholar]
- 16.Sgouras DN, Trang TTH, Yamaoka Y. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2015;20:8–16. doi: 10.1111/hel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Doorn L-J, Schneeberger P, Nouhan N, Plaisier A, Quint W, De Boer W. Importance of Helicobacter pylori cagA and vacAstatus for the efficacy of antibiotic treatment. Gut. 2000;46(3):321–326. doi: 10.1136/gut.46.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miftahussurur M, Waskito LA, Syam AF, Nusi IA, Siregar G, Richardo M, Bakry AF, Rezkitha YAA, Wibawa IDN, Yamaoka Y. Alternative eradication regimens for Helicobacter pylori infection in Indonesian regions with high metronidazole and levofloxacin resistance. Infect Drug Resist. 2019;12:345. doi: 10.2147/IDR.S187063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Brea M, Martínez MJ, Domingo D, Sánchez I, Alarcón T. Metronidazole resistance and virulence factors in Helicobacter pylori as markers for treatment failure in a paediatric population. FEMS Immunol Med Microbiol. 1999;24(2):183–188. doi: 10.1016/S0928-8244(99)00024-3. [DOI] [PubMed] [Google Scholar]
- 20.Niu S, Yang F. Relationship between lansoprasoi triple therapy effect and alleles of vacuolating cytotoxin genotype in patients with gastric ulcer. Chinese J Pract Med. 2014; 2(41).
- 21.De Magalhaes AN, Carvalhaes A, Natan-Eisig J, Paraiso-Ferraz J, Trevisan M, Zaterka S. CagA status and Helicobacter pylori eradication among dyspeptic patients. Gastroenterol Hepatol. 2005;28(8):441–444. doi: 10.1157/13078993. [DOI] [PubMed] [Google Scholar]
- 22.Baryshnikova N, Uspenskiy Y, Suvorov A, Suvorova M. Helicobacter. USA: Wiley-Blackwell; 2012. Efficacy of eradication in patients infected with caga (+) and caga (−) strains of Helicobacter pylori; pp. 105–105. [Google Scholar]
- 23.Broutet N, Marais A, Lamouliatte H, de Mascarel A, Samoyeau R, Salamon R, Mégraud F. cagA Status and eradication treatment outcome of anti-Helicobacter pylori triple therapies in patients with nonulcer dyspepsia. J Clin Microbiol. 2001;39(4):1319–1322. doi: 10.1128/JCM.39.4.1319-1322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solca NM, Bernasconi MV, Valsangiacomo C, Van Doorn L-J, Piffaretti J-C. Population genetics of Helicobacter pylori in the southern part of Switzerland analysed by sequencing of four housekeeping genes (atpD, glnA, scoB and recA), and by vacA, cagA, iceA and IS605 genotypingThe GenBank accession numbers for the sequences reported in this paper are AY004351–AY004662. Microbiology. 2001;147(6):1693–1707. doi: 10.1099/00221287-147-6-1693. [DOI] [PubMed] [Google Scholar]
- 25.Toro C, García-Samaniego J, Alarcón T, Baquero M. Relación entre detección de anticuerpos anti-CagA, sensibilidad antibiótica y úlcera péptica en pacientes con infección por Helicobacter pylori. Enferm Infect Microbiol Clin. 2003;21(3):137–141. doi: 10.1016/S0213-005X(03)72902-4. [DOI] [PubMed] [Google Scholar]
- 26.Elviss NC, Owen RJ, Xerry J, Walker AM, Davies K. Helicobacter pylori antibiotic resistance patterns and genotypes in adult dyspeptic patients from a regional population in North Wales. J Antimicrob Chemother. 2004;54(2):435–440. doi: 10.1093/jac/dkh343. [DOI] [PubMed] [Google Scholar]
- 27.Elviss NC, Owen RJ, Breathnach A, Palmer C, Shetty N. Helicobacter pylori antibiotic-resistance patterns and risk factors in adult dyspeptic patients from ethnically diverse populations in central and south London during 2000. J Med Microbiol. 2005;54(6):567–574. doi: 10.1099/jmm.0.45896-0. [DOI] [PubMed] [Google Scholar]
- 28.Chihu L, Ayala G, Mohar A, Hernández A, Herrera-Goepfert R, Fierros G, Gonzalez-Marquez H, Silva J. Antimicrobial resistance and characterization of Helicobacter pylori strains isolated from Mexican adults with clinical outcome. J Chemother. 2005;17(3):270–276. doi: 10.1179/joc.2005.17.3.270. [DOI] [PubMed] [Google Scholar]
- 29.Francesco VD, Margiotta M, Zullo A, Hassan C, Valle ND, Burattini O, D'Angel R, Stoppino G, Cea U, Giorgio F. Claritromycin resistance and Helicobacter pylori genotypes in Italy. J Microbiol. 2006;44(6):660–664. [PubMed] [Google Scholar]
- 30.Lai C-H, Kuo C-H, Chen P-Y, Poon S-K, Chang C-S, Wang W-C. Association of antibiotic resistance and higher internalization activity in resistant Helicobacter pylori isolates. J Antimicrob Chemother. 2006;57(3):466–471. doi: 10.1093/jac/dki479. [DOI] [PubMed] [Google Scholar]
- 31.Boyanova L, Markovska R, Yordanov D, Marina M, Ivanova K, Panayotov S, Gergova G, Mitov I. High prevalence of virulent Helicobacter pylori strains in symptomatic Bulgarian patients. Diagn Microbiol Infect Dis. 2009;64(4):374–380. doi: 10.1016/j.diagmicrobio.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Taneike I, Nami A, O’connor A, Fitzgerald N, Murphy P, Qasim A, O’Connor H, O’Morain C. Analysis of drug resistance and virulence-factor genotype of Irish Helicobacter pylori strains: is there any relationship between resistance to metronidazole and cagA status? Alimentary Pharmacol Therapeut. 2009;30(7):784–790. doi: 10.1111/j.1365-2036.2009.04095.x. [DOI] [PubMed] [Google Scholar]
- 33.Hu C-T, Chiou P-Y, Wu C-C, Tseng Y-H, Chang Y-J, Lin N-T. Analysis of resistance to clarithromycin and virulence markers in Helicobacter pylori clinical isolates from Eastern Taiwan. Tzu Chi Med J. 2009;21(2):123–128. doi: 10.1016/S1016-3190(09)60023-9. [DOI] [Google Scholar]
- 34.Trespalacios AA, Regino WO, Reyes MM. Resistencia de Helicobacter pylori a metronidazol, claritromicina y amoxicilina en pacientes colombianos. Rev Colomb Gastroenterol. 2010;25(1):31–38. [Google Scholar]
- 35.Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48(10):3703–3707. doi: 10.1128/JCM.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vega AE, Cortiñas TI, Puig ON, Silva HJ. Molecular characterization and susceptibility testing of Helicobacter pylori strains isolated in western Argentina. Int J Infect Dis. 2010;14:e85–e92. doi: 10.1016/j.ijid.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 37.Ayala G, Galván-Portillo M, Chihu L, Fierros G, Sánchez A, Carrillo B, Román A, López-Carrillo L, Silva-Sanchez J, Study Group J Resistance to antibiotics and characterization of Helicobacter pylori strains isolated from antrum and body from adults in Mexico. Microb Drug Resist. 2011;17(2):149–155. doi: 10.1089/mdr.2010.0154. [DOI] [PubMed] [Google Scholar]
- 38.Baba S, Oishi Y, Watanabe Y, Oikawa R, Morita R, Yoshida Y, Hiraishi T, Maehata T, Nagase Y, Fukuda Y. Gastric wash-based molecular testing for antibiotic resistance in Helicobacter pylori. Digestion. 2011;84(4):299–305. doi: 10.1159/000332570. [DOI] [PubMed] [Google Scholar]
- 39.Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and cagA gene correlation: a looming crisis of Helicobacter pylori. World J Gastroenterol WJG. 2012;18(18):2245. doi: 10.3748/wjg.v18.i18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yula E, Nagiyev T, Kaya ÖA, İnci M, Çelik MM, Köksal F. Detection of primary clarithromycin resistance of Helicobacter pylori and association between cagA+ status and clinical outcome. Folia Microbiol. 2013;58(2):141–146. doi: 10.1007/s12223-012-0192-8. [DOI] [PubMed] [Google Scholar]
- 41.Ghotaslou R, Milani M, Akhi MT, Hejazi MS, Nahaei MR, Hasani A, Sharifi Y. Relationship between drug resistance and cagA Gene in Helicobacter pylori. Jundishapur J Microbiol. 2013, 6(10).
- 42.Alfizah H, Rukman AH, Norazah A, Hamizah R, Ramelah M. Ethnicity association of Helicobacter pylori virulence genotype and metronidazole susceptibility. World J Gastroenterol WJG. 2013;19(8):1283. doi: 10.3748/wjg.v19.i8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustamante-Rengifo JA, Matta AJ, Pazos A, Bravo LE. In vitro effect of amoxicillin and clarithromycin on the 3’region of cagA gene in Helicobacter pylori isolates. World J Gastroenterol WJG. 2013;19(36):6044. doi: 10.3748/wjg.v19.i36.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karabiber H, Selimoglu MA, Otlu B, Yildirim O, Ozer A. Virulence factors and antibiotic resistance in children with Helicobacter pylori gastritis. J Pediatr Gastroenterol Nutr. 2014;58(5):608–612. doi: 10.1097/MPG.0000000000000273. [DOI] [PubMed] [Google Scholar]
- 45.Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, Pritchard DM. Analysis of clinical isolates of Helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter. 2014;19(5):387–399. doi: 10.1111/hel.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hussein N, Tunjel I, Majed H, Yousif S, Aswad S, Assafi M. Duodenal ulcer promoting gene 1 (dupA1) is associated with A2147G clarithromycin-resistance mutation but not interleukin-8 secretion from gastric mucosa in Iraqi patients. N Microb N Infect. 2015;6:5–10. doi: 10.1016/j.nmni.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyanova L, Markovska R, Yordanov D, Gergova G, Mitov I. Clarithromycin resistance mutations in Helicobacter pylori in association with virulence factors and antibiotic susceptibility of the strains. Microb Drug Resist. 2016;22(3):227–232. doi: 10.1089/mdr.2015.0199. [DOI] [PubMed] [Google Scholar]
- 48.Fasciana T, Calà C, Bonura C, Di Carlo E, Matranga D, Scarpulla G, Manganaro M, Camilleri S, Giammanco A. Resistance to clarithromycin and genotypes in Helicobacter pylori strains isolated in Sicily. J Med Microbiol. 2015;64(11):1408–1414. doi: 10.1099/jmm.0.000163. [DOI] [PubMed] [Google Scholar]
- 49.Liou J-M, Chang C-Y, Chen M-J, Chen C-C, Fang Y-J, Lee J-Y, Wu J-Y, Luo J-C, Liou T-C, Chang W-H. The primary resistance of Helicobacter pylori in Taiwan after the national policy to restrict antibiotic consumption and its relation to virulence factors—a nationwide study. PLoS ONE. 2015;10(5):e0124199. doi: 10.1371/journal.pone.0124199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alarcón-Millán J, Fernández-Tilapa G, Cortés-Malagón EM, Castañón-Sánchez CA, De Sampedro-Reyes J, Cruz-del Carmen I, Betancourt-Linares R, Román-Román A. Clarithromycin resistance and prevalence of Helicobacter pylori virulent genotypes in patients from Southern México with chronic gastritis. Infect Genet Evol. 2016;44:190–198. doi: 10.1016/j.meegid.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 51.Miftahussurur M, Syam AF, Nusi IA, Makmun D, Waskito LA, Zein LH, Akil F, Uwan WB, Simanjuntak D, Wibawa IDN. Surveillance of Helicobacter pylori antibiotic susceptibility in Indonesia: different resistance types among regions and with novel genetic mutations. PLoS ONE. 2016;11(12):e0166199. doi: 10.1371/journal.pone.0166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zollner-Schwetz I, Leitner E, Plieschnegger W, Semlitsch G, Stepan V, Reiter L, Reicht G, Mörth E, Pavek J, Parsché P. Primary resistance of Helicobacter pylori is still low in Southern Austria. Int J Med Microbiol. 2016;306(4):206–211. doi: 10.1016/j.ijmm.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Bachir M, Allem R, Tifrit A, Medjekane M, Drici A-M, Diaf M, Douidi KT. Primary antibiotic resistance and its relationship with cagA and vacA genes in Helicobacter pylori isolates from Algerian patients. Brazilian J Microbiol. 2018;49:544–551. doi: 10.1016/j.bjm.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farzi N, Yadegar A, Sadeghi A, Asadzadeh Aghdaei H, Marian Smith S, Raymond J, Suzuki H, Zali MR. High prevalence of antibiotic resistance in Iranian Helicobacter pylori isolates: importance of functional and mutational analysis of resistance genes and virulence genotyping. J Clin Med. 2019;8(11):2004. doi: 10.3390/jcm8112004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imkamp F, Lauener FN, Pohl D, Lehours P, Vale FF, Jehanne Q, Zbinden R, Keller PM, Wagner K. Rapid characterization of virulence determinants in Helicobacter pylori isolated from non-atrophic gastritis patients by next-generation sequencing. J Clin Med. 2019;8(7):1030. doi: 10.3390/jcm8071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khani S, Abadi ATB, Mobarez AM. Clarithromycin-susceptible but virulent Helicobacter pylori strains infecting Iranian patients’ stomachs. Infect Drug Resist. 2019;12:3415. doi: 10.2147/IDR.S223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdollahi H, Hashemzadeh M, Khoshnood S, Savari M. Characterization of Helicobacter pylori genotypes from Iranian patients with gastric clinical diseases: predominance of vacA s1a and cagA EPIYA-ABC genotypes. Gene Reports. 2019;16:100458. doi: 10.1016/j.genrep.2019.100458. [DOI] [Google Scholar]
- 58.Farzi N, Yadegar A, Aghdaei HA, Sadeghi A, Zali MR. High prevalence of antibiotic resistance in Helicobacter pylori isolates from Iran: importance of functional and mutational analysis of resistance genes and virulence genotyping. BioRxiv. 2019;1:569814. doi: 10.3390/jcm8112004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang D, Guo Q, Yuan Y, Gong Y. The antibiotic resistance of Helicobacter pylori to five antibiotics and influencing factors in an area of China with a high risk of gastric cancer. BMC Microbiol. 2019;19(1):1–10. doi: 10.1186/s12866-018-1372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korona-Glowniak I, Cichoz-Lach H, Siwiec R, Andrzejczuk S, Glowniak A, Matras P, Malm A. Antibiotic resistance and genotypes of Helicobacter pylori strains in patients with gastroduodenal disease in Southeast Poland. J Clin Med. 2019;8(7):1071. doi: 10.3390/jcm8071071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamidi S, Badmasti F, Sadeghpour Heravi F, Safapoor MH, Mohammad Ali Tabrizi A, Ghorbani M, Azizi O. Antibiotic resistance and clonal relatedness of Helicobacter pylori strains isolated from stomach biopsy specimens in northeast of Iran. Helicobacter. 2020;25(2). [DOI] [PubMed]
- 62.Haddadi M-H, Negahdari B, Asadolahi R, Bazargani A. Helicobacter pylori antibiotic resistance and correlation with cagA motifs and homB gene. Postgrad Med. 2020;132(6):512–520. doi: 10.1080/00325481.2020.1753406. [DOI] [PubMed] [Google Scholar]
- 63.Oktem-Okullu S, Cekic-Kipritci Z, Kilic E, Seymen N, Mansur-Ozen N, Sezerman U, Gurol Y. Analysis of correlation between the seven important Helicobacter pylori (H. pylori) virulence factors and drug resistance in patients with gastritis. Gastroenterol Res Pract. 2020;2020:1–7. doi: 10.1155/2020/3956838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 65.DerSimonian R, Laird N. Meta-analysis in clinical trials. Contr Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 66.Karbalaei M, Keikha M. Potential association between the hopQ alleles of Helicobacter pylori and gastrointestinal diseases: a systematic review and meta-analysis. Meta Gene. 2020;26:100816. doi: 10.1016/j.mgene.2020.100816. [DOI] [Google Scholar]
- 67.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 68.Rothstein HR, Sutton AJ, Borenstein M. Publication bias in meta-analysis: prevention, assessment and adjustments. USA: Wiley; 2006. [Google Scholar]
- 69.Shiota S, Matsunari O, Watada M, Hanada K, Yamaoka Y. Systematic review and meta-analysis: the relationship between the Helicobacter pylori dupA gene and clinical outcomes. Gut Pathogens. 2010;2(1):1–6. doi: 10.1186/1757-4749-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahara S, Sugimoto M, Vilaichone R-K, Mahachai V, Miyajima H, Furuta T, Yamaoka Y. Role of Helicobacter pylori cagA EPIYA motif and vacA genotypes for the development of gastrointestinal diseases in Southeast Asian countries: a meta-analysis. BMC Infect Dis. 2012;12(1):1–13. doi: 10.1186/1471-2334-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matos JI, de Sousa HA, Marcos-Pinto R, Dinis-Ribeiro M. Helicobacter pylori CagA and VacA genotypes and gastric phenotype: a meta-analysis. Eur J Gastroenterol Hepatol. 2013;25(12):1431–1441. doi: 10.1097/MEG.0b013e328364b53e. [DOI] [PubMed] [Google Scholar]
- 72.Maeda S, Yoshida H, Ikenoue T, Ogura K, Kanai F, Kato N, Shiratori Y, Omata M. Structure of cag pathogenicity island in Japanese Helicobacter pyloriisolates. Gut. 1999;44(3):336–341. doi: 10.1136/gut.44.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furuta T, Shirai N, Takashima M, Xiao F, Sugimura H. Effect of genotypic differences in interleukin-1 beta on gastric acid secretion in Japanese patients infected with Helicobacter pylori. Am J Med. 2002;112(2):141–143. doi: 10.1016/S0002-9343(01)01036-1. [DOI] [PubMed] [Google Scholar]
- 74.Zhao JJ, Wang JB, Yang L, Li Y. Influence of Helicobacter pylori genotype on triple eradication therapy. J Gastroenterol Hepatol. 2007;22(12):2251–2555. doi: 10.1111/j.1440-1746.2007.04836.x. [DOI] [PubMed] [Google Scholar]
- 75.Atherton JC, Tham KT, Peek RM, Jr, Cover TL, Blaser MJ. Density of Helicobacter pylori infection in vivo as assessed by quantitative culture and histology. J Infect Dis. 1996;174(3):552–556. doi: 10.1093/infdis/174.3.552. [DOI] [PubMed] [Google Scholar]
- 76.Chisholm SA, Owen RJ. Mutations in Helicobacter pylori rdxA gene sequences may not contribute to metronidazole resistance. J Antimicrob Chemother. 2003;51(4):995–999. doi: 10.1093/jac/dkg192. [DOI] [PubMed] [Google Scholar]
- 77.Kim SY, Joo YM, Lee HS, Chung I-S, Yoo Y-J, Merrell DS, Cha J-H. Genetic analysis of Helicobacter pylori clinical isolates suggests resistance to metronidazole can occur without the loss of functional rdxA. J Antibiot. 2009;62(1):43–50. doi: 10.1038/ja.2008.6. [DOI] [PubMed] [Google Scholar]
- 78.Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77(1):53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mera R, Fontham ET, Bravo LE, Bravo JC, Piazuelo MB, Camargo MC, Correa P. Long term follow up of patients treated for Helicobacter pylori infection. Gut. 2005;54(11):1536–1540. doi: 10.1136/gut.2005.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Argent RH, Thomas RJ, Aviles-Jimenez F, Letley DP, Limb MC, El-Omar EM, Atherton JC. Toxigenic Helicobacter pylori infection precedes gastric hypochlorhydria in cancer relatives, and H. pylori virulence evolves in these families. Clin Cancer Res. 2008;14(7):2227–2235. doi: 10.1158/1078-0432.CCR-07-2022. [DOI] [PubMed] [Google Scholar]
- 81.Jin Yong Y, Jing Y, Ming Yi W, Xiao Zhong G. CagA status and genetic characterization of metronidazole resistant strains of H. pylori from a region at high risk of gastric cancer. 2014. [DOI] [PMC free article] [PubMed]
- 82.Prazeres Magalhães P, de Magalhães Queiroz DM, Campos Barbosa DV, Aguiar Rocha G, Nogueira Mendes E, Santos A, Valle Corrêa PR, Camargos Rocha AM, Martins Teixeira LC, Affonso de Oliveira C. Helicobacter pylori primary resistance to metronidazole and clarithromycin in Brazil. Antimicrob Agents Chemothera. 2002;46(6):2021–2023. doi: 10.1128/AAC.46.6.2021-2023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, Takahashi N, Yoshida M, Azuma T, Hattori M. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11(1):1–28. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong EHJ, Ng CG, Chua EG, Tay ACY, Peters F, Marshall BJ, Ho B, Goh KL, Vadivelu J, Loke MF. Comparative genomics revealed multiple Helicobacter pylori genes associated with biofilm formation in vitro. PLoS ONE. 2016;11(11):e0166835. doi: 10.1371/journal.pone.0166835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kyrillos A, Arora G, Murray B, Rosenwald AG. The presence of phage orthologous genes in Helicobacter pylori correlates with the presence of the virulence factors CagA and VacA. Helicobacter. 2016;21(3):226–233. doi: 10.1111/hel.12282. [DOI] [PubMed] [Google Scholar]
- 86.Ayibatari A, Galleh RP, Ogo AC, Anzaku AA, Akyala AI. Prevalence of virulence genes and associated risk factors of Helicobacter pylori infection among adults in gastric cancer risk region of North Central, Nigeria. Eur J Clin Biomed Sci. 2021;7(6):118–125. [Google Scholar]
- 87.Correa P, Piazuelo MB. Natural history of Helicobacter pylori infection. Digest Liver Dis . 2008;40(7):490–496. doi: 10.1016/j.dld.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krzyżek P, Biernat MM, Gościniak G. Intensive formation of coccoid forms as a feature strongly associated with highly pathogenic Helicobacter pylori strains. Folia Microbiol. 2019;64(3):273–281. doi: 10.1007/s12223-018-0665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yaqub M, Ghezzi P. Adding dimensions to the analysis of the quality of health information of websites returned by Google: cluster analysis identifies patterns of websites according to their classification and the type of intervention described. Front Public Health. 2015;3:204. doi: 10.3389/fpubh.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendoza-Elizalde S, Arteaga-Resendiz NK, Valencia-Mayoral P, Luna RC, Moreno-Espinosa S, Arenas-Huertero F, Zúñiga G, Velázquez-Guadarrama N. Diversification of the vacAs1m1 and vacAs2m2 Strains of Helicobacter pylori in Meriones unguiculatus. Front Microbiol. 2016;7:1758. doi: 10.3389/fmicb.2016.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brennan DE, Dowd C, O’Morain C, McNamara D, Smith SM. Can bacterial virulence factors predict antibiotic resistant Helicobacter pylori infection? World J Gastroenterol. 2018;24(9):971. doi: 10.3748/wjg.v24.i9.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Di Giulio M, Traini T, Trubiani O. Characterization of an Helicobacter pylori environmental strain. J Appl Microbiol. 2008;105(3):761–769. doi: 10.1111/j.1365-2672.2008.03808.x. [DOI] [PubMed] [Google Scholar]
- 93.McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, Cover TL. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J Bacteriol. 2001;183(22):6499–6508. doi: 10.1128/JB.183.22.6499-6508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boehnke KF, Valdivieso M, Bussalleu A, Sexton R, Thompson KC, Osorio S, Reyes IN, Crowley JJ, Baker LH, Xi C. Antibiotic resistance among Helicobacter pylori clinical isolates in Lima, Peru. Infect Drug Resist. 2017;10:85. doi: 10.2147/IDR.S123798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feliciano O, Gutierrez O, Valdés L, Fragoso T, Calderin AM, Valdes AE, Llanes R. Prevalence of Helicobacter pylori vacA, cagA, and iceA genotypes in Cuban patients with upper gastrointestinal diseases. BioMed Res Int. 2015; 2015. [DOI] [PMC free article] [PubMed]
- 96.Pajavand H, Alvandi A, Mohajeri P, Bakhtyari S, Bashiri H, Kalali B, Gerhard M, Najafi F, Abiri R. High frequency of vacA s1m2 genotypes among Helicobacter pylori isolates from patients with gastroduodenal disorders in Kermanshah, Iran. Jundishapur J Microbiol. 2015, 8(11). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.