Abstract
Objectives
The characterization of asymptomatic and mildly symptomatic patients with COVID-19 by observing changes in gene expression profile and possible bacterial coinfection is relevant to be investigated. We aimed to identify transcriptomic and coinfection profiles in both groups of patients.
Methods
A ribonucleic acid (RNA) sequence analysis on nasopharyngeal swabs were performed using a shotgun sequencing pipeline. Differential gene analysis, viral genome assembly, and metagenomics analysis were further performed using the retrieved data.
Results
Both groups of patients underwent a cilia modification and mRNA splicing. Modulations in macroautophagy, epigenetics, and cell cycle processes were observed specifically in the asymptomatic group. Modulation in the RNA transport was found specifically in the mildly symptomatic group. The mildly symptomatic group showed modulation in the RNA transport and upregulation of autophagy regulator genes and genes in the complement system. No link between viral variants and disease severity was found. Microbiome analysis revealed the elevation of Streptococcus pneumoniae and Veillonella parvula proportion in symptomatic patients.
Conclusion
A reduction in the autophagy influx and modification in the epigenetic profile might be involved in halting the disease progression. A global dysregulation of RNA processing and translation might cause more severe outcomes in symptomatic individuals. Coinfection by opportunistic microflora should be taken into account when assessing the possible outcome of SARS-CoV-2 infection.
Keywords: COVID-19, Differential gene expression, Gene ontology, SARS-CoV-2, Transcriptomics
Introduction
The SARS-CoV-2 infection has a variety of clinical manifestations (Koh et al., 2020; Liao et al., 2020), which are affected by multiple host factors, including its genetics (Tavasolian et al., 2020), epigenetics (Schäfer and Baric, 2017), gene expression (Islam et al., 2021), age, sex (Lieberman et al., 2020; Undurraga et al., 2021), and so on. The prevalence of COVID-19 in asymptomatic patients is about 40-45% of the total cases (Oran and Topol, 2020). In the other half of the patients in their first stage of infection, symptoms such as fever, dry cough, and malaise may occur. These symptoms may be followed by dyspnea and could be accompanied by more severe syndromes, which cause their condition to deteriorate (He et al., 2020). Hence, it is important to understand pathophysiologic differences in asymptomatic and mildly symptomatic patients. One of the many methods to study this is through gene expression profiling.
Mapping of gene expression profiles through transcriptomic analysis is one of the most advanced methods to study disease pathophysiology. By studying their expression characteristics, it would be possible to identify biological markers of the disease's pathology and target it for therapeutical intervention (Casamassimi et al., 2017). Consequently, a transcriptomic analysis would be useful to understand SARS-CoV-2 pathogenesis and host responses and may permit the identification of therapeutic targets.
The expression of multiple genes has been used to predict infectious disease severity in viral infections (Calzavara-Silva et al., 2009; Kieboom et al., 2015). Hence, we designed this study to characterize important biological processes and gene expression modulated in asymptomatic and mildly symptomatic patients using diagnostic nasopharyngeal specimens. Our ribonucleic acid (RNA) sequence data would also provide information about the infecting SARS-CoV-2 strain and possible coinfection through metagenomic analysis.
Materials and methods
Selection of samples and RNA extraction
Nasopharyngeal swab samples were collected from diagnostic specimens in Laboratorium Kesehatan Jawa Barat. Patients were categorized into two groups: asymptomatic patients and patients with mild illness on the basis of the guidelines from the United States National Institute of Health (NIH) on the severity of illness (NIH, 2021).
Two nasopharyngeal swab samples from each group were collected from samples that tested positive for SARS-CoV-2, with a cycle threshold value lower than 30. RNA isolation was performed with the QIAamp Viral RNA Mini Kit (QIAGEN, Germany), using factory protocols. Measurement of total RNA quality was performed using the RNA 6000 Pico Kit on Agilent Bioanalyzer 2100 (Agilent Technologies, USA). Measurement of RNA concentration was performed using the Qubit® RNA High Sensitivity Assay on Qubit® 2.0 Fluorometer (Life Technologies, USA).
cDNA synthesis, preparation of DNA library, and DNA sequencing
cDNA library was constructed using 1-10 ng of RNA with the Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero™ Plus (Illumina, USA), using factory protocols. This kit allowed the degradation of cytoplasmic rRNA, human mitochondria, bacterial rRNA, and human β-globin mRNA transcripts. Quantification of DNA library concentration was performed using the Qubit® DNA Broad Range Assay on Qubit® 2.0 Fluorometer (Life Technologies, USA). The cDNA libraries were sequenced using the Illumina NextSeq 550, producing 101 bp paired-end DNA reads. Four technical replicates were generated from each sample, resulting in 16 sets of FASTQ files in total.
SARS-CoV-2 genome analysis
Raw FASTQ reads obtained from each sample were uploaded into the DRAGEN RNA Pathogen Detection 3.5.15 web-based application on the BaseSpace Illumina webpage (https://basespace.illumina.com/apps/12030018/DRAGEN-RNA-Pathogen-Detection), to be aligned onto the genome of SARS-CoV-2 and other respiratory viruses on the Illumina (Yang et al., 2018) and Seattle Study Flu virus panels, producing aligned FASTA sequence files. Finally, these whole genome sequences were uploaded onto the GISAID page (https://gisaid.org).
Differential expression analysis
Raw FASTQ reads obtained from the sequencing were quality-processed using the BBDuk plugin in Geneious Prime v.2020.2, according to the following parameters: exclusion of reads below Q30 and exclusion of reads below 30 bp. Mapping of the processed reads was performed on the annotated human GRCh38 genome, using the Geneious RNA mapper plugin. The reads, which were mapped to multiple locations, were excluded from the expression level calculation. The level of gene expression was measured in raw read counts and exported for the differential expression analysis. Differential expression analysis was performed using the DESeq2 package on R (Love et al., 2014). A raw read count matrix of gene expression from healthy controls was acquired from a previous publication by Lieberman et al (Lieberman et al., 2020). These data were obtained from the NCBI Gene Expression Omnibus, with the accession number GSE154770. The threshold for a differentially expressed gene was set as follows: P-value <0.01, and |log2 fold change| >2. The plots were visualized using the R package ggplot2 (Wickham, 2016).
Gene ontology analysis
The list of differentially expressed genes was used to analyze enriched gene ontology terms. Analysis was performed using clusterProfiler 4.0 to identify gene ontology terms within the three gene ontology categories (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (Yu et al., 2012). We extracted the top 25 most enriched biological process categories only for a more relevant discussion.
Coinfection analysis
Total RNA extraction with QIAamp Viral RNA Mini Kit yielded sufficient bacterial reads for a metagenomic analysis (Zhang et al., 2018). Raw FASTQ files were trimmed using the BBDuk+ plugin in Geneious Prime v.2020.2 software, with the removal of short reads (<70 nucleotides). The processed reads were then individually mapped with KRAKEN2 to the minikraken_8GB_20200312 bacterial database (Wood et al., 2019; Wood and Salzberg, 2014), with the output of bacterial abundance from each taxon, as done previously (Ma et al., 2021). Visualization was performed with multiple R packages: alpha diversity (Shannon index) and beta diversity (Bray-Curtis), with R package ggplot2 (Wickham, 2016) and DESeq2 (Love et al., 2014) for differential analysis and pheatmap (Kolde, 2019) for hierarchical clustering analysis.
Results
Nasopharyngeal specimens were collected during a 2-month period (September-October 2020) from SARS-CoV-2 qualitative reverse transcriptase-polymerase chain reaction-positive diagnostic samples of Laboratorium Kesehatan Jawa Barat, with cycle threshold value less than 30 to ensure high coverage of the viral genome assembly. Patient demographic data were collected as well as their symptoms during specimen collection. Table 1 summarizes the patient's characteristics, symptoms, and co-morbidities. The notation assignment for technical replicates and the number reads (raw, quality-processed, and mapped) are presented in S1 File. The principal component plot (Fig. 1 ) and hierarchical clustering (S1 Figure) of the samples show a close correlation of gene expression between samples from similar groups.
Table 1.
Patients Metadata and Co-morbidities.
| Characteristics | Patient |
|||
|---|---|---|---|---|
| AS1 | AS2 | SY1 | SY2 | |
| Age (yr) | 51 | 15 | 33 | 54 |
| Sex | F | M | F | M |
| Cough | No | No | Yes | Yes |
| Fever | No | No | Yes | Yes |
| Breathing difficulties | No | No | Yes | No |
| Joint pain | No | No | No | Yes |
| Runny nose | No | No | No | Yes |
| Co-morbidities | No | No | No | Chronic heart disease |
| Ct value (mean) | 14.66 | 18.27 | 17.97 | 27.52 |
| SARS-CoV-2 Pango Lineage | C.1.1.398 | C.1.1 | C.1.1.398 | C.1.1.398 |
Fig. 1.
Quality control analysis of differential gene expression. A principal component analysis plot of the top 500 genes.
Viral genome assembly
We noticed that all but one samples belong to the C.1.1.398 lineage (Table 2 ). However, these variants belong to the GR clade with no known mutations affecting the clinical manifestation. The only notable mutation is Spike D614G, which has been proven to increase transmissibility but not severity (Plante et al., 2021). Nonetheless, this mutation was observed in all of our samples, regardless of their disease severity.
Table 2.
Genome characteristics of the infecting SARS-CoV-2.
| Characteristics | Sample |
|||
|---|---|---|---|---|
| AS1 | AS2 | SY1 | SY2 | |
| Accesion ID (GISAID) | EPI_ISL_747235 | EPI_ISL_747239 | EPI_ISL_747236 | EPI_ISL_745033 |
| PANGO lineage (Pango v.4.0.5 PANGO-v1.3) | C.1.1.398 | C.1.1 | C.1.1.398 | C.1.1.398 |
| Clade | GR | GR | GR | GR |
| Category | Non-Variant of Concern, Non-Variant of Interest, Non-Variant Under Monitoring | Non-Variant of Concern, Non-Variant of Interest, Non-Variant Under Monitoring | Non-Variant of Concern, Non-Variant of Interest, Non-Variant Under Monitoring | Non-Variant of Concern, Non-Variant of Interest, Non-Variant Under Monitoring |
| Median assembly coverage | 343x | 11,306x | 929x | 5,512x |
| Amino acid mutations | Spike D614G, N G204R, N R195S, N R203K, NSP1 L149R, NSP2 I273T, NSP3 N1220S, NSP3 T181I, NSP4 G309C, NSP10 A32V, NSP12 P323L | Spike D614G, N G204R, N R203K, NSP1 F143del, NSP1 K141del, NSP1 S142del, NSP12 P323L, NSP15 T33I | Spike D614G, E L21F, N G204R, N R203K, NSP3 T1036I, NSP12 D109N, NSP12 P323L | Spike D614G, N G204R, N P67S, N R203K, NSP2 V628I, NSP4 P187S, NSP12 P323L |
Global gene expression analysis
We used a total of 36,248 sets of genes in this study. We found that a total of 10,489 genes were differentially expressed in asymptomatic patients (478 genes were upregulated, 10,011 genes were downregulated) and 8178 genes in mildly symptomatic patients (2164 genes were upregulated, 6014 genes were downregulated). Differential gene expression analysis showed that a larger set of genes were downregulated in asymptomatic patients than those in mildly symptomatic patients (Fig. 2 ) that might contribute to the halting or progressing the illness. We identified multiple genes within the category of rRNAs and mitochondrial mRNAs that were significantly downregulated in both asymptomatic and symptomatic samples (Fig. 2). This is due to the usage of a different set of healthy control from a different study as mentioned previously, of whom the two types of RNA were not depleted.
Fig. 2.
Global gene expression analysis. Volcano plots showing differentially expressed genes (orange) in asymptomatic and mildly symptomatic samples.
We found that there is a significant upregulation of angiotensin-converting enzyme 2 (ACE2) expression in mildly symptomatic patients but not in asymptomatic patients (Fig. 3 ). However, transmembrane protease serine 2 expression was significantly downregulated in all groups.
Fig. 3.
Gene expression of several SARS-CoV-2 entry proteins. Dot plot displaying the normalized count of ACE2 and TMPRSS2 expression. *P <0.01
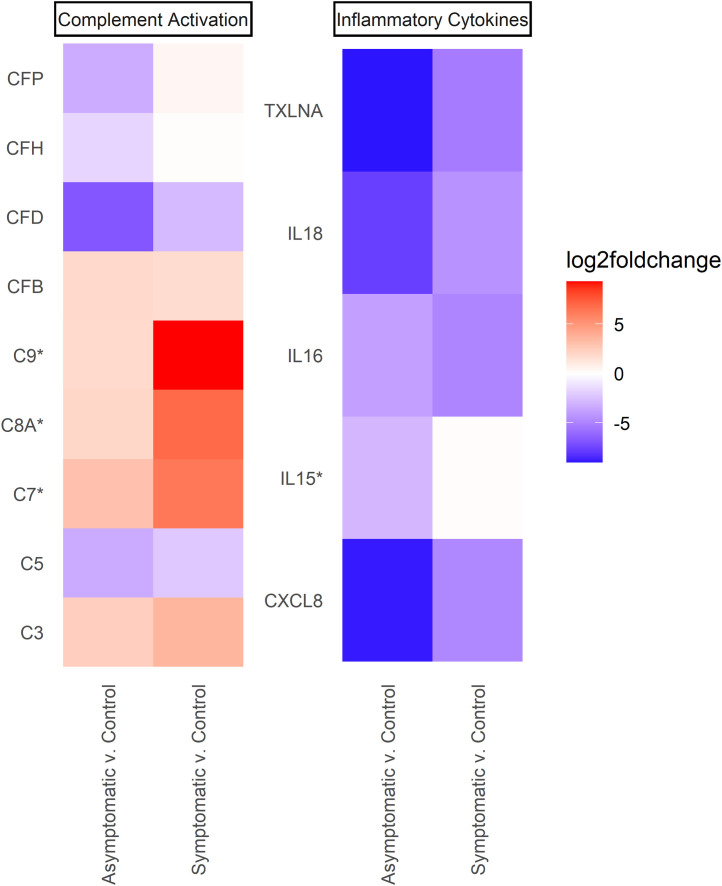
To better assess the inflammatory and immune response differences between the two groups of patients, we visualized a heatmap on the basis of the log2 fold change value of differentially expressed complement system genes (Fig. 4 ) and proinflammatory cytokines (Fig. 4). Interestingly, interleukin (IL)-6 and IL-10 were not differentially expressed, contrasting a previous study (Dhar et al., 2021; Jain et al., 2021; Zhang et al., 2020).
Fig. 4.
The expression level of multiple genes within several immunity categories. Log2 fold change value of several differentially expressed genes within the complement system and proinflammatory cytokines category. The membrane attack complex genes (C7, C8A, and C9) were significantly upregulated in mildly symptomatic patients (P <0.01) but not in asymptomatic ones. A set of proinflammatory cytokines (CXCL8, IL16, IL18, and TXLNA) were downregulated (P <0.01) in both groups. Proinflammatory cytokine interleukin-15 (IL15) was specifically downregulated in asymptomatic samples only (P <0.01; log2 fold change = −2.899).
Gene ontology analysis
We performed gene ontology analysis using DESeq2 differentially expressed genes as input. For more relevant analysis, we generated only gene ontology terms in the biological process category (Fig. 5 ). Asymptomatic and mildly symptomatic patients shared common deregulated biological processes (i.e., cilium modification/assembly, RNA splicing, and nuclear transport). Those processes might be the direct consequences of SARS-CoV-2 infection and may explain the universal pathology of non-severe COVID-19. Cilia regulatory gene RFX3 and cilia component protein DNAH7 (Robinot et al., 2021) were among the genes that were downregulated in our study (P <0.01).
Fig. 5.
Top 25 most enriched biological processes on the basis of the adjusted P-value. Dot plot displaying enriched biological process term in asymptomatic and mildly symptomatic samples. Color magnitude represents adjusted P-value, dot size represents the count of the enriched gene within each category, and the x-axis represents gene ratio (percentage of the enriched genes within that category to the total of assigned genes within this study). The biological process term/category is arranged on the basis of the value of gene ratio, not on their adjusted P-value, for easier visualizing purposes.
Macroautophagy flux reduction in asymptomatic patients
Our study shows expression level differences among genes related to macroautophagy machinery. Moreover, macroautophagy is included in the top 25 most enriched biological processes of asymptomatic patients, indicating its importance in modulating disease severity. Hence, we further analyzed the expression level of genes (log2 fold change) within this ontology term (Fig. 6 ). Generally, we observed a large set of downregulated autophagy-related genes in asymptomatic patients (Fig. 5). Autophagy reduction in asymptomatic patients was featured by a complete downregulation of autophagy-related genes (ATGs). These genes are affiliated with the autophagosomes and DMVs formation (Yin et al., 2016). We found an upregulation of MAPILC3C in the symptomatic group (Fig. 6). Those genes are positive regulators in the autophagosome formation process (Bonam et al., 2020; Yu et al., 2008). The MAP1LC3C protein is a member of the MAP1LC3 protein family, which plays part in the autophagosome initiation (Bonam et al., 2020).
Fig. 6.
Log2 fold change of genes within the macroautophagy ontology term. Autophagy-related genes (ATG13, ATG4D, and ATG9B) were specifically downregulated in asymptomatic patients. Autophagy regulator gene MAP1LC3C was significantly downregulated in the asymptomatic group, yet upregulated in the mildly symptomatic group.
mRNA catabolism dysregulation in mildly symptomatic patients
We found that some biological processes related to mRNA processing (mRNA catabolic process, RNA transport, etc) are particularly enriched in mildly symptomatic patients (Fig. 5). RNA splicing was the only ontologic term within this category that was enriched in asymptomatic patients. This showcases the central role of mRNA processing in the progression of the disease. Hence, we built a heatmap showing the log2 fold change value of differentially expressed genes within this category (Fig. 7 ).
Fig. 7.
Log2 fold change of genes within the mRNA catabolic process term. A downregulation of K-homology splicing regulatory protein (KHSRP) gene expression among mildly symptomatic patients (P <0.01) was detected. Our study also found the upregulation of ZC3H12D, an mRNA regulator zinc finger gene, in asymptomatic patients (P <0.01) but not in mildly symptomatic patients.
Coinfection with local opportunistic bacteria
Taxonomic mapping from the KRAKEN2 result (using minikraken_8GB_20200312 databases) was plotted to a bar plot on the basis of the abundance data for each species. On the basis of our DESeq2 analysis result, we found 30 most differentially abundant species that were higher in symptomatic or asymptomatic patients. We visualized the relative abundance of the 30 species with a heatmap on the basis of Euclidean distance. We found that the Streptococcus pneumoniae dominated the SY1 sample, with an average percentage of 74% and 0.22% in the SY2 sample. S. pneumoniae was also found in asymptomatic samples, with a percentage of 0.01% in AS1 and 0.03% in AS2. Streptococcus cohnii (5.5% in SY1 patient and less than 0.1% in other patients) and Haemophilus influenzae (3.2% in SY1 patient and less than 0.1% in other patients) also had a higher relative abundance in SY1 samples. We found that only Veillonella parvula had a higher abundance in both symptomatic patients (Fig. 8 ).
Fig. 8.
Heatmap plot of species abundance within asymptomatic and mildly symptomatic patients. It is shown that Veillonella parvula abundance is significantly higher in the symptomatic group. ***the highlighted V. parvula.
There is no intrasample variability for each technical replicate and among asymptomatic samples (Fig. 9 a). We also found that there is a great dissimilarity in beta diversity (Fig. 9b). Another research with a bigger sample size also showed diversity variation among samples from the same level of disease severity (Ma et al., 2021). SY1’s alpha diversity was lower than the others (Fig. 9a) because of the overgrowth of S. pneumoniae (Fig. 9c), whereas SY2’s alpha diversity was high. This difference between symptomatic patients might be due to the use of a cross-sectional study with different types of symptoms present (Table 1).
Fig. 9.
Diversity and Relative Abundance Analysis. (A) Alpha diversity analysis with the Shannon index showed that there were no significant differences in intrasample variation. (B) Beta diversity analysis with Bray-Curtis dissimilarity showed no diversity variation among each sample from the same category. (C) Relative abundance of each species to species with >1% relative abundance.
Discussion
Our study aims to identify differences in biological processes and alteration in the microflora community of nasopharyngeal samples from asymptomatic and mildly symptomatic patients with COVID-19. We collected two samples for each group and analyzed their RNA sequence data. According to clinical output categorization by NIH (NIH, 2021), patients AS1 and AS2 were categorized as asymptomatic, whereas patients SY1 and SY2 were mildly symptomatic, possessing at least one common symptom of COVID-19. Patient SY2 possessed a chronic heart disease condition. Although this finding may affect the outcome of the study, Table 1 shows no particular symptoms related to this condition. Hence, its influence over the gene expression of the nasopharynx tissue would be minimal. On the other hand, the infecting SARS-CoV-2 variants are not listed as variants of concern, variants of interest, or variants under monitoring by WHO (who.int). Mutations found in our samples were not known to influence the manifestation of the disease, except for the presence of Spike D614G, which was only known to rise the transmissibility and fitness of the virus (Plante et al., 2021).
Our study confirms a previous study that reported the loss of the ciliary layer due to SARS-CoV-2 infection (Robinot et al., 2021). We observed the downregulation of RFX3, a positive regulator of ciliogenesis, and DNAH7, a component of cilia structure, as identical to that previous study (Robinot et al., 2021) and implies a disruption in the ciliated basal body along the upper respiratory tract, regardless of its clinical manifestation. Although the previous study highlighted the downregulation of ciliogenesis regulator FOXJ1 at the later stage of infection (Robinot et al., 2021), we found no significant differences in the expression of FOXJ1 in both groups of patients (P >0.05). This may suggest that FOXJ1 may still be expressed at the basal level but its downregulation occurs post-translationally due to viral infection, as proven in a study with ciliated EC cells (Abdi et al., 2018). The direct consequence of ciliary impairment is loss of smell (anosmia), one of the most common symptoms of SARS-CoV-2 infection (Meng et al., 2020), considering that the olfactory sensors lie along with the ciliated cells of the nasal cavity (Li et al., 2020). The cilia may also act as an entry point for SARS-CoV-2 along the upper respiratory tract, as previously studied in other coronaviruses (Afzelius, 1994), allowing a high load of virion in the nasal cavity and pharynx (Zou et al., 2020) regardless of case severity.
Another body of biological processes that share similar dysregulation in both groups of the patient lies within the nucleocytoplasmic transport and RNA splicing category. Multiple pieces of evidence demonstrated that the SARS-CoV-2 proteins’ interference over the components of nucleocytoplasmic transport with ORF6 protein performs a vital role in this process (Addetia et al., 2021; Kato et al., 2021; Miyamoto et al., 2021) by binding to nucleoporin protein (Kato et al., 2021; Miyamoto et al., 2021), dislocating them from the nuclear membrane (Kato et al., 2021), and blocking mRNA transport from nucleus to cytoplasm (Addetia et al., 2021) and STAT1 transport into the nucleus (Miyamoto et al., 2021), hindering a proper antiviral interferon response. Although these findings suggest a dysregulation of nucleocytoplasmic transport component at the protein level, our transcriptomic study implies a more systematic dysregulation at their transcriptional level. We found a large set of genes were downregulated within this category of biological process. This provides a hint of intervention in the nucleocytoplasmic gene expression system by yet unknown components of SARS-CoV-2. However, the blocking of mRNA nuclear transport by the viral components would halt multiple cell regulation and pathways (Ren et al., 2010). Moreover, SARS-CoV-2 NSP16 protein binds to U1 and U2 small non-coding RNA region of the spliceosome, inhibiting it from splicing premature mRNA (Banerjee et al., 2020). This action disrupts an array of mRNA modifications and alters multiple pathways and cellular functions (Srivastava et al., 2020).
Specifically, we noticed the deregulation of processes related to epigenetic and cell cycle modification in asymptomatic patients. These findings suggest the involvement of the two modifications in halting the progression of the disease. We have not been able to identify which specific action contributes to this phenomenon because of the complex and unpredicted outcomes of epigenetic regulation. However, we hypothesized that the host cell's response to viral infection by modifying its histone and chromatin structure allows efficient viral elimination through a proper immune response as described before (Menachery et al., 2014).
Interestingly, we found the specific downregulation of IL-15 in asymptomatic patients (P < 0.01) but not in symptomatic patients. In general, overexpression of inflammatory genes and reduction of activated T cells are the hallmark of betacoronavirus infection, including during SARS-CoV-2 infection (Lau et al., 2013; Ryabkova et al., 2021). Multiple reports showcased the “cytokine storm” phenomenon as the main cause of poor prognosis among severe and critically ill patients (Guo et al., 2020; Kumar et al., 2019; Ramasamy and Subbian, 2021; Trougakos et al., 2021). However, we found no other upregulation of inflammatory cytokines in our samples. The discrepancy between our findings and the previous studies might be because the use of nasopharyngeal tissue is not the proper anatomical location to observe this phenomenon. Nonetheless, our result is interesting because IL-15 overexpression leads to excessive activation and exhaustion of neutrophils around the local tissue (Ratthé and Girard, 2004). Activation of neutrophils itself is used as a prognostic factor in the management of patients with COVID-19. A high neutrophils-to-lymphocytes ratio marks the possible severe outcome of the disease (Liu et al., 2020).
Our study found the dysregulation of the mRNA metabolic process, including its translation. We observed this effect explicitly in mildly symptomatic patients. Fig. 5 shows that 12 of the 25 dysregulated biological processes are related to this category. This might benefit the viral infection by halting native mRNA translation and promoting viral genomic RNA translation and replication, which is a very well-known pathogenesis among RNA viruses (Bushell and Sarnow, 2002; Jaafar and Kieft, 2018; Toribio and Ventoso, 2010). Remarkably, a previous single-cell transcriptomic study found consistent dysregulation of the host transcription system in multiple tissues during SARS-CoV-2 infection among symptomatic individuals (Bass et al., 2021). The hijacking of the mRNA translation process includes genes related to viral infection response, specifically innate interferon response (Astuti and Ysrafil, 2020; Banerjee et al., 2020). A delayed interferon response is commonly known as the hallmark of SARS-CoV-2 infection (Astuti and Ysrafil, 2020; Guo et al., 2020; Kasuga et al., 2021; Lau et al., 2013). A more systematic disruption in mRNA metabolism in our symptomatic cohort might need to occur for a poor clinical outcome to occur. Significant downregulation of KHSRP—a negative regulator of retinoic acid-inducible gene I-mediated signaling (Onomoto et al., 2021)—in symptomatic patients might reduce the excessive induction of interferon responses (Onomoto et al., 2021) by halting the activation of retinoic acid-inducible gene I-mediated receptors. We also found the specific upregulation of ZC3H12D gene expression in asymptomatic samples (P <0.01) but not in the symptomatic group. The expression of ZC3H12D negatively correlates with the inflammatory reaction by acting as a destabilizing regulator of several inflammatory gene mRNAs (Zhang et al., 2015), and its expression is high during the case of acute lung injury (Zhang et al., 2015), one of the complication syndrome of patients with COVID-19 (Parasher, 2021).
Other studies also reported aberrant activation of complement system proteins in the blood due to SARS-CoV-2 infection. This activation contributes to the elevation of proinflammatory cytokine release and intravascular thrombosis (Chouaki Benmansour et al., 2021; Bruin et al., 2021; Holter et al., 2020; Java et al., 2020; Noris et al., 2020; Perico et al., 2020). We observed an elevation of complement activation genes (C7, C8A, and C9) within the mildly symptomatic samples, suggesting its involvement in the progression of the disease. The excessive activation of the complement system in the blood causes a high release of inflammatory cytokines and induces vascular thrombosis (Holter et al., 2020; Li and Chen, 2021), the two main causes of the severity of COVID-19 (Parasher, 2021).
We also found a significantly higher expression of the ACE2 gene in mildly symptomatic compared with the control and asymptomatic groups (Fig. 3). It is found that ACE2 expression is a function of interferon expression (Ziegler et al., 2020). Hence, this might contribute to an extreme inflammatory profile among symptomatic patients (Ramasamy and Subbian, 2021). A high level of ACE2 expression in the nasopharyngeal tissue of symptomatic individuals was also observed in a previous transcriptomic study (Islam et al., 2021). Transmembrane protease serine 2 was downregulated in patients with COVID-19, a similar finding to a previous study, where its expression was not differentially expressed among multiple degrees of severity (Jain et al., 2021) or even downregulated among patients with COVID-19 (Islam et al., 2021; Lieberman et al., 2020; Rossi et al., 2021).
We specifically noted an enrichment in the protein catabolic process and macroautophagy/autophagy process in this subset of patients (Fig. 5). Multiple genes with prominent roles in autophagosome influx were significantly downregulated within this group of patients. The most evident ones are the ATGs and MAP1LC3C (P <0.01), suggesting their role in the progression of the disease. The protein products of these ATG genes are central to the formation of autophagosomes and double-membrane vesicles (Andaloussi et al., 2017; Bonam et al., 2020; Rubinsztein et al., 2012; Webber and Tooze, 2010; Xie and Klionsky, 2007; Yun et al., 2020). Coronaviruses and other positive-sense RNA viruses, including SARS-CoV-2, use the DMV compartment to aid their genome replication process, protected from pathogen recognition receptors (Cottam et al., 2014; Netherton and Wileman, 2011; Shroff and Nazarko, 2022; Wolff et al., 2020b, 2020a). Moreover, some autophagy inhibitor compounds have been tested to modulate inflammation in patients with COVID-19. The most notable one is azithromycin, a macrolide antibiotic with an inflammatory regulation property, whose activity in suppressing the autophagosome influx has been well studied (Renna et al., 2011; Venditto et al., 2021).
Our study found that S. pneumoniae, H. influenzae, and S. cohnii had a higher relative abundance in symptomatic patients, specifically in the SY1 patient and only a few in other patients (Fig. 9). The high relative abundance of pathogenic bacteria in SY1 patient affected the lower alpha diversity index in SY1 patient than in others. Besides, the great dissimilarity of beta diversity might be caused by the different individual profiles of bacteria. Pneumococcal infections have been reported in many COVID-19 cases and worsen the patients’ outcomes (Amin-Chowdhury et al., 2021). Although this species is reported as a commensal opportunistic pathogen, residing mostly in the upper airways (Weiser et al., 2018), the colonization of this bacteria leads to respiratory infection. This might happen during the loss of mucociliary clearance due to viral infection (Sender et al., 2021), which is reported in our study and is induced by the depletion of ciliated cells (Li et al., 2020; Robinot et al., 2021). S. pneumoniae had many virulence factors, such as toxin pneumolysin, that cause pore-forming. The effect of pore-forming is to influence inflammatory responses and internalization of other bacteria, such as opportunistic pathogen H. influenzae (Neill et al., 2015). In patients with pneumonia, the normal microflora S. cohnii colonizes and causes bacteremia (Sender et al., 2021). S. pneumoniae colonization can interact with other commensal bacteria through cooperative or competitive relationships. S. pneumoniae cooperatively interacts with H. influenzae and S. cohnii (Sender et al., 2021; Weiser et al., 2018). The clinical data showed that SY1 patients had breathing difficulty symptoms that might be due to the overgrowth of S. pneumoniae, H. influenzae, and S. cohnii in the respiratory tract. Interestingly, Veillonella parvulla had a higher relative abundance in both symptomatic patients (SY1 and SY2) than in asymptomatic patients (Fig. 9). This suggested that the presence of V. parvula might be related to the symptoms of patients with COVID-19. Previously, V. parvula was also found to increase its abundance in patients with COVID-19 compared with the normal patients (Ma et al., 2021).
CRediT authorship contribution statement
Miftahul Faridl: Formal analysis, Investigation, Visualization, Writing – original draft. Karlina Mellyani: Formal analysis, Visualization, Writing – original draft. Karimatu Khoirunnisa: Investigation. Popi Septiani: Methodology, Validation. Ernawati Arifin Giri-Rachman: Writing – review & editing, Validation. Husna Nugrahapraja: Validation. Ema Rahmawati: Funding acquisition, Resources. Cut Nur Cinthia Alamanda: Funding acquisition, Supervision, Resources. Ryan Bayusantika Ristandi: Funding acquisition, Resources. Rifky Walujayati Rachman: Funding acquisition, Resources. Rini Robiani: Funding acquisition, Resources. Azzania Fibriani: Conceptualization, Investigation, Methodology, Supervision, Writing – review & editing, Project administration.
Acknowledgments
Declaration of Competing Interest
The authors have no competing interests to declare.
Funding
This work was supported by the Institute of Research and Community Services, Bandung Institute of Technology, grant number 293/IT1.B07.1/TA.00/2022.
Ethical approval
This study was approved by the ethical committee of Department of Medicine, Padjadjaran University, with approval number 320/UN6.KEP/EC/2021. The informed consent was waived for this study because no potentially identifying information was obtained.
Acknowledgments
We would like to thank Rebecca Stephanie for her input in the writing of this manuscript.
Data availability
All raw reads, gene counts, and bacterial abundance data files are available from the NCBI Gene Expression Omnibus, with accession number GSE186651. The SARS-CoV-2 full genome sequences can be accessed through the GISAID database, with the accession numbers EPI_ISL_747235, EPI_ISL_747239, EPI_ISL_747236, and EPI_ISL_745033.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.06.035.
Appendix. Supplementary materials
References
- Abdi K, Lai CH, Paez-Gonzalez P, Lay M, Pyun J, Kuo CT. Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat Commun. 2018;9:1655. doi: 10.1038/s41467-018-03812-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addetia A, Lieberman NAP, Phung Q, Hsiang TY, Xie H, Roychoudhury P, et al. Sars-cov-2 orf6 disrupts bidirectional nucleocytoplasmic transport through interactions with rae1 and nup98. mBio. 2021:12. doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius BA. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 1994;424:295–300. doi: 10.1007/BF00194614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin-Chowdhury Z, Aiano F, Mensah A, Sheppard CL, Litt D, Fry NK, et al. Impact of the coronavirus disease 2019 (COVID-19) pandemic on invasive pneumococcal disease and risk of pneumococcal coinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): prospective national cohort study, England. Clin Infect Dis. 2021;72:e65–e75. doi: 10.1093/cid/ciaa1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaloussi AE El, Habib S, Soylemes G, Laknaur A, Elhusseini H, Al-Hendy A, et al. Defective expression of ATG4D abrogates autophagy and promotes growth in human uterine fibroids. Cell Death Discov. 2017;3:17041. doi: 10.1038/cddiscovery.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I, Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress Host Defenses. Cell. 2020;183:1325–1339. doi: 10.1016/j.cell.2020.10.004. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass A, Liu Y, Dakshanamurthy S. Single-cell and bulk RNAseq profiling of COVID-19 patients reveal immune and inflammatory mechanisms of infection-induced organ damage. Viruses. 2021;13 doi: 10.3390/v13122418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam SR, Bayry J, Tschan MP, Muller S. Progress and challenges in the use of MAP1LC3 as a legitimate marker for measuring dynamic autophagy in vivo. Cells. 2020;9 doi: 10.3390/cells9051321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin S de, Bos LD, Roon MA van, Tuip-de Boer AMT, Schuurman AR, Koel-Simmelinck MJA, Bogaard HJ, Tuinman PR, van Agtmael MA, Hamann J, Teunissen CE, Wiersinga WJ, Koos Zwinderman AH, Brouwer MC, van de Beek D, Vlaar APJ. Amsterdam UMC COVID-19 Biobank Investigators, et al. Clinical features and prognostic factors in Covid-19: a prospective cohort study. EBiomedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzavara-Silva CE, Gomes ALV, Maia RCC, Acioli-Santos B, Gil LHVG, Marques ETA. Early molecular markers predictive of dengue hemorrhagic fever. An Acad Bras Cienc. 2009;81:671–677. doi: 10.1590/s0001-37652009000400006. [DOI] [PubMed] [Google Scholar]
- Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A. Transcriptome profiling in human diseases: new advances and perspectives. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18081652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaki Benmansour NC, Carvelli J, Vivier E. Complement cascade in severe forms of COVID-19: recent advances in therapy. Eur J Immunol. 2021;51:1652–1659. doi: 10.1002/eji.202048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, K V, Damodar S, Gujar S, Das M. IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: results from meta-analysis and regression. Heliyon. 2021;7:e06155. doi: 10.1016/j.heliyon.2021.e06155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Cheng X, Feng X, Wan H, Chen S, Xiong M. Clinical symptom differences between mild and severe COVID-19 patients in China: a meta-analysis. Front Public Health. 2020;8 doi: 10.3389/fpubh.2020.561264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holter JC, Pischke SE, Boer E de, Lind A, Jenum S, Holten AR, et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci U S A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ABMMK, Khan MAAK, Ahmed R, Hossain MS, Kabir SMT, Islam MS, et al. Transcriptome of nasopharyngeal samples from COVID-19 patients and a comparative analysis with other SARS-CoV-2 infection models reveal disparate host responses against SARS-CoV-2. J Transl Med. 2021;19:32. doi: 10.1186/s12967-020-02695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Kieft JS. Viral RNA structure-based strategies to manipulate translation. Nat Rev Microbiol. 2018;17:110–123. doi: 10.1038/s41579-018-0117-x. 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Ramaswamy S, Harilal D, Uddin M, Loney T, Nowotny N, et al. Host transcriptomic profiling of COVID-19 patients with mild, moderate, and severe clinical outcomes. Comput Struct Biotechnol J. 2021;19:153–160. doi: 10.1016/j.csbj.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Java A, Apicelli AJ, Liszewski MK, Coler-Reilly A, Atkinson JP, Kim AHJ, et al. The complement system in COVID-19: friend and foe? JCI Insight. 2020;5 doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga Y, Zhu B, Jang KJ, Yoo JS. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53:723–736. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ikliptikawati DK, Kobayashi A, Kondo H, Lim K, Hazawa M, et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem Biophys Res Commun. 2021;536:59–66. doi: 10.1016/j.bbrc.2020.11.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom CH van den, Ahout IML, Zomer A, Brand KH, de Groot R, Ferwerda G, et al. Nasopharyngeal gene expression, a novel approach to study the course of respiratory syncytial virus infection. Eur Respir J. 2015;45:718–725. doi: 10.1183/09031936.00085614. [DOI] [PubMed] [Google Scholar]
- Koh J, Shah SU, Chua PEY, Gui H, Pang J. Epidemiological and clinical characteristics of cases during the early phase of COVID-19 pandemic: a systematic review and meta-analysis. Front Med (Lausanne) 2020;7:295. doi: 10.3389/fmed.2020.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. Pheatmap: pretty heatmaps. R package version. 2012;1(2):p.726. [Google Scholar]
- Kumar S, Nyodu R, Maurya VK, Saxena SK. Immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus. Disorders. 2019;2020:43–53. [Google Scholar]
- Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- Li Q, Chen Z. An update: the emerging evidence of complement involvement in COVID-19. Med Microbiol Immunol. 2021;210:101–109. doi: 10.1007/s00430-021-00704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li M, Ou G. COVID-19, cilia, and smell. FEBS Journal. 2020;287:3672–3676. doi: 10.1111/febs.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Fan S, Chen J, Wu J, Xu S, Guo Y, et al. Epidemiological and clinical characteristics of COVID-19 in Adolescents and Young Adults. Adolescents and Young Adults. Innov States. Innovation (Camb) 2020;1 doi: 10.1016/j.xinn.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLOS Biol. 2020;18 doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zhang F, Zhou F, Li H, Ge W, Gan R, et al. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct Target Ther. 2021;6:191. doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Eisfeld AJ, Schäfer A, Josset L, Sims AC, Proll S, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5:e01114–e01174. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Deng Y, Dai Z, Meng Z. COVID-19 and anosmia: a review based on up-to-date knowledge. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Itoh Y, Suzuki T, Tanaka T, Sakai Y, Koido M, et al. SARS-CoV-2 ORF6 disturbs nucleocytoplasmic trafficking to advance the viral replication. bioRxiv. 2021 doi: 10.1038/s42003-022-03427-4. 02.24.432656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health. Clinical spectrum | COVID-19 treatment guidelines 2021, https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/; (accessed June 6, 2021). [PubMed]
- Neill DR, Mitchell TJ, Pneumolysin KA. Streptococcus pneumoniae. Mol Mech Host-Pathogen Interact. 2015:257–275. [Google Scholar]
- Netherton CL, Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol. 2011;1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris M, Benigni A, Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98:314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol. 2021;18:539–555. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasher A. COVID-19: current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad Med J. 2021;97:312–320. doi: 10.1136/postgradmedj-2020-138577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico L, Benigni A, Casiraghi F, Ng LFP, Renia L, Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol. 2020;(17):46–64. doi: 10.1038/s41581-020-00357-4. 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S, Subbian S. Critical determinants of cytokine storm and type i interferon response in COVID-19 pathogenesis. Clin Microbiol Rev. 2021;34 doi: 10.1128/CMR.00299-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratthé C, Girard D. Interleukin-15 enhances human neutrophil phagocytosis by a Syk-dependent mechanism: importance of the IL-15Rα chain. J Leukoc Biol. 2004;76:162–168. doi: 10.1189/jlb.0605298. [DOI] [PubMed] [Google Scholar]
- Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A. 2010;107:10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna M, Schaffner C, Brown K, Shang S, Tamayo MH, Hegyi K, et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest. 2011;121:3554–3563. doi: 10.1172/JCI46095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinot R, Hubert M, Melo GD de, Lazarini F, Bruel T, Smith N, et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun. 2021;12:4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ÁD, de Araújo JLF, de Almeida TB, Ribeiro-Alves M, de Almeida Velozo C, Almeida JM de, et al. Association between ACE2 and TMPRSS2 nasopharyngeal expression and COVID-19 respiratory distress. Sci Rep. 2021;11:9658. doi: 10.1038/s41598-021-88944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Ryabkova VA, Churilov LP, Shoenfeld Y. Influenza infection, SARS, MERS and COVID-19: cytokine storm – the common denominator and the lessons to be learned. Clin Immunol. 2021;223 doi: 10.1016/j.clim.2020.108652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Baric RS. Epigenetic landscape during coronavirus infection. Pathogens. 2017;6 doi: 10.3390/pathogens6010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender V, Hentrich K, Henriques-Normark B. Virus-induced changes of the respiratory tract environment promote secondary infections with Streptococcus pneumoniae. Front Cell Infect Microbiol. 2021;0:199. doi: 10.3389/fcimb.2021.643326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff A, Nazarko TY. The molecular interplay between human coronaviruses and autophagy. Cells. 2022;2021:10. doi: 10.3390/cells10082022. Page;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Daulatabad SV, Srivastava M, Janga SC. Role of SARS-CoV-2 in altering the RNA binding protein and miRNA directed post-transcriptional regulatory networks in humans. bioRxiv. 2020 doi: 10.3390/ijms21197090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavasolian F, Rashidi M, Hatam GR, Jeddi M, Hosseini AZ, Mosawi SH, et al. HLA, immune response, and susceptibility to COVID-19. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio R, Ventoso I. Inhibition of host translation by virus infection in vivo. Proc Natl Acad Sci U S A. 2010;107:9837–9842. doi: 10.1073/pnas.1004110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trougakos IP, Stamatelopoulos K, Terpos E, Tsitsilonis OE, Aivalioti E, Paraskevis D, et al. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J Biomed Sci. 2021;28:9. doi: 10.1186/s12929-020-00703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga EA, Chowell G, Mizumoto K. COVID-19 case fatality risk by age and gender in a high testing setting in Latin America: Chile, March–August 2020. Infect Dis Pover. 2021;10:11. doi: 10.1186/s40249-020-00785-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditto VJ, Haydar D, Abdel-Latif A, Gensel JC, Anstead MI, Pitts MG, et al. Immunomodulatory effects of azithromycin revisited: potential applications to COVID-19. Front Immunol. 2021;0:285. doi: 10.3389/fimmu.2021.574425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16:355–367. doi: 10.1038/S41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2, 2016.
- Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Melia CE, Snijder EJ, Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/J.TIM.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang Y, Walls SD, Gross SM, Schroth GP, Jarman RG, Hang J. Targeted sequencing of respiratory viruses in clinical specimens for pathogen identification and genome-wide analysis. Methods Mol Biol. 2018;1838:125–140. doi: 10.1007/978-1-4939-8682-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Wang L, Lv B, Lu Y, Zeng L, Chen Y, et al. TMEM74, a lysosome and autophagosome protein, regulates autophagy. Biochem Biophys Res Commun. 2008;369:622–629. doi: 10.1016/j.bbrc.2008.02.055. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMI A. J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun EJ, Kim S, Hsieh JT, Baek ST. Wnt/β-catenin signaling pathway induces autophagy-mediated temozolomide-resistance in human glioblastoma. Cell Death Dis. 2020;11:771. doi: 10.1038/s41419-020-02988-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Lou X, Yan H, Pan J, Mao H, Tang H, et al. Metagenomic analysis of viral nucleic acid extraction methods in respiratory clinical samples 11 Medical and Health Sciences 1108 Medical Microbiology. BMC Genomics. 2018;19:1. doi: 10.1186/s12864-018-5152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang WC, Chen JK, Zhou L, Wang M, Wang ZD, Yang B, Xia YM, Lei S, Fu EQ, Jiang T, dong Wang Z, et al. ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol. 2015;67:206–212. doi: 10.1016/j.molimm.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hao Y, Ou W, Ming F, Liang G, Qian Y, et al. Serum interleukin-6 is an indicator for severity in 901 patients with SARS-CoV-2 infection: a cohort study. J Transl Med. 2020;18:406. doi: 10.1186/s12967-020-02571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw reads, gene counts, and bacterial abundance data files are available from the NCBI Gene Expression Omnibus, with accession number GSE186651. The SARS-CoV-2 full genome sequences can be accessed through the GISAID database, with the accession numbers EPI_ISL_747235, EPI_ISL_747239, EPI_ISL_747236, and EPI_ISL_745033.