Abstract
Shigella flexneri, Salmonella enterica serotype Typhimurium, and Listeria monocytogenes were applied to FTA filters, and the filters were used directly as templates to demonstrate their sensitivity and applicability in PCR-based detection assays. With pure cultures, the sensitivities of detection by FTA filter-based PCR were 30 to 50 and 200 CFU for the gram-negative enterics and Listeria, respectively. Different numbers of S. flexneri cells were used in controlled contamination experiments with several different foods (produce, beef, and apple cider). Aliquots from concentrated food washes subsequently spotted onto FTA filters and assayed by PCR gave consistently positive results and detection limits similar to those observed with pure-culture dilutions. This universal method for PCR template preparation from bacterial cells is rapid and highly sensitive and reduces interference from food-associated inhibitors of PCR. In addition, its broad applicability eliminates the need for multiple methods for analysis of food matrices.
Food-borne illnesses caused by pathogenic bacteria still occur at unacceptably high frequencies in industrialized nations and developing countries. Recently, a report from the Centers for Disease Control and Prevention presented estimates and causes of food-borne outbreaks for known illnesses in the United States from 1983 to 1992 and for passive and active surveillance for 1992 to 1997 and 1996 to 1997, respectively (13). The estimates ranged from 6 to 81 million cases of food-borne illnesses per year, with 5,000 deaths occurring annually. Although food safety initiatives have since been introduced to reduce contamination by food handlers and to improve sanitary conditions at the sites where foods are grown, harvested, and processed, food-borne illnesses attributed to these potential sources of contamination continue. This is illustrated by periodic reports of major outbreaks, such as those recently linked to contaminated sprouts in which Salmonella spp. or Escherichia coli O157:H7 was identified as the causative pathogen (18).
Increased public awareness of the health-related and economic impact of food-borne contamination and illness has resulted in greater efforts to develop more sensitive methods of pathogen detection and identification. Advances in molecular biology technology, particularly the PCR, have allowed for more reliable microbial identification and surveillance. PCR has also become a valuable tool for investigating food-borne outbreaks and identifying the responsible etiological agents. PCR techniques have provided increased sensitivity, allowed for more rapid processing times, and enhanced the likelihood of detecting bacterial pathogens. In addition to the analysis of foods, PCR has also been successfully applied to the detection and identification of pathogenic organisms in clinical and environmental samples (16, 21).
The reliability of PCR detection methods depends, in part, on the purity of the target template and the presence of sufficient numbers of target molecules. With such complex matrices as foods, steps must be taken to limit the effects of any potentially inhibitory compounds present that may limit PCR amplification of the intended target (4, 11, 17). This is in addition to enrichment steps that are frequently required to enhance PCR detection sensitivities and overcome problems of low pathogen numbers. Other means must be employed, however, when selective enrichment methods are not possible or do not exist. These other means include immunomagnetic separation (2, 7) and filter (19) formats, both of which are designed to concentrate microorganisms and remove potential PCR inhibitors.
In this study, we developed a protocol that uses FTA filters to prepare bacterial DNA templates derived from pure cultures and from artificially contaminated foods without arduous processing, preenrichment, and purification steps. The FTA filter is a fibrous matrix impregnated with chelators and denaturants that effectively traps and lyses microorganisms on contact (3). Released DNA is sequestered and preserved intact within the membrane. Following a series of brief washes to remove cell debris and other nonbinding contaminants, filters can be used directly in PCR assays or as a solid medium to store samples for later use. With this filter-based technology, enhanced detection sensitivities have been observed (14) compared to the detection sensitivities obtained with conventional template preparations. Templates from low numbers of target cells can be efficiently and rapidly prepared with minimal handling and sample loss. Previously, FTA filters have been used as a blood storage medium (3, 5), for bacterial ribotyping (15), and for preparation of plant genomic DNA (12). The uses of these filters have also been extended to include preparation of PCR templates from parasitic organisms isolated from foods and clinical specimens (14). In the present study, gram-negative and gram-positive bacteria were tested to determine the applicability of FTA filters in PCR detection. Furthermore, we examined the limits of detection of Shigella flexneri cells when they were seeded onto different foods (bean sprouts, alfalfa sprouts, cilantro, lettuce, tomatoes, beef, and apple cider). In addition, a comparison was made between PCR amplification from templates prepared directly from washed produce and PCR amplification from filters. Whereas several investigators have described different methods for preparing template DNA for PCR (20), including the use of other filter types (1), the protocol described here is a more efficient, sensitive, and uniform method for PCR template preparation.
Sensitivity of the PCR assay with pure cultures.
S. flexneri 2457T and Salmonella enterica serotype Typhimurium phage type DT-104 were grown overnight in Luria broth at 37°C in a shaking water bath. Cells were diluted 1:100 in 5 ml of Luria-Bertani broth and incubated as described above for approximately 4 h to obtain a cell count of 109 to 1010 CFU/ml. Listeria monocytogenes cells were grown overnight at 37°C either in brain heart infusion broth or on agar (1.5%) medium. Bacterial cells from cultures grown for either 4 h or overnight (Listeria cultures) were serially diluted with Butterfield's phosphate buffer. PCR detection sensitivities were then compared by using templates prepared either from boiled lysates or by FTA filter application. Aliquots (10 μl) were transferred to thin-wall PCR tubes (Perkin-Elmer), covered with a few drops of mineral oil (Sigma), boiled for 5 min, and then placed on ice until needed. From the same dilutions, 10-μl aliquots were also applied to FTA filters (Fitzco, Inc.). After application of the bacterial cells, the filters were air dried on a heating block at 56°C (15 to 20 min). The FTA filters were washed twice with FTA purification buffer (GIBCO/BRL) for 2 min and twice in 10 mM Tris (pH 8.0) containing 0.1 mM EDTA for 2 min. The filters were air dried as described above, and either the filters were stored at −20°C or the spotted areas were removed with a 6-mm-diameter hole puncher and directly used as templates for PCR. For PCR templates prepared by boiling, the reaction mixtures (total volume, 25 μl) contained the following: 1× PCR buffer (Qiagen), 200 μM (each) dATP, dCTP, dGTP, and dTTP, and each PCR primer at a concentration of 0.2 μM. The mixtures were overlaid with mineral oil. Enzyme (Qiagen Taq DNA polymerase; 1.5 U) was added after the reaction vessels reached 80°C. For reaction mixtures applied onto FTA filters, the conditions were the same, except that the total reaction volume was 100 to 200 μl and the buffer and DNA polymerase (2.5 U/100 μl of reaction mixture) were purchased from Promega. The total number of cycles was 30, and the annealing temperature was 60°C; the denaturing and extension temperatures were 94 and 72°C, respectively. Each step of the cycle was 1 min long. The PCR primers used for amplifying the ipaH gene of Shigella (10) were 5′-GTTCCTTGACCGCCTTTCCGATACCGTC-3′ and 5′-GCCGGTCAGCCACCCTCTGAGAGTAC-3′; the primers used for amplifying the invA gene of salmonellae (9) were 5′-ACCACGCTCTTTCGTCTGG3′ and 5′-GAACTGACTACGTAGACGCTC-3′; and the primers used for amplifying the hemolysin gene of L. monocytogenes were 5′-CGGAGGTTCCGCAAAAGATG-3′ and 5′-CCTCCAGAGTGATCGATGTT-3′ (8). The expected size of the ipaH PCR product is 600 bp; the expected size of the invA PCR product is 941 bp; and a 234-bp product results from targeting the hemolysin gene of L. monocytogenes. PCR-amplified products were visualized in 1% agarose gels in 0.5× Tris-acetate–EDTA (pH 7.8) buffer with ethidium bromide (0.2 μg/ml) by using a Run-one gel system (Embi-Tec).
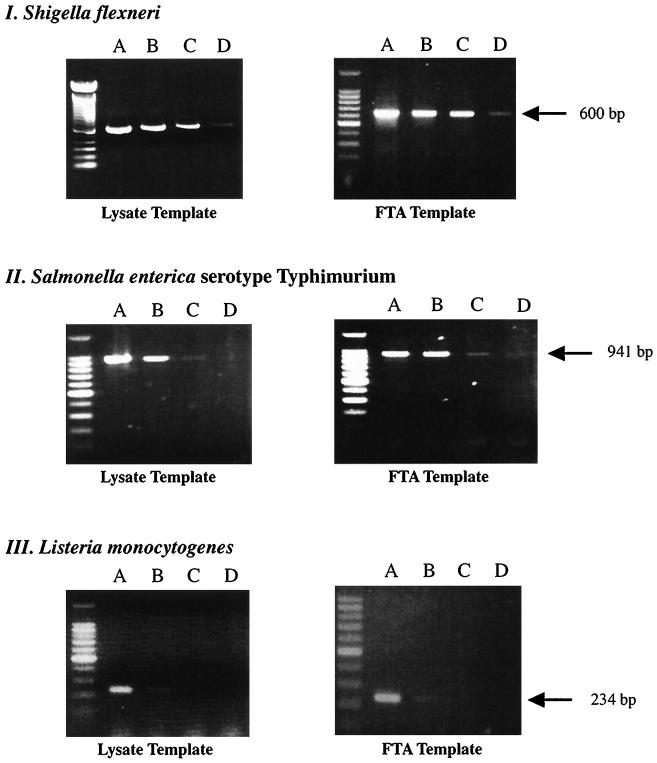
When a dilution series prepared from pure cultures was used, the detection limits were independent of the preparation method (Fig. 1). Templates prepared by boiling or from FTA filters gave similar PCR results. Amplification of the ipaH gene of Shigella, which is present in multiple copies, was observed from as few as 40 CFU. Primers targeting the invA gene in S. enterica serotype Typhimurium phage type DT-104 successfully amplified a template from 30 CFU. The minimum number of CFU required to yield a positive result for L. monocytogenes was 2 × 102 CFU. Boiling easily disrupts gram-negative bacteria, such as shigellae and salmonellae. However, gram-positive organisms, such as Listeria, are impervious to lysis by boiling. An additional reagent to enhance lysis, such as lyphostatin, may be needed to increase the sensitivity of PCR-based analyses.
FIG. 1.
Comparison of detection limits of PCR-based assays with lysates and FTA filters as templates. (I) PCR products generated from 10-fold serially diluted S. flexneri cells (lane A, 5 × 104 CFU; lane B, 5 × 103 CFU; lane C, 5 × 102 CFU; lane D, 50 CFU) when boiled lysates and FTA filters were used as PCR templates. (II and III) Same comparison performed with S. enterica serotype Typhimurium and L. monocytogenes, respectively. The arrows indicate the expected size of each amplicon.
PCR data from unseeded and seeded foods.
Table 1 shows the background microbial population that was enumerated for each food tested. Foods with moderate to large microbial floras were selected in order to assess their impact on detecting low levels of the targeted bacteria (Shigella) when they were applied to FTA filters. Untreated food samples were also tested by using the PCR-based assay for Shigella detection to determine if the microbial content yielded any nonspecific amplicons. The samples chosen for analysis included samples of bean sprouts, alfalfa sprouts, lettuce, tomato, cilantro, ground beef, and apple cider. Solid samples (10 g) were placed in 250-ml beakers and washed once with 10 ml of 1× phosphate-buffered saline with gently agitation for 15 to 20 min at room temperature. The wash buffer was decanted from each food sample, and serial dilutions were then made to 10−3; 10 μl from the 10-ml wash and 10 μl of each dilution were used as PCR templates. Apple cider was assayed directly and by using serial dilutions as described above. Each sample was evaluated directly with the PCR assay or spotted onto FTA filters. No amplicons were observed with either template preparation method in the absence of the appropriate target (data not shown).
TABLE 1.
Detection limits for S. flexneri in various artificially contaminated foods
| Food sample | Background count (CFU/ml) | Detection limit (CFU/ml)
|
|
|---|---|---|---|
| Nonfiltered | FTA filter | ||
| Cilantro | 1.0 × 108 | 5.0 × 104 | 50 |
| Tomato | 9.0 × 103 | 5.0 × 102 | 5.0 × 102 |
| Beef | 4.0 × 104 | NDa | 40 |
| Lettuce | 4.0 × 104 | ND | 50 |
| Alfalfa sprouts | 2.0 × 108 | 50 | 50 |
| Bean sprouts | 5.0 × 106 | 50 | 50 |
| Apple cider | 0 | ND | 50b |
ND, not detected.
Represents 1 μl of suspension spotted onto an FTA filter.
The same foods were artificially contaminated with Shigella. Bacterial cultures were grown as described above and were serially diluted with Butterfield's phosphate buffer. As described above, 10 g of bean sprouts, alfalfa sprouts, lettuce, tomato, cilantro, or ground beef or 10 ml of apple cider was placed in a 250-ml beaker and seeded with 100 μl of the corresponding bacterial dilution; the inoculation levels ranged from 102 to 105 CFU. The wash buffer was then decanted into Poly-prep chromatography columns (Bio-Rad) with glass wool added to remove large particulates. Apple cider was directly added to the columns. The filtrate was collected in 12.5-ml polypropylene tubes and centrifuged at 8,000 × g for 5 min, the supernatant was discarded, and the resulting pellet was suspended in 100 μl of 1× phosphate-buffered saline. Ten microliters of this suspension was then either transferred to PCR tubes and boiled for 5 min or applied to FTA filters and processed for PCR as described previously.
As shown in Table 1, suspensions that were boiled and used directly in the PCR assay gave mixed results. Shigella was not detected in all foods seeded and tested. PCR products were observed only for tomato, cilantro, alfalfa sprouts, and bean sprouts. Beef, lettuce, and apple cider samples were negative. In contrast, application of 10-μl portions of seeded food washes to FTA filters gave very consistent results when the PCR-based assay was used. Table 1 shows that as little as 50 CFU was detected in seeded cilantro and lettuce, whereas 5 × 102 CFU was detected in seeded tomato. PCR amplification using filters was also able to detect Shigella seeded in alfalfa and bean sprouts at levels ranging from 50 to 5 × 102 CFU (Table 1). A PCR amplicon was similarly seen in washes from ground beef seeded with 40 to 4 × 103 CFU of Shigella. With apple cider, application of 10 μl of seeded apple cider to a filter did not produce a PCR product. However, when 1 μl of material was placed on a filter, PCR products were detected in samples containing 50 to 5 × 102 CFU.
There are two reasons for our primary focus on detection of shigellae: these human pathogens are extremely difficult to detect in complex matrices such as foods and the infectious dose of shigellae is reported to be in the range from 10 to 200 organisms (6). Shigella is also noted for its high transmission rates through person-to-person contact. At present, foods are not routinely screened for the presence of human pathogenic bacteria such as shigellae; rather, they are screened only after clinical information and epidemiological information identify an outbreak caused by the consumption of contaminated food. This results in additional difficulties in isolating and identifying the pathogen from a source; after 7 to 10 days, the physical state of the food sample and the overwhelming bacterial background make recovery of the etiological agent with current bacteriological methods a challenge. To fully explore the utility of FTA filters as a component of a PCR detection method for surveillance and epidemiology, the filters must be capable of detecting low numbers of bacteria amid a considerable background of competing microflora while remaining largely unaffected by matrix-derived factors that may inhibit PCR. Our results indicated that the FTA filter format has the potential to fulfill these requirements.
The consumption of fresh vegetables is a common cause of food-borne illnesses, and therefore, artificial contamination of alfalfa and bean sprouts, lettuce, tomato, and cilantro with Shigella was appropriate for our study. Although weak, highly variable results were observed when we used aliquots obtained directly from washes of foods as templates, PCR products were reliably generated from FTA filters spotted with food wash suspensions. The FTA filters were particularly successful at minimizing interference from high counts of indigenous microbial flora in foods, such as alfalfa sprouts. Also, the use of a multicopy gene, such as ipaH, as the PCR target can provide additional sensitivity to the overall assay. For detection of Salmonella, we also used a single-copy target, the invA gene, and demonstrated similar levels of detection. In one experiment, we used a multiplex assay (three genes) for detection of Salmonella and observed a detection limit of 4 × 102 CFU (data not shown). In all cases, applying food washes to the filters was better than, or as good as, boiling the washes and using them directly in the PCR assay. Amplification from shigellae seeded in apple cider was more problematic irrespective of filter use. The amount of extraneous material centrifuged concurrently with bacterial cells was very significant, and this reduced the sensitivity of the PCR assay. Whether this was due to material that blocked binding of the bacteria to the filter, thus preventing lysis, or due to introduction of PCR inhibitors into the reaction mixture is not clear. Additional preparation prior to application of the material to the FTA filters may be required to overcome diminished sensitivity. Further improvements in this format and its application to food testing, particularly with sprouts and cider, are ongoing.
With the development of real-time PCR detection, analysis and identification of suspected food-borne contaminants might be performed in as little as 1 to 2 h. With its shorter analysis times and greater detection sensitivities, this improved template PCR preparation format can be easily adapted to surveillance studies. With the rapid increase in global commerce, variations of this protocol may be particularly useful for international tracking of food-borne outbreaks. As a means of ensuring food safety, this method can be easily adapted to a hazard analysis and critical control point program to prevent contaminated foods from reaching the consumer. The hazard analysis and critical control point approach is aimed at reducing the number of incidents of human illnesses from food-borne agents, such as those attributed to Salmonella in meats and poultry.
Preparation of PCR templates with FTA filters requires fewer steps and less handling time with considerably less likelihood of sample loss and decreased sensitivity compared to other methods. This should improve detection of food-borne pathogens such as those used in this study, as well as emerging pathogens such as E. coli O157:H7, Campylobacter, and other established bacterial agents that cause food-borne illnesses. FTA filters can contribute greatly to elimination of public health concerns for detecting pathogens in foods as it is necessary to have a sufficient amount of template that is free of PCR inhibitors. Passing food isolates through these filters can effectively concentrate targeted organisms, even in the presence of high levels of indigenous microflora, and can also eliminate potential inhibitors of PCR-based assays. The results of this study demonstrate that this extractionless, filter-based method can be used to detect food-borne bacterial pathogens regardless of food type.
Acknowledgments
We thank Dan Levy for his critical reading of the manuscript and his comments.
REFERENCES
- 1.Bej A K, Mahbudani M H, Atlas R M. Polymerase chain reaction-gene probe detection of microorganisms by using filter-concentrated samples. Appl Environ Microbiol. 1991;57:3529–3534. doi: 10.1128/aem.57.12.3529-3534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bej A K, Mahbubani M H, Miller R, DiCesare J L, Haff L, Atlas R M. Multiplex PCR amplification and immobilized capture probes for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 3.Belgrader P, Del Rio S A, Turner K A, Marino M A, Weaver K R, Williams P E. Automated DNA purification and amplification from blood-stained cards using a robotic workstation. BioTechniques. 1995;19:426–432. [PubMed] [Google Scholar]
- 4.Bhaduri S, Cottrell B. A simplified sample preparation method from various foods for PCR detection of pathogenic Yersinai enterocolitica: a possible model for other food pathogens. Mol Cell Probes. 1998;12:79–83. doi: 10.1006/mcpr.1998.0155. [DOI] [PubMed] [Google Scholar]
- 5.Burgoyne, L. A. March 1996. Solid medium and method for DNA storage. U.S. patent 5,496,562.
- 6.DuPont H L, Levine M M, Hornick R B, Formal S B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 7.Fluit A C, Torensma R, Visser M J C, Aarsman C J M, Poppelier M J J G, Keller B H I, Klapwijk P, Verhoef J. Detection of Listeria monocytogenes in cheese with the magnetic immuno-polymerase chain reaction assay. Appl Environ Microbiol. 1993;59:1289–1293. doi: 10.1128/aem.59.5.1289-1293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furrer B, Candrian U, Juethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 9.Galan J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman A B, Venkatesan M, Oaks E V, Buysee J M. Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of Shigella flexneri. J Bacteriol. 1990;172:1905–1915. doi: 10.1128/jb.172.4.1905-1915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinneman K C, Trost P A, Hill W E, Weagant S D, Bryant J L, Kaysner C A, Wekell M M. Comparison of template preparation methods from foods for amplification of Escherichia coli O157 Shiga-like toxins type I and II DNA by multiplex polymerase chain reaction. J Food Prot. 1995;58:722–726. doi: 10.4315/0362-028X-58.7.722. [DOI] [PubMed] [Google Scholar]
- 12.Lin J-J, Fleming R, Kuo J, Matthew B F, Saunders J A. Detection of plant genes using a rapid, nonorganic DNA purification method. BioTechniques. 2000;28:346–350. doi: 10.2144/00282pf01. [DOI] [PubMed] [Google Scholar]
- 13.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlandi P A, Lampel K A. Extraction-free, filter-based template preparation for the rapid and sensitive PCR detection of pathogenic parasitic protozoa. J Clin Microbiol. 2000;38:2271–2277. doi: 10.1128/jcm.38.6.2271-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers C, Burgoyne L. Bacterial typing: storing and processing of stabilized reference bacteria for polymerase chain reaction without preparing DNA—an example of an automatable procedure. Anal Biochem. 1997;247:223–227. doi: 10.1006/abio.1997.2031. [DOI] [PubMed] [Google Scholar]
- 16.Simon T. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 1999;33:3545–3556. [Google Scholar]
- 17.Soumet C, Ermel G, Fach P, Colin P. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett Appl Microbiol. 1994;19:294–298. doi: 10.1111/j.1472-765x.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 18.Taormina P J, Beuchat L R, Slutsker L. Infections associated with eating seed sprouts: an international concern. Emerg Infect Dis. 1999;5:626–634. doi: 10.3201/eid0505.990503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls I, Sheridan J J, Welsh R W, McDowell D A. Separation of microorganisms from meat and their rapid enumeration using a membrane filtration-epifluorescent microscopy technique. Lett Appl Microbiol. 1990;10:23–26. doi: 10.1111/j.1472-765x.1990.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang R-F, Cao W-W, Cerniglia C E. A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. J Appl Microbiol. 1997;83:727–736. doi: 10.1046/j.1365-2672.1997.00300.x. [DOI] [PubMed] [Google Scholar]
- 21.White T J, Madej R, Persing D H. The polymerase chain reaction: clinical applications. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]