Abstract
We looked at the diversity of NO2− oxidizers at field scale by examining isolates at clump scale and in microsamples of soil (diameter, 50 μm). The genetic distances (as determined by amplified ribosomal DNA restriction analysis performed with Nitrobacter-specific primers) in a small clump of soil were as large as those between reference strains from large geographical areas. Diversity in individual microsamples was shown by serotyping.
Bacterial diversity is now accepted as one of the components of soil function, particularly its sustainability. Endemism and clonality, which have an intrinsic spatial dimension, are studied (7, 13, 16, 17), as spatial patterns may have significant implications in soil function. In the case of bacteria, in contrast to other organisms in soil for which hierarchical models of spatial organization have been proposed (1), the spatial arrangement in soil is not well understood yet (11). It was recently shown that only 1 g of soil was needed to represent the prominent bacteria in large homogeneous grassland areas (5).
The distribution of microorganisms at the microscale is rarely explored, although due to the size of bacteria, such distribution obviously represents a functional scale where major interactions take place within the soil structure. Soil is a typical highly heterogeneous medium from both textural and structural standpoints (18), and small-scale heterogeneity of physicochemical characteristics probably results in the patchy arrangement of bacteria observed by Hattori and Hattori (12).
At present, most diversity studies of soil target community changes that follow stress (20); more rarely, a functional or taxonomic group (e.g., Rhizobium) is studied (29). This is probably linked to methodological limits: no specific media have succeeded in providing a reliable means for targeting a defined group among the numerous groups in the soil, and only recently have molecular tools done so. However, the study of spatial heterogeneity of bacterial distribution seems to require the use of a defined bacterial group, as suggested by Felske and Akkermans (5), for interpretation. The genus Nitrobacter, the genus of chemilithotrophic bacteria responsible for the majority of NO3− formation from NO2− in soil, is a good candidate for such studies because there is little redundancy in soil (15) and its presence in a culture is easily detected (14). The rrs-rrl intergenic spacer (IGS) of Nitrobacter seems to be appropriate for studying diversity at the subspecies level (19). Nitrobacter-specific primers, which can be used in complex DNA mixtures (10), were designed for the rrs-rrl IGS and for the rrl gene. It has been suggested (5, 6) that fingerprints of rrs ribosomal DNA extracted from soil were indicative of the numerically dominant bacteria (even with 1-g soil samples) but did not take into account the microheterogeneity, particularly the diversity of small specific communities, which requires appropriate sampling techniques (16). It is, however, recognized that the degree of diversity generally depends on sample size (23). The sample size relevant to Nitrobacter spatial arrangement at a microhabitat scale has been studied elsewhere (unpublished data). It was shown that some microsamples less than 500 μm in diameter did not harbor any nitrifiers.
The objective of this work was to evaluate the diversity of the model genus Nitrobacter at different spatial scales down to a spatial scale close to the microhabitat size. Genetic distances, obtained by amplified ribosomal DNA restriction analysis (ARDRA) of Nitrobacter-specific IGS-rrl amplicons, were compared by using “patterns” obtained from samples of soil taken at various levels of spatial organization, including microsamples, clumps, a field, and Nitrobacter species reference strains from large geographical areas.
Soil sampling.
The agricultural soil studied was an Alfisol cultivated with maize. The surface soil was a sandy loam (bulk density, 1.3 mg · m−3; 17% clay, 39.2% silt, and 40.4% sand; organic carbon content, 1.4%; pH(H2O) 6.4 [8]) and was weakly structured (24). A 2- to 3-cm3 coherent soil clod (clump a) was gradually subdivided under a binocular microscope into microsamples called volumetric units (VU) by using a sterile scalpel blade. A calibrated grid was used to spot VU that fit into squares that were 50, 100, and 250 μm on each side; these VU are referred to below as size 50, size 100, and size 250, respectively. Each VU, sampled randomly because three-dimensional coordinates could not be used due to the lack of an appropriate technique, was taken up with a sterile plugged glass capillary that had been dipped into sterile, biologically inert silicone oil (SV40) and was transferred to 2 ml of defined culture medium by swirling the tip of the capillary in the medium. About 30 VU of each size were sampled. They were subsequently cultured in NO2− oxidizer culture medium (final concentration of NO2−, 1 mM) at 28°C in the dark in the wells of 24-well microculture plates (27), until the NO2− disappeared (about 1 to 8 weeks). The same experiment was carried out with another 2- to 3-cm3 clump of soil (clump b) that was obtained 1 year later at the same spot in the field.
Culturing and analysis of soil microsamples.
After NO2− disappeared from the culture medium (in the presence of NO2− oxidizers), as determined by the Griess-Ilosvay spot test (14), VU positive for the presence of NO2− oxidizers were selected from clump a. Six size 250 replicates, seven size 100 replicates, and four size 50 replicates from clump a were transferred into 100 ml of mineral medium and incubated again until NO2− disappeared in order to obtain enough biomass for DNA extraction. For clump b, four size 250 replicates, five size 100 replicates, and seven size 50 replicates were treated similarly. Total DNA from mixed soil cultures was extracted as described by Rouvier et al. (25). The reference organisms included Nitrobacter winogradskyi, Nitrobacter sp. strain DE30, N. winogradskyi agilis, Nitrobacter sp. strain DE11, Nitrobacter vulgaris Z, 269, and nevada, Nitrobacter sp. strain LL, and Nitrobacter hamburgensis X14. These organisms represented three of the four Nitrobacter species (10, 19, 27), N. winogradskyi, N. vulgaris, and N. hamburgensis.
Nitrobacter-specific PCR was performed with primers FGPI149-457 (5′-CGGCGCTTGTGCTCA-3′) and FGPL420′-458 (5′-CCTGACAGGTTCTGG-3′) for the rrs and rrl genes, respectively (10). The PCR were carried out as described by Grundmann et al. (10). Distances were computed by using the Neighbor-Joining algorithm (26).
Isolation and serotyping.
Isolates were obtained from 5-g fresh soil subsampled from a 3-kg sample of soil taken at different places in the same field and subsequently sieved (mesh size, 2 mm) and mixed. Serial 10-fold dilutions of soil were prepared, and 0.5-ml aliquots of each dilution were inoculated into six wells of 24-well microtiter plates filled with 1.5 ml of mineral medium containing NO2− at a final concentration of 1 mM. The microtiter plates were incubated for 90 days at 28°C. Total disappearance of NO2− from a well indicated the presence of NO2−-oxidizing bacteria (27). The contents of five wells positive for the presence of NO2−-oxidizing bacteria were transferred to new medium, and pure isolates were obtained as described by Soriano and Walker (28).
Seven sera were raised against reference strains corresponding to seven serotypes. These sera were designated by using the strain designations (see above), as follows: LL, Z, DE11, DE30, W, Ag, and X14. Most organisms were grown at 28°C in mineral medium with NO2− as the energy source (27); the exceptions were N. hamburgensis and N. vulgaris, which were grown as described by Bock et al. (3) and Bock et al. (4), respectively. Each serum was tested against each microsample culture, each isolate, and each reference strain. Samples (25 μl) of each culture were fixed on microprint 12-well multitest slides and treated for fluorescent analysis (27). The presence of fluorescent cells in each culture was determined with a Zeiss microscope.
Genetic distances between ARDRA patterns for clumps of soil.
The detection threshold of ARDRA gels, as tested by mixing DNA of two different strains in various proportions, indicated that the ARDRA pattern of each strain could be recognized if the quantities of DNA were in range of ratios down to 1/20 (data not shown). At larger ratios, the fragments of the minority strain were indistinguishable from the background smear. Besides, as the sample sizes were multiples of one another and the larger samples did not produce complex ARDRA patterns, the patterns observed in microsamples probably were the dominant patterns.
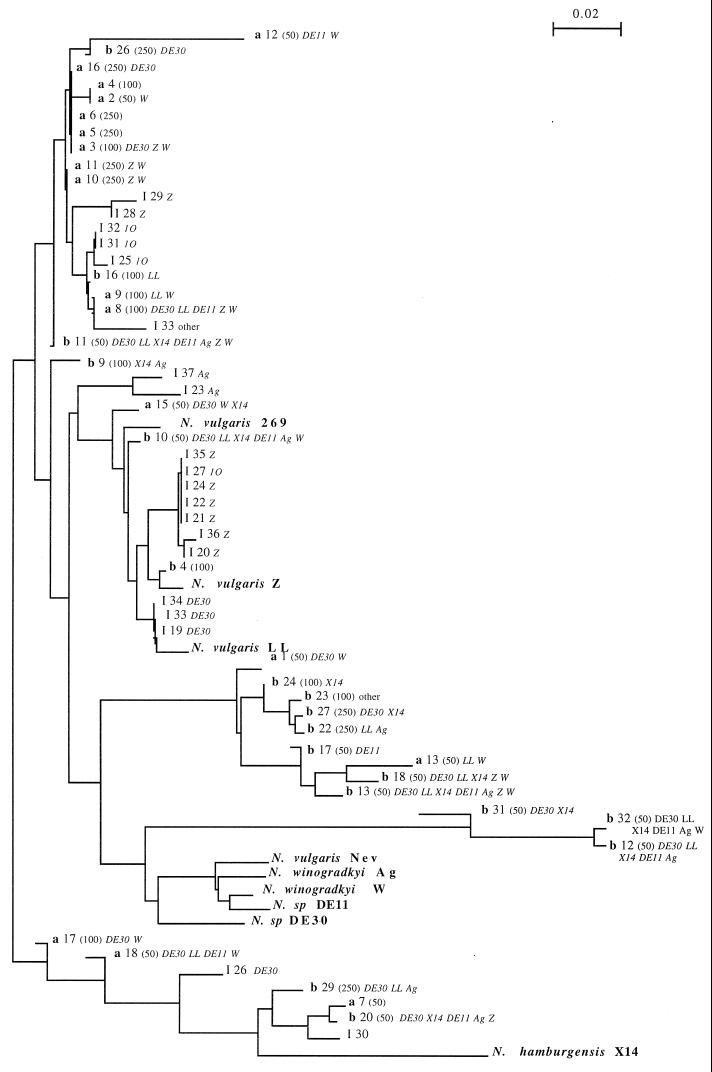
For the 17 VU sampled in clump a (VU designated a in Fig. 1), 11 ARDRA patterns were obtained, all of which were different from the patterns of the reference strains, as indicated in the dendrogram of genetic distances (Fig. 1). The patterns for a few microsamples (the a3, a5, a6, a16 patterns, the a8 and a9 patterns, and the a10 and a11 patterns) were identical. The genetic distances between genotypes from the microsamples were as large (Fig. 1) as the genetic distances between the genotypes of Nitrobacter reference species, the genetic distances between genotypes from the microsamples and genotypes of the reference species were also as large as the genetic distances between the genotypes of the reference species (the smallest genetic distance between two species was 5.5%). The results obtained for clump b (VU designated b in Fig. 1) are similar, except that no identical genotypes were found among the 18 VU.
FIG. 1.
ARDRA distance dendrogram of the genotypes (as calculated by the method of Saitou and Nei [26]). a, microsamples from clump a; b, microsamples from clump b. The numbers are sample numbers. The values in parentheses are the sizes of the UV (in micrometers). I, isolate. Reference organisms are indicated by boldface type. Serotyping results for microsamples and isolates are indicated when available. Bar = 0.02 nucleotide substitution per site.
Genetic distances between isolates.
The 20 isolates obtained from 5 g of homogenized soil obtained at the field site yielded patterns different from the patterns of the reference organisms and from the patterns of the microsamples of soil (Fig. 1). Some isolates were as distant from the reference organisms as the microsamples were. Some isolates came from the same culture well of the most-probable-number count analysis and yielded identical patterns; isolates I21, I22, I24, I27, and I35 produced identical patterns, as did isolates I34, I33, and I19, and isolates I31 and I32.
Serotyping in microsamples.
The detailed serotyping results are indicated on the dendrogram in Fig. 1; both serotype analysis and ARDRA were performed for VU. The serotyping data clearly showed that several serotypes coexisted closely in a single VU (Fig. 1). The sera did not cross-react with the reference strains although the possibility that they cross-reacted with non-Nitrobacter autochtonous cells in the soil sample cannot be eliminated. Furthermore, sera were checked against 40 isolates from the same soil on Luria-Bertani medium (data not shown). Only one isolate yielded an equivocal reaction yet did not have a typical Nitrobacter shape. Although the level of discrimination of serotyping, which corresponds to phenotypes, has not been clearly established yet (3, 19), there is at least large phenotypic diversity.
Even in the smallest samples, several serotypes (up to a maximum of seven serotypes) could be detected. The number of serotypes detected per VU was clearly larger for smaller samples (Fig. 1), as described by Grundmann and Gourbière (9). Thus, diversity may be hidden by a culturing artifact (9) or the ARDRA detection threshold with larger samples.
Spatial limit of identical ARDRA patterns.
We showed that the diversity of Nitrobacter, in terms of genetic distances based on rrs-rrl studies, was as large in a small clump of soil as the diversity of reference strains from different geographical areas. It was possible to find identical ARDRA patterns, considered in the scope of this study a clone, in microsamples from a clump of soil but not at a larger scale. Our results point out that the size of a clone is difficult to delineate considering the low numbers of VU showing identical patterns (4 and 2) and that quantification of the extent of an individual clone's habitat requires thorough spatial investigation at a smaller scale. The spatial coordinates of microsamples would be needed to prove the existence of a relationship between the spatial distance and the genetic distance for the range of distances explored in this work.
Evolutionary implication and soil status.
It is tempting to bring together our results and their evolutionary and soil function implications. Mutations are undoubtedly responsible for diversity. Nitrobacter species are slow growers, but mutations happen between divisions (21). Based on rrs gene analysis, genetic distances indicate that Nitrobacter emerged about 50 to 100 million years ago (22). Considering that the rrs-rrl IGS accumulates 10 times more mutations than the 16S rRNA (10) and given that a universal substitution rate rules bacteria evolution (21), 1% divergence in the IGS would represent 5 million years. The time estimate yielded by such calculations based on the present study (Fig. 1), about 60 million years, corresponds to the lower value for the period mentioned above. This time estimate seems inconsistent with the age of the soil, which is about 200,000 years old (i.e., Quaternary [Riss era]). Besides, the diversity results cannot be separated from the physical status of the soil. The soil studied is of glacial origin, which means that substantial mixing of imported particles occurred. The soil is also cultivated and weakly structured (24). Previous results indicated that this soil has high percolating values, as shown by the mercury porosimetry intrusion method; pores as small as 1.8 μm in diameter were connected. This is much larger than the size of the cells in the soil (2) and allows transportation by water movement for example. These conditions should provide a high potential for migration of cells during rain events and ploughing. These remarks lead to the hypothesis that migration of cells happens at different time and spatial scales on a regular basis.
Our study of the spatial diversity at a submillimetric scale in which ARDRA of the rrs-rrl genes of the genus Nitrobacter was used revealed diversity at the microhabitat scale close to that found for the whole genus. We showed that evaluation of spatial clonality is complex and requires refined sampling strategies. Furthermore, the large genetic distances observed in relation to local Nitrobacter sampling distances suggest that biological and physical processes (mutations and migrations) were involved in the small-scale diversity observed.
REFERENCES
- 1.Beare M H, Coleman D C, Crossley D A, Jr, Hendrix P F, Odum E P. A hierarchical approach to evaluating the significance of soil biodiversity to biogeochemical cycling. Plant Soil. 1995;170:5–22. [Google Scholar]
- 2.Bock E, Koops H P, Arms H. Nitrifying bacteria. In: Schlegel H G, Bowien D, editors. Autotrophic bacteria. Madison, Wis: Science Technique Publishers; 1989. pp. 80–96. [Google Scholar]
- 3.Bock E, Koops H P, Möller U C, Rudert M. A new facultative nitrite-oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol. 1990;153:105–110. [Google Scholar]
- 4.Bock E, Sundermeyer-Klinger H, Stackebrand E. New facultative lithotrophic nitrite-oxidizing bacteria. Arch Microbiol. 1983;136:281–284. [Google Scholar]
- 5.Felske A, Akkermans A D L. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb Ecol. 1998;36:31–36. doi: 10.1007/s002489900090. [DOI] [PubMed] [Google Scholar]
- 6.Führ A. Untersuchungen zu der Biodiversität natürlicher Bakterienpopulationen im Boden mit der denaturierenden Gradientengelelektrophorese (DGGE) von 16S rDNA-Sequenzen. Ph.D. thesis. Kaiserlautern, Germany: Universität Kaiserslautern; 1996. [Google Scholar]
- 7.Fulthorpe R R, Rhodes A N, Tiedje J M. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl Environ Microbiol. 1998;64:1620–1627. doi: 10.1128/aem.64.5.1620-1627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann G L, Renaud P, Ross L, Bardin R. Differential effects of soil water content and temperature on nitrification and aeration. Soil Sci Soc Am J. 1995;59:1342–1349. [Google Scholar]
- 9.Grundmann G L, Gourbière F. A micro-sampling approach to improve the inventory of bacterial diversity in soil. Appl Soil Ecol. 1999;387:1–4. [Google Scholar]
- 10.Grundmann, G. L., M. Neyra, and P. Normand. High resolution phylogenic analysis of NO2−-oxidizing Nitrobacter species using the rrs, rrs-rrl IGS sequence and rrl genes. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 11.Harris P J. Consequences of spatial distribution of microbial communities in soil. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass. New York, N.Y: John Wiley and Sons; 1994. pp. 239–246. [Google Scholar]
- 12.Hattori T, Hattori R. The physical environment in soil microbiology: an attempt to extend principles of microbiology to soil microorganisms. CRC Crit Rev Microbiol. 1976;4:423–460. doi: 10.3109/10408417609102305. [DOI] [PubMed] [Google Scholar]
- 13.Haubold B, Rainey P B. Genetic and ecotypic structure of a fluorescent Pseudomonas population. Mol Ecol. 1996;5:747–761. [Google Scholar]
- 14.Keeney D R, Nelson D W. Nitrogen—inorganic forms. In: Page A L, et al., editors. Methods of soil analysis, part 2, 2nd ed. Agronomy Monographs. Vol. 9. Madison, Wis: Soil Science Society of America; 1982. pp. 643–693. [Google Scholar]
- 15.Marilley L, Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. [Google Scholar]
- 16.Mau M, Timmis K N. Use of subtractive hybridization to design habitat-based oligonucleotide probes for investigation of natural bacterial communities. Appl Environ Microbiol. 1998;64:185–191. doi: 10.1128/aem.64.1.185-191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynard Smith J, Smith N J, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metting F B., Jr . Structure and physiological ecology of soil microbial communities. In: Metting F B Jr, editor. Soil microbial ecology. Applications in agricultural and environmental management. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 3–25. [Google Scholar]
- 19.Navarro E, Fernandez M P, Grimont F, Clays-Josserand A, Bardin R. Genomic heterogeneity of the genus Nitrobacter. Int J Syst Bacteriol. 1992;42:554–560. [Google Scholar]
- 20.Nüsslein K, Tiedje J M. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl Environ Microbiol. 1999;65:3622–3626. doi: 10.1128/aem.65.8.3622-3626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 22.Orso S, Gouy M, Navarro E, Normand P. Molecular phylogenetic analysis of Nitrobacter spp. Int J Syst Bacteriol. 1994;44:83–86. doi: 10.1099/00207713-44-1-83. [DOI] [PubMed] [Google Scholar]
- 23.Pielou E C. An introduction to mathematical ecology. 3rd ed. New York, N.Y: Wiley Interscience; 1977. [Google Scholar]
- 24.Ranjard L, Richaume A, Jocteur-Monrozier L, Nazaret S. Response of soil bacteria to Hg(II) in relation to soil characteristics and cell location. FEMS Microbiol Ecol. 1997;24:321–331. [Google Scholar]
- 25.Rouvier C, Prin Y, Reddell P, Normand P, Simonet P. Genetic diversity among Frankia strains nodulating members of the family Casuarinaceae in Australia revealed by PCR and restriction fragment length polymophism analysis with crushed root nodules. Appl Environ Microbiol. 1996;62:979–985. doi: 10.1128/aem.62.3.979-985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou R R, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Ecol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt E L, Belser L W. Autotrophic nitrifying bacteria. In: Weaver R W, Angle J S, Bottomley P S, editors. Methods of soil analysis, part 2. Microbiogical and biochemical properties. SSSA Book series no. 5. Madison, Wis: Soil Science Society of America; 1994. pp. 159–177. [Google Scholar]
- 28.Soriano S, Walker N. Isolation of ammonia-oxidizing autotrophic bacteria. J Appl Bacteriol. 1968;31:493–497. doi: 10.1111/j.1365-2672.1968.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 29.Souza V, Nguyen T T, Hudson R R, Pinero D, Lenski R E. Hierarchical analysis of linkage disequilibrium in Rhizobium populations: evidence for sex? Proc Natl Acad Sci USA. 1992;89:8389–8393. doi: 10.1073/pnas.89.17.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]