Abstract
Drug-inducible suicide systems may help to minimize risks of human induced pluripotent stem cell (hiPSC) therapies. Recent research challenged the usefulness of such systems since rare drug-resistant subclones were observed. We have introduced a drug-inducible Caspase 9 suicide system (iCASP9) into the AAVS1 safe-harbor locus of hiPSCs. In these cells, apoptosis could be efficiently induced in vitro. After transplantation into mice, drug treatment generally led to rapid elimination of teratomas, but single animals subsequently formed tumor tissue from monoallelic iCASP9 hiPSCs. Very rare drug-resistant subclones of monoallelic iCASP9 hiPSCs appeared in vitro with frequencies of ∼ 3 × 10-8. Besides transgene elimination, presumably via loss of heterozygosity (LoH), silencing via aberrant promoter methylation was identified as a major underlying mechanism. In contrast to monoallelic iCASP9 hiPSCs, no escapees from biallelic iCASP9 cells were observed after treatment of up to 0.8 billion hiPSCs. The highly increased safety level provided by biallelic integration of the iCASP9 system may substantially contribute to the safety level of iPSC-based therapies.
Keywords: suicide gene, induced pluripotent stem cells, Caspase 9, gene editing, silencing, loss of heterozygosity
Graphical abstract

Drug-inducible suicide systems may help to minimize risks of hiPSC therapies. Monoallelic integration of a Caspase 9 system allowed formation of drug-resistant iPSC clones due to loss of heterozygosity or promoter methylation. In contrast, no escapees were observed after treatment of high numbers of cells with biallelic integration of the Caspase9 system.
Introduction
Pluripotent stem cell (PSC) technologies have come of age, and a number of clinical trials applying embryonic stem cell (ESC)- or induced PSC (iPSC)-based cell products are ongoing or in preparation. However, the tumorigenic potential of PSC-derived cell products either in terms of teratoma formation by contaminating undifferentiated cells or due to malignant transformation caused by mutations that were acquired and enriched during reprogramming,1 or culture expansion,2 is considered a major safety concern.
While improved protocols for targeted differentiation of PSCs into the therapeutic derivatives of interest substantially reduce the risk for teratoma formation, introduction of synthetic fail-safe systems with drug-inducible suicide genes have been proposed to further decrease tumor risks. Such suicide systems include the inducible expression of the herpes simplex virus-thymidine kinase (HSV-TK), which has already been clinically applied more than 25 years ago,3 and the inducible Caspase 9 (iCASP9) safety switch system,4 which has been clinically applied more recently, in T cells using a retroviral approach.5
In a first study that attempted to quantitatively define the safety level of PSC transplants with integrated fail-safe systems, ESCs carrying an HSV-TK system controlled by one allele of the endogenous cyclin-dependent kinase 1 (CDK1) locus were applied.6 Despite general functionality of the HSV-TK suicide system concerning the elimination of cycling ESCs, rare proliferating and ganciclovir-resistant subclones of mouse ESCs were observed in vivo and in vitro with an average frequency of 6.6 × 10-8. The major underlying mechanism for failure of the integrated drug-inducible suicide system in mouse ESCs was attributed to transgene loss, probably occurring as loss of heterozygosity (LoH) caused by mitotic recombination, gene conversion, or chromosomal nondisjunction. Furthermore, the authors demonstrated that in case of ESCs with biallelic (homozygous) integration of HSV-TK in the CDK1 locus, no resistant clones appeared among a total of 1.2 × 108 cells, pointing to a substantially higher safety level.
One potential disadvantage of the applied HSV-TK system is that HSV-TK is a viral transgene. Therefore, it may be immunologically recognized in vivo, leading to the killing of HSV-TK-expressing cells even without the application of ganciclovir. It should be stressed that placing an HSV-TK transgene under control of a cell-cycle-dependent locus such as CDK16 indeed restricts the targeted elimination to only cycling cells, such as tumor cells. This concept, though, is probably not applicable if the iPSC-derived graft is required to proliferate in order to fulfill its therapeutic function. Since all cycling cells will express the HSV-TK gene, they may become a target of the host’s immune system. In addition, the HSV-TK fail-safe system is most effective in fast-dividing cells and may fail to eliminate populations of slowly dividing tumor cells. Furthermore, the use of the prodrug ganciclovir as potent antiviral drug, e.g., for the treatment of serious herpes virus infections, may not be possible in patients that received cellular grafts that contain an HSV-TK suicide system.
Because of these limitations and the lack of data for human iPSCs (hiPSCs), we aimed at quantitatively defining the safety level of hiPSC transplants with another integrated drug-inducible suicide system, the iCASP9 safety switch. In this system, the caspase activation and recruitment domain of human Caspase 9, a gene expressed in fetal and adult tissue that is upregulated during induction of apoptosis, has been replaced by a modified dimerizer-binding domain of the FK506-binding protein 12.5 This system requires caspase dimerization after application of an otherwise bioinert small molecule (chemical inducer of dimerization [CID]) for the activation of intracytoplasmic caspase-3/7 and induction of apoptosis4,7 (Figure 1A). This system is considered nonimmunogenic, as Caspase 9 is an endogenous gene.
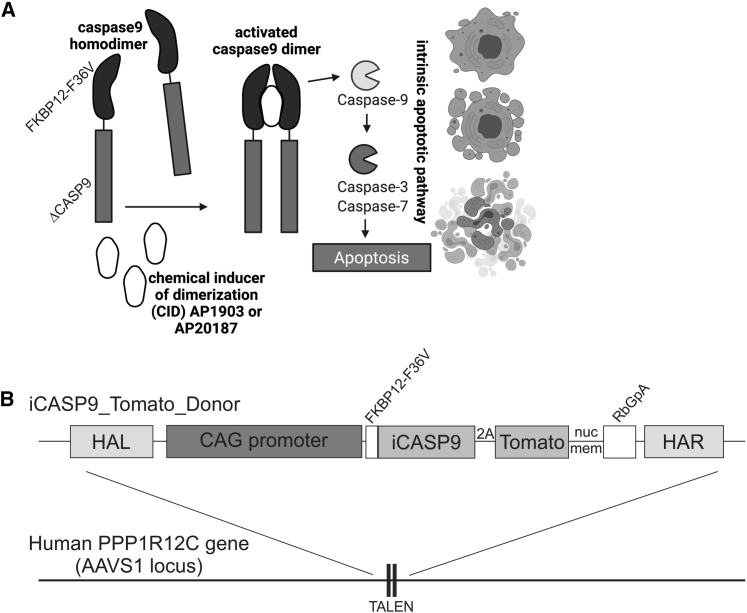
Figure 1.
The iCASP9 suicide switch and TALEN-based integration into the safe-harbor locus AAVS1 in human iPSCs
(A) Function of the iCASP9 suicide system. The CASP9 gene is deleted for its endogenous caspase activation and recruitment domain (ΔCASP9) and is coupled to the sequence of a mutated FK506-binding protein (FKBP12) with an F36V mutation that confers increased affinity for chemical inducers of dimerization (CIDs). FKBP12-F36V binds to otherwise bioinert small-molecule dimerizing agents AP1903 and AP20187, which are lipid-permeant tacrolimus analogs. In the presence of the drugs, the iCASP9 pro-molecule dimerizes and activates the intrinsic apoptotic pathway, leading to cell death via apoptosis (scheme adapted from Gargett and Brown8). (B) Schematic illustration of iCASP9 donor construct and the AAVS1 target site. The iCASP9 donor construct consists of left- and right-handed homology arms (HAL and HAR, respectively), a CAG promoter, and an iCASP9 gene coupled via a 2A site to a dTomatonucmem fluorescence marker with a nuclear membrane localization signal. Integration of the iCASP9 donor construct was achieved via TALEN-based gene editing into the AAVS1 located in the PPP1R12C gene on human chromosome 19.
In order to ensure the safe application of the safety switch and efficient cell killing in both fast- and slow-dividing hiPSC derivatives, we have integrated the iCASP9 construct into the PPP1R12C/adeno-associated-virus site 1 (AAVS1) safe-harbor locus.9 iCASP9 was placed under control of a synthetic promoter consisting of the cytomegalovirus early-enhancer element and chicken beta-actin (CAG) promoter, known for robust expression in undiffentiated hiPSCs and their differentiated progeny10,11 (Figure 1B). Methylation of the endogenous PPP1R12C and the transgenic CAG promoters was analyzed to explore transgene silencing as a potential additional mechanism for the appearance of CID-resistant cell clones. In consideration of the results of Liang et al.6 we have generated hiPSC clones either heterozygous or homozygous for the integrated iCASP9 safety switch. Potential emergence of cell clones escaping the induced suicide was monitored in vivo and in vitro using therapeutically relevant cell numbers.
Results
Generation of hiPSC lines with mono- or biallelic integration of an iCASP9 suicide gene coupled to a Tomato fluorescent reporter
In order to enable inducible induction of apoptosis, we applied transcription-activator-like effector nuclease (TALEN)-mediated gene editing to integrate an iCASP9 safety switch into the AAVS1 safe-harbor locus. The iCASP9 gene was place under control of the CAG promoter. A dTomato-fluorescent protein fused to a nuclear membrane localization signal (dTomatonucmem) was utilized to facilitate indirect monitoring of iCASP9 expression and discrimination from autofluorescence and was coupled to the iCASP9 gene via a 2A site (Figure 1B). Allele-specific PCR analysis confirmed site-specific integration into one or both alleles of the AAVS1 locus of Phoenix12 and hCBPSC213 hiPSC lines (Figures S1A and S1B). For all further procedures and analyses, one correctly targeted monoallelic (heterozygous) and biallelic (homozygous) clone per cell line was chosen. These clones showed considerable levels of dTomatonucmem expression (Figure S1C), absence of chromosomal aberrations as demonstrated by karyotyping (Figure S1D), expressed typical pluripotency markers, differentiated into derivatives of all three germ layers, and formed teratomas upon injection into NODSCID mice (Figure S2). For clarity, all selected iCASP9 cell lines that were applied thereafter are abbreviated according to their origin and genotype: monoallelic iCASP9 Phoenix, biallelic iCASP9 Phoenix, monoallelic iCASP9 hCBiPS2, and biallelic iCASP9 hCBiPS2.
Chemical inducers of dimerization efficiently induce apoptosis in monoallelic and biallelic iCASP9 iPSC clones
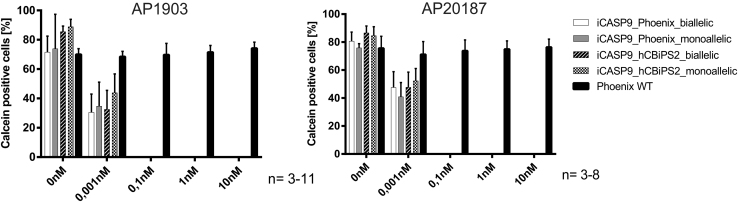
Undifferentiated cells of monoallelic and biallelic iCASP9 Phoenix and iCASP9 hCBiPS2 clones were cultivated as monolayers on Geltrex and were treated for 24 h with different concentrations of two CID variants, AP1903 and AP20187, before the staining of viable cells with Calcein. Intercalating DNA-binding live/dead dyes such as 7-AAD or propidium iodide that were used in previous studies11,14 were found to be less useful for this purpose since (1) dead cell bodies after DNA degradation were obviously detected by flow cytometry as false-negative “viable” cells, and (2) the number of such false-negative viable cells, to a large extent, depends on the duration of CID treatment and the period between CID treatment and fluorescence-activated cell sorting (FACS) analysis (data not shown). In contrast, Calcein provided much more reliable results. Flow-cytometry analysis revealed very efficient killing of cells in all four mono- and biallelic cell lines after treatment with either CID variants at concentrations ≥0.1 nM (Figure 2).
Figure 2.
CID efficiently induce apoptosis in monoallelic and biallelic iCASP9 iPSC clones
FACS analysis of undifferentiated iCASP9 clones cultivated on Geltrex with or without 24 h CID AP1903/AP20187 treatment. As demonstrated by loss of Calcein viability staining, efficient cell killing of iCASP9 iPSCs is achieved with 0.1 nM AP1903 or AP20187 (mean ± SEM; n = 3–11). 300,000 cells per experiment were treated.
In vivo CID treatment against preformed teratomas from monoallelic iCASP9 hiPSCs eliminates human cells and teratomas in most, but not all, mice
In vivo experiments involving NODSCID mice were conducted to confirm the above results in vivo (Figure 3A). All mice after the injection of iPSCs under the kidney capsule showed massive increase of girth, the typical sign for teratoma formation after 8 weeks. Mice that received nontransgenic Phoenix hiPSCs and subsequent CID treatment, as well as mice after the injection of monoallelic iCASP9 Phoenix iPSCs followed by vehicle treatment, all developed teratomas that stained positive for human nuclear antigen (Figure 3B). In contrast, treatment with CID resulted in almost complete elimination of cystic teratoma structures in three out of five mice injected with transgenic monoallelic iCASP9 Phoenix iPSCs (Table 1; Figure 3B). In these mice, only some abnormal puffy mouse tissue with eosinophilic infiltration around the kidney could be detected (Figure 3B, lower middle panel), probably representing fibrotic mouse tissue that formed in response to the massive CID-induced cell death and the resulting infiltration of phagocytes and granulocytes, followed by subsequent pro-fibrotic cytokine release. Also, in the remaining two out of five mice, extensive elimination of frequently cystic teratoma structures was observed. However, in these mice, other tumor-like tissue was detected around the kidney that stained positive with an anti-human nucleoli antibody, suggesting that despite killing of the vast majority of engrafted cells, re-growths of escapees occurred that were apparently resistant to CID induction of iCASP9-mediated suicide.
Figure 3.
AP1903 eliminates preformed teratoma in the majority of NODSCID mice after injection of monoallelic iCASP9 Phoenix iPSCs but did not prevent formation of human tumor-like tissue in all animals
(A) Schematic illustration of the workflow for teratoma induction and CID treatment. (B) NODSCID mice were sacrificed and sectioned 9 days after CID/vehicle treatment subsequent to injection of undifferentiated Phoenix iPSCs under the kidney capsule. (Immuno-)histology demonstrated growth of teratoma in all mice that received wild-type (WT) iPSCs and in monoallelic iCASP9 iPSCs without CID treatment (top and upper middle panels). In 3/5 mice that received iCASP9 iPSCs, only some abnormal puffy mouse tissue with eosinophilic infiltration around the kidney could be detected (lower middle panel), probably representing fibrotic mouse tissue that formed in response to the massive CID-induced cell death and the resulting infiltration of phagocytes and granulocytes followed by pro-fibrotic cytokine release. Human tumor-like tissue was detected in 2/5 mice, suggesting growth of CID-resistant cell clones (bottom panel; see also Table 1). intraperitoneal, i.p. (scale bars: 500 μm for H&E, 100 μm for other images).
Table 1.
CID treatment did not reliably eliminate teratoma formed in NODSCID mice after transplantation of monoallelic iCASP9 Phoenix iPSCs
| iPSC clone | CID treatment (50 μg/day/animal i.p.) | Number of mice treateda | Number of mice with detectable human cell mass after CID treatment |
|---|---|---|---|
| Monoallelic iCASP9 | AP1903 | 5 | 2 |
| Monoallelic iCASP9 | Vehicle | 3 | 3 |
| Wild type | AP1903 | 5 | 5 |
| Wild type | Vehicle | 3 | 3 |
106 cells/mouse have been transplanted under the kidney capsule of NOD.CB17-PrkdcScid/J mice.
In vitro CID treatment led to selection of rare CID-resistant cell subclones from monoallelic, but not biallelic, iCASP9 iPSC clones
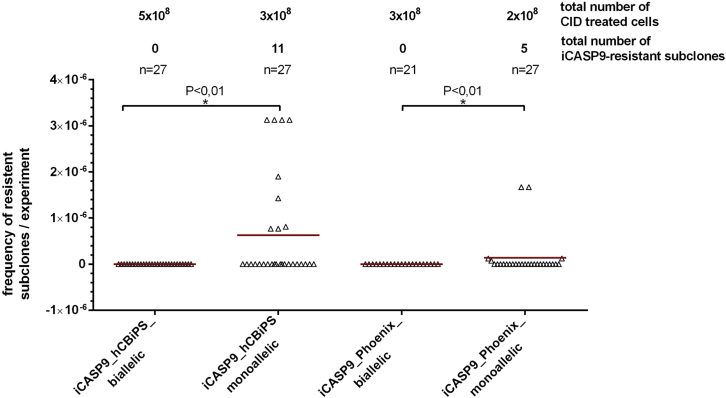
In order to further explore the frequency of the observed CID resistance in monoallelic versus biallelic iPSCs, iCASP9 hCBiPS2 and iCASP9 Phoenix cells were treated with CID (either AP1903 or AP20187, at concentrations ranging from 0.5 to 10 nM). The cells were seeded onto irradiated feeder cells and were cultivated for 3 weeks to promote the propagation of potentially surviving cells. In accordance with the in vivo experiments, the survival of extremely rare CID-resistant cell clones was observed in several independent experiments, with overall frequencies of 3.6 × 10-8 for hCBiPS2 and 2.5 × 10-8 for Phoenix iPSCs. We never observed surviving clones in biallelic iCASP9 iPSCs despite the high number of 0.8 billion treated cells (Figure 4; Table 2).
Figure 4.
CID treatment of mono- and bialleic iPSCs led to selection of rare CID-resistant cell subclones from monoallelic, but not biallelic, iCASP9 iPSC clones
Frequencies of rare CID-resistant cell subclones/CID-treated cells in individual experiment are shown. Monoallelic and biallelic iPSCs were treated with CID (concentrations from 0.5 to 10 nM AP1903/AP20187), seeded onto irradiated feeder cells, and cultivated for 3 weeks to promote the propagation of potentially surviving cells. Survival of apparently CID-resistant extremely rare cell clones was observed in several independent experiments (1 × 105 to 1 × 108 cells/experiment) with overall frequencies of 3.6 × 10-8 (hCBiPS2) or 2.5 × 10-8 (Phoenix). We never observed surviving clones in biallelic transgenic iPSCs despite the high number of treated cells. Data are presented as mean ± SD (n = 21–27). D'Agostino-Pearson omnibus normality test. Kruskal-Wallis test (p < 0.01).
Table 2.
Rare monoallelic iCASP9 iPSC subclones become CID resistant in vitro via transgene loss or methylation of the CAG promoter
| iPSC line | Number of CID-treated cells/experiment | Total number of CID-treated cells | CID conc., nM | Total number of CID-resistant subclones | Frequency of CID- resistant cell subclones | Subclones further cultivated and analyzed | Subclones with eliminated transgene (confirmed/tested) | Subclones with CAG promoter methylated (confirmed/tested) |
|---|---|---|---|---|---|---|---|---|
| iCASP9 hCBiPS2 monoallelic | 1 × 105–1 × 108 | 3 × 108 | 0.5–10 | 11 | 3.6 × 10-8 | 7 | 0/7 | 2/2a |
| iCASP9 Phoenix monoallelic | 2 × 105–8 × 107 | 2 × 108 | 1–10 | 5 | 2.5 × 10-8 | 4 | 3/4 | 0/1b |
| iCASP9 hCBiPS2 biallelic | 2 × 105–5 × 108 | 5.2 × 108 | 0.5–10 | 0 | 0 | 0 | 0 | 0 |
| iCASP9 Phoenix biallelic | 1 × 105–3 × 108 | 3.14 × 108 | 1 –10 | 0 | 0 | 0 | 0 | 0 |
No further clones tested.
Mechanism for CID resistance remained unclear.
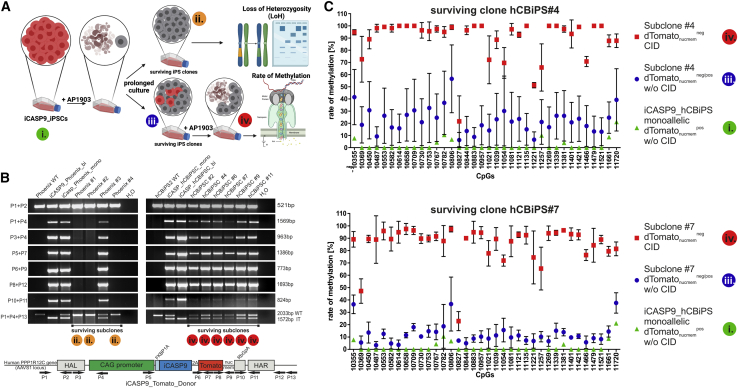
Rare monoallelic iPSC subclones become CID resistant due to transgene elimination, probably via LoH or silencing via CAG and PPP1R12C promoter methylation
Eleven CID-resistant subclones derived from monoallelic iCASP9 iPSCs (Table 2) were further cultivated to investigate mechanisms of the acquired CID resistance. Three out of four subclones derived from iCASP9 Phoenix did not show any dTomatonucmem expression. PCR-based analysis of the genomic sequence of the transgene cassette in the AAVS1 locus of these iCASP9 Phoenix subclones (Figure 5B, (#1, #2, and #4) revealed elimination of the complete transgene, very likely via LoH. FACS analysis of dTomatonucmem expression in these clones after 18–20 passages post CID treatment confirmed the absence of the transgene (data not shown). While the remaining Phoenix subclone #3 showed diminished dTomatonucmem expression, analysis by Nanopore sequencing indicated that neither transgene elimination (Figure 5B) nor increased methylation of CpGs in the CAG promoter or adjacent AAVS1 (PPP1R12C) locus (Figures S4 and S5) had occurred. Instead, a point mutation in the iCASP9 gene was detected that led to a replacement of an Alanine to a Threonine at position 81 of the FKBP1A component of the suicide system (data not shown). Since this point mutation could not explain the observed decreased Tomato expression in Phoenix subclone #3, the underlying mechanism of CID resistance remained elusive.
Figure 5.
Rare monoallelic iPSC subclones become CID resistant due to transgene elimination probably via loss of heterozygosity (LoH) or silencing via CAG promoter methylation
(A) Scheme illustrating appearance and selection for Tomatonucmemneg cells resistant to CID-induced apoptosis and reappearance of Tomatonucmempos CID-sensitive cells during culture expansion, and analysis of different stages for LoH and methylation of promoter elements (and surrounding genomic DNA; data not shown). Colored circles mark stages that have been further analyzed for LoH and promoter methylation. (B) LoH occurred in three CID-resistant monoallelic iCASP9 Phoenix iPSC subclones but not in analyzed CID-resistant monoallelic iCASP9 hCBiPS2 subclones. PCR-based analysis of the genomic sequence of the transgene cassette in the AAVS1 locus revealed elimination of the transgene in Phoenix subclones #1, #2, and #4, very likely via LoH. Primer combinations and locations are depicted in the scheme below. (C) Nanopore sequencing showed methylation of the CAG promoter in 2/2 analyzed CID-resistant monoallelic dTomatonucmemneg hCBiPS2 subclones that did not show transgene elimination. Analysis of CpG islands in the CAG promoter of surviving subclones #4 and #7 indicate a strong correlation between cell survival and loss of dTomatonucmem expression after CID treatment and a high methylation rate in the CAG promoter. Data are presented as mean ± SD (n = 3–4). A scheme of CpG islands in the AAVS1 locus and the integrated iCASP9 donor construct is shown in Figure S4. IT, integrated transgene; WT, wild type.
Loss of the transgene could not be demonstrated in any of the seven analyzed iCASP9 hCBiPS2 subclones. Interestingly, a second CID treatment led to elimination of dTomatonucmem-expressing cells in all these CID-resistant subclones; however, dTomatonucmem-expressing cells re-appeared in all subclones several days after CID treatment (Figure S3), suggesting that reversible epigenetic mechanisms are responsible for transgene silencing.
Exemplarily, two of these phenotypically similar subclones (#4 and #7) were analyzed for iCASP9 and dTomatonucmem mRNA expression and for increased methylation of CpGs in the AAVS1 locus or the CAG promoter via Nanopore sequencing (Figures 5 and S6). The observed high methylation rate in the CAG promoter in these subclones (Figure 5C) correlates with loss of dTomatonucmem expression (Figure S3) and their resistance toward CID treatment. This was accompanied by a strong reduction of dTomatonucmem expression in both clones and by a reduction of iCASP9 expression as well (Figure S6B). For hCBiPS2#7, the reduction of iCASP9 expression is less pronounced than for hCBiPS2#4, a finding that may correlate with the observation that the PPP1R12C was heavily methylated after CID treatment in hCBiPS2#4 (Figure S6C).
Discussion
The availability of human ESCs and iPSCs with their far-reaching potential for proliferation and differentiation offers novel opportunities for the development of tailored cellular therapies. PSC-based therapies have their associated therapeutic risks, including ones that are related to abnormal cell functions such as arrhythmia in the case of cardiomyocytes or dysregulation of hormones in pancreatic islands; however, it is tumor formation that is especially concerning. Integration of synthetic fail-safe systems into PSCs have been proposed as an additional safety measure, enabling induced suicide in cases of transplant-associated tumor formation. In view of the relatively high levels of retroviral and lentiviral vector silencing, especially in pluripotent cells, it is, however, not surprising that suicide-resistant cells are commonly observed after retro- and lentiviral delivery of such systems. In contrast, targeted integration into genomic loci known for their robust expression is considered an approach that is more appropriate to achieve reliable induced elimination of transplanted cells.
A recent study, however, challenged that assumption by detection of rare suicide-resistant cells: with a total of eight ganciclovir-resistant escapee clones among a total of 120 million cells for a monoallelic HSV-TK suicide gene integrated into the endogenous CDK1 locus, Liang et al. observed an overall frequency of 6.7 × 10-8 escapees.6
With a total of 16 escapee clones among 500 million cells (3.2 × 10-8), we have now observed a comparable frequency for the CAG promoter-controlled iCASP9 safety switch inserted in the AAVS1 locus. While Liang et al.6 observed transgene loss and hypothesized LoH via mitotic recombination or chromosomal nondisjunction as main mechanism behind the appearance of clonal escapees among mouse ESCs, we were able to demonstrate that transgene silencing due to aberrant promoter methylation in rare hiPSC subclones apparently contributes to a similar extent to the development of CID-resistant subclones. Interestingly, the observed aberrant subclonal promoter methylation was unstable, and reversion of the methylation always led to the re-appearance of Tomatonucmem-expressing CID-sensitive cells within the escapees after withdrawal of CID. In both analyzed clones, the Tomatonucmem reporter expression correlated perfectly with the observed strong methylation of the CAG promoter.
Whether the lack of methylation of the endogenous PPP1R12C promoter in hCBiPS2#7 may have contributed to the less pronounced decrease in iCASP9 expression compared with subclone #4 remains speculative. Actually, promoter methylation should influence both transgenes that are coupled by a 2A (protease recognition) site and does not explain the observed stronger decrease of dTomato expression. The observed level of iCASP9 expression observed in clone #7, however, seems to be sufficient to turn cells into CID-resistant ones. Apparently, a certain threshold level of iCASP9 expression is necessary for induction of apoptosis.
Using another synthetic suicide system integrated at another genomic site than described by Liang et al.,6 our data suggest that CID-resistant escapees are a general phenomenon that can occur with low frequency upon use of different suicide genes, different promoters controlling the suicide gene, and at different integration sites. In consideration of the observed contribution of transgene silencing to the emergence of rare CID-resistant subclones, the choice of integration site, promoter, and the transplanted cell type will presumably influence the frequency of CID-resistant clones.
Nonetheless, similar to Liang et al., we never observed resistant cell clones that develop from iPSCs carrying a homozygous (biallelic) safety switch. This finding further argues for LoH as the underlying mechanism for transgene elimination in monoallelic subclones, as observed by us and Liang et al.6 since elimination of the suicide gene via LoH can be excluded in cells homozygous for the integrated safety switch.
In case of the observed aberrant promoter methylation as the responsible mechanism for safety-switch inactivation, the question remains whether such rare events depend on the status of the cellular methylation machinery, which would imply that in the event of methylation of one allele, there is also an increased likelihood for similar methylation of the second allele.
Since we did not observe any escapees from two iPSC lines with the integrated biallelic iCASP9 safety switch, even among the huge number of 0.8 billion cultured iPSCs, it is much more likely that the very rare observation (∼3 × 10-8) of aberrant methylation represents a random event that can occur on one allele, independent of the second allele. If this theory applies, a simultaneous methylation of both alleles would be extremely unlikely (3 × 10-8 × 3 × 10-8 = 9 × 10-16).
In theory, individual loss-of-function mutations may also lead to CID resistance. Homologous recombination may even lead to transmission of the mutation to the second allele. While such events are also considered very rare, Liang et al.6 identified a large deletion within the suicide gene. Even we observed a point mutation in Phoenix subclone #3 that led to an exchange of one amino acid; however, the relevance of this mutation remained unclear. In this subclone, further mechanisms such as closed chromatin due to histone modifications have to be considered.
Our results indicate that even targeted integration of a monoallelic safety switch into a safe-harbor site known for robust transgene expression is insufficient to exclude development of CID-resistant subclones. Although a biallelic approach obviously offers much higher safety levels, the question remains how to calculate the safety level, not only for a specific cell product but also for a given clinical scenario. Addressing that question, Liang et al. developed the term safe-cell level (SCL) as the number of therapeutic batches in which there is expected to be one none-safe batch.6 This SCL, which is actually reflecting the frequency of escapees, was then put in relation to clinically relevant cell numbers in terms of the therapeutically applied cell doses. These doses will often be substantially higher than typical cell numbers handled in conventional cell culture systems and may range from 105 to 1010 cells, e.g., it is estimated that ∼109 cardiomyocytes would have to be replaced after myocardial infarction.
It is debatable, however, whether relation of a calculated SCL to the size of a required therapeutic cell dose is the most appropriate approach to estimate the safety level of a given cell product. Integrated suicide genes are mainly considered as a safety measure to eliminate proliferating tumor cells that either arise as teratomas, from contaminating undifferentiated PSCs, or develop from rare cell clones that carry mutations or epigenetic aberrations, which lead to altered expression or function of oncogenes. It is, however, unlikely that a very rare cell clone within the therapeutic cell batch that acquired CID resistance during the production phase becomes enriched during culture expansion because such an event should not lead to any selection advantage. And it is even more unlikely that such a clone belongs to a tiny population of undifferentiated cells with teratoma-forming potential or undergoes another very rare event, which is tumor transformation.
More relevant are other scenarios, where after therapeutic application, single contaminating undifferentiated PSCs among the cell transplant form a teratoma or an individual cell clone acquires genetic aberration(s) and undergoes tumor transformation, in both cases followed by massive proliferation. Among the generated large number of tumor cells, clonal CID resistance may develop in vivo as a second, independent, and again, very rare, event.
Therefore, instead of calculating a SCL in consideration of the size of a produced cell batch or the therapeutic cell dose, it seems more reasonable to estimate the safety of a cell product by putting the frequency of suicide-resistant escapee clones in relation to the number of cells in a tumor mass, which are detectable during routine tumor screening. In the case of bone marrow or blood cell transplantation, the minimal number of circulating leukemic cells that are detectable by routine screening procedures in clinical hematology should be considered. If the frequency of suicide-resistant escapee clones is low enough, it should be possible to detect cancer formation by routine screening procedures prior to development of rare CID-resistant subclones.
Recent estimations for cell numbers in tumors tissue range from 107–108 cells for a tumor of 1 cm3, a size that should be reliably detectable by modern imaging approaches.15 Large tumors may even contain more than 109 to 1010 cells. Provided a frequency of ∼5 × 10-8 for clonal suicide escapees in cells with a monoallelic safety switch, and 108 cells in a tumor, it is obvious that a tumor, when it becomes large enough to become detected clinically, may already contain CID-resistant tumor cell clones.
Future studies are required to further explore to what extent the frequency of clonal escapees depends on the utilized suicide system, applied promoter, integration site, and cell type. Monoallelic integration of synthetic safety switches, however, clearly appears insufficient to provide an increased safety level for cell therapy products. In contrast to monoallelic iCASP9 hiPSCs, we could not detect any drug-resistant escapees from biallelic iCASP9 cells among 0.8 billion iPSCs. This cell number is already ∼10× higher than the estimated cell number in a tumor of 1 cm3 in size, which should be reliably detectable by regular tumor screening, e.g., via magnetic resonance imaging (MRI), before further increased tumor cell numbers may lead to escapees. Large mass-production settings would be required to further define the actual risk for appearance of clonal escapees from cells with integrated biallelic safety switches. Experimental proof, however, of the theoretical independency of suicide transgene inactivation on both alleles, which would imply extremely low frequencies of clonal escapees in a range of 10-15, is almost impossible. Since even with most advanced mass culture technologies, production of not more than 107 cells/mL is possible,16 a culture volume of approximately 100,000 L would be required to generate the huge number of 1015 cells.
Materials and methods
Plasmid construction
AAVS1 TALENs were generated via the Golden Gate assembly method17 and contain wild-type FokI nuclease domains. AAVS1 locus-specific TALEN expression cassettes18 were placed under control of a CAG promoter.
The AAVS1.iCaspase 9 donor was based on the vector SFG.iCasp9.2A.ΔCD19, described in Stasi et al.5 For generation of the AAVS1 donor plasmid, standard cloning technologies were used. The AAVS1-targeting vector contains CAG-iCasp9-2A-dTomatonucmem-RbGpA flanked by two arms of ∼700 bp AAVS1 locus homology sequences in a pUC19 expression vector backbone (Thermo Fisher Scientific).
As described in Stasi et al.5 the transgene iCASP 9 consists of the sequence of the human FK506-binding protein 12 (FKBP12; GenBank: AH002818) with an F36V mutation, connected through a Ser-Gly-Gly-Gly-Ser linker to the gene encoding human CASP9 (GenBank number: NM_001229).
Cell culture
We used two hiPSC lines generated in house. hCBiPS2 (officially registered in hPSCreg as MHHi009-A) is based on the lentiviral transduction of cord-blood-derived endothelial cells,13 and Phoenix (MHHi001-A) is based on cord-blood-derived CD34+ cells transduced with Sendai virus vectors.12 The hiPSC lines were cultured and expanded on irradiated mouse embryonic fibroblasts (MEFs) in iPSC medium (knockout Dulbecco’s modified Eagle’s medium [DMEM] supplemented with 20% knockout serum replacement [KSR], 1 mM L-glutamine, 0.1 mM β-mercaptoethanol, 1% nonessential amino-acid stock [all from Thermo Fisher Scientific], and 10 ng/mL basic fibroblast growth factor [bFGF], supplied by the Institute for Technical Chemistry, Leibniz University, Hannover, Germany19).
Transfection and clone establishment
For Nucleofection, hiPSCs were expanded as monolayer cultures on Geltrex (Thermo Fisher Scientific), cultivated in mTESR1 (STEMCELL Technologies), and harvested by Accutase (Thermo Fisher Scientific). The transfection was performed with the Neon Transfection system (Thermo Fisher Scientific). 1×106 cells were resuspended in 105 μL Neon buffer, electroporated with 5 μg of each plasmid encoding for AAVS1- specific-TALEN and 15 μg donor plasmid with two pulses at 1,000 V for 20 ms, and plated onto Geltrex-coated dishes with MEF conditioned medium (CM) (DMEM/F12 supplemented with 15% KSR, 100 μM β-mercaptoethanol, 1% nonessential amino acid stock [all from Thermo Fisher Scientific], 10 ng/mL bFGF and 10 mM Y-27632 [both from the Institute for Technical Chemistry, Leibniz University Hannover]).
Transfected cells were cultivated as a monolayer prior to FACS. For clone generation, cells were harvested from the monolayer culture by Accutase on day 10 after transfection and were sorted on the FACSAria IIu (BD Bioscience) or XDP (Beckman-Coulter) for dTomatonucmem-positive cells. Sorted populations were plated at low density onto Geltrex-coated dishes. Colonies were picked manually and transferred into feeder-based culture conditions resulting in single-cell clones, which were also analyzed via PCR screening (Figure S1, schematic illustration).
PCR screening and characterization of transgenic clones
Genomic DNA was prepared using the QIAamp DNA Blood Mini Kit (QIAGEN) according to the manufacturer’s instructions, and 100 ng of gDNA was amplified by PCR with GoTaq DNA polymerase (Promega). Sequences and specifications of the primers are shown in Table S1.
Karyotype analysis
After treatment of adherent hiPSCs for 30 min with KaryoMAX Colcemid (Invitrogen), cells were trypsinized, and metaphases were prepared according to standard procedures. Fluorescence R banding using chromomycin A3 and methyl green was performed as previously described in detail 20 . At least 15–20 metaphases were analyzed per clone. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN).
Immunohistochemistry for characterization of transgenic clones
Cells were fixed with 4% paraformaldehyde (w/v), permeabilized with Triton X-100 blocking solution and stained by standard protocols using primary antibodies, listed in Table S2, and appropriate secondary antibodies. Incubation with primary antibodies and corresponding isotype controls was performed overnight at 4°C. Staining of living cells for extracellular markers was performed for 1 h. Secondary antibody staining was performed afterward. Cells were counterstained with DAPI (Sigma) for the analysis with an Axio Observer A1 fluorescence microscope and Axiovision software 4.71 (Zeiss) or propidium iodide (PI) (final concentration 1 μg/mL) (Thermo Fisher Scientific) for the analysis using a MACSQuant Analyzer 10 (Miltenyi Biotech). Flow cytometric data evaluation was performed with FlowJo 7.6.5 software (Celeza).
In vitro differentiation
hiPSCs cultured on MEFs in iPSC medium were detached with collagenase IV (Thermo Fisher Scientific), dispersed in small clumps, and cultured in differentiation medium (80% IMDM supplemented with 20% fetal calf serum, 1 mM L-glutamine, 0.1 mM β-mercapthoethanol, and 1% nonessential amino acid) in ultra-low-attachment plates (Corning) for 7 days. On day 7, the formed embryoid bodies (EBs) were plated onto 0.1% gelatin-coated tissue culture dishes and cultured for a further 13 days before fixation and immunostaining.
Induction of apoptosis with the CID AP20187 and AP1903
iPSCs were seeded on Geltrex-coated tissue culture plates, cultivated in mTESR1, and grown 48 h before functionality of the suicide gene was assessed by adding the CID (AP20187, B/B Homodimerizer, Clontech Laboratories) or AP1903. AP1903 was synthesized in house at the Institute for Organic Chemistry, Leibniz University, Hannover, Germany. CIDs were applied for 24 h with concentrations of 0.001, 0.1, 1, and 10 nM. For flow cytometric analysis, iCASP9 hiPSCs not treated with CID or the nontransgenic hiPSCs were dissociated into single cells using Accutase. Samples based on iCASP9 hiPSCs treated with CID in different concentrations were already in suspension.
For detection of living cells, undifferentiated hiPSCs were stained with 1 μM Calcein AM (Thermo Fisher Scientific) for 45 min, and additionally, all samples were analyzed in parallel using PI. Data were acquired on MACSQuant Analyzer 10 and analyzed using FlowJo software.
Teratoma assay
For in vivo testing of iCASP9 suicide switch functionality, the iCASP9 hiPSCs were cultivated as colonies on MEFs in iPSC medium. After detachment with collagenase IV, the cell suspension was mixed in ratio 1:1 with Matrigel (Thermo Fisher Scientific) to increase cell survival. Teratomas were induced by injection of the cell/Matrigel suspension (∼1 million cells per injection) under the kidney capsule of NOD/SCID mice. After 8–10 weeks, the animals showed increased abdominal girth and body weight.
Then, the mice were administrated 50 μg/day/animal (2.5 mg/kg) AP1903 (4% AP1903 stock solution [3.125 mg/mL], 10% PEG 400 [100%], and 86% Tween [2% in water]) or the same amount of PEG400/Tween solution only as vehicle. The injection was given intraperitoneally (i.p.) for 3 days. Nine days after the last injection, mice were sacrificed. During all experiments, the “Principles of laboratory animal care” (NIH publication no. 86-23, revised 1985) were followed. The animal studies were approved by the responsible authority according to the Animal Welfare Law of Lower Saxony (33.12-42502-04-15/1979).
Histological analysis and confocal microscopy
For histological analyses, the samples were fixed in formalin and processed for paraffin embedding according to standard protocols. Histological staining was conducted on paraffin cuts using hematoxylin and eosin (H&E).
For immunofluorescence analysis of human cells, tissue slides of paraffin were used. Antigen retrieval was achieved by preheated sodium citrate buffer according to standard protocols. After antigen retrieval, tissue slides were stained using the protocol described before for immunohistochemistry.
Confocal images were acquired with a Zeiss LSM 780 and ZEN black 2.3. To separate the immunostaining signal from autofluorescence background, online-fingerprinting mode was used. For that reference, spectra were taken from single fluorescence staining. Several autofluorescence reference spectra were taken from specific regions of nonstained samples and used for unmixing. Images were obtained with a 20x/0.8 Plan-Apochromat (405 and 488 nm excitation at zoom 0.6 [image dimensions 1,024 × 1,024 pixels, 700 × 700 μm]).
To correctly compare immunostainings, all samples were prepared in parallel and imaged at identical acquisition settings. Images were displayed at the same brightness settings to emphasize the specificity of the conditions. A faint Gaussian blur was applied to the images.
In vitro testing for CID-treatment-surviving subclones
iPSCs were seeded on Geltrex-coated tissue culture plates or flasks, cultivated in mTESR1, and grown until a confluent monolayer was reached. Medium was changed to medium containing the chemical inducer AP20187 or AP1903 in concentrations from 0.5 up to 10 nM. After 24 h, the complete medium containing all treated cells were centrifuged and transferred into feeder-based culture conditions. The treated hiPSCs were cultured in iPSC medium (described in Cell culture). In the first 24 h, the medium was supplemented with 10 mM Y-27632 to allow the survival of single cells. After 3 weeks of cultivation, surviving colonies were picked and cultivated further under feeder-based culture conditions. After prolonged cultivation, surviving subclones were transferred to feeder-free-conditions described before and were treated again with 10 nM AP1903 (Figure 5, illustration). The achieved population was analyzed for rate of methylation via Nanopore sequencing and as described before (PCR screening and characterization of transgenic clones) for correct integration of the iCASP9 donor construct (Figure 5, illustration). Sequences and specifications of the primers are shown in Table S1.
Nanopore sequencing
Genomic DNA samples were collected before and after the treatment with AP1903 and stored as a pellet at -80°C. Isolation of genomic DNA was done using the NucleoBond HMW DNA Kit (Macherey Nagel) according to the manufacturer’s instructions. For Cas-mediated PCR-free enrichment of the construct integrated between base positions 55,115,763 and 55,115,769 on chromosome 19 of hCBIPS2 and Phoenix iPSCs, guide RNAs were designed by means of the “Alt-R Custom Cas9 crRNA Design Tool” (IDT). For this purpose, the sequences of the regions, in which the potential guide RNAs should be located in order to obtain fragments of approximately 5,000 bp in length, were pasted into the dialogue box. The program outputs alternative guide RNAs in the inserted region and indicates the respective on-target potential (values from 0 to 100, the higher the better) and off-target risk (values from 0 to 100, the higher the value, the lower the off-target risk). Only guides with an on-target potential ≥60 and an off-target risk ≥75 were accepted for our sequencing experiments and were ordered from IDT (Integrated DNA Technologies, Coralville, IA, USA). During establishment of the best guide RNA strategy, seven guide RNAs were tested. However, according to the criteria on how guide RNAs should be positioned in relation to the target (for more detail, see also https://biorxiv.org/cgi/content/short/2021.09.17.460763v1), a combination of guide RNA nos. 2,134, 2,136, 55, and 2,139 (Table S3) proved to be most effective as they yielded the highest number of calls per CpG side. It is of utmost importance to obtain a sufficient number of calls per CpG site (https://biorxiv.org/cgi/content/short/2021.09.17.460763v1) in order to be able to make a valid statement about the methylation rates of the CpGs. The respective sequences of the guides and their position in the construct are shown in Figure S4. To enable sequencing of this CG-rich and therefore difficult-to-sequence area, the DNA had to be cut into shorter pieces of about 5,000 base pairs each in order to avoid the formation of secondary structures. Before sequencing, the DNA quality was assessed in order to ensure that only high-molecular-weight DNA was used in the subsequent library preparation for the Nanopore run. For this purpose, a pulsed-field gel analysis was carried out using a Pippin Pulse electrophoresis power supply (Sage Science, Beverly, MA USA). Five μg of DNA was used per sample for the library preparation by the Cas-mediated PCR-free enrichment protocol. The latter includes the usage of the Ligation Sequencing (SQK-LSK109) Kit (Oxford Nanopore Technologies, Oxford, UK) and the Native Barcoding Expansion 1–12 (PCR-free) (EXP-NBD104) Kit (Oxford Nanopore Technologies). The entire process consists of the following steps: dephosphorylation of genomic DNA, preparation of the Cas9 ribonucleoprotein complexes (RNPs), cleavage and dA tailing of target DNA, native barcode ligation with appropriate purification by beads and final pooling of samples, adapter ligation (due to barcoding, AMII has been used instead of AMX), and purification by AMPure XP beads (0.3 × volume). The pellet was resuspended in 14 μL of preheated elution buffer at 37°C for 20 min, and 12 μL of the elution was added to the sequencing buffer and loading beads for the final library-preparation step. Afterward, the SpotON flow cell FLO-MIN106D (Oxford Nanopore Technologies) was primed and loaded, and the sequencing run was started on a MinION device (Oxford Nanopore Technologies).
For basecalling and demultiplexing data, guppy basecaller v.5.0.11 (Oxford Nanopore Technologies) was used with standard settings and config file “dna_r9.4.1_450bps_hac”. The basecalled reads were aligned to the reference genome using minimap2 v.2.20 (https://doi.org/10.1093/bioinformatics/bty191). For methylation calling, we used nanopolish v.0.13.2 (https://doi.org/10.1038/nmeth.4184).
PCR-based analysis of LoH
Genomic DNA was prepared using the QIAamp DNA Blood Mini Kit (QIAGEN) according to the manufacturer’s instructions, and 100 ng of gDNA was amplified by PCR with GoTaq DNA polymerase (Promega). Sequences and specifications of the primers are shown in Table S1.
Statistical analyses
Data are presented as mean ± SD (n = 21–27) and were tested for normal distribution using the D'Agostino-Pearson omnibus normality test. As data points were not normally distributed, group comparison was performed by means of the nonparametric Kruskal-Wallis test (p < 0,01) for independent samples.
Acknowledgments
The authors thank Jennifer Beier, Janina Zöllner, Nicole Cleve, Annika Franke, Alexandra Lipus, and Viktor Lutscher for technical assistance and helpful discussion and Wladimir Solodenko for chemical synthesis of AP1903. We are also thankful to Ian Shum for proofreading. Illustrations in Figures 1, 3, and 5 were created with BioRender.com. The plasmid SFG.iCasp9.2A.ΔCD19 was kindly provided by G. Dotti (Baylor College of Medicine). This work received funding from the German Research Foundation (DFG; grants: Cluster of Excellence REBIRTH EXC 62/2, 62/3, and 62/4), “Förderung aus Mitteln des Niedersächsischen Vorab” (grant: ZN3340) supported by the REBIRTH - Research Center for Translational Regenerative Medicine (until 2019: REBIRTH - Cluster of Excellence), and from the German Center for Lung Research (DZL, BREATH 82DZL002A1).
Author contributions
S.W. designed, performed, and analyzed experiments. S.M. and A.H. was involved in the cloning procedure of the donor and design of guide RNAs. K.J., M.D., and H.F. designed, performed, and analyzed Nanopore sequencing. S.G. analyzed the H&E stainings and provided expertise and feedback. W.K. and A.M. performed and analyzed the in vivo experiments (teratoma assay). A.K. provided Y-27632 and AP1903. A.Z. and E.P. were involved in analyzing of remaining human cells and editing the resulting pictures (confocal microscopy). G.G. performed and analyzed the karyotype analysis. M.A. provided expertise and feedback. N.L. helped with data interpretation. T.M. contributed to experimental design. R.Z. contributed expertise and helpful discussion. U.M., S.W., and A.H. wrote the manuscript.
Declaration of interests
The authors have no commercial, proprietary, or financial interest in the products or companies described in this article.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.05.011.
Supplemental information
References
- 1.Kosanke M., Osetek K., Haase A., Wiehlmann L., Davenport C., Schwarzer A., Adams F., Kleppa M.J., Schambach A., Merkert S., et al. Reprogramming enriches for somatic cell clones with small scale mutations in cancer-associated genes. Mol. Ther. 2021;29:2535–2553. doi: 10.1016/j.ymthe.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews P.W., Ben-David U., Benvenisty N., Coffey P., Eggan K., Knowles B.B., Nagy A., Pera M., Reubinoff B., Rugg-Gunn P.J., Stacey G.N. Assessing the safety of human pluripotent stem cells and their derivatives for clinical applications. Stem Cell Rep. 2017;9:1–4. doi: 10.1016/j.stemcr.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oldfield E.H., Ram Z., Culver K.W., Blaese R.M., DeVroom H.L., Anderson W.F. Gene therapy for the treatment of brain tumors using intra-tumoral transduction with the thymidine kinase gene and intravenous ganciclovir. Hum. Gene Ther. 1993;4:39–69. doi: 10.1089/hum.1993.4.1-39. [DOI] [PubMed] [Google Scholar]
- 4.Fan L., Freeman K.W., Khan T., Pham E., Spencer D.M. Improved artificial death switches based on caspases and FADD. Hum. Gene Ther. 1999;10:2273–2285. doi: 10.1089/10430349950016924. [DOI] [PubMed] [Google Scholar]
- 5.Di Stasi A., Tey S.K., Dotti G., Fujita Y., Kennedy-Nasser A., Martinez C., Straathof K., Liu E., Durett A.G., Grilley B., et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Q., Monetti C., Shutova M.V., Neely E.J., Hacibekiroglu S., Yang H., Kim C., Zhang P., Li C., Nagy K., et al. Linking a cell-division gene and a suicide gene to define and improve cell therapy safety. Nature. 2018;563:701–704. doi: 10.1038/s41586-018-0733-7. [DOI] [PubMed] [Google Scholar]
- 7.Budihardjo I., Oliver H., Lutter M., Luo X., Wang X. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 8.Gargett T., Brown M.P. The inducible caspase-9 suicide gene system as a "safety switch" to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 2014;5:235. doi: 10.3389/fphar.2014.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J.R., Maguire S., Davis L.A., Alexander M., Yang F., Chandran S., ffrench-Constant C., Pedersen R.A. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 10.Merkert S., Wunderlich S., Bednarski C., Beier J., Haase A., Dreyer A.K., Schwanke K., Meyer J., Göhring G., Cathomen T., Martin U. Efficient designer nuclease-based homologous recombination enables direct PCR screening for footprintless targeted human pluripotent stem cells. Stem Cell Rep. 2014;2:107–118. doi: 10.1016/j.stemcr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Z.D., Tchao J., Wu L., Carman A.J. Precision installation of a highly efficient suicide gene safety switch in human induced pluripotent stem cells. Stem Cells Transl. Med. 2020;9:1378–1388. doi: 10.1002/sctm.20-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase A., Göhring G., Martin U. Generation of non-transgenic iPS cells from human cord blood CD34(+) cells under animal component-free conditions. Stem Cell Res. 2017;21:71–73. doi: 10.1016/j.scr.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Haase A., Olmer R., Schwanke K., Wunderlich S., Merkert S., Hess C., Zweigerdt R., Gruh I., Meyer J., Wagner S., et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Lipus A., Janosz E., Ackermann M., Hetzel M., Dahlke J., Buchegger T., Wunderlich S., Martin U., Cathomen T., Schambach A., et al. Targeted integration of inducible caspase-9 in human iPSCs allows efficient in vitro clearance of iPSCs and iPSC-macrophages. Int. J. Mol. Sci. 2020;21:2481. doi: 10.3390/ijms21072481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Monte U. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell Cycle. 2009;8:505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 16.Manstein F., Ullmann K., Kropp C., Halloin C., Triebert W., Franke A., Farr C.M., Sahabian A., Haase A., Breitkreuz Y., et al. High density bioprocessing of human pluripotent stem cells by metabolic control and in silico modeling. Stem Cells Transl. Med. 2021;10:1063–1080. doi: 10.1002/sctm.20-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morbitzer R., Elsaesser J., Hausner J., Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holkers M., Maggio I., Liu J., Janssen J.M., Miselli F., Mussolino C., Recchia A., Cathomen T., Gonçalves M.A.F.V. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R., John J., Lavrentieva A., Müller S., Tomala M., Zhao Y., Zweigerdt R., Beutel S., Hitzmann B., Kasper C., et al. Cytokine production using membrane adsorbers: human basic fibroblast growth factor produced by Escherichia coli. Eng. Life Sci. 2012;12:29–38. doi: 10.1002/elsc.201100045. [DOI] [Google Scholar]
- 20.Schlegelberger B., Metzke S., Harder S., Zühlke-Jenisch R., Zhang Y., Siebert R. Classical and molecular cytogenetics of tumor cells. Diagnostic cytogentics. 1999:151–185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.