Abstract
Treatment-induced tumor dormancy is a state in cancer progression where residual disease is present but remains asymptomatic. Dormant cancer cells are treatment-resistant and responsible for cancer recurrence and metastasis. Prostate cancer treated with androgen-deprivation therapy (ADT) often enters a dormant state. ADT-induced prostate cancer dormancy remains poorly understood due to the challenge in acquiring clinical dormant prostate cancer cells and the lack of representative models. In this study, we aimed to develop clinically relevant models for studying ADT-induced prostate cancer dormancy. Dormant prostate cancer models were established by castrating mice bearing patient-derived xenografts (PDX) of hormonal naïve or sensitive prostate cancer. Dormancy status and tumor relapse were monitored and evaluated. Paired pre- and postcastration (dormant) PDX tissues were subjected to morphologic and transcriptome profiling analyses. As a result, we established eleven ADT-induced dormant prostate cancer models that closely mimicked the clinical courses of ADT-treated prostate cancer. We identified two ADT-induced dormancy subtypes that differed in morphology, gene expression, and relapse rates. We discovered transcriptomic differences in precastration PDXs that predisposed the dormancy response to ADT. We further developed a dormancy subtype-based, predisposed gene signature that was significantly associated with ADT response in hormonal naïve prostate cancer and clinical outcome in castration-resistant prostate cancer treated with ADT or androgen-receptor pathway inhibitors.
Implications:
We have established highly clinically relevant PDXs of ADT-induced dormant prostate cancer and identified two dormancy subtypes, leading to the development of a novel predicative gene signature that allows robust risk stratification of patients with prostate cancer to ADT or androgen-receptor pathway inhibitors.
Introduction
Prostate cancer survival and growth are largely dependent on androgens (1). Androgen-deprivation therapy (ADT) has been a cornerstone of treatment for locally advanced and metastatic prostate cancer (2). While ADT initially leads to rapid apoptotic or atrophic responses of prostate cancer (3), more than 50% of ADT-treated cases experience recurrence within 2 years (4, 5), indicating the presence of residual cancer cells that are either insensitive to treatment or in a temporarily dormant stage.
In the clinical context, tumor dormancy is the extensive period of time in which tumors remain in a growth arrest in the primary site or in metastatic dissemination, and patients remain asymptomatic before relapse (6–10). During tumor dormancy, the residual tumor cells are often undetectable by routine diagnostic methods such as medical imaging and biomarkers. The dormant tumor cells are thought to contribute to therapeutic resistance, evade immune surveillance, and lead to future recurrence. Tumor dormancy can be further subdivided into various categories/subtypes at different levels, including the clinical level (e.g., metastatic dormancy, treatment-induced dormancy), cell population level (e.g., cellular dormancy, tumor mass dormancy), and mechanistic level (e.g., angiogenic dormancy, immunologic dormancy; refs. 6–10).
ADT-induced prostate cancer dormancy remains poorly understood. To study ADT-induced prostate cancer dormancy, cancer cells must be characterized during the dormant state. However, it is challenging to acquire sufficient clinical dormant prostate cancer tissues and establish representative experimental models. As a result, the effects of ADT-induced tumor dormancy on disease progression remain unknown, as are clinically useful predictive markers for patients treated with ADT. Therefore, mechanistic understanding of ADT-induced prostate cancer dormancy is critically important, and reliable biomarkers that can predict ADT response and stratify patients for recurrence risks are urgently needed.
We have previously established a large collection of prostate cancer patient-derived xenograft (PDX) models (www.livingtumorlab.com), mostly from primary hormone-naïve prostate cancer. These models retain major characteristics of the patients' original tumors, including gene expression and treatment response (11–13). Therefore, these models are clinically relevant platforms for prostate cancer discovery (14–18), personalized cancer therapy (19–21), predictive marker development (19, 22), and drug development (23–26).
In this study, we aimed to model ADT-induced dormant prostate cancer, to explore ADT-induced dormant prostate cancer at biological, cellular, and gene expression levels, to study the clinical association of the findings in PDXs, and to develop a predictive gene signature for risk-stratification in patients treated with ADT. Herein, using a panel of hormonal naïve or sensitive prostate cancer PDXs, we established 11 ADT-induced dormant prostate cancer PDX models, including nine from hormone-naïve prostate cancer. Our PDX models revealed two ADT-induced prostate cancer dormancy subtypes differing greatly in morphology, gene expression, and biological recurrence. We discovered transcriptomic predispositions in the precastration PDXs of the two dormancy subtypes. We further developed a dormancy subtype-based, predisposed gene signature that was strongly associated with ADT treatment response in hormone-naïve prostate cancer. This gene signature was also an independent risk factor for patients with castration-resistant prostate cancer (CRPC) treated with androgen-receptor pathway inhibitors. These findings provide unique insights of prostate cancer dormancy and will help improve the management of the disease.
Materials and Methods
Materials and animals
Chemicals, solvents and solutions were obtained from Sigma-Aldrich, unless otherwise indicated. Six- to 8-week-old non-obese diabetic Rag1null IL2rgnull (NRG) mice were purchased from the Animal Resource Centre of the BC Cancer Research Institute, Vancouver, Canada. Animal experiments were conducted according to a protocol (A17–0165) approved by the University of British Columbia (UBC) Animal Care Committee (Vancouver, Canada). Human prostate cancer specimens used for PDXs were obtained from patients with informed written consent, according to a protocol (H09–01628) approved by the Institutional Review Board of the UBC.
PDXs and castration-induced prostate cancer dormancy
Tissues from 11 PDX models were grafted at the subrenal capsule grafting sites (SRC) of NRG mice (n = 55) as previously described (13, 27). In brief, cryopreserved PDX seeds were recovered with 6- to 8-week-old NRG mice by SRC implantation (13, 20). Expanded PDX tissues were then regrafted into NRG mice supplemented with testosterone to ensure a human-equivalent serum testosterone level was reached. For each tumor line, when tumor volume reached an approximate volume of 150 mm3 by palpation and microultrasound imaging, the mice were randomly assigned into three subgroups: Precastration (PRE), Dormancy (CX), and Long-term Monitoring (LM) groups. PRE PDX tissues were harvested without host castration. Mice in the CX and LM groups were surgically castrated by removing testes and testosterone pellets. PDX tissues of the CX group were harvested at week 12 postcastration. This time point was determined by a pilot study where both tumor volume and serum levels of PSA remained stable at a lower or undetectable level. The remaining 33 mice (3/tumor line) were monitored for postcastration relapse for up to 42 weeks. Relapsed tumors were collected when tumor volumes reached 300 mm3 or when hosts reached humane endpoint due to aging or sickness. All mice bearing PDXs without a sign of relapse were terminated at week 42 postcastration.
Histology, IHC, and quantification
Harvested xenografts were bisected through the longest dimension, fixed in 10% neutral-buffered formalin, and embedded in paraffin. Preparation of formalin-fixed, paraffin-embedded tissue sections, routine hematoxylin and eosin (H&E) staining, and IHC were conducted as previously described (20). Residual tumor volumes in dormant prostate cancer (12 weeks postcastration time point) were determined by microscopic imaging analyses as previously described (20).
For IHC, a rabbit monoclonal anti—androgen receptor (AR) antibody (1:100, Abcam), a rabbit monoclonal anti-PSA antibody (1:200, Abcam), a monoclonal mouse anti-human Ki-67 antibody (1:25, Dako), and a rabbit monoclonal anti—caspase-3 antibody (1:50, Cell Signaling Technology) were used as primary antibodies. For quantification of Ki-67 or caspase-3 IHC stains, positive cells per 5,000 cancer cells were counted by two independent investigators.
RNA microarray gene profiling
RNA from frozen PDX tissues were extracted and purified according to standard protocols. Total RNA samples were processed using Agilent's One-Color Microarray-Based Gene Expression Analysis Low Input Quick Amp Labeling v6.0. An input of 100 ng of total RNA was used to generate cyanine-3–labeled cRNA. Samples were hybridized on Agilent SurePrint G3 Human GE 8 × 60K Microarray. Arrays were scanned with the Agilent DNA Microarray Scanner at a 3-mm scan resolution and data were processed with Agilent Feature Extraction 10.10. Processed signal was quantile normalized with Agilent GeneSpring 11.5.1. The RNA microarray data generated in this study are publically available on Gene Expression Omnibus (GEO) at GSE193500.
Bioinformatics and statistical analysis
Student t test was used to detect differences in abundance levels among the groups studied. Hierarchical clustering of gene expression data was performed using distance metrics calculated from pairwise correlation coefficients.
Gene Set Enrichment Analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp) was used as previously described (15) to determine whether a defined set of genes (Hallmark Gene Sets) showing significant and consistent differences between two biological phenotypes (e.g., PRE vs. CX, or tumor mass dormancy vs. cellular dormancy). All GSEA analyses in this study used whole transcriptomic data without expression level cut-off as expression datasets. False discovery rate (FDR) q values were calculated using 1,000 permutations. A gene set was considered significantly enriched if its normalized enrichment score (NES) has an FDR q below 0.25.
Ingenuity pathway analysis (IPA) was performed as previously described (11, 17). Differentially-expressed gene list was generated using fold change (FC; ≥2) and P values (P < 0.05) between samples of two groups as cut-offs.
Clinical cohorts
Data from High-low clinical trial cohort and Takeda clinical trial cohort (28) were acquired from C. Collins. Data from The Cancer Genome Atlas (TCGA) cohort (PanCancer Atlas) and SU2C cohort (PNAS 2019) were accessed through cBioPortal (29, 30). Data of Sowalsky neoadjuvant-hormonal-therapy (NHT) cohort (31) were acquired from A.G. Sowalsky. Data of the Alumkal cohort (32) were acquired from J.J. Alumkal. In all cohorts, only patients that received preoperative or postsurgery/postbiopsy ADT or androgen-receptor inhibitors were included in this study.
Gene signature development and validation
Candidate genes that are differentially expressed in precastration PDXs of different dormancy subtypes were identified by applying stringent filters to ensure the reliability of results. The PDX-based candidate genes were developed into a predictive gene signature by using the TCGA cohort as a training cohort. The predictive gene signature was further validated in the Sowalsky NHT cohort for its predictive power in predicting ADT response. Another two independent cohorts, SU2C and Alumkal, were used to validate the prognostic value of the predictive gene signature in predicting clinical outcome for patients with castration refractory prostate cancer treated with AR inhibitors. Methods used to develop and validate the predictive gene signature were explained below in details.
The gene read counts from TCGA cohort were converted by log-transformation. Values of zeros were replaced with the minimal value of the cohort to avoid negative infinity. Based on availabilities in clinical follow-ups, the primary outcome for each cohort was as follows: for TCGA the primary outcome was 5-year progression-free survival (PFS); for SU2C the primary outcome was 3-year overall survival (OS); and for Alumkal, 3-year PFS. The gene candidates were ranked based on the contribution to the patient stratification in TCGA training cohort using the method of Information Gain and Signal Noise Ratio. Top ranked gene candidates were selected for fusion-cross validation. A balanced-mode logistic regression model was used to build a predictive gene signature. The binary prediction results were presented as “1” and “0”. Patients indicated as “1” were predicted to have a high risk in developing recurrence compared with ones indicated as “0”. The performance of gene signature was evaluated by area under the ROC curve (AUC). Kaplan–Meier and Cox proportional hazard regression model were employed to validate the prediction in survival outcome and to perform multivariate analyses. All statistical calculations were performed using R (version 3. 4. 1; http://www.rproject.org/), GraphPad Prism version 8 (GraphPad Software), and SPSS (IBM SPSS Statistics for Windows, Version 25.0.). All P values were two-sided, and a value of less than 0.05 was considered statistically significant.
Results
Eleven dormant prostate cancer PDX models were established and resembled the clinical course of ADT treatment response
Eleven castration-induced dormant prostate cancer PDX models were established from 11 parental hormone-naïve or -sensitive PDX tumor lines. The biological and genetic background of the 11 parental PDX tumor lines is summarized in Supplementary Table S1. All PDXs exhibited a marked response to host castration by 12 weeks postcastration: average tumor volume decreased by 88.35% and host serum PSA levels were at an undetectable level (Fig. 1A and B). Consistent with our pilot study, the low tumor volume were held at a low but a steady state with undetectable serum PSA—a state mimicking the clinical tumor dormancy period seen in patients with prostate cancer after ADT treatment. The PSA nadir was consistent with the low tumor volumes of the dormant xenografts, indicating the attainment of dormant state in prostate cancer PDXs as compared with the acute castration effects where serum PSA is still detectable and tumor volumes constantly decreasing. The castration-induced prostate cancer dormancy in PDX was further evaluated at morphologic and genetic levels, as described in the following sections.
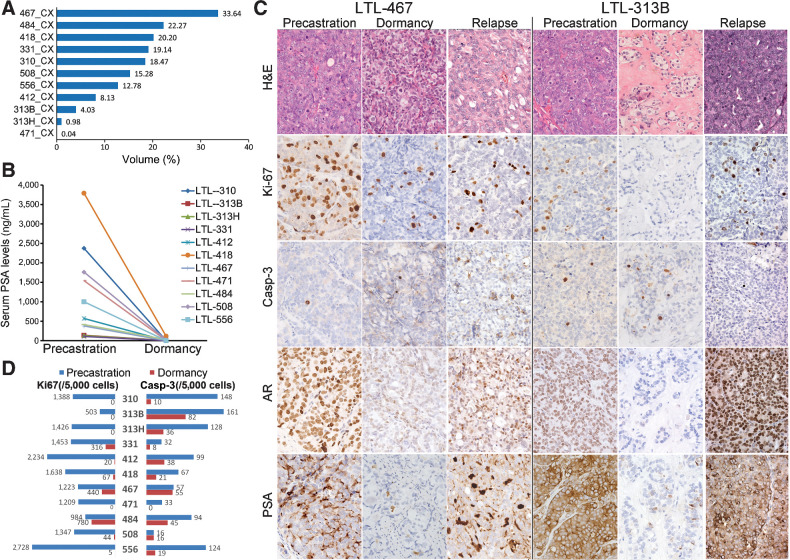
Figure 1.
At week 12 after host castration, dormant prostate cancers were induced in PDXs. A, Percentage volumes of dormant (CX) PDXs at 12 weeks postcastration relative to the PDX volumes before host castration (mean ± SD = 11.65% ± 11.18%). B, Serum PSA of the hosts dropped to undetectable levels on day 84 (week 12) after castration. C, Representative images of H&E and IHC in precastration, dormancy, and relapsed prostate cancer PDXs. D, Ki-67 and caspase-3 (cleaved) percentages in precastration and dormant (at 12 weeks postcastration) PDXs. Casp-3, cleaved caspase-3.
PDXs recapitulated morphologic characteristics of ADT-induced dormant prostate cancer in the clinic
Light microscopy revealed residual carcinoma in all postcastration PDXs. The residual prostate cancer cells demonstrated morphologic changes previously reported in clinical ADT-treated samples (e.g., reductions of glandular structures, hyalinised stroma, cytoplasmic vacuolization, and nuclear pyknosis; Fig. 1C; ref. 33, 34). Mitotic figures were rarely seen. AR nuclear protein expression was decreased, and PSA protein expression was undetectable or weakly expressed in tumor foci (Fig. 1C). Compared with precastration PDXs, cell proliferation and apoptosis in postcastration dormant PDXs, as measured by anti-Ki67 or anti-cleaved caspase-3 immunostaining, were significantly reduced (P = 0.0001 and P = 0.0013, respectively; Fig. 1D), indicating decreased cellular activities in both proliferation and apoptosis in the PDXs 12 weeks after castration, as opposed to the presence of high proliferative and apoptotic activities in the precastration PDXs. The low level of apoptotic activity in the 12-week postcastration PDXs differentiates the dormant tumors from those in acute castration response that involves prominent degree of cell death.
Gene expression analyses revealed broad changes of dormant prostate cancer PDXs in AR signaling and cell proliferation
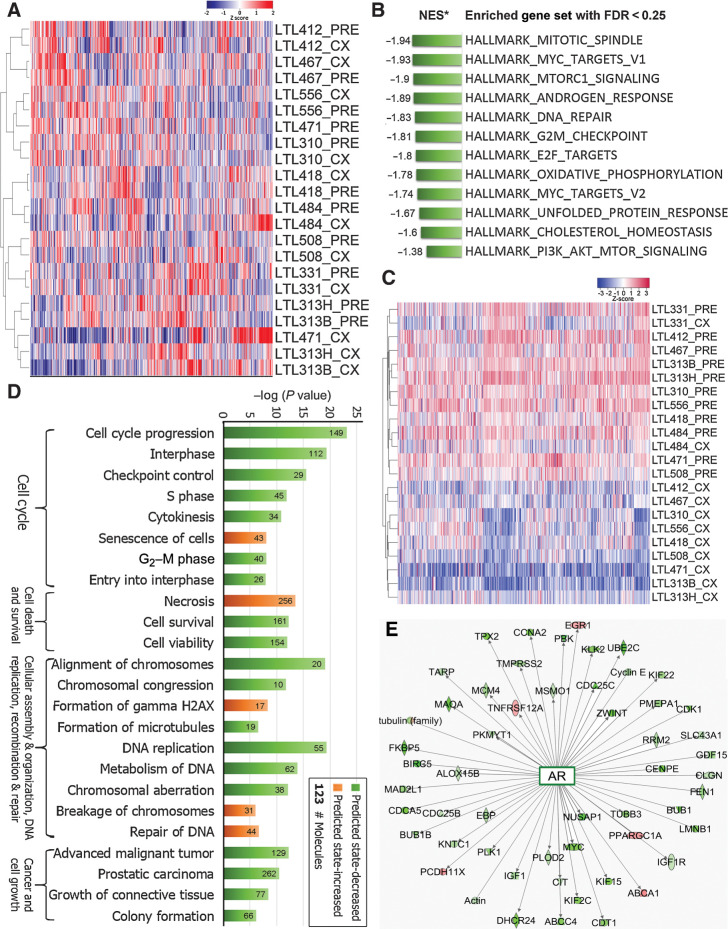
Global transcript profiling of 11 pairs of precastration and postcastration (dormant) PDXs demonstrated that each dormant PDX clustered closely with its paired precastration PDXs (Fig. 2A). GSEA using whole transcriptomic data showed that 12 hallmark gene sets related to androgen response, DNA repair, cell cycle progression, and protein synthesis were significantly enriched in precastration PDXs (FDR < 0.25; Fig. 2B), suggesting these gene sets are downregulated in dormant PDXs. An unsupervised hierarchical clustering analysis based on GSEA Leading Edge Genes demonstrated a split between the precastration and the postcastration dormant PDXs (Fig. 2C). IPA on differential-expression genes (FC of ≥2, P < 0.05) between precastration and dormant PDXs showed that, consistent with GSEA, the most significantly downregulated pathways in dormant PDXs were typically related to cell cycle progression (435 genes), cell survival (315 genes), cellular assembly (205 genes), and cell growth (739 genes; Fig. 2D). Of 58 differentially-expressed genes that were identified by IPA to have direct interactions with AR, 53 were downregulated in dormant PDXs (Fig. 2E). These results were in line with the morphologic findings, indicating prostate cancer dormancy in postcastration PDXs.
Figure 2.
Compared with the precastration PDXs, dormant PDXs showed specific changes in gene expressions. A, genome-wide unsupervised hierarchical cluster showed most dormant (CX) PDXs clustered with their parental precastration (PRE) PDXs. B, GSEA based on whole transcripts without preranking identified 12 significantly enriched gene sets in PRE PDXs (FDR < 0.25). C, Unsupervised hierarchical clustering based on the Leading Edge genes generated by GSEA showed a distinct separation between PRE and most CX PDXs. D, IPA revealed that deregulated pathways were mainly associated with cell cycle, cell death and survival, cellular assembly, DNA repair, and cell growth. E, Fifty-three of 58 differentially-expressed genes that were indicated by IPA to have a direct interaction with AR were downregulated in PDXs of dormant prostate cancer.
Two biologically different ADT-induced prostate cancer dormancy subtypes were associated with disease progression
We observed two dormancy subtypes in postcastration dormant PDXs. As revealed by morphologic analysis, mitotic figures were absent in 8 of 11 postcastration dormant PDX models, with Ki67 less than 5% (Fig. 1C and D), indicating a status of cellular dormancy that involves a state of temporary cell cycle arrest in tumor cells (7, 10). In contrast, the other three dormant PDX models (LTL-467CX, LTL-331CX, and LTL-484CX) showed mitotic figures, with Ki67 percentages at 5% to 15% (Fig. 1C and D), indicating a state of tumor mass dormancy where overall tumor growth is stopped due to likely an equilibrium between cell death and cell proliferation (7, 10). Since quantification using Ki67 alone has limitations in uniformity and consistency (35), we adopted a quantified histopathologic scoring system for ADT-treated clinical prostate cancer samples (36) to evaluate dormant prostate cancer comprehensively (Supplementary Table S2). Three PDX models with score 0 to 2 were considered as tumor mass dormancy, and five with score 7 to 10 as typical cellular dormancy (Table 1). Three PDX models with score 3 to 6 appeared to be less responsive than the typical cellular dormancy PDXs, with the former harboring more residual cancer cells and demonstrating cytologic hints of active biosynthesis (e.g., large conspicuous nucleoli and abundant eosinophilic cytoplasm). However, these three models were still considered as cellular dormancy since mitotic figures were absent and Ki-67 positivity was low (<5%).
Table 1.
Histopathologic quantification results of dormant prostate cancer in PDXs.
| Contributing factors | ||||||
|---|---|---|---|---|---|---|
| PDX ID | Pyknosis | Cytoplasmic vacuolization | Mitoses | Ki-67 positivity | Reduction in cell density and stromal changes | Final score |
| LTL-313B-CX | 2 | 2 | 2 | 2 | 1 | 9 |
| LTL-313H-CX | 2 | 2 | 2 | 2 | 1 | 9 |
| LTL-471-CX | 2 | 1 | 2 | 2 | 2 | 9 |
| LTL-556-CX | 2 | 1 | 2 | 2 | 1 | 8 |
| LTL-310-CX | 2 | 1 | 2 | 2 | 0 | 7 |
| LTL-508-CX | 1 | 1 | 2 | 2 | 0 | 6 |
| LTL-412-CX | 0 | 0 | 2 | 2 | 0 | 4 |
| LTL-418-CX | 0 | 0 | 2 | 1 | 0 | 3 |
| LTL-331-CX | 0 | 0 | 0 | 0 | 0 | 0 |
| LTL-467-CX | 0 | 0 | 0 | 0 | 0 | 0 |
| LTL-484-CX | 0 | 0 | 1 | 0 | 0 | 1 |
GSEA using transcriptomic profiles of three tumor mass dormancy and five typical cellular dormancy PDXs demonstrated that 10 hallmark gene sets were significantly enriched in tumor mass dormancy (FDR < 0.25), including those related to cell cycle, glycolysis, Wnt/β-catenin, DNA repair, and PI3K–AKT–mTOR signaling (Supplementary Table S3). These results coincided with the relatively higher degree of activities of the residual tumor cells in tumor mass dormancy.
We further analyzed GSEA Leading Edge genes and generated a 42-gene set (Fig. 3A; Supplementary Table S4). Based on this gene set, principal component analysis (PCA) exerting 11 pairs of precastration and dormant PDXs showed that the three tumor mass dormancy PDXs were separated from the rest of the dormant PDXs, grouping closely with the proliferative precastration PDXs (Fig. 3B). The result suggested that this gene set, similar to the histopathologic score system, reflected dormant status in postcastration PDXs.
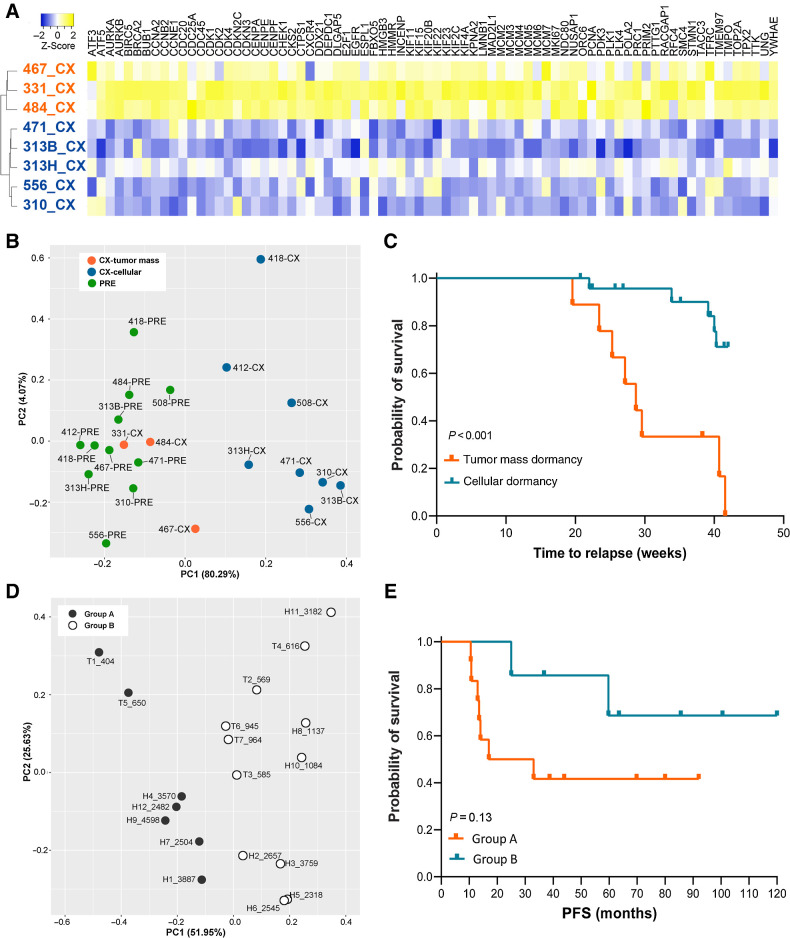
Figure 3.
Two subtypes of castration-induced prostate cancer dormancy were associated with disease progression. A, A 42-gene set generated by GSEA Leading Edge analysis (tumor mass dormancy, orange; versus cellular dormancy, blue). B, PCA using the Leading Edge gene set demonstrated a clear split between eight PDXs in cellular dormancy and three in tumor mass dormancy. The latter were grouped closely to precastration samples. C, Kaplan–Meier estimate of PFS in PDXs showed that PDXs displaying tumor mass dormancy had a significantly shorter time to relapse (P < 0.001). D, PCA based on the same Leading Edge gene set separated 18 patients treated with preoperative ADT (NHT) into two subgroups (groups A and B; n = 7 and n = 11, respectively). E, Kaplan–Meier of patients in the two subgroups (P = 0.13). PC, principal component.
To investigate whether the dormancy status functionally impacted disease progression in PDXs, we castrated male mouse hosts bearing 11 PDX tumor lines (3 mice/line) and monitored for tumor relapse. Seven of the 11 PDX lines developed relapsed tumors within 42 weeks postcastration (Supplementary Table S5), including 6 AR+/PSA+ CRPC and one neuroendocrine prostate cancer. The proliferative status of relapsed tumors was also confirmed by H&E and Ki67 staining (Fig. 1C). PDXs displaying tumor mass dormancy had a significantly shorter time to relapse (P = 0.0001; Fig. 3C) and had much higher relapse frequency than those exhibiting cellular dormancy [8 of 9 mice (88.8%), and 5 of 24 mice (20.83%), respectively; P = 0.0001; Supplementary Table S5].
To determine if the Leading Edge gene set that segregated dormancy subtypes can be applied to clinical prostate cancer samples and distinguished the dormancy subtypes post NHT, we further analyzed transcriptomic profiles from 18 patients who were exposed to preoperative NHT (High-low and Takeda clinical cohorts; Supplementary Table S6; ref. 28). Based on the same Leading Edge gene set used for PDX analysis, PCA showed that the 18 cases were separated into two subgroups (Fig. 3D). One group tended to have a shorter PFS compared with the other group, although the Kaplan–Meier survival analysis did not reach statistical significance due to small sample size (P = 0.13; Fig. 3E).
Transcriptomic differences in precastration PDXs predisposed the dormancy response to ADT
Since dormancy is a resultant of acute cellular injury brought about by androgen ablation, we hypothesized that there was genomic and/or transcriptomic predisposition that enabled the tumors to enter one of the two dormancy subtypes after ADT. Upon analyzing the genomic profiling data of precastration PDXs (11), we did not observe significant differences in genetic configuration (Supplementary Table S1). Transcriptomic analyses showed that a total of 1,619 genes were differentially expressed between precastration PDXs of tumor mass dormancy and typical cellular dormancy [cut-offs: FC ≥ 2 and logbaseline expression intensity ≥ 6]. IPA showed that the precastration PDXs entering tumor mass dormancy after ADT had significantly decreased expression in Wnt/β-catenin, Myc-mediated apoptosis, and p38-MARK signaling and increased expression in integrin and epithelial–mesenchymal transition (EMT) signaling (Supplementary Table S7). These results suggested a presence of transcriptomic predisposition before host castration, which dictated the type of dormancy in response to ADT.
A dormancy subtype-based, predisposed gene signature predicted ADT response and clinical outcomes in independent hormone-naïve or CRPC clinical cohorts
Reliable biomarkers are urgently needed for prediction of ADT response and risk stratification of patients with prostate cancer receiving ADT. Based on the aforementioned findings, we hypothesized that a predictive gene signature derived from the dormancy subtype-related, transcriptomic predisposition in precastration PDXs could be used for predicting ADT response in patients prior to the administration of treatment and for risk stratification. To this end, top 234 differentially-expressed genes between the precastration PDXs of tumor mass dormancy and typical cellular dormancy (cut-offs: P < 0.01, logbaseline gene expression intensity ≥ 6) were used as candidate genes for signature development. Four clinical cohorts (Supplementary Table S8) were employed for development and validation of the gene signature. Using the TCGA cohort (29, 30) as a training cohort (n = 69, only primary hormone-naïve prostate cancers subsequently treated with ADT were included), we developed the PDX candidate genes into a 34-gene signature [hereafter referred to as, Predisposed Gene Signature (PGS); Supplementary Table S9] that separated patients with or without recurrence.
We next validated the PGS using the Sowalsky cohort (ref. 31; n = 37; Supplementary Table S8), in which 37 patients with localized primary hormone-naïve prostate cancer were treated with preoperative ADT (6 months of Lupron plus enzalutamide). Pre-ADT treatment biopsies were profiled for RNA expression, and ADT response (nonresponders, incomplete, and exceptional responders) was defined by posttreatment residual cancer volumes (31, 37). We performed single-sample GSEA using the PGS and generated a PGS enrichment score for each pre-ADT tumor. We observed a strong proportional relationship between the PGS score and residual cancer volumes [RCB, r(35) = 0.74, P < 0.00001; Fig. 4A]. The nonresponders had a significantly higher PGS score compared with the exceptional responders (P < 0.0001) and incomplete responders (P < 0.0001; Fig. 4B). These results strongly suggest that PGS is signficantly enriched in intrinsically resistant tumors in primary prostate cancer.
Figure 4.
PGS predicted ADT response or clinical outcomes in independent hormone-naïve or CRPC clinical cohorts. A, Single-sample gene set enrichment score of PGS is proportionally related to the residual tumor volumes [RCB, r(35) = 0.74, P < 0.00001] in primary hormone-naïve prostate cancer treated with preoperative ADT (Sowalsky clinical cohort, n = 37). B, In the Sowalsky cohort, the PGS gene set enrichment score is significantly higher in nonresponders (NR) compared with exceptional responders (ER; P < 0.0001) and incomplete responders (IR; P < 0.0001). C and D, Kaplan–Meier estimate of OS or PFS segregated according to the PGS score in SU2C (A,n = 60) and Alumkal (B,n = 25) clinical cohorts. In both cohorts, patients with PGS = 1 had worse outcome than those with PGS = 0 (P = 0.0002 and 0.006, respectively). E, Panther Protein Classification of the PGS genes revealed that the genes were involved in various biological processes.
Since the predisposed PGS reflects intrinsic resistances to ADT, we further investigated whether this signature could predict clinical outcomes for patients with more advanced stages of prostate cancer. The PGS was applied to patients with prostate cancer in two independent CRPC cohorts: SU2C (refs. 29, 30, 38; n = 60) and Alumkal (ref. 32; n = 25), separately (Supplementary Table S8). All patients received postbiopsy/-surgery AR inhibitors. The binary prediction results were presented as “1” and “0”. In agreement with the initial discovery TCGA cohort, patients with PGS = 1 had significantly shorter OS or PFS on AR inhibitors compared with those with PGS = 0 (SU2C: P = 0.0002; Alumkal: P = 0.006; Fig. 4C and D). Multivariate Cox regression analysis revealed that the PGS was an independent risk factor in both SU2C and Alumkal cohorts (Table 2, P = 0.003 and 0.004, respectively). Panther Protein Classification and IPA analysis of the PGS revealed that the 34 genes mainly associated with cell growth, proliferation, and transcriptional regulation (Fig. 4E).
Table 2.
Multivariate Cox regression analysis of PGS in SU2C and Alumkal clinical cohorts.
| Cohort | Covariates | Coefficients | SE | P | HR (95% CI) |
|---|---|---|---|---|---|
| SU2C | Age (<65 vs. ≥65 y/o) | 0.791 | 0.41 | 0.054 | 2.205 (0.987–4.923) |
| Gleason score (<7 vs. ≥7) | 0.581 | 0.421 | 0.168 | 1.787 (0.783–4.078) | |
| PSA at diagnosis (<20 vs. ≥20 μg/mL) | 0.001 | 0.001 | 0.095 | 1.001 (1.000–1.002) | |
| PGS (PGS = 1 vs. PGS = 0) | 1.326 | 0.449 | 0.003 | 3.767 (1.561–9.088) | |
| Alumkal | Age (<65 vs. ≥65 y/o) | 0.994 | 0.706 | 0.159 | 2.702 (0.678–10.772) |
| Gleason score (<7 vs. ≥7) | 0.339 | 0.522 | 0.516 | 1.404 (0.505–3.905) | |
| PSA at diagnosis (<20 vs. ≥20 μg/mL) | 0.557 | 0.596 | 0.350 | 1.746 (0.543–5.613) | |
| PGS (PGS = 1 vs. PGS = 0) | 1.741 | 0.61 | 0.004 | 5.702 (1.726–18.842) |
Discussion
In this study, we study ADT-induced dormancy in prostate cancer using highly clinically-relevant PDX models. Treatment-induced tumor dormancy is a common phenomenon after systemic treatment, such as chemotherapy and hormonal therapy, and has been used to explain posttreatment tumor relapse owing to the regrowth of drug-resistant, dormant cancer cells (10). Modeling ADT-induced prostate cancer dormancy has been a major challenge in studying post-ADT dormancy in primary prostate cancer. Experimental cellular dormancy in cell lines is usually achieved by highly artificial culture conditions (39). Prostate cancer PDX models are usually developed from highly aggressive prostate cancer, mostly CRPC tissues, instead of primary hormone-naïve prostate cancer (13). We therefore established 11 PDX models of castration-induced dormant prostate cancer, nine of which were from hormone-naïve prostate cancer. One arresting benefit of using these PDXs to model prostate cancer dormancy is that the dormant prostate cancer PDX models simplify intricate clinical scenarios (e.g., treatment duration and tolerance, and various collecting time points) during the collection of dormant clinical samples.
We adopted the original clinical definition for tumor dormancy: the extensive period of time in which tumors remain in a growth arrest and patients remain asymptomatic before relapse (6–10). In the context of our PDX models, we used serum PSA and tumor volume as clinically relevant indicators for the castration-induced dormancy. The attainment of postcastration tumor dormancy was determined by tumor volumes reaching the lowest point without further change and the serum PSA reaching nadir. In addition, the dormant prostate cancer PDXs demonstrated characteristics similar to those observed in post-ADT clinical prostate cancer samples, including changes in morphology and marker expressions, and biological recurrence (33, 34). Moreover, these dormancy PDX models demonstrated various ki67 index after host castration, which is consistent with the observation in clinical NHT cohorts (40). In view of these, these dormancy models provide unique clinically relevant research tools for ADT-induced prostate cancer dormancy, recapitulating different features of ADT-induced prostate cancer dormancy in the clinic.
In 1970s to 80s, experimental models of dormancy have revealed that tumor dormancy may also result from a balance between cell proliferation and death so that the tumor mass maintains at a constant size (41–43). Tumor dormancy is thus divided into two subtypes at the cell population level: cellular dormancy (nonproliferative tumor cells persisting over a long period of time without dying) and tumor mass dormancy (populations of cancer cells with cell proliferation balanced by cell death; refs. 6–10, 44, 45). Our results demonstrated that these two ADT-induced prostate cancer dormancy subtypes were significantly different in morphology and gene expressions. PDXs typically in cellular dormancy were characterized by no proliferative activities (no mitoses figures, negative for Ki67 stains); and those in tumor mass dormancy might be achieved by a low, but balanced, levels of cell proliferation and apoptosis (indicated by positive Ki67 and caspase-3 stains in limited number of residual cancer cells). The two dormancy subtypes showed signficant difference at the transcriptomic level in hallmark gene sets such as cell cycle, glycolysis, PI3K-AKT-MTOR, and DNA repair. We futher demostrated that the dormancy subtypes functionally impacted recurrence in PDXs, indicating that post-ADT prostate cancer dormancy status is associated with tumor latency, and could be a useful indicator for ADT response and outcome prediction. Currently, large collections of post-ADT prostate cancer samples with domrancy-subtype characterization and follow-up information are not available. Future validation with such cohorts will help better understand the relationship between dormant subtypes and disease progression in ADT-treated prostate cancer.
Notably, the dormancy period investigated in this study is different from the acute castration effects (e.g., drastic cell death, rapid tumor shrinkage, and microenvironment changes). In our pilot study, we found that the acute castration effects are most drastic during the first week after castration. After 4 to 6 weeks postcastration, host PSA generally reached nadir, and tumor volumes remained unchanged. Our study revealed that at 12 weeks postcastration, the residual dormant cancer cells had re-established a stable, nonprogressing state either by cell cycle arrest or by the balance between cell death and proliferation. The cellular activities including both proliferation and apoptosis are decreased in dormant cancer cells, as indicated by low levels of Ki67 and caspase-3 expressions, respectively. The low level of apoptotic activity in the 12-week postcastration PDXs differentiates the dormant tumors from those in acute castration response that involves prominent cell death. This dormant period in PDX models can last months before proliferative, relapsed tumors re-emerge. This period of tumor dormancy could be considered as part of the chronic castration effect because the androgen levels stayed at nadir through this period. However, given the later development of relapsed tumors, it suggests that, at the cellular levels, cancer cells in the dormancy period are undergoing dynamic changes that will enable them to resist the chronic suppressive effect of androgen deprivation. These changes perhaps cannot be attributed solely to the chronic castration effect.
In addition to tumor dormancy, other terms have been used to describe the growth arrest state of cancer cells, such as cellular quiescence. Quiescence is a reversible, nonproliferative state of cells at G0 phase in response to environmental stress and changes (46). Although quiescent cells share many similar characteristics with the cells in tumor dormancy (e.g., growth stagnation, decreased metabolism, survival advantage, and resistance to adjuvant therapies), cancer cell quiescence differs from tumor dormancy in a number of aspects. First, although quiescence is often used to describe the state of G0–G1 cell cycle arrest in tumor cells in cellular dormancy, it does not reflect the cells in tumor mass dormancy where growth equilibrium resulted from a balance between cellular proliferation and death is present. Second, tumor dormancy is primarily derived from clinical observations, describing the tumor as a whole, while quiescence describes the G0-arrest state of cancer cells at cellular level. Third, quiescent cells are in a delay of growth or differentiation and can immediately resume growth or differentiation once the environmental stress is removed. Dormant cells, however, may need to develop new mechanisms in order to exit the dormant state and grow again in a persistently unfavorable environment (47). In this study, although cancer cells in the xenografts in cellular dormancy demonstrate a quiescent, nonproliferate state, the cells in tumor mass dormancy remain certain levels of proliferation activities and therefore are not in a quiescent state. Moreover, in spite of the consistently low androgen level in the castrated hosts, a significant number of PDX models that exhibited cellular dormancy or tumor mass dormancy have conquered the low-androgen environment and developed recurrent tumors within a period of 42 weeks. One of the PDX models, LTL-331, even undergone a neuroendocrine transdifferentiation and re-emerged as a neuroendocrine prostate cancer that recapitulated the donor patient's clinical disease progression. The development of recurrent tumors in a continuously androgen-suppressed environment, especially in the PDX models in cellular dormancy, suggest that cancer cells might have developed adaptive mechanisms enabling them to grow in unfavorable environment. In view of this, tumor dormancy is a more accurate description for this study than quiescence, reflecting dynamic changes of the ever-evolving dormant cancer cells.
Predicting prostate cancer patients' response to ADT has remained a major challenge in clinical management. We herein revealed that predisposed transcriptomic differences in the precastration PDXs might contribute to the two distinctive dormancy subtypes. These transcriptomic predispositions were linked to Wnt/β-catenin, apoptosis, p38-MAPK, integrin, and EMT signaling. These findings suggest that de novo gene expression signatures may lead to tumor mass dormancy after ADT, thus impact survival. We further developed and validated a 34-gene PGS for predicting ADT response in primary prostate cancer. The 34-gene PGS was unbiasedly generated using a mathematic model, based on the top gene candidates differentially expressed between precastration PDXs that gave rise to tumor mass dormancy and those gave rise to cellular dormancy after host castration. The reason for using precastration PDXs was to mimic the potential availability of the patient samples before receiving ADT. The generated PGS will then allow us to predict the ADT response prior to the administration of treatment. We further validated the PGS in multiple independent clinical cohorts. In the Sowalsky cohort (hormone-naïve prostate cancer), the PGS was proportionally correlated with the residual tumor volume, being significantly higher in nonresponders compared with exceptional responders. The results indicate that the PGS is strongly enriched in intrinsically resistant tumors in primary prostate cancer and is a strong indicator for de novo response to ADT treatment in primary hormone-naïve prostate cancer, suggesting a causal relationship between predisposition to ADT as a cause of tumor dormancy and the development of treatment-refractory disease. Moreover, we demonstrated that PGS was predictive of clinical outcome and was an independent risk factor in two independent CRPC cohorts (SU2C and Alumkal) treated with AR pathway inhibitors. These results suggest that PGS is an important determinant of patient outcome and response to subsequent AR pathway inhibitor therapy even after development of castration resistance. Taken together, these findings are of immense clinical significance, as it has the potential to aid in the clinical management of prostate cancer by predicting therapeutic response in patients with cancer. For patients with high PGS, there might be a necessity in combining other adjuvant therapies with ADT or AR pathway inhibitors to eliminate proliferative cells in the tumor mass dormancy. Further prospective study with patients harboring hormone-naïve localized high-risk and advanced prostate cancer will validate the clinical significance of the studies. In addition, understanding factors that mediate this gene signature may lead to new approaches to target the disease.
In conclusion, we have successfully established highly clinically relevant PDX models of ADT-induced dormant prostate cancer. We uncovered two dormancy subtypes that showed significant difference in morphology, transcriptomic predispostion, and disease progression. We developed and validated a novel predisposed gene signature for robust risk-stratification for patients with prostate cancer receiving ADT or androgen-receptor pathway inhibitors. Future in-depth studies focusing on mechanisms underlying tumor dormancy, clinical impact of the predicative gene signature, and therapeutic target development will help improve understanding of dormant prostate cancer. We may also develop new therapeutic approaches to target dormant prostate cancer.
Supplementary Material
Acknowledgments
The authors thank Fang Zhang, James Killam, Peter Gout, members of Y.Z. Wang Laboratory for their great support during various scientific discussions, Robert Bell, Sonia Kung, and Ladan Fazli for their assistance in clinical data management. Portions of this work used the computational resources of the NIH HPC Biowulf cluster. This research was supported in part by the Canadian Institutes of Health Research (grants nos. 141635, 144159, 153081, 173338), Terry Fox Research Institute (grant no.1062), Mitacs Accelerate Program (grant nos. IT10125, IT06414, IT12387, IT14958), the NCI grant pilot project award (P50CA097186), the Canadian Foundation for Translational Immunology to Y. Wang; Pacific Northwest Prostate Cancer (SPORE/NCI P50 CA097186), NCI Drug Resistance and Sensitivity Network (NCI P50 CA186786-07S1, NCI R01 CA251245), Rogel Cancer Center Innovation Award (P30 CA046592) to J.J. Alumkal; The CDMRP Prostate Cancer Research Program (Impact Award to S.P. Balk, M.-E. Taplin, and A.G. Sowalsky; Early Investigator Research Award to S.Wilkinson); the Prostate Cancer Foundation (Young Investigator Award to S.Wilkinson; Challenge Award to S.P. Balk and Y.Wang); and the Intramural Research Program of the NIH, NCI (to A.G. Sowalsky).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is featured in Highlights of This Issue, p. 671
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
M.-E. Taplin reports personal fees from Epizyme, Pfizer, AstraZeneca, Janssen, Research to Practice, Clovis, Wolters Kluwer Health, Celgene, Bayer HealthCare, Myovant, Roviant, Astellas Pharma, Abbvie; and personal fees from Arcus Biosciences during the conduct of the study. S.P. Balk reports grants from Prostate Cancer Foundation, NIH; and grants from U.S. Department of Defense during the conduct of the study. J.J. Alumkal reports personal fees from Astellas Pharma; and other support from Astellas Pharma during the conduct of the study; personal fees from Dendreon, Bristol-Myers Squibb, Merck; and other support from Beactica outside the submitted work. A.G. Sowalsky reports grants from U.S. Department of Defense during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
X. Dong: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft. H. Xue: Data curation, formal analysis, investigation, methodology, project administration, in vivo animal works. F. Mo: Data curation, software, formal analysis, methodology. Y. Lin: Formal analysis, methodology. D. Lin: Formal analysis, investigation, writing–review and editing. N.K.Y. Wong: Methodology, writing–review and editing. Y. Sun: Software, formal analysis, methodology. S. Wilkinson: Resources, formal analysis, methodology. A.T. Ku: Resources, formal analysis, methodology. J. Hao: Software, formal analysis, investigation, methodology, writing–review and editing. X. Ci: Resources, methodology, writing–review and editing. R. Wu: Resources, methodology. A. Haegert: Resources, methodology. R. Silver: Resources, formal analysis. M.-E. Taplin: Resources, formal analysis. S.P. Balk: Resources, formal analysis. J.J. Alumkal: Resources, writing–review and editing. A.G. Sowalsky: Resources, formal analysis, methodology, writing–review and editing. M. Gleave: Resources, writing–review and editing. C. Collins: Resources, formal analysis, writing–review and editing. Y. Wang: Conceptualization, supervision, funding acquisition, investigation, project administration, writing–review and editing.
References
- 1. Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol 1999;26:407–21. [PubMed] [Google Scholar]
- 2. Shahinian VB, Kuo YF, Freeman JL, Orihuela E, Goodwin JS. Increasing use of gonadotropin-releasing hormone agonists for the treatment of localized prostate carcinoma. Cancer 2005;103:1615–24. [DOI] [PubMed] [Google Scholar]
- 3. Ruckle HC, Klee GG, Oesterling JE. Prostate-specific antigen: concepts for staging prostate cancer and monitoring response to therapy. Mayo Clin Proc 1994;69:69–79. [DOI] [PubMed] [Google Scholar]
- 4. Crawford ED, Eisenberger MA, McLeod DG, Spaulding JT, Benson R, Dorr FA, et al. A controlled trial of leuprolide with and without flutamide in prostatic carcinoma. N Engl J Med 1989;321:419–24. [DOI] [PubMed] [Google Scholar]
- 5. Denis LJ, Carnelro de Moura JL, Bono A, Sylvester R, Whelan P, Newling D, et al. Goserelin acetate and flutamide versus bilateral orchiectomy: a phase III EORTC trial (30853). EORTC GU Group and EORTC Data Center. Urology 1993;42:119–29; discussion 29–30. [DOI] [PubMed] [Google Scholar]
- 6. Yeh AC, Ramaswamy S. Mechanisms of cancer cell dormancy–another hallmark of cancer? Cancer Res 2015;75:5014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007;7:834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hensel JA, Flaig TW, Theodorescu D. Clinical opportunities and challenges in targeting tumour dormancy. Nat Rev Clin Oncol 2013;10:41–51. [DOI] [PubMed] [Google Scholar]
- 9. Recasens A, Munoz L. Targeting cancer cell dormancy. Trends Pharmacol Sci 2019;40:128–41. [DOI] [PubMed] [Google Scholar]
- 10. Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer 2020;20:398–411. [DOI] [PubMed] [Google Scholar]
- 11. Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 2014;74:1272–83. [DOI] [PubMed] [Google Scholar]
- 12. Lin D, Xue H, Wang Y, Wu R, Watahiki A, Dong X, et al. Next generation patient-derived prostate cancer xenograft models. Asian J Androl 2014;16:407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Revelo MP, Sudilovsky D, Cao M, Chen WG, Goetz L, et al. Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate 2005;64:149–59. [DOI] [PubMed] [Google Scholar]
- 14. Lin D, Watahiki A, Bayani J, Zhang F, Liu L, Ling V, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res 2008;68:4352–9. [DOI] [PubMed] [Google Scholar]
- 15. Choi SYC, Ettinger SL, Lin D, Xue H, Ci X, Nabavi N, et al. Targeting MCT4 to reduce lactic acid secretion and glycolysis for treatment of neuroendocrine prostate cancer. Cancer Med 2018;7:3385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat Commun 2019;10:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ci X, Hao J, Dong X, Choi SY, Xue H, Wu R, et al. Heterochromatin protein 1alpha mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res 2018;78:2691–704. [DOI] [PubMed] [Google Scholar]
- 18. Herberts C, Murtha AJ, Fu S, Wang G, Schonlau E, Xue H, et al. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur Urol 2020;78:834–44. [DOI] [PubMed] [Google Scholar]
- 19. Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 2015;1:466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong X, Guan J, English JC, Flint J, Yee J, Evans K, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res 2010;16:1442–51. [DOI] [PubMed] [Google Scholar]
- 21. Collins CC, Volik SV, Lapuk AV, Wang Y, Gout PW, Wu C, et al. Next generation sequencing of prostate cancer from a patient identifies a deficiency of methylthioadenosine phosphorylase, an exploitable tumor target. Mol Cancer Ther 2012;11:775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mo F, Lin D, Takhar M, Ramnarine VR, Dong X, Bell RH, et al. Stromal gene expression is predictive for metastatic primary prostate cancer. Eur Urol 2018;73:524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hao J, Ci X, Xue H, Wu R, Dong X, Choi SYC, et al. Patient-derived Hormone-naive prostate cancer xenograft models reveal growth factor receptor bound protein 10 as an androgen receptor-repressed gene driving the development of castration-resistant prostate cancer. Eur Urol 2018;73:949–60. [DOI] [PubMed] [Google Scholar]
- 24. Lin D, Wang X, Choi SYC, Ci X, Dong X, Wang Y. Immune phenotypes of prostate cancer cells: Evidence of epithelial immune cell-like transition? Asian J Urol 2016;3:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010;17:535–46. [DOI] [PubMed] [Google Scholar]
- 26. Tung WL, Wang Y, Gout PW, Liu DM, Gleave M, Wang Y. Use of irinotecan for treatment of small cell carcinoma of the prostate. Prostate 2011;71:675–81. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y, Wang JX, Xue H, Lin D, Dong X, Gout PW, et al. Subrenal capsule grafting technology in human cancer modeling and translational cancer research. Differentiation 2016;91:15–9. [DOI] [PubMed] [Google Scholar]
- 28. Wyatt AW, Mo F, Wang K, McConeghy B, Brahmbhatt S, Jong L, et al. Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer. Genome Biol 2014;15:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilkinson S, Ye H, Karzai F, Harmon SA, Terrigino NT, VanderWeele DJ, et al. Nascent prostate cancer heterogeneity drives evolution and resistance to intense hormonal therapy. Eur Urol 2021;80:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Nat Acad Sci USA 2020;117:12315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montironi R, Diamanti L, Santinelli A, Galetti-Prayer T, Zattoni F, Selvaggi FP, et al. Effect of total androgen ablation on pathologic stage and resection limit status of prostate cancer. Initial results of the Italian PROSIT study. Pathol Res Pract 1999;195:201–8. [DOI] [PubMed] [Google Scholar]
- 34. Tetu B. Morphological changes induced by androgen blockade in normal prostate and prostatic carcinoma. Best Prac Res Clin Endocrinol Metab 2008;22:271–83. [DOI] [PubMed] [Google Scholar]
- 35. Tang LH, Gonen M, Hedvat C, Modlin IM, Klimstra DS. Objective quantification of the Ki67 proliferative index in neuroendocrine tumors of the gastroenteropancreatic system: a comparison of digital image analysis with manual methods. Am J Surg Pathol 2012;36:1761–70. [DOI] [PubMed] [Google Scholar]
- 36. Wang X, Qi M, Zhang J, Sun X, Guo H, Pang Y, et al. Differential response to neoadjuvant hormonal therapy in prostate cancer: Predictive morphological parameters and molecular markers. Prostate 2019;79:709–19. [DOI] [PubMed] [Google Scholar]
- 37. Karzai F, Walker SM, Wilkinson S, Madan RA, Shih JH, Merino MJ, et al. Sequential prostate magnetic resonance imaging in newly diagnosed high-risk prostate cancer treated with neoadjuvant enzalutamide is predictive of therapeutic response. Clin Cancer Res 2021;27:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bui AT, Laurent F, Havard M, Dautry F, Tchenio T. SMAD signaling and redox imbalance cooperate to induce prostate cancer cell dormancy. Cell Cycle 2015;14:1218–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sowalsky AG, Ye H, Bhasin M, Van Allen EM, Loda M, Lis RT, et al. Neoadjuvant-intensive androgen deprivation therapy selects for prostate tumor Foci with diverse subclonal oncogenic alterations. Cancer Res 2018;78:4716–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gimbrone MA Jr, Leapman SB, Cotran RS, Folkman J. Tumor dormancy in vivo by prevention of neovascularization. J Exp Med 1972;136:261–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weinhold KJ, Goldstein LT, Wheelock EF. The tumor dormant state. Quantitation of L5178Y cells and host immune responses during the establishment and course of dormancy in syngeneic DBA/2 mice. J Exp Med 1979;149:732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wheelock EF, Weinhold KJ, Levich J. The tumor dormant state. Adv Cancer Res 1981;34:107–40. [DOI] [PubMed] [Google Scholar]
- 44. Enderling H, Hahnfeldt P, Hlatky L, Almog N. Systems biology of tumor dormancy: linking biology and mathematics on multiple scales to improve cancer therapy. Cancer Res 2012;72:2172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wells A, Griffith L, Wells JZ, Taylor DP. The dormancy dilemma: quiescence versus balanced proliferation. Cancer Res 2013;73:3811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Coller HA. Cell biology. The essence of quiescence. Science 2011;334:1074–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Manjili MH. Tumor dormancy and relapse: From a natural byproduct of evolution to a disease state. Cancer Res 2017;77:2564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 4. PGS predicted ADT response or clinical outcomes in independent hormone-naïve or CRPC clinical cohorts. A, Single-sample gene set enrichment score of PGS is proportionally related to the residual tumor volumes [RCB, r(35) = 0.74, P < 0.00001] in primary hormone-naïve prostate cancer treated with preoperative ADT (Sowalsky clinical cohort, n = 37). B, In the Sowalsky cohort, the PGS gene set enrichment score is significantly higher in nonresponders (NR) compared with exceptional responders (ER; P < 0.0001) and incomplete responders (IR; P < 0.0001). C and D, Kaplan–Meier estimate of OS or PFS segregated according to the PGS score in SU2C (A, n = 60) and Alumkal (B, n = 25) clinical cohorts. In both cohorts, patients with PGS = 1 had worse outcome than those with PGS = 0 (P = 0.0002 and 0.006, respectively). E, Panther Protein Classification of the PGS genes revealed that the genes were involved in various biological processes.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/7661/9355619/5eae3b434c70/782fig4.jpg)