Abstract
Exosomes are a sub-population of extracellular vesicles. It is released from all types of cells and are observed to be involved in cellular communications. It contains DNA, RNA, proteins and lipids. Tumor-derived exosomes can modify the tumor micro-environment and promote tumor development. Exosomal miRNAs are functionally linked with cancer progression, metastasis, and aggressive tumor phenotypes. In this review, we initially discuss on the fundamental biology of exosomes and then summarize the recent understanding of the exosomal miRNAs in oral cancer with various biological events. Moreover, the dynamic impact of exosomal miRNAs in the oral cancer micro-environment and their multiple parameter alterations can lead to (i) increased uncontrolled cell proliferation, (ii) oral cancer angiogenesis, (iii) oral cancer metastasis, (iv) drug resistance in oral cancer, (v) reprogramming of the immune system in oral cancer, and (vi) clinical significance of exosomal miRNA in oral cancer detection. Exosomes research can pave way to identify early detection tools in future and personalized medicine development for oral cancer. Thus, our review provides an informative biological insight into exosomal miRNAs in oral cancer, which can benefit the researchers working in the corresponding domain.
Keywords: Exosomes, miRNAs, Oral cancer
Introduction
Oral cancer is one of the most challenging disease which is the sixth most common health problem globally among other types of cancer (Kumar et al. 2016). Oral cavity and oropharynx-related cancers are called oral cancers or mouth cancers which include epithelial cancers, especially squamous cells, salivary gland cancers, soft-tissue cancers, hematolymphoid cancers, and odontogenic carcinomas (Feller and Lemmer 2012). Among various types of oral cancers, the most common (90%) oral cancer type is the Oral Squamous Cell Carcinoma (OSCC) (Feller and Lemmer 2012; Lewis et al. 2016) which has a survival rate of only 5 years (Turner et al. 2013). According to the current statistics, in the year 2022, it is estimated that 54,000 new (males 38,700 and females 15,300) oral cancer cases will be identified in the US and the mortality associated with the disease would be 11,230 (males 7870 and females 3360) (Siegel et al. 2022). Taking into consideration the WHO statistics in 2020, the prevalence of Oral Cancer in India is predicted as one of the leading cancers leading to morbidity and death, and it is ranked as the number one fatal disease in males and the fourth one in females. In this global scenario, the demand for a dependable clinical screening biomarker and efficient treatment tools for oral cancer prevention is indispensable. Recently, exosomes, a type of extracellular vehicles (EVs), have been identified and included in the list of promising cancer screening tools in liquid biopsies (Lopez et al. 2018). The endosomal originated exosomes (40–200 nm) (Shao et al. 2018) contain several biomolecules such as proteins, lipids, and large amounts of nucleic acids including mRNA, microRNAs, circular RNAs, and long non-coding RNAs which play roles in cellular communications in cancer (Behera and Tyagi 2018).
It is experimentally observed that in exosomes, the secreted rate of oncogenic cells is ten times higher than non-oncogenic cells, and the cancer cell-derived exosomes promote cellular signaling via mRNAs, non-coding miRNAs (ncRNAs), and miRNAs (Mao et al. 2018; Akers et al. 2013); ncRNAs which are 19–25 nucleotides long miRNAs were considered to be the dark matter of genetic, since the exact functional role of ncRNAs in biology was not determined for a long time. However, recent scientific research progress has opened up the mystery of ncRNAs and enlisted the role of dynamic gene expression regulation of ncRNAs in many cells (Morris and Mattick 2014; Esteller 2011). The total ncRNAs are of three types, and their classification is based on the variable sequence length of the ncRNAs. In this classification, miRNAs are considered as short sequences containing (~ 21 nucleotides) acting as a major regulatory factor in cell biology controlling genomic expression. In the study of cancer, miRNAs control several oncogenic developmental episodes by creating genetic instability (Berindan-Neagoe et al. 2014). Besides, scientists have observed that exosomes derived from different cells maintain their uniqueness in mRNAs and miRNA levels even if their origins are different from each other (Chaput and Théry 2011) Additionally, collected pieces of evidence also confirm that, cancer cells released exosomal miRNAs play a significant and effective role in maintaining the tumor environment (Tkach and Théry 2016). Moreover, exosomal miRNAs are clinically significant as they can serve as early clinical diagnostic markers for cancer and they can also contribute to cancer therapeutic development. To date, there are numerous articles available regarding exosomal miRNAs and oral cancer independently (Aqil et al. 2014; Aqil et al. 2015; Rajguru et al. 2020; Mallik and Zhao 2020), while only a few research articles have been published on both exosomal miRNAs and oral cancer together (Shoff et al. 2020; Sakha et al. 2016; Kulkarni et al. 2017). Also, the research articles cover only a specific portion of the related topic. Hence, to provide more biological insight, we provide a comprehensive review of the association of exosomal miRNAs and oral cancer in the maintenance and regulation of various biological events, viz., oral cancer cell proliferation, angiogenesis, metastasis and EMT, drug resistance, and immunity in oral cancer. Interestingly, our review work also highlights the participation of exosomal miRNAs in oral cancer progression and metastasis which would be beneficial to cancer researchers.
Exosomes biogenesis
Exosomes are the transitional by-products of early-to-late endosomes (Huotari and Helenius 2011) originating from the plasma membrane (Shao et al. 2018). Early endosomes (EEs) are processed through two different pathways similar to that of the formation of the lysosomes, one in which the “recycling endosomes” play a role and the other which involves the “late endosomes” (LEs), also called a multivesicular bodies (MVBs). MVBs carry several membrane-bound intraluminal vesicles (ILVs), and are a subset of endosomes and fusion of these with the plasma membrane releases its contents outside of the cells and these extracellular vesicles are called exosomes (Li et al. 2019). Exosomes biogenesis occurs by two different pathways, one, which is dependent on the “Endosomal Sorting Complexes Required for Transport” (ESCRT) called as ESCRT-dependent, while the other is ESCRT-independent (Li et al. 2019). In the initial stages of the ESCRT-dependent pathway, ILVs are synthesized by the developing endosomes. ESCRT, which includes a group of proteins, generates ILVs through a complex networking cascade (Colombo et al. 2013). This complex ESCRT was identified in early 2000 and comprises four types (viz., ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III). In the pathway initiated by ESCRT-0, the ESCRT-0 connects with high phosphatidylinositol 3-phosphate (PI3P) containing part of the membrane and binding occurs through Zinc Finger Domains (ZFDs) and Ubiquitin-interacting Motifs (UIMs) (Schmidt and Teis 2012). ESCRT-0 have two subunits called hepatocyte growth factor regulated tyrosine kinase substrate (HRS) and signal-transducing adaptor molecule 1/2 (STAM-1/2), and this dimer can bind through interactions with eight multiple ubiquitination moieties. ESCRT-0 containing HRS C-terminal activates ESCRT-I (Schmidt and Teis 2012; Henne et al. 2011), and ESCRT-I and ESCRT-II together play a major role in endosomal cytoplasmic budding from the plasma membrane (Wollert and Hurley 2010). During budding, ESCRT-0 guides the cytoplasmic packaging component, and the cargo selection process is regulated by ESCRT-II and ESCRT-III (Wollert and Hurley 2010). ESCRT-III containing proteins such as oligomerized sucrose-nonfermenting (Snf7), tumor susceptibility gene (TSG101), and ALG-2-interacting Protein X (Alix) are involved in classical vesicle budding (Teis et al. 2010). Of these, the TSG101 and Alix are the key components of the ESCRT system and are used as exosomes markers for screening in case of ESCRT-dependent processes (Kowal et al. 2014).
However, the scientific explanation regarding ESCRT-independent pathway is not entirely clear. Multiple cargo sorting and budding mechanisms are observed that are related to ceramide-mediated membrane budding (Niel et al. 2011). Ceramides are produced by the disruption of sphingomyelin via neutral sphingomyelinase, creating a raft-like construct because of its self-organizing property. This type of structure formation enhances membrane budding and CD9, CD63, and CD81 are markers of the ESCRT-independent pathway (Niel et al. 2011; Verweij et al. 2011; Choi et al. 2015; Thakur et al. 2014; Huang et al. 2013; Eirin et al. 2014).
Biosynthesis of miRNAs and packaging in exosomes
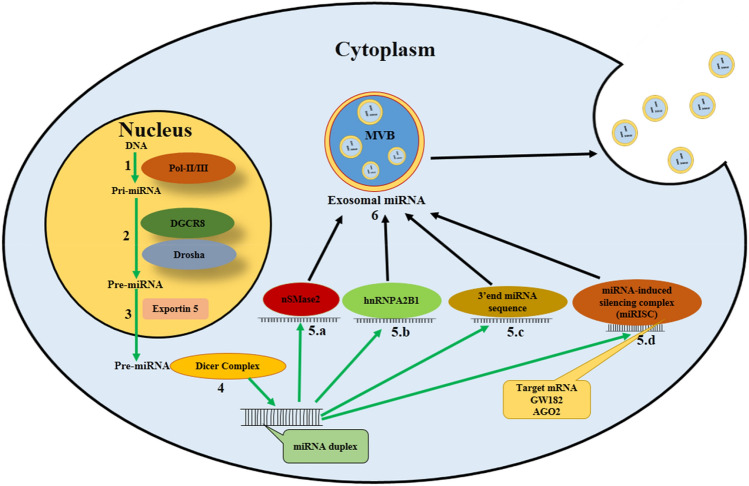
miRNAs are the outcomes of the transcription processes of central dogma and, with the help of DNA polymerases II/III, synthesize primary miRNAs (pri-miRNAs) initially in the nucleus. The combination of DiGeorge syndrome chromosomal region 8 (DGCR8) and Drosha (it is a Class 2 ribonuclease III enzyme, in humans DROSHA gene is encoded by it) in the nucleus converts primary miRNAs to precursor miRNAs (pre-miRNAs). Exportin 5, a RanGTP-dependent dsRNA-binding protein, plays an important role in the transport of pre-miRNAs in the cellular cytoplasm. The Dicer complex influences the processing of pre-miRNAs and converts them into double-strand miRNAs. After that, the exosomal miRNA sorting process starts, and four powerful pathways are responsible for sorting miRNAs into exosomes (Fig. 1). The first one discovered was the neural sphingomyelinase 2 (nSMase2)-dependent pathway, which is responsible for exosomal miRNA packaging (Zhang et al. 2015), and high expression of nSMase2 is attributed to miRNA enrichment present in the exosomes (Kosaka et al. 2013). The second packaging mechanism is related to heterogeneous nuclear ribonucleoprotein (hnRNP)-dependent pathway. The hnRNP has three subtypes of protein family, viz., hnRNPA1, hnRNPA2B1, and hnRNPC. These are involved in the packaging of the miRNA in exosomes and hnRNPA2B1 regulates the sorting of exosomal miRNA by identifying the GGAG motifs in the miRNA sequences (Villarroya-Beltri et al. 2013). The third pathway is 3′ end Gen Script end miRNA sequence-dependent pathway which has an important contribution in packaging the signal process and guiding it into the exosomes (Koppers-Lalic et al. 2014a, b). The fourth pathway is related to miRNA-induced silencing complex (miRISC) which are found in MVBs. Furthermore, conversion of MVBs into lysosomes causes high aggregation of miRISCs. It plays a crucial role in MVBs development and exosomal miRNAs-based gene silencing. One of the main components of miRISC is Argonaute 2 (Ago2), and knockout of Ago2 alters the quantity of miRNAs in exosomes (Guduric-Fuchs et al. 2012; Momen-Heravi and Bala 2018). In this way, the four cellular signaling pathways are involved in miRNAs’ packaging in exosomes.
Fig. 1.
Biosynthesis of miRNA and its incorporation into exosomes. (1) Primary miRNA (Pri-mRNA) synthesis occurs in nucleus via transcription process and involvement of DNA polymerase II/III. (2) Pri-miRNA to precursor mRNA (Pre-miRNA) conversion by nuclear complex of DGCR8 and Drosha. (3) Exportin 5 transports Pre-miRNA from the nucleus to the cytoplasm. (4) Cytoplasmic Dicer complex modifies pre-miRNA converting it to double-stranded miRNA. Mature miRNA packaging by (5.a) nSMase2 pathway (5.b) hnRNP-dependent pathway in which the hnRNPA2B1 protein identifies the GGAG sequence of 3’ end of miRNA and plays a role in miRNA packaging in exosomes. (5.c) 3’ end sequence dependent pathway which provides the guidance for miRNA sorting in exosomes. (5.d) miRNA-induced silencing complex (miRISC) contain miRNA, GW182, Argonaute 2(Ago2), and miRNA targeted mRNA; this complex destabilizes and suppresses the translation of miRNA. (6) All pathways (5.a, 5.b, 5.c and 5.d) assist in packaging of miRNA into multivesicular bodies (MVBs) and miRNA containing exosomes are released from MVBs.
Role of exosomal miRNAs in oral cancer
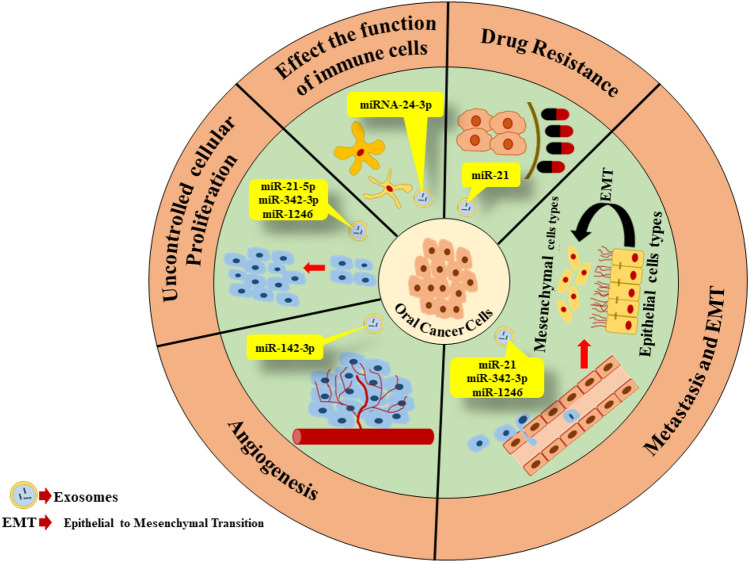
Exosomal miRNAs are smart influencers and modifiers of the oral tumor micro-environment. As a result, different processes of cancer cells are altered such as tumor growth, angiogenesis, metastasis, drug resistance, and immune responses (Fig. 2). The list of important exosomal miRNAs and their role in oral cancer is described in Table 1.
Fig. 2.
The dynamic role of exosomal miRNAs in oral cancer. Exosomal miRNAs in oral cancer regulates several episodes of oral cancer development. It alters natural cellular proliferation activity, intiates secondary tumor developmental cascade via angiogenesis, metastasis, epithelial-to-mesenchymal transition, and reprogramming of immune responses. Exosomal miRNAs play a role in development of drug resistance in oral cancer cells population in tumor microenvironmment. Oral tumor-derived exosomal miRNAs also establish complex intercellular communications which influences aggressive cancer progression and development
Table 1.
List of important exosomal miRNAs in oral cancer which are involved in oral tumor growth, angiogenesis metastasis, drug resistance, and immune responses
| Oral cancer features | Exosomal miRNAs | Functions | References |
|---|---|---|---|
| Tumor growth |
miR-21-5p miR-342-3p miR-1246 |
Activation of the nuclear factor kappa B (NF-κB) inflammatory pathway | (Momen-Heravi and Morvan 2018; Sakha et al. 2016.) |
| Angiogenesis | miR-142-3p | Elevated expression of Type I TGFβ receptor (TβRI) in the donor cancer cells and increase of TβRI action in recipient endothelial cells | (Dickman et al. 2017) |
| Metastasis |
miR-21 miR-342-3p miR-1246 |
Down-regulation of Snail, Vimentin, and E-cadherin | (Shan et al. 2018; Sakha et al. 2016; Li et al. 2016a, b) |
| Drug resistance | miR-21 | Activation of phosphatidylinositol 3 kinase (PTEN) and Pyruvate Dehydrogenase Deficiency (PDCD4) | (Harmati et al. 2017; Liu et al. 2017) |
| Immune response modifications | miRNA-24-3p | Down-regulation of fibroblast growth factor (FGF-11), which inhibits phosphorylation of the signal transducer and activator of transcription (STAT) and extracellular signal-regulated kinases (ERK) protein of T cells | (Ye et al. 2016) |
Contribution of exosomal miRNAs in oral cancer cell proliferation
Cellular proliferation disorderliness is one of the hallmarks of cancer development and initiation. This phase happens via mutations of cell division regulatory proteins (Shan et al. 2018). The exosomes released from Oral Squamous Cell Carcinoma (OSCC) cells cause activation of some signaling pathways such as Jun N-terminal Kinase (JNK), Protein kinase B (AKT), and Mitogen-activated protein kinase (MAPK)/extracellular-signal-regulated kinase (ERK) (Sento et al. 2016). In case of oral cancer, the oncogenic miR-21-5p from the exosomes activate the NF-κB inflammatory pathway (Momen-Heravi and Bala 2018).
Role of exosomal miRNAs in oral cancer angiogenesis
Angiogenesis is a tumor development stage where the tumor forms a network of new blood vessels from the existing blood vessels which provides all the cell growth essential nutrients for the developing tumor. This concept regarding angiogenesis was first hypothesized by Folkman in 1971 (Wang et al. 2010). During the past decade, scientific research had actively reported that exosomes could help tumor angiogenesis which in turn could initiate the development and progression of oral cancer (De Andrade et al. 2018; Dickman et al. 2017). TGF‐β pathway has two transmembrane receptors, viz., transforming growth factor-beta receptors I/II (TβRI/TβRII), both of which play a key role in exosomes-based angiogenesis in oral cancer. OSCC cells secrete exosomes that contain miR-142-3p which can help in cancer progression and angiogenesis, and also have elevated expression of TβRI in the donor cancer and recipient endothelial cells (Dickman et al. 2017).
Role of exosomal miRNAs in oral cancer metastasis and EMT
Metastasis accounts for a great majority of cancer-associated deaths, and in this critical mechanism, oncogenic cells lose their cellular addition property. As a result, the cancer cells migrate and enter the blood or lymph vessels, reach different parts of the body, and construct a fresh cluster of cancer cells (called a secondary tumor) (Santos et al. 2018). Epithelial-to-mesenchymal transition (EMT) encompasses dynamic alterations of the cellular system to alteration of epithelial cellular contact property and gaining a cell motile mesenchymal nature with invasive phenomena (Greening et al. 2015a, b). This cellular modulation demands for cancer growth and metastasis (Blackwell et al. 2017). Oral cancer cells release exosomal-related molecules contributing to dynamic cellular alterations and accelerating oncogenic growth. However, the mechanism involved in this process remains unsolved. Based on the proteomic analysis-related pieces of evidence, it has been demonstrated that some tumor-associated proteins are observed in OSCC-derived exosomes, viz., Matrix Metalloproteinase-13 (MMP-13), Heat Shock Protein-90 (HSP-90), Tumor Necrosis Factor Receptor-Associated Protein 1 (TRAP1), and Epidermal Growth Factor Receptor (EGFR). Researchers predict that these proteins may play a key factor in several stages of oral oncogenic propagation, and it may also be used as an OSCC clinical diagnosis biomarker (Ono et al. 2018; Shan et al. 2018). Exosomal hypoxia-inducible factor-1 (HIF-1α) and latent membrane protein 1 (LMP1) positive exosomes influence high motility and invasiveness of head and neck cancer (HNC) via EMT (Aga et al. 2014). HNC-derived exosomes contain a high percentage of Desmoglein-2 (Dsg-2) (Overmiller et al. 2017) which may regulate the tumor progression by damaging matrix metalloproteinase and caveolins through promoting EVs’ biogenesis and mitogenic effects (Overmiller et al. 2017; Vered et al. 2015). Extreme metastatic oral squamous cell carcinoma cells secrete exosomes containing miR-1246 and miR-342-3p that boost up the oncogenic growth, metastasis, and invasion of recipient cells (Sakha et al. 2016). Hence, poor metastatic cells transform to aggressive metastatic cells because of the downregulation of the Multiple Acyl-CoA Dehydrogenase Deficiency (MADD)/DENN domain consisting of protein 2D (DENN2D) by the influence of exosomal miR-1246. miRNA-21 containing exosomes also show an increased expression of mesenchymal markers such as vimentin and snail and downregulation of E-cadherin. This scientific evidence suggests that OSCC malignant cell clusters migrate to distant organs via EMT (Wang et al. 2019; Shan et al. 2018; Li et al. 2016a, b).
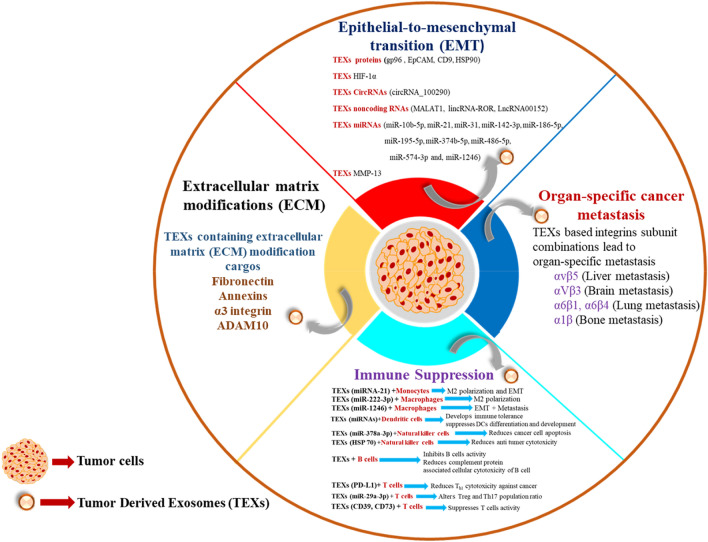
Recent studies have highlighted the association between exosomes and metastasis, and it has been demonstrated that multiple events like immune suppression, organ-specific metastasis, extracellular matrix (ECM)-remodeling, and EMT have been observed to play a role in modifying the interaction between the exosomes and metastasis (Fig. 3).
Fig. 3.
Interrelation between exosomes and metastasis. In cancer metastasis, multiple vital events (immune suppression, organ-specific metastasis, extracellular matrix (ECM)-remodeling, and EMT) regulate via exosomes and its intracellular components play a vital role in the cellular transformation.
Immune suppression: Immune suppression is one of the events which occurs during cancer progression and metastasis. In this process, immune cells are reprogrammed via multiple tumor-inducing factors and the tumor-derived exosomes (TEXs) are the vital components that induce immune suppression. Myeloid-derived suppressor cells (MDSCs) are pathologically activated neutrophils and monocytes with potent immunosuppressive activity. TEXs HSP72 reprogrammes MDSCs for the promotion of cancer and immune suppression (Gao et al. 2020). Monocytes are a group of sub-population of leukocytes, later transforming into macrophages and dendritic cells (DCs). TEXs-mediated increased expression of arginase and ROS influences monocytes activity (Javeed et al. 2016). TEXs miRNA-21 cargo enhances macrophage 2(M2) polarization and EMT (Hsieh et al. 2018) and these events encourage cancer development. Macrophages are antigen-presenting cells (APCs) which form a bridge between innate and adaptive immunity. TEXs down-regulate phosphatase and tensin homolog (PTEN), and enhances signal transducer and activator of transcription 3 (STAT3), and M2 polarization via miR-222-3p (Yang et al. 2018; Zhou et al. 2020; Wang et al. 2020; Kugeratski and Kalluri 2021), and TEXs-derived miR-1246 also directs macrophage-based EMT and metastasis. Dendritic cells (DCs) are classified as professional antigen-presenting cells (APCs). The miRNA cargos of TEXs suppressed DCs differentiation and development while promoting immunological tolerance (Ding et al. 2015). Nature killer cells (NK cells) are the players of innate immunity and it also plays a vital role against cancer cells via cytolytic activity (Morvan and Lanier 2016). This activity is reprogramed via through TEXs-mediated HSP70, which reduces NK cell-mediated cancer cell apoptosis (Gastpar et al. 2005) and miR-378a-3p cargo of TEXs acts on NK cells by mediating anti-tumor cytotoxicity (Briand et al.2020). B cells are key players of humoral immunity of the adaptive immune system. TEXs inhibit B-cell activity and their surface protein reduces cytotoxicity of B cells (Yang et al.2012; Capello et al. 2019). T cells are a type of lymphocyte that is responsible for the cellular immunity component of the adaptive immune system. Cytotoxic T (TC) and Helper T cells 1 (Th1) cells are the important components of the anti-tumor response in the immune system (Borst et al. 2018). T helper 17 (Th17) cells have a dual role in cancer; on one side, it suppresses the immune response for angiogenesis, and on another side, it creates an anti-tumor response (Guéry and Hugues 2015). TEXs cargos CD39 and CD73 suppresses T-cells activity (Clayton et al. 2011), and miR-29a-3p alters the ratio of regulatory T cells (Tregs) and Th17 population which promotes cancer development (Zhou et al. 2018). TEXs also mediate Programmed death-ligand 1 (PD-L1)-based signaling and reduces Th1-mediated cytotoxicity against cancer cells (Chen et al. 2018).
Organ-specific metastasis: TEXs play a major role in organ-specific metastasis through integrins. Integrins are one of the major binding receptors of the extracellular matrix (ECM), and based on these, EMT regulation occurs. The same mechanism takes place in TEXs-mediated cancer cell migration with organ specificity. In general, some specific patterns (α6β1, α6β4, αVβ3, αvβ5) of integrins are expressed in TEXs. The integrins associated with liver metastasis (αvβ5), brain metastasis (α2β), lung metastasis (α6β1, α6β4), and bone metastasis (αvβ3, α4β1) are involved in metastasis (Tian et al. 2019; Wortzel et al. 2019).
Extracellular matrix (ECM)-remodeling: Extracellular matrix (ECM) remodeling is related to several cellular events like cell morphology maintenance, cell growth, cell proliferation, and cell migration. In the tumor micro-environment, multiple factors influence ECM, and TEXs are one of them. TEXs containing fibronectin is involved in ECM remodeling. The proteomic analysis revealed that TEXs cargo annexins, α3 integrin, and A Disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) are involved in cellular migration and invasion (Becker et al. 2016). These complex cellular ECM modifications play a crucial role in cancer metastasis.
EMT and exosomes: EMT is the most dynamic event thatoccurs in cancer metastasis and the involvement of tumor-derived exosomes (TEXs) becomes more complex. Current scientific pieces of evidence suggests that TEXs biological active cargo like proteins (gp96, EpCAM, CD9, HSP90) (Han et al. 2018; Huang et al. 2018), HIF-1α (Nonaka and Wong 2018), CircRNAs (circRNA_100290) (Egea-Jimenez and Zimmermann 2020), non-coding RNAs (MALAT1, lincRNA-ROR, LncRNA00152) (Lebastchi and Callender 2014, Lee and Roberts 2013; Li et al. 2016a, b; Li et al. 2017), miRNAs (miR-10b-5p, miR-21, miR-31, miR-142-3p, miR-186-5p, miR-195-5p, miR-374b-5p, miR-486-5p, miR-574-3p, and miR-1246) (Jeck and Sharpless 2014; Kim et al. 2018; Kiyota et al. 2015; Langevin et al. 2017; Latifkar et al. 2019), and MMP-13 promote EMT and enhances metastasis.
Role of exosomal miRNAs in drug resistance oral cancer
Drug resistance is a major factor that plays a crucial role in the unsuccessful chemotherapy-based oncogenic treatment of oral cancer. Oral cancer cells secrete exosomes that carry some oncogenic molecules, because of which the cancer cells become resistant to the effect of the anti-proliferation and anti-metastatic chemotherapeutic drugs, viz., doxorubicin, cisplatin, and ROS-related drugs (Jelonek et al. 2015). Extreme chemoresistant OSCC cells exhibit drug resistance and the DNA damage is decreased by exosomes containing miRNA-21 which in turn triggers phosphatase and tensin homolog (PTEN) and programmed cell death protein 4 (PDCD4) (Harmati et al. 2017; Liu et al. 2017). Exosomes secretion is also increased by radiation, stress, and uptake of cancer-derived exosomes by normal cells, and at the same time the tumor cells become radiation-tolerant by the influence of the AKT signaling pathway and hence play a key role in repairing double-strand DNA damage (Mutschelknaus et al. 2016, 2017).
Exosomal miRNAs and immunity in oral cancer
The efficient proliferation and metastasis of cancer cells are not possible if cells are not capable of escaping from immune surveillance or treatment-based immune surveillance. Exosomes released from tumor cells create complex cellular signaling networks that build tumor immunity (Gao et al. 2018). Tumor cell-derived exosomes initiate the Treg (regulatory T cells) and tumor micro-environment-related macrophages, and then modulate the anti-tumor immune responses, thus preparing the tumor cell for immune escape and tolerance (Webber et al. 2015; Greening et al. 2015a, b). HNC-derived exosomes inhibit the T lymphocytes division and prevent T-cell subsets, Th-1 and Th-17 reproduction and influences the transformation of all of them to myeloid-derived suppressor cells (MDSCs) and Trag (Whiteside 2013). Treg cells become more susceptible compared to the subset of other T cells for immune suppression by tumor-released exosomes (Muller et al. 2016). Cytotoxic T cells (CD8 + T cells) are the major players in anti-cancer immune response, while HNC-derived exosomes carry high galectin-1 that controls low-level phosphorylation of STAT-1/-3 and high-level phosphorylation of STAT-5 by ERK/MAPK signaling pathway or low expression of CD27/28-induced cytotoxic T cells, promoting a suppressor phenotype (Maybruck et al. 2017). Hypoxic conditions of HNC tumor micro-environment up-regulated exosomes release and they contained miRNA-24-3p which down-regulated FGF-11, inhibiting phosphorylation of the STAT and ERK proteins of T cells (Ye et al. 2016). HNC-derived exosomes also regulates the translation of surface protein of CD8 + T cells such as major histocompatibility complex I (MHC-I) and Fas ligand L (Fas-L) which are playing a role in endocytosis and apoptosis (Ye et al. 2014). Oral cancer cell-derived exosomes also motivates the conversation of the human monocytic cell line (THP-1) cells to tumor-related macrophages M2 subtypes. On the other hand, these alterations do not have any symbolic impact on primary human macrophages (Al-Samadi et al. 2017).
Clinical significance of exosomal miRNAs in oral cancer
Exosomal miRNA-based liquid biopsy is the beginning of a new clinical diagnosis era. Generally, exosomes show strong superiority with unique biological active signatures in oral cancer biopsy. Some dynamic facts that requires attention for detailed investigation are discussed here. First, exosomes exist in all parts of the body and it is highly stable because of their lipid bilayer capsule shield. It is usually stored at 4 °C for 24 hrs and long time stored at − 80 °C in a pH 7 solution (Cheng et al. 2019). Second, exosomes are released from living cells, which contains abundant information about the parental cells. Third, exosomes are identified via specific membrane surface proteins like CD63, HSP70, TS101, and ALIX (Xu et al. 2016) which confers a uniqueness to these vesicles and marks them different from other vesicles. It can also be characterized by electron microscopy because of its specific cup-shaped sized appearance (Xu et al. 2016); fourth, exosomes contain specific parental protein signatures that assists to identify the source of the specific organs. Fifth, exosomes carry several biomarkers which indicate the cell’s normal or pathological status. Thus, these detailed information supports researchers and helps them to analyze and understand multiple pathological conditions of the human body (Sun et al. 2019). Sixth, exosomes provide clues about circulating tumor cells (CTCs) (Avgeris et al. 2019; Tovar-Camargo et al. 2016). All of these pieces of evidence prove that diagnostic accuracy once again can be developed in oral cancer by research on exosomes. In OSCC, miR-27a-3p, miR-223 (Tachibana et al. 2016), miR-302b-3p, miR-365, miR-412-3p, miR-494-3p, miR-512-3p, and miR-517b-3p are detected as biomarkers of oral cancer (Gai et al. 2018). The miR-21, miR-34, and miR-155 are found specific to oral cancer stem cells (Shoff et al. 2020). miR-24-3p has been reported to demonstrate high expression in oral cancer patients (Ye et al. 2014, 2016). miR-21 regulates hypoxic conditions in the tumor micro-environment and promotes EMT via (hypoxia-inducible factor) HIF1a/HIF2a-dependent pathway (Li et al. 2016a, b). In the current scenario of cancer research, exosomal microRNA profiling data have helped us to improve treatment efficiency and predict drug resistance in cancer patients. Extraexosomal microRNAs analysis also has paved the way to discover more precise specific biomarkers and proper therapeutic solution for treatment. Thus, exosomal miRNA-based oral cancer profiling can assist in early detection of cancers and may play a role in the near future for the development of personalized medicine and develop promising cancer diagnostic approaches to alleviate the suffering of oral cancer patients.
Conclusion
Research on exosomes opens a new orientation for a better understanding of cancer biology. Exosomes play a dynamic regulatory role in cancer growth, angiogenesis, metastasis, and immunity of oral cancer. Exosomal miRNAs alter genetic material and create genetic instability in cancer cells. Profiling of exosomal miRNAs databases helps to develop advanced diagnostic tools. However, some challenges associated with exosomal research cannot be avoided. First, we always follow International Society for Extracellular Vesicles (ISEV) guidelines which gives information on nomenclature, separation, characterization update protocols, and procedures for clinical application of exosomes. However, exosomal heterogeneity in size and sub-population indicates that we must develop reliable technologies which can help us in the isolation of exosomes and examination of exosomal miRNAs. Second, our present expertise is limited to exosome synthesis and how miRNA changes to exosomes as well as how exosomal miRNA works in recipient cells. In this scenario, a proper molecular mechanism pathway of exosomal miRNA transport for the next level of research exploration is imminent. Third, the destiny of exosomes and exosomal miRNAs is still not a clear understandable story. There are still many contradictions. Therefore, large magnification and tracking technologies as well as new in vivo models should be developed for better analysis. Fourth, substantial research combined with clinical safety and patient databases is required for proving the exosomal miRNA therapeutic efficiency. Moreover, the next level of research work is required for making a clear concept of how miRNAs of exosomes create genetic instability in oral cancer. Therefore, extensive research is recommended to understand the role of exosomal miRNA in oral cancer in terms of the development and progression, while the detailed study of the exosomal miRNAs profiling contributes to the innovation of exosomal miRNA-based clinical diagnosis tools and cancer therapeutic development against oral cancer in the future.
Data availability
Data sharing is not applicable—no new data are generated. Data sharing does not apply to this article as no new data were created or analyzed in this study.
Declarations
Conflict of interest
The authors report no conflict of interest.
References
- Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, Yoshizaki T, Pagano JS, Shackelford J. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. 2014;33:4613–4622. doi: 10.1038/onc.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Samadi A, Awad SA, Tuomainen K, Zhao Y, Salem A, Parikka M, Salo T. Crosstalk between tongue carcinoma cells, extracellular vesicles, and immune cells in in vitro and in vivo models. Oncotarget. 2017;8:60123–60134. doi: 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J Extracell Vesicles. 2014;25:3. doi: 10.3402/jev.v3.23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqil M, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. Transcriptomic analysis of mRNAs in human monocytic cells expressing the HIV-1 Nef protein and their exosomes. Biomed Res Int. 2015;2015:492395. doi: 10.1155/2015/492395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgeris M, Panoutsopoulou K, Papadimitriou MA, et al. Circulating exosomal miRNAs: clinical significance in human cancers. Expert Rev Mol Diagn. 2019;19:979–995. doi: 10.1080/14737159.2019.1673732. [DOI] [PubMed] [Google Scholar]
- Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera J, Tyagi N. Exosomes: mediators of bone diseases, protection, and therapeutics potential. Oncoscience. 2018;5:181–195. doi: 10.18632/oncoscience.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell RH, Foreman KE, Gupta GN. The role of cancer-derived exosomes in tumorigenicity & epithelial-to-mesenchymal transition. Cancers (basel) 2017;9:105. doi: 10.3390/cancers9080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- Briand J, Garnier D, Nadaradjane A, et al. Radiotherapy-induced overexpression of exosomal miRNA-378a-3p in cancer cells limits natural killer cells cytotoxicity. Epigenomics. 2020;12:397–408. doi: 10.2217/epi-2019-0193. [DOI] [PubMed] [Google Scholar]
- Capello M, Vykoukal JV, Katayama H, et al. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat Commun. 2019;10:254. doi: 10.1038/s41467-018-08109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput N, Théry C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol. 2011;33:419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zeng Q, Han Q, Xia W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell. 2019;10:295–299. doi: 10.1007/s13238-018-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015;4:474–490. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- Clayton A, Al-Taei S, Webber J, et al. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol. 2011;187:676–683. doi: 10.4049/jimmunol.1003884. [DOI] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- de Andrade A, de Oliveira CE, Dourado MR, Macedo C, Winck FV, Paes Leme AF, Salo T, Coletta RD, de Almeida FR, Galvão HC. Extracellular vesicles from oral squamous carcinoma cells display pro- and anti-angiogenic properties. Oral Dis. 2018;5:725–731. doi: 10.1111/odi.12765. [DOI] [PubMed] [Google Scholar]
- Dickman CT, Lawson J, Jabalee J, MacLellan SA, LePard NE, Bennewith KL, Garnis C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget. 2017;9:15252–15266. doi: 10.18632/oncotarget.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Zhou L, Qian Y, et al. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget. 2015;6:29877–29888. doi: 10.18632/oncotarget.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Jimenez AL, Zimmermann P. Lipids in exosome biology. Handb Exp Pharmacol. 2020;259:309–336. doi: 10.1007/164_2019_220. [DOI] [PubMed] [Google Scholar]
- Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O'Brien D, van Wijnen AJ, Lerman LO. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;1:55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Feller L, Lemmer J. Oral squamous cell carcinoma: epidemiology, clinical presentation and treatment. J Cancer Ther. 2012;3:263–268. doi: 10.4236/jct.2012.34037. [DOI] [Google Scholar]
- Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, Molinaro L, Carossa S, Camussi G, Arduino PG. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer. 2018;1:439. doi: 10.1186/s12885-018-4364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wang L, Dai T, Jin K, Zhang Z, Wang S, Xie F, Fang P, Yang B, Huang H, van Dam H, Zhou F, Zhang L. Tumor-derived exosomes antagonize innate antiviral immunity. Nat Immunol. 2018;3:233–245. doi: 10.1038/s41590-017-0043-5. [DOI] [PubMed] [Google Scholar]
- Gao Y, Xu H, Li N, et al. Renal cancer-derived exosomes induce tumor immune tolerance by MDSCs-mediated antigen-specific immunosuppression. Cell Commun Signal. 2020;18:106. doi: 10.1186/s12964-020-00611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastpar R, Gehrmann M, Bausero MA. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening DW, Gopal SK, Mathias RA, Liu L, Sheng J, Zhu HJ, Simpson RJ. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60–71. doi: 10.1016/j.semcdb. [DOI] [PubMed] [Google Scholar]
- Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Guduric-Fuchs J, O'Connor A, Camp B, O'Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéry L, Hugues S. Th17 cell plasticity and functions in cancer immunity. Biomed Res Int. 2015;2015:314620. doi: 10.1155/2015/314620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: emerging roles in systemic disease. Int J Biol Sci. 2018;14:633–643. doi: 10.7150/ijbs.25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmati M, Tarnai Z, Decsi G, Kormondi S, Szegletes Z, Janovak L, Dekany I, Saydam O, Gyukity-Sebestyen E, Dobra G, Nagy I, Nagy K, Buzas K. Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J Oral Pathol Med. 2017;4:259–266. doi: 10.1111/jop.12486. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;1:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hsieh CH, Tai SK, Yang MH. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering MiR-21-abundant exosomes. Neoplasia. 2018;20:775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Yang J, Zheng J, et al. Characterization of selective exosomal microRNA expression profile derived from laryngeal squamous cell carcinoma detected by next-generation sequencing. Oncol Rep. 2018;40:2584–2594. doi: 10.3892/or.2018.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;17:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javeed N, Gustafson MP, Dutta SK, et al. Immunosuppressive CD14+HLA-DRlo/neg monocytes are elevated in pancreatic cancer and "primed" by tumour-derived exosomes. Oncoimmunology. 2016;6:e1252013. doi: 10.1080/2162402X.2016.1252013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelonek K, Wojakowska A, Marczak L, Muer A, Tinhofer-Keilholz I, Lysek-Gladysinska M, Widlak P, Pietrowska M. Ionizing radiation affects protein composition of exosomes secreted in vitro from head and neck squamous cell carcinoma. Acta Biochim Pol. 2015;2:265–272. doi: 10.18388/abp.2015_970. [DOI] [PubMed] [Google Scholar]
- Kim BK, Lee EM, Kim JH, et al. Relationship between ultrasonographic and pathologic calcification patterns in papillary thyroid cancer. Medicine (baltimore) 2018;41:e12675. doi: 10.1097/MD.0000000000012675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyota N, Tahara M, Fujii M. Adjuvant treatment for post-operative head and neck squamous cell carcinoma. Jpn J Clin Oncol. 2015;45:2–6. doi: 10.1093/jjco/hyu195. [DOI] [PubMed] [Google Scholar]
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Würdinger T, Meijer GA, Pegtel DM. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;6:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;15:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Kugeratski FG, Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288:10–35. doi: 10.1111/febs.15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni V, Uttamani JR, Naqvi AR, Nares S. microRNAs: emerging players in oral cancers and inflammatory disorders. Tumour Biol. 2017;5:1010428317698379. doi: 10.1177/1010428317698379. [DOI] [PubMed] [Google Scholar]
- Kumar M, Nanavati R, Modi TG, Dobariya C. Oral cancer: etiology and risk factors: a review. J Cancer Res Ther. 2016;2:458–463. doi: 10.4103/0973-1482.186696. [DOI] [PubMed] [Google Scholar]
- Langevin S, Kuhnell D, Parry T, et al. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget. 2017;8:82459–82474. doi: 10.18632/oncotarget.19614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifkar A, Ling L, Hingorani A, et al. Loss of sirtuin 1 alters the secretome of breast cancer cells by impairing lysosomal integrity. Dev Cell. 2019;49:393–408.e7. doi: 10.1016/j.devcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebastchi AH, Callender GG. Thyroid cancer. Curr Probl Cancer. 2014;38:48–74. doi: 10.1016/j.currproblcancer.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Lee RS, Roberts CW. Linking the SWI/SNF complex to prostate cancer. Nat Genet. 2013;45:1268–1269. doi: 10.1038/ng.2805. [DOI] [PubMed] [Google Scholar]
- Lewis AG, Tong T, Maghami E. Diagnosis and management of malignant salivary gland tumors of the parotid gland. Otolaryngol Clin North Am. 2016;49:343–380. doi: 10.1016/j.otc.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, Li X, Chen J, Liu K, Li C, Zhu G. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Wang S, et al. Exosomes derived from hypoxic oral squamous cell carcinoma Cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- Li X, Cao Y, Gong X, et al. Long noncoding RNAs in head and neck cancer. Oncotarget. 2017;8:10726–10740. doi: 10.18632/oncotarget.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, Daugaard M, Guns E, Hoorfar M, Li ITS. Challenges and opportunities in exosome research-perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3:011503. doi: 10.1063/1.5087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Chen G, Sun D, Lei M, Li Y, Zhou C, Li X, Xue W, Wang H, Liu C, Xu J. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim Biophys Sin (shanghai) 2017;49:808–816. doi: 10.1093/abbs/gmx078. [DOI] [PubMed] [Google Scholar]
- Lopez A, Harada K, Mizrak Kaya D. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev Anticancer Ther. 2018;18:19–38. doi: 10.1080/14737140.2018.1403320. [DOI] [PubMed] [Google Scholar]
- Mallik S, Zhao Z. Graph- and rule-based learning algorithms: a comprehensive review of their applications for cancer type classification and prognosis using genomic data. Brief Bioinform. 2020;21:368–394. doi: 10.1093/bib/bby120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Li X, Gong S, Yuan H, Jiang Y, Huang W, Sun X, Dang X. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018;9–10:248–259. doi: 10.1038/s41417-018-0032-3. [DOI] [PubMed] [Google Scholar]
- Maybruck BT, Pfannenstiel LW, Diaz-Montero M, Gastman BR. Tumor-derived exosomes induce CD8+ T cell suppressors. J Immunother Cancer. 2017;5:65. doi: 10.1186/s40425-017-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F, Bala S. Extracellular vesicles in oral squamous carcinoma carry oncogenic miRNA profile and reprogram monocytes via NF-κB pathway. Oncotarget. 2018;9:34838–34854. doi: 10.18632/oncotarget.26208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. the rise of regulatory RNA. Nat Rev Genet. 2014;6:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep. 2016;6:20254. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschelknaus L, Peters C, Winkler K, Yentrapalli R, Heider T, Atkinson MJ, Moertl S. Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. PLoS ONE. 2016;11:e0152213. doi: 10.1371/journal.pone.0152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschelknaus L, Azimzadeh O, Heider T, Winkler K, Vetter M, Kell R, Tapio S, Merl-Pham J, Huber SM, Edalat L, Radulović V, Anastasov N, Atkinson MJ, Moertl S. Radiation alters the cargo of exosomes released from squamous head and neck cancer cells to promote migration of recipient cells. Sci Rep. 2017;7:12423. doi: 10.1038/s41598-017-12403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Wong DTW. Liquid biopsy in head and neck cancer: promises and challenges. J Dent Res. 2018;97:701–708. doi: 10.1177/0022034518762071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Eguchi T, Sogawa C, Calderwood SK, Futagawa J, Kasai T, Seno M, Okamoto K, Sasaki A, Kozaki KI. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J Cell Biochem. 2018;119:7350–7362. doi: 10.1002/jcb.27039. [DOI] [PubMed] [Google Scholar]
- Overmiller AM, Pierluissi JA, Wermuth PJ, Sauma S, Martinez-Outschoorn U, Tuluc M, Luginbuhl A, Curry J, Harshyne LA, Wahl JK, 3rd, South AP, Mahoney MG. Desmoglein 2 modulates extracellular vesicle release from squamous cell carcinoma keratinocytes. FASEB J. 2017;31:3412–3424. doi: 10.1096/fj.201601138RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajguru JP, Mouneshkumar CD, Radhakrishnan IC, Negi BS, Maya D, Hajibabaei S, Rana V. Tumor markers in oral cancer: a review. J Family Med Prim Care. 2020;9:492–496. doi: 10.4103/jfmpc.jfmpc_1036_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. doi: 10.1038/srep38750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JMO, Peixoto da Silva S, Costa NR, Gil da Costa RM, Medeiros R. The role of microRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers (basel) 2018;10:493. doi: 10.3390/cancers10120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sento S, Sasabe E, Yamamoto T. Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell-derived exosomes. PLoS ONE. 2016;11:e0148454. doi: 10.1371/journal.pone.0148454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, You B, Shi S, Shi W, Zhang Z, Zhang Q, Gu M, Chen J, Bao L, Liu D, You Y. Hypoxia-induced matrix metalloproteinase-13 expression in exosomes from nasopharyngeal carcinoma enhances metastases. Cell Death Dis. 2018;9:382. doi: 10.1038/s41419-018-0425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoff M, Booker T, Leavitt B, Harmon D, Kingsley K, Howard KM. Differential exosome miRNA expression in oral cancer stem cells. ExRNA. 2020;2:1. doi: 10.1186/s41544-019-0045-6. [DOI] [Google Scholar]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q, Gu X, Xiang J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234:1416–1425. doi: 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- Tachibana H, Sho R, Takeda Y, Zhang X, Yoshida Y, Narimatsu H, Otani K, Ishikawa S, Fukao A, Asao H, Iino M. Circulating miR-223 in oral cancer: its potential as a novel diagnostic biomarker and therapeutic target. PLoS ONE. 2016;11:e0159693. doi: 10.1371/journal.pone.0159693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicleformation. EMBO J. 2010;29(5):871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Liu S, Li B. Potential role of exosomes in cancer metastasis. Biomed Res Int. 2019;2019:4649705. doi: 10.1155/2019/4649705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA biomarkers: emerging frontiers in colorectal and other human cancers. Expert Rev Mol Diagn. 2016;16:553–567. doi: 10.1586/14737159.2016.1156535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L, Mupparapu M, Akintoye SO. Review of the complications associated with treatment of oropharyngeal cancer: a guide for the dental practitioner. Quintessence Int. 2013;44:267–279. doi: 10.3290/j.qi.a29050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vered M, Lehtonen M, Hotakainen L, Pirilä E, Teppo S, Nyberg P, Sormunen R, Zlotogorski-Hurvitz A, Salo T, Dayan D. Caveolin-1 accumulation in the tongue cancer tumor microenvironment is significantly associated with poor prognosis: an in-vivo and in-vitro study. BMC Cancer. 2015;15:25. doi: 10.1186/s12885-015-1030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 2011;30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin P, Han C, Cai W, Zhao X, Sun B. Vasculogenic mimicry contributes to lymph node metastasis of laryngeal squamous cell carcinoma. J Exp Clin Cancer Res. 2010;29:60. doi: 10.1186/1756-9966-29-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yu F, Ding H, Wang Y, Li P, Wang K. Emerging function and clinical values of exosomal microRNAs in cancer. Mol Ther Nucleic Acids. 2019;7:791–804. doi: 10.1016/j.omtn.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang X, Si M. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- Webber J, Yeung V, Clayton A. Extracellular vesicles as modulators of the cancer microenvironment. Semin Cell Dev Biol. 2015;40:27–34. doi: 10.1016/j.semcdb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41:245–251. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Xu R, Greening DW, Zhu HJ, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Chalasani G, Ng YH, et al. Exosomes released from Mycoplasma infected tumor cells activate inhibitory B cells. PLoS ONE. 2012;7:e36138. doi: 10.1371/journal.pone.0036138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, McKay D, Pollard JW, et al. diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018;19:5492–5503. doi: 10.1158/0008-5472.CAN-18-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He J, Peng JY, Chen QY, Mo HY, Jun-Cui ZXS, Zeng YX, Li J. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. 2016;240:329–340. doi: 10.1002/path.4781. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li X, Wu X, et al. Exosomes released from tumor-associated macrophages Transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6:1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang Z, Gao S. Tumor-associated macrophages: recent insights and therapies. Front Oncol. 2020;10:188. doi: 10.3389/fonc.2020.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable—no new data are generated. Data sharing does not apply to this article as no new data were created or analyzed in this study.