Summary
Characterizing resident immune cells in human skin using single-cell assays provides insight into their role in infections, inflammation, and cancer. We describe an optimized protocol to rapidly isolate viable cells from 6-mm skin punch-biopsies. We provide an example in which we coupled single-cell RNA sequencing (scRNA-seq) with single-cell T-cell receptor sequencing (scTCR-seq) of skin and blood cells to study transcriptional profiles and clonotypes of skin resident and peripheral circulating, memory, and effector T cells. This is an improved protocol based on Saluzzo et al. (2021).
For complete details on the use and execution of this protocol, please refer to Saluzzo et al. (2021).
Subject areas: Cell isolation, Single Cell, Health Sciences, Sequencing, RNAseq, Immunology
Graphical abstract
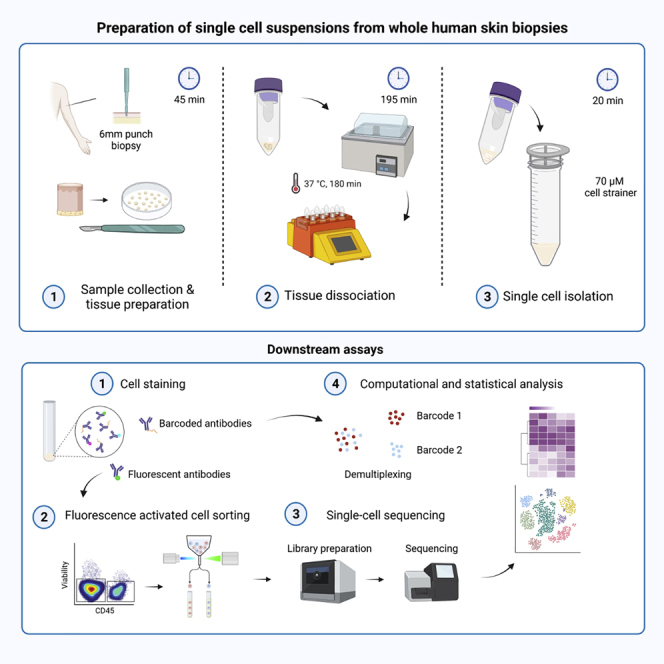
Highlights
-
•
Preparation of a high-quality single-cell suspension from human skin punch biopsies
-
•
Suitable for single-cell RNA and TCR sequencing to study skin and blood T cells
-
•
Cost minimization by hash-tagging and pooling blood and skin cells of one individual
-
•
Overview of the downstream bioinformatic analysis for the T cell characterization
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Characterizing resident immune cells in human skin using single-cell assays provides insight into their role in infections, inflammation, and cancer. We describe an optimized protocol to rapidly isolate viable cells from 6-mm skin punch-biopsies. We provide an example in which we coupled single-cell RNA sequencing (scRNA-Seq) with single-cell T-cell receptor sequencing (scTCR-Seq) of skin and blood cells to study transcriptional profiles and clonotypes of skin resident and peripheral circulating, memory, and effector T cells. This is an improved protocol based on Saluzzo et al. (2021).
Before you begin
The protocol below describes specific steps for obtaining highly viable skin single-cell suspension from one 6 mm single human skin punch-biopsy. We provide an example, in which we coupled scRNA-Seq with scTCR-Seq of skin and blood from the same individuals. We believe that coupling scRNA-Seq with scTCR-Seq from both skin and blood is fundamental when studying skin diseases, as the skin hosts more memory T cells compared with the circulation (Clark, 2015). The identification of T cell receptor via scTCR-Seq allows tracking of specific TCR clones between skin and blood to analyze their phenotype and distribution in tissue (Saluzzo et al., 2021). We provide evidence of the suitability of this protocol to study structural cells in the human skin and suggest statistical tools that can be used for quantification and bioinformatics analysis of their interaction with immune cells. This is an improved protocol, modified from Saluzzo et al. (2021).
A skin punch biopsy is a safe and minimally invasive diagnostic procedure, routinely performed by dermatologists for diagnostic work-up. If working on healthy skin, we recommend to take a biopsy from a non-sun exposed area and, to allow interindividual comparisons, from the same area of the body. We chose the inner upper arm and stick to the same area for all the patients. Biopsies are performed after injecting a local anesthesia, which is usually composed of Xilocain 2% or Lidocain 2%. We often perform 6 mm diameter punch-biopsy, which results in a reasonable number of isolated cells. Before taking the biopsy, it is mandatory to make sure that the all the reagents are in place for the next steps of processing, which needs to be performed as quickly as possible to obtain high-quality viable cells. Tissue samples should be transferred to a small tube containing RPMI medium stored at 4°C and processes immediately. Carefully read the instructions in the data sheet of the Miltenyi Whole Skin Dissociation Kit (130-101-540) and prepare enzyme aliquots accordingly. Ensure adequate supply of stock solutions as described in materials and equipment. We recommend the preparation of single aliquots for the enzymatic reaction, adapted for one biopsy. Have aliquoted single use enzymes for the digestion buffer prepared (stored at −20°C) and ready to be defrosted (refer to materials and equipment for recipe tables). All preparation steps for tissue samples and stock solutions are carried out in a biosafety cabinet. Pre-heat a water bath with shaker to 37°C and pre-cool a centrifuge with 50cc tube rotor to 4°C. Have a gentleMACS dissociator with program h_skin_01 (Duration: 36 s, rpr: 116) ready for use.
Institutional permissions
Human skin biopsies and blood samples should be obtained under an IRB (Institutional Review Board)-approved clinical protocol with informed consent from patients. All experiments illustrated in this protocol involved the voluntary participation of patients with fully-informed written consent for the collection of samples and necessary personal data. The study was performed according to the Declaration of Helsinki and approved by the local ethics committee of the Medical University of Vienna (ECS 1087/2016).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human BD Fc Block | BD Biosciences | Cat#: 564219 |
| κPE/CF 594 anti-human CD45, 1:100 | BD Biosciences | Cat#: 562279 |
| PE anti-human HLA-ABC, 1:200 | BioLegend | Cat#: 311405 |
| BV421 anti-human CD3, 1:200 | BioLegend | Cat#: 300434 |
| PE-Cy7 anti-human CD90, 1:100 | BioLegend | Cat#: 328124 |
| AF488 anti-human CD34, 1:50 | BD | Cat#: 345801 |
| 7-AAD Viability Staining Solution, 1:500 | eBioscience | Cat#: 00-6993-50 |
| TotalSeq-C0251 anti-human Hashtag 1 Antibody | BioLegend | Cat#: 394661 |
| TotalSeq-C0252 anti-human Hashtag 2 Antibody | BioLegend | Cat#: 394663 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum (FBS) | Gibco | Cat#: 10500-064 |
| EDTA | Thermo Fisher Scientific | Cat#: 15575-020 |
| Bovine serum albumin (BSA) | Sigma | Cat#: A8412-100mL |
| PBS | Gibco | Cat#: 10010-023 |
| RPMI 1640 Medium | Thermo Fisher Scientific | Cat#: 11875093 |
| Dimethyl sulfoxide (DMSO) | Sigma | Cat#: D2650 |
| Critical commercial assays | ||
| Whole Skin Dissociation Kit, human | Miltenyi Biotec | Cat#: 130-101-540 |
| Software and algorithms | ||
| FlowJo | Tree Star | https://www.flowjo.com/solutions/flowjo |
| Seurat v4.0.1 | Stuart et al. (2019) | https://github.com/satijalab/seurat |
| CITE-seq software | Stoeckius et al., 2017 | https://cite-seq.com/ |
| Other | ||
| gentleMACS™ C Tubes | Miltenyi Biotec | Cat#: 130-093-237 |
| gentleMACS™ Octo Dissociator | Miltenyi Biotec | Cat#: 130-095-937 |
| 70 micron cell strainer | Falcon | Cat#: 352350 |
| Scalpel No.11 | Medi-Safe Surgicals | Cat#: BYDMSS11 |
Materials and equipment
Digestion mix from Miltenyi Whole Skin Dissociation Kit (see key resources table)
| Reagent | Aliquots preparation | Amount per reaction |
|---|---|---|
| Enzyme P | 25 μL aliquots | 12.5 μL |
| Enzyme D | Dissolved in Buffer L, 50 μL aliquots | 50 μL |
| Enzyme A | Dissolved in Buffer L, 10 μL aliquots | 2.5 μL |
| Buffer L | 30 mL, do not aliquot | 435 μL |
| Total | 50 reactions per Kit | 500 μL |
Store Buffer L at 4°C and enzymes at −20°C until Kit expiration date, avoid freeze/thaw cycles.
Wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS ×1 | n/a | 45 mL |
| Fetal calf serum (FCS), heat-inactivated | 10% | 5 mL |
| Total | 50 mL |
Store at 4°C for one month.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS ×1 | n/a | 50 mL |
| Ultra pure EDTA (0.5 M), pH 8 | 2 mM | 200 μL |
| Bovine serum albumin (BSA) | 1% | 500 mg |
| Total | n/a | 50 mL |
Sterile filter and store at 4°C for one month.
FACS-Sort buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS ×1 | n/a | 48.98 mL |
| Bovine serum albumin (BSA) | 0.08% | 40 mg |
| Total | n/a | 50 mL |
Sterile filter and store at 4°C for one month.
Tissue culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI | n/a | 47.5 mL |
| FCS | 5% | 2.5 mg |
| Total | n/a | 50 mL |
Sterile filter and store at 4°C for one month.
Biopsies and PBMCs freezing media
| Reagent | Final concentration | Amount |
|---|---|---|
| Dimethyl sulfoxide (DMSO) | 10% | 5 mL |
| Fetal bovine serum (FBS) | 90% | 45 mL |
| Total | n/a | 50 mL |
Sterile filter and store at 4°C for one month.
Step-by-step method details
Sample collection
Timing: 30 min
Skin 6 mm punch biopsies are obtained by qualified clinicians under local anesthesia. Collected tissue samples are placed in a 15 mL Falcon tube containing 2 mL of RPMI 1640, kept on ice and transferred immediately to the laboratory for further processing.
CRITICAL: The tissue should be kept on ice and processed right after collection, in any case within 15–30 min.
Prepare tissue for digestion
Timing: 15 min
The purpose of this step is to dissect the biopsy into smaller pieces to allow optimal enzymatic digestion. Dissection and subsequent steps are performed at 20°C–24°C) unless otherwise stated. All steps have to be performed under sterile conditions.
-
1.Prepare digestion mix in a gentleMACS C tube according to instructions in Miltenyi Whole Skin Dissociation Kit (https://www.miltenyibiotec.com/upload/assets/IM0007590.PDF).
-
a.Thaw one aliquot of each Enzyme (P, A, D).
-
b.Transfer 435 μL of Buffer L into the gentleMACS C tube.
-
c.Add 12.5 μL of Enzyme P and resuspend carefully.
-
d.Add 50 μL of Enzyme D and resuspend carefully.
-
e.Add 2.5 μL of Enzyme A and resuspend carefully.
-
a.
Note: As suggested by the manufacturer, adding Enzyme P to the digestion mix will increase the yield, but not preserve surface epitopes like CD4 or CD8. Therefore, be sure to skip the use of this enzyme in case of downstream assays that rely on surface markers, like CITE-Seq for example.
-
2.Rinse tissue with PBS to remove remaining blood.
-
a.Remove skin biopsy from the tube, place on a 60 mm petri dish (Figure 1A) and add 1–2 mL of sterile PBS.
-
b.Gently shake the petri dish, then aspirate the PBS. Repeat 2–3 time, if necessary, until clear.
-
c.Completely aspirate PBS and discard.
-
a.
-
3.Dissect the skin biopsy:
-
a.Remove subcutaneous fat from the biopsy by carefully scraping it using a curved cutting-edge scalpel.
-
b.Dissect tissue by fixating the biopsy with forceps and gently cutting it with a curved scalpel (Methods video S1). Continue until obtaining pieces of 1 mm2 dimension (Figure 1B).
 CRITICAL: Wet the scalpel with digestion mix in between to prevent drying of tissue pieces. All dissected pieces have to be maintained moist all the time. Troubleshooting problem 1.Methods video S1. Rinsing the cell strainer, related to step 10Download video file (94.4MB, mp4)
CRITICAL: Wet the scalpel with digestion mix in between to prevent drying of tissue pieces. All dissected pieces have to be maintained moist all the time. Troubleshooting problem 1.Methods video S1. Rinsing the cell strainer, related to step 10Download video file (94.4MB, mp4) -
c.Collect the tissue pieces by scraping them off the petri dish with the scalpel (Figure 1C).
-
d.Transfer dissected tissue into the gentleMACS C tube with the prepared digestion mix (Figure 1D) and tightly close the lid.Note: The procedure is the same also in case of smaller size biopsies, see “expected outcomes”.
-
a.
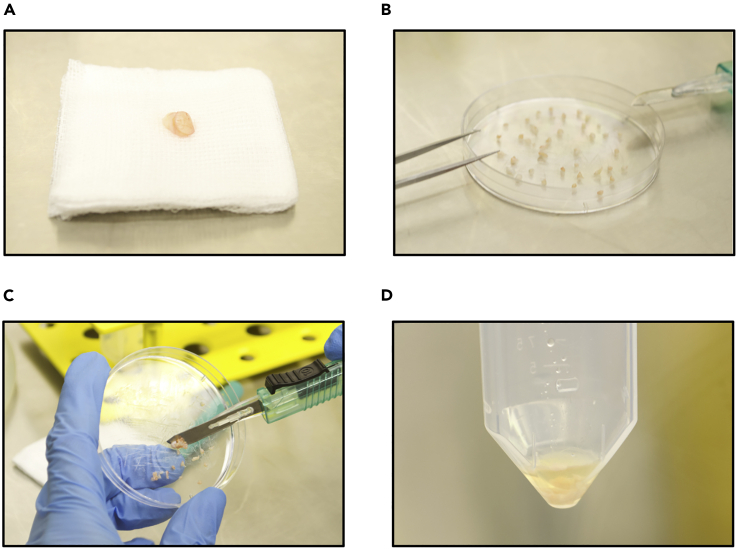
Figure 1.
Key steps of skin processing
(A) Representative skin biopsy on a saline-soaked compress immediately after excision.
(B) Biopsy in petri dish during dissection.
(C) Collection of minced tissue using a scalpel.
(D) Minced biopsy in gentleMACS C tube before attachment to dissociator.
Enzymatic and mechanical tissue dissociation
Timing: 195 min (with 15 min for step 5)
The purpose of this step is to digest the tissue first enzymatically in a water bath and then mechanically using the gentleMACS dissociator.
-
4.Perform the enzymatic digestion:
-
a.Transfer the gentleMACS C tube with the minced biopsy to a pre-heated water bath with shaker at 37°C at 220 RPM for 3 h.
-
b.Take out the gentleMACS C tube from the water bath, add 500 μL of cold tissue culture media (see materials and equipment) to the tube containing the digestion mix with the biopsy and close the lid tightly beyond the first resistance.
-
a.
Note: We use cold RPMI with 5% FCS, but also other culture media are appropriate, for instance also RPMI 10% FCS. This step is important to increase the volume of liquid for the dissociation step. It stops the digestions and also starts cooling down the cells. After dissociation, all further steps will be performed at 4°C.
Note: The digestion time is optimized for healthy normal skin, independently of biopsy size. Digestion time can be reduced in case of biopsy taken from lesional skin of patient suffering from inflammatory skin diseases (like psoriasis, atopic dermatitis, active cutaneous lupus and others) or cutaneous skin lymphomas. In some cases, the digestion time can be even reduced to 30 min, but the protocol has to be optimized according to the expected cellular density and the inflammatory stage of the disease, as well as the cell types of interest.
Optional: The incubation time in the water bath, especially when omitting Enzyme P, can be prolonged to 12–18 h to increase the cell yield. We strongly discourage prolonging the digestion time for single-cell RNA sequencing experiments.
-
5.Perform the mechanical digestion in the gentle MACS dissociator:
-
a.Place the tube upside down into the slot of the dissociator and check that the tube is correctly attached.
-
b.Select the program h_skin_01 and run it one time. Troubleshooting problem 2.
-
c.Carefully detach the gentleMACS C tube from the dissociator, the content should look opaque.
-
d.Place the gentleMACS C tube in the pre-cooled centrifuge and spin down the tissue for 1–2 min, 300 g).
-
a.
CRITICAL: This quick final centrifugation step is important to make sure that all tissue pieces and liquid droplets on the walls of the tube and on the lid are collected to the bottom of the tube. It is specially needed in case of smaller biopsies.
Note: The wide surface of the bottom part of the gentleMACS tubes allows a good exposure of all tissue to the digestion media. If the tubes and the dissociator are not available, then consider using a 50 mL Falcon tube with 1 ml digestion media for the enzymatic digestion in the water bath.
Prepare single-cell suspension
Timing: 20 min
The purpose of this step is to finalize the tissue preparation and obtain the skin-derived single cell suspension.
-
6.
Place a sterile 70 μM cell strainer on a fresh 50 mL Falcon tube.
-
7.
Take the gentleMACS C tube from the centrifuge, the content will look opaquer and more homogeneous (Figure 2A compared to Figure 1D). Briefly vortex it to loosen the tissue (5 s.
-
8.
Transfer the content of the gentleMACS C tube to the prepared cell strainer by quickly pouring the tube and tapping the tube over the cell strainer to transfer remaining tissue pieces.
-
9.
Transfer 10 mL of ice-cold wash buffer (PBS with 10% FCS) into the empty gentleMACS C tube to collect remaining cells.
-
10.
Rinse the cell strainer ten times with 1 mL of PBS 10% FCS wash buffer from the gentleMACS C tube. Apply rotatory movements to the strainer and the tube in order to make sure to wash the strainer throughout (Methods video S1, Rinsing the cell strainer) Troubleshooting problem 3.
CRITICAL: While rinsing the cell strainer, pay attention to rinse thoroughly in the edges as well.
-
11.
After all liquid has passed through, discard the cell strainer and close the Falcon tube.
Note: Connective tissue will remain inside the cell strainer (Figure 2B). This can be discarded. Further washing does not improve the cell yield.
-
12.
Spin down the cells in the pre-cooled centrifuge for 10 min at 300 g.
-
13.
Carefully aspirate the supernatant with a suction pump Troubleshooting problem 4.
CRITICAL: In our experience, the pellet is always hard to see at the bottom of the 50 mL Falcon tube. Therefore, when aspirating with the suction pump one has to be extra careful to aspirate only on the side of the tube and leave 10–20 μL of buffer.
Note: This protocol does not require a red blood cell lysis step, as in our experience the pellet (if visible) never looks red. Furthermore, the use of subcutaneous lidocaine and epinephrine as local anesthetic before taking the biopsy, minimizes the presence of red-blood cells in the tissue. Finally, small cells are excluded during the sorting in the lymphocytes gating.
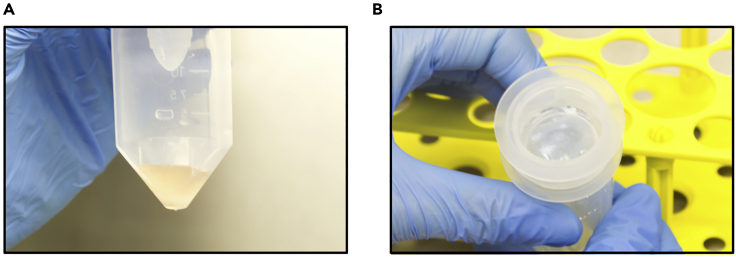
Figure 2.
Key steps of single-cell suspension preparation
(A) GentleMACS tube containing digested pieces of tissue after 3 h digestion processing through the dissociator and short spinning.
(B) White material remaining in the cell strainer after rinsing.
Staining of the cells and isolation of live cells
Timing: 45 min
The purpose of this step is to stain the single cell suspension with fluorochrome-labeled antibodies for downstream FACS-purification before 10× Genomics sequencing. The identification and sorting of viable cells are crucial to the protocol, whereas the quantification of the structural cells versus immune cells is facultative. We provide an example in which we used two different TotalSeq hashtag antibodies for skin cells and for PBMCs in order to isolate both cell types from the same individual in a single 10× Genomics reaction. Moreover, we provide an example of a FACS panel that can be applied to the skin cells during the sorting, to make sure that the cells of interest are present in the sorted population.
-
14.
Dislodge the pellet and resuspend cells in 50 μL of FACS-Sort Buffer.
-
15.
Add 1 μL of Fc-Block (see key resources table) to the 50 μL pellet and live 5 min at 20°C–24°C.
-
16.
Add 50 μL of the antibody mix prepared in FACS buffer.
Note: Considering that there was already 10–20 μL of liquid left before adding the 50 μL of Fc-block and the 50 μL of antibody mix, the final volume of the staining mix will be slightly more than 100 μL. This is not an issue, as the panel is designed to have an excess of antibodies and the staining is therefore not compromised.
Optional: The antibody mix does not necessarily have to be prepared before, as the antibodies can also be added freshly to the cells. In this case, the cells can be resuspended directly in 100 μL of FACS staining buffer and the Fc-Block can be added for first (2 μL instead of 1 μL to maintain the dilution). After 5 min (in case of using CITE-seq antibodies, we recommend to extend this incubation time to 15 min), all the rest of the antibodies can be added separately. Quickly vortex the cells after each antibody is added.
-
17.
Transfer the whole content into a new FACS-Sort tube and stain for 30 min at 4°C in the dark.
-
18.
Add 1 mL of FACS buffer, and centrifuge at 300 g for 5 min at 4°C.
-
19.
Resuspend in 200 μL of FACS buffer and keep the tube on wet ice until sorting.
Optional: We recommend to hashtag the skin or PBMCs cells simultaneously with the antibody staining to reduce the time until sample collection. We use 2 μL of Total-Seq Antibody (see key resources table) per 100 μL of skin single-cell suspension. If blood from the same donor is processed side by side, we recommend to isolate PBMCs on a Ficoll gradient, stain 1 million PBMCs for CD45+, add a different hashtag (see key resources table) and leave for 30 min at 4°C in the dark simultaneously to skin cells. For blood we use 1 μL of a different Total-Seq antibody in 100 μL of FACS-Sort buffer. Before sorting we exclude non-viable PBMCs with 7-AAD as for skin cells.
Note: If the PBMCs are processed together with the skin, we suggest to perform the PBMCs isolation at 20°C–24°C. We avoid red blood cell lysis for PBMCs, as we usually have a clear pellet after the Ficoll gradient. Moreover, due to the lymphocyte gate during FACS-purification, the small red blood cells are excluded from the sorting.
Note: We reduced the washing steps after antibody stain to a minimum of one. We prioritize cell yield to antibody specificity for single-cell analysis.
-
20.
Shortly before sorting the cells add 2 μL of 7-AAD dye directly to the cell suspension and briefly vortex the tube (5 s) Troubleshooting problem 5.
CRITICAL: The cells should be sorted as soon as possible.
Note: Before sorting the skin cells it is recommended to strain them again through a 40 μL cell-strainer. We skip this step, as we already strained the cells in point 9.-12. and in our experience, we do not need a second straining step.
Note: The manufacturers protocol recommends to wash cells 2 times after the hashtag. In our experience, a single wash-step is sufficient to obtain an almost perfect separation of skin and blood cells (see Figure 3C) and we therefore prefer to avoid a second centrifugation step as we prioritize timing and yield to antibody specificity. Moreover, further washing of the antibody happens while sorting itself, so once the different hashtag-labeled cells enter the same tube, there should not be free hashtag label antibodies in the mix and no further cell labeling should occur.
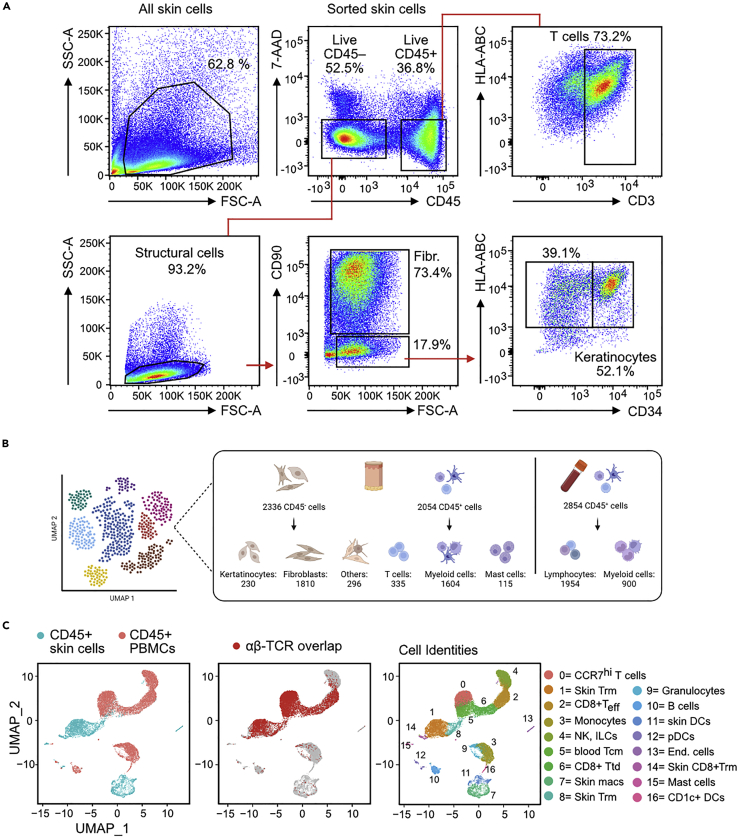
Figure 3.
Expected outcome of FACS-Sort and single-cell analysis of CD45+ skin and PBMCs
(A) FACS plot showing the gating of skin cells applied for sorting viable CD45+ or CD45– cells. The figure also shows how we assure that all main cell population of interest are included in the respective gates. For CD45+ cells we check the presence of T cells and non-T cells with CD3. For CD45– cells we check the presence of fibroblasts (CD90+) and other structural cells (like keratinocytes and endothelial cells) which are CD90+HLA-ABC+. In the sample shown, we also checked for CD34 expression to make sure to have follicular keratinocytes).
(B) Example of the number of single cell population sorted and sequenced after quality control upon a 10× Genomics run. The initial cell input according to the sorter was 10,000 CD45+, 10,000 CD45– and 10,000 PBMC.
(C) Left: UMAP visualization of CD45+ cells sequenced from the skin (light blue) and PBMCs (light red) after demultiplexing with the hashtags. Middle: single cells expressing αβ-TCR were mapped on the UMAP plot. Right: example of cell identity assignments for skin and blood cells. Figure reprinted with permission from Saluzzo et al. (2021).
Flow cytometry and cell sorting for 10× Genomics
Timing: 30 min
The purpose of this step is to quantify and sort viable cells into a small collection tube that can be directly subjected to further volume reduction to optimize the viability of the single cells. We perform our FACS-Sort purification with FACSAria III cytometer (BD Biosciences, Vienna, Austria). We sort 30,000 cells in total. Here we provide an example in which we sorted 15,000 CD45+ skin cells and 15,000 CD45+ PBMCs. Note that it is crucial that your sorting machine is running already before initiation of this step. We recommend to sort cells only once you have experience with flow cytometry of the skin biopsies. It is also crucial to perform the compensation setting for the FACS-Sort with multicolor labeled antibodies separately, to avoid further waiting time for the cells and immediately sort after antibody labeling. This protocol assumes a good knowledge of flow cytometry and cell sorting.
-
21.
Set the speed of cell/min as low as possible. Acquire 1000 cells and plot the cells on a forward scatter (FSC) versus side scatter (SSC) plot (Figure 3A).
-
22.
Set the gate on all lymphocytes and exclude doublets with FSC-height vs FSC-width.
Optional: further exclude duplets with the SSC-height vs SSC-area gate.
-
23.
Further plot the cells for viability using the 7AAD marker. Viable cells are 7-AAD negative (Figure 3A). Troubleshooting problem 6.
-
24.
Set a sorting gate for CD45+ cells and a second for CD45- cells (Figure 3A).
-
25.
Increase sorting speed at roughly 1000 cells/s. Collect sorted cells directly into PCR-tubes containing 20 μL sterile PBS with 0.08% BSA.
Note: Sorting into a PCR tube increases cells yield by avoiding a further centrifugation step. The combination of this protocol with sorting into the tube guarantees a very high yield and we highly recommend it.
Optional: To compare skin and circulating cells, hashtag-labeled CD45+ PBMCs from the same patient, can be sorted into the same tube as CD45+ cells from skin. Sorting of CD45+ PBMCs is critical to allow selection of viable cells and exclude erythrocytes and duplets via the lymphocytic gate. We provide an example in Figure 3B in which we sorted 10,000 CD45+ PBMCs, 10,000 CD45+ skin cells and 10,000 CD45- skin cells and in Figure 3C in which we sorted 15,000 CD45+ PBMCs and 15,000 CD45+ skin cells.
-
26.
Spin down the cells and resuspend them at a concentration between 700 to 1200 cells/μL to efficiently load the cells onto the chromium chip (10× Genomics) Troubleshooting problem 7.
CRITICAL: Run the cells through the chromium chip (10× Genomics) as soon as possible, as timing is crucial to guarantee cell viability at this point. We recommend to rather invest additional chromium chips for immediate runs and not wait for more samples to be sort-purified. Table 2 provides an example of how fast the number of captured cells after quality control is reduced if the samples are not immediately processed through the chromium chip after sorting.
Optional: After FACS-purification the buffer volume can reliably be reduced using the volume reduction device VR-NxT (Silicon Biosystems, Cat #: DA0650). Cells sorted into the appropriate PCR tube (Applied Biosystems MicroAmp Reaction tube, Cat #: N8010612) can be centrifuged for 10 min at 500 g in a centrifuge with swing out rotor. The tube can then be transferred into the VR-Nxt adaptor, closed with the cap (Silicon Biosystems VR-NxT Cap V1.0.0 Cat #: DA0665) following manufacturer’s instructions and volume reduction can be performed using the high-volume setup resulting in a final total volume of 15 μL supernatant, in which the cell pellet is resuspended before loading onto the chromium chip.
Note: If you wish to additionally sort stromal skin cells, consider that the ratio of CD45+ to CD45– cells in the healthy skin of the inner upper arm is roughly 1:3. In other regions of the body, or in lesional skin, this ratio will change. It is fundamental to perform flow cytometry and get familiar with the composition of the skin tissue of interest before proceeding with cell sorting and single cell sequencing. In the healthy skin we sort CD45+ and CD45– cells separately in order to achieve 15,000 CD45+ cells in a tube and 15,000 in a second tube, then we pool volumes together in order to maintain the desired proportion of cells. However, such calculations will of course depend on the aims of each specific experiment. An example of percentage of viable and sorted cells can be found in Figure 3A. An example of cellular yield with initial input of 10,000 CD45+, 10,000 CD45– and 10,000 PBMCs viable sorted cells can be found in Figure 3B.
Note: Our protocol is optimized for high quality cellular yield also thanks to the sorting step. In fact, this step allows to get rid of non-viable cells, which are still quite a high percentage (around 20%), exclude doublets, debris and erythrocytes. We therefore strongly recommend sorting before single cell sequencing. The sorting on CD45+ cells we suggest when studying inflammatory skin diseases or in general tissue resident memory T cells. However, of course, a “natural” CD45+/CD45– ratio is also important to maintain, for instance to study cell-cell interactions.
Table 2.
The yield of cells for single-cell analysis according to time
| Sample | Original counts of FACS-sorted cells | Single cells yield for analysis | Time after sorting |
|---|---|---|---|
| Number 1 | 15.000 | 6635 | 15 min |
| Number 2 | 15.000 | 1002 | 45 min |
| Number 3 | 15.000 | 243 | 120 min |
Expected outcomes
This single-cell isolation protocol is optimized to obtain all the skin cell types in healthy, non-inflamed skin. If dealing with healthy individuals, we highly recommend taking the skin biopsy always from the same and non-UV exposed part of the body to balance the experimental cohort. We use the inner upper arm. This will reduce the probability of individual factors to affect the results. Also keep in mind that the ratio of CD45+ to CD45– skin cells is in general around 1:3 for healthy skin, but it can be the opposite for inflamed tissue, like psoriasis or sarcoidosis (Ho and Kupper, 2019). Moreover, the yield can vary with age and gender of the individuals, so we recommend to select individuals accordingly.
With this protocol, the expected yield is > 20 000 viable CD45+ cells (30%–40% of total skin cells) from a single 6 mm diameter skin punch biopsy from healthy human skin (Figure 3A) (see also Troubleshooting problem 7). The use of multicolor flow cytometry during the sorting allows to check that the major population of interested are present in the final sorting tube and it is highly recommended (Table 1; Figure 3A) Troubleshooting problem 8.
Table 1.
Suggested flow cytometry panel to distinguish immune cells from stromal cells
| Antibody/Target | Conjugate/Dye | Amount used | Dilution |
|---|---|---|---|
| Anti- human CD45+ | PE-CF594 | 1 μL | 1:100 |
| Anti-human HLA-ABC | PE | 0.5 μL | 1:200 |
| Anti-human CD3 | BV421 | 0.5 μL | 1:200 |
| Anti-human CD90 | PE-Cy7 | 1 μL | 1:100 |
| Anti-human CD34 | AF-488 | 2 μL | 1:50 |
| FACS Buffer | 45 μL |
Total Volume of 50 μL Store at 4°C until use
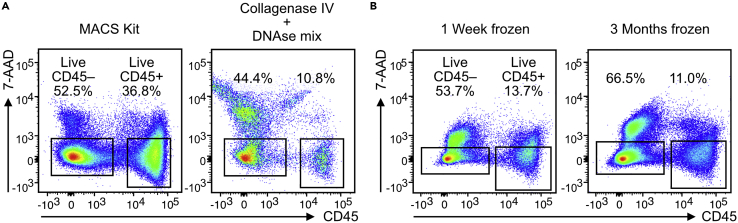
This protocol is superior to the one based on Collagenase IV/DNAse mixture for skin digestion often used in the literature (Strobl et al., 2021) (Figure 4A). We do not separate epidermis from dermis in this protocol, as we do not specifically focus on one compartment. Moreover, we think that other techniques are more useful to study epidermal cells specifically, like TissueFAXS (Tissuegnostics) or special transcriptomics. If one is interested in Langerhans cells only, we recommend other specific protocols for isolation (van den Berg et al., 2015).
Figure 4.
FACS-Sort outcomes with different protocols
(A) Comparison between MACS Kit and classical Collagenase IV/DNAse digestion mix for 6 mm biopsies.
(B) Cell yield after digestion of DMSO-Frozen biopsies at two time points, using the MACS Kit.
Another important feature of our protocol is that it conveniently allows to freeze the whole biopsy in DMSO-based freezing media (10% DMSO in FBS) using a cryogenic container that allows gradual temperature decrease as is the standard protocol for freezing single-cell suspensions. This allows storage of the biopsy at −80°C for months, although this might reduce the percentage of CD45+ viable cells (Figure 4B). After thawing, the biopsy can be processed as described in this protocol.
In case of processing smaller punch biopsies (like 3- or 4-mm diameter or half of a 6 mm biopsy) we follow the same procedures as for the 6 mm biopsy. During tissue dissection the size of the minced pieces does not change for smaller biopsies, they will just be reduced in number. As they can be easily seen by eye and collected with the scalpel, it is important to make sure to collect all pieces of dissected tissue. The digestion of skin biopsies from healthy non-inflamed tissue should also be kept of the same length as for 6 mm biopsies. Even if the use of 70 μm strainers of smaller diameter can be convenient for steps 6–13, the digested biopsy will have to be rinsed with less PBS to fit the 15 mL tube and this might reduce the yield. In our experience, carefully following this protocol for smaller biopsies results in a relatively comparable cell yield. However, we do not recommend to use smaller biopsies for scRNA-Seq from healthy non-inflamed skin, and limit smaller it for flow cytometry or bulk RNA-Seq. In case of inflamed tissue, the size of the biopsy can be reduced also for sc-RNA-Seq. As mentioned in the protocol, the digestion time can also be reduced in case of highly cellular inflammatory skin diseases (like in psoriasis, active cutaneous lupus lesions, atopic dermatitis etc) or cutaneous lymphomas etc. It is possible to use this protocol as orientation and optimize it for skin size and digestion length according not only to the disease type but also to its stage (for example active or quiescent lupus skin lesions).
This protocol allows not only an unbiased representation of all cell types in the skin, but also to modify the yield of the single cell population according to interest. For example, by pooling hashtagged skin and PBMCs together and performing scRNA-Seq coupled with scTCR-Seq we can obtain an optimal visualization of the single TCR clones shared between skin and blood in the same individual and identify their phenotype in health and disease as in Figure 3C, modified from Saluzzo et al., 2021.
Quantification and statistical analysis
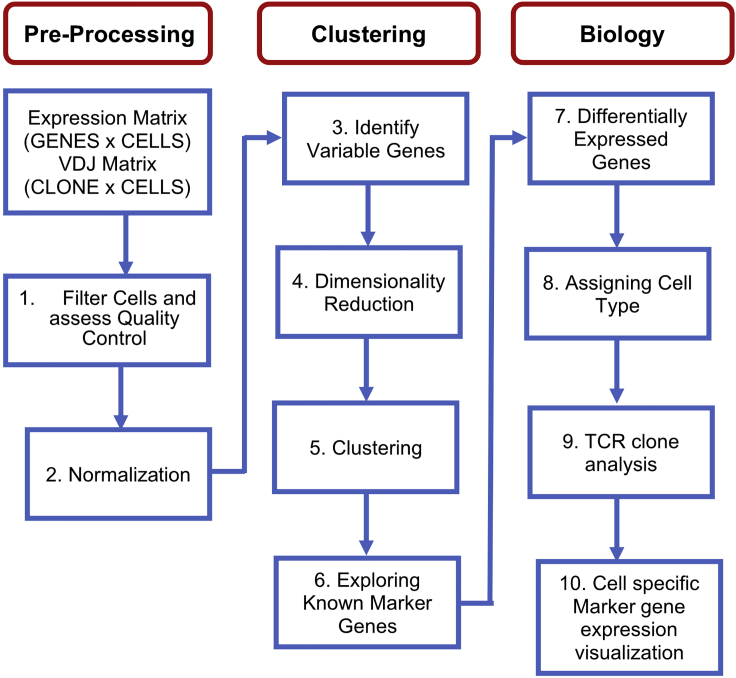
ScRNA-Seq analysis coupled with scTCR-analysis requires specialized statistical and computational methods. Here we suggest a short overview of the crucial steps to assess viability and originate a UMAP that overlay RNA and TCR sequencing data. Moreover, we provide the methods to track and display T cell clones trafficking between two different compartments, in this case skin and blood. Here we provide details for a typical bioinformatics workflow for 10× Genomics scRNA-Seq and scTCR-Seq analysis (Figure 5).
-
1.
Pre-processing: The hashtag oligo identification in order to separate skin and blood cells can be performed using the CITE-seq software (Stoeckius et al., 2017). For downstream 10× scRNA-Seq transcriptome and TCR data analysis, Seurat (v4.0.1) can be used (Stuart et al., 2019). For assessing the cell viability, we estimate the mitochondria gene fraction, and set the cut off for mitochondria genes to <15% to exclude low quality cells (apoptotic or lysing cells). We exclude doublets by using a maximum number of expressed genes, in this case > 3000 detected genes. We also excluded cells with <200 genes detected and those genes detected in < 3 cells as lowly expressed cells and genes respectively. The raw unique molecular identifier counts were scaled and normalized through log transformation.
Optional: An alternative or additional way to exclude doublets is by using hashing information and exclude those cells that are double positive for both hashtags.
Note: It is potentially possible that hashing de-multiplexing fails. In this case, it is still possible to re-assign the reads back to individual patients SNPs calling by using a SNP calling tools such as Freebayes (https://github.com/freebayes/freebayes).
-
2.
Clustering: for both skin and PBMCs we chose to select the top 2000 variable genes for principle-component analysis. Then, the top 15 principal components are selected for UMAP (Uniform Manifold Approximation and Projection) and clustering analysis with resolution 0.5. The Seurat function FindAllMarkers (logFC threshold = 0.25) can be used to find cluster- and sample-specific marker genes. At this point the UMAP clusters can be annotated by using the known marker genes and marker genes detected by function FindAllMarkers.
-
3.
Biology: Before the cluster (UMAPs) analysis, batch correction was performed with the ComBat function from sva (v3.32.1) (Leek et al., 2012) in the R package. To compute and create feature plots, violins plots and dot plots we usually adopt the Seurat functions FeaturePlot, VlnPlot, and DotPlot respectively, using default parameters. To analyze the single cell T cell receptor (TCR) clones from Skin and blood we suggest to use the scRepertoire (1.2.0) (Borcherding et al., 2020) R package. Shared clones between Skin and Blood can be visualized by using the alluvialClonotypes function from scRepertoire. A set of examples for all these graphical representations is available in our latest publication (Saluzzo et al., 2021).
Figure 5.
Bioinformatics workflow for 10× scRNA-seq and scTCR-seq data analysis
Limitations
Single-cell sequencing could be expensive to perform, in particular when using separate sequencing of multiple replicates from different donors. However, cells from different replicates can be hash-tagged and pooled after separately isolating single cells prior to 10× Genomics single-cell library preparation, therefore significantly reducing the sequencing costs. However, this approach has limitations, as it reduces the number of cells captured for each sample in the same pool. This is why this method has to be adopted in studies with clearly defined patients’ population and is best suitable for comparing healthy donors to diseased individuals (due to elimination of technical batch-effects). For studies focusing on specific cell types, as the example provided on tissue resident T cells, single-cell suspension generated in this protocol can be used to enrich specific cell types by sorting. Naturally, the information on the spatial organization of the cell types within the tissues is lost during cell preparation. Immunohistochemistry or Immunofluorescence staining can be used to validate the findings and observe the cells of interests in situ. Additionally, the combination of spatial transcriptomics (Marx, 2021) with single cell sequencing can be a powerful way of restoring a spatial resolution for skin samples and integrate it with single cell findings.
Troubleshooting
Problem 1
A high number of dead cells after the final single-cell suspension.
Potential solution
The possible reason can be letting the biopsy dry after collection or during dissection, not maintaining appropriate temperatures, slow execution of the protocol, long waiting time before sorting. Handle the pipetting steps gently, maintain the correct temperature and centrifugation speeds and follow the timings indicated in the protocol. During tissue preparation, make sure that the sample does not dry out by wetting the scalpel while cutting the biopsy and keeping a moist environment in the petri dish at any moment.
Problem 2
The gentleMACS C tube was not properly attached to the dissociator and the lid did not turn during running the program.
Potential solution
Re-attach the tube or use a different slot and run the program again.
Problem 3
The digested tissue is clogging the cell strainer and the liquid does not pass through.}.
Potential solution
In order to allow all the liquid to pass through, lift the cell strainer slightly, tilt it and push the edges to the inside of the Falcon tube to allow liquid to run through the sides of the strainer. Use a 1000 mL pipette to detach the connective tissue from the bottom of the strainer and then wash again with the washing buffer. Remaining liquid in the strainer can be aspirated by placing the 1000 mL pipette to the downside of the cell strainer and take up the liquid trough the strainer wall. A little amount of liquid will always remain in the strainer, which can then be discarded.
Problem 4
There is no visible pellet after centrifugation of the single-cell suspension.
Potential solution
The pellet is usually not well visible, since the expected yield is in the range of approximately 1–2 × 10ˆ5 cells. Be careful while aspirating the supernatant and resuspend the pellet well in the FACS staining buffer. Pay attention to transfer the complete liquid to the fresh FACS tube. If you want to double-check that cells are present in the FACS tube, take 1–2 μL of re-suspended cells, place the droplet on a cover slip and check under the microscope for the presence of cells.
Problem 5
The sample line clogs during sample acquisition.
Potential solution
The first potential problem is larger tissue pieces are present in the sample. Pass the sample through a 70 μM cell strainer and continue the acquisition. Another possibility is a too high concentration of the cell suspension. Add an appropriate volume (e.g., 100 μL) of cold FACS Buffer to the sample and proceed with acquisition.
Problem 6
Low yield of cells in the final single-cell suspension.
Potential solution
The possible reason could be errors in centrifugation, discarding of supernatant or resuspending the cell pellet. Maintain the correct centrifugation speed, make sure that the cell pellet is not disturbed while removing the supernatant (we suggest to live 10–20 μL of buffer if unsure about the pellet), and take care of resuspending the pellet gently in an appropriate buffer volume.
Problem 7
A low number of cells identified from single-cell sequencing after step 23.
Potential solution
The possible reason is an error in the cell counting leading to the loading of too few cells onto the microfluidic chip. Maintain the single-cell stock concentration between 700 to 1200 cells / μL before loading onto the 10× Chromium controller. Further, the target cell recovery can vary with tissue and cell types.
Problem 8
Not all expected cell types are present in the single-cell sequencing dataset.
Potential solution
The possible reason can be a wrong gating strategy during the cell sort. Carefully check test your gating strategy to ensure that all desired cell types are included in the sorting gates (see Figure 3). Be aware that especially structural cells like keratinocytes or fibroblasts are larger and require adequate gating in the forward and side scatter channels.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Georg Stary (georg.stary@meduniwien.ac.at).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank the Biomedical Sequencing Facility (BSF) at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences for assistance with next-generation sequencing. This research was funded in part by the Clinician Scientist Grant of the Austrian Society of Dermatology and Venereology (AP00871OFF) and by the Medical Scientific Fund of the Mayor of the City of Vienna (project number 18127). This work was further supported by funds from Oesterreichische Nationalbank (Austrian Central Bank, Anniversary Fund, project number 17872), by the Vienna Science and Technology Funds (LS18-058), and by the Austrian Science Fund (FWF; P31494).
Author contributions
S.S., L.M.G., M.F., and G.S. developed the protocol. S.S., L.M.G., and T.N. carried out experiments. S.S. made the illustrations and L.M.G. made the graphical abstract. R.V.P. analyzed single-cell sequencing data. S.S. and G.S. wrote the manuscript, with the help of all co-authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101470.
Contributor Information
Simona Saluzzo, Email: simona.saluzzo@meduniwien.ac.at.
Georg Stary, Email: georg.stary@meduniwien.ac.at.
Data and code availability
This study did not generate any unique datasets or code. The scRNA Seq data reported in Saluzzo et al. (2021) have been deposited at GEO and are publicly available as of the date of publication. NCBI GEO: GSE184320.
References
- van den Berg L.M., Cardinaud S., van der Aar A.M.G., Sprokholt J.K., de Jong M.A.W.P., Zijlstra-Willems E.M., Moris A., Geijtenbeek T.B.H. Langerhans cell–dendritic cell cross-talk via langerin and hyaluronic acid mediates antigen transfer and cross-presentation of HIV-1. J. Immunol. 2015;195:1763–1773. doi: 10.4049/jimmunol.1402356. [DOI] [PubMed] [Google Scholar]
- Borcherding N., Bormann N.L., Kraus G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:1–17. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.A. Resident memory T cells in human health and disease. Sci. Transl. Med. 2015;7:269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.W., Kupper T.S. T cells and the skin: from protective immunity to inflammatory skin disorders. Nat. Rev. Immunol. 2019;19:490–502. doi: 10.1038/s41577-019-0162-3. [DOI] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Method of the Year: spatially resolved transcriptomics. Nature Methods. 2021;18:9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- Saluzzo S., Pandey R.V., Gail L.M., Dingelmaier-Hovorka R., Kleissl L., Shaw L., Reininger B., Atzmüller D., Strobl J., Touzeau-Römer V., et al. Delayed antiretroviral therapy in HIV-infected individuals leads to irreversible depletion of skin- and mucosa-resident memory T cells. Immunity. 2021;54:2842–2858.e5. doi: 10.1016/j.immuni.2021.10.021. [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl J., Gail L.M., Kleissl L., Pandey R.V., Smejkal V., Huber J., Puxkandl V., Unterluggauer L., Dingelmaier-Hovorka R., Atzmüller D., et al. Human resident memory T cells exit the skin and mediate systemic Th2-driven inflammation. J. Exp. Med. 2021;218:e20210417. doi: 10.1084/jem.20210417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code. The scRNA Seq data reported in Saluzzo et al. (2021) have been deposited at GEO and are publicly available as of the date of publication. NCBI GEO: GSE184320.