Abstract
RNA extracts obtained from environmental samples are frequently contaminated with coextracted humic substances and DNA. It was demonstrated that the response in rRNA-targeted oligonucleotide probe hybridizations decreased as the concentrations of humic substances and DNA in RNA extracts increased. The decrease in hybridization signal in the presence of humic substances appeared to be due to saturation of the hybridization membrane with humic substances, resulting in a lower amount of target rRNA bound to the membrane. The decrease in hybridization response in the presence of low amounts of DNA may be the result of reduced rRNA target accessibility. The presence of high amounts of DNA in RNA extracts resulted in membrane saturation. Consistent with the observations for DNA contamination, the addition of poly(A) to RNA extracts, a common practice used to prepare RNA dilutions for membrane blotting, also reduced hybridization signals, likely because of reduced target accessibility and membrane saturation effects.
Hybridizations with oligonucleotide probes targeting rRNA extracted from environmental samples are commonly used to characterize microbial communities (e.g., references 6, 10, 11, 17, and 21). When membrane hybridization assays are used for the quantification of populations in complex microbial communities, in particular for the quantification of low-abundance populations, experimental conditions need to be optimized for detectability and precision (13, 14). Since RNA extracted from environmental samples is frequently contaminated with coextracted humic substances and DNA (3, 10, 11), it is necessary to evaluate the effects of the presence of these contaminants on quantitative membrane hybridizations. We present here results of experiments designed to accomplish this objective.
Humic substances are naturally occurring, heterogeneous organic substances that are yellow to black in color, of relatively high molecular weight, and resistant to degradation (1). They contain anionic functional groups (e.g., partially deprotonated phenolic and carboxylic groups), as well as hydrophobic components (aromatic and aliphatic moieties) (23). The coextraction of humic substances is particularly problematic when isolating RNA from soils, sediments, and water (10, 11). Humic substances have been shown to interfere with enzymatic digestion of DNA, with PCR amplification of DNA (18, 22, 26) and, to a lesser extent, with dot blot hybridization of DNA (24). Recent developments in nucleic acid extraction methods for environmental samples have focused on purification strategies to remove humic substances (7, 10, 27, 28). The effects of humic substances on RNA-targeted hybridizations have not, to our knowledge, been thoroughly addressed.
DNA can be present in RNA extracts in excess of the RNA recovered (3). Contaminating DNA is often digested from RNA extracts with DNase (10, 11). A concern with this approach is that some DNase preparations contain residual RNase activity and cause partial degradation of the RNA (24). Nucleases affect different regions of rRNA molecules to various degrees, and partial degradation of the rRNA molecules may have a serious influence on quantitative hybridization results (14). For example, hybridization results with universal probes are generally used to normalize the responses obtained with specific probes. Samples with partial rRNA degradation of target sites for universal probes (e.g., the 1390 region in the small-subunit rRNA, Escherichia coli numbering) may result in elevated responses (11, 14, 17).
Coextracted humic substances and DNA can be retained on hybridization membranes by two possible mechanisms: (i) by binding to the membrane directly or (ii) by interacting with RNA bound to the membrane. In the first scenario, hybridization signals may be decreased since humic substances and DNA occupy binding sites on the membrane, thus resulting in fewer binding sites for rRNA (this phenomenon is referred to as membrane saturation in this study). On the other hand, hybridization signals may be increased due to nonspecific binding of the oligonucleotide probes to the membrane-bound contaminants. In the second scenario, humic substances and DNA bound to membrane-immobilized rRNA may also result in an increased hybridization response due to nonspecific binding of the oligonucleotide probes. It is also possible, however, that they interfere with the hybridization of probes to target rRNA, thus reducing hybridization signals.
To evaluate the effects of the presence of coextracted humic substances and DNA on quantitative membrane hybridizations targeting rRNA, we first determined the hybridization response with uncontaminated RNA. In a previous study (13), it was demonstrated that Magna Charge, a 0.45-μm-pore-size charge-modified nylon membrane from Micron Separation, Inc. (Westboro, Mass.), exhibited better detectability and lower variability for oligonucleotide probe hybridizations compared to three other membrane types tested. Therefore, Magna Charge membranes were used for all experiments in this study.
Hybridization response with uncontaminated RNA.
In a first experiment, the hybridization response for immobilized RNA in the absence of DNA or humic substances was evaluated for Magna Charge membranes by hybridization with a 32P-labeled oligonucleotide probe. RNA was extracted from a pure culture of E. coli (harvested during exponential growth) using a low-pH, hot-phenol method (16, 21). It was determined that DNA was not coextracted with RNA using polyacrylamide gel electrophoresis (PAGE) (3) (data not shown). The RNA was denatured, diluted to different concentrations (using double-distilled water [ddH2O] containing 0.02 μl of 2% bromophenol blue per ml), and applied in triplicate to a Magna Charge membrane by slot blotting (15). Subsequently, the membrane was baked for 2 h at 80°C, hybridized with a 5′-end 32P-labeled universal probe (S-*-Univ-1390-a-A-18 [30]), and washed as previously described (15). The hybridized membrane was exposed to a phosphor storage screen (Molecular Dynamics, Sunnyvale, Calif.), which was scanned using a PhosphorImager (Molecular Dynamics). The image was quantified with the software package ImageQuant (Molecular Dynamics). Figure 1a shows the hybridization signals (expressed as the sum above background [SAB], the output obtained using ImageQuant) for the various amounts of RNA applied. The relationship between hybridization response and the amount of RNA applied is not linear, especially for the higher amounts of RNA applied. Figure 1b shows the change in slope between each one of two sequential data points in Fig. 1a. The slope starts to decrease between the datum points corresponding to 160 and 320 ng of RNA applied per slot. Two mechanisms may be responsible for this observation. First, the RNA binding capacity of the membrane may have been reached, suggesting that the decrease in slope was due to membrane saturation. Second, the accessibility of the target may have decreased when large amounts of RNA were applied. Regardless of the mechanism(s) responsible for the decrease in slope, a decrease in hybridization response appears to take place between applications of 26.7 and 53.5 ng of RNA/mm2 (the area of one slot is 6 mm2). For the lower amounts of RNA applied (< 10 ng of RNA per slot), the slope increases sharply. This behavior at low levels of radiation (“detectable reciprocity failure”) is an artifact of the phosphor imaging technology (20).
FIG. 1.
(a) Hybridization response for increasing amounts of RNA (nanograms of E. coli RNA applied per slot), expressed as SAB. (b) Change in slope between two sequential datum points in panel a.
For the experiments presented below, samples were diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml. Thus, since a sample volume of 50 μl was applied to each slot, 50 ng of poly(A) was added in addition to RNA, DNA, or humic acid. However, since the total amount of nucleic acid applied per slot was <160 ng, membrane saturation and/or target accessibility due to the presence of poly(A) should not be a concern for the experiments described below.
Effects of humic substances.
To evaluate the effect of the presence of humic substances in RNA extracts on membrane hybridizations, RNA samples extracted from pure cultures of Methanosarcina acetivorans or E. coli (harvested during exponential growth) were mixed with various amounts of humic acids (catalog no. h1,675-2; Aldrich, Milwaukee, Wisc.). The RNA-humic acids samples were denatured, diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml, and applied to Magna Charge membranes (10 ng of RNA per slot) as described above. The membranes were baked, hybridized, and washed using the oligonucleotide probes listed in Table 1 (except for probe S-D-Euca-0502-a-A-16). The hybridization signals were quantified as described above, and the results were expressed as a percentage of the hybridization response for RNA samples that did not contain humic acids (unamended).
TABLE 1.
Oligonucleotide probes, target group, pure culture RNA, and posthybridization wash temperature (TW)
| Probea | Target group | Pure culture RNA | TW (°C) | Reference |
|---|---|---|---|---|
| S-*-Univ-1390-a-A-18 | Virtually all organisms | 44 | 30 | |
| S-D-Euca-0502-a-A-16 | Domain Eucarya | Saccharomyces cerevisiae | 52 | 4 |
| S-D-Bact-0338-a-A-18 | Domain Bacteria | Escherichia coli K-12 | 54 | 4 |
| S-D-Arch-0915-a-A-20 | Domain Archaea | Methanosarcina acetivorans C2A | 56 | 5 |
| S-G-Msar-0821-a-A-24 | Genus Methanosarcina | Methanosarcina acetivorans C2A | 58 | 15 |
Oligonucleotide probe names are standardized according to the method of Alm et al. (2).
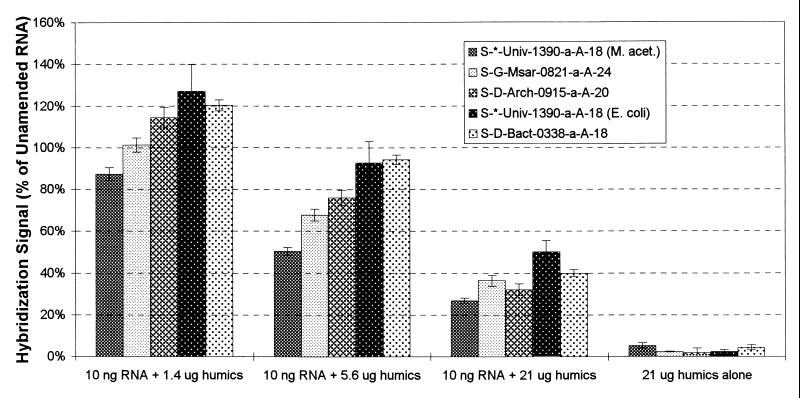
To estimate the amounts of soil humic substances that might be coextracted with 10 ng of RNA, the following assumptions were made: (i) 1 g of soil contains 1010 cells (8, 10, 25), (ii) a typical soil bacterium contains 5.7 × 10−15 g RNA (19), (iii) 80% of the organic carbon content in soils consists of humic substances (12), and (iv) all humic substances present in a sample are recovered during the extraction procedure. Amounts of humic acids corresponding to organic carbon contents of 1% (freshwater sediment [K. Nealson, personal communication], 1.4 μg of humic acids per 10 ng of RNA), 4% (coastal marine sediment [9], 5.6 μg of humic acids per 10 ng of RNA), and 15% (soil [A. Ogram, personal communication], 21 μg of humic acids per 10 ng of RNA) were used in this study. Note that the same four assumptions developed for soils were used for calculating the amounts of humic acids to be added to stimulate RNA extracts from freshwater and marine sediments. The results of this experiment were expressed as a percentage of the hybridization response for unamended RNA samples and are shown in Fig. 2.
FIG. 2.
Hybridization response for RNA amended with humic acids and for humic acids alone. The hybridization response is expressed as a percentage of the hybridization response obtained with RNA only.
The hybridization signals obtained for the 21-μg humic acid (without RNA) application ranged from 1.9 to 5.4% (average, 3.0%) of the hybridization responses obtained with 10 ng of unamended RNA, indicating that the oligonucleotide probes bind nonspecifically to humic acids to a low extent. The presence of 1.4 μg of humic acids did not have a significant effect on the hybridization signals as determined by a two-way analysis of variance (ANOVA) (P < 0.05). For humic acids amendments of >1.4 μg, the hybridization responses decreased (a 23.8% average decrease in the presence of 5.6 μg of humic acids and a 62.9% average decrease for 21 μg of humic acids). These decreases were significant as tested by a fixed two-way ANOVA (P < 0.05) (factor 1 = probe, factor 2 = humic acid level).
As discussed above, the decrease in hybridization signal due to the presence of humic substances may be caused by two mechanisms, membrane saturation or interactions between humic substances and RNA that affect probe hybridization. To determine whether the decrease in hybridization signal was due to membrane saturation, increasing concentrations of humic acids were added to radiolabeled RNA. Ten nanograms of radiolabeled RNA, amended with various amounts of humic acids, was denatured, diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml, and applied to membranes as described above. The membranes were baked, incubated with hybridization solution (without probe), and washed, and the radioactivity was quantified as presented above. The results of this experiment (Fig. 3) showed that radioactive signals were reduced by 18.0, 36.6, and 63.0% for humic acids amendments of 1.4, 5.6, and 21 μg, respectively. Since the levels of signal reduction for the humic acid amendments of 5.6 and 21 μg were similar to those observed in the previous experiment, the signal decreases appeared to be due to membrane saturation. It is unclear why the amendment of 1.4 μg of humic acids to 10 ng of RNA did not have a significant effect on the hybridization response (Fig. 2), whereas the addition of the same amount of humic acids to 10 ng of radiolabeled RNA resulted in an 18% decrease in radioactive signal (Fig. 3). It is possible that the nonspecific binding of probes to humic acids partially compensated the effects of membrane saturation.
FIG. 3.
Radioactive signal for increasing amounts of humic acids added to 10 ng of radiolabeled RNA and for humic acids alone. The signal is expressed as the SAB.
In summary, the presence of humic substances in RNA extracts lowers hybridization signals due to membrane saturation, i.e., by preventing the total amount of RNA applied from binding to the membrane. This effect is less significant for humic acid amounts of ≤1.4 μg per 10 ng of RNA applied to a membrane surface of 6 mm2 (i.e., one slot). On the other hand, small amounts of oligonucleotide probes can bind nonspecifically to humic substances, resulting in a slight increase in hybridization signals.
Effects of DNA.
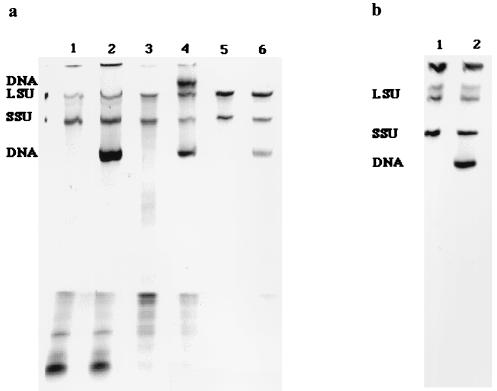
RNA extracts from environmental samples often contain high levels of coextracted DNA relative to the amounts of RNA recovered. For example, we determined that significant amounts of DNA were coextracted during a low-pH, hot-phenol RNA extraction of samples obtained from a coastal microbial mat (29), an anaerobic sewage sludge digester (16), and a solid waste digester (6). The different nucleic acid fractions were visualized after separation using PAGE (3) before and after exposure to DNase (FPLCpure DNase 1; Pharmacia, Piscataway, N.J. [catalog no. 27-0514-01]) (Fig. 4a). The presence of DNA in RNA extracts has not been considered during development of rRNA-targeted hybridization techniques because DNA is generally absent from pure culture RNA extracts, obtained using cells harvested during exponential growth. However, when hybridization protocols are used to quantify populations in environmental samples, in which cells are often in their stationary growth phase (high DNA/RNA ratio), the effect of the presence of DNA needs to be evaluated. To illustrate the difference between DNA levels in RNA extracts obtained from cells harvested in the exponential and stationary growth phases, we extracted RNA using a low-pH, hot-phenol method from Methanosaeta concilii cells harvested in exponential and stationary growth phases. The different nucleic acid fractions were visualized after separation with PAGE (Fig. 4b).
FIG. 4.
PAGE gels of nucleic acid samples extracted from various environmental samples. (a) Lanes 1 and 2, coastal marine microbial mat; lanes 3 and 4, anaerobic sewage sludge digester; lanes 5 and 6, solid waste digester. Samples in lanes 1, 3, and 5 were treated with FPLCpure DNase 1; samples in lanes 2, 4, and 6 are undigested controls. (b) Lane 1, Methanosaeta concilii 11 days after transfer into fresh medium (exponential growth phase); lane 2, M. concilii 25 days after transfer into fresh medium (stationary growth phase). LSU and SSU represent bands for the large rRNA of the large ribosomal subunit (23S and 23S-like rRNA) and for the small-subunit rRNA, respectively.
To evaluate the effect of the presence of DNA in RNA extracts on membrane hybridizations, RNA was removed from E. coli DNA (catalog no. D-2001; Sigma, St. Louis, Mo.) by RNase digestion (Ribonuclease 1 A; Pharmacia). Subsequently, the RNase was removed by one phenol-chloroform extraction, one chloroform extraction, and ethanol precipitation. Ten nanograms of RNA (without DNA) obtained using a low-pH, hot-phenol extraction from E. coli harvested during exponential growth was amended with 1, 10, or 100 ng of the RNase-treated DNA; denatured; diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml; applied to membranes; hybridized; and quantified as described above. In addition, hybridization results were obtained for 10 ng of RNA (unamended) and 10 and 100 ng of DNA. The results were expressed as a percentage of the hybridization response for unamended RNA samples and are presented in Fig. 5.
FIG. 5.
Hybridization response for RNA amended with DNA and for DNA alone. The hybridization response is expressed as a percentage of the hybridization response obtained with RNA only.
In contrast to the hybridization signals obtained with humic acids alone, the hybridization signals obtained with 10 and 100 ng DNA were low (values ranged from 0.0 to 2.1% [average, 0.8%] of the signal obtained with 10 ng of unamended RNA), indicating that oligonucleotide probes did not bind appreciably to DNA for the hybridization and wash conditions used in this experiment.
The effects of DNA contamination on hybridization signals were similar to those observed for humic substances. The presence of 1 ng of DNA did not have a statistically significant effect on the hybridization signals as determined by a two-way ANOVA (P < 0.05). As the DNA amendments increased to 10 and 100 ng, the average hybridization response decreased by 2.2 and 49.7%, respectively. These decreases were significant (P < 0.05) as tested by a fixed two-way ANOVA (factor 1 = probe, factor 2 = DNA level).
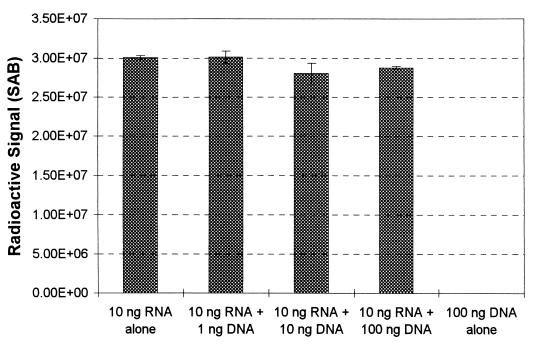
The possibility that the presence of DNA would result in membrane saturation was examined in a manner similar to the experiment described above for humic substances. Increasing amounts of DNA were added to 10 ng of 32P-labeled E. coli RNA, and the nucleic acids were denatured, diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml, and applied to membranes, which were processed as described above. The results of this experiment are shown in Fig. 6 and confirm that DNA (at least for the levels used here) did not prevent the binding of radiolabeled RNA to the membrane (the maximum reduction in signal was observed for an amendment of 10 ng of DNA and was only 6.7%). Since the level of signal reduction due to the addition of DNA to 10 ng of radiolabeled RNA was much lower than the effect of DNA contamination on hybridization signals, the mechanism by which DNA inhibits the hybridization response does not appear to be related to membrane saturation. It is possible that the presence of DNA reduces RNA target accessibility due to interactions between DNA and RNA, but further studies are needed to confirm this hypothesis.
FIG. 6.
Radioactive signal for increasing amounts of DNA added to 10 ng of radiolabeled RNA and for DNA alone. The signal is expressed as the SAB.
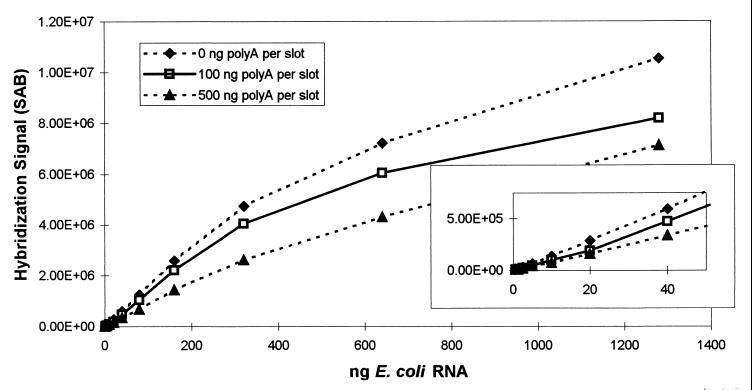
Our observations that the presence of low levels of DNA decreased hybridization signals and that membrane saturation and/or reduced target accessibility resulted in lower hybridization responses for RNA applied between 27 and 53 ng per mm2 prompted us to evaluate the effect of the addition of poly(A) to the dilution water used to prepare RNA samples. Historically, poly(A) has been included in dilution water to provide an alternative, irrelevant target for residual RNase activity. When RNA samples are diluted before blotting, the dilution water generally contains 1 ng of poly(A) per μl (15). Thus, since a 50- to 100-μl sample volume is blotted, 50 to 100 ng of poly(A) is applied with each sample in addition to the target RNA. To evaluate the effect of the presence of poly(A), dilution waters with three different concentrations of poly(A) (0, 1, and 5 ng/μl) were prepared. Various samples with different amounts of E. coli RNA were denatured, diluted with dilution water, applied in 100-μl volumes to membranes, and hybridized with probe S-*-Univ-1390-a-A-18. Hybridization signals were obtained as described above and are plotted in Fig. 7. The inset in Fig. 7 shows that the application of 100 ng of poly(A) per slot, together with low amounts of E. coli RNA (≤40 ng), resulted in lower hybridization signals than those obtained with E. coli RNA without poly(A). Since the total nucleic acid amounts in these samples were below the earlier determined amount of RNA that exhibited reduced hybridization responses (about 160 ng per slot or 27 ng per mm2), a mechanism other than membrane saturation must have been responsible for the decreased response. As hypothesized above, it is possible that the presence of DNA [poly(A)] reduced RNA target accessibility. For higher amounts of E. coli RNA (>40 ng) and for the poly(A) amendment of 500 ng per slot, the reduction in signal was more pronounced and may be explained by membrane saturation effects.
FIG. 7.
Hybridization response for increasing amounts of RNA, with or without poly(A) in the dilution water. The hybridization response is expressed as the SAB for increasing amounts of RNA applied to the membranes (nanograms of E. coli RNA applied per slot).
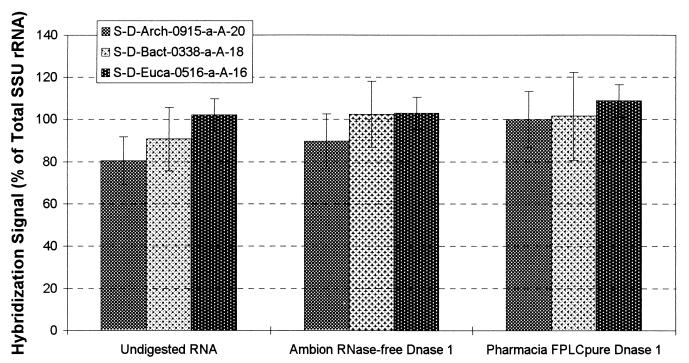
A logical solution to problems associated with high levels of coextracted DNA would be removal of DNA from RNA extracts by DNase digestion. Because of earlier reported concerns with residual RNase activity in DNase preparations, we evaluated the effect of a DNase digestion on hybridization response. Aliquots of pure culture RNA (Table 1) were exposed to two commercial RNase-free DNase preparations (RNase-free DNase 1, catalog no. 2222 [Ambion, Austin, Tex.]; FPLCpure DNase 1 [Pharmacia]). The DNase was removed by one phenol-chloroform extraction, one chloroform extraction, and ethanol precipitation. Then, 10 ng of DNase-exposed RNA was denatured, diluted with ddH2O containing 0.02 μl of 2% bromophenol blue and 1 μg of poly(A) per ml, applied to membranes, and hybridized as described above. Hybridization results obtained with domain-specific probes were expressed as a percentage of the hybridization response obtained with the universal probe. The relative hybridization responses obtained with the RNA samples exposed to the DNase preparations were increased by 3.5 to 14.2% compared to controls not exposed to DNase (Fig. 8). This result may be explained if the presence of small amounts of RNase caused partial degradation of the RNA (e.g., the target site of the universal probe). If so, this explains the increase in relative hybridization response observed in Fig. 8, since the universal probe was used to normalize the responses obtained with the domain-specific probes.
FIG. 8.
Hybridization response for RNA exposed to DNase. The hybridization response is expressed as a percentage of the response obtained with probe S-*-Univ-1390-a-A-18.
Discussion and concluding remarks.
Contaminating humic substances and DNA alter the results of quantitative membrane hybridizations by lowering the expected hybridization response. The presence of humic substances or high levels of DNA in RNA extracts likely results in saturation of hybridization membranes, which decreases the amount of target rRNA that is able to bind to the membranes. The presence of low levels of DNA does not contribute much to saturation but still reduces hybridization responses. This may be due to interactions between contaminating DNA and target rRNA, rendering target sites less accessible to oligonucleotide probes. Removing contaminating DNA by digestion with DNase should not be performed in most cases since DNase digestion may cause site-specific degradation, resulting in elevated specific responses. If large amounts of DNA contaminate an RNA sample, so that membrane saturation becomes a problem, then DNase treatment may be necessary. However, the consequences of site-specific degradation need to be taken into account. The addition of poly(A) to dilution water can also decrease hybridization signals, apparently due to mechanisms similar to those observed for native DNA, and is not recommended if the total amount of nucleic acids [target plus poly(A)] exceeds the binding capacity of the membrane.
When performing quantitative membrane hybridizations of environmental samples, normalizations of specific probe results with universal probe results are necessary. The data presented to date that have not been normalized should be viewed with caution. Since the decreases of hybridization signals due to membrane saturation by humics or DNA are fairly uniform for different target sites, this normalization approach should help to reduce biases caused by contamination by humic substances and DNA. In situations where target RNA has been subjected to site-specific degradation by RNase, however, normalizations will result in falsely elevated specific responses. Instead, greater care should be taken in obtaining RNA extractions without contamination by DNA, rather than relying on subsequent DNase treatment. Methods for removing contaminating humic substances and DNA from environmental RNA extracts, without causing (partial) degradation of rRNA, are needed to yield high-quality quantitative membrane hybridization results.
Acknowledgments
We thank Katherine McMahon for help with preparing Fig. 4, Scott McNaught for help with statistical analyses, and Dominic Frigon and Daniel Oerther for critical review of the manuscript.
This work was supported by grants from the U.S. National Science Foundation (BES 9410476) and from the U.S. Department of Agriculture (95-37500-1911).
REFERENCES
- 1.Aiken G R, McKnight D M, Wershaw R L, MacCarthy P. Humic substances in soil, sediment, and water. New York, N.Y: John Wiley & Sons; 1985. Appendix A. [Google Scholar]
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm E W, Stahl D A. Critical factors influencing the recovery and integrity of rRNA extracted from environmental samples: use of an optimized protocol to measure depth-related biomass distribution in freshwater sediments. J Microbiol Methods. 2000;40:153–162. doi: 10.1016/s0167-7012(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R I, Binder B J, Olson R J, Chisolm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin M E, McMahon K D, Mackie R I, Raskin L. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol Bioeng. 1998;57:342–355. doi: 10.1002/(sici)1097-0290(19980205)57:3<342::aid-bit11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Herrick J B, Madsen E L, Batt C A, Ghiorse W C. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl Environ Microbiol. 1993;59:687–694. doi: 10.1128/aem.59.3.687-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holben W E. Methods of soil analysis, part 2: microbiological and biochemical properties. Madison, Wis: SSSA; 1994. Isolation and purification of bacterial DNA from soils. [Google Scholar]
- 9.Martens C S, Klump J V. Biogeochemical cycling in an organic-rich coastal marine basin-I. Methane sediment-water exchange processes. Geochim Cosmochim Acta. 1980;44:471–490. [Google Scholar]
- 10.Moran M, Torsvik V L, Torsvik T, Hodsen R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogram A, Sun W, Brockman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashid M A. Geochemistry of marine humic compounds. N.Y: Springer-Verlag, New York, Inc.; 1985. [Google Scholar]
- 13.Raskin L, Capman W C, Kane M D, Rittmann B E, Stahl D A. Critical evaluation of membrane supports for use in quantitative hybridizations. Appl Environ Microbiol. 1996;62:300–303. doi: 10.1128/aem.62.1.300-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raskin L, Capman W C, Sharp R, Poulsen L K, Stahl D A. Molecular ecology of gastrointestinal ecosystems. In: Mackie R I, White B A, Isaacson R E, editors. Ecology and physiology of gastrointestinal microbes. 2. Gastrointestinal microbiology and host interactions. London, England: Chapman & Hall, Ltd.; 1997. pp. 243–398. [Google Scholar]
- 15.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raskin L, Zheng D, Griffin M E, Stroot P G, Misra P. Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Leeuwenhoek. 1995;68:297–308. doi: 10.1007/BF00874140. [DOI] [PubMed] [Google Scholar]
- 17.Risatti J, Capman W C, Stahl D A. Community structure of a microbial mat: the phylogenetic dimension. Proc Natl Acad Sci USA. 1994;91:10173–10177. doi: 10.1073/pnas.91.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochelle P A, Fry J C, Parkes R J, Weightman A J. DNA extraction for 16S rRNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett. 1992;100:59–66. doi: 10.1111/j.1574-6968.1992.tb14019.x. [DOI] [PubMed] [Google Scholar]
- 19.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 20.Southern E M. Methods for imaging radioactive samples in molecular biology. Oxford, England: British Electrophoresis Society; 1993. [Google Scholar]
- 21.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steffan R J, Goksøyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stumm W, Morgan J J. Aquatic chemistry, chemical equilibria and rates in natural waters. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 24.Tijssen P. Hybridization with nucleic acid probes, part I: theory and nucleic acid precipitation. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1993. [Google Scholar]
- 25.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai Y-L, Olson B H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young C C, Burghoff R L, Keim L G, Minak-Bernero V, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]