Abstract
Our purpose was to develop a rapid, inexpensive method of diagnosing the source of fecal pollution in water. In previous research, we identified Bacteroides-Prevotella ribosomal DNA (rDNA) PCR markers based on analysis. These markers length heterogeneity PCR and terminal restriction fragment length polymorphism distinguish cow from human feces. Here, we recovered 16S rDNA clones from natural waters that were close phylogenetic relatives of the markers. From the sequence data, we designed specific PCR primers that discriminate human and ruminant sources of fecal contamination.
The inability to identify the source of fecal contamination is partly to blame for the persistent problem of fecal pollution in coastal and inland waters. Although methods exist to quantify fecal pollution, none quickly and accurately identifies the animal source. Antibiotic resistance patterns of fecal streptococci (8, 16, 17) and Escherichia coli ribosomal DNA (rDNA) tracking (14; D. Akre and J. Wilcox, Northwest Algal Symp. Pacific Estuarine Res. Soc. Joint Meet., 1998) have recently emerged as potentially useful, but labor-intensive, solutions to the problem. Their reliability, however, may be considerably less than 100% (16, 17).
Unlike these methods, which require culturing indicator organisms, detection of host-specific molecular markers does not require culturing and holds promise as a precise, rapid method for identifying sources of fecal contamination. The Bacteroides-Prevotella group is one of several noncoliform bacterial groups that has been proposed as an alternative fecal pollution indicator (1, 5, 10), partly because of its abundance in feces. The use of molecular methods makes it more feasible to use anaerobic bacteria that are potentially difficult to grow, such as members of the Bacteroides-Prevotella group, as indicators.
We recently identified host-specific Bacteroides-Prevotella 16S rDNA markers for humans and cows by screening fecal DNAs by length heterogeneity PCR (LH-PCR) (15) or terminal restriction fragment length polymorphism (T-RFLP) (11) analysis (2). Cloning and sequencing experiments revealed that each marker comprised multiple sequences forming host-specific gene clusters. Here, we have identified additional clones, recovered from water samples, that cluster with the fecal clones. Using the sequences from fecal and water clones, we developed cluster-specific primers that can discriminate between human and ruminant feces.
Clones recovered from water samples.
To identify fecal Bacteroides-Prevotella rDNA markers in water, we collected six 1-liter water samples from areas in Tillamook Bay, Oreg., that are frequently contaminated with fecal pollution. We processed the samples as previously described (2). DNAs from each water sample were amplified with Bacteroides-Prevotella-specific primers (Bac32F and Bac708R) as described previously (2). Equal portions of PCR products from all water samples were pooled and cloned into pGEM T-Easy vectors according to the manufacturer's directions (Promega, Madison, Wis.).
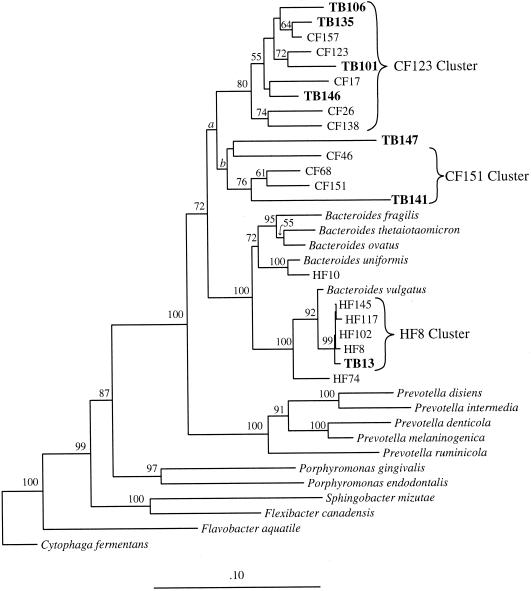
To locate marker clones, we screened 192 clones for LH-PCR and T-RFLP host-specific patterns, and we found 7 unique clones that corresponded to human or cow genetic markers previously identified (2). Clones with host-specific LH-PCR or T-RFLP patterns were sequenced as described elsewhere (2). All sequences were checked for chimeric structure with CHECK_CHIMERA of the Ribosomal Database Project (12) and by comparisons to other clones in our study. Similarities were calculated using the distance function in GCG, version 10 (Genetics Computer Group, Madison, Wis.), with the Kimura two-parameter correction. Sequence analysis of clones recovered from water samples revealed that they were all very similar, but not identical, to clones recovered from human and cow fecal samples (Fig. 1) (2).
FIG. 1.
Phylogenetic relationships among partial 16S rDNA sequences (558 positions) of clones recovered from Tillamook Bay water samples (TB). HF and CF are host-specific genetic markers identified from human and cow fecal clone libraries, respectively. The tree was inferred by neighbor joining. Numbers above the internal branches are percentages of bootstrap replicates that support the branching order. Bootstrap values below 50% are not shown. Bootstrap values for branches a and b dropped from 68 to 47 and 76 to 40, respectively, when TB147 was added to the analysis. The sequence from Cytophaga fermentans was used to root the tree.
Although previous analyses confirmed that noncontaminated water does not contain detectable Bacteroides-Prevotella DNA (2), we performed additional experiments to confirm that the clones recovered from water samples were fecal in origin. We designed primers specific to two of the water clones, TB141 and TB147, and amplified 16S rRNA genes from cow fecal DNAs. The methods for cow fecal sample collection and processing are presented elsewhere (2). Sequence analysis of the PCR products confirmed that the sequences were the same as the sequences of the two clones.
We aligned these clones with the fecal clones from our previous study and inferred a phylogenetic tree with the neighbor-joining algorithm (13) in PHYLIP, version 3.5c (4). Six of the seven clones recovered from water samples clustered with human- or cow-specific sequences identified in our earlier study (Fig. 1). TB13 corresponded to the human-specific cluster HF8 and was greater than 99% similar to other clones in this cluster. The TB13 sequence differed by only one or two bases from HF8, HF117, and HF145; these differences could be attributed to PCR or sequencing errors. The remaining clones corresponded to the cow-specific markers. TB141 had the same T-RFLP pattern as CF46, CF68, and CF151 and was 84.7 to 90.4% similar to the other CF151 clones. TB101, TB106, TB135, and TB146 had the same T-RFLP pattern as the other clones in the CF123 cluster and were 93.3 to 96.1% similar. TB147 had the same T-RFLP pattern as the clones in the CF123 cluster, but the sequence grouped with the CF151 cluster. Additionally, TB147 had the highest similarity with CF17 (88.2%), which is in the CF123 cluster. Bootstrap values for the CF151 cluster dropped considerably when TB147 was included in the analysis, suggesting that the branching order of TB147 is not strongly supported. It is unlikely that TB147 is a chimeric sequence since the same sequence was recovered from fecal and water samples independently.
Primer design.
To develop a PCR assay for identifying sources of fecal bacteria in water, we designed primers specific for each cluster and for clone HF10 (Table 1). We established specificity and optimal annealing temperatures for all primer pairs by using plasmid DNAs from target and closely related nontarget sequences as well as Bacteroides DNA from cultures (B. distasonis, B. fragilis, B. ovatus, B. thetaiotaomicron, B. uniformis, and B. vulgatus; all were gifts from A. Salyers). Additional confirmation of specificity was obtained through PROBE_MATCH of the Ribosomal Database Project. PCR mixtures were described by us previously (2). A thermal minicycler (MJ Research, Watertown, Mass.) was used for all reactions, with the following conditions: 25 cycles of 94°C for 30 s, appropriate annealing temperature (Table 1) for 30 s, and 72°C for 1 min followed by a final 6-min extension at 72°C. To increase the sensitivity of detection, 1 μl of each PCR product was reamplified using the same conditions. PCR products were visualized in a 1% agarose gel stained with 1 μg of ethidium bromide/ml.
TABLE 1.
Primers used in this studya
| Primer | Sequence (5′–3′) | Target | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| Bac32F | AACGCTAGCTACAGGCTT | Bacteroides-Prevotella | 53 | 2 |
| Bac708R | CAATCGGAGTTCTTCGTG | Bacteroides-Prevotella | 2 | |
| CF128F | CCAACYTTCCCGWTACTC | CF123 cluster | 58 | This study |
| CF193F | TATGAAAGCTCCGGCC | CF151 cluster | 55 | This study |
| HF134F | GCCGTCTACTCTTGGCC | HF10 | 61 | This study |
| HF183F | ATCATGAGTTCACATGTCCG | HF8 cluster, HF74 | 59 | This study |
| HF654R | CCTGCCTCTACTGTACTC | HF10 | 61 | This study |
Bac, Bacteroides-Prevotella; HF, human-specific; CF, cow-specific. Numbers correspond to the numbers of the E. coli 16S rRNA gene. All forward primers except HF134F were paired with Bac708R. HF134F was paired with HF654R. Annealing temperatures were empirically determined for each primer pair as described in the text.
Host-specific primers were further tested by amplifying fecal DNAs from target hosts (Table 2). DNAs from human and cow feces and sewage were collected and processed according to methods described elsewhere (2). We detected genes corresponding to the HF8 cluster in 11 of 13 human fecal samples, all of the sewage samples, and none of the cow fecal samples. Using the HF10-targeted primers, we detected PCR product in less than half of the sewage and human fecal samples and in one cow fecal sample. Because HF8 genes were more widely distributed among the humans and primers for HF10 were not as specific as desired, we tested only for HF8 genes in subsequent analyses. Genes from the CF151 and CF123 clusters were detected in all cow samples but in none of the human or sewage samples.
TABLE 2.
Distribution of host-specific genetic markers in feces from targeted hosts
| Target | No. of samples tested | No. of positive PCR resultsa
|
|||
|---|---|---|---|---|---|
| Human markers
|
Cow markers
|
||||
| HF8 cluster | HF10 cluster | CF123 cluster | CF151 cluster | ||
| Human | 13 | 11 | 6 | 0 | 0 |
| Sewage | 3 | 3 | 1 | 0 | 0 |
| Cow | 19 | 0 | 1 | 19 | 19 |
PCR results are from two rounds of 25 cycles each.
To determine the host specificity of these primers, we tested fecal samples collected from other animals (Table 3). Samples were collected with sterile utensils and placed in sterile 50-ml tubes or plastic bags, kept on ice for transport to the lab, and immediately stored at −80°C. Fecal DNAs were extracted using the Fast DNA kit for soil (Bio 101, Vista, Calif.), by following the manufacturer's directions. Samples were tested for marker genes by PCR. HF8 sequences were not detected in any samples (Table 3). CF123 and CF151 sequences, however, were detected in all ruminant animals and in llamas, which are members of the same order (Artiodactyla) but are considered pseudoruminants (3). A positive PCR result for CF123 or CF151, therefore, does not rule out wildlife sources, such as deer and elk, but land use evaluation could determine the likelihood of an agricultural or wildlife source.
TABLE 3.
Distribution of host-specific genetic markers in feces from nontarget animals
| Animal | No. of samples tested | No. of positive PCR resultsa
|
||
|---|---|---|---|---|
| Human marker
|
Cow markers
|
|||
| HF8 cluster | CF123 cluster | CF151 cluster | ||
| Cat | 3 | 0 | 0 | 0 |
| Deerb | 3 | 0 | 2 | 3 |
| Dog | 3 | 0 | 0 | 0 |
| Duck | 3 | 0 | 0 | 0 |
| Elkb | 3 | 0 | 3 | 3 |
| Goatb | 1 | 0 | 1 | 1 |
| Llamac | 1 | 0 | 1 | 1 |
| Pig | 3 | 0 | 0 | 0 |
| Seagull | 3 | 0 | 0 | 0 |
| Sheepb | 4 | 0 | 4 | 4 |
PCR results are from two rounds of 25 cycles each.
Ruminant.
Pseudoruminant.
PCR sensitivity.
Sensitivity of the PCRs was evaluated by amplifying marker genes from serial dilutions of plasmid DNAs from the clones CF123, CF68, and HF145. Detection limits were approximately 10−12 g of DNA (105 gene copies) for all three plasmid DNAs.
We also tested the sensitivity of our host-specific primers using serial dilutions of cow feces or raw sewage. Sensitivity assays were carried out as described elsewhere (2). DNAs from each dilution were tested for the markers by PCR. We measured fecal coliforms in each dilution according to standard methods (7).
Detection of CF123 genes was as sensitive as detection of fecal coliforms (Table 4). Detection of fecal coliforms, however, was 10- to 100-fold more sensitive than detection of CF151 and HF8 genes. The sensitivity assay using cow fecal dilutions was repeated with feces from different cows, and similar results were obtained (Table 4). Although the results varied slightly, we believe that these differences are not significant. Some of the variability may be due to uneven dispersion of cells during fecal suspension and dilution. In addition, because we are not currently able to measure the exact number of the marker genes in a fecal sample and there may be individual variability, these limits of detection represent approximations.
TABLE 4.
Detection limits of host-specific genetic markers and fecal coliformsa
| Source of DNA | Detection limit (g of dry feces/liter)
|
|||
|---|---|---|---|---|
| HF8 cluster | CF123 cluster | CF151 cluster | Fecal coliforms | |
| Cow feces A | ND | 2.8 × 10−7 | 2.8 × 10−5 | 2.8 × 10−7 |
| Cow feces B | ND | 3.6 × 10−6 | 3.6 × 10−5 | 3.6 × 10−6 |
| Sewage | 1.4 × 10−6 | ND | ND | 1.4 × 10−7 |
Results are from dilution assays using either cow feces or raw sewage. Each cow sample combined feces from four cows. The sensitivity of detection of cow feces was measured twice, with two independent samples (A and B). Sewage dilutions were not replicated. Results for detection of the genetic markers are from two rounds (25 cycles each) of PCR. ND, not determined.
If the detection limit of 105 gene copies using plasmid DNAs is extrapolated to the detection results from the serial dilutions of feces, then we must assume that 2 × 10−6 g of cow feces (the average sensitivity for cow feces samples A and B in Table 4) contains at least 105 gene copies. This translates to 5 × 1010 copies/g of feces. Assuming an average of 3 × 1011 bacterial cells/g of feces (6) and an average of five 16S rDNA operons per Bacteroides cell (rRNA Operon Copy Number Collection [http://rdp.cme.msu.edu/rrn/]), then 5 × 1010 copies/g of feces represents 3% of the total bacteria. If Bacteroides cells comprise 30% of the total fecal bacteria (9), we estimate a density of 1011 Bacteroides cells/g of feces; based on this estimate, the host-specific markers would represent 10% of the Bacteroides cells. This estimate seems reasonable, especially considering potential errors associated with pipetting fecal slurries.
These detection limits are similar to other estimates of the contribution of host-specific marker genes to total Bacteroides cells. We calculated the relative abundance of the host-specific LH-PCR peak for the CF151 cluster (2), compared to the relative abundance of total Bacteroides PCR amplicons. The relative fluorescence of the host-specific peak (the area under the peak relative to the total area) was approximately 7% of the total Bacteroides PCR products (data not shown). Additionally, marker sequences recovered from the Tillamook Bay clone library comprised 4% of all Bacteroides clones, which is consistent with the percentages of marker sequences found in our human and cow fecal clone libraries (3.1 and 6.3%, respectively) (2).
Although extensive field testing is required to determine the efficacy of the assays and the geographic distribution of the host-specific markers before these markers can be used for routine water quality monitoring, we believe that these PCR assays provide a promising diagnostic tool for identifying nonpoint sources of fecal pollution. Additionally, our approach for the identification of diagnostic markers can be easily applied to find markers for animals besides humans and ruminants.
Nucleotide sequence accession numbers.
The sequences described in this paper have been submitted to GenBank with accession numbers AF294903, Af294904, AF294905, AF294906, AF294907, AF294908, and AF294909.
Acknowledgments
We are grateful for assistance from Weerathep Pongprasert, Mike Rappé, Nancy Ritchie, and Kevin Vergin.
This work was partially supported by grant NA76RG0476 (project no. R/ECO-04) from the National Oceanic and Atmospheric Administration to the Oregon State University Sea Grant College Program, by appropriations made by the Oregon State legislature, and by grant R827639-01-0 from the U.S. Environmental Protection Agency. This work was also supported by the Research Council of Oregon State University.
REFERENCES
- 1.Allsop K, Stickler J D. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J Appl Bacteriol. 1985;58:95–99. doi: 10.1111/j.1365-2672.1985.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard A E, Field K G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol. 2000;66:1587–1594. doi: 10.1128/aem.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehority B A. Foregut fermentation. In: Mackie R I, White B A, editors. Gastrointestinal microbiology. Vol. 1. New York, N.Y: Chapman & Hall, International Thomson Publishing; 1997. p. 658. [Google Scholar]
- 4.Felsenstein J. PHYLIP—phylogeny inference package (v3.5) Cladistics. 1989;5:164–166. [Google Scholar]
- 5.Fiksdal L, Maki J S, LaCroix S J, Staley J T. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl Environ Microbiol. 1985;49:148–150. doi: 10.1128/aem.49.1.148-150.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 8.Hagedorn C, Robinson S L, Filtz J R, Grubbs S M, Angier T A, Reneau R B., Jr Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl Environ Microbiol. 1999;65:5522–5531. doi: 10.1128/aem.65.12.5522-5531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdeman L V, Good I J, Moore W E C. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl Environ Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Samadpour M, Chechowitz N. Little Soos Creek microbial source tracking: a survey. Seattle: Department of Environmental Health, University of Washington; 1995. [Google Scholar]
- 15.Suzuki M, Rappé M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl Environ Microbiol. 1996;62:3997–4002. doi: 10.1128/aem.62.11.3997-4002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiggins B A, Andrews R W, Conway R A, Corr C L, Dobratz E J, Dougherty D P, Eppard J R, Knupp S R, Limjoco M C, Mettenburg J M, Rinehardt J M, Sonsino J, Torrijos R L, Zimmerman M E. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl Environ Microbiol. 1999;65:3483–3486. doi: 10.1128/aem.65.8.3483-3486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]