Abstract
Shear wave elastography (SWE) is an ultrasound‐based stiffness quantification technology that is used for noninvasive liver fibrosis assessment. However, despite widescale clinical adoption, SWE is largely unused by preclinical researchers and drug developers for studies of liver disease progression in small animal models due to significant experimental, technical, and reproducibility challenges. Therefore, the aim of this work was to develop a tool designed specifically for assessing liver stiffness and echogenicity in small animals to better enable longitudinal preclinical studies. A high‐frequency linear array transducer (12‐24 MHz) was integrated into a robotic small animal ultrasound system (Vega; SonoVol, Inc., Durham, NC) to perform liver stiffness and echogenicity measurements in three dimensions. The instrument was validated with tissue‐mimicking phantoms and a mouse model of nonalcoholic steatohepatitis. Female C57BL/6J mice (n = 40) were placed on choline‐deficient, L‐amino acid‐defined, high‐fat diet and imaged longitudinally for 15 weeks. A subset was sacrificed after each imaging timepoint (n = 5) for histological validation, and analyses of receiver operating characteristic (ROC) curves were performed. Results demonstrated that robotic measurements of echogenicity and stiffness were most strongly correlated with macrovesicular steatosis (R2 = 0.891) and fibrosis (R2 = 0.839), respectively. For diagnostic classification of fibrosis (Ishak score), areas under ROC (AUROCs) curves were 0.969 for ≥Ishak1, 0.984 for ≥Ishak2, 0.980 for ≥Ishak3, and 0.969 for ≥Ishak4. For classification of macrovesicular steatosis (S‐score), AUROCs were 1.00 for ≥S2 and 0.997 for ≥S3. Average scanning and analysis time was <5 minutes/liver. Conclusion: Robotic SWE in small animals is feasible and sensitive to small changes in liver disease state, facilitating in vivo staging of rodent liver disease with minimal sonographic expertise.
Shear wave elastography (SWE) is an ultrasound‐based stiffness quantification technology that is used routinely for noninvasive liver fibrosis assessment in the clinic, but is not widely utilized in rodent models. The aim of this work was to develop a new robotic SWE platform designed specifically for use in small animals to better enable longitudinal preclinical studies. The instrument was validated in a mouse model of nonalcoholic steatohepatitis (NASH), and demonstrated strong correlation with histologic scores of fibrosis and steatosis.

Ultrasound‐based shear wave elastography (SWE) is a revolutionary imaging technique that noninvasively measures the mechanical stiffness of biological tissue, such as liver, in vivo.( 1 ) In SWE, a high‐intensity ultrasonic pulse is used to generate transverse mechanical waves in the tissue (i.e., shear waves), whose speed is directly proportional to the underlying tissue stiffness.( 2 ) Over the past decade, clinical research has demonstrated that SWE and other ultrasound readouts (such as echogenicity, backscatter coefficient, and/or attenuation) are safe and effective at diagnosing a multitude of liver pathologies in humans, including steatosis,( 3 ) fibrosis/cirrhosis,( 4 ) and cancer,( 5 ) and have greatly reduced the reliance on invasive biopsy.( 6 ) While the integration of SWE, and corollary technologies such as transient elastography, in clinical practice has progressed at a robust pace, there has been a notable lack of adoption of these technologies in preclinical research, despite a growing list of publications demonstrating the feasibility of the approach in rodent models (mice( 7 , 8 , 9 ) and rats( 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 )) and relevancy of these models to human liver stiffness measurements.( 24 ) Basic science laboratories still mostly rely on histology to assess hepatic injury and response to therapy,( 25 , 26 , 27 , 28 ) which hampers longitudinal studies because it requires subject endpoints, thus increasing the individuals enrolled and costs.
Several challenges present themselves when considering “reverse translation” of SWE technology from humans to small animals. First, in small animals, higher ultrasonic frequencies (15 MHz or greater) are necessary to achieve adequate resolution to visualize the liver, and generating SWE pulses at these frequencies becomes nontrivial due to transducer heating challenges and bandwidth requirements. As a point of comparison, human livers, which are hundreds of times larger than rodent livers, are typically scanned with ultrasonic frequencies of 2‐3 MHz to achieve appropriate penetration depths. Second, SWE measurements can be susceptible to variations in the environment and operator skill (e.g., errors arising from precompression of the tissue, incorrect or inconsistent transducer placement, and breathing artifacts) that can bias measurements and introduce error.( 29 ) Precompression, in particular, is difficult to control in small animals due to the very short distance of organs from the skin surface and small size of organs. Finally, typical SWE configurations are limited to small two‐dimensional (2D) areas of the liver and can be susceptible to sampling error. Even expert radiographers can display substantial data variance when scanning small animals by hand without extensive training and standard operating procedures.( 22 ) Therefore, the optimal SWE instrument for small‐animal research should include automated (i.e., hands‐free), noncontact (i.e., without operator precompression), and three‐dimensional (3D) whole‐organ sampling to ensure the highest degree of reproducibility and accuracy.
The purpose of this work is to address these limitations and increase the accessibility of SWE to the liver‐disease research community by developing a robotically controlled SWE imaging platform for small animals. In this system, the ultrasound scanning direction is inverted, and hands‐free SWE is achieved with rodents in the prone position using a robotically actuated high frequency transducer. Validation is performed with studies in tissue‐mimicking phantoms, as well as in vivo in a diet‐induced nonalcoholic steatohepatitis (NASH) mouse model scanned longitudinally with histological endpoints.
Materials and Methods
Imaging System Design
The imaging system used was a Vega robotic ultrasound imager (SonoVol).( 30 ) Multiple track location SWE was implemented on a broadband linear array (12‐24 MHz) mounted in the translation carriage that allowed wide‐range lateral and elevation travel (15.25 × 15.25 cm). The translation carriage and transducers were submersed in a proprietary dielectric coolant, which served both as a heat sink and conduit for sound propagation into the animal’s abdomen. The animal was separated from the fluid chamber through an acoustically transmissive membrane (Fig. 1A). Two stepper motors controlling a pulley system were connected to high‐resolution encoders to allow for precise positioning information to be collected simultaneously with ultrasound data. Robotic movement of the transducers and 3D B‐mode data capture were coordinated with SonoEQ v1.5.2 (SonoVol), which is built on top of 3D Slicer.( 31 , 32 ) The data‐acquisition workflow proceeded as follows: (1) Capture a wide‐field B‐mode “scout scan”; (2) reconstruct a 3D volume; (3) visualize the liver; and (4) place fiducial markers in 3D space that indicate the position of the desired SWE capture. After placing multiple fiducials, the software would then jog the linear array to the appropriate position in space, allowing the user to collect SWE data at the desired locations (Fig. 1B,C). SWE data capture was then initiated through a separate custom graphical user interface written in MATLAB (The MathWorks, Natick, MA) to activate SWE pulse sequences on the Vega imager and render SWE images to the screen.
FIG. 1.
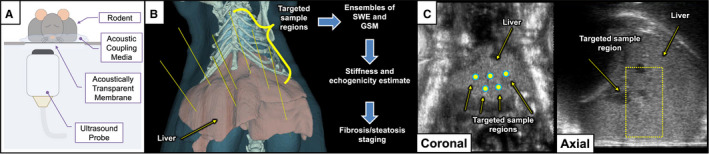
Illustrations of data acquisition system and SWE protocol. (A) Experimental setup depicting anesthetized rodent in prone position on acoustically transparent membrane with robotic ultrasound transducer imaging from below. (B) SWE/GSM measurements are made at any number of targeted sample regions to measure liver fibrosis and steatosis. (C) B‐mode images (coronal and axial) reconstructed from 3D volumetric data are shown depicting the liver with manually placed SWE positions. Extent of SWE range‐gate is indicated with a dashed yellow outline.
SWE excitation and tracking pulses were centered at 12 and 17 MHz, respectively, with 4‐times parallel receive beamforming, 70V transmit voltage, and 3‐kHz multiplexed pulse repetition frequency. Raw beamformed 16‐bit radiofrequency (RF) lines sampled at 60 MHz were saved to disk for off‐line shear wave speed (SWS) estimation. RF data were processed in MATLAB to calculate SWS using several previously published techniques, including normalized cross‐correlation‐based motion tracking,( 33 ) time‐to‐peak linear least squares group SWS estimation,( 34 ) and reflected‐wave filtering.( 35 ) Finally, SWS was converted to Young’s modulus (YM) following standard equations( 34 ) and rendered as a 2D colored overlay on the B‐mode image. In addition, grayscale median (GSM) values were computed from the first RF line in the SWE ensembles to evaluate spatially matched liver echogenicity. GSM processing included envelope detection of RF lines (absolute value of Hilbert transform) followed by a square‐root operation for compression.
Phantom Experiments
Measurements of YM using the system were first validated in tissue‐mimicking phantoms by comparing against a third‐party commercial SWE‐enabled ultrasound system, the Vantage 256 (Verasonics, Inc., Redmond, WA). Two experiments were conducted: one in commercially available tissue‐mimicking phantoms (Model 039; CIRS, Norfolk, VA) and the other in custom fabricated gelatin phantoms. The commercial phantom set included four phantoms with nominal YM stiffnesses of 1.9 kPa, 7.8 kPa, 18.1 kPa, and 38.1 kPa, as reported by the manufacturer. Custom gelatin phantoms (n = 10) were fabricated following well‐established recipes( 36 ) to span a YM range of 1 to 12 kPa and congealed in 50‐mL syringes to better simulate the cylindrical geometry of a mouse abdomen as compared with a large geometry of the commercially available phantoms. After removal from the syringe body, measurements were captured from 10 unique locations per phantom, and the median YM value was computed using custom processing code mentioned previously. Subsequently, SWE measurements with the Vantage system were made in a similar spatial location with an L7‐4 linear array (Philips ATL, Bothell, WA) using plane‐wave imaging, and processed using open‐source SWE code developed by the Radiological Society of North America’s Quantitative Imaging Biomarker Alliance.( 37 )
Animal Experiments
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of North Carolina at Chapel Hill and followed the National Institutes of Health guidelines for animal care. A total of 40 wild‐type mice (11‐week‐old, C57BL/6J females) were purchased from the Jackson Laboratory (Bar Harbor, ME) and were allowed a 2‐week acclimation period before the start of any imaging or experimental diet initialization. For the duration of the study, mice were maintained on a 12‐hour light/dark schedule with ad libitum access to food and water.
Experimental Diet
After the 2‐week acclimation period, and initial control imaging timepoint, mice were switched from standard chow to a choline‐deficient, L‐amino acid‐defined, high‐fat diet (CDAHFD) to induce NASH.( 38 ) The CDAHFD consisted of pellets formulated for 60 kcal% fat, 0.1% methionine by weight, and no added choline (A06071302; Research Diets, New Brunswick, NJ). Food pellets were placed in a Petri dish on the cage floor, as opposed to the standard top‐loaded food hopper, to prevent pellet crumbling and waste. All animals were fed the same CDAHFD formulation, with all animals serving as their own controls, as they were switched to CDAHFD at the same time.
Imaging Timepoints
Mice were imaged at baseline (week 0) before starting the experimental diet, and then seven times more after the start of diet (week 1, 2, 3, 6, 9, 12, and 15) for a total of eight imaging timepoints. Imaging was performed by anesthetizing the mice with vaporized isoflurane (3% induction, 1.5% maintenance for duration of imaging), shaving and depilating the abdomen, and orienting in the prone position on the acoustic membrane of the imaging device with a thin layer of water for coupling (Fig. 1A). Water was used instead of ultrasound gel due to improved air bubble clearance and ease of cleanup. Mouse body temperature was maintained by an overhead heat lamp. Between 6‐8 2D SWE captures were acquired per liver at various positions in the left and right lateral lobes below the xiphoid cartilage with five repeated acquisitions at each location for averaging (for a total of 30‐40 SWE images per liver). After each imaging timepoint, one cage of animals (i.e., n = 5 mice) was sacrificed for histological validation of noninvasive ultrasound measures.
Histological Assessment
Following necropsy (performed within 6 hours of each imaging session), sections of liver lobes were submersion fixed in 10% neutral buffered formalin at room temperature for 48 hours. Fixed tissues were processed, paraffin embedded, sectioned at 5‐µm thickness, and stained with hematoxylin and eosin (H&E) and picrosirius red. Pathology slides were digitized (ScanScope XT; Aperio, Sausalito, CA) at 20× magnification and scored by a board‐certified veterinary pathologist blinded to the SWE results on semi‐quantitative scales for fibrosis, steatosis (both microvesicular and macrovesicular), hypertrophy, and lobular inflammation (see Table 1 for pathology scoring rubric). Fibrosis scoring was based on an adapted Ishak scheme,( 39 ) whereas steatosis and inflammation was based on the rodent NAFLD scheme.( 40 ) Steatosis (S) score was based on percent of image area made up of fat vesicles, not the percent of cells that have fat vesicles. Additionally, a zonation criterion was added to the steatosis score system (i.e., periportal vs. diffuse),( 41 ) which added a further classification category referred to as “S4” in this manuscript. For inflammation score, foci of inflammation were calculated from an average of four 100× fields (where foci contained at least five inflammatory cells).
TABLE 1.
Pathology Grading Criteria
| Fibrosis Stage | F Score | Inflammation | L Score |
|---|---|---|---|
| No fibrosis | 0 | <0.5 | 0 |
| Early fibrous expansion emanating from some portal areas ± short fibrous septa; increased short, disorganized fibrous septa along some sinusoids | 1 | 0.5 ‐ 1.0 | 1 |
| Fibrous expansion emanating from most portal areas; network of loose fibrous septae coalescing along numerous sinusoids | 2 | 1.0 ‐ 2.0 | 2 |
| Fibrous expansion of portal areas with occasional early portal‐portal bridging, which often coalesces with sinusoidal fibrous network (most prominent in liver lobule zone I) | 3 | >2.0 | 3 |
| Fibrous expansion of portal areas with common portal‐portal bridging and some portal‐central vein bridging, which coalesces with sinusoidal fibrosis most prominent in liver lobule zones I and II | 4 | Steatosis (micro/macro) Hypertrophy | S score |
| More prominent bridging, occasional nodule formation (incomplete cirrhosis) | 5 | <5% | 0 |
| Cirrhosis with marked bridging and nodules | 6 | 5%‐33% (periportal) | 1 |
| 34%‐66% (periportal) | 2 | ||
| >66% (periportal) | 3 | ||
| >66% (diffuse) | 4 |
Statistical Methods
All statistical analyses were carried out with MATLAB 2017a. Continuous variables were expressed as mean ± SD or median values with interquartile range. Linear regression and Bland‐Altman analysis were used to examine associations between variables, including computations of Pearson correlation (r) and coefficient of determination (R2). Pairwise Mann‐Whitney U tests were used to compare continuous variables. Empirical receiver operating characteristic (ROC) curves and area under the ROC (AUROCs) curves were used to evaluate the ability of GSM and YM to stage disease pathologies based on histological scores. The optimal operating point values were selected as the point of the ROC curve closest to (0,1). The level of significance was set at P < 0.05 for all tests.
Results
Phantom Validation
A total of 14 phantoms were imaged to validate the robotic SWE measurements spanning a YM range of 0.50‐38.1 kPa (SWS: 0.41‐3.56 m/s). Excellent agreement was observed between the robotic and conventional measurements of median YM (R2 = 0.998). Bland‐Altman analysis demonstrated no statistically significant bias (−0.14 kPa; P = 0.33), low coefficient of variation of 5.7%, and limits of agreement of 1.0 kPa. Phantom validation results are summarized in Supporting Fig. S1.
Longitudinal In Vivo Imaging
Mice on CDAHFD were successfully imaged longitudinally on the robotic system for 15 weeks with both 3D B‐mode and SWE modes to track changes in liver echogenicity (i.e., brightness) and stiffness, respectively, arising from the experimental diet. Representative images and histology are shown in Figs. 2 and 3, respectively. In general, mice tolerated the imaging well with no noticeable adverse side effects. The average time mice were on the imaging system under anesthesia was 3.5 minutes. During the study, one mouse had to be euthanized unexpectedly on recommendation of the veterinarian due to deteriorating health after 7 weeks on diet. This mouse was excluded from histological correlation and ROC analyses because the time of necropsy was not matched with an imaging timepoint.
FIG. 2.
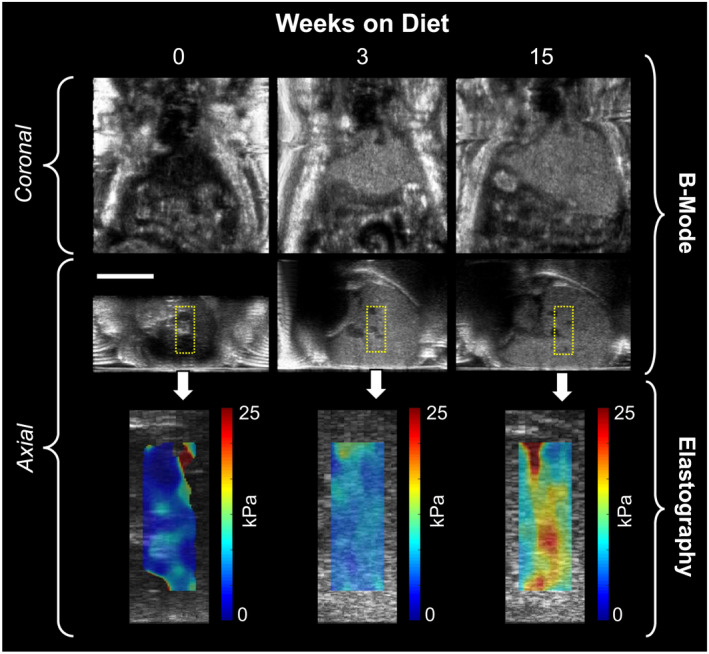
Representative ultrasound images from a single mouse at three timepoints (0, 3, and 15 weeks) on CDAHFD. The echogenicity, stiffness, and size of the liver all increase with time on diet. Scale bar indicates 1 cm.
FIG. 3.
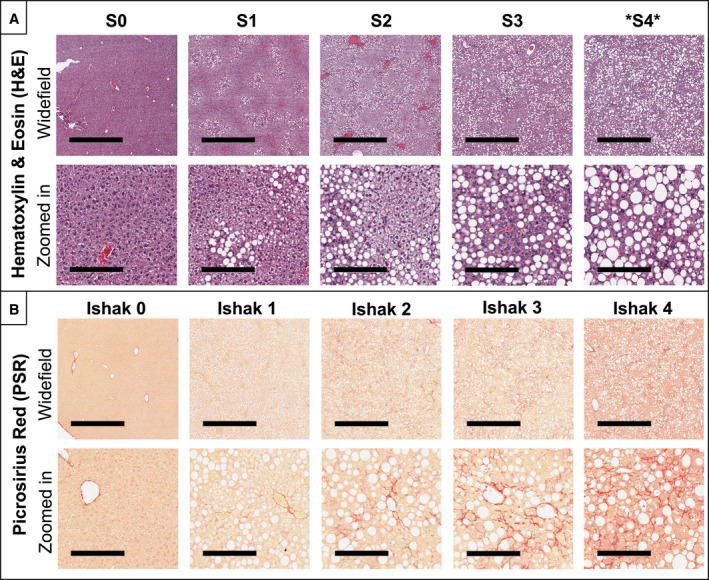
Representative histological images with disease severity category indicated above the columns. Steatosis and inflammation were evaluated using H&E staining (A), while fibrosis was evaluated using picrosirius red staining (B). For each stain, both a widefield (top) and magnified (bottom) view are presented with scale bars of 1 mm and 200 µm, respectively. Note that an additional steatosis category, “S4,” was added to the traditional rodent NALFD system to represent >66% diffuse steatosis. Macrovesicular steatosis rapidly reached S4 by 3‐6 weeks after start of diet, while fibrosis onset was a slower progression, requiring 12‐15 weeks to reach Ishak 4. The different temporal dynamics for these disease progressions can be observed in the longitudinal data presented in Fig. 5.
The longitudinal progression of ultrasound imaging–derived metrics, including liver stiffness and liver echogenicity, is shown in Fig. 4. Statistically significant changes in liver echogenicity were observed as early as week 1 (1.4‐fold higher than baseline) and continued to rise sharply during the first 6 weeks on diet (2.5‐fold higher than baseline). After 6 weeks, echogenicity reached a maximum and plateaued for the remainder of the time course. Liver stiffness, on the other hand, exhibited a more gradual and steadier rise through the course of the study. At baseline, liver stiffness was measured to be 5.5 ± 0.75 kPa for the n = 40 mouse cohort. After 1 week on diet, liver stiffness increased slightly to 6.6 ± 1.1 kPa, representing a 1.2‐fold increase. However, by 6, 12, and 15 weeks, the liver stiffness had increased to 10.2 ± 1.5 kPa, 15.3 ± 1.9 kPa, and 19.1 ± 1.6 kPa, representing 1.9, 2.8, and 3.4‐fold increases, respectively.
FIG. 4.
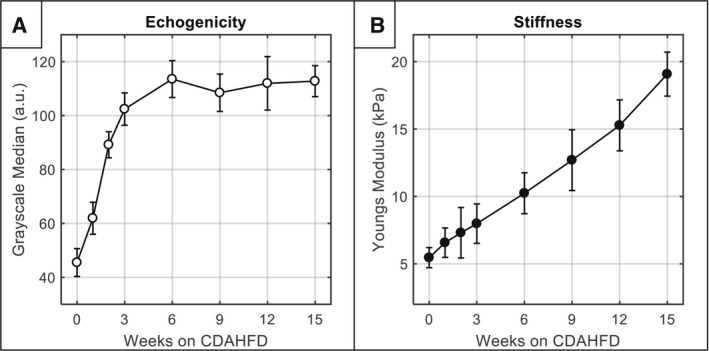
Longitudinal progression of noninvasive imaging parameters (mean ± SD). (A) Echogenicity over time measured by GSM. (B) Liver stiffness over time measured by SWE. Note that the full sample size is included at each timepoint, and sample size decreased over time by 5 mice due to sacrifice for histology (e.g., n = 40 mice at week 0, n = 35 mice at week 1, etc.).
Correlation of Noninvasive Ultrasound Imaging With Histology
At each time point, one cage of mice (n = 5) was sacrificed for histology. Histology demonstrated that as early as 1 week on CDAHFD, mice exhibited abnormal steatosis (both microvesicular and macrovesicular phenotypes S1‐S2) and the beginning of lobular inflammation (L1), indicating borderline NASH. By 3 weeks, periportal macrovesicular steatosis was dominant (S3), while inflammation was midrange (L2), indicating definite NASH; and by 6 weeks, maximum diffuse macrovesicular steatosis and inflammation scores (S4/L3) were recorded in nearly all sampled mice. Fibrosis, on the other hand, progressed more slowly and with a relatively linear trend. By 2‐3 weeks, fibrosis had reached Ishak 1; by 6 weeks, it reached Ishak 2; by 9 weeks, it reached Ishak 3; and by 12‐15 weeks, it reached Ishak 4. No mice in the cohort reached Ishak levels 5 or 6, which correspond to “incomplete” or “definite” cirrhosis. Of note, the pathologist indicated that hypertrophy of cells was not a prominent feature in this model, nor was it a strong differentiator across the time course. Histologic results for macrosteatosis, inflammation, and fibrosis are shown in Fig. 5, whereas results for microsteatosis and hypertrophy are shown in Supporting Fig. S2.
FIG. 5.
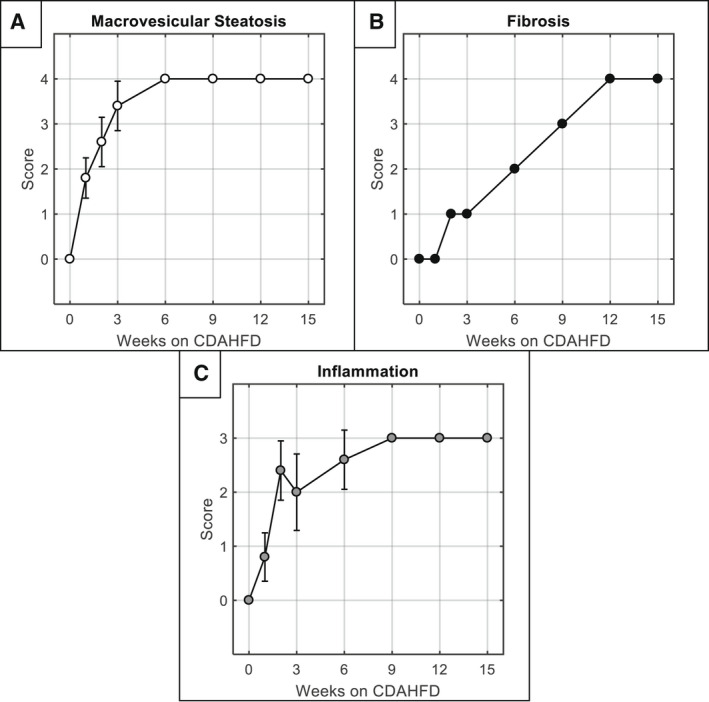
Longitudinal histologic grading (mean ± SD). (A) Macrovesicular steatosis score over time measured from H&E histological stains. (B) Fibrosis score (Ishak) over time measured from picrosirius red histological stains. (C) Lobular inflammation score over time measured from H&E histological stains. Each timepoint includes n = 5 mice. For fibrosis plot in (B), all five mice at each timepoint received the same score; therefore no error bars are plotted.
Correlation analysis comparing in vivo ultrasound imaging and histology pairs revealed the strongest associations were between stiffness/fibrosis, and echogenicity/macrosteatosis, with R2 values of 0.839 and 0.891, respectively (Fig. 6). Pairwise Mann‐Whitney U tests revealed that all grades of steatosis (S0‐S4) and nearly all grades of fibrosis (Ishak 0‐4) were statistically significantly different than all others (Fig. 6). Associations were also observed between all other imaging/histology pairs to lesser degrees (in descending order): echogenicity/inflammation, R2 = 0.755; echogenicity/fibrosis, R2 = 0.623; stiffness/inflammation, R2 = 0.542; and stiffness/steatosis, R2 = 0.482 (Table 2). Raw histologic scores are provided in Supporting Table S3.
FIG. 6.
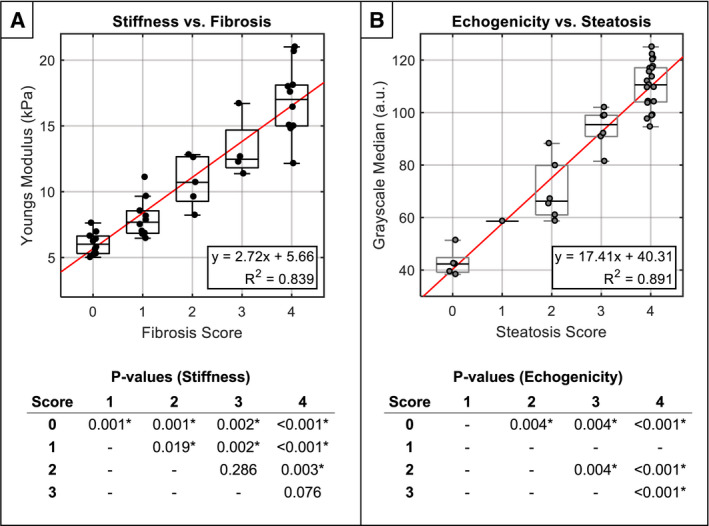
Selected comparison between noninvasive imaging and histology pairs with highest respective correlation. (A) Liver stiffness versus Ishak fibrosis score. (B) Liver echogenicity versus macrovesicular steatosis score. For clarity, P values from pairwise Mann‐Whitney U tests are provided in the tables below (α = 0.05). Note that P values for steatosis grade “1” were not computed, because only one animal was identified with this score. For full comparison matrix showing correlation plots of all imaging/histology pairs, see Supporting Fig. S2.
TABLE 2.
Correlation Analysis Between Imaging and Histology
| Histologic Parameter | Stiffness | Echogenicity | ||||
|---|---|---|---|---|---|---|
| Pearson R | R2 | P Value | Pearson R | R2 | P Value | |
| Macrosteatosis | 0.694 | 0.482 | 9.55E‐07 | 0.944 | 0.891 | 2.30E‐19 |
| Microsteatosis | −0.130 | 0.017 | 0.430 | 0.113 | 0.013 | 0.494 |
| Hypertrophy | 0.431 | 0.186 | 6.13E‐03 | 0.728 | 0.530 | 1.50E‐07 |
| Lobular inflammation | 0.736 | 0.542 | 9.15E‐08 | 0.869 | 0.755 | 7.25E‐13 |
| Fibrosis | 0.916 | 0.839 | 3.06E‐16 | 0.790 | 0.623 | 2.31E‐09 |
Sensitivity Analyses
Sensitivity analysis was conducted on the imaging/histology pairs with the highest correlation, namely, stiffness/fibrosis and echogenicity/macrosteatosis. The AUROCs for classification of Ishak ≥ 1, Ishak ≥ 2, Ishak ≥ 3, and Ishak ≥ 4 fibrosis using liver stiffness measurements were 0.969, 0.984, 0.980, and 0.969, respectively (Fig. 7, top row). The AUROCs for prediction of ≥S2, ≥S3, and ≥S4 steatosis using liver echogenicity measurements were 1.00, 0.997, and 0.976, respectively (Fig. 7, bottom row). Note that only one animal was indicated with a steatosis score of “1”; therefore the “≥S1” category was omitted from the analysis. The optimal cutoff values, as well as measurements of sensitivity and specificity, are shown in Fig. 7. More ROC analysis parameters (e.g., positive predictive value, negative predictive value) are provided in Supporting Tables S1 and S2.
FIG. 7.
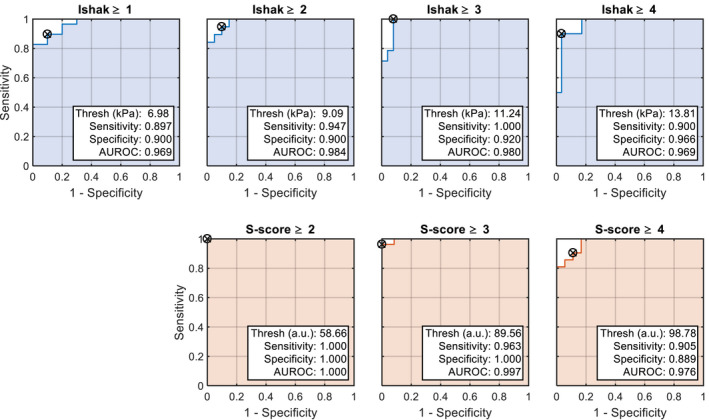
ROC curves for SWE (top row) and GSM (bottom row) at different fibrosis and steatosis thresholds, respectively. Note that ROC analysis for steatosis grade “1” was not computed, because only one animal was identified with this score.
Discussion
In this study, a hands‐free ultrasound instrument for measuring liver stiffness and echogenicity in mouse models of liver disease was built and validated. Imaging metrics were observed to correlate with numerous histologic parameters of NASH including steatosis, inflammation, and fibrosis. The highest histologic correlates for stiffness and echogenicity were fibrosis and macrosteatosis, respectively, with imaging metrics closely tracking the temporal dynamics of the histological scores. Liver stiffness measurements, additionally, were in good agreement with other SWE studies in CDAHFD rodents( 23 ) as well as human SWE imaging in patients with NASH.( 4 ) Importantly, these noninvasive measurements could be taken in a highly repeatable and simple manner, eliminating many of the user‐dependent challenges faced by traditional handheld ultrasound scanning (e.g., consistent beam placement, precompression of tissue biasing stiffness). Furthermore, because this robotic system uses acoustic radiation force to generate shear waves, an invasive actuator need not be inserted into the liver, as is necessary for murine magnetic resonance elastography (MRE),( 42 ) making it a fully noninvasive method with extremely low risk of instrument‐induced animal loss.
Although these results are highly encouraging, it should be noted that liver echogenicity and stiffness are not direct measurements of steatosis and fibrosis. Echogenicity, for example, can be increased by cirrhosis and is not strictly reflective of steatosis.( 43 ) Likewise, liver stiffness can be increased independently under various nonfibrotic conditions including inflammation (edematous tissue reaction and cell infiltrate), obstructive cholestasis, deposition of amyloid, and blood pressure fluctuations (portal, venous, arterial).( 44 ) In contrast, liver stiffness can be decreased by steatosis in the absence of concomitant inflammation.( 44 ) In our study, we did not observe any softening during establishment of steatosis, even in very early timepoints that preceded the onset of histologically identified fibrosis. At the 1‐week timepoint, mean stiffness increased from baseline by 1.1 kPa (5.5 kPa vs. 6.6 kPa; P = 1.7e‐6) and mean echogenicity increased from baseline by 16.4 a.u. (45.5 a.u. vs. 61.9 a.u.; P = 2.2e‐20). This suggests that any softening from lipid sequestration may have been counterbalanced by stiffening from the aggressive inflammatory processes in the CDAHFD model.( 45 ) Furthermore, the coexistence of steatosis and inflammation in this model likely explains the high level of correlation between echogenicity and inflammation score (R2 = 0.76), which even exceeded that of stiffness (R2 = 0.54). Given these observations, diagnostic cutoff values, such as those presented in this manuscript, should be applied carefully and will likely need to be determined on a model‐specific basis, which mirrors what has been done in the clinic with etiology‐specific and inflammation‐adapted cutoff values.( 46 )
Some experimental limitations were noted during the study and highlight areas for future work. First, cirrhotic phenotypes (i.e., Ishak 5 and Ishak 6) were never achieved; therefore, the performance of the robotic imaging system for these levels of pathology is unknown. However, given the substantial literature showing efficacy of SWE for detecting cirrhosis clinically,( 4 ) we believe the system would be similarly sensitive to these advanced forms of the disease. Second, only female mice were used due to the lower prevalence of aggression, in‐cage fighting, and injury; therefore, sex‐based differences could not be analyzed. Male mice can be more susceptible to hepatic fibrosis with certain diets( 47 ) and are slightly larger in size, which may influence imaging penetration depth, particularly in obesity models, and will need to be explored in future studies. Third, the impact of subcapsular depth was not explored in this study. It is recommended that clinical SWE measurements in humans be taken below the liver capsule to avoid reverberation artifact and the stiff subcapsular parenchyma, which can bias measurements.( 1 ) In this study, all SWE images were taken at a depth greater than or equal to 4 mm, which was beyond the subcapsular region and consistent with established preclinical protocols,( 24 ) suggesting bias should have been mitigated. Fourth, the impact of pressure and pulse was not evaluated in this study. Arterial, venous, and portal pressures have all been demonstrated as independent modulators of liver stiffness,( 12 , 13 , 14 ) and sinusoidal pressure (which is closely linked to liver stiffness) has been hypothesized as the primary driving factor behind fibrosis progression at the cellular level.( 48 ) Given this, we expect that pressure differences could explain some intra‐timepoint variability in liver stiffness measurements; however, the magnitude of these effects will need to be explored in future studies. Fifth, mice were not fasted before imaging. In clinical settings, fasting has been shown to be critical, because postprandial hyperemia to the abdominal organs can cause transient increases in liver stiffness.( 1 ) However, feeding behavior between humans and mice is different (mice are nocturnal and feed mostly at night( 49 )), and some anesthetics used in preclinical research for chemical restraint (namely pentobarbital) have been shown to suppress hyperemia.( 50 ) Therefore, fasting may not be as impactful during rodent studies, insofar as the animals naturally will not have consumed food immediately before imaging done during the day, or the effects of abdominal hyperemia will be suppressed by anesthesia. More studies are needed to evaluate these questions and optimize preclinical experimental protocols further.
Finally, in addition to experimental limitations, several instrumental limitations were noted as well. First, while the robotic SWE scanning was performed in 3D, the spatial sampling was coarse (~1‐mm elevational step sizes), and the entire extent of the liver was not comprehensively scanned due to time constraints imposed by slow RF data transfer rate of the prototype system. Spatial heterogeneity was observed in the SWE estimates, particularly around areas of the large hepatic vasculature. We expect that scanning the entire 3D liver volume with SWE will likely improve the accuracy of the final YM estimate (akin to MRE) and should be possible with ongoing improvements to the software architecture (e.g., graphics processing unit techniques( 51 )) that enable real‐time SWE framerates. Second, echogenicity measurements were made without calibration to a reference phantom or normalization to another organ (e.g., hepato‐renal index). This approach was successful because no system parameter was changed throughout the entire study (e.g., gain, frequency, focal zone); however, in the future, more robust and quantitative measurements of backscatter (and/or attenuation) should be explored.( 3 ) Third, no breath or cardiac gating was used. Breathing has been shown to significantly corrupt SWE measurements, and clinical scanning uses breath‐holding during imaging.( 1 ) Effects of breathing and heart motion were mitigated by taking multiple successive SWE captures in each location (n = 5) and averaging the data. Physiological gating would slow acquisition time; however, measurement variance would likely decrease if used. An alternative approach could be retrospective physiological gating, which would not slow down acquisition, but would rely on a method to detect physiological motion within the SWE captures to remove these corrupted samples before averaging the ensemble for that liver.
Supporting information
Supplementary Material
Acknowledgment
Histological services were provided by the Histology Research Core Facility in the Department of Cell Biology and Physiology at the University of North Carolina, Chapel Hill.
References
Author names in bold designate shared co‐first authorship.
- 1. Barr RG. Shear wave liver elastography. Abdom Radiol (New York) 2018;43:800‐807. [DOI] [PubMed] [Google Scholar]
- 2. Sarvazyan AP, Rudenko OV, Swanson SD, Fowlkes JB, Emelianov SY. Shear wave elasticity imaging: a new ultrasonic technology of medical diagnostics. Ultrasound Med Biol 1998;24:1419‐1435. [DOI] [PubMed] [Google Scholar]
- 3. Tada T, Nishimura T, Yoshida M, Iijima H. Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: new trends and role of ultrasonography. J Med Ultrason 2001;2020:511‐520. [DOI] [PubMed] [Google Scholar]
- 4. Herrmann E, de Lédinghen V, Cassinotto C, Chu W‐W, Leung V‐F, Ferraioli G, et al. Assessment of biopsy‐proven liver fibrosis by two‐dimensional shear wave elastography: an individual patient data‐based meta‐analysis. Hepatology 2018;67:260‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu X, Huang X, Chen H, Zhang T, Hou J, Song A, et al. Diagnostic effect of shear wave elastography imaging for differentiation of malignant liver lesions: a meta‐analysis. BMC Gastroenterol 2019;19:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tapper EB, Lok AS‐F. Use of liver imaging and biopsy in clinical practice. N Engl J Med 2017;377:756‐768. [DOI] [PubMed] [Google Scholar]
- 7. Barry CT, Hah Z, Partin A, Mooney RA, Chuang K‐HH, Augustine A, et al. Mouse liver dispersion for the diagnosis of early‐stage fatty liver disease: a 70‐sample study. Ultrasound Med Biol 2014;40:704‐713. [DOI] [PubMed] [Google Scholar]
- 8. Yeh C, Chen B, Tseng L, Jao P, Su T, Li P. Shear‐wave elasticity imaging of a liver fibrosis mouse model using high‐frequency ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control 2015;62:1295‐1307. [DOI] [PubMed] [Google Scholar]
- 9. Bastard C, Bosisio MR, Chabert M, Kalopissis AD, Mahrouf‐Yorgov M, Gilgenkrantz H, et al. Transient micro‐elastography: a novel non‐invasive approach to measure liver stiffness in mice. World J Gastroenterol 2011;17:968‐975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang MH, Palmeri ML, Guy CD, Yang L, Hedlund LW, Diehl AM, et al. In vivo quantification of liver stiffness in a rat model of hepatic fibrosis with acoustic radiation force. Ultrasound Med Biol 2009;35:1709‐1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin S‐H, Ding H, Mao F, Xue L‐Y, Lv W‐W, Zhu H‐G, et al. Non‐invasive assessment of liver fibrosis in a rat model: shear wave elasticity imaging versus real‐time elastography. Ultrasound Med Biol 2013;39:1215‐1222. [DOI] [PubMed] [Google Scholar]
- 12. Piecha F, Peccerella T, Bruckner T, Seitz H‐K, Rausch V, Mueller S. Arterial pressure suffices to increase liver stiffness. Am J Physiol Liver Physiol 2016;311:G945‐G953. [DOI] [PubMed] [Google Scholar]
- 13. Piecha F, Paech D, Sollors J, Seitz H‐K, Rössle M, Rausch V, et al. Rapid change of liver stiffness after variceal ligation and TIPS implantation. Am J Physiol Liver Physiol 2018;314:G179‐G187. [DOI] [PubMed] [Google Scholar]
- 14. Piecha F, Mandorfer M, Peccerella T, Ozga A‐K, Poth T, Vonbank A, et al. Pharmacological decrease of liver stiffness is pressure‐related and predicts long‐term clinical outcome. Am J Physiol Gastrointest Liver Physiol 2018;315:G484‐G494. [DOI] [PubMed] [Google Scholar]
- 15. Shi X, Li J, Min B, Yang R, He C, Yang Y. Application of ultrasound elastography for monitoring the effects of TβR1 shRNA therapy on hepatic fibrosis in a rat model. PLoS One 2021;16:e0253150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang B‐K, Lee SS, Cheong H, Hong SM, Jang K, Lee M‐G. Shear wave elastography for assessment of steatohepatitis and hepatic fibrosis in rat models of non‐alcoholic fatty liver disease. Ultrasound Med Biol 2015;41:3205‐3215. [DOI] [PubMed] [Google Scholar]
- 17. Ogawa S, Moriyasu F, Yoshida K, Oshiro H, Kojima M, Sano T, et al. Relationship between liver tissue stiffness and histopathological findings analyzed by shear wave elastography and compression testing in rats with non‐alcoholic steatohepatitis. J Med Ultrason 2001;2016:355‐360. [DOI] [PubMed] [Google Scholar]
- 18. Kazemirad S, Zhang E, Nguyen BN, Bodson‐Clermont P, Destrempes F, Trudel D, et al. Detection of steatohepatitis in a rat model by using spectroscopic shear‐wave US elastography. Radiology 2017;282:726‐733. [DOI] [PubMed] [Google Scholar]
- 19. Cao W, Zhou Y, Niu Y, Zhu X, Song Y, Guo R. Quantitative analysis of hepatic toxicity in rats induced by inhalable silica nanoparticles using acoustic radiation force imaging. J Ultrasound Med 2017;36:1829‐1839. [DOI] [PubMed] [Google Scholar]
- 20. Langdon JH, Elegbe E, Gonzalez RS, Osapoetra L, Ford T, McAleavey SA. Measurement of liver stiffness using shear wave elastography in a rat model: factors impacting stiffness measurement with multiple‐ and single‐tracking‐location techniques. Ultrasound Med Biol 2017;43:2629‐2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carbonell G, Berná‐Serna JDD, Oltra L, Martínez CM, Garcia‐Carrillo N, Guzmán‐Aroca F, et al. Evaluation of rat liver with ARFI elastography: in vivo and ex vivo study. PLoS One 2019;14:e0217297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin Y, Huh J, Ham SJ, Cho YC, Choi Y, Woo D‐C, et al. Test‐retest repeatability of ultrasonographic shear wave elastography in a rat liver fibrosis model: toward a quantitative biomarker for preclinical trials. Ultrasonography 2021;40:126‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morin J, Swanson TA, Rinaldi A, Boucher M, Ross T, Hirenallur‐Shanthappa D. Application of ultrasound and shear wave elastography imaging in a rat model of NAFLD/NASH. J Vis Exp 2021. 10.3791/62403 [DOI] [PubMed] [Google Scholar]
- 24. Elshaarawy O, Alquzi S, Piecha F, Sandrin L, Bastard C, Mueller S. Liver Stiffness Measurements in Small Animals. In: Elastography L, ed. Cham, Switzerland: Springer International Publishing; 2020:95‐102. [Google Scholar]
- 25. Wang D, Luo Y, Wang X, Orlicky D, Myakala K, Yang P, et al. The sodium‐glucose cotransporter 2 inhibitor dapagliflozin prevents renal and liver disease in Western diet induced obesity mice. Int J Mol Sci 2018;19:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang J, Li Y, Xu D, Zhang X, Zhou X. RUNX1 regulates SMAD1 by transcriptionally activating the expression of USP9X, regulating the activation of hepatic stellate cells and liver fibrosis. Eur J Pharmacol 2021;903:174137. [DOI] [PubMed] [Google Scholar]
- 27. Cheong KO, Shin D‐S, Bak J, Lee C, Kim KW, Je NK, et al. Hepatoprotective effects of zingerone on carbon tetrachloride‐ and dimethylnitrosamine‐induced liver injuries in rats. Arch Pharm Res 2016;39:279‐291. [DOI] [PubMed] [Google Scholar]
- 28. Ikenaga N, Peng Z‐W, Vaid KA, Liu SB, Yoshida S, Sverdlov DY, et al. Selective targeting of lysyl oxidase‐like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut 2017;66:1697‐1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barr RG, Zhang Z. Effects of precompression on elasticity imaging of the breast: development of a clinically useful semiquantitative method of precompression assessment. J Ultrasound Med 2012;31:895‐902. [DOI] [PubMed] [Google Scholar]
- 30. Czernuszewicz TJ, Papadopoulou V, Rojas JD, Rajamahendiran RM, Perdomo J, Butler J, et al. A new preclinical ultrasound platform for widefield 3D imaging of rodents. Rev Sci Instrum 2018;89:075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: a platform for subject‐specific image analysis, visualization, and clinical support. In: Jolesz F, ed. Intraoperative Imaging and Image‐Guided Therapy. New York, NY: Springer; 2014:277‐289. [Google Scholar]
- 32. Lasso A, Heffter T, Rankin A, Pinter C, Ungi T, Fichtinger G. PLUS: open‐source toolkit for ultrasound‐guided intervention systems. IEEE Trans Biomed Eng 2014;61:2527‐2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinton GF, Dahl JJ, Trahey GE. Rapid tracking of small displacements with ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control 2006;53:1103‐1117. [DOI] [PubMed] [Google Scholar]
- 34. Palmeri ML, Wang MH, Dahl JJ, Frinkley KD, Nightingale KR. Quantifying hepatic shear modulus in vivo using acoustic radiation force. Ultrasound Med Biol 2008;34:546‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deffieux T, Gennisson J‐L, Bercoff J, Tanter M. On the effects of reflected waves in transient shear wave elastography. IEEE Trans Ultrason Ferroelectr Freq Control 2011;58:2032‐2035. [DOI] [PubMed] [Google Scholar]
- 36. Hall TJ, Bilgen M, Insana MF, Krouskop TA. Phantom materials for elastography. IEEE Trans Ultrason Ferroelectr Freq Control 1997;44:1355‐1365. [Google Scholar]
- 37. Palmeri ML, Milkowski A, Barr R, Carson P, Couade M, Chen J, et al. Radiological society of North America/quantitative imaging biomarker alliance shear wave speed bias quantification in elastic and viscoelastic phantoms. J Ultrasound Med 2021;40:569‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsumoto M, Hada N, Sakamaki Y, Uno A, Shiga T, Tanaka C, et al. An improved mouse model that rapidly develops fibrosis in non‐alcoholic steatohepatitis. Int J Exp Pathol 2013;94:93‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol 1995;22:696‐699. [DOI] [PubMed] [Google Scholar]
- 40. Liang W, Menke AL, Driessen A, Koek GH, Lindeman JH, Stoop R, et al. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One 2014;9:e115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwen LO, Homeyer A, Schwier M, Dahmen U, Dirsch O, Schenk A, et al. Zonated quantification of steatosis in an entire mouse liver. Comput Biol Med 2016;73:108‐118. [DOI] [PubMed] [Google Scholar]
- 42. Yin M, Glaser KJ, Manduca A, Mounajjed T, Malhi H, Simonetto DA, et al. Distinguishing between hepatic inflammation and fibrosis with MR elastography. Radiology 2017;284:694‐705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu M, Sharma PG, Grajo JR. The echogenic liver: steatosis and beyond. Ultrasound Q 2020;37:308‐314. [DOI] [PubMed] [Google Scholar]
- 44. Mueller S, Lackner C. Histological confounders of liver stiffness. In Liver Elastography. Cham, Switzerland: Springer International Publishing; 2020:233‐242. [Google Scholar]
- 45. Sugasawa T, Ono S, Yonamine M, Fujita S‐I, Matsumoto Y, Aoki K, et al. One week of CDAHFD induces steatohepatitis and mitochondrial dysfunction with oxidative stress in liver. Int J Mol Sci 2021;22:5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mueller S. Introduction to fibrosis assessment by liver stiffness in different etiologies. In: Liver Elastography. Cham, Switzerland: Springer International Publishing; 2020:105‐111. [Google Scholar]
- 47. Yanguas SC, Cogliati B, Willebrords J, Maes M, Colle I, van den Bossche B, et al. Experimental models of liver fibrosis. Arch Toxicol 2016;90:1025‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mueller S. Role of sinusoidal pressure and arterialization in driving fibrosis progression. In: Liver Elastography. Cham, Switzerland: Springer International Publishing; 2020:671‐683. [Google Scholar]
- 49. Ellacott KLJ, Morton GJ, Woods SC, Tso P, Schwartz MW. Assessment of feeding behavior in laboratory mice. Cell Metab 2010;12:10‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anzueto Hernandez L, Kvietys PR, Granger DN. Postprandial hemodynamics in the conscious rat. Am J Physiol 1986;251:G117‐G123. [DOI] [PubMed] [Google Scholar]
- 51. Rosenzweig S, Palmeri M, Nightingale K. GPU‐based real‐time small displacement estimation with ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control 2011;58:399‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


