Abstract
Aerobic degradation of phenylacetic acid in Pseudomonas putida U is carried out by a central catabolism pathway (phenylacetyl-coenzyme A [CoA] catabolon core). Induction of this route was analyzed by using different mutants specifically designed for this objective. Our results revealed that the true inducer molecule is phenylacetyl-CoA and not other structurally or catabolically related aromatic compounds.
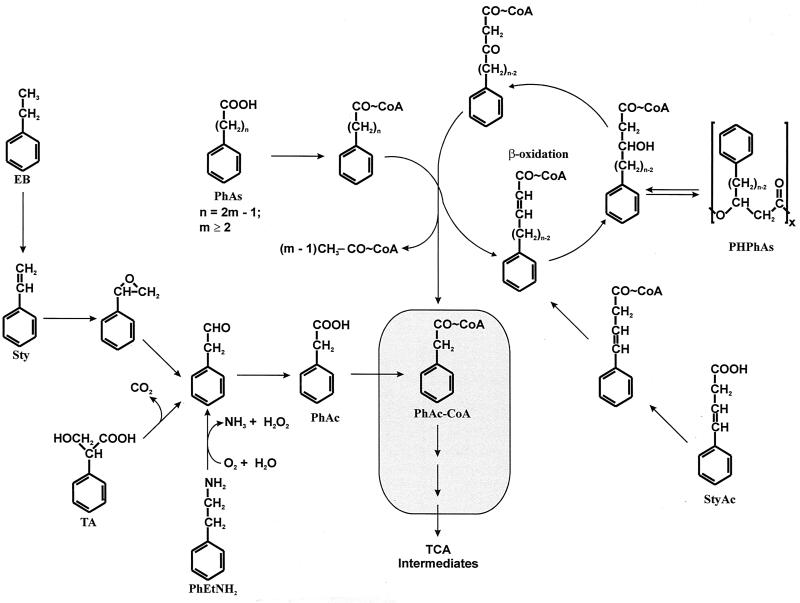
Phenylacetyl-coenzyme A (CoA) catabolon (PhAc-CoAC) is a term used for a complex catabolic unit integrated by different degradative pathways involved in the assimilation of certain aromatic compounds (phenylacetic acid [PhAc], phenylethylamine, ethylbenzene, styrene, tropic acid, trans-styrylacetic acid, and n-phenylalkanoic acids [n-PhAs] containing an odd number of carbon atoms) which converge in a central route, namely, the PhAc-CoAC core (PhAc-CoACC) (2, 13), which converts these molecules into general metabolites (Fig. 1).
FIG. 1.
Organization of the PhAc-CoAC. EB, ethylbenzene; Sty, styrene; TA, tropic acid; PhEtNH2, phenylethylamine; StyAc, trans-styrylacetic acid; PhAs, n-phenylalkanoic acids; TCA, tricarboxylic acid cycle intermediates. The box indicates the PhAc-CoACC.
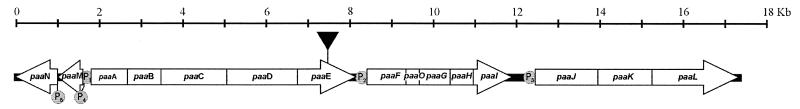
The genetic information required for the PhAc-CoACC in Pseudomonas putida U is contained in a piece of DNA (18 kb) organized in three consecutive operons (Fig. 2). We have previously reported that this route appeared when P. putida U was cultured in chemically defined media containing some of the aromatic compounds indicated above as carbon sources (4, 10, 11) and that other similar, structurally related molecules (2-OH-PhAc, 3-OH-PhAc, 4-OH-PhAc, benzoic acid, and n-PhAs with an even number of carbon atoms) did not have this effect (13, 14). Bearing in mind the diverse chemical structure of the aromatic compounds able to induce the pathway, it seems feasible to assume that a common intermediate (phenylacetyl-CoA) is the true inducer of the convergence pathway. Despite this, the possibility that other molecules generated from the aromatic compounds (PhAc or n-phenylalkanoyl-CoA or their β-oxidation derivatives) might also be involved in this induction cannot be ruled out.
FIG. 2.
Genetic organization of the paa catabolic pathway, P1, P2, P3, P4, and P5 are promoters; paaA to paaL and paaO are catabolic genes; and paaM and paaN are regulatory genes which are divergently transcribed. The genes encode the following proteins: paaA, enoyl-CoA hydratase I; paaB, enoyl-CoA hydratase II; paaC, 3-hydroxyacyl-CoA dehydrogenase; paaD, ketothiolase; paaE, PCL; paaF to paaI and paaO, ring hydroxylation system; paaJ, permease; paaK, porine; paaL, ring-opening enzyme; paaM, possible regulator; and paaN, transcriptional repressor. The solid triangle indicates the site of insertion of the disruption element used to truncate the desired gene.
In order to clarify this point, we designed, obtained, and characterized by different methods (6, 13–17, 19) several mutants of P. putida U affected in the catabolism of certain aromatic compounds (see below). Using these strains, we analyzed induction of the PhAc-CoACC by measuring two target enzymatic activities: the activity of phenylacetyl-CoA ligase (PCL) (encoded by the paaE gene, belonging to the first operon) (10, 11) and the activity of a permease (encoded by the paaJ gene [the third operon in Fig. 2]) involved in the uptake of [1-14C]PhAc (18).
P. putida U was maintained and cultured as previously reported (10). The medium used for growth of P. putida and different mutants was a chemically defined medium (10) containing PhAc, 4-OH-PhAc, 6-phenylhexanoic acid (PhH) (or 8-phenyloctanoic acid [PhO]), other aromatic compounds, or combinations of these compounds as carbon sources (Table 1). In the experiments in which mutants were employed, 4-OH-PhAc and the carbon source used to test induction were added to the media together. The final concentration of each aromatic compound was 5 mM.
TABLE 1.
Evaluation of PCL activity, the [1-14C]PhAc transport system in P. putida U (wild type) and in different mutants (types I, II, and III), and PHPhA accumulation when the bacteria were cultured in a chemically defined medium
| Strain | Carbon source(s) (5 mM) | PCL sp act (U/mg of protein)a | [1-14C]PhAc uptake (pmol/min)b | PHPhA content (%)c | Significance |
|---|---|---|---|---|---|
| P. putida U (wild type) | 4-OH-PhAc | NDd | ND | ND | 4-OH-PhAc does not induce PhAc-CoACC, 4-OH-PhAc is not polymerized |
| Wild type | 4-OH-PhAc + PhAc | 43 | 2,420 | ND | PhAc-CoACC induction occurs, PhAc is not polymerized |
| Type I | 4-OH-PhAc + PhAc | ND | ND | ND | PhAc-CoACC induction does not occur, PhAc is not activated to PhAc-CoA by other ligases, PhAc is not an inducer of PhAc-CoACC |
| Wild type | 4-OH-PhAc + PhH (or PhO) | 38 | 2,725 | 18–20 | PhAc-CoACC induction occurs, PHPhAs accumulate |
| Type II | 4-OH-PhAc + PhH (or PhO) | ND | ND | ND | PhAc-CoACC induction does not occur, PHPhAs do not accumulate, n-PhAs are not inducers of PhAc-CoACC |
| Type II | 4-OH-PhAc + PhAc | 37 | 2,740 | ND | PhAc-CoACC induction occurs, PHPhAs not produced |
| Type III | 4-OH-PhAc + PhH (or PhO) | ND | ND | 50 | PhAc-CoACC induction does not occur, n-PhAs and β-oxidation derivatives are not inducers, strong PHPhA accumulation is observed |
| Type III | 4-OH-PhAc + PhAc | 30 | 2,300 | ND | PhAc-CoACC induction occurs, PHPhAs are not produced |
Since the PhAc-CoACC is required for degradation of PhAc, ethylbenzene, styrene, tropic acid, phenylethylamine, trans-styrylacetic acid, and n-PhAs with an odd number of carbon atoms, it is possible that any of these compounds could induce the PhAc-CoACC. However, although a priori this possibility cannot be ruled out, it seems more reasonable to assume that a single compound is the true inducer. If this is indeed the case, two different molecules, PhAc or PhAc-CoA, could be involved in this process.
PhAc is a catabolite generated from styrene (20), ethylbenzene (8) tropic acid (9), and phenylethylamine (3, 5), whereas PhAc-CoA is obtained from trans-styrylacetic acid, from n-PhAs containing an odd number of carbon atoms, from β-oxidation derivatives of these acids, or from polymer derivatives of these compounds (4). However, we have previously shown that P. putida U mutants that are defective in assimilation of all these compounds due to the existence of a mutation in some of the genes belonging to the second operon (the ring oxidation system) (Fig. 2) accumulates PhAc extracellularly when the bacteria are cultured in a chemically defined medium (4) containing molecules which could be catabolically converted into PhAc-CoA (13). These results suggested that this thioester is hydrolyzed to PhAc (probably by nonspecific thioesterases) before being released into the culture broth. It is thus difficult to establish which of these compounds (PhAc or PhAc-CoA) is the true inducer of the PhAc catabolic pathway, since the two molecules are interconvertible. In order to establish the nature of the molecule that acts as the true inducer of the PhAc-CoACC, three different types of mutants were obtained.
The first group (type I [Fig. 2]) includes mutants lacking PCL activity. These strains were obtained by disrupting the paaE gene, which encodes a protein having this enzymatic activity, with plasmid pK18::mob (17). These mutants are unable to grow in chemically defined media containing PhAc, ethylbenzene, styrene, or phenylethylamine as the sole carbon source, since all of them generate PhAc as a catabolic intermediate, which has to be activated to PhAc-CoA for further degradation (10, 11, 13). However, they grow well in a similar medium containing as carbon sources compounds that produce (through β-oxidation) PhAc-CoA as an intermediate (trans-styrylacetic acid or n-PhAs with an odd number of carbon atoms or derivatives of these compounds) and that, therefore, do not require the presence of PCL activity for total degradation of the molecules.
Type I mutants (PCL−) were used to analyze whether PhAc, ethylbenzene, styrene, or phenylethylamine is able to induce the PhAc-CoACC. Since PCL was not synthesized in these mutants, induction of the PhAc-CoACC was carried out by measuring [1-14C]PhAc uptake (permease activity) (Table 1).
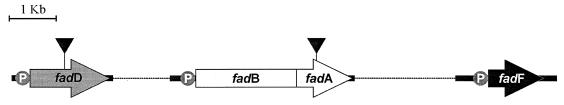
The second group (type II) includes mutants disrupted with transposon Tn5 (6, 19) in the fadD gene, which encodes an acyl-CoA synthetase that catalyzes activation of n-PhAs (containing an acyl chain with more than four carbon atoms), as well as medium- or long-chain aliphatic fatty acids, to their CoA thioesters (4). These mutants were used to establish whether n-PhAs with an odd number of carbon atoms are able to induce the PhAc-CoACC.
The third group of mutants (type III) comprises mutants in which disruption of the fadA gene (encoding the 3-ketoacyl-CoA thiolase of the β-oxidation pathway) was produced with plasmid pK18::mob (17) (Fig. 3). These mutants were unable to synthesize phenylacetyl-CoA from the phenylalkanoyl-CoA β-oxidation derivatives but were able to transport and activate these compounds since they, in contrast to the wild type, accumulate large amounts of poly(3-hydroxyphenylalkanoates) (PHPhAs) when they are cultured in a chemically defined medium containing 4-OH-PhAc and PhH or PhO as carbon sources (Table 1). The presence of PHPhAs as reserve material inside the bacteria was determined by direct microscopic observation (4). The amount of polymer that accumulated was quantified as previously reported (4, 7), and the PHPhA content was recorded as a percentage of the bacterial dry weight. The structures of the polymers synthesized were analyzed by nuclear magnetic resonance as indicated by García et al. (4). For this type of experiment, the wild type (P. putida U) or the different mutants were cultured in the same chemically defined media containing PhAc, 4-OH-PhAc, PhH, PhO, or combinations of these compounds as carbon sources (Table 1). In all cases the final concentration of each aromatic molecule was 5 mM. The data reported above indicate that all the enzymes required for transport, activation, and β-oxidation (except the 3-ketoacyl-CoA thiolase) (Fig. 3) are functional in these mutants. These mutants were used to analyze whether different n-phenylalkanoyl-CoAs (or their β-oxidation derivatives) might be directly involved in induction of the PhAc-CoACC.
FIG. 3.
Genetic organization of the β-oxidation genes in P. putida U. fadD, gene encoding an acyl-CoA synthetase; fadF, gene encoding an acyl-CoA dehydrogenase; fadB, two fused genes encoding several enzymatic activities including enoyl-CoA hydratase, 3-OH-acyl-CoA dehydrogenase, cis-Δ3-trans-Δ2-enoyl-CoA isomerase, and 3-OH-acyl-CoA epimerase; and fadA, gene encoding the 3-ketoacyl-CoA thiolase. Dashed lines indicate that some of the genes are not contiguous in the bacterial chromosome. The solid triangle indicates the site of insertion of the disruption element used to truncate the desired gene.
In all three types of mutants, PCL and PhAc permease activities (see above) were assayed. In these experiments, all the strains analyzed were cultured in the same medium containing the inducer molecule to be analyzed and a different carbon source (4-OH-PhAc, which does not induce the PhAc-CoACC [Table 1]) (14) to support bacterial growth.
The absence of [1-14C]PhAc uptake in the mutants belonging to the first group (those lacking a functional PCL) revealed that PhAc does not induce the PhAc-CoACC (Table 1). It is surprising that PhAc was unable to induce the PhAc-CoACC since PCL, the product of the paaE gene, is required for activation of PhAc to PhAc-CoA and for further catabolism of PhAc-CoA. It should therefore be assumed that a certain quantity of PCL or other acyl-CoA activating enzymes that nonspecifically activate PhAc to PhAc-CoA must be present in P. putida U and that the trace amounts of PhAc-CoA are sufficient to open the whole pathway. However, the results obtained with type I mutants (Table 1) indicated that no uptake of [1-14C]PhAc occurred, suggesting that PCL alone and not nonspecific acyl-CoA synthetases are involved in activation of PhAc. These results also reinforce our previous observations about the absence of PhAc passive diffusion in this bacterium (18) and show that this aromatic compound does not induce (at least in the absence of a functional PCL activity) other permeases which more or less nonspecifically could be involved in the uptake of PhAc.
Analysis of the results obtained with the mutants belonging to the second group (type II) (those lacking acyl-CoA synthetase activity which activates n-PhAs to their CoA thioesters) indicated that neither 6-phenylalkanoic acid nor 8-phenylalkanoic acid, two compounds that generated PhAc-CoA by β-oxidation, act as an inducer of PhAc-CoACC since neither PCL activity nor [1-14C]PhAc uptake was observed (Table 1). Furthermore, these mutants were unable to accumulate PHPhAs, suggesting that acyl-CoA synthetase is the only enzyme that activates n-PhAs to their CoA derivatives. The lack of n-phenylalkanoyl-CoAs implies that the monomers of PHPhAs (3-OH-n-phenylalkanoyl-CoAs) cannot be synthesized.
However, when these mutants were cultured in chemically defined medium containing 4-OH-PhAc and PhAc as carbon sources, both PCL activity and [1-14]PhAc uptake were observed (Table 1), supporting the hypothesis that PhAc-CoA is the true inducer. The fact that the mutants in the third group (those lacking 3-ketoacyl-CoA thiolase activity) were also unable to induce either PCL activity or [1-14]PhAc uptake further supports this hypothesis. Moreover, type III mutants accumulated PHPhAs (Table 1), suggesting that they are able to transport, activate to CoA thioesters, and synthesize the β-oxidation intermediates required for polymerization of these compounds (3-hydroxy-phenylalkanoyl-CoA). These data show that neither phenylalkanoyl-CoA nor its β-oxidative intermediates are able to induce the central pathway involved in degradation of PhAc.
The data in Table 1 indicate that 4-OH-PhAc does not induce the PhAc-CoACC and that, as previously reported by us (4), PhAc or 4-OH-PhAc cannot be polymerized by the enzymatic system involved in synthesis of PHPhAs because of the impossibility of production of the 3-OH-n-PhA monomers required for polymerization.
All the above observations allow us to conclude that the true inducer of the PhAc-CoACC is indeed phenylacetyl-CoA, the first common intermediate of all the pathways involved in degradation of ethylbenzene, styrene, phenylethylamine, tropic acid, trans-styrylacetic acid, and n-PhAs containing an odd number of carbon atoms. Recently, Ferrández et al. (1), using a different approach, have shown that binding of the repressor which controls the catabolic pathway involved in aerobic degradation of PhAc in Escherichia coli W to the DNA promoter region is avoided when PhAc-CoA is present, suggesting (as also shown here) that PhAc-CoA, an intermediate which until recently was considered a catabolic intermediate produced under anaerobic conditions (12), is the true inducer of the PhAc catabolic pathway.
It therefore seems reasonable to propose that each of the independent aromatic compounds indicated above induces its own partial degradative route and that once PhAc-CoA has been synthesized, this molecule opens the common catabolic pathway (the PhAc-CoACC).
Acknowledgments
This work was supported by grant AMB97-0603-C02-01 from Comisión Interministerial de Ciencia y Tecnología (Madrid, Spain) and by grant 1FD97-0245 from Fondo Europeo de Desarrollo Regional. B.G. and E.R.O. are recipients of fellowships from Fondo Europeo de Desarrollo Regional; B.M. is a recipient of a fellowship from CICYT; and D.C. is a recipient of a fellowship from Formación de Personal Investigador.
REFERENCES
- 1.Ferrández A, García J L, Díaz E. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J Biol Chem. 2000;275:12214–12222. doi: 10.1074/jbc.275.16.12214. [DOI] [PubMed] [Google Scholar]
- 2.Ferrández A, Miñambres B, García B, Olivera E R, Luengo J M, García J L, Díaz E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J Biol Chem. 1998;273:25974–25986. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 3.Ferrández A, Prieto M A, García J L, Díaz E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 4.García B, Olivera E R, Miñambres B, Fernández-Valverde M, Cañedo L M, Prieto M A, García J L, Martínez M, Luengo J M. Novel biodegradable aromatic plastics from a bacterial source. Genetic and biochemical studies on a route of the phenylacetyl-CoA catabolon. J Biol Chem. 1999;274:29228–29241. doi: 10.1074/jbc.274.41.29228. [DOI] [PubMed] [Google Scholar]
- 5.Hanion S P, Hill T K, Flavell A, Stringfellow J M, Cooper R. Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology. 1997;143:513–518. doi: 10.1099/00221287-143-2-513. [DOI] [PubMed] [Google Scholar]
- 6.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lageveen R G, Huisman G W, Preusting H, Katelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, Gibson D T. Toluene and ethylbenzene oxidation by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl Environ Microbiol. 1996;62:3101–3106. doi: 10.1128/aem.62.9.3101-3106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long M T, Bartholomew B A, Smith M J, Trudgill P W, Hopper D J. Enzymology of oxidation of tropic acid to phenylacetic acid in metabolism of atropine by Pseudomonas sp. strain AT3. J Bacteriol. 1997;179:1044–1050. doi: 10.1128/jb.179.4.1044-1050.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Blanco H, Reglero A, Rodríguez-Aparicio L B, Luengo J M. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J Biol Chem. 1990;265:7084–7090. [PubMed] [Google Scholar]
- 11.Miñambres B, Martínez-Blanco H, Olivera E R, García B, Barredo J L, Díez B, Moreno M A, Schleissner C, Salto F, Luengo J M. Molecular cloning and expression in different microbes of the DNA encoding Pseudomonas putida U phenylacetyl-CoA ligase. Use of this gene to improve the rate of benzylpenicillin biosynthesis in Penicillium chrysogenum. J Biol Chem. 1996;271:33531–33538. doi: 10.1074/jbc.271.52.33531. [DOI] [PubMed] [Google Scholar]
- 12.Mohamed M E, Fuchs G. Purification and characterization of phenylacetyl-CoA ligase from a denitrifying Pseudomonas sp., an enzyme involved in the anaerobic degradation of phenylacetate. Arch Microbiol. 1993;159:554–562. doi: 10.1007/BF00249035. [DOI] [PubMed] [Google Scholar]
- 13.Olivera E R, Miñambres B, García B, Muñiz C, Moreno M A, Ferrández A, Díaz E, García J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivera E R, Reglero A, Martínez-Blanco H, Fernández-Medarde A, Moreno M A, Luengo J M. Catabolism of aromatics in Pseudomonas putida U. Formal demonstration that phenylacetic acid and 4-hydroxyphenylacetic acid are catabolized by two unrelated pathways. Eur J Biochem. 1994;221:375–381. doi: 10.1111/j.1432-1033.1994.tb18749.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pk18 and pk19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 18.Schleissner C, Olivera E R, Fernández-Valverde M, Luengo J M. Aerobic catabolism of phenylacetic acid in Pseudomonas putida U: biochemical characterization of a specific phenylacetic acid transport system and formal demonstration that phenylacetyl-CoA is a catabolic intermediate. J Bacteriol. 1994;176:7667–7676. doi: 10.1128/jb.176.24.7667-7676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selvaraj G, Iyer V N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983;156:1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]