Abstract
Breast Cancer is the most predominant form of cancer among women worldwide. It has been rigorously studied for biomarker identifications and therapeutic targets. However, various potential genes and their clinical relevance to breast cancer remain unexplored. The heterogeneity of breast cancer is one of the major challenges in early detection. Several studies have reported the significant role of alkaline phosphate (ALP) in the regulation of tumor growth and overall free survival in the pathogenesis of different cancer, including breast cancer which may offer unique therapeutic targets. Therefore, these findings demand a comprehensive study for the biogenesis of ALP genes. This study aims to expression profiling of alkaline phosphate genes in breast cancer and to identify the key pathways and molecular mechanisms underlying breast cancer proliferation and progression. In this study, the transcriptome profiling of invasive breast carcinoma samples was performed and analyzed. We identified that all the ALP genes were downregulated in both Invasive Lobular and Invasive Ductal Carcinoma patients. To understand the underlying molecular mechanism and the clinical significance for these genes in breast cancer, the expression values of genes were measured in adjacent normal and tumor tissues of patients followed by network analysis and functional enrichment analysis. The overall analysis revealed the highly aberrant expression of ALPL gene among all four ALP genes. We identified the functional significance of RUNX2 and WNT3A in deregulating ALPL. Therefore, our findings suggests that downregulation of ALPL could be a potential marker gene for invasive breast carcinoma progression towards bone metastasis.
Keywords: Breast cancer, Invasive lobular carcinoma, Invasive ductal carcinoma, Biomarkers, Alkaline phosphate biomineralization associated, Therapeutic targets
Abbreviations: BRCA, Breast Cancer; ALP, Alkaline Phosphate; ALPL, Alkaline Phosphate Biomineralization associated; DCIS, Ductal carcinoma in situ; DEGs, Differentially Expressed Genes; IDC, Invasive Ductal Carcinoma; ILC, Invasive Lobular Carcinoma
1. Introduction
Breast cancer has been the most frequently observed cancer among women, remains the leading cause of mortality worldwide and the female population of Saudi Arabia is no exception. This makes breast cancer of great interest among scientists and researchers to understand the molecular basis of this dreadful disease to identify the precise therapeutic targets. According to the Saudi Cancer Registry (SCR), the reported number of BRCA cases in 2014 was 28.7% of all female-related cancer cases in Saudi Arabia. The estimated number of breast cancer diagnosed cases according to WHO (World Health Organization) in 2020 was 2,261,419 (11.71%) among all cancer types surpassing lung cancer (Sung et al., 2021) and alone accounts for 3,954 (29%) in Saudi specific population (Xiao et al., 2017). With the advancement in diagnosis approaches, focused therapy and supportive care, the survival rates for breast cancer have continued to increase (DeSantis et al., 2019). However, a 0.3% increase in reported cases within six years states that besides numerous studies on Breast Cancer, other underlying functional aspects remain unnoticed and need to be elucidated as earliest.
Breast carcinoma is majorly classified into two types Ductal Carcinoma In Situ and Lobular Carcinoma In Situ. In breast cancer, at the initial stages, the abnormal cells are restricted to the lining of breast ducts, which may lead to invasive cancer if not treated (Feng et al., 2018). The Invasive Ductal Carcinoma (IDC) and Invasive Lobular Carcinoma (ILC) together account for about 90% of breast cancer, where only IDC accounts for almost 80% of the diagnoses of breast cancer cases, while ILC comprises another 5%-15% (Aiderus et al., 2018). Breast cancer is frequently associated with mutations in genes like BRCA1/2, p53, ATM, and CHD1 (Sultan et al., 2019). In breast cancer, the p53 signaling pathway is activated in response to cellular stresses by other pathways dependent on regulatory kinases and aggressive alteration in p53 often results in worse overall survival. Previous studies have reported the significant role of fatty acid oxidative metabolism in cancer progression by reprogramming their signaling pathways (Koundouros and Poulogiannis, 2020). The gene expression-based biomarker identification has been thoroughly studied and >6,304 breast cancer-associated genes have been identified so far. However, the understanding of the complete underlying mechanism for all the BRCA related genes is still limited. Huge amount of data generated by transcriptomic analysis has been majorly contributing to cancer based research in the identification of marker genes with considerable clinical significance for precise therapeutic targets (Sultan and Zubair, 2019).
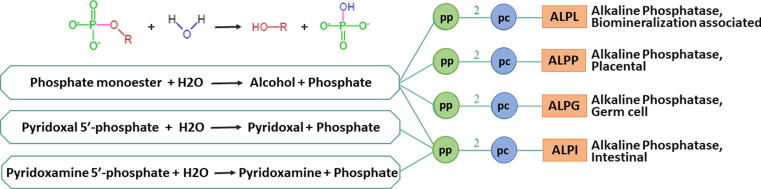
ALP (Alkaline Phosphatase) genes encode for alkaline phosphatase protein members (Sharma et al., 2014). The ALP family includes four groups, namely biomineralization associated/tissue non-specific form (ALPL), intestinal (ALPI), placental (ALPP) and germ-cell (ALPG). ALPL is located on chromosome 1, while the other 3 is located on chromosome 2. The schematic representation of ALP family genes and their constituents is represented in (Fig. 1). Alkaline phosphatase-derived enzyme activities have been reported in different cancer types, but very limited studies are present on their role in breast cancer (Usoro et al., 2010, Yu et al., 2019, Singh et al., 2013, Lou et al., 2021). In a recent study, ALPL gene modulated by retinoic acid has been experimentally observed in breast cancer-specific MCF-7 cell (Fushimi et al., 2020). The physiological functions of ALPL include its effects on vascular endothelial cell growth factor, chondrocyte growth, apoptosis and MAPK signaling (Nam et al., 2017). The biochemical interactions between ALPL and MAP4K1, MAP4KAPK2 and PIK3C2b reflect the evident role of ALPL in MAPK and PI3K-Akt signaling pathways and these pathways also have well-established studies in breast cancer pathogenesis (Han et al., 2017, Bai et al., 2019, Paplomata and O'Regan, 2014, Jiang et al., 2020, Khoury et al., 2020). Therefore, this study aimed to understand the role of ALP genes in relevance with breast cancer pathogenicity.
Fig. 1.
Gene-Reaction Schematic for ALP family genes, where circles represent polypeptides (green) and protein complexes (blue). The lines indicate relationships between them, line from gene (blue circle) indicates that gene codes for that polypeptide (green), line from polypeptide to protein complex circle (blue) indicates that the polypeptide is a subunit of the complex, number over the line indicates the number of copies of the polypeptide in the complex.
2. Material and methods
2.1. Mining of differentially expressed genes (DEGs)
In the present study, breast cancer dataset GSE36295 was retrieved from the Gene Expression Omnibus database in NCBI (National Center for Biotechnology Information) and analyzed in R (v.4.0.0) to identify the differentially expressed genes (DEGs). The dataset encompasses 50 Saudi Arabian subjects of invasive breast carcinoma subtypes. Expression profiling was conducted on Human Gene 1.0 ST GeneChip arrays (Affymetrix) testing platform. In order to identify DEGs in Control vs. ILC and Control vs. IDC group, we have used Bioconductor packages in R, namely affy, limma, RMA, oligo in our customized pipeline. Benjamin and Hochberg (FDR) and t-test methods were utilized to filter the significantly expressed genes. The set of DEGs was curated with the cutoff criteria of p < 0.05 and | logFC | ≥ 1, so genes with logFC ≥ 1 were considered as upregulated and genes with logFC ≤ -1 were considered as downregulated genes.
2.2. Network analysis and target gene identification
The network analysis tool STRING was used to develop the (GGIN) Gene–Gene Interaction Network (Snel et al., 2000). The parameters used for network generation were; all prediction sources enabled medium confidence score ≥ 0.40 and no interactors in the first and second shell. The final GGIN was retrieved for further analysis and visualization in GeneMANIA and Cytoscape (v3.8.0). GeneMANIA (https://www.genemania.org) is a web interface to model protein/gene-protein/gene interactions, protein-DNA interactions, physiological and biochemical responses and signaling pathways. GeneMANIA identified the relevance of DEGs with each other and other neighboring genes. Additionally, the enrichment analysis for the target genes' involvement in biological processes, molecular functions and cellular components was carried out using ClueGO and CluePedia (Bindea et al., 2009, Bindea et al., 2013) in Cytoscape. Additionally, comprehensive gene-pathway interactions were obtained from SIGNOR v2.0 (SIGnaling Network Open Resource) database (Licata et al., 2020).
2.3. Functional classification and pathway enrichment analysis
DAVID (https://www.david.niaid.nih.gov), a database for annotation, visualization and integrated discovery, was used to identify the functional association of the target genes and enriched pathways, a. It is an open-source database that identifies enriched biological processes, molecular functions, and cellular components for the gene of interest. Additionally, the target genes were subjected to pathway enrichment analysis in the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database, representing the biochemical reaction between interacting genes.
2.4. Overall survival analysis
The survival analysis for the significant genes was evaluated by KM plotter (https://kmplot.com/) (Dudley et al., 2016). The primary purpose of the KM Plotter tool is a meta-analysis based discovery and validation of survival biomarkers. It estimates the survival evidence for individual genes in breast cancer based on expression profile cohort of 4929 patients with overall and relapse-free survival from TCGA database. The median survival is computed for 20 target genes and filtered the genes with a probability significance level less than equal to 0.01.
3. Results
3.1. Data normalization and DEGs identification
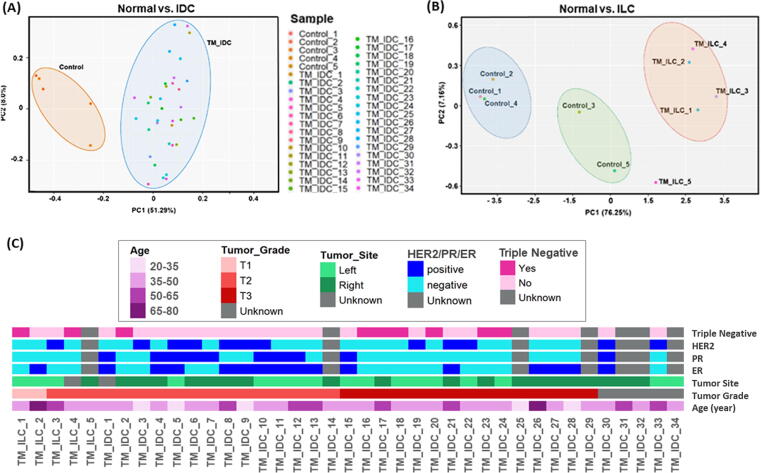
The transcriptome sample array in the dataset GSE36295 was obtained from GEO (NCBI) database. The dataset comprised of 5 control (adjacent normal), 5 ILC, 34 IDC and 6 histologically poorly differentiated samples from female breast cancer patients. The six poorly differentiated samples were not included in this study. The histopathological details, age, tumor grade, site, state of ER (Estrogen Receptor), PR (Progesterone Receptor), HER2 (Human Epidermal growth factor Receptor 2) and Triple Negative is represented in (Fig. 2b). The gene expression was normalized using RMA normalization approach and DEGs were identified using Bioconductor packages (https://www.bioconductor.org/) in R (v4.0.0). DEG analysis was performed with the threshold of |logFC| ≥ 1 and Benjamini and Hochberg adjusted p-value cut-off of 0.05 for Control vs. ILC and Control vs. IDC. We identified a total of 1476 and 786 DEGs in Control vs. IDC and Control vs. ILC samples, respectively (Supplementary Table 1). The Principal Component Analysis revealed the relatedness between the samples of each group for control and cancerous samples (IDC and ILC), as shown in (Fig. 2a) and (Fig. 2b). The samples with similar features are clustered together to differentiate the samples based on aggregate gene expression in normal breast tissue to the invasive breast cancer samples. The identified DEGs common in IDC and ILC was taken further for comprehensive analysis to understand the underlying molecular mechanism and interaction significance of these genes in breast cancer. The annotation based on the clinical description of samples (Fig. 2c) may help clinicians to map the pathological condition of the invasive breast carcinoma patients and the lower expression level of ALPL gene (which could be identified through genetic testing of invasive breast carcinoma patient tumor tissue) may help to consider the urgent need of treatment as our study suggests the deregulated ALPL gene is potential to promote breast calcification and bone metastasis.
Fig. 2.
Principal components analysis of the microarray dataset: Sample distribution in PCA (a) IDC and (b) ILC samples with Normal tissue samples. (c) The clinical details for breast cancer samples, where age, tumor grade, tumor site, HER2, ER, PR and Triple Negative status is represented through a heatmap.
3.2. Gene-Gene interaction network (GGIN) and target gene identification
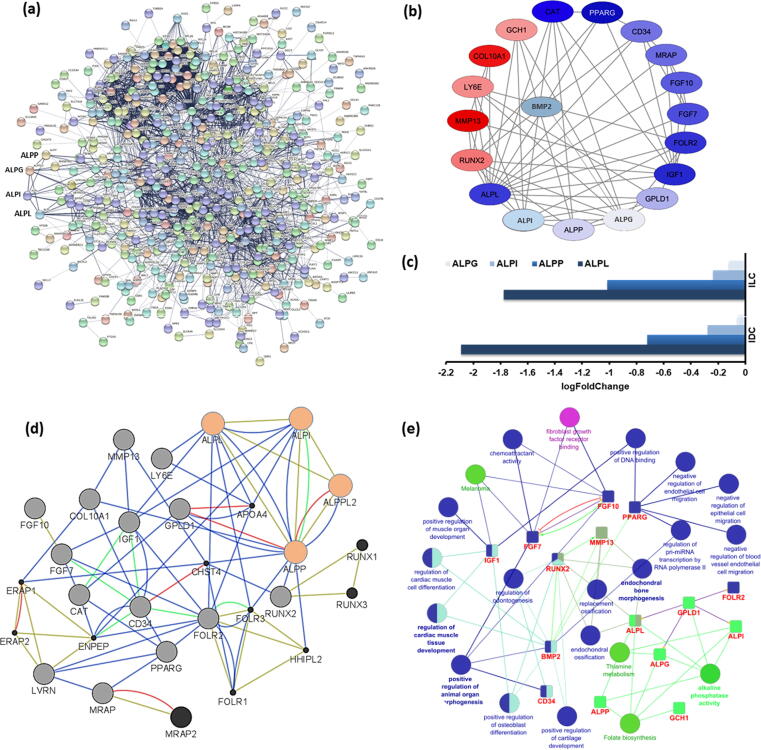
The network analysis was performed to obtain the interaction of the identified differentially expressed genes at the protein level based on average local clustering coefficient of 0.46, average node degree of 11 and enrichment p-value less than 1.0 e−16. A gene-gene interactions network (GGIN) was obtained consisting of 544 genes (nodes) and 2989 interactions (edges) (Fig. 3a). From the GGIN, we observed that ALP genes were highly (highest node degree > 15) interacting with other genes, therefore the genes interacting with the ALP family genes were considered as target genes including ALPL, ALPI, ALPG and ALPP. Among the identified DEGs, PPAR, CD34, COL10A1 were having a well-established role in the pathogenesis of multiple cancer types based on co-expression, co-localization, and pathways. The STRING network was further analyzed in Cytoscape v3.8.0. All the genes interacting with the ALP family genes were considered as target genes (ALPL, ALPI, ALPG/ALPPL2, ALPPP, AQPEP/LVRN, CD34, COL10A1, FGF10, FGF7, GCH1, FOLR2, IGF1, GPLD1, LY6E, MRAP, MMP13 and PPARG) and the final network was plotted with respect to the logFC value of all the genes (Fig. 3b). After that, the interaction attributes between the genes in the network were identified by GeneMANIA (Fig. 3d). Furthermore, ClueGO and CluePedia annotated the target genes and added their interaction types (Fig. 3e). The identified target genes' description and their logFC values from Control vs. IDC and Control vs. ILC analysis are added in Table 1.
Fig. 3.
(a) The Protein-Protein Interaction Network for the differentially expressed genes common in IDC and ILC. (b) The interaction network of target genes is visualized, where node color represents their expression logFC value in cancer samples. The nodes' color shows the expression intensity of each gene (red-downregulation, blue-upregulated genes) and edges represent the interaction between genes. (c) The bar graph showing the expression values of ALP genes in IDC and ILC samples. (d) The interaction attributes obtained from GeneMANIA, where ALP genes are highlighted in orange color, other target genes in grey and additional neighboring genes in black color nodes, color of edges represent interaction types, blue - co-expression, red - physical interactions, green - pathway sharing and yellow - shared protein domain. (e) The Biological Processes (blue), Molecular Functions (purple) and enriched pathways (green) association network are shown. Each group of ontology terms and pathways are represented with different colors based on their functional specificity. The small square nodes represent the target DEGs.
Table 1.
The top 20 differentially expressed hub genes interacting with ALP genes and their expression in invasive ductal (IDC) and invasive lobular (ILC) carcinoma.
| Gene | Gene name | Chrom No.: location (bp): Strand | ILC |
IDC |
||
|---|---|---|---|---|---|---|
| pValue | logFC | pValue | logFC | |||
| ALPL | alkaline phosphatase, biominera-lization associated | chr1: 21509397-21578410:1 | 4.42 × e−3 | −1.776 | 5.47 × e−8 | −2.089 |
| ALPG | alkaline phosphatase, placental | chr2: 232406844-232410714:1 | 5.40 × e−1 | −0.122 | 7.69 × e−1 | −0.060 |
| ALPI | alkaline phosphatase, intestinal | chr2: 232456125-232460753:1 | 1.28 × e−1 | −0.237 | 2.50 × e−2 | −0.278 |
| ALPP | alkaline phosphatase, germ cell | chr2: 232378724-232382889:1 | 5.58 × e−3 | −0.122 | 1.49 × e−3 | −0.722 |
| AQPEP | Laeverin | chr5: 115962454-116027619:1 | 1.98 × e−3 | −1.866 | 3.41 × e−11 | −2.141 |
| BMP2 | Bone Morphogenetic Protein 2 | chr20: 6767686-6780246:1 | 4.94 × e−2 | −0.716 | 3.25 × e−4 | −0.723 |
| CAT | Catalase | chr11: 34438934-34472060:1 | 7.02 × e−5 | −1.801 | 4.27 × e−9 | −1.833 |
| CD34 | CD34 molecule | chr1: 207880972-207911402:-1 | 4.70 × e−3 | −1.239 | 7.15 × e−7 | −1.689 |
| COL10A1 | collagen type X alpha 1 chain | chr6:116118909-116158747:-1 | 9.27 × e−4 | 2.467 | 4.78 × e−3 | 2.208 |
| FGF10 | fibroblast growth factor 10 | chr5:44300247-44389706:-1 | 6.28 × e−4 | −1.849 | 4.85 × e−2 | −1.443 |
| FGF7 | fibroblast growth factor 7 | chr15:49423237-49488775:1 | 4.42 × e−2 | −1.050 | 6.98 × e−6 | −1.944 |
| FOLR2 | folate receptor beta | chr11:72216601-72221950:1 | 7.78 × e−3 | −2.164 | 2.96 × e−6 | −1.980 |
| GCH1 | GTP cyclohydrolase 1 | chr14:54842008-54902826:-1 | 3.13 × e−4 | 1.001 | 1.66 × e−5 | 1.194 |
| GPLD1 | glycosylphosphatidylinositol specific phospholipase D1 | chr6:24424565-24495205:-1 | 9.95 × e−3 | −1.038 | 2.42 × e-6 | −1.294 |
| IGF1 | insulin like growth factor 1 | chr12:102395874-102481744:-1 | 1.28 × e−4 | −1.335 | 2.28 × e−5 | −2.280 |
| LY6E | lymphocyte antigen 6 family member E | chr8:143017982-143023832:1 | 3.08 × e−4 | 1.272 | 1.74 × e−3 | 1.284 |
| MMP13 | matrix metallopeptidase 13 | chr11:102942995-102955732:-1 | 7.13 × e−4 | 3.069 | 7.63 × e-3 | 2.327 |
| MRAP | melanocortin 2 receptor accessory protein | chr21:32291813-32314784:1 | 8.79 × e−3 | −1.552 | 2.34 × e−10 | −1.736 |
| PPARG | peroxisome proliferator activated receptor gamma | chr3:12287368-12434356:1 | 6.33 × e−4 | −2.374 | 5.13 × e−11 | −2.730 |
| RUNX2 | RUNX family transcription factor 2 | chr6:45328157-45664349:1 | 2.39 × e−3 | 1.445 | 2.72 × e−2 | 1.115 |
3.3. Target genes enriched in biological process, molecular functions, cellular components and pathways
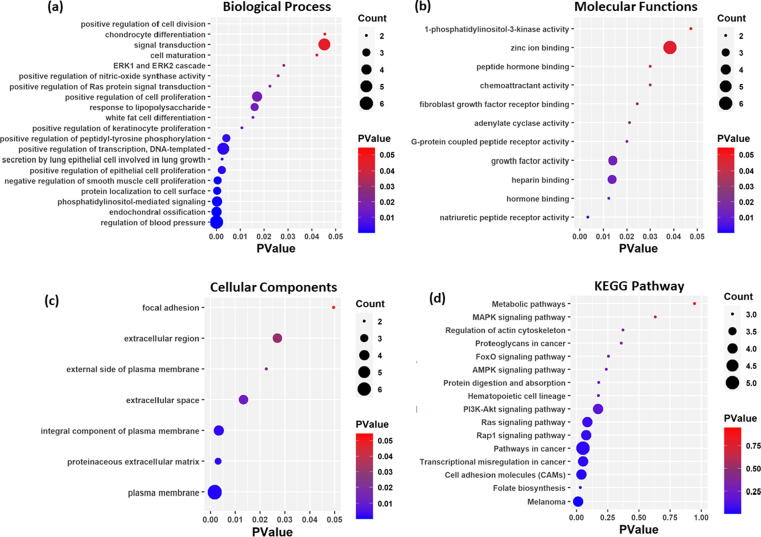
The significantly enriched Gene Ontology terms and the involved genes are presented in Fig. 4(a,b,c). The enrichment analysis of target genes revealed that they are majorly associated with endochondral ossification, cyclic nucleotide biosynthesis, white fat cell differentiation, lipopolysaccharide response, positive regulation of cell proliferation, natriuretic peptide receptor activity, hormone binding, G-protein coupled peptide receptor activity, fibroblast growth factor receptor binding and chemoattractant activity molecular functions. The gene enrichment analysis also identified the DEGs enriched pathways including folate biosynthesis, cell adhesion molecules and regulation of actin cytoskeleton. We observed that ALP genes were enriched in transcriptional misregulation in cancer pathway, pathways in cancer, Rap1 signaling pathway, Ras signaling pathway, PI3K-Akt signaling pathway, AMPK signaling pathway, FoxO signaling pathway, Proteoglycans in cancer, and MAPK signaling pathway, which has been comprehensively studied for their activities in cancer pathogenesis including breast cancer (Fig. 4d).
Fig. 4.
The enriched (a) Biological Process, (b) Molecular Functions, (c) Cellular Components and (d) KEGG pathways involving the target genes. The size of nodes varies based on the number of genes and the color of nodes represents the pvalue (level of significance).
3.4. Overall survival analysis
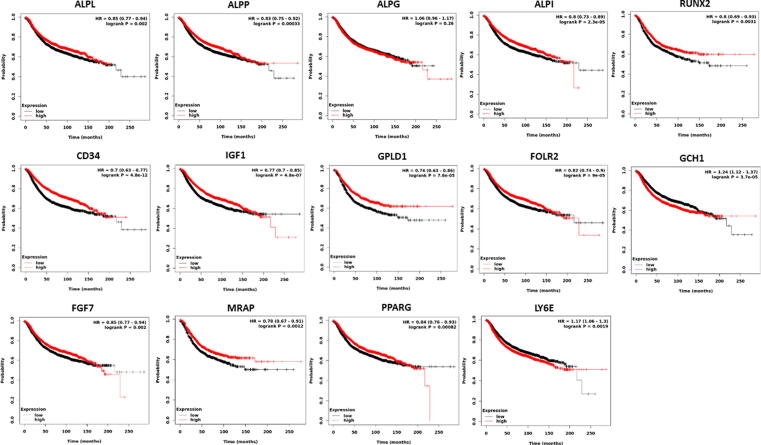
Among the target genes, ALPL, ALPI, ALPP, ALPG, BMP2, GPLD1, IGF1, CD34, FGF7, FOLR2, AQPEP, MRAP, PPARG, CAT, FGF10 were downregulated and LY6E, RUNX2, MMP13, GCH1, COL10A1 were identified as upregulated in both IDC and ILC patients. Survival analysis curves revealed the low expression of ALPL, ALPP, GPLD1, CD34, LY6E is associated with poor survival, while upregulation of RUNX2, LY6E, GCH1 and downregulation of ALPI, ALPG, IGF1, FGF7, FOLR2 and PPARG is associated with overall good survival (Fig. 6). The other target gene was not showing significant survival estimates (logrank Pvalue > 0.01). The survival curves shows poor survival over time for both ALPL and ALPP gene. However, through various studies we found that ALPP gene is primarily expressed in the placenta and is closely related to the intestinal related enzyme; while ALPL plays important role in the growth and development of bones but mutation in ALPL gene may lead to lack of bone calcification (Sharma et al., 2014). Additionally, the expression level of ALPL gene is 2-folds lower than ALPP in both the invasive subtypes. Therefore, the ALPL gene is considered as a targeted gene that is found highly associated with breast cancer calcification and bone metastasis.
Fig. 6.
Kaplan-Meier survival curves estimated by Pearson correlation for the target genes.
4. Discussion
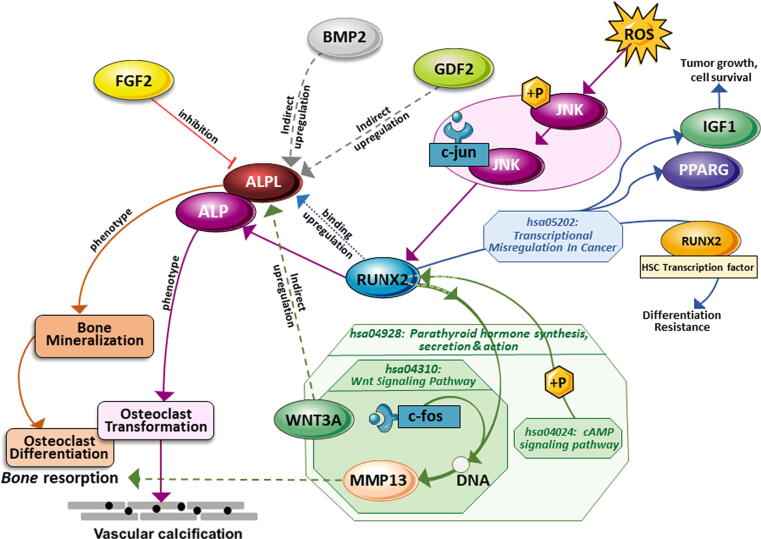
The network analysis was performed based on average local clustering coefficient of 0.46, average node degree of 11 and enrichment p-value less than 1.0 e−16. In the obtained network, the ALP family genes were majorly interacting with other genes, therefore all the genes interacting with ALP family genes were taken as target genes. Furthermore, to identify the functional enrichment specification of the final target genes, they were annotated. The comparative expression analysis for all the four ALP family genes identified that ALPL was highly deregulated than other alkaline phosphate genes including ALPI, ALPG and ALPP in both IDC and ILC (Fig. 3c). Therefore, ALPL gene was further studied to elucidate its role in breast cancer pathogenesis. Pathway enrichment analysis identified the major role of ALPL in thiamine metabolism, folate biosynthesis, metabolic pathways and biosynthesis of cofactors. Additionally, the SIGNOR network revealed that ALPL derived isozyme plays a key role in bone mineralization by regulating diphosphate levels. The ALPL gene is upregulated by RUNX2, WNT3A, BMP2, GDF2, while the transcription factor FGF2 is involved in its inhibition. The inhibition of ALPL by FGF2 supports the finding of our study where we have identified the highly downregulated ALPL expression level. In our study, we found that FGF2 was significantly expressed only in Invasive ductal carcinoma patients but not in Invasive Lobular carcinomas. Therefore, we found that the downregulation of ALPL inhibited by FGF2 reflects the hindrance in bone mineralization which may lead to abnormal cellular functioning resulting in bone metastasis in IDC females. On the other hand, RUNX2 upregulates ALPL gene expression via different oncogenic pathways, including transcriptional misregulation in cancer, Wnt signaling, JNK signaling pathway (Fig. 5). RUNX2 is commonly known as the principal protein-encoding gene for osteogenic differentiation that shows a decrease in bone formation and an increase in fat accumulation (Hu et al., 2018). The functional involvement of RUNX2 includes cell proliferation, epithelial-mesenchymal transition, apoptosis and cancer metastasis. Previous studies have also reported the regulatory roles of RUNX2 in metastatic tumor and cancer cell interactions with bones in various cancer types, including breast cancer and lung cancer (Vishal et al., 2017, Xiao et al., 2021). Moreover, common metastasis of tumors to bones and its prevalence has been frequently observed in prostate, lung, and breast cancers (Wu et al., 2010, Chen et al., 2017). Therefore, the expression alteration of ALPL gene could be a potential biomarker that is directly associated with bone mineralization and differentiation.
Fig. 5.
Schematic diagram of enriched pathways involving the target gene ALPL. The enhanced expression of ALPL due to alteration in RUNX2 via different pathways leads to osteogenesis.
5. Conclusion
Breast Cancer has continued to be one of the global concerns responsible for morbidity and mortality among the female section of the population irrespective of region and caste. However, various biomarkers have been identified and reported for their clinical significance as potential therapeutic targets; few genes have also been observed specific to regional differences. There are various factors defined to be possible reasons for breast cancer, but our knowledge is still limited. In this study, we identified a total of 595 common DEGs from 45 invasive breast carcinoma samples of Invasive Ductal Carcinoma and Invasive Lobular Carcinoma with paired normal breast tissue. A cluster of 20 significantly expressed genes was curated from network analysis. The network analysis aided in understanding the interaction and possible impact of any gene on other respective genes in the same network. We targeted ALP family genes to understand their functional specifications in invasive breast carcinoma. Among the four ALP genes (ALPL, ALPI, ALPL, ALPG), ALPL gene was found highly downregulated in both IDC and ILC patients' tumor tissue which was taken forward for further analysis. To understand the pathogenesis of gene regulation in estimating the cascade of processes associated with disease pathology of breast cancer, enrichment analysis and pathway analysis were performed. Functional enrichment analysis elucidates the function of genes at the molecular level and the biological process in which they are involved along with the molecular pathways in which the gene of interest is actively participating which overall helps in comprehensive analysis and understanding of pathogenesis of the disease on individual gene level. The functional annotation analysis revealed the enriched pathways. We found that ALPL gene was activated by RUNX2 while FGF2 inhibits the functioning of ALPL, which results in abnormal bone mineralization. Also, RUNX2 and BMP2 were involved in transcriptional misregulation in cancer, where the HSC (hematopoietic transcription factor) enhanced RUNX2 was found involved in differentiation resistance and the uncontrolled fusion of hematopoietic transcription factors changes RUNX2 further contributes to tumorigenesis. At the same time, BMP2 acts as a signaling mediator in transcriptional misregulations in cancer. RUNX2 is also identified to be involved in Parathyroid hormone synthesis, secretion and action pathways where the cAMP-induced phosphorylation of RUNX2 activates MMP13 gene, which results in osteoblastic differentiation. Additionally, ROS activation induces RUNX2 and ALP gene via JNK signaling pathway, which has resulted in apoptosis, osteogenic differentiation and vascular calcification. The gene alteration in the observed pathways has been extensively studied for its role in breast cancer pathogenesis. Furthermore, the survival analysis revealed the deregulation of ALPL gene is associated with poor overall survival probability, while the upregulated RUNX2 shows good survival rate over time. These survival curves estimated the survival rate of patient based on high or low individual gene expression levels. From the previous studies, we observed that upregulation of ALPL gene via RUNX2 transcription factor involves normal bone mineralization, while in this study, we found the ALPL gene highly downregulated in Saudi-based females with invasive ductal carcinoma and invasive lobular carcinoma. Moreover, bone metastasis has been frequently observed in various late-stage cancer types, including breast cancer; therefore, our findings suggest that the downregulated ALPL could be a potential biomarker for invasive breast carcinoma bone metastasis progression and demands further analysis with a larger sample size.
Consent for publication
All authors consent to publish this manuscript in the Saudi Journal of Biological Sciences.
Availability of data and material
Data will be available on request by the corresponding or the first author.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This Project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. G: 534- 830- 1439. The authors, therefore, acknowledge with thanks DSR for technical and financial support.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2022.103340.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Aiderus A., Black M.A., Dunbier A.K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer. 2018;18(1):805. doi: 10.1186/s12885-018-4626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z., Yao Q., Sun Z., Xu F., Zhou J. Prognostic Value of mRNA Expression of MAP4K Family in Acute Myeloid Leukemia. Technol. Cancer. Res. Treat. 2019;18:1. doi: 10.1177/1533033819873927. 533033819873927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. J. Bioinform. 2013;29(5):661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. J. Bioinform. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Z., Shen J.F., Zhou Y., Chen X.Y., Liu J.M., Liu Z.L. Clinical characteristics and risk factors for developing bone metastases in patients with breast cancer. Sci. Rep. 2017;7(1):11325. doi: 10.1038/s41598-017-11700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis C.E., Ma J., Gaudet M.M., Newman L.A., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Breast cancer statistics. CA Cancer J. Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- Dudley W.N., Wickham R., Coombs N. An Introduction to Survival Statistics: Kaplan-Meier Analysis. J. Adv. Pract. Oncol. 2016;7(1):91–100. doi: 10.6004/jadpro.2016.7.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Spezia M., Huang S., Yuan C., Zeng Z., Zhang L., Ji X., Liu W., Huang B., Luo W., Liu B., Lei Y., Du S., Vuppalapati A., Luu H.H., Haydon R.C., He T.C., Ren G. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5(2):77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi A., Takeyama H., Tachibana T., Manome Y. Osteogenic cocktail induces calcifications in human breast cancer cell line via placental alkaline phosphatase expression. Sci. Rep. 2020;10(1):12669. doi: 10.1038/s41598-020-69622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Jeng E.E., Hess G.T., Morgens D.W., Li A., Bassik M.C. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017;35(5):463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Yin C., Zhao F., Ali A., Ma J., Qian A. Mesenchymal Stem Cells: Cell Fate Decision to Osteoblast or Adipocyte and Application in Osteoporosis Treatment. Int. J. Mol. Sci. 2018;19(2):360. doi: 10.3390/ijms19020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Dai Q., Su X., Fu J., Feng X., Peng J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol. Biol. Rep. 2020;47(6):4587–4629. doi: 10.1007/s11033-020-05435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury K., Tan A.R., Elliott A., Xiu J., Gatalica Z., Heeke A.L., Isaacs C., Pohlmann P.R., Schwartzberg L.S., Simon M., Korn W.M., Swain S.M., Lynce F. Prevalence of Phosphatidylinositol-3-Kinase (PI3K) Pathway Alterations and Co-alteration of Other Molecular Markers in Breast Cancer. Front. Oncol. 2020;10:1475. doi: 10.3389/fonc.2020.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundouros N., Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer. 2020;122(1):4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata L., Lo Surdo P., Iannuccelli M., Palma A., Micarelli E., Perfetto L., Peluso D., Calderone A., Castagnoli L., Cesareni G. SIGNOR 2.0, the SIGnaling Network Open Resource 2.0: 2019 update. Nucleic Acids Res. 2020;48(D1):D504–D510. doi: 10.1093/nar/gkz949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., Lin W., Zhao H., Jiao X., Wang C., Zhao H., Liu L., Liu Y., Xie Q., Huang X., Huang H., Zhao L. Alkaline phosphatase downregulation promotes lung adenocarcinoma metastasis via the c-Myc/RhoA axis. Cancer Cell Int. 2021;21(1):217. doi: 10.1186/s12935-021-01919-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H.K., Sharma M., Liu J., Hatch N.E. Tissue Nonspecific Alkaline Phosphatase (TNAP) Regulates Cranial Base Growth and Synchondrosis Maturation. Front. Physiol. 2017;8:161. doi: 10.3389/fphys.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paplomata E., O'Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther. Adv. Med. Oncol. 2014;6(4):154–166. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Pal D., Prasad R. Alkaline phosphatase: an overview. Indian J. Clin. Biochem. 2014;29(3):269–278. doi: 10.1007/s12291-013-0408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Pandey A., Tewari M., Kumar R., Sharma A., Singh K.A., Pandey H.P., Shukla H.S. Advanced stage of breast cancer hoist alkaline phosphatase activity: risk factor for females in India. 3 Biotech. 2013;3(6):517–520. doi: 10.1007/s13205-012-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan G., Zubair S., Tayubi I.A., Dahms H.U., Madar I.H. Towards the early detection of ductal carcinoma (a common type of breast cancer) using biomarkers linked to the PPAR(γ) signaling pathway. J. Bioinform. 2019;15(11):799–805. doi: 10.6026/97320630015799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan G., Zubair S. Bioinformatics approaches for big data analytics in precision medicine: An overview. J. Appl. Anal. Comput. 2019:12–15. ISSN 0973-2861. [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer. J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Usoro N.I., Omabbe M.C., Usoro C.A., Nsonwu A. Calcium, inorganic phosphates, alkaline and acid phosphatase activities in breast cancer patients in Calabar, Nigeria. Afr. Health Sci. 2010;10(1):9–13. [PMC free article] [PubMed] [Google Scholar]
- Vishal M., Swetha R., Thejaswini G., Arumugam B., Selvamurugan N. Role of Runx2 in breast cancer-mediated bone metastasis. Int. J. Biol. Macromol. 2017;99:608–614. doi: 10.1016/j.ijbiomac.2017.03.021. [DOI] [PubMed] [Google Scholar]
- Wu Y.Y., Janckila A.J., Ku C.H., Yu C.P., Yu J.C., Lee S.H., Liu H.Y., Yam L.T., Chao T.Y. Serum tartrate-resistant acid phosphatase 5b activity as a prognostic marker of survival in breast cancer with bone metastasis. BMC Cancer. 2010;10:158. doi: 10.1186/1471-2407-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Liu K., Chen J., Gong Y., Zhou X., Huang J. RUNX2 as a Potential Prognosis Biomarker and New Target for Human Lung Cancer. Explor. Res. Hypothesis Med. 2021;12 doi: 10.14218/ERHM.2021.00009. [DOI] [Google Scholar]
- Xiao Y., Ma D., Ruan M., Zhao S., Liu X.Y., Jiang Y.Z., Shao Z.M. Mixed invasive ductal and lobular carcinoma has distinct clinical features and predicts worse prognosis when stratified by estrogen receptor status. Sci. Rep. 2017;7(1):10380. doi: 10.1038/s41598-017-10789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zheng Q., Ding X., Zheng B., Chen X., Chen B., Shen C., Zhang Y., Luan X., Yan Y., Chen W., Xie B., Wang M., Liu J., Fang J., Hu X., Li H., Qiao C., Yang P. Systematic re-analysis strategy of serum indices identifies alkaline phosphatase as a potential predictive factor for cervical cancer. Oncol. Lett. 2019;18(3):2356–2365. doi: 10.3892/ol.2019.10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- Liu Q., Xiang P., Chen M., Luo Y., Zhao Y., Zhu J., Jing W., Yu H. Nano-Sized Hydroxyapatite Induces Apoptosis and Osteogenic Differentiation of Vascular Smooth Muscle Cells via JNK/c-JUN Pathway. Int. J. Nanomed. 2021;16:3633–3648. doi: 10.2147/IJN.S303714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Song W.X., Luo J., Luo X., Chen J., Sharff K.A., Bi Y., He B.C., Huang J.Y., Zhu G.H., Su Y.X., Jiang W., Tang M., He Y., Wang Y., Chen L., Zuo G.W., Shen J., Pan X., Reid R.R., Luu H.H., Haydon R.C., He T.C. BMP-9-induced osteogenic differentiation of mesenchymal progenitors requires functional canonical Wnt/beta-catenin signalling. J. Cell Mol. Med. 2009;13(8B):2448–2464. doi: 10.1111/j.1582-4934.2008.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available on request by the corresponding or the first author.