Abstract
The accumulation of protein inclusions is linked to many neurodegenerative diseases that typically develop in older individuals, due to a combination of genetic and environmental factors. In rare familial neurodegenerative disorders, genes encoding for aggregation-prone proteins are often mutated. While the underlying mechanism leading to these diseases still remains to be fully elucidated, efforts in the past 20 years revealed a vast network of protein–protein interactions that play a major role in regulating the aggregation of key proteins associated with neurodegeneration. Misfolded proteins that can oligomerize and form insoluble aggregates associate with molecular chaperones and other elements of the proteolytic machineries that are the frontline workers attempting to protect the cells by promoting clearance and preventing aggregation. Proteins that are normally bound to aggregation-prone proteins can become sequestered and mislocalized in protein inclusions, leading to their loss of function. In contrast, mutations, posttranslational modifications, or misfolding of aggregation-prone proteins can lead to gain of function by inducing novel or altered protein interactions, which in turn can impact numerous essential cellular processes and organelles, such as vesicle trafficking and the mitochondria. This review examines our current knowledge of protein–protein interactions involving several key aggregation-prone proteins that are associated with Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, or amyotrophic lateral sclerosis. We aim to provide an overview of the protein interaction networks that play a central role in driving or mitigating inclusion formation, while highlighting some of the key proteomic studies that helped to uncover the extent of these networks.
Keywords: amyloid, ALS, Alzheimer, Huntington, misfolding, neurodegenerative disease, Parkinson, protein aggregation, protein interaction network, proteomics
Abbreviations: β-CTF, APP carboxy-terminal fragment; Aβ, amyloid-β; AD, Alzheimer’s disease; AICD, APP intracellular C-terminal domain; AP-MS, affinity purification mass spectrometry; APP, amyloid precursor protein; BioID, proximity-dependent biotin identification; CMA, chaperone-mediated autophagy; ER, endoplasmic reticulum; fALS, familial ALS; HD, Huntingtin’s disease; LB, Lewy body; ND, neurodegenerative disease; NFT, intraneuronal neurofibrillary tangle; PD, Parkinson's disease; PIN, PPI network; polyQ, polyglutamine; PPI, protein–protein interaction; ROS, reactive oxygen species; SN, substantia nigra; Y2H, yeast two-hybrid
Neurodegenerative diseases (NDs) are complex disorders, with multifactorial pathology, that result in progressive damage to neuronal cells and loss of neuronal connectivity, ultimately leading to impaired mobility and/or cognition. Protein aggregation due to misfolding and oligomerization gives rise to extracellular or intracellular inclusions that are a common hallmark for many NDs. Further spreading of these amyloid aggregates in the nervous system–similar to prion-based infections, hence referred to as a prion-like mechanism–is often thought to be a major element in the etiology of NDs (1). In the past few decades, many of the genetic and biochemical causes underlying NDs associated with protein aggregation were uncovered, leading to the distinction between rarer familial forms, where disease-causing mutations are genetically inherited, and the more common sporadic forms, where genetic and environmental risk factors drive the pathogenesis (2). In both cases, the affected proteins are found enriched in pathological aggregates, which highlights their importance in the manifestation of the disease. However, despite the knowledge accumulated and the many clinical trials in which attempts were made to alleviate protein aggregation, to date no therapeutic strategy has been broadly accepted to cure any of the NDs. This led many scientists to question whether protein aggregation is really central to ND etiology or a mere manifestation of other underlying causes (3, 4). Nonetheless, collectively, the work of the past decades generated a more complex understanding of how each aggregation-prone protein engages with many key cellular pathways. In this review, we aim to provide an overview of these intricate connections by bringing together core findings and more recent discoveries.
For each ND, different sets of genes are typically found mutated in the familial forms, and different brain regions and cell types are initially affected. For example, Huntington’s disease (HD) and spinocerebellar ataxia type 1 (SCA1) are linked to the expansion of the CAG repeat of the huntingtin (HTT) and ataxin 1 (ATXN1) genes, respectively, resulting in proteins with an unusually long polyglutamine (polyQ) tract that is very prone to aggregation and causes intracellular deposits in striatal neurons (5, 6). In Alzheimer’s disease (AD), two different types of deposits are observed. The aberrant cleavage products of the transmembrane protein amyloid-β (Aβ) precursor protein (APP) form extracellular plaque deposits in the temporal and parietal brain regions, while the protein tau (MAPT) accumulates intracellularly, in neurofibrillary tangles (7). In Parkinsonś disease (PD), the primarily affected brain area is the substantia nigra (SN), where α-synuclein (α-syn; SNCA) aggregates are found to accumulate in dopaminergic neurons (8). In ALS, cellular aggregates of superoxide dismutase 1 (SOD1), RNA-binding protein FUS (FUsed in Sarcoma), and TAR-DNA–binding protein 43 (TDP-43) have been identified in motor neurons of the primary motor cortex, brainstem, and spinal cord (9). It is therefore important to consider each of these proteins independently and in the context of the cells that are most affected. Note, while we will mostly use short protein names in this review, whenever a gene is linked to ND, the corresponding italicized gene name will also be indicated in brackets if it is different from the protein name.
Protein misfolding and aggregation of disease-associated proteins is facilitated by mutations and posttranslation modifications (e.g., phosphorylation and protein cleavage) that avert formation of the native protein structure, while in some other cases misfolding can also seemingly occur sporadically, without yet a clear explanation. Aggregation is first typically initiated by a seed or/and an oligomer, in which sequence-specific elements of the misfolded protein interact to adopt a non-native conformation, which can then convert other proteins into the toxic form. In many cases, the oligomerization of misfolded proteins leads to the formation of amyloid fibrils with a distinctive β-sheet structure that arise when soluble oligomers begin to assemble into small protofibrils (10). When more proteins are converted into the non-native forms, these protofibrils become longer fibrils that can then form larger cellular inclusions visible by light microscopy. Recently, it has been proposed that oligomerization may be favored by liquid–liquid phase separation of aggregation-prone proteins (11) (Box 1). Moreover, it is now also evident that there are different polymorphs for most amyloid fibrils in vitro and in vivo (polymorph is a term used to indicate the capacity of a polypeptide to generate fibrils with different structures) (12, 13).
Box 1. Liquid–liquid phase separation.
Conventional mechanisms for aggregation of many disease-related proteins proceed through protein misfolding and oligomerization. Recently, increasing attention has been given to the role of liquid–liquid phase separation (LLPS) in this process. Phase-separated droplets provide a concentrated environment where the aggregation process may be accelerated. LLPS occurs when the interactions between molecules in solution are stronger than their interactions with solution to the extent that the entropic cost of demixing is overcome and a condensed phase is formed (393). Beginning with P granules, many cellular membraneless organelles (MLOs) have been shown to exhibit liquid-like properties such as exchanging components with the surrounding environment, deforming under sheer force and fusing (394, 395, 396). This led to the hypothesis that MLOs form through LLPS. While there is a crucial role of LLPS in cellular processes, changes in the properties of phase-separated granules have been linked to NDs. The ability to phase separate in vitro is emerging as a common property of disease-associated proteins. Over time, phase separated granules can mature into more solid, glass-like states (397). The vitrified state consists of thioflavin-positive aggregates for many of the disease-associated proteins discussed here. Phase separation of tau occurs in the presence of polyanionic molecules or RNA-binding proteins such as T-cell intracellular antigen 1 (TIA1) (398, 399). Phosphorylation or the presence of disease-associated mutations in tau promote its phase separation and accelerate the conversion from the liquid-like to solid state eventually forming thioflavin-T–positive fibrils (398, 399, 400, 401). The phase-separation of α-syn requires nonphysiological conditions or long periods of time in vitro (402). However, α-syn localizes to droplets formed by tau through interaction of its negatively charged C-terminal domain with the positively charged proline-rich region of tau (403). The link between phase-separation and neurodegeneration is particularly clear in the case of ALS, where mutations in several RNA-binding proteins have been implicated. RNA-binding proteins are components of many MLOs, and it is thought that their structural properties such as intrinsically disorder regions (IDRs) are important for driving the phase separation of these granules. The interaction between IDRs and RNA can drive phase separation and influence the morphology and dynamics of the resulting protein–RNA granules (404). ALS-associated mutations in IDRs or low-complexity domains of the RNA-binding proteins FUS, TIA1, heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), and TDP-43 accelerate the liquid to solid transition of phase-separated granules in vitro with many eventually forming thioflavin-T–positive inclusions (405, 406, 407, 408, 409, 410). These transitions may influence pathology by disrupting granule functions and trapping RNA-binding proteins and elements of the translational machinery (405). Low-complexity domains are characteristic of HTT that phase separates through weak hydrophobic interactions between its proline-rich region and the polyQ expansion (411, 412). With increasing polyQ length, HTT phase separates more quickly and at lower concentration. The phase separated inclusions eventually convert to solid and irreversible structures. Interestingly, the protein profilin interacts with HTT at its proline-rich domain and reduces its ability to phase separate, as well as reduced the rate of fibril formation (413). This highlights how PPIs and phase separation behavior of disease-associated proteins are intertwined and potentially play a major role in neurodegeneration.
Protein aggregation can cause a series of deleterious events in the cell. First, it will typically lead to a loss of function of the aggregated protein and can then also affect other cellular components that normally form protein–protein interactions (PPIs) with the natively folded protein. In some cases, these interacting proteins will also coaggregate. For each aggregation-prone protein discussed later, we review some of the main known PPIs and briefly discuss their physiological relevance. In several cases, we also underline how some disease-specific mutations may impact these PPIs, especially if this is also linked to aggregation.
In addition, aggregation-prone species can mediate a toxic gain of function due to their ability to engage in aberrant and nonphysiological interactions with different cellular components that would not normally occur (14). Notably, acute or chronic exposure to stress conditions can lead to the unraveling of buried, hydrophobic, and aggregation-prone regions, even in unrelated proteins, thereby inducing their coaggregation with pathogenic polypeptides (15). These gained PPIs can often perturb the normal function of affected proteins. For instance, all the reviewed proteins, in their aggregated form, can interact with mitochondrial proteins resulting in disrupted mitochondrial function. These mitochondrial perturbations are reviewed in more detail in each chapter due to their evident centrality in ND. In addition to interfering with other proteins, aggregated proteins and small protofibrils can also interact with and disrupt cell membranes, which will then exacerbate cytotoxicity (15, 16). As the aggregating proteins can be arranged in different polymorphs, each form may impact the cell differentially, depending on their ability to propagate and to interact with other cellular components. Interestingly, the cellular environment may dictate which conformation is favored, which in turn could impact how the disease will spread (17).
To deal with the challenges posed by aggregation-prone proteins, human cells developed coping mechanisms that largely rely on the protein homeostasis network. Molecular chaperones are central components of this network that maintain protein homeostasis (a.k.a. proteostasis) by facilitating protein folding and disaggregation, as well as by targeting misfolded products for degradation (18). Different cellular pathways can be used to eliminate toxic misfolded proteins, directing them either to the ubiquitin–proteasome or the lysosomal system. Furthermore, the cell displays a protective mechanism that can drive the formation of larger noncytotoxic, or less toxic, inclusions, hence lowering the cytotoxicity of smaller protofibrils via sequestration (19). Nevertheless, the aberrant interactions of aggregating proteins we described previously can also extend to the protein quality control network itself. These can cause deleterious effects by trapping chaperones and reducing the pool of chaperones available for other critical functions, thereby impairing proteasome function and potentially exacerbating the accumulation of aggregated proteins and their cytotoxicity (20, 21). Understanding how the components of the proteostasis network are affected during disease progression could reveal strategies for the treatment of NDs.
The functional characterization of many aggregation-prone proteins associated with NDs has so far proven to be a major challenge. In some cases, the function of the aggregation-prone protein remains to be fully elucidated. Furthermore, the contributing role of the genetic modifiers of ND-causing genes is often poorly understood. Few studies have systematically analyzed the similarities between affected proteins causing the main types of NDs. Therefore, we decided to review past work, first by interrogating the PPI networks (PINs) around the proteins associated with NDs and then by carefully examining how each of these aggregation-prone proteins interacts with the protein homeostasis network and the mitochondria, in order to gain a general view of how the progression of neurodegeneration may impact the cells and to determine any commonalities between these diseases.
Building networks
Identification of PPIs of proteins associated with neurodegeneration was in first guided by knowledge of genes associated with the diseases and the initial characterization of components enriched in protein aggregates, often using immunochemistry. The elucidation of PPIs using targeted approaches—customarily by assessing binding using coimmunoprecipitation—still represents a large portion of our knowledge related to aggregation-prone proteins in neurodegeneration. In most cases, the first unbiased searches to gain better understanding of the role of these aggregation-prone proteins or their binding partners were driven by the yeast two-hybrid (Y2H) method, a technique that emerged over 25 years ago. Advances in PPI identification methodologies in the past 2 decades, including improvement of mass spectrometry instrumentation, have further unraveled a large portion of human PINs, providing additional insights into the functions of proteins involved in NDs. Major contributions to PINs associated with NDs have come from proteomics studies that rely on protein coimmunoprecipitation experiments, which are commonly referred as to affinity purification mass spectrometry (AP-MS) (Box 2), while proximity-labeling approaches (Box 3) are becoming increasingly more popular.
Box 2. Affinity purification mass spectrometry.
Affinity purification mass spectrometry (AP-MS) typically refers to the enrichment and isolation of a particular protein (i.e., bait) and its interaction partners (i.e., prey) using a coimmunoprecipitation method (414, 415). This can be done by the exogenous expression of the bait fused to an affinity tag or by the immunoprecipitation of the endogenous protein with a specific antibody. Antibodies are immobilized on resins such as magnetic or agarose beads and exposed to cellular lysates followed by their retrieval and washing, prior to the elution and digestion of coimmunoprecipitated proteins for shotgun proteomics. Exogenous expression of a bait protein with an epitope tag such as FLAG or HA tag has the advantage of not requiring an antibody for each specific protein. Therefore, the approach can be applied to multiple tagged baits and allow a better control of nonspecific interactions, which are different for each antibody. However, the overexpression of the bait can also lead to false identifications that would not occur at endogenous levels and the tag itself could perturb the bait function. Isotopic antibodies or transfection of the epitope tag alone or fused to other unrelated proteins are typically used as negative controls. Initial efforts to decrease nonspecific background focused on a dual purification scheme (416), but it tends to favor the identification of more stable protein complexes. Alternatively, large scale studies have taken advantage of the AP-MS of other unrelated baits to determine high-confidence interactors (417, 418). These approaches, alongside with more sensitive instruments, have helped to greatly expand PPIs.
Box 3. Proximity labeling.
Proximity labeling of interacting proteins relies on the fusion of the bait protein to an enzyme that generates a reactive, diffusible molecule, which quickly labels lysine residues of nearby proteins in living cells. The main methods used in combination with mass spectrometry are proximity-dependent biotin identification (BioID) and engineered ascorbate peroxidase (APEX), which rely on biotin-AMP reactive molecules and biotin-phenoxyl radicals, respectively. After cell lysis, biotinylated proteins can then be enriched using streptavidin beads. Due to the strength of the interaction between biotin and streptavidin, on bead digestion is typically used (419). The BioID approach is based on a modified form of the Escherichia coli biotin ligase BirA that reduces affinity for biotin-AMP, making it a more promiscuous biotin ligase (420). Later, a smaller biotin ligase was identified (BioID2) and directed evolution was used to generate the TurboID and miniTurbo (28 kDa) (421, 422). Both BioID and BioID2 are typically used with labeling times of 12 to 24 h, while TurboID and miniTurbo produce strong biotinylation after 10 to 20 min. Ascorbate peroxidase is a plant-derived peroxidase that catalyzes the formation of substrate radicals from H2O2 (423). The APEX protein was originally developed for electron microscopy (424). Subsequently, it was adapted for proteomic studies by using biotin phenol as a substrate for labeling proteins that are in close proximity to the bait (425). Using directed evolution Lam et al. identified APEX2 that has an enhanced activity (426). Labeling with APEX is typically carried out for 1 min or less providing much better temporal resolution than BioID, but it may lead to higher nonspecific signal if not carefully controlled.
AP-MS has contributed greatly to the mapping of PPIs and several large-scale studies have significantly expanded our repositories. For instance, the laboratories of Drs Steve Gygi and Wade Harper have now completed AP-MS experiments for 10,128 baits in a series of landmark studies (22, 23, 24). In addition to these monumental efforts, it is important to identify how PINs may potentially adapt in the presence of disease-associated mutations, especially in the context of the NDs that are examined here. Hosp et al. performed an extensive characterization of the interactome of several NDs using quantitative AP-MS to identify interactors of WT and disease-associated mutations from PD, AD, HD, and SCA (25). Notably, they identified a small number of interactors specific to each bait in the presence of a disease-associated mutation that were not found with their WT counterparts. Application of this approach to more cell types and disease-associated proteins will be crucial in uncovering how impacted PINs within affected cells contribute to neurodegeneration.
Proximity labeling techniques offer complementary approaches to AP-MS, especially when subcellular compartments cannot be maintained or insoluble components cannot be immunoprecipitated. By using the engineered ascorbate peroxidase, Markmiller et al., not only determined the composition of stress granules in neuronal cells but also identified changes in stress granule composition in induced motor neurons with ALS-associated mutations in C9orf72 and HNRNPA2B1 ((26)). This study demonstrates the power of proximity labeling in identifying potential mechanisms associated with NDs. Similarly, proximity-dependent biotin identification (BioID) has been used to identify changes in interactions with aggregation-prone proteins. Chou et al. used BioID to identify interactors of TDP-43, including interactors specific to the aggregated form (27). Additionally, BioID has been applied to other disease-associated proteins such as cyclin F (also associated with ALS) and α-syn (28, 29). Importantly, interaction studies have also been applied in the absence of disease models to map the underlying proteostasis network. Piette et al. used a combination of BioID and AP-MS to map the interactors of J-domain proteins and heat shock protein (Hsp) 70s (30). This builds on previous work identifying interactors of chaperones by AP-MS (31) and demonstrates the importance of using these complementary techniques in characterizing PINs.
In addition to protein mass spectrometry, Y2H and other protein complementation assays continue to be used for specifically identifying binary interactions. Haenig et al. recently performed an extensive Y2H screen using nearly 500 bait proteins related to NDs (32). Focusing on inherited ND-causing proteins, they constructed disease-specific networks and validated hits from these networks in patient samples. The ability to study interactions of protein fragments in Y2H has led to specific understanding of differences in PPIs between various disease-associated mutations in α-syn, HTT, and ataxias (33, 34, 35). Proximity ligation assays are also often used to verify that a given PPI occurs in a specific location in the cell or when one of the components cannot be easily assessed using other biochemical approaches (e.g., coimmunoprecipitation). Protein arrays can also be used to probe PPIs and were used to identify interactors for oligomerized α-syn and Aβ (36, 37). While these techniques are not currently widely employed, they offer some unique abilities to probe PPIs in the context of NDs.
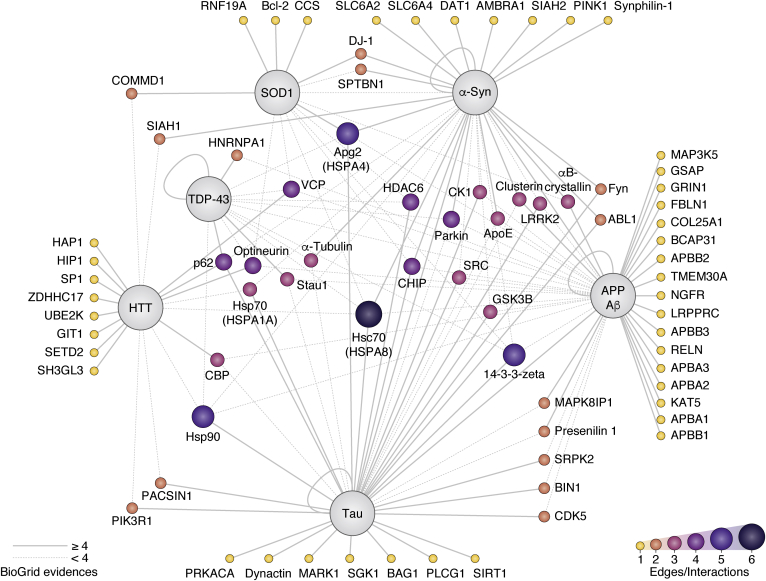
In this review, we focused on six well-known disease-associated proteins that drive aggregation in AD, PD, HD, and ALS (Table S1). For each of the selected protein, we first examined the interactomes (i.e., the ensemble of proteins forming PPIs) that were available in the BioGRID database that included over 714,000 nonredundant human PPIs at the time of writing this review (38). Because a large number of PPIs are reported for a given protein, we initially emphasized mostly PPIs that were reported in multiple independent studies or assessed using different methods, to gain confidence of their potential significance and reduce unspecific hits (Table S1). For instance, there are 554 unique interactors reported for α-syn, but only 18 interactors were characterized in at least four independent observations. We also referenced additional PPIs that were either not yet annotated in BioGRID or placed low in the aforementioned ranking, whenever the PPI association to ND was particularly relevant to a specific area examined in this review. For this analysis, we considered work done in either human or mice. Some proteins known to aggregate such as the major prion protein (PrP) and poly-GA polypeptides translated from C9orf72 with expanded GGGGCC repeats were omitted from this analysis due the relative paucity of information in comparison to other aggregation-prone proteins. For each main ND, we described PPIs with selected aggregation-prone proteins with an emphasis on the elements and events triggering pathogenesis. Whenever possible, we compared both the physiological role of the WT protein and the features distinguishing it from the disease-associated variants. Moreover, we emphasize in each section the PPIs with the proteostasis network and mitochondria. To conclude, we reflect upon the challenges associated with our efforts and outline possible future directions aimed to better handle and integrate the ever-growing PPI databases in the context of NDs.
Alzheimer's disease
AD is the leading cause of dementia and the most frequent ND that is characterized by amyloidogenic proteins in the brain (7). To date, there are no treatments that prevent or slow down the disease. The progression of the disease leads to neuronal loss over time. A potential cause is the accumulation of extracellular deposits of Aβ in plaques and intraneuronal neurofibrillary tangles (NFTs) rich in hyperphosphorylated microtubule-associated protein tau. An initial spike of Aβ levels and a loss of its cellular catabolism is postulated to be the triggering event that leads to the formation of senile plaques and results in the development of the first clinical symptoms within years or decades (7). Recent findings suggest that, although misfolded tau and Aβ are deposited in different brain regions, both phenomena are synergetic in AD (39). However, the exact molecular pathways linking tau and Aβ to neurodegeneration are yet to be fully elucidated.
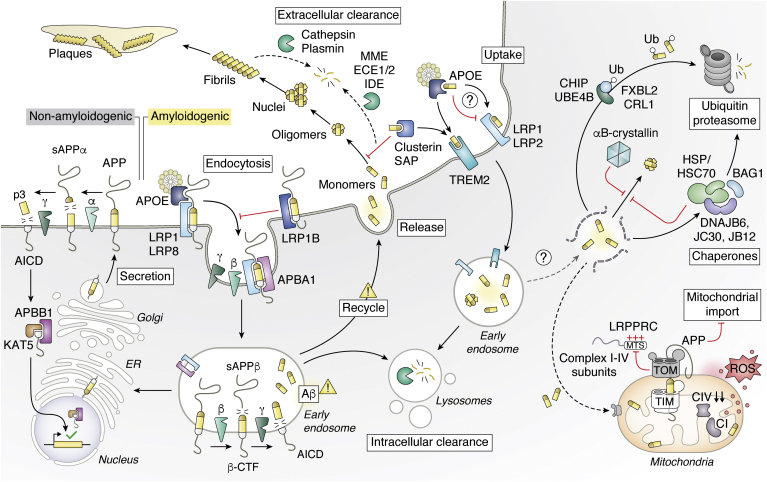
APP is a cell surface receptor and a type I transmembrane precursor protein proteolytically cleaved into a variety of shorter peptides by the transmembrane proteases α-, β-, and γ-secretases. The majority of peptides are nonamyloidogenic products of APP cleavage by α-secretase (ADAM10) and γ-secretase: sAPPα and p3 are released extracellularly and the APP intracellular C-terminal domain (AICD) is released in the cytosol (Fig. 1). AICD then forms a transcriptionally active complex with the adapter protein Aβ precursor protein–binding family B member 1 (APBB1) and the histone acetyltransferase KAT5 (lysine (K) acetyltransferase 5, TIP60) to enhance transcriptional activation (24, 40, 41). Conversely, a small fraction of the precursor protein APP undergoes a different sequential enzymatic processing, generating short amyloidogenic peptides (Aβ), which tend to oligomerize and deposit forming the base of amyloid plaques observed in AD patients' brains. The first step in the formation of the Aβ peptide from APP is catalyzed by the β-secretase 1 (BACE1) or its paralog β-secretase 2 (BACE2) (Fig. 1). These cleave APP after residue 671 generating sAPPβ, a long amino-terminal soluble fragment for extracellular release and a corresponding cell-associated carboxy-terminal fragment (β-CTF). β-CTF is further processed by the γ-secretase, which catalyzes its intramembrane cleavage and yields Aβ for secretion and AICD for intracellular release (42, 43, 44).
Figure 1.
Amyloid β and Alzheimer's disease. At the cell membrane, APP is predominantly cleaved into soluble fragments by α-, β-, and γ-secretases. In the presence of disease-associated mutations, APP cleavage into Aβ fragments increases. Full-length APP also undergoes endocytosis, which influence its processing. Aβ monomers can aggregate to form small soluble oligomers, then amyloid fibrils, and ultimately insoluble amyloid plaques. The extensive processing of APP to Aβ in different subcellar localizations can impact the cell, such as disruption of mitochondrial function. In contrast, interaction with chaperones can modulate oligomerization and aggregation and several proteolytic systems can promote proteolysis of Aβ extracellularly or of related proteins in the cell. Aβ, amyloid-β.
AD is categorized in two subclasses according to the age of onset of the disease: early onset forms of the disease are mostly caused by mutations in the APP or presenilin genes (PSEN1 and PSEN2), while late onset is linked to other genes such as APOE, discussed later, and other environmental factors (45). Increased production of the amyloidogenic form of APP is observed in familial AD-linked mutations of PSEN1 or PSEN2, which encode the catalytic subunit of the γ-secretase complex (46, 47). Because PSEN1 and its homolog PSEN2 directly affect the protease activity of the γ-secretase required for the production of Aβ, they represent major candidates for the development of inhibitory or modulating drugs aimed at preventing AD progression (48). Other components of the γ-secretase complex affecting Aβ levels include presenilin enhancer 2 (PEN2) and anterior pharynx-defective 1 (APH1), both essential for the maturation of the presenilins, as well as nicastrin, necessary for binding β-CTF (49, 50, 51). Mutations in APP have been linked to the autosomal dominant form of AD, with most of its mutations clustered in regions encoding cleavage sites, which favor the generation of Aβ by faulty processing of APP by β- or γ-secretase (52, 53). Other APP mutations within the Aβ sequence increase the self-aggregation tendency favoring amyloid fibril formation (54, 55).
According to the most favored model of Aβ fibrillization, once released, the Aβ monomers can undergo a conformational change shifting from a helical conformation to an abnormal β-sheet structure. In this conformation, monomers start assembling in small oligomers of 3 to 50 Aβ units, which have an increased cytotoxic potential compared to monomeric Aβ and larger insoluble fibrils (56). Some oligomers have seeding activity, which can trigger polymerization of more Aβ into protofibrils. Although protofibrils are still in equilibrium with smaller oligomers, they can rapidly evolve into thermodynamically stable fibrils, which can finally deposit in larger insoluble aggregates, forming plaques (57, 58). The extracellular accumulation of Aβ in neuritic plaques together with the binding of soluble oligomeric Aβ to different exposed cellular receptors has been proposed as a cause of neuronal toxicity in AD. Such toxicity is proposed to derive from the disruption of neuronal homeostasis, the internalization of Aβ leading to cellular defects, and the induction of neurotoxic signals, which can trigger mitochondrial dysfunction or endoplasmic reticulum (ER) stress response (58).
There is a key interplay between plasma membrane proteins that interact with APP or Aβ that impacts AD (Fig. 1). Numerous studies aimed at characterizing the transport of Aβ through the membrane identified low-density lipoprotein receptor-related protein 1, 1B, 2, and 8 (LRP1, 1B, 2, and 8) as major players in AD (59, 60, 61, 62). The aforementioned LRPs interact with a wide range of ligands, including Aβ, the nontoxic precursor APP, and apolipoprotein E (also known as apoE) (63, 64). By interacting with precursor APP, different LRPs play opposing roles in its endocytosis. Whereas, LRP1B retains APP at the cell surface and reduces Aβ peptide production, LRP1 promotes fast endocytosis that results in an increase of APP processing into Aβ (65). Binding of lipid-carrying apoE to LRP8 was shown to recruit the adapter protein Aβ A4 precursor protein-binding family A member 1 (APBA1) and APP, thereby inducing the endocytosis of APP in neuroblastoma cells and leading to increased production of Aβ (Fig. 1) (60). ApoE also plays a central role in AD by directly binding to Aβ. Initially, apoE was shown to bind Aβ and to promote fibrillization of Aβ, particularly isoform 4, which is a major genetic risk factor for AD (66, 67). However, searches for Aβ-binding proteins by affinity chromatography by Calero et al. led to the identification of serum amyloid P component (SAP) and apolipoprotein J (also known as clusterin) as the main plasma interactors of Aβ, while apoE was only marginally enriched (68). Subsequent work confirmed that only a small amount of apoE was bound to Aβ in physiological conditions but that does not preclude the importance of this interaction (69). In fact, apoE has a role in Aβ clearance. Notably, presence of LRP1 on brain microvascular endothelial cells plays a major role to mediate the clearance of Aβ from the brain tissue through the blood–brain barrier via transcytosis (70). Notably, apoE interaction mediates delivery of Aβ to LRP1 and drives its internalization (71). Nevertheless, other reports show that apoE competes with Aβ for the interaction with LRP1, resulting in the suppression of Aβ cellular uptake and clearance (69). One important element is that different cell types may have different ability to uptake Aβ. For instance, the triggering receptor expressed on myeloid cells 2 (TREM2) receptor on microglia cells that binds to apoE and clusterin has been recently found to be involved in Aβ uptake (72). Interestingly, clusterin is a small extracellular chaperone molecule capable of inhibiting Aβ primary and secondary nucleation by suppressing the elongation step of Aβ aggregation in vitro (73, 74). The interplay with plasma membrane proteins and APP or Aβ is very complex and a major challenge is to properly evaluate the importance of each PPI. This is particularly convoluted because multiple genes associated with AD interact with the same protein (e.g., LRP1), the animal models do not perfectly mimic human pathology, and the perturbation of a gene can have different effect in different cell types. For instance, isoform 4 of apoE that is major genetic risk factor for AD in humans is not present in mouse, and the disruption of a LRP can affect APP catabolism in neurons, while it can impact transcytosis in endothelial cells.
Interactions between APP or Aβ with the proteostasis network also play a major role in regulating levels of the different components involved in Aβ production and its oligomerization. Alongside the binding of Aβ to plasma transport proteins, extracellular proteolysis of Aβ prevents its accumulation in the brain (Fig. 1). Aβ can be degraded by four major zinc metalloendopeptidases; including neprilysin (MME), endothelin converting enzymes 1 and 2 (ECE1/2), or insulysin (IDE) (75, 76, 77); or the serine protease plasminogen/plasmin, which can degrade both monomeric and fibrillar Aβ, thereby reducing its toxicity (78). Other proteases with Aβ-degrading activity are the matrix metalloproteases 2 and 9 and the ADAM30-activated cathepsin D (79, 80). In addition to generating Aβ, regulation of APP levels by different cellular proteolytic machineries also plays a major role in AD. For example, ubiquitin ligase ubiquitin-conjugation factor E4 B (UBE4B) can help mediate endocytosis of APP via its ubiquitination, thereby impacting Aβ levels (81). While UBE4B levels are not affected in AD, elements of the endosomal sorting complex required for transport (ESCRT) machinery that further direct endosome trafficking downstream of UBE4B are found expressed at lower levels in AD patients. Importantly, downregulation of ESCRT components and UBE4B leads to higher levels of Aβ production in primary neurons. F-BoX LRR-repeat protein 2 (FBXL2), which is one of the substrate adapters of the Cullin RING ligase 1 (CRL1) ubiquitin ligase, was also shown to interact with APP, and its overexpression in mice reduced levels of Aβ (82). C terminus of Hsc70-interacting protein (CHIP, also known as STUB1) is a E3 ligase that has been shown to target a variety of misfolded proteins for proteasomal degradation. Notably, CHIP binds to both APP and Aβ (83). In addition, CHIP also influences Aβ metabolism by targeting the β-secretase for degradation (84). Interestingly, knockdown of CHIP does not lead to higher levels of APP but reduces the ability of Hsc70 or Hsp70 to interact with APP (83). Earlier proteomic work had shown that several chaperones can bind to cytosolic portion of APP and Aβ, including Hsc70, Hsp90, and especially the small Hsp αB-crystallin that was strongly enriched following affinity purification from brain lysates (85). Interestingly, αB-crystallin interacts with the Aβ fibrils and aggregation-prone misfolded proteins using two different interfaces, indicating that this chaperone has developed distinctive functions (86). A recent study using AP-MS and proximity labeling with molecular chaperones led to the identification of multiple APP interactors such as several components of the Hsp40 family (DnaJB12, DnaJC25, DnaJC30, and DnaJC5) and Hsp70s (HSPA13 and HSPA5) (30). Note that there are 11 human genes encoding for HSP70, and whenever possible we also provide their gene names. This work provides a greater view of the extent of possible PPIs, and together with additional studies it portrays a more refined view on how APP is handled by the protein homeostasis network (Fig. 1). For instance, DnaJC30 and DnaJB6 were also identified as interactors of soluble Aβ oligomers in two different studies using human protein microarray technology (37, 87). Notably, DnaJB6 can capture Aβ oligomers and prevent primary nucleation (88, 89). In contrast, DnaJB12, which was also identified as an interactor of APP-K670N/M671L, the so-called Swedish mutation linked to familial AD, acts as a cochaperone with Hsc70 (HSPA8)—also shown to interact with APP—to promote protein folding, trafficking, and prevention of aggregation (25, 90, 91). Interaction with the proteostasis network within the cell may be key to alleviating Aβ cytotoxicity, especially in relation to mitochondria and perhaps to tau aggregation (see later).
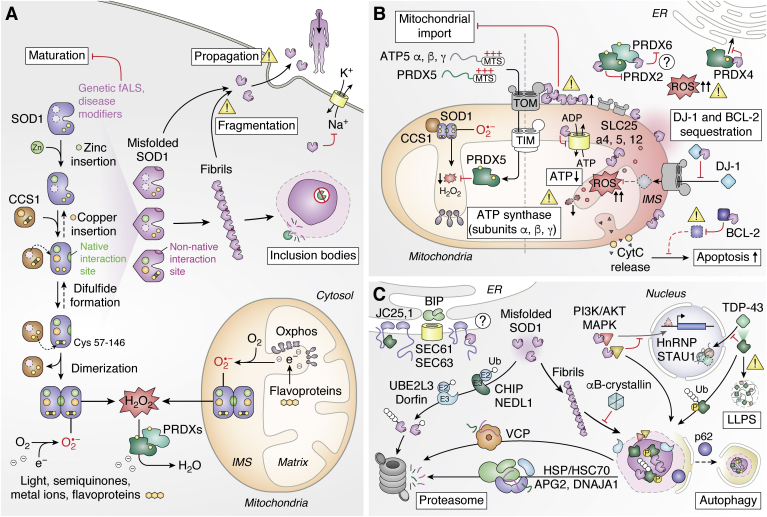
Mitochondrial dysfunction is considered a hallmark of many NDs and AD is no exception. Precursor APP is mainly localized to plasma membrane and ER via its ER-targeting sequence in amino acids 1 to 35; however, as it contains a cryptic mitochondrial targeting sequence between residues 36 to 67, it can also be recruited to the mitochondria (Fig. 1) (92). There it becomes stuck with the cytosolic carboxy terminus associated to the mitochondrial translocase of the outer membrane 40 (TOM40), and the amino terminus is fully imported in the matrix (93, 94). In this conformation, it interacts with other mitochondrial membrane proteins and obstructs the TOM channel, impairing mitochondrial protein import (95). An example of affected mitochondrial proteins is leucine-rich PPR-motif–containing protein (LRPPRC), which is linked to multiple cellular functions including maturation and export of nuclear mitochondrial mRNA (25). Analysis of amyloid plaques from AD patients confirmed that LRPPRC is not found extracellularly, whereas proximity ligation assays in cells and cell fractionation indicated a distinctive mitochondrial colocalization for both APP and LRPPRC (25, 96). Furthermore, LRPPRC has been found to interact intracellularly with early onset AD variants of APP and to induce mitochondrial dysfunction by the dysregulation of mitochondrial gene expression, specifically genes encoding subunits of complex I and complex IV (25, 97, 98). A similar but more direct effect on inner-membrane complexes is observed with internalized toxic Aβ, which can accumulate within the inner mitochondrial membrane and affect complex IV activity (99). Mitochondrial dysfunction is further confirmed in various cellular, transgenic, and human AD models where mitochondrial localization of WT or mutant APP disrupts mitochondrial dynamics, reactive oxygen species (ROS) generation, and energy state, as well as causes a loss of membrane potential (95, 100, 101).
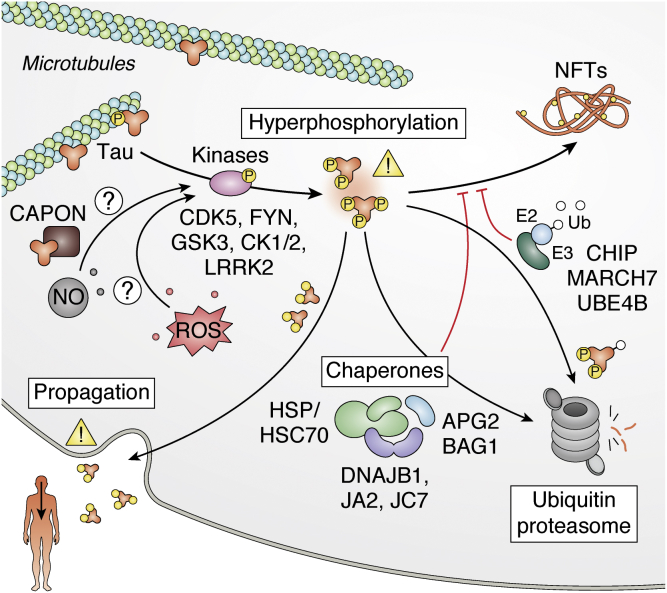
In addition to the major contribution of Aβ in AD, microtubule-associated protein tau is another key player that aggregates. Tau is an intrinsically disordered protein that regulates microtubule stability in neurons. In addition to AD, aggregation of tau is observed in numerous NDs communally termed tauopathies. Notably, numerous tau autosomal dominant mutations that are located near the microtubule-binding domain and promote aggregation have been associated with frontotemporal dementia (102, 103). While phosphorylation plays a role in regulating tau function, tau is found hyperphosphorylated in NFTs (Fig. 2). Notably, phosphorylation events can modulate both PPI and the aggregation propensity of tau in a complex manner. For instance, whereas 14-3-3ζ interacts with unphosphorylated tau and promotes its aggregation, tau phosphorylation on S134 increases 14-3-3ζ affinity but, in this case, reduces tau aggregation (104, 105). Other phosphorylation events have been shown to promote aggregation, and several of these posttranslation modifications are only identified in the context of neurodegeneration (106). Especially, several kinases have been proposed to promote tau aggregation like glycogen synthase kinase-3 (GSK3), casein kinase 1/2 (CK1/2), leucine-rich repeat kinase 2 (LRRK2, PARK8) and Fyn, and many have been considered as candidates for potential AD therapeutics (107, 108, 109, 110). For instance, inhibition of cyclin-dependent kinase 5 (CDK5) that interacts with tau restores long-term potentiation in mice indicating it may mitigate some AD traits (111). However, an effective compound has yet to be identified, and other approaches may need to be taken in consideration to successfully treat AD by taking in account other PPIs. Screens for tau-interacting proteins in transgenic mice expressing WT human tau led to the identification of new interactors like the adapter protein of neuronal nitric oxide (NO) synthase CAPON (carboxyl-terminal PDZ ligand of neuronal NO synthase protein), which can induce tau aggregation and links tau pathology to the production of NO (112). Lowering CAPON levels could suppress tau pathology. Moreover, cell survival could be achieved by ensuring prevention of tau deposition or clearance of hyperphosphorylated tau intracellularly. Components of the ubiquitin system like E3 ligases CHIP, membrane-associated ring-CH-type finger 7 (MARCH7), and the E2 enzyme ubiquitin-conjugating enzyme E2 W (UBE2W) are in place to promote ubiquitination of tau and the removal of its oligomers by the proteasome (113, 114, 115). Hsp70/Hsc70s (HSPA8, HSPA4, HSPA1A) and Hsp90s (HSP90AA1) interact with amyloid structures to, in cooperation with DnaJ proteins or BAG family molecular chaperone regulator 1 (BAG1), modulate their assembly and disassembly or promote client tau degradation (116, 117, 118, 119, 120, 121). Notably, Hsp70 prevents formation of tau aggregates alongside the DnaJA2 and DnaJB1 cochaperones (121, 122, 123). Interestingly, recent work shows that, while DnaJA2, DnaJB1, and Hsc70 all bind to the same tau region, Hsc70 binds preferentially to the monomeric tau, DnaJB1 binds to the oligomerized form, and DnaJA2 binds to both (124). Another recent proteomic study revealed that DnaJC7 is yet another cochaperone that interacts with tau (125). In this case, the cochaperone specifically binds with higher affinity to the natively folded tau, and this interaction is thought to stabilize the native conformation to prevent aggregation. In addition, the small HSP Hsp27 (HSPB1) can also delay formation of tau aggregates via a weak interaction with the small fibrils (126). These studies illustrate how different elements of the proteostasis network exert distinct but complementary roles to prevent protein aggregation in the cell.
Figure 2.
Tau and Alzheimer's disease. Tau associates to the cytoskeleton components. However, in a hyperphosphorylated state, tau can oligomerize and lead to the deposition in NFTs. Chaperones and components of the ubiquitin proteasome system help prevent the aggregation process. NFT, intraneuronal neurofibrillary tangle.
The “relationship” between Aβ and tau in AD is still a matter of debate. Earlier work showed that Aβ can directly induce tau aggregation in vitro and that it may enhance tau phosphorylation (127, 128). However, it is still unclear how Aβ promotes the formation of NFTs, especially since aggregate deposition occurs in different brain regions. There are two important elements: (1) the pattern of NFT deposition in AD has a clear correlation with the disease severity and is actually used to define the six different Braak stages to describe the disease progression (Braak stages are also used in PD, where it follows the spread of α-syn aggregation rather than NFT) and (2) there is now increasing evidence that Aβ formation precedes tau aggregation. For instance, specific phosphorylation events on tau coincide with Aβ formation up to 2 decades before other sites are phosphorylated and tau aggregation begins (129). In addition, Aβ plaque favor an environment for rapid spreading of tau aggregates (130). Recent work also suggests that microglial cells activated by Aβ may play an important role in promoting phosphorylation of tau, notably via the activation of the p38 mitogen-activated protein (MAP) kinase (131). In addition, isoform 4 of apoE, associated with increased risk of AD, also aggravates tau aggregation (132). We will likely continue to refine our understanding of the interplay between Aβ and NFTs in the upcoming years. This relationship is key to a better comprehension of AD etiology and strategies for therapeutics.
Parkinson's disease
PD is the second most common ND and is characterized by motor abnormalities and neuropsychiatric disturbances derived from selective degeneration of dopaminergic neurons in the SN (8). Approximately, 85% of PD cases are sporadic, whereas the remaining 15% represent autosomal dominant or recessive familial PD cases linked to genetic factors (133). At present, mutations in over 19 genes have been associated with PD development, including those encoding α-syn (SNCA/PARK1), parkin (PRKN/PARK2) E3 ubiquitin ligase, PTEN-INduced kinase 1 (PINK1, PARK6), deglycase DJ-1 (PARK7), and LRKK2 (PARK8) (133, 134). Among them, α-syn is the most extensively studied, followed by parkin. However, as none of these PD-associated genes has a 100% penetrance, one of the prevalent models involves a synergistic action between multiple factors causing both familial and sporadic PD (133). We will mostly discuss PPIs with α-syn, as well as summarize parkin/PINK1 function in this review.
The presence of Lewy bodies (LBs) in some neuronal tissues is a key feature of PD. LBs are protein-rich inclusions that contain α-syn among potentially more than 100 different proteins (135). Despite the high concentration of α-syn in neurons, these toxic inclusion bodies do not occur in healthy individuals. Several SNCA mutations and gene duplication events are linked to rare cases of familial PD and promote α-syn fibril formation. In addition to PD, dementia with LBs and multiple system atrophy are the main synucleinopathies characterized by the presence of α-syn inclusions. Identifying and understanding the cellular mechanisms in place to reduce accumulation and propagation of misfolded α-syn in the brain will likely play a major role in the design of therapeutics to prevent synucleinopathies and slow down the progression of PD. Recent structural analyses provided new insights into α-syn fibril structures (136, 137). Interestingly, PD-linked point mutations in α-syn are all predicted to impact the fibril structure. Notably, α-syn A53T fibrils have a smaller interface in comparison to fibrils generated with WT α-syn, which may explain why these fibrils are less stable thereby promoting seeding propagation (138). Similarly, E46K causes a distinct conformation that is less stable in some experimental conditions (139, 140). One key element will be to distinguish whether some of these polymorphs may cause a gain of cytotoxicity by forming new protein interactions.
While the exact physiological function of α-syn is still debated, there is increasing evidence that it plays a role in regulating the homeostasis in synaptic vesicles owing to its ability to bind to multiple synaptic proteins and the cell membrane (Fig. 3A). α-Syn is a small intrinsically disordered protein of 140 amino acids that is localized in presynaptic termini. In its monomeric form, it regulates synaptic vesicle trafficking and subsequent neurotransmitter release, by increasing local calcium release from microdomains to enhance ATP-induced exocytosis (141). In its multimeric membrane-bound state, it supports the assembly of the SNAREs synaptic fusion components at the presynaptic plasma membrane, a function that becomes crucial during aging (142). α-Syn interacts with several cytoskeleton components. Both α/β tubulins have been found to bind α-syn in AP-MS experiments with human brain samples from PD patients and to colocalize with α-syn-positive pathological structures (143). As well, α-syn binds tau, and both proteins have a synergetic effect toward their polymerization into fibrils (144, 145). In a recent study, in which PPIs of a subset of LB-localizing proteins were assessed using a combination of nanobead luminescence and two-color coincidence detection assays, tau was shown to bind more strongly to preformed α-syn fibrils rather than to monomeric and oligomeric α-syn (146). In addition to its association with the microtubular network, α-syn has also been found to associate with dopamine transporter 1 (DAT1), and may thereby regulate dopamine neurotransmission by modulating DAT1 levels at the cell surface by tethering the transporter to microtubules (147, 148, 149). The interaction of DAT1 with the familial PD-linked α-syn A30P mutant is enhanced compared to the WT, thereby decreasing DAT1 surface expression and activity (150). In addition to mutations, posttranslation modifications can also impact PPIs. Notably, α-syn phosphorylated on S129 is found enriched in LB where it accumulates upon progression of the disease (151, 152). While the impact of S129 on PD is not yet fully elucidated, it can clearly induce multiple effects (153). In a comparative proteomic study using different α-syn peptides, McFarland et al. showed that different subsets of interactors are affected depending on whether α-syn is phosphorylated (154). Nonphosphorylated α-syn coprecipitates with protein complexes enriched for mitochondrial proteins (many in complex I), whereas phosphorylation on either S129 or Y125 leads α-syn to interact with more cytoskeletal and vesicular trafficking proteins. In addition, oligomerization of α-syn can also lead to novel nonphysiological PPIs. For instance, Tanudjojo et al. recently showed that upon formation of fibrils, there is a gain of interaction with DJ-1 (155). Notably, KO of DJ-1 also increases the aggregate-induced cytotoxicity, which illustrates how a gain of interaction may aggravate the outcome at the cellular level.
Figure 3.
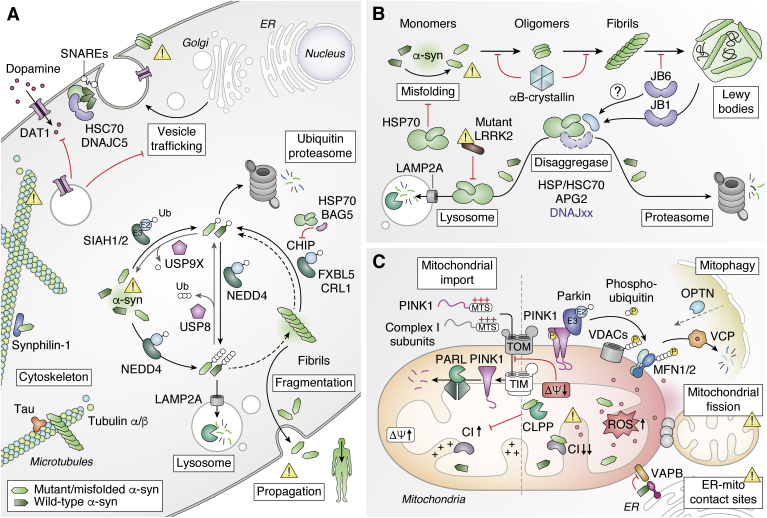
α-Synuclein and Parkinson's disease.A, α-syn regulates synaptic vesicles and neurotransmitter release, which is disrupted upon oligomerization and aggregation. Higher levels of α-syn promotes oligomerization and several pathways control α-syn proteolysis (see also panel B). B, accumulation of misfolded α-syn leads to oligomerization and formation of insoluble amyloid fibrils and Lewy bodies. Several chaperone proteins can modulate α-syn aggregation or promote degradation. C, PD-related mutations alter mitochondrial function. Disruption of normal parkin or PINK1 activity by familial PD-associated mutations leads to impaired clearance of defective mitochondria. Direct interactions between mutant α-syn and mitochondrial proteins drive the formation of ROS and alter membrane potential. PD, Parkinsonś disease; ROS, reactive oxygen species.
The interaction between α-syn and the proteostasis network plays a major role both to promote normal function of α-syn and to prevent toxic gain of function interactions. Interactions with Hsc70/Hsp70, Hsp90, and small HSP αB-crystallin have been shown to reduce oligomerization of α-syn in vitro (Fig. 3B) (156, 157, 158, 159). In addition to preventing oligomerization, other chaperones can also display disaggregase activity and disassemble α-syn fibrils. This is the case with DnaJB1, Hsp70, and the Apg2 nucleotide exchange factor that convert cytotoxic fibrils to nontoxic monomeric α-syn in an ATP-dependent manner (160). Another molecular cochaperone, DnaJB6, has been shown to suppress the aggregation of seeded α-syn in cells and in animal models of PD (146, 161, 162, 163). Different chaperone proteins can bind to different populations of misfolded α-syn. Notably, while the small HSP αB-crystallin preferably binds to seeding oligomers, DnaJB6 interacts instead with assembled fibrils (146). Distinctly, recent work using in-cell nuclear magnetic resonance indicates that a majority of monomeric α-syn could be bound to chaperone proteins inside the cell (164). Interestingly, the binding of α-syn to chaperones and cell membranes is mutually exclusive, as it is mediated via the same N-terminal region of α-syn. Protein mass spectrometry confirms that a large portion of PPIs with that N-terminal region of α-syn are with chaperone proteins. Therefore, inhibition of chaperone interactions increases localization of α-syn at cellular membranes, including with mitochondria, and the aggregation of α-syn. The equilibrium between free-state, membrane-bound, and chaperone-bound α-syn is likely to play a major role in PD etiology (Fig. 3B). Notably, phosphorylation of α-syn Y39, a posttranslation modification that increases with the severity of the PD and is found in LB, perturbs both the interactions with chaperones and cell membranes, and could therefore greatly accelerate α-syn aggregation (164, 165, 166).
As SNCA gene duplication is linked to PD, regulation of α-syn levels in the cell likely plays a major role in the etiology. A subset of α-syn is ubiquitinated in LBs, indicating that ubiquitination may play an important role in regulating α-syn levels (167). Several ubiquitin ligases have been found to mediate α-syn ubiquitination (Fig. 3A). Parkin E3 ligase was first shown to coimmunoprecipitate with and ubiquitinate glycosylated α-syn (168). Parkin was also subsequently shown to ubiquitinate synphilin-1 (SNCAIP), a protein that interacts with α-syn and is enriched in LBs (169). However, subsequent work on parkin has been more focused on its role with PINK1 in regulating mitophagy, a specialized autophagy process in which defective mitochondria are targeted to lysosomes (see later) (170). Instead, more attention was given to other E3 ubiquitin ligases that may potentially conjugate α-syn. The Seven In Absentia Homolog (SIAH) 1 and 2 E3 ubiquitin ligases were shown to ubiquitinate α-syn, which also promotes its aggregation (171). In contrast, the deubiquitinating enzyme ubiquitin-specific peptidase 9 X-linked (USP9X) interacts with ubiquitinated α-syn, and downregulation of USP9X accelerates turnover of α-syn in a proteasome-dependent manner in tissue culture cells indicating that ubiquitin, in this case, promotes turnover—and not aggregation—of α-syn (172). Similarly, ubiquitin-specific peptidase 8 (USP8) is another deubiquitinating enzyme that interacts and reduces turnover of α-syn and is enriched in LBs with less ubiquitinated α-syn in the SN (173). Downregulation of USP8 promotes the accumulation of K63 polyubiquitin chains on α-syn and its lysosomal degradation (K63 polyubiquitin chains are typically not associated with proteasomal degradation). The neuronal precursor cell-expressed developmentally downregulated 4 (NEDD4) E3 ligase that binds to the C-terminal region of α-syn promotes its targeting to the lysosomal pathway, a pathway that was found “druggable” as it mitigates α-syn cytotoxicity (174, 175). In addition to the ubiquitination of the monomeric α-syn, other E3s have been shown to target or affect α-syn fibrils. The CHIP E3 ligase, which also targets tau and accumulates in LBs, can reduce the accumulation of oligomerized α-syn in tissue culture cells (176). CHIP is recruited to preformed fibrils internalized in tissue culture cells. Crosslinking coupled to mass spectrometry experiments show that the interaction with the fibrils is at least mediated by CHIP U-box that recruits the E2 ubiquitin-conjugating enzyme (177). The potential role of this E3 enzyme in regulating α-syn is convoluted, as CHIP also binds Hsp70 via its tetratricopeptide repeat domain. For instance, while CHIP can ubiquitinate α-syn in vitro and in transfected cells, this activity is inhibited by BAG5, which is recruited via Hsp70 (178). The impact of CHIP on α-syn regulation would need to be carefully re-evaluated as many initial experiments relied on transient transfection and overexpression in tissue culture cells. More recently, the CRL1 E3 ligase with F-box/LRR repeat protein 5 (FBXL5) was shown to interact with α-syn fibrils that were taken up by cells (179). Different components of CRL1 colocalize with internalized α-syn fibrils that form inclusions, especially after rupture of the lysosome. Cullin-1, the main scaffold component of CRL1, is required to mediate α-syn ubiquitination and downregulation of different components of CRL1, including the substrate adapter FBXL5, leads to an increased accumulation of internalized α-syn in tissue culture cells. Remarkably, downregulation of FBXL5 or Skp1 (another component of CRL1) in mice allows more spreading of LB-like pathology upon administration of exogenous α-syn fibrils in the ipsilateral brain region, where expression of the E3 components was targeted by a lentivirus. Collectively, past and emerging work indicate that there are multiple E3 ubiquitin ligases that can regulate cellular α-syn levels, whether it is in its monomeric form or assembled in fibrils.
Several additional pathways have been implicated in the clearance of α-syn in a ubiquitin-independent manner. Chaperone-mediated autophagy (CMA) is a cellular mechanism ensuring normal cellular function by degrading misfolded proteins via the lysosomes (180). It is of particular relevance in cells that have limited or no division capacity, such as neurons, and is therefore incapable of diluting accumulated, damaged, and toxic intracellular components. α-syn and LRRK2, another gene linked to familial forms of PD, both contain the CMA-specific recognition motif (KFERQ), which is recognized by Hsc70 (HSPA8) and targets the substrate to lysosomes via the lysosome-associated membrane protein 2 (LAMP2A) membrane receptor prior to the degradation by intralysosomal proteases (Fig. 3B) (181, 182, 183). Importantly, while A30P and A53T mutations of SNCA lead to an increase in binding with LAMP2A, these α-syn mutants are not properly cleared by CMA (181). Similarly, LRRK2 R1441G PD-linked mutation induced accumulation of α-syn oligomers in aged mutant brains by impairing CMA protein clearance (183, 184). These inhibitory effects on CMA could cause toxicity in PD by hindering the degradation of α-syn. In addition to cellular clearance, it is also becoming apparent that cells can expel protein aggregates via exosomes or other specialized mechanisms (185). Recent work has highlighted the contribution of the misfolding-associated protein secretion for secretion of several ND-associated proteins, such as tau and α-syn, that relies on the DnaJC5 cochaperone (186). Interestingly, Trepte et al. report that DnaJC5 binds to α-syn using a double bioluminescence-based two-hybrid technology (termed LuTHy) (187). Both α-syn and DnaJC5 localize to neuronal synaptic vesicles, where they support the folding or assembly of SNAREs and promote synaptic function (188, 189). This interaction may also suggest that, together with Hsc70, the role of DnaJC5 could extend to triaging misfolded proteins associated with NDs. Secretion of α-syn could both protect cells from cytotoxicity, as well as mediate spreading of the pathology in a prion-like manner.
Mitochondrial dysfunction has emerged as a common thread in PD pathophysiology, which has been also highlighted in a recent review (190). We will therefore only emphasize a few key points by first summarizing how other PD-associated genes impacts mitochondria homeostasis before highlighting contributions from α-syn (Fig. 3C). Earlier work characterizing brain tissues from PD patients noted the presence of mitochondrial defects, oxidative stress, and a predominant decrease in complex I activity in the SN (191). Similar PD-like phenotypes can be achieved upon exposure of mouse brains to mitochondrial drugs, which either directly inhibit complex I activity (e.g., the rotenone herbicide) or hamper mitochondrial import (e.g., CCCP or carbonyl cyanide m-chlorophenyl hydrazine, an uncoupler of oxidative phosphorylation) (192). Moreover, several nuclear genes encoding familial PD-linked mutant proteins regulate mitochondria homeostasis, such as CHCHD2, DJ-1, parkin, and PINK1. Notably, the PINK kinase and the parkin ubiquitin ligase play a major role in the regulation of mitophagy, in which defective mitochondria can be targeted for lysosomal degradation via macroautophagy. While PINK1 is normally imported and degraded in mitochondria by presenilins-associated rhomboid-like (PARL), low membrane potential impairs its import, thereby allowing the kinase to then phosphorylate ubiquitin and parkin, which is in turn activated (193, 194, 195, 196). Ubiquitination of a first subset of mitochondrial proteins, including mitofusins, leads to their proteasomal degradation and promotes fission (197, 198). Conjugation of other mitochondrial proteins (e.g., VDAC1; voltage-dependent anion-selective channel 1) then mediates the recruitment of autophagy receptors, such as optineurin, that guide the formation of the autophagosome membrane to eliminate the damaged mitochondria (199, 200).
Mitochondria homeostasis is also impacted by α-syn (Fig. 3C). Earlier work demonstrated that overexpression of α-syn in neuronal cells caused a change in mitochondrial morphology, leading to shorter, wider, and more fragmented mitochondria, which is accompanied by increased levels of free radicals (201, 202). Similar effects on mitochondria were also observed in mouse models (203). A cryptic mitochondrial-targeting signal in the first 32 amino acids in α-syn confers mitochondrial localization in human dopaminergic neurons (204). The mitochondrial localization was enhanced for the A53T mutant in comparison WT α-syn. Similarly, there was more mitochondrial α-syn in the SN of patients with PD in comparison to controls. Subsequent work confirmed that α-syn can directly associate with mitochondrial membrane in neuronal cells to drive mitochondrial fission (202, 205). PD-associated α-syn mutants also reduce mitochondria ER contact sites (also known as MAM or mitochondria-associated ER membranes) owing to its PPI with vesicle-associated membrane protein (VAMP)–associated protein B (VAPB), which is enhanced with the A30P and A53T mutations (206, 207). In mitochondria from PD brains, as well as cultured cells, WT and A53T α-syn interact with complex I subunits and increase the production of ROS (204). In addition, α-syn interacts with the mitochondrial matrix peptidase CaseinoLytic protease P (ClpP), which is directly involved in complex I maintenance and mitochondrial energy metabolism (208). α-syn suppresses ClpP protease activity thus triggering mitochondrial oxidative damage and neurotoxicity. Aggregating α-syn drives additional PPIs that impact mitochondria. Oligomeric, but not monomeric or fibrillar, α-syn interacts with TOM20 and impairs mitochondrial import (209). In addition, α-syn oligomers are located in close proximity to the ATP synthase and cause its oxidation, which is then linked to the observed increased transition pore opening that triggers neuronal cell death (210).
The convergence of parkin, PINK1, and α-syn on mitochondrial dynamics as well as the interconnectivity of all these genes related to familial PD reveals an intricate network in which it is hard to disentangle the cause and effect of the disease. An apparent common involvement of these PARK genes in mitochondrial stress response provides a potential physiological basis for the prevalence of α-syn pathology in PD.
Huntington's disease
HD is an autosomal dominant inherited neurodegenerative condition that presents with progressive motor, cognitive, and psychiatric impairments due to neuronal dysfunction and extensive cell loss mainly in the cerebral cortex and the striatum (211). After the onset of symptoms, the average survival time is about 15 years, and there is currently no effective treatment for HD. A pathological expansion of the trinucleotide CAG in the first exon of the HTT gene is the main cause of the disease, encoding a mutant HTT (mutHTT) with expanded polyQ stretches. In healthy individuals, the CAG sequence is repeated 9 to 35 times, whereas it exceeds 35 repeats in the HD population (212). HTT is ubiquitously expressed and localizes in the cytosol and nucleus where it is essential for development and is involved in diverse functions, including vesicle transport along axons and transcription (211). The etiology of HD is determined by the combined result of the emerging toxic properties of mutHTT protein, and potentially its mRNA, and the associated loss of normal HTT functions, with an inverse relationship between the expanded repeat length and the age of disease onset that can be modulated by additional gene modifiers (213). PolyQ expansions in mutHTT have been shown to drive the formation of pathological amyloid fibrils that are able to perturb the intracellular proteostasis network or deplete factors crucial for the basic neuronal cell functionality (214, 215). Although mutHTT is typically thought to accumulate and exert its toxicity from within the cell, there is also recent evidence for an extracellular localization and transfer to neighboring cells or other tissues via the blood stream, supporting the idea that mutHTT can propagate in a prion-like fashion (216, 217). This may contribute to the HD pathology or its worsening, as blood can serve as a vehicle of propagation for mutHTT outside of the brain, enabling peripheral pathological conditions that were previously observed such as changes in protein metabolism or mitochondrial function in skeletal muscle, hepatocytes, or immune system cells (218, 219, 220).
HTT is a large 3146 amino acid protein (if containing a 23-glutamine polyQ, or 23Q) that is ubiquitously expressed and is hypothesized to act as a scaffold for binding multiple protein assemblies (221). Recent work shows that HTT mainly consists of α-helices arranged in three domains that are stabilized by the HTT-associated protein 40 (HAP40)–interacting protein: the N- and C-terminal domains consisting of multiple HEAT (HTT, Elongation factor 3, phosphatase 2A, and kinase TOR) repeats that are linked by the bridge domain, which contains tandem α-helical repeats (222). However, large segments of the N-terminal region remain poorly defined structurally, such as the polyQ, as they are highly dynamic. Preceding the variable polyQ stretch, whose function in normal HTT remains poorly understood, the HTT amino terminus consists of 17 residues arranged in an amphipathic α-helix, which has a nuclear export signal. This region may interact with the cell membrane and is prone to posttranslational modifications (223). Following the polyQ stretch, a proline-rich domain drives the binding of different WW or SH3 domain–containing proteins, involved in nucleic acid binding and processing or cellular dynamics (25, 224, 225, 226), in addition to maintaining the solubility of mutHTT in vitro and in vivo (227, 228). Interaction with cellular proteases, such as caspases and calpains, promotes cleavage of both WT HTT and mutHTT amino termini, with enhanced proteolysis being specifically observed in HD patient brain extracts (229). This processing has been linked to the generation and accumulation of small HTT fragments with the polyQ portion, which can translocate into the nucleus, associate with membranes, cause toxicity, and accelerate HTT aggregation (229, 230, 231, 232). Aberrant splicing can also lead to the translation of the exon 1 isoform with the pathogenic polyQ segment (233). Following seeding, pathological elongated polyQ stretches form ordered and β-sheet-rich amyloid fibrils (234, 235). The β-hairpin-containing β-sheets are connected through hydrogen bond interactions that provide a critical monomeric core fundamental to trigger the aggregation and the formation of amyloid fibrils (236). Due to the high affinity of the β-sheets, higher molecular weight aggregates can be formed by recruiting not only pathogenic polyQ protein but also other endogenous nonpathogenic proteins, thereby disrupting other cellular networks and their functions (221).
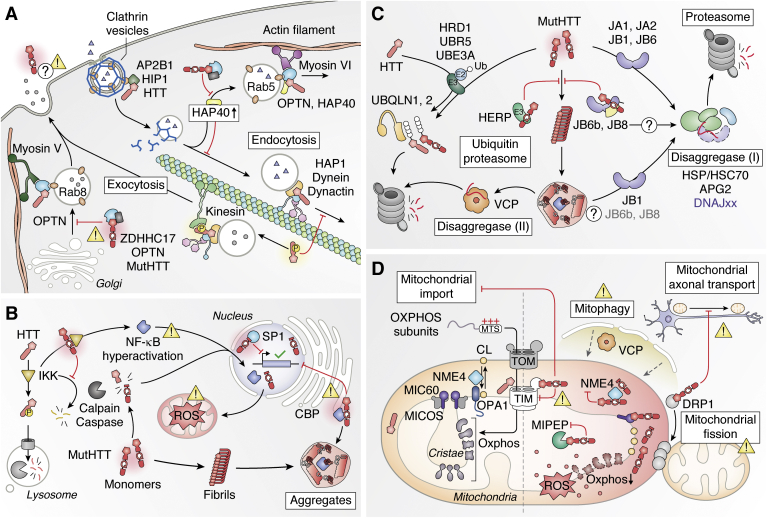
One hypothesis is that HTT acts as a hub to tether multiple partners to promote several major processes in the cell such as vesicle trafficking and transcription (Fig. 4, A and B). A range of studies have shown that several PPIs can be perturbed by extended polyQ in mutHTT and several high-throughput experiments have vastly expanded the list of candidate interactors using Y2H screening and AP-MS (225, 237, 238, 239). For instance, Shirasaki et al. employed AP-MS to define a spatiotemporally resolved HTT-interactome in transgenic mice expressing mutHTT (97Q) and identified interactions that are more specific to some brain regions (cortex, striatum, and cerebellum) and specific ages (2 and 12 months) (238). HTT forms a network of PPIs with motor and motor-associated proteins such as HAP1 and HAP40, HTT-interacting protein 1 (HIP1), optineurin, and Zinc finger DHHC-type palmitoyltransferase 17 (ZDHHC17; HIP14) consistent with HTT being an essential integrator of intracellular vesicular trafficking and transport in the cell (224, 225, 238, 239, 240) (Fig. 4A). HAP1 serves as adapter protein recruiting and stabilizing the anterograde microtubule motor kinesin and retrograde motor dynactin–dynein complex (225, 241, 242, 243). Dynein, which requires dynactin for its activation, constitutively interacts with HTT (25, 225), whereas interaction with kinesin to recruit vesicle occurs only when HTT is phosphorylated on serine 421 (244, 245). Therefore, the dynein–dynactin–mediated retrograde motility can occur when HTT is not phosphorylated (246). Interactions with other proteins also determine the association to the actin cytoskeleton. High cytoplasmic levels of HAP40 promote the formation of a complex with HTT and optineurin on Rab5-positive endocytic vesicles (247). Through optineurin, the actin-based motor myosin VI associates the cargo to the actin cytoskeleton for its transport (224, 248). Interestingly, a complex with HTT, optineurin, and Rab8 has been shown to regulate exocytic membrane trafficking from the Golgi (248, 249). MutHTT leads to mislocalization of optineurin and irregular cargo transport, leading to impaired palmitoylation of ZDHHC17 substrates, with implications in HD pathogenesis (250). HTT interaction with the palmitoyl transferase is also reduced in presence of an extended polyQ (251). Through the interaction with HIP1 and adapter-related protein complex 2 subunit beta 1 (AP2B1), which also binds HTT at the amino terminus, HTT may play a role also in clathrin-mediated endocytosis and trafficking, although mutHTT does not seem to directly inhibit clathrin-dependent endocytosis (25, 225, 252, 253). Nonetheless, the sequestration of the Hsc70 chaperone in mutHTT aggregates affects clathrin-mediated endocytosis (254). This phenomenon was also observed when other proteins aggregate, like mutated SOD1 and ataxin-1. Numerous studies have also shown that HTT binds to transcription factors such as specificity protein 1 (SP1) and other components regulating transcription like CREB-binding protein (CBP) (Fig. 4B) (255, 256, 257). Presence of extended polyQ increased SP1 binding, and CBP is recruited to HTT inclusions, which can then lead to transcriptional dysregulation (255, 256, 257). Several groups have also integrated multiple omics approaches to identify interactors that can act as gene modifiers. For instance, knockdowns of the vacuolar protein sorting 35 (Vps35) and the brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2) Rho GTPase that both interact with HTT reduce HTT toxicity in a drosophila model and tissue culture cell–based model, respectively (238, 258). The drosophila model also indicates that fly HTT genetically interacts with autophagy genes, and work in mammalian cells shows that HTT is required for selected autophagy, notably by acting as a scaffold for several autophagy-regulating proteins, such as p62 (SQSTM1) (259).
Figure 4.
Huntington and Huntington's disease.A, HTT participates in vesicle trafficking along microtubules or actin filaments. mutHTT may disrupt these processes through mislocalization or disruption of interactors. B, cleavage or mis-splicing of HTT accelerates fibril formation that leads to a loss of HTT function as well as sequestration of other factors. In the presence of mutHTT, IKK causes hyperaction of NF-κB and modulated SP1 activity. C, soluble and aggregated mutHTT can be targeted for degradation. Aggregated mutHTT can be disrupted by the disaggregases VCP or DnaJB1 allowing for subsequent degradation. D, mitochondrial function becomes disrupted in HD. mutHTT disrupts import via its interaction with TIM23. The interaction of mutHTT with Drp1 disrupts its functions in anterograde mitochondrial transport and mitochondrial fission. HD, Huntingtin’s disease.
Multiple elements of the chaperone system interact with HTT, and several components can reduce mutHTT aggregation or regulate HTT clearance (Fig. 4C). The main components of the human HSP70-based disaggregase system have been identified in diverse interactome studies of WT and mutant HTT. Members of the Hsp70/Hsc70 family have been shown to interact with mutHTT, including genes encoded by HSPA8 and HSPA1A, alongside Hsp40 cochaperones from the family A (DNAJA1 and DNAJA2) and family B (DNAJB1 and DNAJB6), as well as small Hsps (HSP27) (25, 260, 261). Notably, soluble mutHTT-53Q and mutHTT-103Q oligomers colocalize and interact with Hsp70 (HSPA8, HSPA1A/B) and DnaJB1, both in vitro and in rat neuronal model cell lines (262). The Hsc70 (HSPA8), DnaJB1, and the nucleotide exchange factor Apg2 cooperatively suppress mutHTT fibrillization in vitro (263). Moreover, their depletion promotes mutHTT aggregation, while overexpression of DnaJB1 alone was shown to be sufficient to strongly reduce aggregation. A previous screen for HD modifiers also identified, DnaJB6 and DnaJB8, as suppressors of mutHTT aggregation (264). These two cochaperones were shown to directly bind to the polyQ stretches of mutHTT-Q119 through a serine/threonine (S/T)–rich region in their carboxy terminus, thereby preventing aggregation independently of the Hsp70 machinery (264, 265). Notably, only cytosolic isoform B and not nuclear isoform A of DNAJB6 is able to suppress aggregation of polyQ-containing and other aggregation-prone proteins (266), making it the major focus for future studies on polyQ aggregation. Indeed, DnaJB6 levels are higher in undifferentiated neuronal cells, which could explain why stem cells may be protected from HTT aggregation (267). Hsp90 also interacts with HTT (HSP90AA1 and HSP90AB1) (260, 268, 269). Interestingly, in one study in which new mouse HD models were generated by expressing HTT fragments that mimic protease cleavages, Hsp90 shows greater interaction with a longer HTT fragment that mimics caspase 6 cleavage than a shorter and more toxic fragment that can be generated by caspase 2 (268). While overexpression of Hsp90 reduces cytotoxicity, downregulation of Hsp90 leads to more cell death alongside lower levels of the exogenous HTT fragments, indicating Hsp90 interaction may regulate mutHTT turnover. In a separate study, Hsp90 was found to specifically interact with the N-terminal amphipathic α-helix of HTT (269). Because Hsp90 also binds to the USP19-deubiquitinating enzyme, it may play a major role in regulating HTT cellular levels by modulating its ubiquitination. Yet another chaperone, the chaperonin T-complex protein ring complex (TRiC, also known as CCT for chaperonin-containing TCP-1), interacts with the N-terminal amphipathic α-helix of HTT, which underlies the importance of this domain in HTT aggregation (270). Previously, TRiC had been shown to promote assembly of nontoxic HTT oligomers in vitro and tissue culture cells (271, 272). In a recent study, levels of TRiC components were shown to be reduced in mouse neural stem and progenitor cells, which could explain why diseases onset occurs in older individuals (273).
Clearance of HTT can be mediated by several components of ubiquitin proteasome system as well as by the lysosome (Fig. 4C). The N-terminal region of mutHTT interacts with the E6-associated protein (E6-AP or UbeE3A) ubiquitin ligase (274, 275). Levels of this E3 ligase are reduced in older mice, which coincide with the accumulation of HTT associated with K63-linked polyubiquitin in a HD mouse model, and which could result from a failure to adequately target mutHTT for proteasomal degradation (275). Accordingly, exogenous expression of E6-AP in tissue culture cells and HD mouse model reduces accumulation of HTT inclusions (274, 275). Interestingly, mutHTT does not aggregate in induced pluripotent stem cells derived from HD patients (276, 277). Koyuncu et al. showed that the Ubr5 E3 ligase, which binds to mutHTT alongside E6-AP, is expressed at higher levels in induced pluripotent stem cells and targets mutHTT for proteasome degradation (278). Recent work also showed that single nucleotide polymorphisms near UBR5 acts as a modifier of the HD age onset, which suggests that Ubr5 may play a major role in regulating mutHTT levels in neuronal cells (213). Ubiquilin 1 and 2 (UBQLN1/2), which functionally link the ubiquitination machinery to the proteasome, are shown bind to polyubiquitin chains on mutHTT (279, 280). Several ER-associated ubiquitin ligases have also been involved in targeting mutHTT for degradation. For instance, the homocysteine-responsive ER-resident protein (Herp, HERPDU1) E3 binds to the N terminus of HTT with an extended polyQ (Q160) and can mediate proteasomal degradation of the mutHTT in neuronal tissue culture cells (281). Interestingly, the expression of HERPDU1 is induced upon ectopic expression of mutHTT and the E3 ligase localizes in the periphery of the HTT inclusions, where it could mediate mutHTT conjugation. Human HMG-CoA reductase degradation 1 (Hrd1) is another ER-associated E3 that interacts with an N-terminal fragment of mutHTT and mediates its degradation in tissue culture cells (282). Interestingly, the turnover of the ectopic HTT fragment also requires the VCP/p97 disaggregase. The ATPase VCP can bind to the first 17 residues of HTT and is capable of disassembling mutHTT toxic aggregates via its disaggregase function both in vitro and in HD cellular models (283). In addition to its disaggregase function, VCP also recruits linear ubiquitin chain assembly complex (LUBAC) an E3 ligase that mediates conjugation of linear ubiquitin chains (i.e., linked via the methionine 1 of ubiquitin instead of a lysine) on aggregated mutHTT in tissue culture cells (284). Notably, increase of linear ubiquitin reduces the recruitment of SP1 to mutHTT inclusions. Markedly, in addition to HTT, VCP interacts with ALS-related SOD1 and AD-related APP, suggesting that human VCP might act as a general disaggregase for several protein aggregates linked to neurodegeneration (25, 285, 286, 287, 288, 289). Several additional posttranslation modifications may regulate HTT clearance. For instance, conjugation of the SUMO ubiquitin-like protein on lysine residues located in the N terminus of HTT—which have also been found ubiquitinated—reduces turnover and promote aggregation of mutHTT (290). In contrast, phosphorylation by TANK-binding kinase 1 (TBK1) suppresses mutHTT aggregation and toxicity (291). Similarly, inhibitor of κB kinase (IKK) activation and the resulting HTT phosphorylation promotes degradation by the proteasome and lysosome (261). Indeed, CMA has been shown to also to regulate mutHTT clearance with the participation of Hsc70 (292). Interestingly, the activation of IKK is induced upon binding of mutHTT leading to a hyperactivation of the transcription factor NF-κB with repercussions for mitochondrial homeostasis, similar for what was observed in PD (293, 294).
Mitochondrial dysfunction plays a critical role also in HD pathogenesis (Fig. 4D). Both full-length mutHTT and its cleavage-associated fragments directly associate with mitochondria, resulting in impaired protein import, membrane permeabilization, impaired energy metabolism, abnormal mitochondrial trafficking, and impaired mitochondrial dynamics (295, 296, 297, 298). As observed in human HD-affected brains, trafficking motors and mitochondrial components become sequestered in HTT inclusions (299). MutHTT interacts with mitochondrial Drp1, a mediator of mitochondrial fission, and abnormally enhances its activity (297). This results in impaired mitochondrial dynamics causing defects in anterograde mitochondrial axonal transport and synaptic deficiencies. In addition, mutHTT also recruits VCP to mitochondria, which induces excessive mitophagy (300, 301). Remarkably, inhibition of the HTT–VCP PPI reduces mitochondrial impairment and cell death in HD cell culture and mouse models. Therefore, the interaction between mutHTT and VCP may have different consequences depending on the localization of both proteins. Furthermore, mutHTT affects both the organization and composition of mitochondria by interacting with additional proteins in the intermembrane space. Yablonska et al. showed that both WT HTT and mutHTT-Q111 localize to the mitochondrial intermembrane space where they interact with translocase of mitochondrial inner membrane 23 (TIM23) (302). In particular, the increased interaction between TIM23 and mutHTT results in reduced TIM23-dependent import of mitochondrial matrix proteins, drastically altering the mitochondrial proteome. Another interesting mitochondrial partner of mutHTT is mitofilin, the main component of the mitochondrial contact site and cristae organizing system (MICOS) that is involved in forming of contact sites to the outer membrane to maintain mitochondrial cristae morphology (25, 303). Notably, compromised membrane architecture has been observed both in the striatum and the cortex of HD mice, which has been associated with impaired oligomerization of optic atrophy 1 (OPA1), which interacts with mitofilin (304). A Y2H study focused on disease-associated HTT mutations revealed mutHTT interactions with other mitochondrial proteins with possible additional relevance for mitochondrial impairment in HD pathology (239). For instance, mutHTT interacts with mitochondrial intermediate peptidase (MIPEP) and NME4, a nucleoside diphosphate kinase, which are both key to mitochondrial function. However, additional studies would be required to evaluate this possibility. Although the mechanisms by which mutHTT mediates mitochondrial pathology in HD are not fully elucidated, numerous studies now provide insight into the consequences of gained mitochondrial PPIs with mutHTT.
The great diversity in novel or altered PPIs with mutHTT together with the variety of their cellular localizations, suggests that multiple pathways are simultaneously affected during the development of HD. One challenge is to identify a therapeutic approach that can alleviate these different effects simultaneously. In comparison to the other NDs discussed in this review, HD has the particularity of being a genetic disorder essentially driven by the mutation of one gene. Therefore, HD is an ideal candidate for a genetic therapeutic approach such as antisense oligonucleotides to reduce HTT expression. Though, the recent failed antisense oligonucleotide trials suggest that the endogenous HTT function may be important and that more refined approaches to target HTT are needed (305).
Amyotrophic lateral sclerosis
ALS is a fatal neurodegenerative disorder causing progressive loss of motor neurons with no known effective treatment. Approximately 90% of ALS cases are sporadic and the remaining 10% are familial ALS (fALS) (306). There are over 30 genes linked to ALS including the SOD1 Cu/Zn superoxide dismutase, TDP-43 (TARPDBP), cyclin F (CCNF), FUS, p62, ubiquitin-like protein ubiquilin 2 (UBQLN2), Matrin 3 (MATR3), or repeat expansions in C9orf72 (307, 308, 309, 310, 311, 312, 313, 314, 315). In this review, we will direct our main focus to SOD1, as approximately 20% of fALS patients carry mutations in the SOD1 gene (mutSOD1), and TDP-43 that is found aggregated in the majority of ALS cases.
There are a large number of SOD1 mutations (over 180) associated with fALS and many are thought to impact protein folding and promote aggregation. SOD1 is an enzyme that detoxifies superoxide anions by converting them to hydrogen peroxide, and it is typically present in the cytosol, nucleus, peroxisomes, and mitochondrial intermembrane space of mammalian cells where it forms a stable homodimer (316). For its maturation, SOD1 generally undergoes four main steps: zinc insertion, copper insertion, disulfide bond formation, and homodimer formation (Fig. 5A). The copper chaperone for SOD1 (CCS) interacts with SOD1 providing it with copper and facilitating the transition to the holoform (317, 318). More specifically, CCS interacts with the zinc-metalated, disulfide-reduced SOD1. Recent work has shown that copper has to be transferred from CCS to SOD1 before SOD1 homodimerization occurs, following disulfide bond formation (319). Nonetheless, absence of CCS in mouse models in which mutSOD1 is overexpressed does not accelerate the disease onset, despite SOD1 remaining mostly inactive in this case (320). These results indicate that failure to complete copper loading on SOD1 and loss of function of SOD1 are not at the basis of the pathology. Instead, it has been proposed that there is a gain of toxicity following conformational changes of monomeric SOD1 (321, 322). In fact, many SOD1 mutations destabilize the protein structure leading to more exposed hydrophobic regions, especially the monomeric metal-free species (323, 324, 325). The reduced capacity to dimerize and the exposure of more hydrophobic regions can lead to new PPI. For instance, gain of PPI were noted for a truncated mutSOD1 that cannot dimerize due to a two base pair deletion at codon 126 (Leu126delTT), notably with several subunits of the Na+/K+ ATPase pump (326). More recently, an independent study that used an antibody specific to misfolded SOD1 for AP-MS confirmed the interaction of the Na+/K+ ATPase pump with fALS-associated SOD1-G93A (327). Importantly, this interaction was detected in early stages and was confirmed for other fALS-associated mutants. Moreover, the levels of Na+/K+ ATPase are reduced in tissues derived from patients with sporadic ALS. Uncoupling the Na+/K+ ATPase by misfolded SOD1 could be highly cytotoxic in motor neurons and play an important role in the ALS etiology.
Figure 5.
Superoxide dismutase 1, TDP-43, and ALS.A, SOD1 is localized to the cytosol or IMS where is converts superoxide to hydrogen peroxide. Normal folding and maturation of SOD1 is disrupted by disease-associated mutations. Misfolded SOD1 becomes incorporated into β-sheet-rich fibrils and inclusion bodies. B, misfolded SOD1 can be degraded by either the proteasome or by the lysosome. Soluble mutSOD1 may be specifically targeted by the E3 ligase dorfin. Larger aggregates or inclusion bodies may be degraded through autophagy or become resolubilized by disaggregases such as Hsc70 or VCP and then be subsequently degraded by the proteasome. C, mutSOD1 may disrupt mitochondrial import and thus energy and redox states. Altered interactions between SOD1 and the components of the antioxidant system may further contribute to increased ROS. Formation of aggregates sequesters soluble BCL-2, disrupting its antiapoptotic activity. TDP-43 physiological interactions and its involvement in ALS. IMS, intermembrane mitochondrial space; ROS, reactive oxygen species.
Since most assessed fALS-associated mutants reduce protein folding, interaction of SOD1 with the proteostasis network is pivotal and may have a major impact in disease progression (Fig. 5B). Proteomic analysis of SOD1 PPIs in a mouse model led to several chief observations (328). First, the mutant protein—but not the WT SOD1—shows a marked interaction with Hsc70 (HSP8A) and Hsp70 (HSPA1B), even in 1-month-old mice prior to the presence of any SOD1 inclusions. Interaction of mutated SOD1 with Hsc70/Hsp70 had also been observed in several other studies (326, 329, 330, 331). Secondly, despite the interaction of mutated SOD1 with Hsc70 and Hsp70 in young mice, motor neuron disease developed in 4- to 12-month-old mice (328). Finally, there is a gain of multiple additional PPIs with other chaperones coinciding with appearance of aggregated SOD1 in later stages, including with Apg2/Hsp110 (HSP4), Hsp105 (HSPH1), BiP/Grp78 (HSPA5), and DnajA1. Association to the ER-associated chaperone is perhaps at first puzzling. However, a recent study by Piette et al. also found WT SOD1 associating to BiP (30). In parallel, C9orf72 was found associated to SEC63. In mammalian cells, transport of signal peptide-containing proteins into the ER is mediated by the Sec61 channel complex, which relies on BiP and SEC63 (332). Association of WT counterparts of fALS-associated proteins to the Sec61 complex and its interactors could hint to a possible involvement of ER dysfunction and protein-folding stress in the progression of sporadic ALS, as previously shown (333). Interestingly, presence of a mutSOD1 can modify the composition of the client protein pool interacting with Hsc70 and Hsp70, even when mildly overexpressed in tissue culture cells (334). These proteomic experiments relied on engineered chaperone proteins to trap normally transient interactions with client proteins (i.e., proteins folded by a given chaperone). The authors of the study noted that in the presence of misfolded mutSOD1, there is a gain of client proteins that are predicted to be more disordered, whereas proteins with more stretches of residues predicted to be aggregation-prone accumulate in the pellet fraction, perhaps due to a competition with Hsp70 binding. These results illustrate how presence of misfolded proteins can shift the equilibrium within the cell by potentially altering the cell folding capacity. It will be important to address how proteostasis is affected in differentiated cells and in physiological conditions in future studies. In another recent study, the binding of two mutSOD1 to Hsp70 and Hsp90 was compared using a modified ELISA (335). In this case, the G41S mutant associated a severe form of fALS shows further increased binding to Hsp70 in comparison to WT SOD1, whereas G37R that is associated with a milder phenotype displays, instead, a higher association to Hsp90 but not Hsp70. While only two SOD1 mutants were compared, other disease-associated mutations with known disease severity were assessed and show the same trend. These results imply that the severity of the disease may in part be determined by how a misfolded protein is triaged by the proteostasis network and confirm that Hsp90 can act as protein-folding buffer in the cell to mitigate the effect of some diseases-associated mutations.
In addition to the SOD1 PPIs with chaperone proteins, levels of SOD1 are regulated by proteolysis. Both the ubiquitin–proteasome system and autophagy are reported to mediate degradation of mutSOD1 when transfected in neuronal cells (336). In agreement with a role for autophagy in SOD1 clearance, mutSOD1 mutants—but not WT—bind to p62 (also linked to fALS), which facilitate autophagy (337). In addition, the α-crystallin chaperone (HSPB8) that is induced in mouse motor neurons that express SOD1-G93A can facilitate the autophagy clearance of aggregated SOD1 and TDP-43 in tissue culture neuronal cells (338). Moreover, several studies report that induction of autophagy increases SOD1 clearance or reduces accumulation of aggregated SOD1 and improves some ALS phenotypes (339, 340, 341). Interestingly, in another study where fibroblast cells were derived from ALS patients, levels of mutSOD1 are increased by proteasome inhibition, but not by autophagy suppression (342). One possibility is that different cell types may have different capacity to triage misfolded SOD1 or that autophagy can only efficiently clear aggregated SOD1. Several E3 ubiquitin ligases have been implicated in regulating SOD1 levels, especially of the fALS-associated mutants (Fig. 5B). As for other ND-associated proteins, CHIP has been shown to interact—together with Hsc70—with mutated but not WT SOD1 (329, 330). Whereas CHIP may be involved in the regulation of SOD1 turnover, several results also indicate that another E3 ligase is involved in ubiquitinating SOD1. Indeed, dorfin, which is a RING between RING (denoted RBR) E3, interacts specifically with mutated but not WT SOD1 and is recruited to SOD1 inclusion bodies (343). The presence of a disulfide crosslink between the cysteine residues at positions 6 and 111 of SOD1 has been suggested to both mediate SOD1 aggregation and the recognition by dorfin (344). However, another study, using a different C6 mutation, also showed that the disulfide bond between these residues is not required for aggregation, indicating that another mechanism may mediate recognition of misfolded SOD1 by dorfin (345). Other ubiquitin ligases have been shown to target misfolded SOD1 for proteolysis. For instance, the NEDD4-Like ubiquitin ligase 1 (NEDL1) E3 that is specifically expressed in neuronal cells interacts with and ubiquitinates several fALS mutants, but not WT SOD1, as well as reduces the half-lives of mutSOD1 when overexpressed (346). In addition, E6-AP (Ube3A), the ER-associated glycoprotein 78 (gp78 or AMFR; autocrine motility factor receptor) and cellular inhibitor of apoptosis protein 1/2 (cIAP 1/2) E3s have all been shown to interact and regulate levels of different mutSOD1 (347, 348, 349). While several ubiquitin ligases have been implicated in regulating SOD1 levels, careful evaluation of each E3 ligase expression levels in different neuronal cells will need to be more precisely quantified in future studies to establish a more comprehensive model of their possible impact in ALS.
Failure to adequately fold or eliminate SOD1 leads to the accumulation of misfolded SOD1 and its aggregation. Inclusion bodies enriched in SOD1 can be observed in the cytoplasm of motor neurons and astrocytes in mouse models and may play a key role in the ALS etiology (321, 350). These inclusion bodies present two distinct areas, an external protease-sensitive coating and a more compact protease-resistant core. The exact structure varies with different mutations (351). Three main regions of SOD1, each with different physiochemical properties, were identified as fibril cores (352). Synthetic peptides corresponding to these three core regions can form fibrils, either alone or in a mixture with the other peptides. Furthermore, lowering structural stability by reduction of the highly conserved intramolecular disulfide bond between cysteines 57 to 146, increased aggregation of mutSOD1-G93A under physiological conditions, suggesting that exposing fibril-forming core regions through structure destabilization can facilitate fibrillar aggregation of SOD1 (351, 352). Une et al. identified proteins that preferentially bind to the different aggregation cores upon normal conditions and upon inhibition of autophagy or of the ubiquitin proteasome system in mouse (331). Proteins interacting with SOD1 cores are involved with PI3K/AKT and MAP kinase cascades, suggesting that mutant SOD1 could indirectly affect cell growth and cell survival through sequestration of such components. In addition to cellular aggregates, there is growing experimental evidence supporting uptake and prion-like propagation of pathological conformations of SOD1, but the precise mechanism underlying the transfer is still unknown (353, 354, 355). In line with this, extracellular matrix proteins or cell surface receptors were also identified as interacting proteins in several studies (356, 357). Interestingly, SOD1 can also form homotrimers and fALS mutants that promote trimer formation and are more cytotoxic in model neuronal cells (358, 359). In contrast, assembly into large insoluble fibrils appear to mitigate neurotoxic effects both in model tissue culture cells and in a mouse model, indicating that large SOD1 aggregates might be cytoprotective (359, 360). It will be important to determine in future work whether small SOD1 oligomers engage in specific PPI that are detrimental to the cell.
As a fraction of SOD1 is localized in the mitochondria, it is perhaps not a surprise that several PPIs with mitochondrial proteins have been reported. In addition, expression of mutSOD1 impacts mitochondrial homeostasis. For instance, overexpression of the cochaperone CCS in mice leads to the accumulation of mutSOD1-G93A in mitochondria, inducing a severe mitochondrial pathology that accelerates neurological deficits and shortens lifespan (Fig. 5C) (361). Different forms of mutSOD1 preferentially associate to the cytoplasmic side of the mitochondrial outer membrane. Proteomic studies by Li et al. showed that expression of different fALS-associated SOD1 variants leads to a decrease in mitochondrial protein import, despite increased levels of some components of the mitochondrial import machinery (362). Interestingly, this defect is more pronounced in organelles purified from motor neurons than from the liver, suggesting a potential mechanism of motor neuron susceptibility. In addition, both WT and mutant SOD1 have been shown to bind the antiapoptotic and mitochondrial-associated protein Bcl-2 in vitro and in vivo in mouse and human spinal cord, providing evidence of a direct link between SOD1 and an apoptotic pathway (363). SOD1 localizes to the intermembrane mitochondrial space, and gain of PPI with one fALS-associated mutant (Leu126delTT) was reported for several intermembrane mitochondrial space proteins such as members of the solute carrier family (SLC25A 4, 5, 12) (326). Interestingly, additional SOD1 interactors that reside in the matrix were also reported in the same study, such as the subunits α, β, and γ of ATP synthase. This may be particularly relevant as subunit β (ATP5B) is reported to be depleted or affected in the mitochondrial fraction of G93A mice spinal cord, suggesting that loss of SOD1 might play a role either in stabilizing the ATPase complex or rather promote efficient mitochondrial import, affecting, when mutated, the energy and redox levels of the organelle (364, 365, 366). In a large proteomic study aimed to generate a high-confidence mitochondrial protein network linked to ND disorders, Malty et al. identified SOD1 interacting with members of the peroxiredoxin antioxidant system (PRDX 2, 4, 5, and 6), as previously reported (288, 367, 368). Disruption of the interaction with PRDX5, a hydrogen peroxide scavenger localizing in the mitochondrial matrix, results in an increase of ROS, which highlights the importance of SOD1–PRDX5 binding (288). Interestingly, DJ-1 (PARK7), has also been found to interact with SOD1 as well as PRDX 3 and 6, which are active in protecting mitochondria by clearing ROS. Whether DJ-1 interaction is related to its antioxidant defense role or its chaperone activity remains unclear, but the shared PPIs between proteins implicated in PD and ALS raises the question whether DJ-1 and SOD1 might have a role in the same antioxidant pathway, which may be commonly affected in these two different NDs.
Despite mutations in TDP-43 accounting for less than 1% of ALS cases, cytoplasmic inclusions of TDP-43 are found in the majority of ALS cases (369). TDP-43 aggregates are not limited to ALS and have been found in frontotemporal dementia, AD, HD, and PD. TDP-43 is an RNA-binding protein, which is predominantly localized to the nucleus, and is involved in RNA processing, such as splicing. The number of reported PPIs is lower than for other disease-associated proteins in this review. However, TDP-43 interacts with a large number of different RNAs, including its own mRNA to reduce, in this case, TDP-43 expression levels (370). In addition, depletion of TDP-43 in mouse brain tissue results in the downregulation of synapse related genes, suggesting a potential mechanism of neuronal vulnerability to changes in TDP-43 levels associated with ALS. If not properly localized, TDP-43 can also impede translation of mitochondrial genes, thereby inducing mitochondrial damages (371). Of the reported PPIs with TDP-43, many are interactions with ribonucleoproteins or RNA-binding proteins. Freibaum et al. identified nuclear and cytosolic interaction networks using AP-MS of tagged TDP-43 (372). In this case, most PPIs are not affected by the presence of disease-associated mutations in TDP-43 (A315T or M337V), but many of these interactions are RNA-dependent suggesting they are indirect. The cytosolic PIN comprises translation initiation and elongation factors and ribosomal subunits, including the stress granule component Stau1, which was later shown to interact with TDP-43 in a RNA-dependent manner (373). The nuclear network of interactors included transcription and splicing factors, like HNRNPA1 and HNRNPH1. Additional evidence for the interaction of TDP-43 with RNA-binding proteins, including HNRNPH1, comes from the BrainMap published by Pourhaghighi et al. (374). Using cofractionation of proteins obtained from mouse brain tissues, the authors identified a complex of RNA-binding proteins that included several ALS-associated proteins such as TIA1, ataxin 2 (ATXN2), Ewing's sarcoma (EWS), and FUS. Additional studies have found that TDP-43 and FUS drive together the expression of Hdac6 and that their PPI is also RNA dependent (375, 376). The identification of several ALS-associated proteins as interactors suggests that a common pathway is affected in the development of ALS.
Regulation of TDP-43 interaction with both RNA and the protein homeostasis network is likely playing a major role in ALS pathology. Yu et al. recently showed that several mutations or posttranslation modifications that reduce RNA binding drive TDP-43 demixing in atypical droplets that contain both liquid spherical shells and cores (377). While TDP-43 is more concentrated in the shell, it is also present at a lower concentration in the core where it interacts with various Hsp70s and Apg2. These interactions are required to maintain phase separation and prevent TDP-43 aggregation. Notably, Hsp70 may directly interact with a more conserved stretch of the C-terminal low complexity domain that plays a key role in LLPS of TDP-43 (378). Alongside preventing aggregation, chaperone proteins also promote the clearance of TDP-43, especially after the activation of the heat shock transcription factor 1 (HSF1) master regulator of the heat shock response (379, 380, 381). For example, DNAB8, a small HSP, inhibits the aggregation of smaller fragment of TDP-43 that is particularly prone to aggregate in the cytosol, which can then be cleared by the proteasome and autophagy (382).
TDP-43 is ubiquitinated in aggregates found in the motor neurons of ALS patients (383), and several E3 ubiquitin ligases have been shown to interact with TDP-43. The Praja ubiquitin ligase was recently shown to be upregulated by HSF1, and it mediates clearance by binding to the C-terminal domain of TDP-43 (384). RING finger 122 (RNF112, Znf179) is another ubiquitin ligase that interacts and ubiquitinates TDP-43 (385). Notably, TDP-43 inclusions are observed in several brain regions of the RNF112 mouse KO, suggesting it may play an important role in regulating TDP-43 turnover. In addition to regulating turnover, ubiquitination could also play a role in TDP-43 localization. For instance, parkin and TDP-43 interaction is reported in several studies, and expression of both human proteins in rat cortex results in an increased ubiquitination of TDP-43 and mislocalization of TDP-43 to the cytosol, which could promote cytosolic aggregation (386, 387, 388). Cyclin F, which is a CRL1 substrate adapter protein encoded by a gene associated to fALS, binds to the N-terminal region of TDP-43 and mediates its ubiquitination in vitro (389). Remarkably, one of cyclin F ALS-associated mutation (S621G) leads to an increase of K48-linked ubiquitin chains conjugated to TDP-43 in vitro, and there is an accumulation of cytosolic inclusions containing both TDP-43 and K48 ubiquitin chains in the spinal cord section of a patient with that mutation. These results are surprising, as K48-chains are mostly thought to mediate proteasome degradation, which should in principle hamper TDP-43 aggregation. One possibility is that the ubiquitination of TDP-43 may increase the affinity for ubiquilin 2—yet another protein encoded by a fALS-associated gene—that contains a ubiquitin-binding domain and binds to the C-terminal region of TDP-43 (390). Ubiquitin 2 itself undergoes phase separation, localizes to stress granules, and helps mediate proteasomal degradation of misfolded proteins (391, 392). In this case, the tightened interaction between TDP-43 and ubiquilin 2 could favor the solidification of elements in the stress granule condensates, which then would undergo a liquid to solid transition and nucleate aggregation.
Outlook and future directions
While clearer views are emerging regarding the role of each aggregation-prone protein, many areas still require further clarification. For example, when more than one function has been attributed to a given aggregation-prone protein, it is not always clear which function(s) may be more important in the context of the pathology and how they may each exacerbate or mitigate misfolding and oligomerization. In addition to the perturbation of normal function, the mutations, posttranslational modifications, and oligomerization of aggregation-prone proteins can also affect cellular processes due to deviant PPIs and other cytotoxic events (e.g., damage to the cell membrane). It remains to be determined which of the affected cellular processes are dominant in the progression of the disease, especially in differentiated neurons. One possibility is that a combination of multiple “system failures” within the cell contributes to cell death. Alternatively, distinct “series of unfortunate events” may occur in different neuronal cells, with some cells being more susceptible to a specific type of injury. For instance, while impaired vesicle trafficking in some cells containing HTT inclusions may be particularly detrimental, misregulated transcription may be more damaging in other cells in which an HTT inclusion has formed. This may be particularly true for human neuronal cells that have an average lifespan of over 10 years, during which time they may get exposed to different sets of stimuli or stresses.
To summarize the numerous PPIs discussed here, we assembled a PIN (Fig. 6). For clarity, we only included proteins for which there are at least four experimental evidence reported in BioGRID for at least one of the six aggregation-prone proteins. Perhaps not surprisingly, multiple components of the proteostasis network interact with several aggregation-prone proteins and are placed at the center of the PIN. In contrast, proteins involved in the physiological function of the examined proteins tend to be located at the periphery of the PIN. Notably, Hsc70 (e.g., HSPA8) and Hsp70 (e.g., Apg2/HSPA4) plays a dominant role in preventing both misfolding and aggregation, by driving the disaggregation of oligomerized and aggregated proteins and the clearance of misfolded proteins. Together with Hsp90, these chaperones occupy a central location in the PIN. Oddly, DNAJ proteins are noticeably absent from the PIN because the number of observations are below our cutoff. Nonetheless, DnaJA1, A2, B1, and B6 each bind to multiple aggregation-prone proteins. Because these cochaperones play a key role in mediating the Hsc70/Hsp70 interactions, it would be important to examine their expression levels and their activity in human neuronal cells upon aging. This would determine whether a lower folding capacity in the cell is really the main triggering factor for aggregation. Other central elements include proteins associated to the ubiquitin proteasome system such as VCP, p62, and CHIP. One striking observation is that each aggregation-prone protein has been found to be targeted for degradation by more than one pathway, yet the presence of inclusions indicates that cells are not able to adequately prevent accumulation of misfolded proteins. For example, several E3s can mediate ubiquitination of a given misfolded protein, but it is not clear whether all these pathways are really simultaneously active. One possible issue is that many of the earlier studies were often based on transient transfection experiments, in which both the E3 and the substrate were vastly overexpressed in comparison to their endogenous levels. This is particularly true for CHIP, as the bulk of studies on the topic was performed over a decade ago when only fewer molecular and cell biology tools were available. Therefore, while there are possibly several redundant quality control pathways at play, not all are necessarily active in a given cell type, especially in differentiated neurons. Side-by-side evaluation of some of these pathways will need to be performed in suitable cell models to better assess the role of each of these E3s in neurodegeneration.
Figure 6.
Protein–protein interaction (PPI) network of the six aggregation-prone proteins analyzed in this review. Source data was obtained from BioGRID on March 11, 2022, and condensed in Table S1. Here are represented only the proteins that display at least one high-confidence interaction (with number of PPI evidence on BioGRID greater or equal to four), which are depicted with solid lines. Interactions with less than four PPI evidence on BioGRID are represented as dotted lines. The size of the nodes is proportional to the number of interaction(s) with the aggregation-prone proteins, and the nodes are colored as indicated.
One challenging issue for evaluating the role of PPIs in disease progression is that there is no common repository for altered PPI due to a disease-associated mutation or upon oligomerization of a protein. For instance, most PPIs reported in BioGRID are assumed to occur between WT proteins. Some PPI specific to ND-associated mutants are included in the main database, but others, such as those involving oligomerized fibrils, are often not included. Perhaps, the creation of PPI databases that are better structured, with datasets being organized by gene and including the ND-associated mutations, could lay the foundation for a greater comprehension of the PINs involved in disease etiology. Another main issue is that most studies rely on coimmunoprecipitation experiments using either overexpressed baits in tissue culture cells or homogenized tissues, which implies nonphysiological conditions or loss of contextual information, respectively. For instance, some PPIs may not occur at lower concentrations or only occur in specific cell types. Therefore, it remains essential to test the role of each candidate PPI using adequate cell or animal models in order to properly determine whether it plays a role in the disease.
Mitochondrial dysfunction can be triggered by all the reviewed aggregation-prone proteins and may therefore play a central role in the etiology of neurological disorders. Neurons critically depend on this organelle and its correct localization to enable brain development, differentiation, neurotransmission, and neuronal activity. Markedly, many mitochondrial PPIs that we cited are not found in our main PIN (Fig. 6). Similar to the Hsp40 cochaperones, the number of reported evidence remains low for many of these interacting mitochondrial proteins. Nonetheless, some clear shared paths are emerging. For instance, impaired mitochondrial protein import and maturation of OXPHOS subunits, including the subsequent increase in ROS production and a lower energy state, are a common signature in all NDs that we reviewed. Additional detrimental defects further diversify the burden on mitochondria for each ND. For instance, accumulation of mutHTT on the outer mitochondrial membrane alters mitochondrial dynamics and promotes mitophagy in addition to disrupting protein import. In the case of ALS, import defects were extended to ROS-scavenging enzymes, thereby favoring ROS accumulation, alongside the inactivation of antiapoptotic factors. Although the exact timing between mitochondrial dysfunction and the onset of NDs are yet to be fully elucidated, it is clear that the impact of the aberrant interactions of mutant and/or misfolded aggregation-prone proteins leads to failure to maintain functional mitochondria. This often results in aberrant mitochondrial fusion/fission dynamics, impaired axonal transport, and ultimately neuronal death, thereby representing one of the major risk factors in the development and progression of neurodegenerative disorders.
Despite numerous clinical trials and the vast expansion of our knowledge in NDs, no cure has yet been established for these disorders. One important aspect is that each aggregation-prone protein engages multiple PPIs with various components of different cellular networks. Therefore, multifactor approaches will likely be required to effectively combat the cytotoxicity associated with protein aggregation. Alongside, we will need to better define which PPIs occur in differentiated cells and which elements are most likely altered upon aging, as they might exacerbate neurodegeneration. Importantly, while mutations are great tools to start investigating how PINs may be altered in diseases, better models will need to be established to understand how PPIs may be affected in sporadic forms of NDs.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We have tried to include as much as possible the relevant literature and we apologize to colleagues whose contributions have been omitted.
Author contributions
G. C. methodology and original figures design; G. C., C. M., and T. M. writing-original draft; G. C., C. M., and T. M. writing-review and editing.
Funding and additional information
Our work is supported by CIHR project scheme grant (PJT-148489). G. C. is supported by a DFG Walter Benjamin fellowship (CA 2559/1-1) and a MSFHR Research Trainee fellowship (RT-2020-0517) and C. M. by a UBC scholarship.
Edited by Paul Fraser
Contributor Information
Gaetano Calabrese, Email: gaetano.calabrese@msl.ubc.ca.
Thibault Mayor, Email: mayor@mail.ubc.ca.
Supporting information
References
- 1.Goedert M. NEURODEGENERATION. Alzheimer's and Parkinson's diseases: the prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 2.Bertram L., Tanzi R.E. The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh D.M., Selkoe D.J. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat. Rev. Neurosci. 2016;17:251–260. doi: 10.1038/nrn.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makin S. The amyloid hypothesis on trial. Nature. 2018;559:S4–S7. doi: 10.1038/d41586-018-05719-4. [DOI] [PubMed] [Google Scholar]
- 5.Ross C.A., Tabrizi S.J. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 6.Orr H.T., Zoghbi H.Y. SCA1 molecular genetics: a history of a 13 year collaboration against glutamines. Hum. Mol. Genet. 2001;10:2307–2311. doi: 10.1093/hmg/10.20.2307. [DOI] [PubMed] [Google Scholar]
- 7.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J., et al. Parkinson disease. Nat. Rev. Dis. Primers. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 9.Foerster B.R., Welsh R.C., Feldman E.L. 25 years of neuroimaging in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2013;9:513–524. doi: 10.1038/nrneurol.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- 11.Mathieu C., Pappu R.V., Taylor J.P. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petkova A.T., Leapman R.D., Guo Z., Yau W.M., Mattson M.P., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., et al. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer M.H., Wanker E.E., Andrade-Navarro M.A. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucl. Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 16.Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C., Cotman C.W., et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 17.Peng C., Gathagan R.J., Covell D.J., Medellin C., Stieber A., Robinson J.L., et al. Cellular milieu imparts distinct pathological alpha-synuclein strains in alpha-synucleinopathies. Nature. 2018;557:558–563. doi: 10.1038/s41586-018-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansbury P.T., Lashuel H.A. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 20.Keck S., Nitsch R., Grune T., Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J. Neurochem. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 21.Guo Q., Lehmer C., Martinez-Sanchez A., Rudack T., Beck F., Hartmann H., et al. In Situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. 2018;172:696–705.e612. doi: 10.1016/j.cell.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huttlin E.L., Bruckner R.J., Navarrete-Perea J., Cannon J.R., Baltier K., Gebreab F., et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184:3022–3040.e3028. doi: 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosp F., Vossfeldt H., Heinig M., Vasiljevic D., Arumughan A., Wyler E., et al. Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 2015;11:1134–1146. doi: 10.1016/j.celrep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e513. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou C.C., Zhang Y., Umoh M.E., Vaughan S.W., Lorenzini I., Liu F., et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018;21:228–239. doi: 10.1038/s41593-017-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner S.L., Cheng F., Hogan A.L., Grima N., Yang S., Ke Y.D., et al. ALS/FTD-causing mutation in cyclin F causes the dysregulation of SFPQ. Hum. Mol. Genet. 2021;30:971–984. doi: 10.1093/hmg/ddab073. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.Y., Kim H., Jo A., Khang R., Park C.H., Park S.J., et al. Alpha-synuclein A53T binds to transcriptional adapter 2-alpha and blocks histone H3 acetylation. Int. J. Mol. Sci. 2021;22:5392. doi: 10.3390/ijms22105392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piette B.L., Alerasool N., Lin Z.Y., Lacoste J., Lam M.H.Y., Qian W.W., et al. Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol. Cell. 2021;81:2549–2565.e2548. doi: 10.1016/j.molcel.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z.Y., Larsen B., et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haenig C., Atias N., Taylor A.K., Mazza A., Schaefer M.H., Russ J., et al. Interactome mapping provides a network of neurodegenerative disease proteins and uncovers widespread protein aggregation in affected brains. Cell Rep. 2020;32:108050. doi: 10.1016/j.celrep.2020.108050. [DOI] [PubMed] [Google Scholar]
- 33.Kruger R. The role of synphilin-1 in synaptic function and protein degradation. Cell Tissue Res. 2004;318:195–199. doi: 10.1007/s00441-004-0953-z. [DOI] [PubMed] [Google Scholar]
- 34.Lim J., Hao T., Shaw C., Patel A.J., Szabo G., Rual J.F., et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 35.Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnack C., Danzer K.M., Hengerer B., Gillardon F. Protein array analysis of oligomerization-induced changes in alpha-synuclein protein-protein interactions points to an interference with Cdc42 effector proteins. Neuroscience. 2008;154:1450–1457. doi: 10.1016/j.neuroscience.2008.02.049. [DOI] [PubMed] [Google Scholar]
- 37.Virok D.P., Simon D., Bozso Z., Rajko R., Datki Z., Balint E., et al. Protein array based interactome analysis of amyloid-beta indicates an inhibition of protein translation. J. Proteome Res. 2011;10:1538–1547. doi: 10.1021/pr1009096. [DOI] [PubMed] [Google Scholar]
- 38.Oughtred R., Rust J., Chang C., Breitkreutz B.J., Stark C., Willems A., et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busche M.A., Hyman B.T. Synergy between amyloid-beta and tau in Alzheimer's disease. Nat. Neurosci. 2020;23:1183–1193. doi: 10.1038/s41593-020-0687-6. [DOI] [PubMed] [Google Scholar]
- 40.Cao X., Sudhof T.C. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J. Biol. Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 41.Endres K., Deller T. Regulation of alpha-secretase ADAM10 in vitro and in vivo: genetic, epigenetic, and protein-based mechanisms. Front. Mol. Neurosci. 2017;10:56. doi: 10.3389/fnmol.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hussain I., Powell D., Howlett D.R., Tew D.G., Meek T.D., Chapman C., et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 43.Lin X., Koelsch G., Wu S., Downs D., Dashti A., Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada H., Zhang W., Peterhoff C., Hwang J.C., Nixon R.A., Ryu S.H., et al. Proteomic identification of sorting nexin 6 as a negative regulator of BACE1-mediated APP processing. FASEB J. 2010;24:2783–2794. doi: 10.1096/fj.09-146357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reitz C., Rogaeva E., Beecham G.W. Late-onset vs nonmendelian early-onset alzheimer disease: a distinction without a difference? Neurol. Genet. 2020;6:e512. doi: 10.1212/NXG.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn K., Shelton C.C., Tian Y., Zhang X., Gilchrist M.L., Sisodia S.S., et al. Activation and intrinsic gamma-secretase activity of presenilin 1. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Strooper B., Iwatsubo T., Wolfe M.S. Presenilins and gamma-secretase: Structure, function, and role in alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mekala S., Nelson G., Li Y.M. Recent developments of small molecule gamma-secretase modulators for Alzheimer's disease. RSC Med. Chem. 2020;11:1003–1022. doi: 10.1039/d0md00196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G., Nishimura M., Arawaka S., Levitan D., Zhang L., Tandon A., et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 50.Luo W.J., Wang H., Li H., Kim B.S., Shah S., Lee H.J., et al. PEN-2 and APH-1 coordinately regulate proteolytic processing of presenilin 1. J. Biol. Chem. 2003;278:7850–7854. doi: 10.1074/jbc.C200648200. [DOI] [PubMed] [Google Scholar]
- 51.Zhou R., Yang G., Guo X., Zhou Q., Lei J., Shi Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science. 2019;363 doi: 10.1126/science.aaw0930. [DOI] [PubMed] [Google Scholar]
- 52.Cai X.D., Golde T.E., Younkin S.G. Release of excess amyloid beta protein from a mutant amyloid beta protein precursor. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- 53.Citron M., Oltersdorf T., Haass C., McConlogue L., Hung A.Y., Seubert P., et al. Mutation of the beta-amyloid precursor protein in familial Alzheimer's disease increases beta-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 54.Wisniewski T., Ghiso J., Frangione B. Peptides homologous to the amyloid protein of Alzheimer's disease containing a glutamine for glutamic acid substitution have accelerated amyloid fibril formation. Biochem. Biophys. Res. Commun. 1991;179:1247–1254. doi: 10.1016/0006-291x(91)91706-i. [DOI] [PubMed] [Google Scholar]
- 55.Hardy J., Selkoe D.J. The amyloid hypothesis of alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 56.Cleary J.P., Walsh D.M., Hofmeister J.J., Shankar G.M., Kuskowski M.A., Selkoe D.J., et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 57.Finder V.H., Glockshuber R. Amyloid-beta aggregation. Neurodegener. Dis. 2007;4:13–27. doi: 10.1159/000100355. [DOI] [PubMed] [Google Scholar]
- 58.Chen G.F., Xu T.H., Yan Y., Zhou Y.R., Jiang Y., Melcher K., et al. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017;38:1205–1235. doi: 10.1038/aps.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammad S.M., Ranganathan S., Loukinova E., Twal W.O., Argraves W.S. Interaction of apolipoprotein J-amyloid beta-peptide complex with low density lipoprotein receptor-related protein-2/megalin. A mechanism to prevent pathological accumulation of amyloid beta-peptide. J. Biol. Chem. 1997;272:18644–18649. doi: 10.1074/jbc.272.30.18644. [DOI] [PubMed] [Google Scholar]
- 60.He X., Cooley K., Chung C.H., Dashti N., Tang J. Apolipoprotein receptor 2 and X11 alpha/beta mediate apolipoprotein E-induced endocytosis of amyloid-beta precursor protein and beta-secretase, leading to amyloid-beta production. J. Neurosci. 2007;27:4052–4060. doi: 10.1523/JNEUROSCI.3993-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lakshmana M.K., Yoon I.S., Chen E., Bianchi E., Koo E.H., Kang D.E. Novel role of RanBP9 in BACE1 processing of amyloid precursor protein and amyloid beta peptide generation. J. Biol. Chem. 2009;284:11863–11872. doi: 10.1074/jbc.M807345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kounnas M.Z., Moir R.D., Rebeck G.W., Bush A.I., Argraves W.S., Tanzi R.E., et al. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 63.Haas J., Beer A.G., Widschwendter P., Oberdanner J., Salzmann K., Sarg B., et al. LRP1b shows restricted expression in human tissues and binds to several extracellular ligands, including fibrinogen and apoE-carrying lipoproteins. Atherosclerosis. 2011;216:342–347. doi: 10.1016/j.atherosclerosis.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pietrzik C.U., Yoon I.S., Jaeger S., Busse T., Weggen S., Koo E.H. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J. Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cam J.A., Zerbinatti C.V., Knisely J.M., Hecimovic S., Li Y., Bu G. The low density lipoprotein receptor-related protein 1B retains beta-amyloid precursor protein at the cell surface and reduces amyloid-beta peptide production. J. Biol. Chem. 2004;279:29639–29646. doi: 10.1074/jbc.M313893200. [DOI] [PubMed] [Google Scholar]
- 66.Yang D.S., Smith J.D., Zhou Z., Gandy S.E., Martins R.N. Characterization of the binding of amyloid-beta peptide to cell culture-derived native apolipoprotein E2, E3, and E4 isoforms and to isoforms from human plasma. J. Neurochem. 1997;68:721–725. doi: 10.1046/j.1471-4159.1997.68020721.x. [DOI] [PubMed] [Google Scholar]
- 67.Ma J., Yee A., Brewer H.B., Jr., Das S., Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372:92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- 68.Calero M., Rostagno A., Ghiso J. Search for amyloid-binding proteins by affinity chromatography. Met. Mol. Biol. 2012;849:213–223. doi: 10.1007/978-1-61779-551-0_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verghese P.B., Castellano J.M., Garai K., Wang Y., Jiang H., Shah A., et al. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1807–1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Storck S.E., Meister S., Nahrath J., Meissner J.N., Schubert N., Di Spiezio A., et al. Endothelial LRP1 transports amyloid-beta(1-42) across the blood-brain barrier. J. Clin. Invest. 2016;126:123–136. doi: 10.1172/JCI81108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruzali W.A., Kehoe P.G., Love S. Influence of LRP-1 and apolipoprotein E on amyloid-beta uptake and toxicity to cerebrovascular smooth muscle cells. J. Alzheimers Dis. 2013;33:95–110. doi: 10.3233/JAD-2012-121336. [DOI] [PubMed] [Google Scholar]
- 72.Yeh F.L., Wang Y., Tom I., Gonzalez L.C., Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 73.Scheidt T., Lapinska U., Kumita J.R., Whiten D.R., Klenerman D., Wilson M.R., et al. Secondary nucleation and elongation occur at different sites on Alzheimer's amyloid-beta aggregates. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aau3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satapathy S., Wilson M.R. The dual roles of clusterin in extracellular and intracellular proteostasis. Trends Biochem. Sci. 2021;46:652–660. doi: 10.1016/j.tibs.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N.P., et al. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 76.Eckman E.A., Reed D.K., Eckman C.B. Degradation of the Alzheimer's amyloid beta peptide by endothelin-converting enzyme. J. Biol. Chem. 2001;276:24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- 77.Edbauer D., Willem M., Lammich S., Steiner H., Haass C. Insulin-degrading enzyme rapidly removes the beta-amyloid precursor protein intracellular domain (AICD) J. Biol. Chem. 2002;277:13389–13393. doi: 10.1074/jbc.M111571200. [DOI] [PubMed] [Google Scholar]
- 78.Tucker H.M., Kihiko M., Caldwell J.N., Wright S., Kawarabayashi T., Price D., et al. The plasmin system is induced by and degrades amyloid-beta aggregates. J. Neurosci. 2000;20:3937–3946. doi: 10.1523/JNEUROSCI.20-11-03937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Letronne F., Laumet G., Ayral A.M., Chapuis J., Demiautte F., Laga M., et al. ADAM30 downregulates APP-linked defects through Cathepsin D activation in alzheimer's disease. EBioMedicine. 2016;9:278–292. doi: 10.1016/j.ebiom.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin K.J., Cirrito J.R., Yan P., Hu X., Xiao Q., Pan X., et al. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J. Neurosci. 2006;26:10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gireud-Goss M., Reyes S., Tewari R., Patrizz A., Howe M.D., Kofler J., et al. The ubiquitin ligase UBE4B regulates amyloid precursor protein ubiquitination, endosomal trafficking, and amyloid beta42 generation and secretion. Mol. Cell Neurosci. 2020;108:103542. doi: 10.1016/j.mcn.2020.103542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watanabe T., Hikichi Y., Willuweit A., Shintani Y., Horiguchi T. FBL2 regulates amyloid precursor protein (APP) metabolism by promoting ubiquitination-dependent APP degradation and inhibition of APP endocytosis. J. Neurosci. 2012;32:3352–3365. doi: 10.1523/JNEUROSCI.5659-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar P., Ambasta R.K., Veereshwarayya V., Rosen K.M., Kosik K.S., Band H., et al. CHIP and HSPs interact with beta-APP in a proteasome-dependent manner and influence Abeta metabolism. Hum. Mol. Genet. 2007;16:848–864. doi: 10.1093/hmg/ddm030. [DOI] [PubMed] [Google Scholar]
- 84.Singh A.K., Pati U. CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of beta-secretase. Aging Cell. 2015;14:595–604. doi: 10.1111/acel.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cottrell B.A., Galvan V., Banwait S., Gorostiza O., Lombardo C.R., Williams T., et al. A pilot proteomic study of amyloid precursor interactors in Alzheimer's disease. Ann. Neurol. 2005;58:277–289. doi: 10.1002/ana.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mainz A., Peschek J., Stavropoulou M., Back K.C., Bardiaux B., Asami S., et al. The chaperone alphaB-crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat. Struct. Mol. Biol. 2015;22:898–905. doi: 10.1038/nsmb.3108. [DOI] [PubMed] [Google Scholar]
- 87.Olah J., Vincze O., Virok D., Simon D., Bozso Z., Tokesi N., et al. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J. Biol. Chem. 2011;286:34088–34100. doi: 10.1074/jbc.M111.243907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mansson C., Arosio P., Hussein R., Kampinga H.H., Hashem R.M., Boelens W.C., et al. Interaction of the molecular chaperone DNAJB6 with growing amyloid-beta 42 (Abeta42) aggregates leads to sub-stoichiometric inhibition of amyloid formation. J. Biol. Chem. 2014;289:31066–31076. doi: 10.1074/jbc.M114.595124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Osterlund N., Lundqvist M., Ilag L.L., Graslund A., Emanuelsson C. Amyloid-beta oligomers are captured by the DNAJB6 chaperone: direct detection of interactions that can prevent primary nucleation. J. Biol. Chem. 2020;295:8135–8144. doi: 10.1074/jbc.RA120.013459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kouchi Z., Sorimachi H., Suzuki K., Ishiura S. Proteasome inhibitors induce the association of Alzheimer's amyloid precursor protein with Hsc73. Biochem. Biophys. Res. Commun. 1999;254:804–810. doi: 10.1006/bbrc.1998.9977. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto Y.H., Kimura T., Momohara S., Takeuchi M., Tani T., Kimata Y., et al. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct. Funct. 2010;35:107–116. doi: 10.1247/csf.10023. [DOI] [PubMed] [Google Scholar]
- 92.Devi L., Anandatheerthavarada H.K. Mitochondrial trafficking of APP and alpha synuclein: relevance to mitochondrial dysfunction in Alzheimer's and Parkinson's diseases. Biochim. Biophys. Acta. 2010;1802:11–19. doi: 10.1016/j.bbadis.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song J., Becker T. Clogging the mitochondrial protein-entry gate promotes obesity. Nat. Metab. 2019;1:1175–1176. doi: 10.1038/s42255-019-0155-3. [DOI] [PubMed] [Google Scholar]
- 94.Pavlov P.F., Wiehager B., Sakai J., Frykman S., Behbahani H., Winblad B., et al. Mitochondrial gamma-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB J. 2011;25:78–88. doi: 10.1096/fj.10-157230. [DOI] [PubMed] [Google Scholar]
- 95.Devi L., Prabhu B.M., Galati D.F., Avadhani N.G., Anandatheerthavarada H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sterky F.H., Ruzzenente B., Gustafsson C.M., Samuelsson T., Larsson N.G. LRPPRC is a mitochondrial matrix protein that is conserved in metazoans. Biochem. Biophys. Res. Commun. 2010;398:759–764. doi: 10.1016/j.bbrc.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 97.Olahova M., Hardy S.A., Hall J., Yarham J.W., Haack T.B., Wilson W.C., et al. LRPPRC mutations cause early-onset multisystem mitochondrial disease outside of the French-Canadian population. Brain. 2015;138:3503–3519. doi: 10.1093/brain/awv291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U. S. A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pavlov P.F., Hansson Petersen C., Glaser E., Ankarcrona M. Mitochondrial accumulation of APP and abeta: significance for alzheimer disease pathogenesis. J. Cell Mol. Med. 2009;13:4137–4145. doi: 10.1111/j.1582-4934.2009.00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X., Su B., Siedlak S.L., Moreira P.I., Fujioka H., Wang Y., et al. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schmidt C., Lepsverdize E., Chi S.L., Das A.M., Pizzo S.V., Dityatev A., et al. Amyloid precursor protein and amyloid beta-peptide bind to ATP synthase and regulate its activity at the surface of neural cells. Mol. Psych. 2008;13:953–969. doi: 10.1038/sj.mp.4002077. [DOI] [PubMed] [Google Scholar]
- 102.Spillantini M.G., Bird T.D., Ghetti B. Frontotemporal dementia and Parkinsonism linked to chromosome 17: a new group of tauopathies. Brain Pathol. 1998;8:387–402. doi: 10.1111/j.1750-3639.1998.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forrest S.L., Kril J.J., Stevens C.H., Kwok J.B., Hallupp M., Kim W.S., et al. Retiring the term FTDP-17 as MAPT mutations are genetic forms of sporadic frontotemporal tauopathies. Brain. 2018;141:521–534. doi: 10.1093/brain/awx328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sadik G., Tanaka T., Kato K., Yanagi K., Kudo T., Takeda M. Differential interaction and aggregation of 3-repeat and 4-repeat tau isoforms with 14-3-3zeta protein. Biochem. Biophys. Res. Commun. 2009;383:37–41. doi: 10.1016/j.bbrc.2009.03.107. [DOI] [PubMed] [Google Scholar]
- 105.Hashiguchi M., Sobue K., Paudel H.K. 14-3-3zeta is an effector of tau protein phosphorylation. J. Biol. Chem. 2000;275:25247–25254. doi: 10.1074/jbc.M003738200. [DOI] [PubMed] [Google Scholar]
- 106.Martin L., Latypova X., Wilson C.M., Magnaudeix A., Perrin M.L., Yardin C., et al. Tau protein kinases: involvement in alzheimer's disease. Ageing Res. Rev. 2013;12:289–309. doi: 10.1016/j.arr.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 107.Toral-Rios D., Pichardo-Rojas P.S., Alonso-Vanegas M., Campos-Pena V. GSK3beta and tau protein in alzheimer's disease and epilepsy. Front. Cell Neurosci. 2020;14:19. doi: 10.3389/fncel.2020.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen C., Gu J., Basurto-Islas G., Jin N., Wu F., Gong C.X., et al. Up-regulation of casein kinase 1epsilon is involved in tau pathogenesis in Alzheimer's disease. Sci. Rep. 2017;7:13478. doi: 10.1038/s41598-017-13791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Briner A., Gotz J., Polanco J.C. Fyn kinase controls tau aggregation in vivo. Cell Rep. 2020;32:108045. doi: 10.1016/j.celrep.2020.108045. [DOI] [PubMed] [Google Scholar]
- 110.Li J.Q., Tan L., Yu J.T. The role of the LRRK2 gene in Parkinsonism. Mol. Neurodegener. 2014;9:47. doi: 10.1186/1750-1326-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seo J., Kritskiy O., Watson L.A., Barker S.J., Dey D., Raja W.K., et al. Inhibition of p25/Cdk5 attenuates tauopathy in mouse and iPSC models of frontotemporal dementia. J. Neurosci. 2017;37:9917–9924. doi: 10.1523/JNEUROSCI.0621-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hashimoto S., Matsuba Y., Kamano N., Mihira N., Sahara N., Takano J., et al. Tau binding protein CAPON induces tau aggregation and neurodegeneration. Nat. Commun. 2019;10:2394. doi: 10.1038/s41467-019-10278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shimura H., Schwartz D., Gygi S.P., Kosik K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 114.van Nerom C., Bormans G., Cleynhens B., de Roo M., Verbruggen A. A convenient configuration for safe and efficient radio-HPLC of 99mTc-radiopharmaceuticals. J. Pharm. Biomed. Anal. 1989;7:1849–1852. doi: 10.1016/0731-7085(89)80202-x. [DOI] [PubMed] [Google Scholar]
- 115.Ye Y., Klenerman D., Finley D. N-terminal ubiquitination of amyloidogenic proteins triggers removal of their oligomers by the proteasome holoenzyme. J. Mol. Biol. 2020;432:585–596. doi: 10.1016/j.jmb.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schirmer C., Lepvrier E., Duchesne L., Decaux O., Thomas D., Delamarche C., et al. Hsp90 directly interacts, in vitro, with amyloid structures and modulates their assembly and disassembly. Biochim. Biophys. Acta. 2016;1860:2598–2609. doi: 10.1016/j.bbagen.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 117.Fontaine S.N., Rauch J.N., Nordhues B.A., Assimon V.A., Stothert A.R., Jinwal U.K., et al. Isoform-selective genetic inhibition of constitutive cytosolic Hsp70 activity promotes client tau degradation using an altered Co-chaperone complement. J. Biol. Chem. 2015;290:13115–13127. doi: 10.1074/jbc.M115.637595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trzeciakiewicz H., Ajit D., Tseng J.H., Chen Y., Ajit A., Tabassum Z., et al. An HDAC6-dependent surveillance mechanism suppresses tau-mediated neurodegeneration and cognitive decline. Nat. Commun. 2020;11:5522. doi: 10.1038/s41467-020-19317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taylor I.R., Ahmad A., Wu T., Nordhues B.A., Bhullar A., Gestwicki J.E., et al. The disorderly conduct of Hsc70 and its interaction with the Alzheimer's-related Tau protein. J. Biol. Chem. 2018;293:10796–10809. doi: 10.1074/jbc.RA118.002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elliott E., Tsvetkov P., Ginzburg I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of tau protein. J. Biol. Chem. 2007;282:37276–37284. doi: 10.1074/jbc.M706379200. [DOI] [PubMed] [Google Scholar]
- 121.Nachman E., Wentink A.S., Madiona K., Bousset L., Katsinelos T., Allinson K., et al. Disassembly of Tau fibrils by the human Hsp70 disaggregation machinery generates small seeding-competent species. J. Biol. Chem. 2020;295:9676–9690. doi: 10.1074/jbc.RA120.013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patterson K.R., Ward S.M., Combs B., Voss K., Kanaan N.M., Morfini G., et al. Heat shock protein 70 prevents both tau aggregation and the inhibitory effects of preexisting tau aggregates on fast axonal transport. Biochemistry. 2011;50:10300–10310. doi: 10.1021/bi2009147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mok S.A., Condello C., Freilich R., Gillies A., Arhar T., Oroz J., et al. Mapping interactions with the chaperone network reveals factors that protect against tau aggregation. Nat. Struct. Mol. Biol. 2018;25:384–393. doi: 10.1038/s41594-018-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Irwin R., Faust O., Petrovic I., Wolf S.G., Hofmann H., Rosenzweig R. Hsp40s play complementary roles in the prevention of tau amyloid formation. Elife. 2021;10 doi: 10.7554/eLife.69601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hou Z., Wydorski P.M., Perez V.A., Mendoza-Oliva A., Ryder B.D., Mirbaha H., et al. DnaJC7 binds natively folded structural elements in tau to inhibit amyloid formation. Nat. Commun. 2021;12:5338. doi: 10.1038/s41467-021-25635-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baughman H.E.R., Clouser A.F., Klevit R.E., Nath A. HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem. 2018;293:2687–2700. doi: 10.1074/jbc.M117.803411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rank K.B., Pauley A.M., Bhattacharya K., Wang Z., Evans D.B., Fleck T.J., et al. Direct interaction of soluble human recombinant tau protein with Abeta 1-42 results in tau aggregation and hyperphosphorylation by tau protein kinase II. FEBS Lett. 2002;514:263–268. doi: 10.1016/s0014-5793(02)02376-1. [DOI] [PubMed] [Google Scholar]
- 128.Guo J.P., Arai T., Miklossy J., McGeer P.L. Abeta and tau form soluble complexes that may promote self aggregation of both into the insoluble forms observed in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1953–1958. doi: 10.1073/pnas.0509386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barthelemy N.R., Li Y., Joseph-Mathurin N., Gordon B.A., Hassenstab J., Benzinger T.L.S., et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer's disease. Nat. Med. 2020;26:398–407. doi: 10.1038/s41591-020-0781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He Z., Guo J.L., McBride J.D., Narasimhan S., Kim H., Changolkar L., et al. Amyloid-beta plaques enhance Alzheimer's brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 2018;24:29–38. doi: 10.1038/nm.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., Lamb B.T. Regulation of tau pathology by the microglial fractalkine receptor. Neuron. 2010;68:19–31. doi: 10.1016/j.neuron.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tran J., Anastacio H., Bardy C. Genetic predispositions of Parkinson's disease revealed in patient-derived brain cells. NPJ Parkinsons Dis. 2020;6:8. doi: 10.1038/s41531-020-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deng H., Wang P., Jankovic J. The genetics of Parkinson disease. Ageing Res. Rev. 2018;42:72–85. doi: 10.1016/j.arr.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 135.Fares M.B., Jagannath S., Lashuel H.A. Reverse engineering Lewy bodies: how far have we come and how far can we go? Nat. Rev. Neurosci. 2021;22:111–131. doi: 10.1038/s41583-020-00416-6. [DOI] [PubMed] [Google Scholar]
- 136.Li B., Ge P., Murray K.A., Sheth P., Zhang M., Nair G., et al. Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 2018;9:3609. doi: 10.1038/s41467-018-05971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li Y., Zhao C., Luo F., Liu Z., Gui X., Luo Z., et al. Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res. 2018;28:897–903. doi: 10.1038/s41422-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun Y., Hou S., Zhao K., Long H., Liu Z., Gao J., et al. Cryo-EM structure of full-length alpha-synuclein amyloid fibril with Parkinson's disease familial A53T mutation. Cell Res. 2020;30:360–362. doi: 10.1038/s41422-020-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boyer D.R., Li B., Sun C., Fan W., Zhou K., Hughes M.P., et al. The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl. Acad. Sci. U. S. A. 2020;117:3592–3602. doi: 10.1073/pnas.1917914117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao K., Li Y., Liu Z., Long H., Zhao C., Luo F., et al. Parkinson's disease associated mutation E46K of alpha-synuclein triggers the formation of a distinct fibril structure. Nat. Commun. 2020;11:2643. doi: 10.1038/s41467-020-16386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang C.C., Chiu T.Y., Lee T.Y., Hsieh H.J., Lin C.C., Kao L.S. Soluble alpha-synuclein facilitates priming and fusion by releasing Ca(2+) from the thapsigargin-sensitive Ca(2+) pool in PC12 cells. J. Cell Sci. 2018;131:jcs213017. doi: 10.1242/jcs.213017. [DOI] [PubMed] [Google Scholar]
- 142.Burre J., Sharma M., Tsetsenis T., Buchman V., Etherton M.R., Sudhof T.C. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alim M.A., Hossain M.S., Arima K., Takeda K., Izumiyama Y., Nakamura M., et al. Tubulin seeds alpha-synuclein fibril formation. J. Biol. Chem. 2002;277:2112–2117. doi: 10.1074/jbc.M102981200. [DOI] [PubMed] [Google Scholar]
- 144.Jensen P.H., Hager H., Nielsen M.S., Hojrup P., Gliemann J., Jakes R. alpha-synuclein binds to Tau and stimulates the protein kinase A-catalyzed tau phosphorylation of serine residues 262 and 356. J. Biol. Chem. 1999;274:25481–25489. doi: 10.1074/jbc.274.36.25481. [DOI] [PubMed] [Google Scholar]
- 145.Giasson B.I., Forman M.S., Higuchi M., Golbe L.I., Graves C.L., Kotzbauer P.T., et al. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300:636–640. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 146.Leitao A.D.G., Rudolffi-Soto P., Chappard A., Bhumkar A., Lau D., Hunter D.J.B., et al. Selectivity of Lewy body protein interactions along the aggregation pathway of alpha-synuclein. Commun. Biol. 2021;4:1124. doi: 10.1038/s42003-021-02624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee F.J., Liu F., Pristupa Z.B., Niznik H.B. Direct binding and functional coupling of alpha-synuclein to the dopamine transporters accelerate dopamine-induced apoptosis. FASEB J. 2001;15:916–926. doi: 10.1096/fj.00-0334com. [DOI] [PubMed] [Google Scholar]
- 148.Wersinger C., Sidhu A. Disruption of the interaction of alpha-synuclein with microtubules enhances cell surface recruitment of the dopamine transporter. Biochemistry. 2005;44:13612–13624. doi: 10.1021/bi050402p. [DOI] [PubMed] [Google Scholar]
- 149.Butler B., Saha K., Rana T., Becker J.P., Sambo D., Davari P., et al. Dopamine transporter activity is modulated by alpha-synuclein. J. Biol. Chem. 2015;290:29542–29554. doi: 10.1074/jbc.M115.691592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wersinger C., Vernier P., Sidhu A. Trypsin disrupts the trafficking of the human dopamine transporter by alpha-synuclein and its A30P mutant. Biochemistry. 2004;43:1242–1253. doi: 10.1021/bi035308s. [DOI] [PubMed] [Google Scholar]
- 151.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S., et al. Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 152.Walker D.G., Lue L.F., Adler C.H., Shill H.A., Caviness J.N., Sabbagh M.N., et al. Changes in properties of serine 129 phosphorylated alpha-synuclein with progression of Lewy-type histopathology in human brains. Exp. Neurol. 2013;240:190–204. doi: 10.1016/j.expneurol.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oueslati A. Implication of alpha-synuclein phosphorylation at S129 in synucleinopathies: what have we learned in the last decade? J. Parkinsons Dis. 2016;6:39–51. doi: 10.3233/JPD-160779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McFarland M.A., Ellis C.E., Markey S.P., Nussbaum R.L. Proteomics analysis identifies phosphorylation-dependent alpha-synuclein protein interactions. Mol. Cell Proteomics. 2008;7:2123–2137. doi: 10.1074/mcp.M800116-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tanudjojo B., Shaikh S.S., Fenyi A., Bousset L., Agarwal D., Marsh J., et al. Phenotypic manifestation of alpha-synuclein strains derived from Parkinson's disease and multiple system atrophy in human dopaminergic neurons. Nat. Commun. 2021;12:3817. doi: 10.1038/s41467-021-23682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rekas A., Adda C.G., Andrew Aquilina J., Barnham K.J., Sunde M., Galatis D., et al. Interaction of the molecular chaperone alphaB-crystallin with alpha-synuclein: effects on amyloid fibril formation and chaperone activity. J. Mol. Biol. 2004;340:1167–1183. doi: 10.1016/j.jmb.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 157.Dedmon M.M., Christodoulou J., Wilson M.R., Dobson C.M. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J. Biol. Chem. 2005;280:14733–14740. doi: 10.1074/jbc.M413024200. [DOI] [PubMed] [Google Scholar]
- 158.Pemberton S., Madiona K., Pieri L., Kabani M., Bousset L., Melki R. Hsc70 protein interaction with soluble and fibrillar alpha-synuclein. J. Biol. Chem. 2011;286:34690–34699. doi: 10.1074/jbc.M111.261321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Falsone S.F., Kungl A.J., Rek A., Cappai R., Zangger K. The molecular chaperone Hsp90 modulates intermediate steps of amyloid assembly of the Parkinson-related protein alpha-synuclein. J. Biol. Chem. 2009;284:31190–31199. doi: 10.1074/jbc.M109.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gao X., Carroni M., Nussbaum-Krammer C., Mogk A., Nillegoda N.B., Szlachcic A., et al. Human Hsp70 disaggregase reverses Parkinson's-linked alpha-synuclein amyloid fibrils. Mol. Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Aprile F.A., Kallstig E., Limorenko G., Vendruscolo M., Ron D., Hansen C. The molecular chaperones DNAJB6 and Hsp70 cooperate to suppress alpha-synuclein aggregation. Sci. Rep. 2017;7:9039. doi: 10.1038/s41598-017-08324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Deshayes N., Arkan S., Hansen C. The molecular chaperone DNAJB6, but not DNAJB1, suppresses the seeded aggregation of alpha-synuclein in cells. Int. J. Mol. Sci. 2019;20:4495. doi: 10.3390/ijms20184495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arkan S., Ljungberg M., Kirik D., Hansen C. DNAJB6 suppresses alpha-synuclein induced pathology in an animal model of Parkinson's disease. Neurobiol. Dis. 2021;158:105477. doi: 10.1016/j.nbd.2021.105477. [DOI] [PubMed] [Google Scholar]
- 164.Burmann B.M., Gerez J.A., Matecko-Burmann I., Campioni S., Kumari P., Ghosh D., et al. Regulation of alpha-synuclein by chaperones in mammalian cells. Nature. 2020;577:127–132. doi: 10.1038/s41586-019-1808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brahmachari S., Ge P., Lee S.H., Kim D., Karuppagounder S.S., Kumar M., et al. Activation of tyrosine kinase c-Abl contributes to alpha-synuclein-induced neurodegeneration. J. Clin. Invest. 2016;126:2970–2988. doi: 10.1172/JCI85456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dikiy I., Fauvet B., Jovicic A., Mahul-Mellier A.L., Desobry C., El-Turk F., et al. Semisynthetic and in vitro phosphorylation of alpha-synuclein at Y39 promotes functional partly helical membrane-bound states resembling those induced by PD mutations. ACS Chem. Biol. 2016;11:2428–2437. doi: 10.1021/acschembio.6b00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tofaris G.K., Razzaq A., Ghetti B., Lilley K.S., Spillantini M.G. Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J. Biol. Chem. 2003;278:44405–44411. doi: 10.1074/jbc.M308041200. [DOI] [PubMed] [Google Scholar]
- 168.Shimura H., Schlossmacher M.G., Hattori N., Frosch M.P., Trockenbacher A., Schneider R., et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 169.Chung K.K., Zhang Y., Lim K.L., Tanaka Y., Huang H., Gao J., et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 170.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee J.T., Wheeler T.C., Li L., Chin L.S. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum. Mol. Genet. 2008;17:906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 172.Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A.J., Shani V., et al. alpha-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Alexopoulou Z., Lang J., Perrett R.M., Elschami M., Hurry M.E., Kim H.T., et al. Deubiquitinase Usp8 regulates alpha-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4688–4697. doi: 10.1073/pnas.1523597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tofaris G.K., Kim H.T., Hourez R., Jung J.W., Kim K.P., Goldberg A.L. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tardiff D.F., Jui N.T., Khurana V., Tambe M.A., Thompson M.L., Chung C.Y., et al. Yeast reveal a "druggable" Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tetzlaff J.E., Putcha P., Outeiro T.F., Ivanov A., Berezovska O., Hyman B.T., et al. CHIP targets toxic alpha-Synuclein oligomers for degradation. J. Biol. Chem. 2008;283:17962–17968. doi: 10.1074/jbc.M802283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bendifallah M., Redeker V., Monsellier E., Bousset L., Bellande T., Melki R. Interaction of the chaperones alpha B-crystallin and CHIP with fibrillar alpha-synuclein: effects on internalization by cells and identification of interacting interfaces. Biochem. Biophys. Res. Commun. 2020;527:760–769. doi: 10.1016/j.bbrc.2020.04.091. [DOI] [PubMed] [Google Scholar]
- 178.Kalia L.V., Kalia S.K., Chau H., Lozano A.M., Hyman B.T., McLean P.J. Ubiquitinylation of alpha-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS One. 2011;6 doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gerez J.A., Prymaczok N.C., Rockenstein E., Herrmann U.S., Schwarz P., Adame A., et al. A cullin-RING ubiquitin ligase targets exogenous alpha-synuclein and inhibits Lewy body-like pathology. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aau6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 182.Dice J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 183.Orenstein S.J., Kuo S.H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ho P.W., Leung C.T., Liu H., Pang S.Y., Lam C.S., Xian J., et al. Age-dependent accumulation of oligomeric SNCA/alpha-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: role for therapeutic activation of chaperone-mediated autophagy (CMA) Autophagy. 2020;16:347–370. doi: 10.1080/15548627.2019.1603545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Stefanis L., Emmanouilidou E., Pantazopoulou M., Kirik D., Vekrellis K., Tofaris G.K. How is alpha-synuclein cleared from the cell? J. Neurochem. 2019;150:577–590. doi: 10.1111/jnc.14704. [DOI] [PubMed] [Google Scholar]
- 186.Fontaine S.N., Zheng D., Sabbagh J.J., Martin M.D., Chaput D., Darling A., et al. DnaJ/Hsc70 chaperone complexes control the extracellular release of neurodegenerative-associated proteins. EMBO J. 2016;35:1537–1549. doi: 10.15252/embj.201593489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Trepte P., Kruse S., Kostova S., Hoffmann S., Buntru A., Tempelmeier A., et al. LuTHy: a double-readout bioluminescence-based two-hybrid technology for quantitative mapping of protein-protein interactions in mammalian cells. Mol. Syst. Biol. 2018;14 doi: 10.15252/msb.20178071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chandra S., Gallardo G., Fernandez-Chacon R., Schluter O.M., Sudhof T.C. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123:383–396. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 189.Calo L., Hidari E., Wegrzynowicz M., Dalley J.W., Schneider B.L., Podgajna M., et al. CSPalpha reduces aggregates and rescues striatal dopamine release in alpha-synuclein transgenic mice. Brain. 2021;144:1661–1669. doi: 10.1093/brain/awab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Trinh D., Israwi A.R., Arathoon L.R., Gleave J.A., Nash J.E. The multi-faceted role of mitochondria in the pathology of Parkinson's disease. J. Neurochem. 2021;156:715–752. doi: 10.1111/jnc.15154. [DOI] [PubMed] [Google Scholar]
- 191.Dexter D.T., Sian J., Rose S., Hindmarsh J.G., Mann V.M., Cooper J.M., et al. Indices of oxidative stress and mitochondrial function in individuals with incidental Lewy body disease. Ann. Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 192.Langston J.W. The MPTP story. J. Parkinsons Dis. 2017;7:S11–S19. doi: 10.3233/JPD-179006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McLelland G.L., Goiran T., Yi W., Dorval G., Chen C.X., Lauinger N.D., et al. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. 2018;7 doi: 10.7554/eLife.32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wong Y.C., Holzbaur E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 201.Hsu L.J., Sagara Y., Arroyo A., Rockenstein E., Sisk A., Mallory M., et al. alpha-synuclein promotes mitochondrial deficit and oxidative stress. Am. J. Pathol. 2000;157:401–410. doi: 10.1016/s0002-9440(10)64553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nakamura K., Nemani V.M., Azarbal F., Skibinski G., Levy J.M., Egami K., et al. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J. Biol. Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Subramaniam S.R., Vergnes L., Franich N.R., Reue K., Chesselet M.F. Region specific mitochondrial impairment in mice with widespread overexpression of alpha-synuclein. Neurobiol. Dis. 2014;70:204–213. doi: 10.1016/j.nbd.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Devi L., Raghavendran V., Prabhu B.M., Avadhani N.G., Anandatheerthavarada H.K. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nakamura K., Nemani V.M., Wallender E.K., Kaehlcke K., Ott M., Edwards R.H. Optical reporters for the conformation of alpha-synuclein reveal a specific interaction with mitochondria. J. Neurosci. 2008;28:12305–12317. doi: 10.1523/JNEUROSCI.3088-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., et al. alpha-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 2014;34:249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Paillusson S., Gomez-Suaga P., Stoica R., Little D., Gissen P., Devine M.J., et al. alpha-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca(2+) homeostasis and mitochondrial ATP production. Acta Neuropathol. 2017;134:129–149. doi: 10.1007/s00401-017-1704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hu D., Sun X., Liao X., Zhang X., Zarabi S., Schimmer A., et al. Alpha-synuclein suppresses mitochondrial protease ClpP to trigger mitochondrial oxidative damage and neurotoxicity. Acta Neuropathol. 2019;137:939–960. doi: 10.1007/s00401-019-01993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Di Maio R., Barrett P.J., Hoffman E.K., Barrett C.W., Zharikov A., Borah A., et al. alpha-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Sci. Transl. Med. 2016;8:342ra378. doi: 10.1126/scitranslmed.aaf3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ludtmann M.H.R., Angelova P.R., Horrocks M.H., Choi M.L., Rodrigues M., Baev A.Y., et al. alpha-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson's disease. Nat. Commun. 2018;9:2293. doi: 10.1038/s41467-018-04422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tabrizi S.J., Flower M.D., Ross C.A., Wild E.J. Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 2020;16:529–546. doi: 10.1038/s41582-020-0389-4. [DOI] [PubMed] [Google Scholar]
- 212.Kremer B., Goldberg P., Andrew S.E., Theilmann J., Telenius H., Zeisler J., et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 213.Genetic Modifiers of Huntington's Disease Consortium CAG repeat not polyglutamine length determines timing of Huntington's Disease Onset. Cell. 2019;178:887–900.e14. doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Riguet N., Mahul-Mellier A.L., Maharjan N., Burtscher J., Croisier M., Knott G., et al. Nuclear and cytoplasmic huntingtin inclusions exhibit distinct biochemical composition, interactome and ultrastructural properties. Nat. Commun. 2021;12:6579. doi: 10.1038/s41467-021-26684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thibaudeau T.A., Anderson R.T., Smith D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018;9:1097. doi: 10.1038/s41467-018-03509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Masnata M., Sciacca G., Maxan A., Bousset L., Denis H.L., Lauruol F., et al. Demonstration of prion-like properties of mutant huntingtin fibrils in both in vitro and in vivo paradigms. Acta Neuropathol. 2019;137:981–1001. doi: 10.1007/s00401-019-01973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rieux M., Alpaugh M., Sciacca G., Saint-Pierre M., Masnata M., Denis H.L., et al. Shedding a new light on huntington's disease: how blood can both propagate and ameliorate disease pathology. Mol. Psych. 2021;26:5441–5463. doi: 10.1038/s41380-020-0787-4. [DOI] [PubMed] [Google Scholar]
- 218.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89:910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 219.Stuwe S.H., Goetze O., Lukas C., Klotz P., Hoffmann R., Banasch M., et al. Hepatic mitochondrial dysfunction in manifest and premanifest Huntington disease. Neurology. 2013;80:743–746. doi: 10.1212/WNL.0b013e318282514e. [DOI] [PubMed] [Google Scholar]
- 220.van der Burg J.M., Bjorkqvist M., Brundin P. Beyond the brain: widespread pathology in huntington's disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- 221.Wanker E.E., Ast A., Schindler F., Trepte P., Schnoegl S. The pathobiology of perturbed mutant huntingtin protein-protein interactions in Huntington's disease. J. Neurochem. 2019;151:507–519. doi: 10.1111/jnc.14853. [DOI] [PubMed] [Google Scholar]
- 222.Guo Q., Bin H., Cheng J., Seefelder M., Engler T., Pfeifer G., et al. The cryo-electron microscopy structure of huntingtin. Nature. 2018;555:117–120. doi: 10.1038/nature25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 224.Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 225.Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 226.Gao Y.G., Yang H., Zhao J., Jiang Y.J., Hu H.Y. Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure. 2014;22:378–386. doi: 10.1016/j.str.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 227.Bhattacharyya A., Thakur A.K., Chellgren V.M., Thiagarajan G., Williams A.D., Chellgren B.W., et al. Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 228.Pigazzini M.L., Lawrenz M., Margineanu A., Kaminski Schierle G.S., Kirstein J. An expanded polyproline domain maintains mutant huntingtin soluble in vivo and during aging. Front. Mol. Neurosci. 2021;14:721749. doi: 10.3389/fnmol.2021.721749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kim Y.J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K.B., et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 231.Tanaka Y., Igarashi S., Nakamura M., Gafni J., Torcassi C., Schilling G., et al. Progressive phenotype and nuclear accumulation of an amino-terminal cleavage fragment in a transgenic mouse model with inducible expression of full-length mutant huntingtin. Neurobiol. Dis. 2006;21:381–391. doi: 10.1016/j.nbd.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 232.Wang C.E., Tydlacka S., Orr A.L., Yang S.H., Graham R.K., Hayden M.R., et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum. Mol. Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 235.Altschuler E.L., Hud N.V., Mazrimas J.A., Rupp B. Structure of polyglutamine. FEBS Lett. 2000;472:166–168. doi: 10.1016/s0014-5793(00)01381-8. [DOI] [PubMed] [Google Scholar]
- 236.Hoop C.L., Lin H.K., Kar K., Magyarfalvi G., Lamley J.M., Boatz J.C., et al. Huntingtin exon 1 fibrils feature an interdigitated beta-hairpin-based polyglutamine core. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1546–1551. doi: 10.1073/pnas.1521933113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Culver B.P., Savas J.N., Park S.K., Choi J.H., Zheng S., Zeitlin S.O., et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J. Biol. Chem. 2012;287:21599–21614. doi: 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shirasaki D.I., Greiner E.R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D.H., et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Suter B., Fontaine J.F., Yildirimman R., Rasko T., Schaefer M.H., Rasche A., et al. Development and application of a DNA microarray-based yeast two-hybrid system. Nucl. Acids Res. 2013;41:1496–1507. doi: 10.1093/nar/gks1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wanker E.E., Rovira C., Scherzinger E., Hasenbank R., Walter S., Tait D., et al. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- 241.McGuire J.R., Rong J., Li S.H., Li X.J. Interaction of huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J. Biol. Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- 242.Li S.H., Gutekunst C.A., Hersch S.M., Li X.J. Interaction of huntingtin-associated protein with dynactin P150Glued. J. Neurosci. 1998;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Engelender S., Sharp A.H., Colomer V., Tokito M.K., Lanahan A., Worley P., et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 244.Caviston J.P., Holzbaur E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Humbert S., Bryson E.A., Cordelieres F.P., Connors N.C., Datta S.R., Finkbeiner S., et al. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 246.Colin E., Zala D., Liot G., Rangone H., Borrell-Pages M., Li X.J., et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Pal A., Severin F., Lommer B., Shevchenko A., Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J. Cell Biol. 2006;172:605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sahlender D.A., Roberts R.C., Arden S.D., Spudich G., Taylor M.J., Luzio J.P., et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hattula K., Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 250.Butland S.L., Sanders S.S., Schmidt M.E., Riechers S.P., Lin D.T., Martin D.D., et al. The palmitoyl acyltransferase HIP14 shares a high proportion of interactors with huntingtin: implications for a role in the pathogenesis of huntington's disease. Hum. Mol. Genet. 2014;23:4142–4160. doi: 10.1093/hmg/ddu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Singaraja R.R., Hadano S., Metzler M., Givan S., Wellington C.L., Warby S., et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 252.Legendre-Guillemin V., Metzler M., Charbonneau M., Gan L., Chopra V., Philie J., et al. HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin. Identification of a novel interaction with clathrin light chain. J. Biol. Chem. 2002;277:19897–19904. doi: 10.1074/jbc.M112310200. [DOI] [PubMed] [Google Scholar]
- 253.Trushina E., Singh R.D., Dyer R.B., Cao S., Shah V.H., Parton R.G., et al. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum. Mol. Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- 254.Yu A., Shibata Y., Shah B., Calamini B., Lo D.C., Morimoto R.I. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1481–1490. doi: 10.1073/pnas.1321811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Li S.H., Cheng A.L., Zhou H., Lam S., Rao M., Li H., et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 257.Nucifora F.C., Jr., Sasaki M., Peters M.F., Huang H., Cooper J.K., Yamada M., et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 258.Tourette C., Li B., Bell R., O'Hare S., Kaltenbach L.S., Mooney S.D., et al. A large scale Huntingtin protein interaction network implicates Rho GTPase signaling pathways in Huntington disease. J. Biol. Chem. 2014;289:6709–6726. doi: 10.1074/jbc.M113.523696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rui Y.N., Xu Z., Patel B., Chen Z., Chen D., Tito A., et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mitsui K., Nakayama H., Akagi T., Nekooki M., Ohtawa K., Takio K., et al. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J. Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Thompson L.M., Aiken C.T., Kaltenbach L.S., Agrawal N., Illes K., Khoshnan A., et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lotz G.P., Legleiter J., Aron R., Mitchell E.J., Huang S.Y., Ng C., et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J. Biol. Chem. 2010;285:38183–38193. doi: 10.1074/jbc.M110.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Scior A., Buntru A., Arnsburg K., Ast A., Iburg M., Juenemann K., et al. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 2018;37:282–299. doi: 10.15252/embj.201797212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hageman J., Rujano M.A., van Waarde M.A., Kakkar V., Dirks R.P., Govorukhina N., et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 265.Kakkar V., Mansson C., de Mattos E.P., Bergink S., van der Zwaag M., van Waarde M., et al. The S/T-Rich motif in the DNAJB6 chaperone delays polyglutamine aggregation and the onset of disease in a mouse model. Mol. Cell. 2016;62:272–283. doi: 10.1016/j.molcel.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 266.Sarparanta J., Jonson P.H., Golzio C., Sandell S., Luque H., Screen M., et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 2012;44:S451–452. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Thiruvalluvan A., de Mattos E.P., Brunsting J.F., Bakels R., Serlidaki D., Barazzuol L., et al. DNAJB6, a key factor in neuronal sensitivity to amyloidogenesis. Mol. Cell. 2020;78:346–358 e349. doi: 10.1016/j.molcel.2020.02.022. [DOI] [PubMed] [Google Scholar]
- 268.O'Brien R., DeGiacomo F., Holcomb J., Bonner A., Ring K.L., Zhang N., et al. Integration-independent transgenic Huntington disease fragment mouse models reveal distinct phenotypes and life span in vivo. J. Biol. Chem. 2015;290:19287–19306. doi: 10.1074/jbc.M114.623561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.He W.T., Xue W., Gao Y.G., Hong J.Y., Yue H.W., Jiang L.L., et al. HSP90 recognizes the N-terminus of huntingtin involved in regulation of huntingtin aggregation by USP19. Sci. Rep. 2017;7:14797. doi: 10.1038/s41598-017-13711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Tam S., Spiess C., Auyeung W., Joachimiak L., Chen B., Poirier M.A., et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kitamura A., Kubota H., Pack C.G., Matsumoto G., Hirayama S., Takahashi Y., et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 272.Behrends C., Langer C.A., Boteva R., Bottcher U.M., Stemp M.J., Schaffar G., et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 273.Vonk W.I.M., Rainbolt T.K., Dolan P.T., Webb A.E., Brunet A., Frydman J. Differentiation drives widespread rewiring of the neural stem cell chaperone network. Mol. Cell. 2020;78:329–345.e329. doi: 10.1016/j.molcel.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mishra A., Dikshit P., Purkayastha S., Sharma J., Nukina N., Jana N.R. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J. Biol. Chem. 2008;283:7648–7656. doi: 10.1074/jbc.M706620200. [DOI] [PubMed] [Google Scholar]
- 275.Bhat K.P., Yan S., Wang C.E., Li S., Li X.J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Jeon I., Lee N., Li J.Y., Park I.H., Park K.S., Moon J., et al. Neuronal properties, in vivo effects, and pathology of a Huntington's disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30:2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 277.Consortium H. D. i. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Koyuncu S., Saez I., Lee H.J., Gutierrez-Garcia R., Pokrzywa W., Fatima A., et al. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington's disease patients. Nat. Commun. 2018;9:2886. doi: 10.1038/s41467-018-05320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang H., Monteiro M.J. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem. Biophys. Res. Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zeng L., Wang B., Merillat S.A., Minakawa E.N., Perkins M.D., Ramani B., et al. Differential recruitment of UBQLN2 to nuclear inclusions in the polyglutamine diseases HD and SCA3. Neurobiol. Dis. 2015;82:281–288. doi: 10.1016/j.nbd.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Luo H., Cao L., Liang X., Du A., Peng T., Li H. Herp promotes degradation of mutant huntingtin: involvement of the proteasome and molecular chaperones. Mol. Neurobiol. 2018;55:7652–7668. doi: 10.1007/s12035-018-0900-8. [DOI] [PubMed] [Google Scholar]
- 282.Yang H., Liu C., Zhong Y., Luo S., Monteiro M.J., Fang S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ghosh D.K., Roy A., Ranjan A. The ATPase VCP/p97 functions as a disaggregase against toxic Huntingtin-exon1 aggregates. FEBS Lett. 2018;592:2680–2692. doi: 10.1002/1873-3468.13213. [DOI] [PubMed] [Google Scholar]
- 284.van Well E.M., Bader V., Patra M., Sanchez-Vicente A., Meschede J., Furthmann N., et al. A protein quality control pathway regulated by linear ubiquitination. EMBO J. 2019;38 doi: 10.15252/embj.2018100730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Imai Y., Soda M., Murakami T., Shoji M., Abe K., Takahashi R. A product of the human gene adjacent to parkin is a component of Lewy bodies and suppresses Pael receptor-induced cell death. J. Biol. Chem. 2003;278:51901–51910. doi: 10.1074/jbc.M309655200. [DOI] [PubMed] [Google Scholar]
- 286.Hirabayashi M., Inoue K., Tanaka K., Nakadate K., Ohsawa Y., Kamei Y., et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- 287.Jang J.K., Park K.J., Lee J.H., Ko K.Y., Kang S., Kim I.Y. Selenoprotein S is required for clearance of C99 through endoplasmic reticulum-associated degradation. Biochem. Biophys. Res. Commun. 2017;486:444–450. doi: 10.1016/j.bbrc.2017.03.060. [DOI] [PubMed] [Google Scholar]
- 288.Malty R.H., Aoki H., Kumar A., Phanse S., Amin S., Zhang Q., et al. A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-kappaB signaling. Cell Syst. 2017;5:564–577.e512. doi: 10.1016/j.cels.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Moutaoufik M.T., Malty R., Amin S., Zhang Q., Phanse S., Gagarinova A., et al. Rewiring of the human mitochondrial interactome during neuronal reprogramming reveals regulators of the respirasome and Neurogenesis. iScience. 2019;19:1114–1132. doi: 10.1016/j.isci.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 291.Hegde R.N., Chiki A., Petricca L., Martufi P., Arbez N., Mouchiroud L., et al. TBK1 phosphorylates mutant Huntingtin and suppresses its aggregation and toxicity in Huntington's disease models. EMBO J. 2020;39 doi: 10.15252/embj.2020104671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Qi L., Zhang X.D., Wu J.C., Lin F., Wang J., DiFiglia M., et al. The role of chaperone-mediated autophagy in huntingtin degradation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Khoshnan A., Ko J., Watkin E.E., Paige L.A., Reinhart P.H., Patterson P.H. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J. Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Laforge M., Rodrigues V., Silvestre R., Gautier C., Weil R., Corti O., et al. NF-kappaB pathway controls mitochondrial dynamics. Cell Death Differ. 2016;23:89–98. doi: 10.1038/cdd.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Choo Y.S., Johnson G.V., MacDonald M., Detloff P.J., Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 296.Giacomello M., Oliveros J.C., Naranjo J.R., Carafoli E. Neuronal Ca(2+) dyshomeostasis in Huntington disease. Prion. 2013;7:76–84. doi: 10.4161/pri.23581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., et al. Mutant huntingtin's interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Bossy-Wetzel E., Petrilli A., Knott A.B. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Trushina E., Dyer R.B., Badger J.D., 2nd, Ure D., Eide L., Tran D.D., et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Guo X., Sun X., Hu D., Wang Y.J., Fujioka H., Vyas R., et al. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington's disease. Nat. Commun. 2016;7:12646. doi: 10.1038/ncomms12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Guo X., Qi X. VCP cooperates with UBXD1 to degrade mitochondrial outer membrane protein MCL1 in model of Huntington's disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:552–559. doi: 10.1016/j.bbadis.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Yablonska S., Ganesan V., Ferrando L.M., Kim J., Pyzel A., Baranova O.V., et al. Mutant huntingtin disrupts mitochondrial proteostasis by interacting with TIM23. Proc. Natl. Acad. Sci. U. S. A. 2019;116:16593–16602. doi: 10.1073/pnas.1904101116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ott C., Dorsch E., Fraunholz M., Straub S., Kozjak-Pavlovic V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hering T., Kojer K., Birth N., Hallitsch J., Taanman J.W., Orth M. Mitochondrial cristae remodelling is associated with disrupted OPA1 oligomerisation in the Huntington's disease R6/2 fragment model. Exp. Neurol. 2017;288:167–175. doi: 10.1016/j.expneurol.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 305.Kingwell K. Double setback for ASO trials in Huntington disease. Nat. Rev. Drug Discov. 2021;20:412–413. doi: 10.1038/d41573-021-00088-6. [DOI] [PubMed] [Google Scholar]
- 306.Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W., et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers. 2017;3:17071. doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 307.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 308.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 310.Deng H.X., Chen W., Hong S.T., Boycott K.M., Gorrie G.H., Siddique N., et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Fecto F., Yan J., Vemula S.P., Liu E., Yang Y., Chen W., et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 314.Johnson J.O., Pioro E.P., Boehringer A., Chia R., Feit H., Renton A.E., et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Williams K.L., Topp S., Yang S., Smith B., Fifita J.A., Warraich S.T., et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat. Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Fischer L.R., Igoudjil A., Magrane J., Li Y., Hansen J.M., Manfredi G., et al. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain. 2011;134:196–209. doi: 10.1093/brain/awq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Casareno R.L., Waggoner D., Gitlin J.D. The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J. Biol. Chem. 1998;273:23625–23628. doi: 10.1074/jbc.273.37.23625. [DOI] [PubMed] [Google Scholar]
- 318.Schmidt P.J., Kunst C., Culotta V.C. Copper activation of superoxide dismutase 1 (SOD1) in vivo. Role for protein-protein interactions with the copper chaperone for SOD1. J. Biol. Chem. 2000;275:33771–33776. doi: 10.1074/jbc.M006254200. [DOI] [PubMed] [Google Scholar]
- 319.Sala F.A., Wright G.S.A., Antonyuk S.V., Garratt R.C., Hasnain S.S. Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Subramaniam J.R., Lyons W.E., Liu J., Bartnikas T.B., Rothstein J., Price D.L., et al. Mutant SOD1 causes motor neuron disease independent of copper chaperone-mediated copper loading. Nat. Neurosci. 2002;5:301–307. doi: 10.1038/nn823. [DOI] [PubMed] [Google Scholar]
- 321.Bruijn L.I., Houseweart M.K., Kato S., Anderson K.L., Anderson S.D., Ohama E., et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 322.Benkler C., O'Neil A.L., Slepian S., Qian F., Weinreb P.H., Rubin L.L. Aggregated SOD1 causes selective death of cultured human motor neurons. Sci. Rep. 2018;8:16393. doi: 10.1038/s41598-018-34759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Tiwari A., Xu Z., Hayward L.J. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2005;280:29771–29779. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- 324.Furukawa Y., O'Halloran T.V. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 325.Kayatekin C., Zitzewitz J.A., Matthews C.R. Disulfide-reduced ALS variants of Cu, Zn superoxide dismutase exhibit increased populations of unfolded species. J. Mol. Biol. 2010;398:320–331. doi: 10.1016/j.jmb.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Watanabe Y., Morita E., Fukada Y., Doi K., Yasui K., Kitayama M., et al. Adherent monomer-misfolded SOD1. PLoS One. 2008;3:e3497. doi: 10.1371/journal.pone.0003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ruegsegger C., Maharjan N., Goswami A., Filezac de L'Etang A., Weis J., Troost D., et al. Aberrant association of misfolded SOD1 with Na(+)/K(+)ATPase-alpha3 impairs its activity and contributes to motor neuron vulnerability in ALS. Acta Neuropathol. 2016;131:427–451. doi: 10.1007/s00401-015-1510-4. [DOI] [PubMed] [Google Scholar]
- 328.Wang J., Farr G.W., Zeiss C.J., Rodriguez-Gil D.J., Wilson J.H., Furtak K., et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Urushitani M., Kurisu J., Tateno M., Hatakeyama S., Nakayama K., Kato S., et al. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J. Neurochem. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 330.Choi J.S., Cho S., Park S.G., Park B.C., Lee D.H. Co-chaperone CHIP associates with mutant Cu/Zn-superoxide dismutase proteins linked to familial amyotrophic lateral sclerosis and promotes their degradation by proteasomes. Biochem. Biophys. Res. Commun. 2004;321:574–583. doi: 10.1016/j.bbrc.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 331.Une M., Yamakawa M., Watanabe Y., Uchino K., Honda N., Adachi M., et al. SOD1-interacting proteins: roles of aggregation cores and protein degradation systems. Neurosci. Res. 2021;170:295–305. doi: 10.1016/j.neures.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 332.Schorr S., Klein M.C., Gamayun I., Melnyk A., Jung M., Schauble N., et al. Co-chaperone specificity in gating of the polypeptide conducting channel in the membrane of the human endoplasmic reticulum. J. Biol. Chem. 2015;290:18621–18635. doi: 10.1074/jbc.M115.636639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Matus S., Valenzuela V., Medinas D.B., Hetz C. ER dysfunction and protein folding stress in ALS. Int. J. Cell Biol. 2013;2013:674751. doi: 10.1155/2013/674751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ryu S.W., Stewart R., Pectol D.C., Ender N.A., Wimalarathne O., Lee J.H., et al. Proteome-wide identification of HSP70/HSC70 chaperone clients in human cells. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Karras G.I., Yi S., Sahni N., Fischer M., Xie J., Vidal M., et al. HSP90 shapes the consequences of human genetic variation. Cell. 2017;168:856–866 e812. doi: 10.1016/j.cell.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kabuta T., Suzuki Y., Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J. Biol. Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 337.Gal J., Strom A.L., Kwinter D.M., Kilty R., Zhang J., Shi P., et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J. Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Crippa V., Sau D., Rusmini P., Boncoraglio A., Onesto E., Bolzoni E., et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum. Mol. Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 339.Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Castillo K., Nassif M., Valenzuela V., Rojas F., Matus S., Mercado G., et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 341.Imamura K., Izumi Y., Watanabe A., Tsukita K., Woltjen K., Yamamoto T., et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf3962. [DOI] [PubMed] [Google Scholar]
- 342.Keskin I., Forsgren E., Lange D.J., Weber M., Birve A., Synofzik M., et al. Effects of cellular pathway disturbances on misfolded superoxide dismutase-1 in fibroblasts derived from ALS patients. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Niwa J., Ishigaki S., Hishikawa N., Yamamoto M., Doyu M., Murata S., et al. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J. Biol. Chem. 2002;277:36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- 344.Niwa J., Yamada S., Ishigaki S., Sone J., Takahashi M., Katsuno M., et al. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J. Biol. Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- 345.Karch C.M., Borchelt D.R. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Miyazaki K., Fujita T., Ozaki T., Kato C., Kurose Y., Sakamoto M., et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004;279:11327–11335. doi: 10.1074/jbc.M312389200. [DOI] [PubMed] [Google Scholar]
- 347.Matentzoglu K., Scheffner M. Ubiquitin ligase E6-AP and its role in human disease. Biochem. Soc. Trans. 2008;36:797–801. doi: 10.1042/BST0360797. [DOI] [PubMed] [Google Scholar]
- 348.Ying Z., Wang H., Fan H., Zhu X., Zhou J., Fei E., et al. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum. Mol. Genet. 2009;18:4268–4281. doi: 10.1093/hmg/ddp380. [DOI] [PubMed] [Google Scholar]
- 349.Choi J.S., Kim K., Lee D.H., Cho S., Ha J.D., Park B.C., et al. cIAPs promote the proteasomal degradation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2016;480:422–428. doi: 10.1016/j.bbrc.2016.10.065. [DOI] [PubMed] [Google Scholar]
- 350.Bruijn L.I., Becher M.W., Lee M.K., Anderson K.L., Jenkins N.A., Copeland N.G., et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 351.Furukawa Y., Kaneko K., Yamanaka K., Nukina N. Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2010;285:22221–22231. doi: 10.1074/jbc.M110.113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Ida M., Ando M., Adachi M., Tanaka A., Machida K., Hongo K., et al. Structural basis of Cu, Zn-superoxide dismutase amyloid fibril formation involves interaction of multiple peptide core regions. J. Biochem. 2016;159:247–260. doi: 10.1093/jb/mvv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Munch C., O'Brien J., Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Thomas E.V., Fenton W.A., McGrath J., Horwich A.L. Transfer of pathogenic and nonpathogenic cytosolic proteins between spinal cord motor neurons in vivo in chimeric mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E3139–E3148. doi: 10.1073/pnas.1701465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ayers J.I., Fromholt S., Koch M., DeBosier A., McMahon B., Xu G., et al. Experimental transmissibility of mutant SOD1 motor neuron disease. Acta Neuropathol. 2014;128:791–803. doi: 10.1007/s00401-014-1342-7. [DOI] [PubMed] [Google Scholar]
- 356.Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J.P. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat. Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 357.Zhao W., Beers D.R., Henkel J.S., Zhang W., Urushitani M., Julien J.P., et al. Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia. 2010;58:231–243. doi: 10.1002/glia.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Proctor E.A., Fee L., Tao Y., Redler R.L., Fay J.M., Zhang Y., et al. Nonnative SOD1 trimer is toxic to motor neurons in a model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:614–619. doi: 10.1073/pnas.1516725113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zhu C., Beck M.V., Griffith J.D., Deshmukh M., Dokholyan N.V. Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4661–4665. doi: 10.1073/pnas.1800187115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Gill C., Phelan J.P., Hatzipetros T., Kidd J.D., Tassinari V.R., Levine B., et al. SOD1-positive aggregate accumulation in the CNS predicts slower disease progression and increased longevity in a mutant SOD1 mouse model of ALS. Sci. Rep. 2019;9:6724. doi: 10.1038/s41598-019-43164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Son M., Puttaparthi K., Kawamata H., Rajendran B., Boyer P.J., Manfredi G., et al. Overexpression of CCS in G93A-SOD1 mice leads to accelerated neurological deficits with severe mitochondrial pathology. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6072–6077. doi: 10.1073/pnas.0610923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Li Q., Vande Velde C., Israelson A., Xie J., Bailey A.O., Dong M.Q., et al. ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21146–21151. doi: 10.1073/pnas.1014862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Pasinelli P., Belford M.E., Lennon N., Bacskai B.J., Hyman B.T., Trotti D., et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 364.Massignan T., Casoni F., Basso M., Stefanazzi P., Biasini E., Tortarolo M., et al. Proteomic analysis of spinal cord of presymptomatic amyotrophic lateral sclerosis G93A SOD1 mouse. Biochem. Biophys. Res. Commun. 2007;353:719–725. doi: 10.1016/j.bbrc.2006.12.075. [DOI] [PubMed] [Google Scholar]
- 365.Lukas T.J., Luo W.W., Mao H., Cole N., Siddique T. Informatics-assisted protein profiling in a transgenic mouse model of amyotrophic lateral sclerosis. Mol. Cell Proteomics. 2006;5:1233–1244. doi: 10.1074/mcp.M500431-MCP200. [DOI] [PubMed] [Google Scholar]
- 366.Casoni F., Basso M., Massignan T., Gianazza E., Cheroni C., Salmona M., et al. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J. Biol. Chem. 2005;280:16295–16304. doi: 10.1074/jbc.M413111200. [DOI] [PubMed] [Google Scholar]
- 367.Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z., et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G., et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Tziortzouda P., Van Den Bosch L., Hirth F. Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat. Rev. Neurosci. 2021;22:197–208. doi: 10.1038/s41583-021-00431-1. [DOI] [PubMed] [Google Scholar]
- 370.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wang W., Wang L., Lu J., Siedlak S.L., Fujioka H., Liang J., et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat. Med. 2016;22:869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Milev M.P., Ravichandran M., Khan M.F., Schriemer D.C., Mouland A.J. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Front. Microbiol. 2012;3:367. doi: 10.3389/fmicb.2012.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Pourhaghighi R., Ash P.E.A., Phanse S., Goebels F., Hu L.Z.M., Chen S., et al. BraInMap elucidates the macromolecular connectivity landscape of mammalian brain. Cell Syst. 2020;10:333–350.e314. doi: 10.1016/j.cels.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Wang G., Yang H., Yan S., Wang C.E., Liu X., Zhao B., et al. Cytoplasmic mislocalization of RNA splicing factors and aberrant neuronal gene splicing in TDP-43 transgenic pig brain. Mol. Neurodegener. 2015;10:42. doi: 10.1186/s13024-015-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Kim S.H., Shanware N.P., Bowler M.J., Tibbetts R.S. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Yu H., Lu S., Gasior K., Singh D., Vazquez-Sanchez S., Tapia O., et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science. 2021;371 doi: 10.1126/science.abb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Gu J., Wang C., Hu R., Li Y., Zhang S., Sun Y., et al. Hsp70 chaperones TDP-43 in dynamic, liquid-like phase and prevents it from amyloid aggregation. Cell Res. 2021;31:1024–1027. doi: 10.1038/s41422-021-00526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wang P., Wander C.M., Yuan C.X., Bereman M.S., Cohen T.J. Acetylation-induced TDP-43 pathology is suppressed by an HSF1-dependent chaperone program. Nat. Commun. 2017;8:82. doi: 10.1038/s41467-017-00088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Lin P.Y., Folorunso O., Taglialatela G., Pierce A. Overexpression of heat shock factor 1 maintains TAR DNA binding protein 43 solubility via induction of inducible heat shock protein 70 in cultured cells. J. Neurosci. Res. 2016;94:671–682. doi: 10.1002/jnr.23725. [DOI] [PubMed] [Google Scholar]
- 381.Chen H.J., Mitchell J.C., Novoselov S., Miller J., Nishimura A.L., Scotter E.L., et al. The heat shock response plays an important role in TDP-43 clearance: evidence for dysfunction in amyotrophic lateral sclerosis. Brain. 2016;139:1417–1432. doi: 10.1093/brain/aww028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Crippa V., Cicardi M.E., Ramesh N., Seguin S.J., Ganassi M., Bigi I., et al. The chaperone HSPB8 reduces the accumulation of truncated TDP-43 species in cells and protects against TDP-43-mediated toxicity. Hum. Mol. Genet. 2016;25:3908–3924. doi: 10.1093/hmg/ddw232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 384.Watabe K., Kato Y., Sakuma M., Murata M., Niida-Kawaguchi M., Takemura T., et al. Praja1 RING-finger E3 ubiquitin ligase suppresses neuronal cytoplasmic TDP-43 aggregate formation. Neuropathology. 2020;40:570–586. doi: 10.1111/neup.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Lee Y.C., Huang W.C., Lin J.H., Kao T.J., Lin H.C., Lee K.H., et al. Znf179 E3 ligase-mediated TDP-43 polyubiquitination is involved in TDP-43- ubiquitinated inclusions (UBI) (+)-related neurodegenerative pathology. J. Biomed. Sci. 2018;25:76. doi: 10.1186/s12929-018-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Chen H., Li Y., Li Y., Chen Z., Xie L., Li W., et al. PARK2 promotes mitochondrial pathway of apoptosis and antimicrotubule drugs chemosensitivity via degradation of phospho-BCL-2. Theranostics. 2020;10:9984–10000. doi: 10.7150/thno.47044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zhu X., Ma X., Tu Y., Huang M., Liu H., Wang F., et al. Parkin regulates translesion DNA synthesis in response to UV radiation. Oncotarget. 2017;8:36423–36437. doi: 10.18632/oncotarget.16855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Hebron M.L., Lonskaya I., Sharpe K., Weerasinghe P.P., Algarzae N.K., Shekoyan A.R., et al. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6) J. Biol. Chem. 2013;288:4103–4115. doi: 10.1074/jbc.M112.419945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Rayner S.L., Yang S., Farrawell N.E., Jagaraj C.J., Cheng F., Davidson J.M., et al. TDP-43 is a ubiquitylation substrate of the SCF(cyclin F) complex. Neurobiol. Dis. 2022;167:105673. doi: 10.1016/j.nbd.2022.105673. [DOI] [PubMed] [Google Scholar]
- 390.Cassel J.A., Reitz A.B. Ubiquilin-2 (UBQLN2) binds with high affinity to the C-terminal region of TDP-43 and modulates TDP-43 levels in H4 cells: characterization of inhibition by nucleic acids and 4-aminoquinolines. Biochim. Biophys. Acta. 2013;1834:964–971. doi: 10.1016/j.bbapap.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Hjerpe R., Bett J.S., Keuss M.J., Solovyova A., McWilliams T.G., Johnson C., et al. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell. 2016;166:935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Dao T.P., Kolaitis R.M., Kim H.J., O'Donovan K., Martyniak B., Colicino E., et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell. 2018;69:965–978.e966. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 395.Weber S.C., Brangwynne C.P. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 397.Shin Y., Berry J., Pannucci N., Haataja M.P., Toettcher J.E., Brangwynne C.P. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017;168:159–171.e114. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ambadipudi S., Biernat J., Riedel D., Mandelkow E., Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 2017;8:275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ash P.E.A., Lei S., Shattuck J., Boudeau S., Carlomagno Y., Medalla M., et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2014188118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37 doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Boyko S., Surewicz K., Surewicz W.K. Regulatory mechanisms of tau protein fibrillation under the conditions of liquid-liquid phase separation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:31882–31890. doi: 10.1073/pnas.2012460117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Sawner A.S., Ray S., Yadav P., Mukherjee S., Panigrahi R., Poudyal M., et al. Modulating alpha-synuclein liquid-liquid phase separation. Biochemistry. 2021;60:3676–3696. doi: 10.1021/acs.biochem.1c00434. [DOI] [PubMed] [Google Scholar]
- 403.Siegert A., Rankovic M., Favretto F., Ukmar-Godec T., Strohaker T., Becker S., et al. Interplay between tau and alpha-synuclein liquid-liquid phase separation. Protein Sci. 2021;30:1326–1336. doi: 10.1002/pro.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Boeynaems S., Holehouse A.S., Weinhardt V., Kovacs D., Van Lindt J., Larabell C., et al. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7889–7898. doi: 10.1073/pnas.1821038116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Murakami T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 407.Lin Y., Protter D.S., Rosen M.K., Parker R. formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Conicella A.E., Zerze G.H., Mittal J., Fawzi N.L. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Mackenzie I.R., Nicholson A.M., Sarkar M., Messing J., Purice M.D., Pottier C., et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816.e809. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Peskett T.R., Rau F., O'Driscoll J., Patani R., Lowe A.R., Saibil H.R. A liquid to solid phase transition underlying pathological huntingtin Exon1 aggregation. Mol. Cell. 2018;70:588–601.e586. doi: 10.1016/j.molcel.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Aktar F., Burudpakdee C., Polanco M., Pei S., Swayne T.C., Lipke P.N., et al. The huntingtin inclusion is a dynamic phase-separated compartment. Life Sci. Alli. 2019;2 doi: 10.26508/lsa.201900489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Posey A.E., Ruff K.M., Harmon T.S., Crick S.L., Li A., Diamond M.I., et al. Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 2018;293:3734–3746. doi: 10.1074/jbc.RA117.000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Gingras A.C., Gstaiger M., Raught B., Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 415.Smits A.H., Vermeulen M. Characterizing protein-protein interactions using mass spectrometry: challenges and opportunities. Trends Biotechnol. 2016;34:825–834. doi: 10.1016/j.tibtech.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 416.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 417.Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Met. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Roux K.J., Kim D.I., Burke B., May D.G. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2018;91 doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Kim D.I., Jensen S.C., Noble K.A., Kc B., Roux K.H., Motamedchaboki K., et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Samavarchi-Tehrani P., Samson R., Gingras A.C. Proximity dependent biotinylation: key enzymes and adaptation to proteomics approaches. Mol. Cell Proteomics. 2020;19:757–773. doi: 10.1074/mcp.R120.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Met. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.