Abstract
Background
Automated brain tumor identification facilitates diagnosis and treatment planning. We evaluate the performance of traditional machine learning (TML) and deep learning (DL) in brain tumor detection and segmentation, using MRI.
Methods
A systematic literature search from January 2000 to May 8, 2021 was conducted. Study quality was assessed using the Checklist for Artificial Intelligence in Medical Imaging (CLAIM). Detection meta-analysis was performed using a unified hierarchical model. Segmentation studies were evaluated using a random effects model. Sensitivity analysis was performed for externally validated studies.
Results
Of 224 studies included in the systematic review, 46 segmentation and 38 detection studies were eligible for meta-analysis. In detection, DL achieved a lower false positive rate compared to TML; 0.018 (95% CI, 0.011 to 0.028) and 0.048 (0.032 to 0.072) (P < .001), respectively. In segmentation, DL had a higher dice similarity coefficient (DSC), particularly for tumor core (TC); 0.80 (0.77 to 0.83) and 0.63 (0.56 to 0.71) (P < .001), persisting on sensitivity analysis. Both manual and automated whole tumor (WT) segmentation had “good” (DSC ≥ 0.70) performance. Manual TC segmentation was superior to automated; 0.78 (0.69 to 0.86) and 0.64 (0.53 to 0.74) (P = .014), respectively. Only 30% of studies reported external validation.
Conclusions
The comparable performance of automated to manual WT segmentation supports its integration into clinical practice. However, manual outperformance for sub-compartmental segmentation highlights the need for further development of automated methods in this area. Compared to TML, DL provided superior performance for detection and sub-compartmental segmentation. Improvements in the quality and design of studies, including external validation, are required for the interpretability and generalizability of automated models.
Keywords: artificial intelligence, brain tumor, machine learning, meta-analysis, segmentation
Key Points.
Human expertise outperformed automated methods in sub-compartmental segmentation.
DL performed superiorly to TML for detection and sub-compartmental segmentation.
Transparency and generalizability of models should be improved.
Importance of the Study.
Despite the increasing research on artificial intelligence techniques in medical imaging, their safe implementation into clinical practice depends on rigorous and generalizable evidence. This study systematically evaluated the performance of automated brain tumor detection and segmentation methods, and assessed the quality of reporting using the Checklist for Artificial Intelligence in Medical Imaging guideline. Although automated and manual methods in whole tumor segmentation performed comparably, manual methods performed better in sub-compartmental segmentation. Within automated methods, deep learning was found to be superior to traditional machine learning in detection and sub-compartmental segmentation, but explaining this was hindered by the paucity in reported methods of model interpretability. Less than a third of studies reported external validation of their automated method. The variability found in study reporting undermines the credibility of automated methods, impacting their benefit for patients and health systems. Hence, there is a need for adherence to international reporting standards and guidelines.
Brain tumors present a significant burden on healthcare worldwide due to the neurological deficits produced and subsequent poor prognosis, with an average 5-year survival of 35% in malignant subtypes.1 MRI is the gold standard modality engendering brain tumor diagnosis and subsequently informing surgical intervention, radiotherapy planning, and chemotherapy. Inevitably, qualitative MRI assessment has always been subject to high inter-rater variability, as well as being a notoriously laborious process.2 However, the emergence of Artificial Intelligence (AI) has sparked the hope of overcoming these limitations.
The advent of Computer-Aided Diagnosis (CAD) using AI can potentially improve brain tumor patient outcomes. Traditional machine learning (TML) techniques have become widely used for image classification but are restricted by a requirement for specifying “feature vectors” for extraction from the raw data.3 Conversely, deep learning (DL) techniques provide effective and automatic representation of complex image features, which has contributed to their increased popularity,3 but the interpretation of automatically identified features remains a problem.4 In addition, both TML and DL techniques are vulnerable to overfitting and selection bias.5 Therefore, to safely use CAD in clinical settings, large robust studies which evaluate their quality and generalizability are crucial.4 Holistic and standardized evaluation of scientific reporting is facilitated by established guidelines, such as the recently proposed Checklist for Artificial Intelligence in Medical Imaging (CLAIM).6
The research on AI in neuro-oncology imaging has been amplified by the introduction of open access image datasets, such as the annual Multimodal Brain Tumor Segmentation Challenge (BRATS).7 This provides the ideal foundation for an in-depth review to identify optimal automated methods. Three former systematic reviews and meta-analyses evaluated performance of AI-related techniques in neuro-oncological imaging.8–10 However, these focused on specific brain tumor types and whole tumor (WT) segmentation, and none have evaluated sub-compartmental segmentation nor addressed performance disparities between CAD and human expert segmentation. Moreover, there remains a paucity in comprehensively assessing the quality of studies in this field. We present the largest systematic review and meta-analysis that objectively evaluates performance of automated detection and segmentation techniques and assesses the reporting quality of included studies.
Materials and Methods
Search Strategy
This systematic review and meta-analysis were conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement11 (PROSPERO; CRD42021247925). We searched PubMed, Web of Science, and Scopus for studies published between January 1, 2000, and May 8, 2021. The search was initially performed on June 19, 2020 and updated on May 8, 2021. The search strategy is found in the Supplementary Appendix. The search was limited to publications written in English. The citations of included articles were hand-searched to identify additional appropriate articles.
Inclusion and Exclusion Criteria
Studies were included if they developed or validated a semi-automatic or fully automatic adult brain tumor detection or segmentation method using MRI. Exclusion criteria: (1) studies reporting tumor classification or tumor grading methods only; (2) studies utilizing MRI spectroscopy only for method development; (3) studies reporting methods on pediatric, pituitary, and/or brainstem tumors only; (4) abstracts or conference proceedings; and (5) no performance metrics reported.
Study Selection and Data Extraction
Extracted citations were imported into the Rayyan systematic review site (https://www.rayyan.ai) for study selection. Following removal of duplicates, titles and abstracts were screened, and full texts of relevant publications reviewed. Study screening was completed by two independent reviewers (O.K., J.D.S.), with disagreements resolved through a consensus-based approach with the wider group.
Two independent reviewers (O.K., A.H.) extracted study characteristics from included studies with disagreements resolved through consensus. Data extracted included: (1) Author; (2) year; (3) dataset(s) utilized with the number of patients/images; (4) type of tumors(s) studied; (5) MRI modality; (6) performance evaluation metrics; (7) type of algorithm utilized; (8) feature extraction; (9) inference time in slice/second for segmentation; (10) user interaction (ie, automatic vs semi-automatic); and (11) validation technique(s).
Reporting and Quality Evaluation
The reporting quality of studies was assessed according to CLAIM.6 The risk of bias and applicability was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) guideline,12 with consideration of some CLAIM items (see Supplementary Appendix). Three reviewers (O.K., A.H., D.B.) independently appraised included studies with any disagreements resolved through consensus. A “good” domain was deemed by its reporting in ≥70% of studies.
Definitions
DL was referred to studies that utilized deep neural networks as their method of choice. TML was referred to methods not classified as DL. Detection studies were defined as those that reported performance results for techniques that identified the presence of a tumor in an image. Segmentation studies were defined as those that reported performance results for techniques that segmented brain tumors, whether it was WT, tumor core (TC), and/or enhancing tumor (ET) segmentations as defined by BRATS.7 Following previous work, dice similarity coefficient (DSC) of ≥0.7 was considered to represent “good” overlap.13
Statistical Analysis
A meta-analysis was conducted for both automated detection and segmentation studies to compare DL with TML methods and to evaluate the segmentation performance of CAD to that of manual experts. Studies providing performance metrics for their method on different datasets were assumed to be independent of each other. This is because we are interested in providing an overview of the two methods rather than exact point estimates.
For detection methods, contingency tables consisting of True Positive, False Positive, False Negative, and True Negative were constructed. For studies that did not directly provide contingency tables, missing data were calculated with Review Manager 5.3 (https://revman.cochrane.org/) using sensitivity, specificity, and number of images. If neither contingency tables nor sufficient data were reported for computation, then the study was excluded from meta-analysis. A unified hierarchical summary receiver operating characteristic model was developed for the detection meta-analysis. Summary estimates of sensitivity and specificity with 95% CIs were derived using the random-effects bivariate binomial model parameters and equivalence equations of Harbord et al.14 The reason for using the hierarchical model is that it considers the correlation between sensitivity and specificity, accounting for within-study variability, as well as variability (also called heterogeneity) in effects between studies (ie, between-study variability). Receiver operating characteristic (ROC) curves were used to plot summary estimates of sensitivity against false positive rate (FPR, ie, 1-specificity). The ROC curve plots also exhibit the uncertainty around the summary estimates via 95% confidence regions, and heterogeneity between accuracy estimates via 95% prediction regions.
Segmentation methods were evaluated using a random effects model, and reported in terms of pooled DSC, a universally used and reported metric. The restricted maximum likelihood estimator was used to calculate the heterogeneity variance (τ 2). The inverse variance method was used to calculate a pooled effect size. Knapp-Hartung adjustments were used to calculate the confidence interval. A prerequisite for study inclusion in the meta-analysis was reporting outcome of interest (ie, DSC), in combination with an SD. Subgroup analysis comparing tumor types was performed where possible. A comparative analysis was conducted to evaluate the performance of CAD versus human experts. Sensitivity analysis was performed looking at studies that only performed out-of-sample external validation. Subgroup or sensitivity analysis was avoided when the number of studies in a group is small (n < 5). Study heterogeneity was formally evaluated using Higgins’ inconsistency index (I2) (I2 > 50% = significant heterogeneity). All analyses were performed in R (version 4.0.2, http://www.r-project.org/) using the tidyverse, metaDTA, dmetar, meta, and ComplexUpset packages.
Results
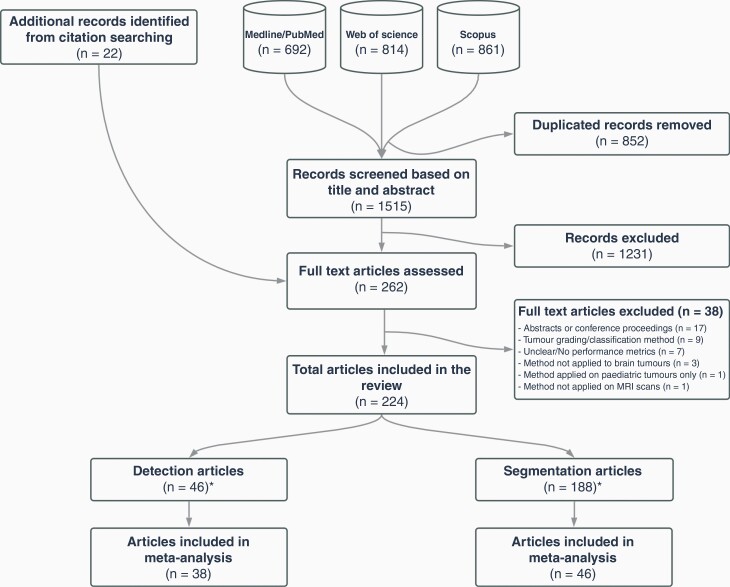
Our search identified 2367 records, of which 1515 records were screened (Figure 1). An additional 22 texts were identified through cross-referencing. Two-hundred and sixty-two full texts were assessed for eligibility and 224 were included in the systematic review: 188 segmentation and 46 detection studies (10 studies reported both detection and segmentation results; see “Eligible Studies” in Supplementary Appendix). Forty-six segmentation15–60 and 38 detection39,42,45,61–95studies were eligible for meta-analysis.
Figure 1.
Study selection flow diagram (*10 studies reported both detection and segmentation results).
Study Characteristics
Study characteristics are shown in Supplementary Table 1 (segmentation) and Supplementary Table 2 (detection). 40.6% (n =95) of studies used DL and 59.4% (n = 139) used TML methods. There was a clear increase in the use of DL from 2018 (Supplementary Figure 1). Most studies utilized a fully automated algorithm (n = 222; 94.9%).
80.7% (n = 189) used data from open-access repositories, with BRATS being the most popular of them (n = 156; 66.7%). 29.0% (n = 68) used local datasets, all of which were retrospectively collected data. 11.9% (n = 28) used both local and public datasets. 2.1% (n = 5) did not specify dataset(s) used (Supplementary Figure 2). Publicly available datasets are detailed in Supplementary Table 3.
The most studied tumors were high-grade gliomas (HGG) (n = 173; 73.9%) and low-grade gliomas (LGG) (n = 171; 73.1%), with 59.0% (n = 138) of studies involving both (Supplementary Figure 3). 9.8% (n = 23) did not report the type of tumor studied. Regarding MRI sequences, T2 (n = 169; 72.2%), fluid-attenuated inversion recovery (FLAIR) (n = 165; 70.5%), T1-contrast enhanced (T1CE) (n = 164; 70.1%), and T1 (n = 143; 61.1%) modalities were the most studied (Supplementary Figure 4). 48.3% (n = 113) of studies combined all these sequences, and 20.1% (n = 47) used just one for the algorithm development. A small minority (n = 19, 8.1%) did not report the type of MRI used.
The most common metrics used for evaluating performance were DSC (n = 168; 71.8%) and sensitivity (n = 168; 71.8%) (Supplementary Figure 5). 55.1% (n = 129) of studies reported internal validation. 31.2% (n = 73) used random split validation and 32.1% (n = 75) used resampling methods (Supplementary Figure 6). Overall, less than a third of studies (n = 70; 30%) performed external validation. Specifically, 49.5% (n = 47/95) of DL and 16.5% (n = 23/139) of TML studies reported external validation (Supplementary Figure 7). Details of algorithm performance and validation techniques of studies are found in Supplementary Table 4 (segmentation) and Supplementary Table 5 (detection). Regarding segmentation inference time, DL methods performed the fastest (median: 0.2 s/MRI slice, interquartile range [IQR]: 0.1–0.9), whereas fully automated TML methods achieved a median of 2.6 s (IQR: 1.1–12.6) and semi-automated techniques achieved 48.16 s (IQR: 6.2–134.9) (P < .001; Kruskal-Wallis test) (Supplementary Figure 5).
Reporting Quality
Detailed CLAIM assessment is presented in Supplementary Table 6 (segmentation) and Supplementary Table 7 (detection). With respect to “good” reported CLAIM items, 95.3% (n = 223) stated the source of the data (CLAIM item 7) and 86.8% (n = 203) clearly reported how ground truths were derived (CLAIM items 14–18). Almost all studies reported detailed model structure and initialization of parameters (CLAIM items 22–24). 83.8% (n = 196) clearly reported training procedures and hyperparameters in sufficient detail (CLAIM item 25) (Supplementary Figure 9).
However, only 1.3% (n = 3) of studies clarified missing data handling. No studies reported sample size calculations (CLAIM item 19). Less than two-thirds (n = 144, 61.5%) specified how data was partitioned (CLAIM item 20). Only 32.5% (n = 76) of studies reported uncertainty around performance metrics (CLAIM item 29). 67.1% (n = 157) studies reported performing internal and/or external validation (CLAIM item 32). Just 2.6% (n = 6) specified inclusion and exclusion flow of participants or images (CLAIM item 33) and only 6% (n = 14) defined demographics and clinical characteristics of cases in each partition (CLAIM item 34). Ten studies made the algorithm source code publicly available (CLAIM item 41; for available links to source codes see Supplementary Table 8).
Risk of bias and applicability assessment
—Detailed QUADAS-2 assessment is presented in Supplementary Table 9 (segmentation) and Supplementary Table 10 (detection). In the patient selection domain of risk of bias, 21.4% (n = 50) studies were considered to have unclear or high risk of bias as they did not express the exclusion criteria in the utilized dataset(s). In the reference standard domain, 13.2% (n = 31) were deemed to have unclear or high risk of bias as they did not clearly define how the ground truth segmentation was derived. In terms of applicability, the main source of concern was in the index test domain; 31.6% (n = 74) had high applicability concerns as they did not validate the algorithm (Supplementary Figure 10).
Meta-analysis
Detection meta-analysis
—Thirty-eight detection studies provided sufficient data to construct contingency tables (69 tables). Only one study performed an external validation. 28.9% (n = 11; 20 tables) of studies utilized DL methods and the remaining 71.1% (n = 27; 49 tables) utilized TML methods (Table 1).
Table 1.
Detection Meta-analysis Results
| Method | Author | Year | TP | FN | FP | TN | Total | Sensitivity | Specificity | Weighted Specificity | Weighted Sensitivity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DL | Çinar and Yildirim80 | 2020 | 147 | 8 | 0 | 98 | 253 | 0.948 | 1 | 5.129 | 4.467 |
| DL | Devanathan and Venkatachalapathy81 | 2020 | 153 | 2 | 3 | 95 | 253 | 0.987 | 0.969 | 4.241 | 4.708 |
| DL | Gurunathan and Krishnan82 | 2020 | 514 | 25 | 21 | 1752 | 2312 | 0.954 | 0.988 | 6.356 | 5.694 |
| DL | Rai et al.65 | 2021 | 1232 | 141 | 135 | 2421 | 3929 | 0.897 | 0.947 | 6.417 | 6.582 |
| DL | Abd-Ellah et al.63 | 2018 | 239 | 1 | 0 | 109 | 349 | 0.996 | 1 | 2.887 | 3.571 |
| DL | Atici et al.79 | 2019 | 1082 | 220 | 110 | 2171 | 3583 | 0.831 | 0.952 | 6.424 | 6.567 |
| DL | Kaur and Ghandi86 | 2020 | 20 | 0 | 0 | 30 | 50 | 1 | 1 | 2.003 | 0.991 |
| DL | Kaur and Ghandi86 | 2020 | 52 | 0 | 0 | 22 | 74 | 1 | 1 | 1.343 | 1.889 |
| DL | Kaur and Ghandi86 | 2020 | 140 | 0 | 0 | 20 | 160 | 1 | 1 | 0.877 | 2.958 |
| DL | Kaur and Ghandi86 | 2020 | 238 | 12 | 18 | 238 | 506 | 0.952 | 0.93 | 5.983 | 6.155 |
| DL | Thangarajan and Chokkalingam87 | 2020 | 159 | 10 | 7 | 93 | 269 | 0.941 | 0.93 | 5.501 | 5.835 |
| DL | Kalaiselvi et al.88 | 2020 | 56 | 7 | 17 | 201 | 281 | 0.889 | 0.922 | 6.145 | 5.163 |
| DL | Rajinikanth et al.89 | 2020 | 388 | 12 | 11 | 589 | 1000 | 0.97 | 0.982 | 6.092 | 5.725 |
| DL | Rajinikanth et al.89 | 2020 | 387 | 13 | 9 | 591 | 1000 | 0.968 | 0.985 | 6.105 | 5.611 |
| DL | Rajinikanth et al.89 | 2020 | 392 | 8 | 8 | 592 | 1000 | 0.98 | 0.987 | 5.948 | 5.45 |
| DL | Rajinikanth et al.89 | 2020 | 395 | 5 | 4 | 596 | 1000 | 0.988 | 0.993 | 5.692 | 4.845 |
| DL | Rajinikanth et al.89 | 2020 | 393 | 7 | 9 | 391 | 800 | 0.983 | 0.978 | 5.727 | 5.771 |
| DL | Rajinikanth et al.89 | 2020 | 395 | 5 | 6 | 194 | 600 | 0.988 | 0.97 | 4.967 | 5.814 |
| DL | Huang et al.90 | 2020 | 4244 | 52 | 34 | 6348 | 10678 | 0.988 | 0.995 | 6.36 | 6.373 |
| DL | Huang et al.90 | 2020 | 397 | 6 | 12 | 480 | 895 | 0.985 | 0.976 | 5.805 | 5.83 |
| TML | Jayachandran and Dhanasekaran67 | 2013 | 10 | 0 | 1 | 4 | 15 | 1 | 0.8 | 0.424 | 1.387 |
| TML | Deepa and Emmanuel72 | 2018 | 68 | 2 | 0 | 11 | 81 | 0.971 | 1 | 1.517 | 1.927 |
| TML | Selvapandian and Manivannan73 | 2018 | 47 | 3 | 3 | 72 | 125 | 0.94 | 0.96 | 3.22 | 1.897 |
| TML | Chen et al.91 | 2021 | 238 | 9 | 3 | 54 | 304 | 0.964 | 0.947 | 2.98 | 2.475 |
| TML | Edalati-rad and Mosleh75 | 2019 | 42 | 0 | 1 | 36 | 79 | 1 | 0.973 | 1.442 | 1.71 |
| TML | Dahshan et al.95 | 2014 | 87 | 0 | 1 | 13 | 101 | 1 | 0.929 | 0.534 | 2.283 |
| TML | Song et al.76 | 2019 | 242 | 7 | 1 | 56 | 306 | 0.972 | 0.982 | 2.853 | 2.346 |
| TML | Johnpeter and Ponnuchamy77 | 2019 | 58 | 2 | 2 | 98 | 160 | 0.967 | 0.98 | 3.142 | 1.729 |
| TML | Amin et al.69 | 2017 | 42 | 4 | 0 | 39 | 85 | 0.913 | 1 | 3.12 | 1.362 |
| TML | Amin et al.69 | 2017 | 60 | 5 | 0 | 35 | 100 | 0.923 | 1 | 3.045 | 1.619 |
| TML | Amin et al.64 | 2019 | 70 | 0 | 2 | 14 | 86 | 1 | 0.875 | 0.64 | 2.333 |
| TML | Amin et al.64 | 2019 | 61 | 1 | 7 | 17 | 86 | 0.984 | 0.708 | 1.725 | 2.463 |
| TML | Amin et al.64 | 2019 | 68 | 6 | 2 | 10 | 86 | 0.919 | 0.833 | 2.433 | 2.375 |
| TML | Amin et al.64 | 2019 | 69 | 0 | 5 | 12 | 86 | 1 | 0.706 | 0.646 | 2.473 |
| TML | Amin et al.64 | 2019 | 69 | 0 | 4 | 15 | 88 | 1 | 0.789 | 0.72 | 2.432 |
| TML | Amin et al.64 | 2019 | 72 | 0 | 4 | 10 | 86 | 1 | 0.714 | 0.543 | 2.469 |
| TML | Amin et al.64 | 2019 | 290 | 0 | 10 | 106 | 406 | 1 | 0.914 | 1.481 | 2.544 |
| TML | Amin et al.64 | 2019 | 296 | 0 | 5 | 105 | 406 | 1 | 0.955 | 1.445 | 2.487 |
| TML | Amin et al.64 | 2019 | 301 | 50 | 5 | 50 | 406 | 0.858 | 0.909 | 3.363 | 2.558 |
| TML | Amin et al.64 | 2019 | 296 | 0 | 10 | 100 | 406 | 1 | 0.909 | 1.422 | 2.55 |
| TML | Amin et al.64 | 2019 | 306 | 11 | 0 | 89 | 406 | 0.965 | 1 | 3.151 | 2.187 |
| TML | Amin et al.64 | 2019 | 295 | 10 | 1 | 100 | 406 | 0.967 | 0.99 | 3.175 | 2.298 |
| TML | Amin et al.64 | 2019 | 70 | 4 | 1 | 11 | 86 | 0.946 | 0.917 | 2.117 | 2.236 |
| TML | Amin et al.64 | 2019 | 70 | 0 | 2 | 14 | 86 | 1 | 0.875 | 0.64 | 2.333 |
| TML | Amin et al.64 | 2019 | 74 | 6 | 0 | 6 | 86 | 0.925 | 1 | 1.798 | 2.045 |
| TML | Amin et al.64 | 2019 | 71 | 0 | 3 | 12 | 86 | 1 | 0.8 | 0.59 | 2.419 |
| TML | Amin et al.64 | 2019 | 74 | 0 | 0 | 12 | 86 | 1 | 1 | 0.518 | 1.969 |
| TML | Amin et al.64 | 2019 | 70 | 3 | 0 | 13 | 86 | 0.959 | 1 | 1.965 | 1.916 |
| TML | Jayachandran and Dhanasekaran66 | 2012 | 4 | 1 | 0 | 5 | 10 | 0.8 | 1 | 1.719 | 0.482 |
| TML | Wang et al.83 | 2020 | 25 | 0 | 1 | 24 | 50 | 1 | 0.96 | 1.271 | 1.525 |
| TML | Kesav and Rajini84 | 2020 | 43 | 1 | 1 | 21 | 66 | 0.977 | 0.955 | 1.91 | 1.891 |
| TML | Alam et al.78 | 2019 | 38 | 1 | 0 | 1 | 40 | 0.974 | 1 | 0.193 | 1.806 |
| TML | Murali and Meena85 | 2020 | 182 | 5 | 0 | 25 | 212 | 0.973 | 1 | 2.234 | 2.225 |
| TML | Arunkumar et al.74 | 2018 | 20 | 0 | 1 | 19 | 40 | 1 | 0.95 | 1.146 | 1.457 |
| TML | Gupta and Khanna62 | 2017 | 600 | 0 | 13 | 488 | 1101 | 1 | 0.974 | 2.358 | 2.511 |
| TML | Gupta and Khanna62 | 2017 | 320 | 0 | 12 | 269 | 601 | 1 | 0.957 | 2.221 | 2.489 |
| TML | Bahadure et al.61 | 2015 | 128 | 3 | 4 | 65 | 200 | 0.977 | 0.942 | 2.835 | 2.371 |
| TML | Dvorák et al.68 | 2013 | 63 | 9 | 9 | 122 | 203 | 0.875 | 0.931 | 3.453 | 2.246 |
| TML | Sriramakrishnan et al.39 | 2019 | 4441 | 70 | 60 | 78 | 4649 | 0.984 | 0.565 | 3.02 | 2.63 |
| TML | Patil and Hamde92 | 2021 | 50 | 0 | 0 | 44 | 94 | 1 | 1 | 1.558 | 1.421 |
| TML | Patil and Hamde92 | 2021 | 50 | 0 | 0 | 44 | 94 | 1 | 1 | 1.558 | 1.421 |
| TML | Anitha and Raja70 | 2017 | 14 | 1 | 2 | 83 | 100 | 0.933 | 0.976 | 3.199 | 0.92 |
| TML | Anitha and Raja70 | 2017 | 14 | 1 | 1 | 59 | 75 | 0.933 | 0.983 | 3.075 | 0.848 |
| TML | Kebir et al.42 | 2019 | 961 | 332 | 805 | 1630 | 3728 | 0.743 | 0.669 | 3.547 | 2.625 |
| TML | Lahmiri71 | 2017 | 20 | 0 | 1 | 29 | 50 | 1 | 0.967 | 1.496 | 1.311 |
| TML | Simaiya et al.93 | 2017 | 504 | 63 | 111 | 843 | 1521 | 0.889 | 0.884 | 3.536 | 2.598 |
| TML | Kalaiselvi et al.45 | 2019 | 1683 | 73 | 154 | 2740 | 4650 | 0.958 | 0.947 | 3.539 | 2.61 |
| TML | Kalaiselvi et al.45 | 2019 | 63 | 0 | 7 | 211 | 281 | 1 | 0.968 | 2.734 | 1.917 |
| TML | Tejas P and Padma94 | 2021 | 74 | 6 | 0 | 20 | 100 | 0.925 | 1 | 2.749 | 1.875 |
DL, deep learning; FN, False Negative; FP, false Positive; TML, Traditional Machine Learning; TN, True Negative; TP, True Positive.
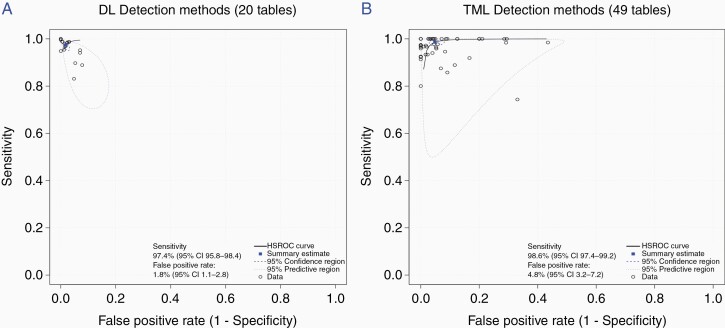
Overall, the pooled sensitivity was 0.98 (95% CI, 0.97 to 0.99) and the FPR was 0.035 (95% CI, 0.025 to 0.048). DL and TML had comparable sensitivity, but DL achieved a lower FPR compared to TML; 0.018 (95% CI, 0.011 to 0.028) and 0.048 (95% CI, 0.032 to 0.072) (P < .001), respectively (Figures 2A and 2B).
Figure 2.
Hierarchical receiving operating curves (ROC) of (A) deep learning (DL) and (B) traditional machine learning (TML) studies included in detection meta-analysis.
Segmentation meta-analysis
—Due to limited numbers of semi-automated studies, segmentation meta-analysis solely focused on fully automated methods. Forty-six fully automated segmentation studies provided sufficient data to be included in the meta-analysis. 34.8% (n = 16) of studies utilized DL and 65.2% (n = 30) utilized TML methods. Less than half (n = 19; 41.3%) of studies performed external validation. 97.8% (n = 45) of studies provided segmentation results for WT, 41.3% (n = 19) for TC and 39.1% (n = 18) for ET.
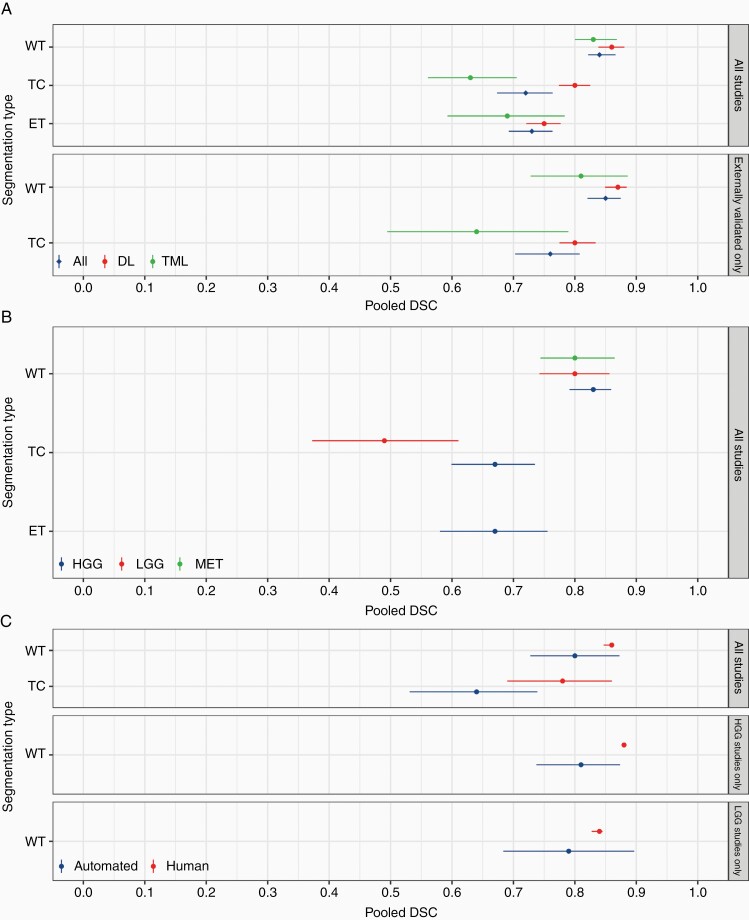
Overall, a DSC of 0.84 (95% CI, 0.82 to 0.87; I2 = 99.99%) for WT, 0.72 (95% CI, 0.67 to 0.76; I2 = 99.99%) for TC, and 0.73 (95% CI, 0.69 to 0.76; I2 = 99.99%) for ET (Figure 3A; Supplementary Table 11) were achieved. This persisted on sensitivity analysis of externally validated studies; a DSC of 0.85 (95% CI, 0.82 to 0.87; I2 = 99.97%) was achieved for WT and 0.76 (95% CI, 0.70 to 0.80; I2 = 99.96%) for TC (Figure 3A; Supplementary Table 12).
Figure 3.
Segmentation meta-analysis for (A) all studies and externally validated only studies, stratified by deep learning (DL) and traditional machine learning (TML), (B) subgroup segmentation meta-analysis by tumor type (high-grade glioma [HGG], low-grade glioma [LGG], and metastatic brain tumor [MET]), and (C) automated versus human segmentation.
TML versus DL segmentation meta-analysis
—DL was comparable to TML for WT segmentation, 0.86 (95% CI, 0.84 to 0.88; I2 = 99.99%) and 0.83 (95% CI, 0.80 to 0.87; I2 = 99.99%; P = .21), respectively (Figure 3A; Supplementary Table 11). This was relatively consistent on sensitivity analysis; 0.87 (95% CI, 0.85 to 0.88; I2 = 100%) and 0.81 (95% CI, 0.73 to 0.89; I2 = 99.94%; P = .10), respectively (Figure 3A; Supplementary Table 12).
In terms of TC segmentation, DL achieved a statistically significant higher DSC compared to TML, 0.80 (95% CI, 0.77 to 0.83; I2 = 99.97%) and 0.63 (95% CI, 0.56 to 0.71; I2 = 100%; P < .001). This remained unchanged on sensitivity analysis; 0.80 (95% CI, 0.77 to 0.83; I2 = 99.97%) and 0.64 (95% CI, 0.49 to 0.79; I2 = 99.87%; P = .009), respectively.
Finally, for ET segmentation, DL methods achieved higher DSC when compared to TML, 0.75 (95% CI, 0.72 to 0.78; I2 = 99.91%) and 0.69 (95% CI, 0.59 to 0.78; I2 = 100%), respectively. However, this did not reach statistical significance (P = .17).
Subgroup analysis by tumor type
—Most studies (91.3%; n = 42/46) applied their segmentation method on gliomas (91.3%; n = 42/46) HGG and 84.78% (n = 39/46; LGG), 10.87% (n = 5/46) on metastatic brain tumors, 4.35% (n = 2/46) on meningiomas, and 1.79% (n = 1/46) on nerve sheath tumors. 58.69% of studies (n = 27/46) sufficiently categorized their segmentation results by tumor type required for subgroup analysis (Supplementary Table 13).
Since few studies applied their segmentation techniques to meningiomas and nerve sheath tumors, they could not be included in subgroup analyses. The subgroup analysis thus compared HGG, LGG, and metastatic brain tumors. Only WT segmentation results for metastatic brain tumors were possible to compute due to limited studies. ET segmentation was predominantly performed on HGG, thereby excluding it from subgroup analysis. It was not possible to compare DL and TML methods in diagnosing different types of tumors due to the small number of studies.
For WT segmentation, no difference was observed between HGG, LGG, and metastatic tumors, 0.83 (95% CI, 0.79 to 0.86; I2 = 99.99%), 0.80 (95% CI, 0.74 to 0.86; I2 = 99.98%) and 0.80 (95% CI, 0.74 to 0.86; I2 = 99.95%; P = .64), respectively (Figure 3B; Supplementary Table 13). For TC segmentation, a higher DSC was achieved for HGG compared to LGG, 0.67 (95% CI, 0.60 to 0.74; I2 = 99.97%) and 0.49 (95% CI, 0.37 to 0.61; I2 = 99.98%; P = .0027), respectively.
Automated versus human expert segmentation
—Only 30.4% (n = 14/46) of studies provided sufficient data for comparison between automated and expert manual segmentation for WT and TC segmentation. All studies included multiple (>1) independent expert operators for generating ground truth segmentations; one study (7.1%) utilized two operators and 13 (92.9%) utilized four operators as part of the BRATS challenge (Supplementary Table 14).
For WT segmentation, both achieved “good” performance, but higher DSC was achieved in the manual group than the automated group 0.86 (95% CI, 0.85 to 0.86; I2 = 99.90%) and 0.80 (95% CI, 0.73 to 0.87; I2 = 99.98%; P = .11), respectively (Figure 3C; Supplementary Table 14). However, for TC segmentation, manual segmentation outperformed automated segmentation, 0.78 (95% CI, 0.69 to 0.86; I2 = 99.94%) and 0.64 (95% CI, 0.53 to 0.74; I2 = 99.98%; P = .014), respectively.
For HGG tumors, manual segmentation outperformed automated 0.88 (95% CI, 0.87 to 0.88; I2 = 28.85%) and 0.81 (95% CI, 0.74 to 0.87; I2 = 99.98%; P = .015), respectively. Conversely, manual was comparable to automated segmentation for LGG; 0.84 (95% CI, 0.83 to 0.85; I2 = 95.68%) and 0.79 (95% CI, 0.68 to 0.90, I2 = 99.96%, P = .33), respectively.
Discussion
To date, this is the largest meta-analysis evaluating automated brain tumor segmentation and detection methods. Automation provides benefits including elimination of human inter-rater variability and reduced inference time2; particularly DL methods, which showed an impressive median inference time of 0.2 seconds/MRI slice.
Previous studies have concluded that, in general, automated methods are comparable to human expertise in terms of performance.10,96 However, our research highlights that this only holds true for WT segmentation in brain tumors. Notably, we found that manual methods outperformed automated techniques for TC segmentation. Sub-compartmental segmentation, including TC, is a major influence on tumor progression monitoring and radiotherapy planning.97 Hence, our finding cautions the application of machine learning in all its potential uses in routine clinical practice and highlights the need for further research on sub-compartmental automated segmentation (TC and ET). Since most methods used conventional MRI scans (ie, T1, T2, T1CE, and FLAIR), future studies could combine these multimodal sequences with other specialized MRI sequences to increase the number of features, assessing for potential enhanced segmentation results. Soltaninejad et al.30 and Durmo et al.98 incorporated features obtained from diffusion-weighted and diffusion tensor imaging and showed promising results in the automated identification of brain tumors. Including other MRI sequences in publicly available datasets, such as BRATS, could facilitate investigations into the diagnostic value of additional features.
Regarding automated detection, we have replicated the findings of Cho et al.’s systematic review on brain tumor metastasis8; DL had a significantly lower FPR than TML, whilst sensitivity between the two methods remained similar. To the best of our knowledge, there has been no previous evaluation of automated sub-compartmental segmentation of brain tumors. Our study extends confidence in DL to tumor segmentation; the DL group achieved “good” (DSC ≥ 0.7) performance for all segmentation types (WT, TC, ET), whereas for TML, “good” performance was limited to WT segmentation. This trend persisted with sensitivity analysis investigating only externally validated studies, reinforcing these results. DL techniques support the automatic identification of complex features unlike TML, which requires hand-crafted feature vectors.3 However, the advantages of DL remain ambiguous, due to its “black box” nature; the interpretability of learned features and the explainability of the model’s decisions could be improved.3,4 Certain methods, such as saliency maps or feature attribution attempt to deduce how these learning algorithms detect complex features.99 However, just 2.1% (n = 5) of studies reported such methods, hindering model interpretation. This highlights the importance of future work reporting DL interpretation to improve comprehension and transparency of algorithmic predictions.
Van Kempen et al.9 reported good performance of machine learning algorithms for glioma WT segmentation, also showing that automated segmentation for both HGG and LGG were comparable. Our subgroup analysis, stratified by tumor type, showed “good” performance, and no statistically significant difference between tumor types for WT segmentation. However, this was not consistent for TC segmentation; both HGG and LGG tumors did not reach “good” performance as was evident for WT. This is clinically pertinent, because of the aforementioned value of reliable automated sub-compartmental segmentation in treatment pathways. HGG TC segmentation performance was found to be significantly better than LGG. This may be due to LGG’s slow growth, lack of surrounding vasogenic edema, and poor enhancement on MRI, making LGGs radiologically more difficult to identify.98 Moreover, HGGs are highly proliferative tumors resulting in higher lesion contrast and enhancement, making them radiologically more noticeable.98 This study shows that although manual WT segmentation statistically outperformed automated segmentation for HGG, both achieved “good” performance (DSC ≥ 0.7). On the other hand, for LGG tumors, manual and automated segmentation were statistically comparable in terms of performance; however, only manual segmentation achieved “good” performance. This could be because LGGs can simply conform to normal anatomy (eg, expanding gyri), making them difficult to diagnose, especially when small. This further highlights the need for future work on improving machine learning performance to segment LGG more accurately to achieve comparable results to that of manual segmentation.
Reporting guidelines reinforce robust evaluation and generalizability of diagnostic models. The recent CLAIM checklist, developed on the foundations of earlier well-established guidelines, is the first to address AI applications in medical imaging. This is the first study to adopt this pertinent guideline for the comprehensive assessment of reporting quality for brain tumor identification. Although over 70% of studies detailed data sources, model design, and ground truth definitions, only a minority reported missing data handling, data partitioning, study participant flow, and external validation. This is consistent with Yusuf et al.’s systematic review5 which found poor reporting of the study participant flow, the distribution of disease severity, and model validation techniques within ML-based diagnosis models. Such findings reiterate the necessity for studies to employ guidelines to aid their interpretation and reusability. This is paramount in ensuring reliable research is the basis of pioneering novel techniques into clinical practice.
The absence of external validation jeopardizes the generalizability of models for clinical use. Our study highlights such a limitation, with only 41.3% (n = 19/46) of segmentation and 2.6% (n = 1/38) of detection studies in the meta-analysis undertaking external validation. To address this, we performed a sensitivity analysis on segmentation models that were externally validated, which showed similar results to the original analysis. To ensure that future studies externally validate their machine learning algorithms, authors should utilize the CLAIM guideline when reporting their study. In addition, journals should encourage authors to provide details about elements of reporting outlined CLAIM for editors and reviewers during the assessment of AI-related manuscripts in medical imaging. Secondly, high heterogeneity was observed which may be due to methodological diversity in machine learning techniques. Thirdly, only a quarter of included studies were eligible for meta-analysis because of inadequate reporting, particularly the uncertainty values of performance metrics, thus compromising data availability. This issue has been recognized by non-neuro-oncology systematic reviews.96 Fourthly, most studies failed to report manual segmentation results, impeding a direct comparison of the techniques. To promote standardization of ground-truth images for training AI algorithms, experts should utilize structured reporting during manual segmentation.100 Finally, most studies tested and trained their algorithms on open-access datasets. We propose that available automated algorithms be applied to prospective, routinely collected MRI data to assess performance and feasibility for use in daily clinical practice.
To conclude, we found promising results for the use of AI algorithms in brain tumor identification and highlight the areas for future research. Further improvements to study design are needed, with adherence to reporting guidelines, which will avail transparent evaluation and generalizability of diagnostic AI models.
Supplementary Material
Contributor Information
Omar Kouli, School of Medicine, University of Dundee, Dundee, UK; NHS Greater Glasgow and Clyde, Glasgow, UK.
Ahmed Hassane, NHS Greater Glasgow and Clyde, Glasgow, UK.
Dania Badran, NHS Greater Glasgow and Clyde, Glasgow, UK.
Tasnim Kouli, School of Medicine, University of Dundee, Dundee, UK.
Kismet Hossain-Ibrahim, Division of Neurosurgery, NHS Tayside, Dundee, UK.
J Douglas Steele, Division of Imaging Science and Technology, School of Medicine, University of Dundee, Dundee, UK.
Funding
This work was supported by the SINAPSE innovation fund. The funders had no role in study design, analysis, and interpretation, or writing.
Conflict of interest statement. The authors declare no conflicts of interest.
Authorship statement. O.K., K.H.-I., and J.D.S. contributed to study conception and design. O.K., A.H., D.B., K.H.-I., and J.D.S. contributed to literature search, data extraction, and quality assessment of studies. All authors had access to the raw data, and contributed to data analysis, interpretation, and writing of the manuscript. O.K., K.H.-I., and J.D.S. guarantee the integrity of the work.
References
- 1. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018; 392(10145):432–446. [DOI] [PubMed] [Google Scholar]
- 2. Porz N, Bauer S, Pica A, et al. . Multi-modal glioblastoma segmentation: man versus machine. PLoS One. 2014; 9(5):e96873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 4. Montavon G, Samek W, Müller KR. Methods for interpreting and understanding deep neural networks. Digit Signal Process. 2018; 73:1–15. [Google Scholar]
- 5. Yusuf M, Atal I, Li J, et al. . Reporting quality of studies using machine learning models for medical diagnosis: a systematic review. BMJ Open. 2020; 10(3):e034568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mongan J, Moy L, Kahn CE Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. [DOI] [PMC free article] [PubMed]
- 7. Menze BH, Jakab A, Bauer S, et al. . The multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2014; 34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho SJ, Sunwoo L, Baik SH, et al. . Brain metastasis detection using machine learning: a systematic review and meta-analysis. Neuro-oncology. 2021; 23(2):214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Kempen EJ, Post M, Mannil M, et al. . Performance of machine learning algorithms for glioma segmentation of brain MRI: a systematic literature review and meta-analysis. Eur Radiol. 2021; 31(12):9638–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng Q, Yang L, Zeng B, et al. . Artificial intelligence performance in detecting tumor metastasis from medical radiology imaging: a systematic review and meta-analysis. EClinicalMedicine. 2021; 31:100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salameh JP, Bossuyt PM, McGrath TA, et al. . Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): explanation, elaboration, and checklist. BMJ. 2020; 370. [DOI] [PubMed] [Google Scholar]
- 12. Whiting PF, Rutjes AW, Westwood ME, et al. . QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011; 155(8):529–536. [DOI] [PubMed] [Google Scholar]
- 13. Zou KH, Warfield SK, Bharatha A, et al. . Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Acad Radiol. 2004; 11(2):178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harbord RM, Deeks JJ, Egger M, et al. . A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2007; 8(2):239–251. [DOI] [PubMed] [Google Scholar]
- 15. Sanjuán A, Price CJ, Mancini L, et al. . Automated identification of brain tumors from single MR images based on segmentation with refined patient-specific priors. Front Neurosci. 2013; 7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu W, Chen AY, Zhao L, et al. . Brain tumor detection and segmentation in a CRF (conditional random fields) framework with pixel-pairwise affinity and superpixel-level features. Int J Comput Assist Radiol Surg. 2014; 9(2):241–253. [DOI] [PubMed] [Google Scholar]
- 17. Dvorak P, Bartusek K, Kropatsch WG, et al. . Automated multi-contrast brain pathological area extraction from 2D MR images. J Appl Res Technol. 2015; 13(1):58–69. [Google Scholar]
- 18. Steed TC, Treiber JM, Patel KS, et al. . Iterative probabilistic voxel labeling: automated segmentation for analysis of The Cancer Imaging Archive glioblastoma images. Am J Neuroradiol. 2015; 36(4):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hasan AM, Meziane F, Aspin R, et al. . Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry. 2016; 8(11):132. [Google Scholar]
- 20. Ilunga-Mbuyamba E, Avina–Cervantes JG, Garcia–Perez A, et al. . Localized active contour model with background intensity compensation applied on automatic MR brain tumor segmentation. Neurocomputing. 2017; 220:84–97. [Google Scholar]
- 21. Thiruvenkadam K, Perumal N. Fully automatic method for segmentation of brain tumor from multimodal magnetic resonance images using wavelet transformation and clustering technique. Int J Imaging Syst Technol. 2016; 26(4):305–314. [Google Scholar]
- 22. Liu Y, Stojadinovic S, Hrycushko B, et al. . Automatic metastatic brain tumor segmentation for stereotactic radiosurgery applications. Phys Med Biol. 2016; 61(24):8440. [DOI] [PubMed] [Google Scholar]
- 23. Li Y, Jia F, Qin J. Brain tumor segmentation from multimodal magnetic resonance images via sparse representation. Artif Intell Med. 2016; 73:1–13. [DOI] [PubMed] [Google Scholar]
- 24. Soltaninejad M, Yang G, Lambrou T, et al. . Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assist Radiol Surg. 2017; 12(2):183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imtiaz T, Rifat S, Fattah SA, et al. . Automated brain tumor segmentation based on multi-planar superpixel level features extracted from 3D MR images. IEEE Access. 2019; 8:25335–25349. [Google Scholar]
- 26. Kaur T, Saini BS, Gupta S. A novel fully automatic multilevel thresholding technique based on optimized intuitionistic fuzzy sets and tsallis entropy for MR brain tumor image segmentation. Australas Phys Eng Sci Med. 2018; 41(1):41–58. [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, Stojadinovic S, Hrycushko B, et al. . A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One. 2017; 12(10):e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Essadike A, Ouabida E, Bouzid A. Brain tumor segmentation with Vander Lugt correlator based active contour. Comput Methods Programs Biomed. 2018; 160:103–117. [DOI] [PubMed] [Google Scholar]
- 29. Pinto A, Pereira S, Rasteiro D, et al. . Hierarchical brain tumour segmentation using extremely randomized trees. Pattern Recognit. 2018; 82:105–117. [Google Scholar]
- 30. Soltaninejad M, Yang G, Lambrou T, et al. . Supervised learning based multimodal MRI brain tumour segmentation using texture features from supervoxels. Comput Methods Programs Biomed. 2018; 157:69–84. [DOI] [PubMed] [Google Scholar]
- 31. Charron O, Lallement A, Jarnet D, et al. . Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med. 2018; 95:43–54. [DOI] [PubMed] [Google Scholar]
- 32. Li Q, Gao Z, Wang Q, et al. . Glioma segmentation with a unified algorithm in multimodal MRI images. IEEE Access. 2018; 6:9543–9553. [Google Scholar]
- 33. Grøvik E, Yi D, Iv M, et al. . Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. J Magn Reson Imaging. 2020; 51(1):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eltayeb EN, Salem NM, Al-Atabany W. Automated brain tumor segmentation from multi-slices FLAIR MRI images. BioMed Mater Eng. 2019; 30(4):449–462. [DOI] [PubMed] [Google Scholar]
- 35. Wang G, Li W, Ourselin S, et al. . Automatic brain tumor segmentation based on cascaded convolutional neural networks with uncertainty estimation. Front Comput Neurosci. 2019; 13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Li A, Wang M. A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput Biol Med. 2019; 108:150–160. [DOI] [PubMed] [Google Scholar]
- 37. Tong J, Zhao Y, Zhang P, et al. . MRI brain tumor segmentation based on texture features and kernel sparse coding. Biomed Signal Proc Control. 2019; 47:387–392. [Google Scholar]
- 38. Dogra J, Jain S, Sood M. Glioma extraction from MR images employing gradient based kernel selection graph cut technique. Vis Comput. 2020; 36(5):875–891. [Google Scholar]
- 39. Sriramakrishnan P, Kalaiselvi T, Rajeswaran R. Modified local ternary patterns technique for brain tumour segmentation and volume estimation from MRI multi-sequence scans with GPU CUDA machine. Biocybern Biomed Eng. 2019; 39(2):470–487. [Google Scholar]
- 40. Mlynarski P, Delingette H, Criminisi A, et al. . 3D convolutional neural networks for tumor segmentation using long-range 2D context. Comput Med Imaging Graph. 2019; 73:60–72. [DOI] [PubMed] [Google Scholar]
- 41. Mallick PK, Ryu SH, Satapathy SK, et al. . Brain MRI image classification for cancer detection using deep wavelet autoencoder-based deep neural network. IEEE Access. 2019; 7:46278–46287. [Google Scholar]
- 42. Tchoketch Kebir S, Mekaoui S, Bouhedda M. A fully automatic methodology for MRI brain tumour detection and segmentation. Imaging Sci J. 2019; 67(1):42–62. [Google Scholar]
- 43. Alagarsamy S, Kamatchi K, Govindaraj V, et al. . Multi-channeled MR brain image segmentation: a new automated approach combining BAT and clustering technique for better identification of heterogeneous tumors. Biocybern Biomed Eng. 2019; 39(4):1005–1035. [Google Scholar]
- 44. Shivhare SN, Kumar N, Singh N. A hybrid of active contour model and convex hull for automated brain tumor segmentation in multimodal MRI. Multimedia Tools Appl. 2019; 78(24):34207–34229. [Google Scholar]
- 45. Kalaiselvi T, Kumarashankar P, Sriramakrishnan P. Three-phase automatic brain tumor diagnosis system using patches based updated run length region growing technique. J Digit Imaging. 2020; 33(2):465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu Y, Zhao Z, Wu W, et al. . Automatic glioma segmentation based on adaptive superpixel. BMC Med Imaging. 2019; 19(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rehman ZU, Naqvi SS, Khan TM, et al. . Fully automated multi-parametric brain tumour segmentation using superpixel based classification. Expert Syst Appl. 2019; 118:598–613. [Google Scholar]
- 48. Zhou C, Ding C, Wang X, et al. . One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Process. 2020; 29:4516–4529. [DOI] [PubMed] [Google Scholar]
- 49. Khan H, Shah PM, Shah MA, et al. . Cascading handcrafted features and Convolutional Neural Network for IoT-enabled brain tumor segmentation. Comput Commun. 2020; 153:196–207. [Google Scholar]
- 50. Xue J, Wang B, Ming Y, et al. . Deep learning–based detection and segmentation-assisted management of brain metastases. Neuro-oncology. 2020; 22(4):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thiruvenkadam K, Nagarajan K. Fully automatic brain tumor extraction and tissue segmentation from multimodal MRI brain images. Int J Imaging Syst Technol. 2021; 31(1):336–350. [Google Scholar]
- 52. Ben naceur M, Akil M, Saouli R, et al. . Fully automatic brain tumor segmentation with deep learning-based selective attention using overlapping patches and multi-class weighted cross-entropy. Med Image Anal. 2020; 63:101692. [DOI] [PubMed] [Google Scholar]
- 53. Aboelenein NM, Songhao P, Koubaa A, et al. . HTTU-Net: hybrid two track U-net for automatic brain tumor segmentation. IEEE Access. 2020; 8:101406–101415. [Google Scholar]
- 54. Hassen OA, Abter SO, Abdulhussein AA, et al. . Nature-inspired level set segmentation model for 3D-MRI brain tumor detection. CMC Comput Mater Contin. 2021; 68(1):961–981. [Google Scholar]
- 55. Kao PY, Shailja S, Jiang J, et al. . Improving patch-based convolutional neural networks for MRI brain tumor segmentation by leveraging location information. Front Neurosci. 2020; 13:1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Debnath S, Talukdar FA, Islam M. Combination of contrast enhanced fuzzy c-means (CEFCM) clustering and pixel based voxel mapping technique (PBVMT) for three dimensional brain tumour detection. J Ambient Intell Hum Comput. 2021; 12(2):2421–2433. [Google Scholar]
- 57. Baid U, Talbar S, Rane S, et al. . A novel approach for fully automatic intra-tumor segmentation with 3D U-Net architecture for gliomas. Front Comput Neurosci. 2020; 14:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitchell JR, Kamnitsas K, Singleton KW, et al. . Deep neural network to locate and segment brain tumors outperformed the expert technicians who created the training data. J Med Imaging. 2020; 7(5):055501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sran PK, Gupta S, Singh S. Integrating saliency with fuzzy thresholding for brain tumor extraction in MR images. J Vis Commun Image Represent. 2021; 74:102964. [Google Scholar]
- 60. Takahashi S, Takahashi M, Kinoshita M, et al. . Fine-tuning approach for segmentation of gliomas in brain magnetic resonance images with a machine learning method to normalize image differences among facilities. Cancers. 2021; 13(6):1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bahadure NB, Ray AK, Thethi HP. Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int J Biomed Imaging. 2017; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gupta N, Khanna P. A non-invasive and adaptive CAD system to detect brain tumor from T2-weighted MRIs using customized Otsu’s thresholding with prominent features and supervised learning. Signal Process Image Commun. 2017; 59:18–26. [Google Scholar]
- 63. Abd-Ellah MK, Awad AI, Khalaf AA, et al. . Two-phase multi-model automatic brain tumour diagnosis system from magnetic resonance images using convolutional neural networks. EURASIP J Image Video Process. 2018; 2018(1):1–0. [Google Scholar]
- 64. Amin J, Sharif M, Raza M, et al. . Brain tumor detection using statistical and machine learning method. Comput Methods Programs Biomed. 2019; 177:69–79. [DOI] [PubMed] [Google Scholar]
- 65. Rai HM, Chatterjee K, Dashkevich S. Automatic and accurate abnormality detection from brain MR images using a novel hybrid UnetResNext-50 deep CNN model. Biomed Signal Proc Control. 2021; 66:102477. [Google Scholar]
- 66. Jayachandran A, Dhanasekaran R. Automatic detection of brain tumor in magnetic resonance images using multi-texton histogram and support vector machine. Int J Imaging Syst Technol. 2013; 23(2):97–103. [Google Scholar]
- 67. Jayachandran A, Dhanasekaran R. Brain tumor severity analysis using modified multi-texton histogram and hybrid kernel SVM. Int J Imaging Syst Technol. 2014; 24(1):72–82. [Google Scholar]
- 68. Dvořák P, Kropatsch WG, Bartušek K. Automatic brain tumor detection in t2-weighted magnetic resonance images. Meas Sci Rev. 2013; 13(5):223–230. [Google Scholar]
- 69. Amin J, Sharif M, Yasmin M, et al. . A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit Lett. 2017; 139:118–127. [Google Scholar]
- 70. Anitha R, Siva Sundhara Raja D. Development of computer-aided approach for brain tumor detection using random forest classifier. Int J Imaging Syst Technol. 2018; 28(1):48–53. [Google Scholar]
- 71. Lahmiri S. Glioma detection based on multi-fractal features of segmented brain MRI by particle swarm optimization techniques. Biomed Signal Proc Control. 2017; 31:148–155. [Google Scholar]
- 72. Deepa AR, Emmanuel WS. An efficient detection of brain tumor using fused feature adaptive firefly backpropagation neural network. Multimedia Tools Appl. 2019; 78(9):11799–11814. [Google Scholar]
- 73. Selvapandian A, Manivannan K. Performance analysis of meningioma brain tumor classifications based on gradient boosting classifier. Int J Imaging Syst Technol. 2018; 28(4):295–301. [Google Scholar]
- 74. Arunkumar N, Mohammed MA, et al. . Fully automatic model-based segmentation and classification approach for MRI brain tumor using artificial neural networks. Concurrency Comput Pract Exp. 2020; 32(1):e4962. [Google Scholar]
- 75. Edalati-rad A, Mosleh M. Improving brain tumor diagnosis using MRI segmentation based on collaboration of beta mixture model and learning automata. Arab J Sci Eng. 2019; 44(4):2945–2957. [Google Scholar]
- 76. Song G, Huang Z, Zhao Y, et al. . A noninvasive system for the automatic detection of gliomas based on hybrid features and PSO-KSVM. IEEE Access. 2019; 7:13842–13855. [Google Scholar]
- 77. Johnpeter JH, Ponnuchamy T. Computer aided automated detection and classification of brain tumors using CANFIS classification method. Int J Imaging Syst Technol. 2019; 29(4):431–438. [Google Scholar]
- 78. Alam MS, Rahman MM, Hossain MA, et al. . Automatic human brain tumor detection in MRI image using template-based K means and improved fuzzy C means clustering algorithm. Big Data Cogn Comput. 2019; 3(2):27. [Google Scholar]
- 79. Atici MA, Sagiroglu S, Celtikci P, et al. . A novel deep learning algorithm for the automatic detection of high-grade gliomas on T2-weighted magnetic resonance images: a preliminary machine learning study. Turk Neurosurg. 2020; 30(2):199–205. [DOI] [PubMed] [Google Scholar]
- 80. Çinar A, Yildirim M. Detection of tumors on brain MRI images using the hybrid convolutional neural network architecture. Med Hypotheses. 2020; 139:109684. [DOI] [PubMed] [Google Scholar]
- 81. Devanathan B, Venkatachalapathy K. Brain tumor detection and classification model using optimal Kapur’s thresholding based segmentation with deep neural networks. IIOABJ. 2020; 11:1–8. [Google Scholar]
- 82. Gurunathan A, Krishnan B. Detection and diagnosis of brain tumors using deep learning convolutional neural networks. Int J Imaging Syst Technol. 2021; 31(3):1174–1184. [Google Scholar]
- 83. Wang J, Shao W, Kim J. Automated classification for brain MRIs based on 2D MF-DFA method. Fractals. 2020; 28(06):2050109. [Google Scholar]
- 84. Kesav OH, Rajini GK. Automated detection system for texture feature based classification on different image datasets using S-transform. Int J Speech Technol. 2021; 24(2):251–258. [Google Scholar]
- 85. Murali E, Meena K. Brain tumor detection from MRI using adaptive thresholding and histogram based techniques. Scalable Comput Pract Exper. 2020; 21(1):3–10. [Google Scholar]
- 86. Kaur T, Gandhi TK. Deep convolutional neural networks with transfer learning for automated brain image classification. Mach Vis Appl. 2020; 31(3):1–6. [Google Scholar]
- 87. Thangarajan SK, Chokkalingam A. Integration of optimized neural network and convolutional neural network for automated brain tumor detection. Sensor Rev. 2021; 41(1):16–34. [Google Scholar]
- 88. Kalaiselvi T, Padmapriya T, Sriramakrishnan P, et al. . Development of automatic glioma brain tumor detection system using deep convolutional neural networks. Int J Imaging Syst Technol. 2020; 30(4):926–938. [Google Scholar]
- 89. Rajinikanth V, Joseph Raj AN, Thanaraj KP, et al. . A customized VGG19 network with concatenation of deep and handcrafted features for brain tumor detection. Appl Sci. 2020; 10(10):3429. [Google Scholar]
- 90. Huang Z, Xu H, Su S, et al. . A computer-aided diagnosis system for brain magnetic resonance imaging images using a novel differential feature neural network. Comput Biol Med. 2020; 121:103818. [DOI] [PubMed] [Google Scholar]
- 91. Chen B, Zhang L, Chen H, et al. . A novel extended Kalman filter with support vector machine based method for the automatic diagnosis and segmentation of brain tumors. Comput Methods Programs Biomed. 2021; 200:105797. [DOI] [PubMed] [Google Scholar]
- 92. Patil DO, Hamde ST. Automated detection of brain tumor disease using empirical wavelet transform based LBP variants and ant-lion optimization. Multimedia Tools Appl. 2021; 80(12):17955–17982. [Google Scholar]
- 93. Simaiya S, Lilhore UK, Prasad D, et al. . MRI brain tumour detection & image segmentation by hybrid hierarchical K-means clustering with FCM based machine learning model. Ann Romanian Soc Cell Biol. 2021; 28:88–94. [Google Scholar]
- 94. Tejas P. A novel hybrid approach to detect brain tumor in MRI images. Turk J Comput Math Educ. 2021; 12(3):3412–3416. [Google Scholar]
- 95. El-Dahshan ES, Mohsen HM, Revett K, et al. . Computer-aided diagnosis of human brain tumor through MRI: a survey and a new algorithm. Expert Syst Appl. 2014; 41(11):5526–5545. [Google Scholar]
- 96. Liu X, Faes L, Kale AU, et al. . A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019; 1(6):e271–e297. [DOI] [PubMed] [Google Scholar]
- 97. Ghaffari M, Samarasinghe G, Jameson M, et al. . Automated post-operative brain tumour segmentation: a deep learning model based on transfer learning from pre-operative images. Magn Reson Imaging. 2022; 86:28–36. [DOI] [PubMed] [Google Scholar]
- 98. Durmo F, Lätt J, Rydelius A, et al. . Brain tumor characterization using multibiometric evaluation of MRI. Tomography. 2018; 4(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shad R, Cunningham JP, Ashley EA, et al. . Designing clinically translatable artificial intelligence systems for high-dimensional medical imaging. Nat Mach Intell. 2021; 3(11):929–935. [Google Scholar]
- 100. Dos Santos DP, Brodehl S, Baeßler B, et al. . Structured report data can be used to develop deep learning algorithms: a proof of concept in ankle radiographs. Insights Imaging. 2019; 10(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.