Abstract
Maternal embryonic leucine zipper kinase (MELK) is of vital importance due to its significant role in cancer development and its association with poor prognosis in different cancers. Here, we employed several computer aided drug design approaches to shortlist potential binding molecules of MELK. For virtual screening, asinex oncology library (containing 6334 drugs) and comprehensive marine natural products database (containing approximately 32,000 drugs) were used. The study identified two drug molecules: Top-2 and Top-3 as high affinity binding MELK molecules compared to the control co-crystalized Top-1 inhibitor. Both the shortlisted compounds and the control showed high stable binding free energy and high GOLD score. The compounds and control also reported stable dynamics with root mean square deviations (RMSD) value ∼ 2 Å in 500 ns. Similarly, the MELK active site residues were observed in good stability with the compounds. Further, it was noticed the compounds/control formed multiple hydrogen bonds with the MELK active pocket residues which is the main reason of high intermolecular stability. Atomic level binding free energies determined van der Waals and electrostatic energies to play vital role in stable complex formation. From drug likeness and pharmacokinetics perspective, the compounds are ideal molecules for further investigation. Overall, the results are promising and might be tested in in vivo and in vitro studies against MELK.
Keywords: Maternal embryonic leucine zipper kinase, Virtual screening, Molecular dynamics simulations, Binding free energies, Pharmacokinetics
1. Introduction
The maternal embryonic leucine zipper kinase (MELK) is also referred as murine protein K38 (MPK38) or pEg3 (Li et al., 2016). The protein is serine/threonine kinase and is important for different biological functions such as tumorigenesis, mitotic progression, apoptosis, mitotic progression and stem cell phenotypes (Maes et al., 2019). The MELK is reported to phosphorylate G2/M proteins in order to regulate G2/M transition (Sun et al., 2021). The protein is able to indirectly drive the expression of proteins such as survivin, cyclin B1, and aurora kinases by phosphorylating and activating the transcription of FOXM1 (Fischer et al., 2015). The Cdc25B is also phosphorylated by MELK, which in turn activates Cdk1 in early mitosis (Boutros et al., 2007). Beside this, the MELK targets several other proteins such as c-Jun, p53, apoptosis signal-regulating kinase 1 (ASK1), enhancer of zeste homolog 2 (EZH2), SOX2, and DEPDC1 (Thangaraj et al., 2020). Many studies have reported the association of between high level of MELK and onset of different types of cancers and poor cancer prognosis (Jiang and Zhang, 2013). Also, the therapeutic target database (TTD) database includes MELK as a drug target for clinical studies. Considering the vital importance of MELK in cancers development and good druggable nature, the protein is considered as an attractive drug target [8,9].
Previously, “OTSSP167”, a potent MELK inhibitor has been described to halt cancer cells growth in myeloma, leukemia, kidney cancer cells, prostate cancer, small cell lung cancer and neuroblastoma (Fakhri and Kahl, 2017). The drug is under different clinical trials and has been observed to show promising activity against solid tumor and relapsed leukemia (Maes et al., 2019). This highlights the potential of drug molecules to modulate the function of MELK for management of cancers, thus warranting for the search of new inhibitory molecules.
Traditional drug discovery is a very slow process and consume significant cost and human efforts (Ahmad et al., 2022, Menchaca et al., 2020). In contrast, computer aided drug designing is fast and is cost effective in screening new drug molecules against any given biological macromolecule (Islam et al., 2022, Yu and MacKerell, 2017). In this regard, virtual screening of drug libraries is an effective way to identify the best binding molecules and understand their conformational dynamics with respect to a biological target (Lionta et al., 2014). Though the chances of false positive results are high, cross validation through different computational techniques can minimize such errors. The molecular docking studies are usually validated by running a control molecule that is proven experimentally for biological potency against the target biomolecule. The control drug binding interactions and binding mode are often reproduced through molecular docking and once the results are same, virtual screening of libraries are performed. Another way of docking validation is to perform comparative docking where the results of one docking software is affirmed by screening the compounds against the target in another docking software. Further validation is achieved by conducing long run of molecular dynamics simulation that only decipher the dynamics of biomolecule-drug complex but also examine the time dependent behavior of the system (Karplus and McCammon, 2020). This is also highlight critical amino acids of proteins that are key in interactions and holding the ligands at the active pocket. The atomic level interactions are further determined via MM/PBSA and MM/GBSA methods (Genheden and Ryde, 2015). Both methods results are comparable and is of significance importance in drug designing (Hou et al., 2011, Tahir ul Qamar et al., 2021a, Tahir ul Qamar et al., 2021b). The findings of the study to deliver new drug molecules to show efficient binding with MELK for experimentalists thus saving time and cost.
2. Materials and methods
2.1. MELK structure retrieval
The crystal structure of MELK was taken from protein data bank (Sussman et al., 1998) using a PDB 4 digit code of 5M5A (Klaeger et al., 2017). The 5M5A structure was determined by X-RAY diffraction method and is the most recent (06–12-2017) structure of the protein in PDB database with good resolution of 1.90 Å. After fetching, the MELK was treated in receptor preparatory phase where the co-crystalized ligands such as water, Na+ and CL- and MELK inhibitor were removed. The protein was analyzed in energy minimization phase in UCSF Chimera 1.15 (Pettersen et al., 2004) to add missing hydrogen bonds and energy minimized the structure using two algorithms. The first algorithm applied is steepest descent for 1000 rounds, followed by the second algorithm of conjugate gradient algorithm for 1000 cycles. The purpose of the first refinement is to remove highly unfavorable steric clashes while the conjugate gradient remove severe clashes and is much slower than steepest descent. One these steps are finished, the MELK is saved in.pdb format for downward processing.
2.2. Inhibitors library preparation
For virtual screening, different drug libraries were used. The libraries used include Asinex oncology library (containing 6334 drugs) (https://www.asinex.com/?page_id=36) and Comprehensive Marine Natural Products Database (containing approximately 32,000 drugs) (Lyu et al., 2021). Both the libraries contain diverse group of chemical and natural molecules and are less explored for their potential as anti-cancer compounds. The libraries after retrieval were imported to PyRx 0.8 software (Dallakyan and Olson, 2015). The libraries drugs were energy minimized using MM2 force field in PyRx software and filtered using Lipinski rule of five (Lipinski, 2004). The rule ensured that only potential drug like molecules are forwarded. The different parameters ensured in the Lipinski rule of five include molecular weight < 500 kDa, <5 hydrogen bond donors and 10 hydrogen bond acceptors, logP value < 5 and topological polar surface area (<101.64 Å2). The filtered compounds were then converted into.pdbqt format to be used in virtual screening against MELK.
2.3. Molecular docking
To shortlist the best binding drug molecules to the MELK active site, a blind molecular docking study was carried out. For accuracy of results, the docking study was carried out using two well-known molecular docking software: AutoDock Vina (Trott and Olson, 2010) and genetic optimization for ligand docking (GOLD) (Iqbal et al., 2016). First, the libraries filtered drug like molecules were screened against MELK using AutoDock Vina and each molecule was assigned with binding energy value. The number of docking iteration set for each molecule is 50. The shortlisted 50 molecules were redocked to the enzyme using GOLD using the same number of iterations. In AutoDock Vina, the selection of compounds was based on lowest binding energy while in GOLD, compounds ranking was done via GOLD fitness score. Top 2 compounds with same conformation in both docking software, and lowest binding energy value and highest GOLD score were selected. Further, to ensure consistency in the results, a control molecule (named as Top-1) was redocked to MELK and examined to reproduce the same binding conformation reported in the crystal structure. The top 3 complexes selected were visualized in UCSF Chimera 1.15 (Pettersen et al., 2004) and Discovery Studio version 2020 (Biovia, 2017).
2.4. Molecular dynamics simulations
For confirmation on the docked conformation stability of the compounds and understand complexes dynamics, molecular dynamics simulation was performed. This was achieved using AMBER20 simulation package (Case et al., 2020). The initial systems libraries were prepared using antechamber program. For MELK, FF14SB force field (Case et al., 2014) was used while for compounds, GAFF (Dickson et al., 2012) was considered. The complexes were submerged into TIP3 water box. The box size was set to 12 Å to make sure the complete submersion of complexes inside the box. The complexes were neutralized by adding adequate number of counter ions. The energy minimization was performed for total of 1500 steps. After that, the complexes were heated to 310 K gradually for 100 ps. Each complex was then equilibrated for 50 ps using periodic boundary conditions. SHAKE algorithm (Kräutler et al., 2001) was used to constrain hydrogen bonds length while Langevin was applied to maintain temperature (Feller et al., 1995). The van der Waal interactions was defined by a cut-off distance of 10 Å. The simulation trajectories were generated in the production phase at time scale of 500 ns and CPPTRAJ module (Roe and Cheatham III, 2013) was used to measure different statistical parameters of complexes. XMGRACE software (Turner, 2005) was used for plotting different systems stability graphs. Additionally, we performed MMPB/GBSA analysis on 1000 frames of simulation trajectories via MMPBSA.py module of AMBER (Miller et al., 2012). Entropy of the systems was determined using AMBER normal mode analysis (Genheden et al., 2012). Hydrogen bonds analysis was performed in visual molecular dynamics software using Hbonds plugin (Humphrey et al., 1996). Only hydrogen bonds that were within 3 Å were considered.
2.5. WaterSwap binding free energies
The overall intermolecular docked stability measurement was revalidated by WaterSwap (Ahmad et al., 2018, Altharawi et al., 2021, Woods et al., 2014). For this purpose, WaterSwap from Sire package was employed for total of 1000 interactions. The binding energy prediction was done through Bennett’s acceptance ratio (BAR), thermodynamic integration (TI) and free energy perturbation (FEP). A system is considered in good stability if the energy value different among the three algorithm is less than 1 kcal/mol.
2.6. Drug likeness and ADMET properties analysis
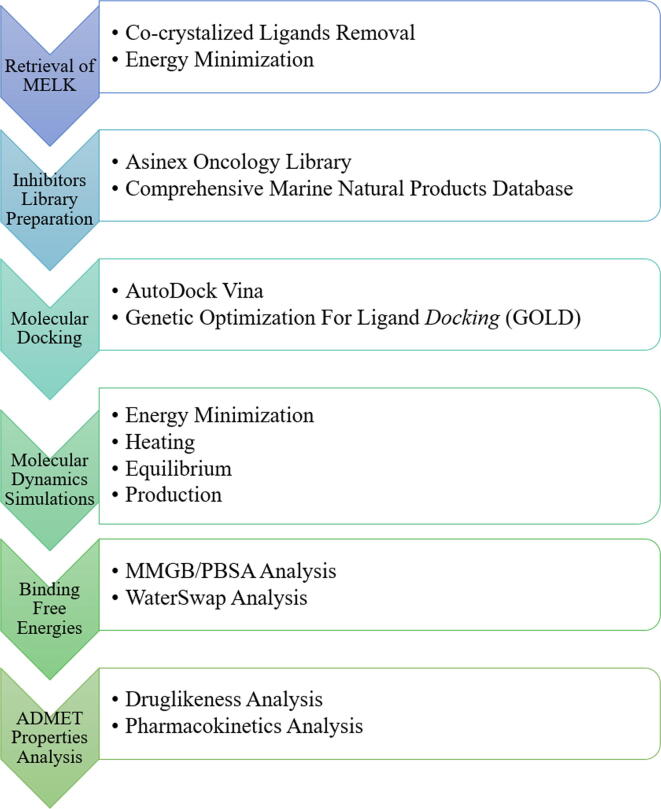
The drug likeness and different pharmacokinetics properties of compounds were evaluated using pkCSM (Pires et al., 2015) and SWISSADME (Daina et al., 2017) servers. The complete flow of steps used herein is presented in Fig. 1.
Fig. 1.
Flow chart of methodology steps employed in the study.
3. Results
3.1. Comparative docking studies
Virtual screening was conducted to filtered compounds that show stable conformation at MELK active pocket. This shortlisted two drug molecules named as: Top-2 (COC1 = CC(=CC = C1OCC[N]2C = CN = C2C)CN3CCC(O)(CC3)COC4 = CC = C(C)C = C4) and Top-3 (COC1 = C(OCC2 = NC = CC = C2)C = C(C = C1)C(=O)N3CCC(CC3)CC4 = CC = CC5 = NC = CC = C45) as strong binders of MELK active pocket. In contrast, Top-1 inhibitor (COC(=O)C1(O)CC2OC1(C)[N]3C4 = CC = CC = C4C5 = C3C6 = C(C7 = C(C = CC = C7)[N]26)C8 = C(O)[NH]C = C58) which was used as a control and co-crystalized molecule. The AutoDock binding affinity of Top-1, Top-2 and Top-3 is −13.81 kcal/mol, −13.68 kcal/mol, and −13.70 kcal/mol, respectively. The GOLD score of the compounds is Top-1 (80.36), Top-2 (81.65) and Top-3 (80.67). The AutoDock vina binding energy and GOLD score of top 10 compounds are shown Table 1.
Table 1.
Comparative molecular docking scores of selected high affinity binders.
| Compounds | Gold fitness score | AutoDock Vina binding energy in kcal/mol |
|---|---|---|
Top-1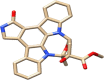
|
80.36 | −13.81 |
Top-2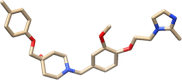
|
81.65 | −13.68 |
Top-3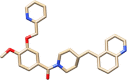
|
80.06 | −13.70 |
Top-4
|
75.69 | −10.28 |
Top-5
|
74.99 | −10.58 |
Top-6
|
75.36 | −11.01 |
Top-7
|
73.26 | −9.62 |
Top-8
|
72.55 | −8.62 |
Top-9
|
73.02 | −8.69 |
Top-10
|
−72.36 | −8.00 |
Top-1 is methyl 6,13-dihydroxy-5-methyl-6,7,8,14-tetrahydro-5H-16-oxa-4b,8a,14-triaza-5,8-methanodibenzo[b,h]cycloocta[jkl]cyclopenta[e]-as-indacene-6-carboxylate while Top-2 is 1-(3-methoxy-4-(2-(2-methyl-1H-imidazol-1-yl)ethoxy)benzyl)-4-((p-tolyloxy)methyl)piperidin-4-ol and Top-3 is (4-methoxy-3-(pyridin-2-ylmethoxy)phenyl)(4-(quinolin-5-ylmethyl)piperidin-1-yl)methanone. All the three compounds were found to interfere with the active cavity of MELK, which might halt interaction with the natural substrate. Also, the size of compounds was important parameter in achieving stable conformation at the MELK active pocket.
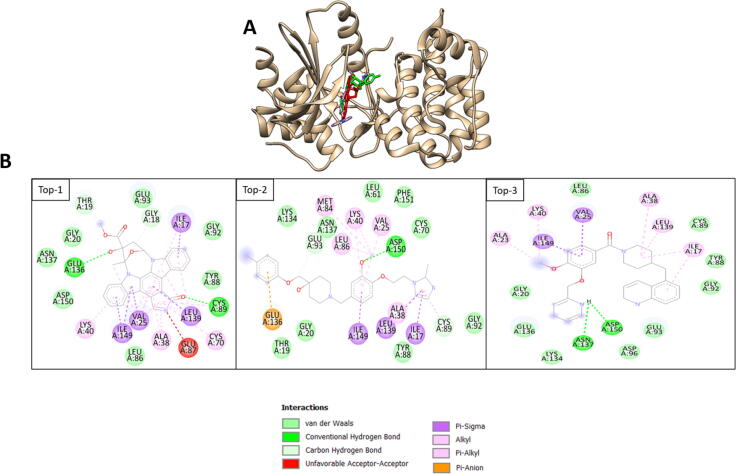
The intermolecular binding conformation are given in Fig. 2A while binding interactions are given in Fig. 2B. All the compounds reported to bind to the same active pocket of MELK present at junction of domain I and domain II. Visualization of the complexes revealed Top-1 hydrogen bond interaction with Cys89 and Glu136. Top-2 was seen in hydrogen bonds with Asp150 while Top-3 in Asn137 and Asp150. Beside hydrogen bonds, multiple van der Waals contacts were reported between the compounds and MELK. From interactions perspective, the compounds binding with the MELK residues is dominated by hydrophobic interactions and less by hydrophilic interactions. This signifies that the intermolecular binding requires a balance interactions of strong hydrophilic interactions and weak hydrophobic contacts. However, further structure of the compounds structure might enhance the compounds deep MELK binding pocket. The common non-bounded interactions observed in the complexes are listed in Table 2.
Fig. 2.
A. Compounds binding at the active site of MELK. The protein is presented in tan new cartoon while the Top-1, Top-2, and Top-3 are shown by red, green and purple, respectively. B. Compounds interactions with MELK. Top-1 is control while Top-2 and Top-3 is the best two filtered docked molecules.
Table 2.
Common non-bounded interactions observed in the complexes.
| Residue | Ligand atom | Distance (Angstrom) |
|---|---|---|
| O Ile17 | C | 3.83–4 |
| O Ile17 | C | 3.25–4 |
| CD1 Ile17 | C | 3.53–4 |
| CD1 Ile17 | C | 3.58–4 |
| CA Gly18 | O | 3.47–4 |
| C Gly18 | O | 3.81–4 |
| N Thr19 | C | 3.69–4 |
| C Thr19 | C | 3.85–4 |
| C Thr19 | C | 3.79–4 |
| N Gly20 | C | 3.63–4 |
| CA Gly20 | C | 3.81–4 |
| CG2 Val25 | C | 3.80–4 |
| CG2 Val25 | C | 3.68–4 |
| CB Ala38 | O | 3.87–4 |
| CB Ala38 | C | 3.76–4 |
| CB Ala38 | N | 3.27–4 |
| CD Lys40 | C | 3.53–4 |
3.2. Molecular dynamics simulation
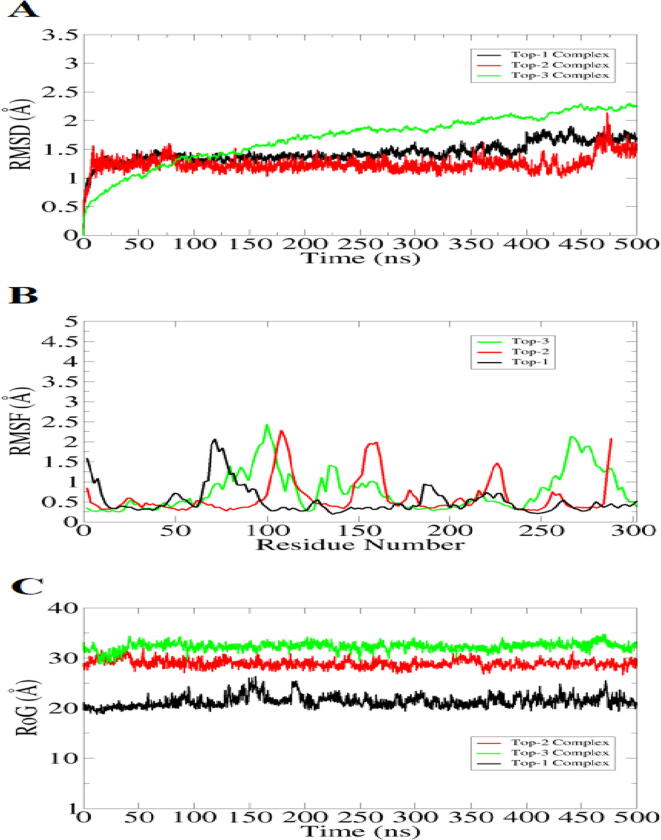
Molecular dynamic simulation was carried out to decipher whether the docked conformation of compounds with respect to MELK is stable or not (Karplus and McCammon, 2020, Tahir ul Qamar et al., 2021a, Tahir ul Qamar et al., 2021b). This was important to underpin as blockage of MELK active pocket by compounds for longer time ensure non-functional MELK enzyme and thus curtail cancer progression. The MELK dynamics in the presence of compounds were checked by root mean square deviation (RMSD) considering carbon alpha atoms (Kuzmanic and Zagrovic, 2010). The complexes conformation in each frame were superimposed and the structural deviations were measured in term of Å. The RMSD of complexes is plotted in Fig. 3A. The systems revealed very stable dynamics with no drastic conformational changes observed. Top-1 and Top-2 were found to be the most stable with some minor structure changes were noticed at the end. These changes were the outcome of flexible loops of MELK. The mean RMSD of Top-1 is 1.3 Å and that of Top-2 is 1.4 Å. Top-3 on the other hand experienced steady RMSD increase touching high RMSD of 2 Å. These changes were due flexible Top-3 conformation and remained in strong and short distance contacts with MELK. Next, root mean square fluctuation (RMSF) of MELK residues was investigated (Fig. 3B). Majority of MELK active site residues were found in stable dynamics with some having RMSF up to 2.5 Å. The MELK structure integrity was additionally studied using radius of gyration (RoG) (Lobanov et al., 2008) as mentioned in Fig. 3C. The RoG findings complement the RMSD and reported stable compactness of MELK. This demonstrate that the MELK is in stable conformation in the presence of compounds. The mean RoG of Top-1, Top-2 and Top-3 is 2.56 Å, 29.87 Å, and 32 Å, respectively. Furthermore, solvent accessible surface area (SASA) analysis was conducted tp confirm complexes stability (Muneer et al., 2021). The SASA analysis of Top-1, Top-2 and Top-3 is given in S-Fig. 1, S-Fig. 2 and S-Fig. 3, respectively. The analysis suggested that the complexes remained in good stability and the water molecules do not affect the binding of compounds to the protein.
Fig. 3.
Investigating structure stability of MELK in the presence of compounds. The conformational stability of MELK was examined considering carbon alpha deviation of the protein. Different statistical analysis were conducted like RMSD (A), RMSF (B) and RoG (C).
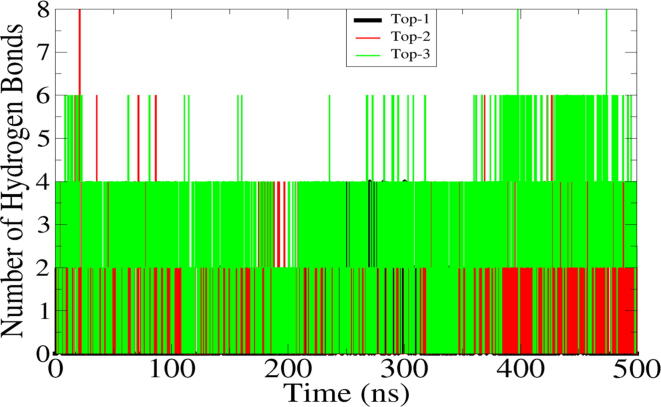
3.3. Hydrogen bonds analysis
Hydrogen bonds are critical in determining specificity of drugs binding to receptor biomolecule (Altharawi et al., 2021, Wade and Goodford, 1989). Hydrogen bonds are formed between highly electronegative atoms and are essential for stable docking of drugs to their targeted biomolecule. Hydrogen bonds analysis between the MELK and compounds is presented in Fig. 4. As can be seen that in contrast to Top-1 control, both Top-2 and Top-3 filtered compounds produced more than 2 hydrogen bonds. This explain the good intermolecular strength of the complexes. For Top-1, majority of frames reported at least 2 hydrogen bonds while some revealed 3 and 4. For Top-1, the average hydrogen bonds are 4 while some reported up to 8. In case of Top-3, the number of hydrogen bonds interactions is 5–6 in each frame.
Fig. 4.
Hydrogen bonds analysis between the MELK and compounds during simulation time. Only hydrogen bonds produced within 5 Ã… are plotted only.
3.4. MM\GBSA and MM\PBSA binding free energies
MM\GBSA and MM\PBSA are powerful and modest approaches to estimate atomic level binding free energies of protein–ligand complex (Ahmad et al., 2017, Alamri et al., 2021, Genheden and Ryde, 2015, Hou et al., 2011). The different binding free energies in kcal/mol of all three complexes are tabulated in Table 3. It can be spotted that van der Waals energy dominates the gas phase energy, followed by electrostatic energy. The van der Waals energy is −38.10 for Top-1, −25.34 for Top-2 and −29.87 for Top-3. Similarly, the electrostatic energy for Top-1, Top-2 and Top-3 is −18.00, −16.80 and −15.20, respectively. From solvation energy, the net polar solvation energy contributed non-significantly in all three systems in contrast to non-polar solvation energy that favors complex formation and remained stable during the analysis. Overall, the net binding free energy of Top-1 is −39.75 in Top-1, −30.55 in Top-2 and –32.51 in Top-3 in MM\GBSA. In MM\PBSA, the net binding free energy of Top-1, Top-2 and Top-3 is −34.61, –33.16 and –32.28, respectively. Further, entropy energy was estimated for each system. The entropy contribution is 1.34, 3.6 and 5.04 for Top-1, Top-2 and Top-3, respectively in MM\GBSA. While in MM\PBSA, the entropy energy is 2.65 (Top-1), −1.54 (Top-2) and 0.15 (Top-3).
Table 3.
Binding free energy analysis of compounds. The analysis is based on 1000 frames from simulation trajectories and presented as net values of complex, receptor and ligand. The unit of energy values given is kcal/mol.
| Energy Parameter | Top-1 Complex | Top-2 Complex | Top-3 Complex |
|---|---|---|---|
| MM\GBSA | |||
| Van der Waal energy | −38.10 | −25.34 | −29.87 |
| Electrostatic energy | −18.00 | −16.80 | −15.20 |
| Polar energy | 21.96 | 17.99 | 19.33 |
| Non polar energy | −5.61 | −6.40 | −6.77 |
| Total gas phase | −56.1 | −42.14 | −45.07 |
| Total solvation | 16.35 | 11.59 | 12.56 |
| Net energy | −39.75 | −30.55 | –32.51 |
| Entropy energy | 1.34 | 3.6 | 5.04 |
| MM\PBSA | |||
| Van der Waal energy | −38.10 | −25.34 | −29.87 |
| Electrostatic energy | −18.00 | −16.80 | −15.20 |
| Polar energy | 28.36 | 15.22 | 17.41 |
| Non polar energy | −6.87 | −6.24 | −4.62 |
| Total gas phase | −56.1 | −42.14 | −45.07 |
| Total solvation | 21.49 | 8.98 | 12.79 |
| Net energy | −34.61 | –33.16 | –32.28 |
| Entropy energy | 2.65 | −1.54 | 0.15 |
3.5. Decomposition of binding free energy per residue of MELK
The active site residues that are engaged in different types of interaction with the compounds were further subjected to residue wise binding free energy analysis (Sanober et al., 2017). In this analysis, the net energy contribution of each active pocket residue to overall net MM\GBSA binding free energy was determined as presented in Table 4. It was revealed that the majority of the active pocket residue contributed significantly in retaining the active conformation of compounds at MELK active pocket. Notably Cys70, Glu87, Tyr88, Cys89, Glu136 Asn137, Lue139, and Asp150 were found to play important role in compound binding during simulation time.
Table 4.
Residues contributing majorly to the net binding energy of compounds with MELK.
| Residue |
Compounds |
||
|---|---|---|---|
| Top-1 | Top-2 | Top-3 | |
| Ile17 | −1.25 | −0.67 | −0.55 |
| Gly18 | −0.80 | −1.12 | −1.0 |
| Thr19 | −0.63 | 0.34 | 0.40 |
| Gly20 | −1.22 | −1.41 | −0.84 |
| Val25 | −0.83 | −0.66 | −0.54 |
| Ala38 | −1.36 | −1.88 | −1.00 |
| Lys40 | −1.22 | −1.28 | −1.30 |
| Cys70 | −2.41 | −2.04 | −2.90 |
| Leu86 | −0.64 | −1.24 | −1.67 |
| Glu87 | −1.25 | −1.63 | −2.70 |
| Tyr88 | −1.05 | −1.35 | −2.47 |
| Cys89 | −3.67 | −2.35 | −1.87 |
| Glu136 | −4.25 | −3.21 | −2.55 |
| Asn137 | −3.54 | −4.01 | −5.01 |
| Leu139 | −2.31 | −1.47 | −1.58 |
| Ile149 | −1.33 | −1.0 | −1.21 |
| Asp150 | −2.61 | −3.67 | −6.31 |
3.6. WaterSwap analysis
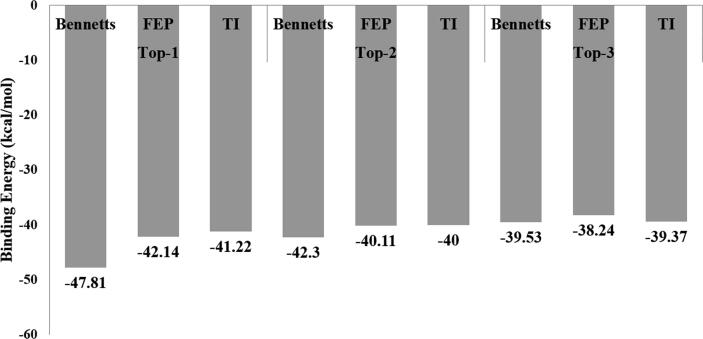
The binding free energies analysis done by MM\GBSA and MM\PBSA were re-estimated by WaterSwap to ensure accurasy in the results. WaterSwap used three methods i. e. BAR, TI and FEP for net binding free energy of complexes and is considered more relaible than MM\GBSA and MM\PBSA as it take into account the interaction energy contribution of water moelcules in protein–ligand interactions. Results of WaterSwap are given in Fig. 5. It can cearly be seen that the three algorithms depiected very stable nature of the complexes with net binding energy value < -41 kcal/mol for Top-1, < −40 kcal/mol for Top-2 and < 38 kcal/mol for Top-3. These findings confirm that the intermolecular binding affinity is very high and compounds binding mode and interactions are very stable.
Fig. 5.
WaterSwap analysis of compounds. All the energy values are presented in kcal/mol.
3.7. Drug likeness and pharmacokinetics
Further investigation of compounds oral bioavailability and absorption, distribution, metabolism, excretion as well as toxicity was conducted (Table 5) (Lombardo et al., 2017). This was vital to understand and extremely important for compounds success to reach the market. For drug likeness, several drug like rules were applied: for example Lipinski rule of five (Lipinski, 2004), Ghose (Ghose et al., 1999), Veber (Veber et al., 2002), Egan (Egan et al., 2000) and Muegge (Muegge et al., 2001). The filtered compounds (Top-2 and Top-3) are revealed to follow all the mentioned rule except Ghose while the control (Top-1) complete the parameters of all drug rules. The drug like rules ensure the selected compounds are good candidates in term of solubility, metabolic stability, permeability, transportations, clearance from the body and in vitro pharmacology. The compounds are moderable soluble, which is important from drug design perspective as it ensure high concentration of drugs can be reached the target site of action. The gastrointestinal absorption of the compounds is also high which ensured that the compound can reach the target organ in good concentration. The compounds also have zero alert for PAINs which mean that the compounds have selective binding to only one biological target and avoid non-specific interactions (Whitty, 2011). The compounds were also unveiled no major toxicity in different assays and are also non mutagenic.
Table 5.
Evaluation of compounds drug likeness and ADMET properties.
| Property |
Compound |
||
|---|---|---|---|
| Physicochemical Properties | Top-1 | Top-2 | Top-3 |
| Formula | C27H21N3O5 | C27H35N3O4 | C29H29N3O3 |
| Molecular weight | 467.47 g/mol | 465.58 g/mol | 467.56 g/mol |
| Heavy atoms | 35 | 34 | 35 |
| Aromatic heavy atoms | 23 |
17 |
22 |
| Fraction Csp3 |
0.22 |
0.44 | 0.28 |
| Rotatable bonds |
2 |
10 |
8 |
| H-bond acceptors | 5 | 6 | 5 |
| H-bond donors | 3 | 1 | 0 |
| Molar Refractivity | 132.11 | 136.45 | 140.42 |
| TPSA | 101.64 Å2 | 68.98 Å2 | 64.55 Å2 |
| Lipophilicity | |||
| Consensus Log Po/w | 3.04 |
3.44 | 4.26 |
| Water Solubility | Moderately soluble | Moderately soluble | Moderately soluble |
| Pharmacokinetics | |||
| GI absorption |
High |
High |
High |
| BBB permeant |
No |
Yes | Yes |
| P-gp substrate |
Yes |
Yes | Yes |
| CYP1A2 inhibitor | No | No | No |
| CYP2C19 inhibitor | Yes | No | Yes |
| CYP2C9 inhibitor | Yes | No | Yes |
| CYP2D6 inhibitor | No | Yes | Yes |
| CYP3A4 inhibitor | No | Yes | Yes |
| Log Kp (skin permeation) | −6.99 cm/s | −6.63 cm/s | −5.78 cm/s |
| Druglikeness | |||
| Lipinski | Yes | Yes; 0 violation | Yes; 0 violation |
| Ghose |
No |
No | No |
| Veber | Yes | Yes | Yes |
| Egan |
Yes |
Yes | Yes |
| Muegge |
No |
Yes | Yes |
| Bioavailability Score | 0.55 | 0.55 | 0.55 |
| Medicinal Chemistry | |||
| PAINS | 0 alert | 0 alert | 0 alert |
| Brenk | 0 alert | 0 alert | 0 alert |
| Synthetic accessibility | 4.77 | 3.96 | 3.66 |
| Toxicity | |||
| Hepatotoxicity | Yes | No | No |
| Skin Sensitisation | No | No | No |
|
T. pyriformis toxicity |
No | No | No |
| AMES toxicity | No | No | No |
| Minnow toxicity | No | No | No |
| Carcino mouse | Negative | Negative | Negative |
| Excretion | |||
| Total Clearance |
−0.067 |
0.078 | 0.082 |
| Renal OCT2 substrate | No | No | No |
4. Discussion
The MELK major role in activation of pathways key for tumor aggressive growth and resistance to treatment in many adult cancers make it a central target for therapeutic intervention (Jiang and Zhang, 2013). Considering the published literature reporting the MELK as promising drug target, many efforts have been done to investigate promising drug molecules to interfere with the biological function of this protein. The protein was targeted by OTS167 molecule in neuroblastoma growth and found that the drug molecule hinder the neuroblastoma xenografts. In another study, it was reported that MELK was evaluated for its role in diffuse large B cell lymphoma and mantle cell lymphoma. The study revealed increase expression of MELK on both of the mentioned cancer types and is associated with worse clinical outcomes. Further, they demonstrated that the treatment of the said cancers with OTSSP157 prolonged survival of the mice model suggesting strong anti-lymphoma activities of the compounds both in vivo and in vitro. Together, findings of the study found MELK as potential new drug target for treating aggressive B cell malignancies (Maes et al., 2019). Another study reported that biological function and molecular events of MELK in esophageal squamous cell carcinoma, which is common gastro malignancy and correlated to high mortality rate around the world. The study found that MELK enhances cancer cells migration, tumorigenesis, invasion and metastasis of esophageal squamous cell carcinoma and activates FOCm1 signaling pathway (Chen et al., 2020). Due to these findings, the study suggest MELK as therapeutic target for esophageal squamous cell carcinoma patients even those suffering form advanced stage cancer. Recently, several computational studies have been conducted against several cancer targets. In one study, β-bourbonene, a compound from a traditional medicinal plant Ficus carica L, has been revealed to show promising biological activity against different cancer targets such as B-cell lymphoma 2 (Bcl-2), cyclin-dependent kinase 2 (CDK-2), cyclin-dependent kinase 6 (CDK-6), and vascular endothelial growth factor receptor 2 (VEGFR-2) (Gurung et al., 2021). In another study, taxifolin natural compound was unveiled as promising binding molecule of protein kinases B (AKT1) protein and thus predicted to inhibit ovarian cancerous cells (Ajjarapu et al., 2021).
In this current study, we have employed computational approaches to identify potential inhibitory molecules that can block the function of MELK. The compounds have good potency in term of binding to the MELK and revealed stable dynamics at the active site of MELK. To the best of our understanding, both these compounds are not reported before and not tested experimentally against MELK. Thus, we believe that findings of our study will speed up drug development against MELK and the screened compounds may serve as lead structure for further derivatives development.
5. Conclusions
In this study, two drug molecules were identified as Top-2 and Top-3 compared to the control co-crystalized Top-1 inhibitor as high affinity binding MELK molecules. The filtered molecules showed binding to the same binding pocket and interact with same set of hotspot residues. The stable conformation of the compounds at MELK binding site is investigated due to a balanced network of multiple hydrogen bonds. The compounds also revealed highly favorable net binding free energies and non-favorable entropy energy contribution. Several active pocket amino acids were revealed that contributed in stable compounds conformation with the MELK. From pharmacokinetics perspective, the compounds are drug like molecules and fulfill the parameters of most drug like rules and also revealed good pharmacokinetic properties. The compounds are also found non-toxic and non-mutagenic. Despite the promising findings of the study, one limitation of the study is the lack of experimental validation. However, we believe the outcomes of the study speed up drug discovery against MELK and thus might provide novel lead molecules that can be structural optimized for derivatives with enhanced biological activities.
Funding
This research received no external funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2022.103335.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data Availability Statement.
The data presented in this study are available within the article.
References
- Ahmad F., Albutti A., Tariq M.H., Din G., Tahir ul Qamar M., Ahmad S. Discovery of Potential Antiviral Compounds against Hendra Virus by Targeting Its Receptor-Binding Protein (G) Using Computational Approaches. Molecules. 2022;27(2):554. doi: 10.3390/molecules27020554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S., Raza S., Uddin R., Azam S.S. Comparative subtractive proteomics based ranking for antibiotic targets against the dirtiest superbug: Acinetobacter baumannii. J. Mol. Graph. Model. 2018;82:74–92. doi: 10.1016/j.jmgm.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Raza S., Uddin R., Azam S.S. Binding mode analysis, dynamic simulation and binding free energy calculations of the MurF ligase from Acinetobacter baumannii. J. Mol. Graph. Model. 2017;77:72–85. doi: 10.1016/j.jmgm.2017.07.024. [DOI] [PubMed] [Google Scholar]
- Ajjarapu S.M., Tiwari A., Taj G., Singh D.B., Singh S., Kumar S. Simulation studies, 3D QSAR and molecular docking on a point mutation of protein kinase B with flavonoids targeting ovarian Cancer. BMC Pharmacol. Toxicol. 2021;22:1–23. doi: 10.1186/s40360-021-00512-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamri M.A., Tahir ul Qamar M., Afzal O., Alabbas A.B., Riadi Y., Alqahtani S.M. Discovery of anti-MERS-CoV small covalent inhibitors through pharmacophore modeling, covalent docking and molecular dynamics simulation. J. Mol. Liq. 2021;330:115699. doi: 10.1016/j.molliq.2021.115699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altharawi A., Ahmad S., Alamri M.A., Tahir ul Qamar M. Structural insight into the binding pattern and interaction mechanism of chemotherapeutic agents with Sorcin by docking and molecular dynamic simulation. Colloids Surf. B Biointerf. 2021;112098 doi: 10.1016/j.colsurfb.2021.112098. [DOI] [PubMed] [Google Scholar]
- Biovia D.S. San Diego; CA, USA: 2017. Discovery studio visualizer. [Google Scholar]
- Boutros R., Lobjois V., Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer. 2007;7(7):495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., et al. The FF14SB force field. Amber. 2014;14:29–31. [Google Scholar]
- Case, D.A., Belfon, K., Ben-Shalom, I., Brozell, S.R., Cerutti, D., Cheatham, T., Cruzeiro, V.W.D., Darden, T., Duke, R.E., Giambasu, G., et al., 2020. Amber 2020.
- Chen L., Wei Q., Bi S., Xie S. Maternal embryonic leucine zipper kinase promotes tumor growth and metastasis via stimulating FOXM1 signaling in esophageal squamous cell carcinoma. Front. Oncol. 2020;10:10. doi: 10.3389/fonc.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Chemical Biology. Springer; 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- Dickson C.J., Rosso L., Betz R.M., Walker R.C., Gould I.R. GAFFlipid: a General Amber Force Field for the accurate molecular dynamics simulation of phospholipid. Soft Matter. 2012;8(37):9617. doi: 10.1039/c2sm26007g. [DOI] [Google Scholar]
- Egan W.J., Merz, K.M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- Fakhri B., Kahl B. Current and emerging treatment options for mantle cell lymphoma. Ther. Adv. Hematol. 2017;8(8):223–234. doi: 10.1177/2040620717719616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller S.E., Zhang Y., Pastor R.W., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103(11):4613–4621. [Google Scholar]
- Fischer M., Quaas M., Nickel A., Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget. 2015;6(39):41402–41417. doi: 10.18632/oncotarget.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S., Kuhn O., Mikulskis P., Hoffmann D., Ryde U. The normal-mode entropy in the MM/GBSA method: effect of system truncation, buffer region, and dielectric constant. J. Chem. Inf. Model. 2012;52(8):2079–2088. doi: 10.1021/ci3001919. [DOI] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M., Farooq S. Molecular docking and dynamics simulation study of bioactive compounds from Ficus carica L. with important anticancer drug targets. PLoS One. 2021;16(7):e0254035. doi: 10.1371/journal.pone.0254035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, T., Wang, J., Li, Y., Wang, W., 2011. Assessing the Performance of the MM_PBSA and MM_GBSA Methods. 1. The Accuracy.pdf 69–82. [DOI] [PMC free article] [PubMed]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iqbal S., Shamim A., Azam S.S., Wadood A. Identification of potent inhibitors for chromodomain-helicase- DNA-binding protein 1-like through moleculardocking studies. Med. Chem. Res. 2016;25(12):2924–2939. doi: 10.1007/s00044-016-1712-x. [DOI] [Google Scholar]
- Islam S., Hosen M.A., Ahmad S., ul Qamar M.T., Dey S., Hasan I., Fujii Y., Ozeki Y., Kawsar S.M.A. Synthesis, antimicrobial, anticancer activities, PASS prediction, molecular docking, molecular dynamics and pharmacokinetic studies of designed methyl α-D-glucopyranoside esters. J. Mol. Struct. 2022;1260:132761. doi: 10.1016/j.molstruc.2022.132761. [DOI] [Google Scholar]
- Jiang P., Zhang D. Maternal embryonic leucine zipper kinase (MELK): a novel regulator in cell cycle control, embryonic development, and cancer. Int. J. Mol. Sci. 2013;14(11):21551–21560. doi: 10.3390/ijms141121551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, M., 2002. Molecular dynamics simulations of biomolecules. [DOI] [PubMed]
- Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.-A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., Schoof M., Canevari G., Casale E., Depaolini S.R., Feuchtinger A., Wu Z., Schmidt T., Rueckert L., Becker W., Huenges J., Garz A.-K., Gohlke B.-O., Zolg D.P., Kayser G., Vooder T., Preissner R., Hahne H., Tõnisson N., Kramer K., Götze K., Bassermann F., Schlegl J., Ehrlich H.-C., Aiche S., Walch A., Greif P.A., Schneider S., Felder E.R., Ruland J., Médard G., Jeremias I., Spiekermann K., Kuster B. The target landscape of clinical kinase drugs. Science. 2017;358(6367) doi: 10.1126/science:aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler V., van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22(5):501–508. [Google Scholar]
- Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010;98(5):861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li Z., Guo T., Xing X.-F., Cheng X., Du H., Wen X.-Z., Ji J.-F. Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget. 2016;7(5):6266–6280. doi: 10.18632/oncotarget.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionta E., Spyrou G., Vassilatis K.D., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr. Top. Med. Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lobanov M.Y., Bogatyreva N.S., Galzitskaya O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008;42(4):623–628. [PubMed] [Google Scholar]
- Lombardo F., Desai P.V., Arimoto R., Desino K.E., Fischer H., Keefer C.E., Petersson C., Winiwarter S., Broccatelli F. In Silico Absorption, Distribution, Metabolism, Excretion, and Pharmacokinetics (ADME-PK): Utility and Best Practices. An Industry Perspective from the International Consortium for Innovation through Quality in Pharmaceutical Development: Miniperspective. J. Med. Chem. 2017;60:9097–9113. doi: 10.1021/acs.jmedchem.7b00487. [DOI] [PubMed] [Google Scholar]
- Lyu C., Chen T., Qiang B., Liu N., Wang H., Zhang L., Liu Z. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021;49:D509–D515. doi: 10.1093/nar/gkaa763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes A., Maes K., Vlummens P., De Raeve H., Devin J., Szablewski V., De Veirman K., Menu E., Moreaux J., Vanderkerken K., De Bruyne E. Maternal embryonic leucine zipper kinase is a novel target for diffuse large B cell lymphoma and mantle cell lymphoma. Blood Cancer J. 2019;9(12) doi: 10.1038/s41408-019-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchaca T.M., Juárez-Portilla C., Zepeda R.C. Drug Discovery and Development-New Advances. IntechOpen; 2020. Past, Present, and Future of Molecular Docking. [Google Scholar]
- Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8(9):3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Muegge I., Heald S.L., Brittelli D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001;44(12):1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- Muneer I., Ahmad Sajjad, Naz A., Abbasi S.W., Alblihy A., Aloliqi A.A., Alkhayl F.F., Alrumaihi F., Ahmad S., El Bakri Y. Discovery of Novel Inhibitors from Medicinal Plants for V-Domain Ig Suppressor of T-Cell Activation (VISTA) Front. Mol. Biosci. 2021:951. doi: 10.3389/fmolb.2021.716735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Sanober G., Ahmad S., Azam S.S. Identification of plausible drug targets by investigating the druggable genome of MDR Staphylococcus epidermidis. Gene Reports. 2017;7:147–153. [Google Scholar]
- Sun H., Ma H., Zhang H., Ji M. Up-regulation of MELK by E2F1 promotes the proliferation in cervical cancer cells. Int. J. Biol. Sci. 2021;17(14):3875–3888. doi: 10.7150/ijbs.62517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J.L., Lin D., Jiang J., Manning N.O., Prilusky J., Ritter O., Abola E.E. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54(6):1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- Tahir ul Qamar M., Ahmad S., Fatima I., Ahmad F., Shahid F., Naz A., Abbasi S.W., Khan A., Mirza M.U., Ashfaq U.A., Chen L.-L. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104389. [DOI] [PubMed] [Google Scholar]
- Thangaraj K., Ponnusamy L., Natarajan S.R., Manoharan R. MELK/MPK38 in cancer: from mechanistic aspects to therapeutic strategies. Drug Discovery Today. 2020;25(12):2161–2173. doi: 10.1016/j.drudis.2020.09.029. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, P.J., 2005. XMGRACE, Version 5.1. 19. Cent. Coast. Land-Margin Res. Oregon Grad. Inst. Sci. Technol. Beaverton, OR.
- Tahir ul Qamar M., Ahmad S., Khan A., Mirza M.U., Ahmad S., Abro A., Chen L.-L., Almatroudi A., Wei D.-Q. Structural probing of HapR to identify potent phytochemicals to control Vibrio cholera through integrated computational approaches. Comput. Biol. Med. 2021;138:104929. doi: 10.1016/j.compbiomed.2021.104929. [DOI] [PubMed] [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Wade R.C., Goodford P.J. The role of hydrogen-bonds in drug binding. Prog. Clin. Biol. Res. 1989;289:433–444. [PubMed] [Google Scholar]
- Whitty A. Growing PAINS in academic drug discovery. Future Med. Chem. 2011;3(7):797–801. doi: 10.4155/fmc.11.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C.J., Malaisree M., Michel J., Long B., McIntosh-Smith S., Mulholland A.J. Rapid decomposition and visualisation of protein-ligand binding free energies by residue and by water. Faraday Discuss. 2014;169:477–499. doi: 10.1039/c3fd00125c. [DOI] [PubMed] [Google Scholar]
- Yu W., MacKerell A.D. Computer-aided drug design methods. Antibiotics. Springer. 2017:85–106. doi: 10.1007/978-1-4939-6634-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- Emmerich C.H., Gamboa L.M., Hofmann M.C.J., Bonin-Andresen M., Arbach O., Schendel P., Gerlach B., Hempel K., Bespalov A., Dirnagl U., Parnham M.J. Improving target assessment in biomedical research: the GOT-IT recommendations. Nat. Rev. Drug Discov. 2021;20(1):64–81. doi: 10.1038/s41573-020-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Jalal S.I., Sledge G.W., Meroueh S.O. Small-molecule binding sites to explore protein–protein interactions in the cancer proteome. Mol. Biosyst. 2016;12(10):3067–3087. doi: 10.1039/c6mb00231e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available within the article.