Abstract
A bioreporter was made containing a tfdRPDII-luxCDABE fusion in a modified mini-Tn5 construct. When it was introduced into the chromosome of Ralstonia eutropha JMP134, the resulting strain, JMP134-32, produced a sensitive bioluminescent response to 2,4-dichlorophenoxyacetic acid (2,4-D) at concentrations of 2.0 μM to 5.0 mM. This response was linear (R2 = 0.9825) in the range of 2.0 μM to 1.1 × 102 μM. Saturation occurred at higher concentrations, with maximal bioluminescence occurring in the presence of approximately 1.2 mM 2,4-D. A sensitive response was also recorded in the presence of 2,4-dichlorophenol at concentrations below 1.1 × 102 μM; however, only a limited bioluminescent response was recorded in the presence of 3-chlorobenzoic acid at concentrations below 1.0 mM. A significant bioluminescent response was also recorded when strain JMP134-32 was incubated with soils containing aged 2,4-D residues.
The herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) is widely used in both agricultural and domestic weed control applications. While it is rapidly degraded in most environments, the initial step in the degradation of 2,4-D is a dioxygenase-mediated attack on the acetic acid moiety, yielding glyoxylate and 2,4-dichlorophenol (DCP) (13). At concentrations ranging from 120 to 250 μM, DCP is known to be toxic to 2,4-D degraders and other microorganisms (8, 28), giving rise to concern over the fate of 2,4-D in the environment. As both 2,4-D and DCP are moderately nonpolar molecules, they have a tendency to partition into organic matter. This reduction in bioavailability is difficult to assess with traditional analytical approaches but is an important factor affecting the longevity of these compounds in the environment.
Bioreporters are being increasingly used as a nondestructive means of assaying gene expression, thereby allowing the assessment of biologically relevant analyte concentrations. Analysis of gene expression typically relies on transcriptional fusions between a promoter of interest and a reporter gene. Commonly used reporter genes include lacZ, gfp, luxAB, and luxCDABE. Use of the entire luxCDABE gene cassette has been extensive (1, 2, 7, 17, 22, 35, 36) because such reporters do not require the addition of an exogenous substrate for signal production. The bioluminescent signal generated by luxCDABE fusions is typically short-lived, thus allowing for repetitive sampling under dynamic conditions. Similar bioreporters have recently been shown to be compatible with emerging signal detection technologies, such as integrated circuits capable of processing and communicating signal input (39). We report here on the development of a bioluminescent reporter for the detection of 2,4-D degradation in aqueous samples and demonstrate its use in slurries containing aged 2,4-D residues.
Strain construction.
Ralstonia (formerly Alcaligenes) (44) eutropha JMP134 contains plasmid pJP4, which encodes all the enzymes involved in the metabolism of 2,4-D. This plasmid and the associated enzymes have been well characterized (10, 26, 27, 30, 41). As transcription of the genes associated with 2,4-D degradation is known to be inducible (12, 23), construction of a functional bioreporter was deemed feasible. To construct such a reporter both promoter and regulatory elements were selected from pJP4 and fused to promoterless lux reporter genes (33).
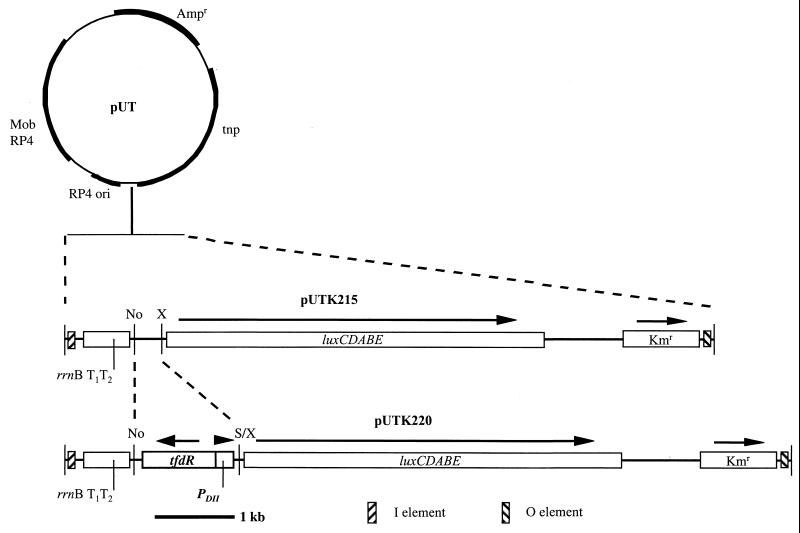
The tfdDII promoter (tfdPDII) was chosen because it has two domains that are identical to those of the promoter of tfdA (27) but does not contain a piece of the ISJP4 insertion element, which has interrupted the tfdA promoter region (24). To fully activate transcription of the genes encoding 2,4-D-degradative enzymes, TfdR, a LysR-type DNA binding protein, is required (25). In order to efficiently incorporate both of these elements into the construct, a 1.1-kb fragment containing both tfdPDII and tfdR was amplified from the plasmid pJP4. Amplification of this fragment was facilitated by two PCR primers made using a DNA 1000 Oligo Synthesizer (Beckman, Fullerton, Calif.). The first primer, GCGGCCGCCTATTTCTGTCCTTTCCCGCG, targeted the region just downstream of the 3′ end of tfdR and contained an introduced NotI site on its 5′ end (underlined). The second primer, ACTAGTCGCAGCGGCAGATCG, was targeted to the 5′ end of tfdDII and contained an SpeI site on its 5′ end (underlined). PCR was achieved using a modified touchdown protocol (34) in a PT200 thermocycler (MJ Research, Watertown, Mass.). After an initial 5-min denaturation at 95°C, 10 cycles were executed in which the denaturing, annealing, and extension times and temperatures were 95°C for 30 s, 65°C for 1 min, and 72°C for 2 min. During these first 10 cycles the annealing temperature was lowered by 1°C per cycle. An additional 20 cycles were then executed with an annealing temperature of 55°C. The resulting amplicon containing the promoter sequence upstream of tfdDII and tfdR in its entirety was cloned into pCR2.1-TOPO using a TOPO TA cloning kit from Invitrogen (Carlsbad, Calif.). The resulting plasmid, pTFDR, was digested with NotI and SpeI, yielding a 1.1-kb fragment which was ligated into pUTK215 (20), a modified mini-Tn5 suicide delivery system containing a promoterless luxCDABE cassette from Vibrio fischeri (33) that was previously digested with NotI and XbaI. The ligation mix was transformed into electrocompetent Escherichia coli S17(λpir) (9) using an Electroporation System Electro Cell Manipulator 600 (BTX, San Diego, Calif.) according to the manufacturer's instructions. Transformants were selected on Luria-Bertani plates containing kanamycin (50 mg/liter). Plasmid DNA was isolated from the transformants and analyzed using restriction endonucleases to confirm the presence of the tfdPDII-tfdR fragment. A plasmid containing this insert was named pUTK220 (Fig. 1). In all cases plasmid and chromosomal DNA were isolated and enzymatically modified according to procedures outlined by Ausubel et al. (4).
FIG. 1.
Construction of mini-Tn5 tfd-lux suicide vector pUTK220 from pUTK215, a pUT derivative (18). No, NotI; X, XbaI; S/X, SpeI-XbaI heterologous cloning sites; tnp, transposase; rrnB T1T2, transcriptional terminators from E. coli rrnB (2); Ampr, ampicillin resistance; Kmr, kanamycin resistance. RP4 ori, replication origin of RP4, Mob RP4, mobilization region of RP4.
E. coli SV17(λpir) pUTK220 was mated with R. eutropha JMP134. Transformants were plated on minimal salts medium (MSM) containing 2.25 mM 2,4-D and kanamycin (50 mg/liter) (40). Colonies able to grow on 2,4-D in the presence of kanamycin were transferred to Luria-Bertani plates containing 2.25 mM 2,4-D and were inspected in the dark for the production of light. Two transformants producing enough light to be visible in a dark room were further analyzed with regard to the kinetics of light production in the presence of 2,4-D at various concentrations (data not shown). Of these two, a transformant designated strain JMP134-32 was chosen for further analyses.
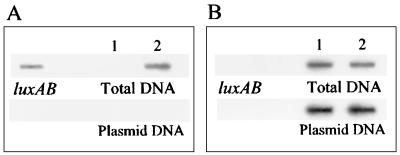
To ascertain whether transposition from pUTK220 had occurred in the chromosome or into the endogenous plasmid pJP4, both total DNA and plasmid DNA were loaded independently onto a Biotrans nylon membrane from ICN (Irvine, Calif.) using a Bioslot apparatus from Bio-Rad (Hercules, Calif.). The membrane was first hybridized with a 32P-labeled, PCR-generated luxAB probe amplified from V. fischeri (2) and was then stripped and reprobed with a 32P-labeled, PCR-generated tfdC probe amplified from pJP4 (31). Blots were hybridized and washed as described previously (1) and were then visualized on a Storm 840 PhosphorImager from Molecular Dynamics (Sunnyvale, Calif.). Results from these slot blot hybridizations (Fig. 2) demonstrated that transposition from pUTK220 resulted in insertion of the tfdRPDII-luxCDABE fusion into the chromosome of JMP134-32.
FIG. 2.
Slot blot analysis of DNA from strain JMP134 (lanes 1) and strain JMP134-32 (lanes 2). The blot was probed with a 32P-labeled luxAB fragment (A), stripped, and then reprobed with a 32P-labeled tfdC fragment (B).
Construct stability was assessed by repeated passages of JMP134-32 on nonselective YEPG medium (1). After 10 passages or approximately 65 generations, the culture was serially diluted and spread onto YEPG plates amended with kanamycin (50 mg/liter). Colony hybridizations (3) were then performed using the luxAB probe mentioned above and visualized using the Storm 840 PhosphorImager. All colonies probed positive for the lux genes, demonstrating maintenance of the construct in recombination-proficient JMP134-32 without selection.
Bioluminescent response.
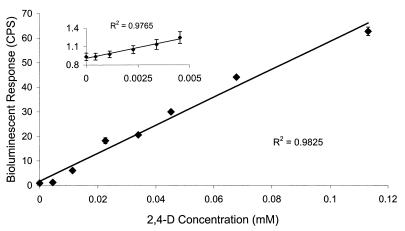
The bioluminescent response to 2,4-D was determined using a growing-cell assay that has been described previously (17). Briefly, JMP134-32 was grown to an optical density at 600 nm of 0.35 in YEPG at 28°C. Strain JMP134-32 was exposed to 2,4-D (98% pure) from Aldrich Chemical Company (Milwaukee, Wis.) by adding 50 μl of the YEPG-grown cells to 50 μl of MSM containing 2,4-D at various concentrations. The 2,4-D additions were serially diluted into MSM from a 5.0 mM stock dissolved in MSM. Light was measured in quadruplicate, using static opaque 96-well plates, in a Wallac 1450 Microbeta Plus liquid scintillation counter (Wallac, Turku, Finland) at room temperature. Preliminary experiments showed that 60- to 100-min incubations were sufficient to provide a consistent light response. The mean light response to 2,4-D concentrations from 0.0 μM to 1.1 × 102 μM was obtained after 100 min of incubation and is plotted (Fig. 3). There was a statistically significant (P < 0.05) linear bioluminescent response by R. eutropha JMP134-32 to increasing concentrations of 2,4-D (from 2.0 μM to approximately 112 μM) (R2 = 0.9825). At concentrations above 112 μM and up to 1.25 mM, the response appeared to follow saturation kinetics, with a maximal bioluminescent response induced by 1.25 mM 2,4-D (Fig. 4).
FIG. 3.
Bioluminescent response of strain JMP134-32 to various concentrations of 2,4-D. Inset shows response to low-level concentrations and is plotted on a different scale. CPS, luminescence counts per second.
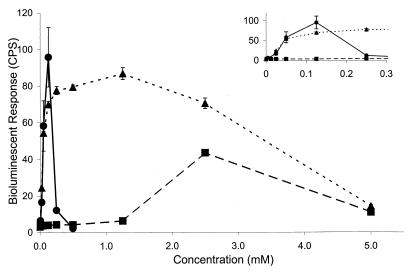
FIG. 4.
Comparison of the bioluminescent response of strain JMP134-32 to those of different compounds known to induce the genes involved in 2,4-D degradation: 3-CB, ■; DCP, ●; and 2,4-D, ▴. Inset shows response to low-level concentrations and is plotted on a different scale.
Toxic effects, as measured by a 10 to 20% reduction in optical density at 600 nm, were observed at 2,4-D concentrations greater than 1.25 mM. The observed decrease in optical density is consistent with published reports which demonstrate that the accumulation of DCP during 2,4-D degradation in more complex media can be toxic (8, 23). The decrease in optical density alone, however, cannot explain the magnitude of the decrease in light output observed at 2,4-D concentrations above 1.25 mM (Fig. 4). The mechanism of DCP toxicity is assumed to be the same as that for other chlorinated phenols which are known to act as uncouplers of electron transport and can make membranes permeable to small molecules, such as ATP (15, 23). It is therefore possible that decreased light output at higher 2,4-D concentrations (Fig. 4) results in part from the rapid loss of ATP to the extracellular milieu when DCP accumulates and disrupts the membrane. ATP is a reactant required for light production (39), and its loss to the supernatant would dramatically reduce both cellular energy level and total light output.
R. eutropha JMP134-32 was also incubated with several other compounds to determine the range of inducing substrates. DCP, 3-chlorobenzoate, and benzoate were 99% pure, while 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) was 97% pure. The chemicals listed above were obtained from Aldrich. 4-Nitrophenoxyacetic acid was obtained from TCI America (Portland, Oreg.) and was 98% pure. Light production was not induced by 2,4,5-T during the same time period (0 to 4 h). Bioluminescence was not observed in the presence of either benzoic acid or 4-nitrophenoxyacetic acid but was induced in the presence of both DCP and 3-chlorobenzoic acid (3-CB). The bioluminescent response to 3-CB was measured in the same manner as was that induced by 2,4-D. However, the volatile nature of DCP precluded its measurement using the Wallac 1450. DCP was therefore measured in triplicate by placing sealed sample vials into a light-tight box connected to a Detection System 7070 photomultiplier (Oriel, Stratford, Conn.) via a liquid light pipe (17). Light from samples containing 100, 75, 50, or 25 μM 2,4-D was also measured using the Oriel System 7070. As responses to 2,4-D measured by both instruments were linear in this range, a conversion factor was derived that allowed the direct normalization of Oriel-derived DCP data to output from the Wallac 1450. The means of the normalized data obtained after 100 min of incubation are plotted (Fig. 4).
Not surprisingly, the bioluminescent response of strain JMP134-32 to low levels of DCP was very similar to its response to 2,4-D. DCP is known to be an intermediate in the metabolism of 2,4-D (11) and has been used as an inducing agent for increasing the production of enzymes involved in 2,4-D degradation (26). This is likely due to its conversion to 2,4-dichloromuconate, a breakdown product of both 2,4-D and DCP and a known inducing agent of the 2,4-D pathway (12). Strain JMP134-32 showed a statistically significant increase in light production concomitant with increasing DCP concentrations from 1.0 μM to 1.1 × 102 μM. However, toxic effects were observed at much lower concentrations of DCP than with 2,4-D. These toxic effects are consistent with published observations concerning the effects of DCP concentrations greater than 200 μM on 2,4-D-degrading organisms (28).
Given the strong bioluminescent response of strain JMP134-32 to 2,4-D and DCP, the limited response to 3-CB, especially at low concentrations, was surprising (Fig. 4). Although light production was statistically greater than the control at 3-CB concentrations of 12 μM, the amount of light was not statistically significantly different from that produced in the presence of 3-CB concentrations as high as 5.0 × 102 μM. More dramatic increases in light production did not occur in the presence of 3-CB until concentrations exceeded 1.0 mM, with maximal measured light production occurring in the presence of 2.5 mM 3-CB (Fig. 4).
Light production in the presence of 1.2 mM 3-CB was approximately twofold greater than background levels. This is consistent with the findings of Leveau and van der Meer (25), who reported a two- to threefold increase in 3,5-dichlorocatechol dioxygenase activity toward 3-chlorocatechol by cells containing a functional regulatory protein (TfdR) grown in the presence of 1.0 mM 3-CB. When knockout mutants of JMP134 unable to degrade the inducer, 2,4-dichloromuconate, were grown in the presence of 2.25 mM 2,4-D, Filer and Harker (12) were also able to detect an approximately twofold increase in transcription of the tfdCDEF operon.
The bioluminescent response of strain JMP134-32 in the presence of approximately 2.25 mM 2,4-D was more than 20-fold higher than the background level and 10-fold higher than that reported by Filer and Harker (12). Comparisons between different reporter systems are difficult to make due to possible differences in the stability of the reporter transcript, the strength of the ribosomal binding site, and the longevity of the protein (43). However, it is unlikely that the observed differences between the reporter constructs are related to the strengths of the different promoters, as Leveau et al. (23) have recently shown that the relative abundance of mRNA transcribed from the tfdC and tfdDII promoters is not dramatically different. Although the reasons for differences in signal strength are unclear, the low signal-to-noise ratio in response to 2,4-D, combined with the easily measured bioluminescent signal, makes JMP134-32 a candidate for use as a bioreporter in more complex substrates than just minimal media.
To assess the usefulness of JMP134-32 in more environmentally relevant samples, bioluminescence assays were performed on soil extracts and soil slurries from Agent Orange-contaminated soils. Soil was obtained from a loading-unloading area at Hardstand 7, Eglin Air Force Base (Niceville, Fla.). To determine the concentrations of the contaminants, soil was extracted by shaking it for 30 min with a 1:1 mixture of hexane and acetone. The extracts were analyzed using a Supelcosil LC-18-T reverse-phase C18 column (Supelco, Bellefonte, Pa.) attached to a binary LC 250 series pump (Perkin-Elmer, Foster City, Calif.) and a Perkin-Elmer model LC-235 diode array detector. Data analysis was achieved using the Turbochrome version 4.1 software package (Perkin-Elmer, PE Nelson Division, San Jose, Calif.). Samples were eluted at a flow rate of 1.0 ml/min with a 0.025% H3PO4–acetonitrile gradient, as follows: 0% acetonitrile (isocratic, 5.0 min), 0 to 50% acetonitrile (5.0-min ramp), 50% acetonitrile (isocratic, 20 min), and 50 to 0% acetonitrile (5.0-min ramp). Absorbance of eluted compounds was monitored at 210 nm.
High-performance liquid chromatography (HPLC) analysis revealed the soil to have 22.4-mg/kg and 73.8-mg/kg concentrations of 2,4-D and 2,4,5-T, respectively, while DCP and 2,4,5-trichlorophenol concentrations were both found to be less than 1 mg/kg (31). Initial experiments involved washing 5.0 g of soil with 5.0 ml of MSM. The slurries were centrifuged at 10,000 × g for 10 min after being shaken for 24, 48, or 120 h. Then 100 μl of the supernatant was added to 100 μl of JMP134-32 that had been grown in YEPG medium to an optical density at 600 nm of 0.35. Cells were also added to a negative control consisting of MSM alone and a positive control consisting of MSM plus 2,4-D (20 mg/liter). No bioluminescence was observed in the soil extracts. HPLC analysis of these soil washings revealed trace levels of 2,4-D and 2,4,5-T that were below the HPLC quantitation limit (2.3 × 102 nM) and below the level required to induce a significant bioluminescent response in MSM (2.0 μM).
Under the assumption that cells added directly to a soil slurry may act as a sink for 2,4-D (5), JMP134-32 was grown to an optical density at 600 nm of 0.5 in YEPG and centrifuged at 10,000 × g for 10 min. The cells were then resuspended in 0.5 volume of MSM to a final optical density at 600 nm of 1.0 and were added to soil (1.0 ml of cells/g of soil). Triplicate 100-μl samples of the slurry were monitored for bioluminescence as described above. To control for nonspecific induction of bioluminescence, the procedure described above was repeated using a previously characterized bioreporter, Pseudomonas fluorescens 5RL, which is insensitive to 2,4-D (19). No bioluminescence was observed when 5RL was added to the contaminated soil (data not shown).
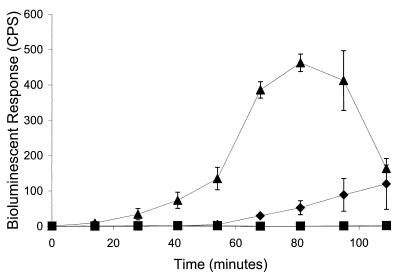
Although there appeared to be a significant lag compared to the performance of the aqueous positive control, a significant bioluminescent response was observed after the addition of JMP134-32 to 2,4-D-contaminated soil (Fig. 5). A similar lag in bioluminescent response was observed when P. fluorescens HK44, a bioluminescent reporter bacterium responsive to naphthalene, was incubated with naphthalene-contaminated soils (21). The authors suggested that the lag was due to the quenching of emitted light by soil particles in the slurry. Since the soil-washing experiment demonstrated that 2,4-D and DCP concentrations in the extracts were below the levels required for significant light induction (>2.0 μM), the bioluminescent signal recorded most likely resulted from metabolism of 2,4-D that had partitioned from the soil to the cells. The observed lag may therefore indicate that desorption of 2,4-D from the soil is a rate-limiting step. This is consistent with the findings of Ogram et al., who demonstrated that only solution-phase 2,4-D was readily degraded in soil slurries (29).
FIG. 5.
Bioluminescent response of strain JMP134-32 to Agent Orange-contaminated soil, ⧫; MSM containing 90 μM 2,4-D, ▴; and MSM control, ■.
The decrease in bioluminescence observed in the aqueous control after 80 min (Fig. 5) was not seen during the initial experiments that used a much lower concentration of cells (optical density at 600 nm, 1.0 versus 0.18). Leveau et al. have shown that challenging actively growing, dense cultures (optical densities at 600 nm of >0.8) with 2,4-D results in the rapid accumulation of DCP and is accompanied by cell death (23). This accumulation may be a result of disproportionate synthesis rates of tfdA and tfdB, resulting in a transient 100-fold relative increase in tfdA transcript abundance (23). As mentioned earlier, the resultant accumulation of DCP may also damage cell membrane integrity, resulting in the rapid loss of ATP (15), thereby limiting bioluminescence.
Conclusions.
The limited responsiveness to 3-CB combined with sensitivity to low concentrations of both 2,4-D and DCP makes strain JMP134-32 a potentially useful bioreporter for the detection of 2,4-D and its breakdown products in aqueous samples. The specific nature of this reporter is a simple alternative to fingerprinting methods which rely on the response of multiple bioreporters, each consisting of the promoter from a general stress gene fused to a reporter gene (6). Such systems relying on multiple reporters could give misleading signals that cannot be controlled for if unknowns contain solvents (16). Producing a positive signal specifically in response to 2,4-D and DCP, strain JMP134-32 also differs from Burkholderia sp. strain RASC c2, a recently described DCP-degrading bacterium that contains a lux cassette expressed from an uncharacterized constitutive promoter (37). Strain RASC c2 has no reported response to 2,4-D and responds to DCP only as one of many potential food sources or toxicants (38).
Having its reporter element chromosomally encoded, strain JMP134-32 is not prone to problems encountered by plasmid-based systems, such as copy number effects (42) or the need for selective pressure in order to ensure that plasmid loss does not occur (32). The inclusion of tfdR also makes for a versatile autonomous reporter that could be placed in other organisms unable to degrade 2,4-D, thereby allowing measurement of extracellular metabolite concentrations.
This reporter construct has been used to successfully detect 2,4-D in aqueous media and also in slurries containing soil with aged 2,4-D residues. Response to aged 2,4-D residues in soil took longer than response to 2,4-D in aqueous media. Although the soil slurry may have quenched some of the bioluminescent signal, more work needs to be done to determine the correlation between this observed lag in bioluminescence and the bioavailability of 2,4-D residues in soil.
Specific applications of this bioreporter must be carefully evaluated, as underestimation of 2,4-D or DCP concentration or false-negative results may occur if 2,4-D or DCP is at toxic levels in the sample. This limitation is easily addressed by performing analyses on samples that have been serially diluted in an appropriate medium (14). Increased bioluminescence in more dilute samples would indicate diminished toxicity and serve as an effective control. Spiking samples that do not emit light with known concentrations of 2,4-D is another means of assessing possible false negatives as a result of toxicity-related issues. Despite these limitations, the work described in this report details the development of a whole-cell bioluminescent reporter for the detection of 2,4-D that is rapid and easy to perform and may be useful in elucidating factors involved in the bioavailability of 2,4-D in environmentally relevant samples.
Acknowledgments
This work was supported in part by a Dow Foundation Sphere award to G.S.S. and in part by the Waste Management Research and Education Institute, University of Tennessee, Knoxville. A.G.H. was supported by an appointment to the Alexander Hollaender Distinguished Postdoctoral Fellowship Program sponsored by the U.S. Department of Energy, Office of Health and Environmental Research, and administered by the Oak Ridge Institute for Science and Education.
REFERENCES
- 1.Applegate B, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn F M, Bienkowski P, Sayler G. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J Ind Microbiol Biotechnol. 1997;18:4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- 2.Applegate B M, Kehrmeyer S R, Sayler G S. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl Environ Microbiol. 1998;64:2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Applegate B M, Matrubutham U, Sanseverino J, Sayler G S. Biodegradation genes as marker genes in microbial ecosystems. 1995. p. 6.1.8. , 1–14. In A. D. L. Akerman, J. D. Van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 5.Baveye P, Bladon R. Bioavailability of organic xenobiotics in the environment: a critical perspective. In: Baveye P, et al., editors. Bioavailability of organic xenobiotics in the environment. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 227–248. [Google Scholar]
- 6.Ben-Israel O, Ben-Israel H, Ulitzur S. Identification and quantification of toxic chemicals by use of Escherichia coli carrying lux genes fused to stress promoters. Appl Environ Microbiol. 1998;64:4346–4352. doi: 10.1128/aem.64.11.4346-4352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burlage R S, Palumbo A V, Heitzer A, Sayler G. Bioluminescent reporter bacteria detect contaminants in soil samples. Appl Biochem Biotechnol. 1994;45:731–740. [Google Scholar]
- 8.Daugherty D D, Karel S F. Degradation of 2,4-dichlorophenoxyacetic acid by Pseudomonas cepacia DBO1(pRO101) in a dual-substrate chemostat. Appl Environ Microbiol. 1994;60:3261–3267. doi: 10.1128/aem.60.9.3261-3267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lorenzo V, Fernandez S, Herrero M, Jakubzik U, Timmis K N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 10.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Don R H, Weightman A J, Knackmuss H-J, Timmis K N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filer K, Harker A R. Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl Environ Microbiol. 1997;63:317–320. doi: 10.1128/aem.63.1.317-320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukumori F, Hausinger R. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J Bacteriol. 1993;175:2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harkey G A, Young T M. Effect of soil contaminant extraction method in determining toxicity using the Microtox assay. Environ Toxicol Chem. 1999;19:276–282. [Google Scholar]
- 15.Heipieper H J, Keweloh H, Rehm H J. Influence of phenols on growth and membrane permeability of free and immobilized Escherichia coli. Appl Environ Microbiol. 1991;57:1213–1217. doi: 10.1128/aem.57.4.1213-1217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitzer A, Applegate B, Kehrmeyer S, Pinkart H, Webb O F, Phelps T J, White D C, Sayler G S. Physiological considerations of environmental applications of lux reporter fusions. J Microbiol Methods. 1998;33:45–57. [Google Scholar]
- 17.Heitzer A, Webb O F, Thonnard J E, Sayler G S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1992;58:1839–1846. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston W. Ph.D. dissertation. Knoxville: University of Tennessee; 1996. [Google Scholar]
- 20.Kehrmeyer S R. Ph.D. dissertation. Knoxville: University of Tennessee; 1998. [Google Scholar]
- 21.King J M H, Digrazia P M, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler G S. Rapid sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 22.Layton A C, Muccini M, Ghosh M M, Sayler G S. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl Environ Microbiol. 1998;64:5023–5026. doi: 10.1128/aem.64.12.5023-5026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leveau J H, Konig F, Fuchslin H, Werlen C, van der Meer J R. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol Microbiol. 1999;33:396–406. doi: 10.1046/j.1365-2958.1999.01483.x. [DOI] [PubMed] [Google Scholar]
- 24.Leveau J H J, van der Meer J R. Genetic characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutropha JMP134. Gene. 1997;202:103–114. doi: 10.1016/s0378-1119(97)00460-5. [DOI] [PubMed] [Google Scholar]
- 25.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leveau J H J, Zehnder A J B, van der Meer J R. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1998;180:2237–2243. doi: 10.1128/jb.180.8.2237-2243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrubutham U, Harker A R. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alcaligenes eutrophus JMP134. J Bacteriol. 1994;176:2348–2353. doi: 10.1128/jb.176.8.2348-2353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mungkarndee P, Bhamidimarri S M R, Mawson A J, Chong R. The role of a metabolic intermediate in the biodegradation of inhibitory substrates. Water Sci Technol. 1997;36:27–36. [Google Scholar]
- 29.Ogram A V, Jessup R E, Ou L T, Rao P S C. Effects of sorption on biological degradation rates of (2,4-dichlorophenoxy)acetic acid in soils. Appl Environ Microbiol. 1985;49:582–587. doi: 10.1128/aem.49.3.582-587.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Pantoja D, Guzmán L, Manzano M, Pieper D, González B. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4) Appl Environ Microbiol. 2000;66:1602–1608. doi: 10.1128/aem.66.4.1602-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice J F. Ph.D. dissertation. Knoxville: University of Tennessee; 1999. [Google Scholar]
- 32.Rice J F, Fowler R F, Arrage A A, White D C, Sayler G S. Effects of external stimuli on environmental bacterial strains harboring an algD-lux bioluminescent reporter plasmid for the study of corrosive biofilms. J Ind Microbiol. 1995;15:318–328. [Google Scholar]
- 33.Rogowsky P M, Close T J, Chimera J A, Shaw J J, Kado C I. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987;169:5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux K H. Using mismatched primer-template pairs in touchdown PCR. BioTechniques. 1994;16:812–814. [PubMed] [Google Scholar]
- 35.Selifonova O, Burlage R, Barkay T. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol. 1993;59:3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selifonova O V, Eaton R W. Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl Environ Microbiol. 1996;62:778–783. doi: 10.1128/aem.62.3.778-783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw L J, Beaton L, Glover K, Killham K, Meharg A A. Development and characterization of a lux-modified 2,4-dichlorophenol-degrading Burkholderia sp. RASC Environ Microbiol. 1999;1:393–399. doi: 10.1046/j.1462-2920.1999.00049.x. [DOI] [PubMed] [Google Scholar]
- 38.Shaw L J, Beaton Y, Glover A, Killham K, Meharg A A. Interactions between soil, toxicant, and a lux-marked bacterium during solid phase-contact toxicity testing. Environ Toxicol Chem. 2000;19:1247–1252. [Google Scholar]
- 39.Simpson M L, Sayler G S, Applegate B M, Ripp S, Nivens D E, Paulus M J, Jellison G E., Jr Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol. 1998;16:332–338. [Google Scholar]
- 40.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;41:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 41.Streber W R, Timmis K N, Zenk M H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Rawlings M, Gibson D T, Labbe D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 43.Wood K V. Marker proteins for gene expression. Curr Opin Biotechnol. 1996;6:50–58. doi: 10.1016/0958-1669(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 44.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]