Abstract
Next-generation COVID-19 vaccines are critical due to the ongoing evolution of SARS-CoV-2 virus and rapid waning duration of the neutralizing antibody response against current vaccines. The mRNA vaccines mRNA-1273 and BNT162b2 were developed using linear transcripts encoding the prefusion-stabilized trimers (S-2P) of the wildtype spike, which have shown a reduced neutralizing activity against the variants of concern B.1.617.2 and B.1.1.529. Recently, a new version of spike trimer, termed VFLIP (five (V) prolines, Flexibly-Linked, Inter-Protomer disulfide) was developed. Based on the original amino acid sequence of the wildtype spike, VFLIP was genetically engineered by using five proline substitutions, a flexible cleavage site amino acid linker, and an inter-protomer disulfide bond. It has been suggested to possess native-like glycosylation, and greater pre-fusion trimeric stability as opposed to S-2P. Here, we report that the spike protein VFLIP-X, containing six rationally substituted amino acids to reflect emerging variants (K417N, L452R, T478K, E484K, N501Y and D614G), offers a promising candidate for a next-generation SARS-CoV-2 vaccine. Mice immunized by a circular mRNA (circRNA) vaccine prototype producing VFLIP-X had detectable neutralizing antibody titers for up to 7 weeks post-boost against SARS-CoV-2 variants of concern (VOCs) and variants of interest (VOIs). In addition, a balance in TH1 and TH2 responses was achieved by immunization with VFLIP-X. Our results indicate that the VFLIP-X delivered by circRNA induces humoral and cellular immune responses, as well as broad neutralizing activity against SARS-CoV-2 variants.
Keywords: VFLIP, Spike, SARS-CoV-2, Vaccine
1. Introduction
Implementing next-generation vaccines and therapeutics is vital to combating and controlling continuous evolution and ongoing global transmission of SARS-CoV-2 variants of concern (VOCs) and variants of interest (VOIs). The first-generation prefusion-stabilized spike engineered by two proline substitutions (S-2P) has served as an initial immunogen in current SARS-CoV-2 vaccines, presenting one or more RBDs (receptor-binding domains) “up” (Kirchdoerfer et al., 2018; Pallesen et al., 2017; Wrapp et al., 2020). As serum neutralizing activity is primarily directed to the prefusion spike, structure-based engineering that better stabilizes the prefusion state could elicit more potent neutralizing antibody responses (Bowen et al., 2021). Importantly, immunodominant neutralizing epitopes are mainly located in the RBD (Bowen et al., 2021; Piccoli et al., 2020; Dejnirattisai et al., 2021a). However, multiple RBD mutations in emerging variants resulted in changes in immunodominance hierarchy and impaired neutralization potency of RBD-directed neutralizing antibodies (Hastie Kathryn et al., 2021; Wilks et al., 2022). Specifically, common mutations, including substitutions at positions 417, 452, 484 and 501, have been identified as major drivers of antigenic differences (Wilks et al., 2022; Greaney et al., 2022). Other RBD-targeted neutralizing monoclonal antibodies are unaffected by mutations in the RBD of emerging variants (Dejnirattisai et al., 2021b; Cerutti et al., 2021; Cameroni et al., 2022). Moreover, the utilization of RBD as an immunogen in several vaccine platforms has shown promising clinical immunogenicity results (Mulligan et al., 2020; Yang et al., 2021; Janssen et al., 2022; Chen et al., 2022). Thus, rationally engineered SARS-CoV-2 RBD and spike proteins could serve as potential immunogens to elicit potent protective immune responses.
Next-generation vaccine design strategies have been suggested to engineer antigens with enhanced stability and native-like quaternary structure, conformational dynamics, and glycosylation profiles (Harvey et al., 2021; Graham et al., 2019). A recently described spike protein, namely VFLIP, has been engineered by using five proline substitutions in the S2 subunit, a flexible S1/S2 linker, and two cysteine substitutions to introduce an inter-protomer disulfide bond formation (Olmedillas et al., 2021). In this work, one proline substitution at residue 986, shared between S-2P and the second-generation HexaPro, was converted back to lysine (K986) restoring native salt bridge formation between K986 and D427. The VFLIP spike displays improved thermostability and native RBD motion, of which its receptor binding can trigger the conformational dynamics from “down” to “up” conformation (Olmedillas et al., 2021). In addition, glycosylation patterns of VFLIP are more similar to the authentic virus than previously engineered spike constructs. Moreover, immunization of VFLIP subunit candidate vaccine in mice results in superior immunogenicity compared to S-2P. When combined with next-generation vaccine platforms, such as mRNA, the antigen design strategy might pave the way for improved, cross-variant neutralization of SARS-CoV-2 (Edwards et al., 2022).
The inherent instability and relatively short half-life of linear mRNA transcripts impede their utilization as next-generation vaccine candidates. Specifically, longer half-life of mRNA can confer improved immunogenicity through a sustained antigen production in target cells. Unlike their linear counterparts, circRNAs impart greater stability due to the covalently closed structure, rendering protection from degradation by exonucleases, and can therefore extend half-life of mRNA. Efficient circularization of long RNAs is achieved through a permuted intron-exon splicing strategy with other assisting elements, including homology arms and spacer sequences (Wesselhoeft et al., 2018a). To produce translatable exogenous circRNAs, these RNAs have been engineered to comprise an IRES (internal ribosome entry site) elements of several viruses (e.g., EMCV, CVB3, PV, etc.) placed prior to a coding sequence as a translation initiation site. Importantly, circRNAs elicit more durable protein expression as compared to their linear cognates. In this work, we utilized the circRNA platform to engineer the spike protein VFLIP-X, which is VFLIP containing six rationally substituted amino acids. We show in mice that the circRNA vaccine prototype producing VFLIP-X elicited neutralizing antibodies for up to 7 weeks post-boost against SARS-CoV-2 VOCs and VOIs. VFLIP-X delivered by circRNA also induces favorable humoral and cellular immune responses. Together, our work highlights the potential of a SARS-CoV-2 circRNA vaccine expressing VFLIP-X spike as a next-generation COVID-19 vaccine.
2. Results
2.1. Expression of the circRNA vaccine prototype producing VFLIP-X
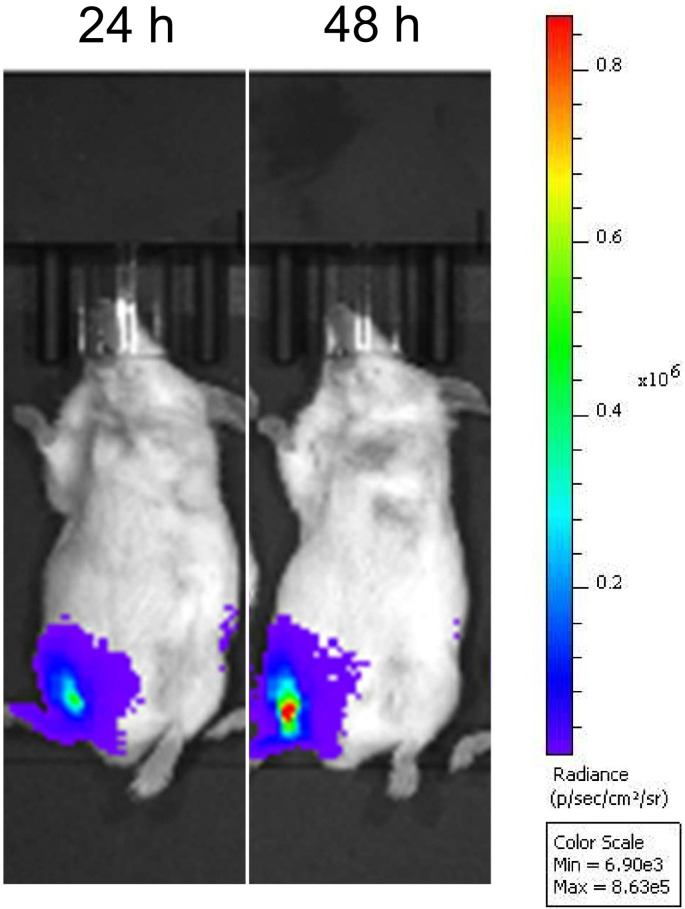
We initially confirmed circRNA-driven in vivo protein expression by intramuscularly injecting BALB/c mice with LNP-formulated circRNA encoding firefly luciferase (FLuc). At 24 and 48 h after the administration, we clearly observed bioluminescence in those mice (Fig. S1), suggesting that the circRNA template is applicable for development of an mRNA vaccine prototype.
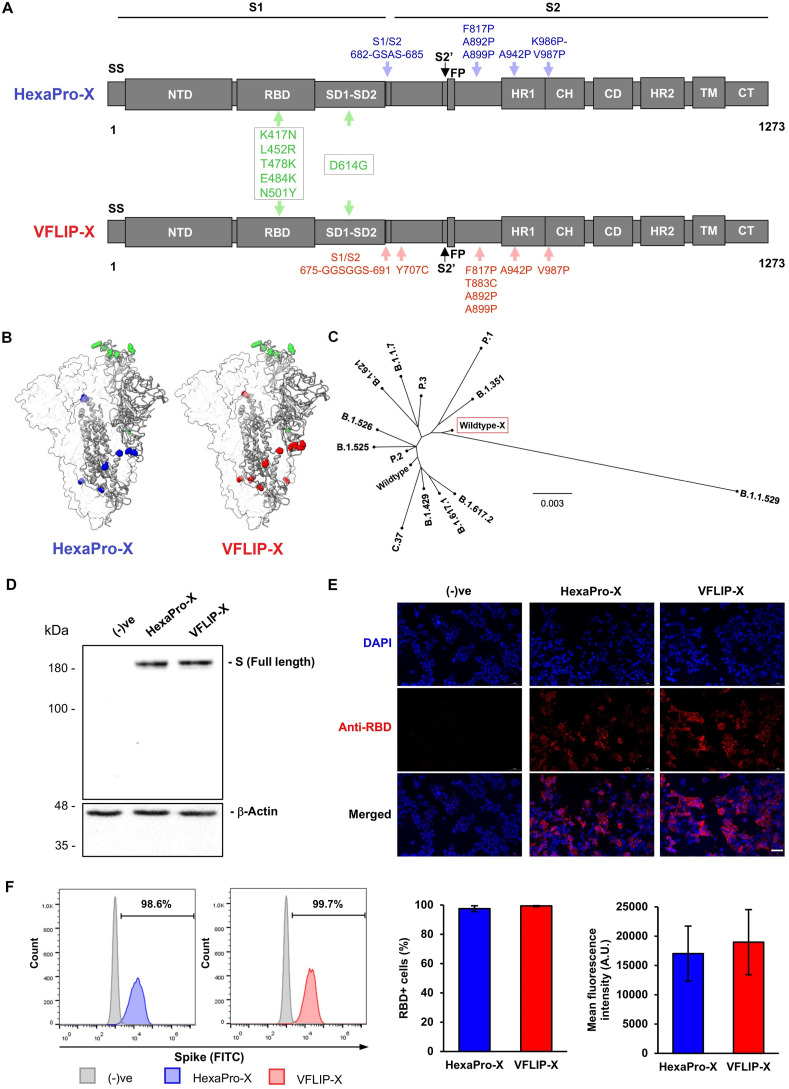
To develop a circRNA expressing SARS-CoV-2 spike as a vaccine prototype capable of neutralizing broad SARS-CoV-2 variants, the full-length, membrane-bound version of the recently engineered VFLIP spike possessing native-like glycosylation was chosen (Olmedillas et al., 2021). Further, a substitution of six amino acids was rationally selected based on the co-mutation (D614G) found in all SARS-CoV-2 variants as well as five mutations (K417N, L452R, T478K, E484K and N501Y) co-identified in several variants of concern (VOCs) and variants of interest (VOIs) (Fig. 1 A). The selected six mutations and the amino acid substitutions pertinent to the originally reported VFLIP spike were structurally presented in the spike trimers (Fig. 1B). We named this spike construct VFLIP-Cross (VFLIP-X). An unrooted phylogenetic tree was constructed to visualize relationships among amino acid sequences derived from the wildtype isolate containing the six rationally substituted amino acids (wildtype-X) and SARS-CoV-2 VOC and VOI spikes. Compared to VOC and VOI spikes, wildtype-X spike is found at a distinct clade among those spike variants, and is more related to Omicron (B.1.1.529) spike variant (Fig. 1C). The result suggests that the six rationally substituted amino acids confer a unique sequence feature, yet interrelated to VOC and VOI spikes.
Fig. 1.
Design strategy and expression of SARS-CoV-2 spike proteins harboring six rationally substituted amino acids. (A) Schematic representation of HexaPro-X and VFLIP-X showing the S1 and S2 subunits. Amino acid positions described in the original HexaPro (Hsieh et al., 2020) and VFLIP (Olmedillas et al., 2021) spikes are shown in blue and red colors, respectively. The positions of six rationally substituted amino acids (K417N, L452R, T478K, E484K, N501Y and D614G) are indicated by green color. (B) Molecular models of HexaPro-X and VFLIP-X spike trimers. The RBD-up protomer is shown in ribbons colored corresponding to (A). The structures were prepared in SWISS-MODEL. (C) An unrooted phylogenetic tree comparing amino acid sequences derived from the wildtype spike containing six rationally substituted amino acids (wildtype-X) and spike sequences derived from SARS-CoV-2 VOCs and VOIs. (D) Western blot analysis of full-length SARS-CoV-2 spike expression in HEK293T cells transfected with circRNAs encoding HexaPro-X and VFLIP-X. The proteins were visualized by an anti-RBD antibody. (E) Immunofluorescence analysis of HexaPro-X and VFLIP-X in HEK293T cells immunostained by an anti-RBD antibody. Scale bar, 50 μm. (F) Flow cytometry analysis of HEK293T cells expressing HexaPro-X and VFLIP-X spikes upon delivery by circRNA-LNP. The proteins were visualized by an anti-RBD antibody. Percentage of RBD-positive cells and mean fluorescence intensity from biological duplicates are shown. The error bars indicate the ±SD.
To ascertain whether the VFLIP-X spike can be expressed as a full-length spike in vitro, protein expression was determined in HEK293T transfected with a circRNA prototype encoding the spike protein. We also compared expression of the VFLIP-X spike to a membrane-bound, prefusion-stabilized spike containing the six rationally substituted amino acids (HexaPro-X). Similar expression levels of full-length HexaPro-X and VFLIP-X were determined using western blot, immunofluorescence staining and flow cytometry analyses using a polyclonal anti-RBD antibody (Fig. 1D–F), indicating that the circRNA prototype is capable of producing the spike antigens.
2.2. VFLIP-X confers neutralization against SARS-CoV-2
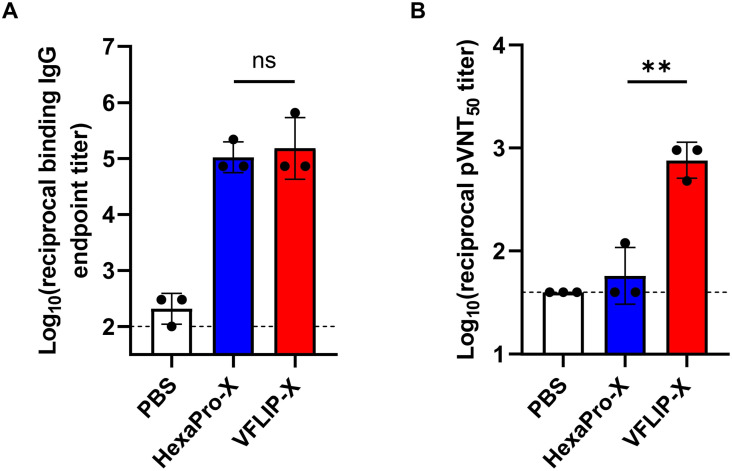
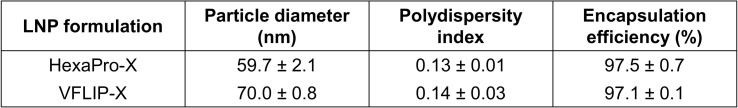
To elucidate immunogenicity of the circRNA vaccine prototype, we formulated circRNAs expressing HexaPro-X or VFLIP-X with LNPs, and determined the physicochemical properties of the circRNA-LNPs. We confirmed similar average size, polydispersity index, and encapsulation efficiency of circRNA-LNPs producing either HexaPro-X or VFLIP-X (Table S1). Next, seven-week-old female BALB/c mice were intramuscularly immunized with 5 μg of the circRNAs using a prime-boost regimen separated by a 3-week interval. Immunization with VFLIP-X induced high B.1.1.529 S-binding serum IgG titers similar to HexaPro-X (Fig. 2 A). Importantly, we found that only sera from mice vaccinated by VFLIP-X elicits high neutralization against B.1.1.529 pseudovirus. In contrast, HexaPro-X induced little to no neutralizing titers for B.1.1.529 pseudovirus (Fig. 2B). Further, similar results were observed for infectious SARS-CoV-2 variant isolates including wildtype, B.1.1.7, B.1.351, B.1.617.2 and B.1.1.529 (Fig. S2), suggesting a very poor immunogenicity of membrane-bound HexaPro-X spike. The result is consistent with a previous finding using the membrane-bound version of wildtype HexaPro spike (Kalnin et al., 2021). Therefore, only the circRNA expressing VFLIP-X was thus chosen for subsequent experiments.
Fig. 2.
Mice immunized with VFLIP-X full length spike produce neutralizing antibodies against B.1.1.529. BALB/c mice (n = 3) were immunized at weeks 0 and 3 with 5 μg of circRNA-LNP encoding HexaPro-X (blue) or VFLIP-X (red). Control mice were administered with PBS (white). (A) IgG levels of two weeks post-boost sera were assessed by enzyme-linked immunosorbent assay (ELISA) using recombinant Omicron (B.1.1.529) spike protein. (B) Neutralization activity of two weeks post-boost sera were determined using B.1.1.529 pseudotyped virus. Data are presented as GMT ± geometric SD. Horizontal dotted lines represent assay limits of detection. Immunized groups were compared by student's t-test (parametric, two-tailed unpaired). ** p < 0.01.
2.3. VFLIP-X elicits cross-neutralizing antibody production against SARS-CoV-2 variants
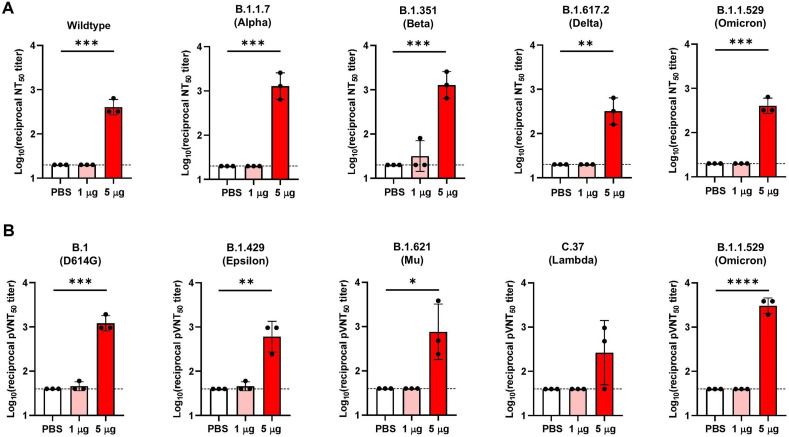
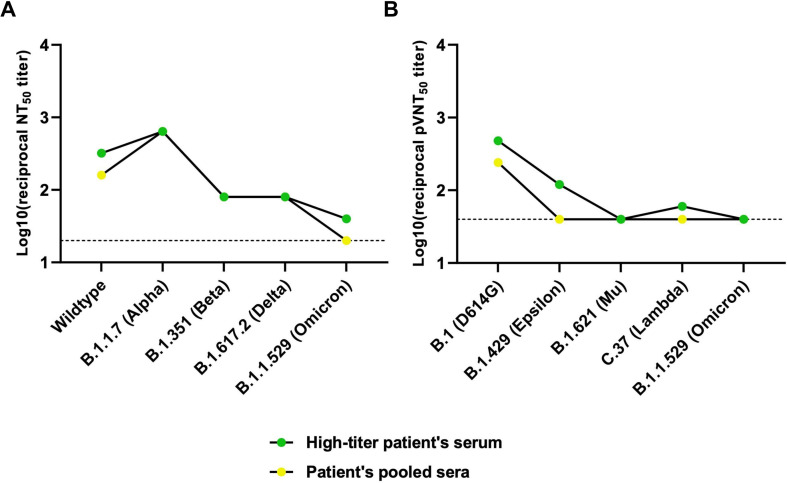
To determine whether VFLIP-X provides a cross-variant neutralizing activity, sera samples from mice administered with 1 or 5 μg of VFLIP-X 7 weeks post-boost were collected and tested for neutralizing activity against infectious SARS-CoV-2 variant isolates (wildtype, B.1.1.7, B.1.351, B.1.617.2, and B.1.1.529) and pseudoviruses (B.1, B.1.429, B.1.621, C.37 and B.1.1.529). We observed little to no neutralizing activity against SARS-CoV-2 VOCs and VOIs in mice using 1 μg dose of VFLIP-X (Fig. 3 A–B). Remarkably, VFLIP-X immunized at 5 μg elicited neutralizing antibody levels against the panel of infectious SARS-CoV-2 variant isolates and pseudotyped SARS-CoV-2 viruses. In animals receiving 5 μg VFLIP-X, neutralizing activity was similar for wildtype, B.1.617.2 and B.1.1.529 as determined by microneutralization assay using infectious SARS-CoV-2 variant isolates (Fig. 3A). A similar neutralizing activity was also observed for B.1.1.7 and B.1.351 variants. Notably, among the pseudoviruses included here, the highest pseudovirus-neutralizing activity was achieved against the variant B.1.1.529, while the lowest neutralizing titer was observed for the variant C.37 (Fig. 3B). We also compared the cross-neutralizing activities of the vaccine candidate VFLIP-X with human convalescent sera collected during the first COVID-19 pandemics in 2020. The human sera demonstrate limited cross-variant neutralizing activity as compared to 5 μg dose of VFLIP-X vaccine candidate (Fig. S3). Specifically, we observed little to no neutralizing activity against B.1.1.529 from human convalescent serum samples. Our results indicate that VFLIP-X demonstrates enhanced neutralizing activity against the heavily mutated SARS-CoV-2 variant B.1.1.529 while maintaining a strong response against other VOCs and VOIs, including the ancestral strain wildtype and B.1.
Fig. 3.
VFLIP-X elicits cross-neutralizing antibodies against SARS-CoV-2 VOCs and VOIs. BALB/c mice (n = 3) were immunized at weeks 0 and 3 with 1 μg (pink) or 5 μg (red) of circRNA-LNP encoding VFLIP-X. Control mice were administered with PBS (white). (A) Infectious virus 50% neutralization titers (NT50) of seven weeks post-boost sera were determined against the indicated infectious SARS-CoV-2 variant isolates. (B) Lentivirus-based pseudovirus 50% neutralization titers (pVNT50) of seven weeks post-boost sera were determined against the indicated SARS-CoV-2 variants. Data are presented as GMT ± geometric SD. Horizontal dotted lines represent assay limits of detection. Control group was compared with 5 μg immunized group by student's t-test (parametric, two-tailed unpaired). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
2.4. VFLIP-X induces humoral and cellular immune responses against B.1.1.529 variant
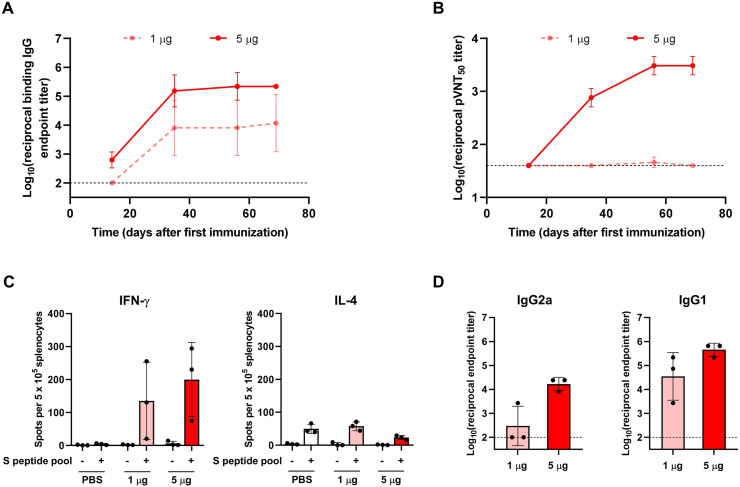
The humoral and cellular immune responses were assessed in BALB/C mice immunized with 1 or 5 μg of VFLIP-X in a series of experiments. Two-dose immunization of VFLIP-X induced high B.1.1.529 S-binding serum IgG titers in a dose-dependent manner (Fig. 4 A). The binding IgG titers were increased one week after the second immunization and sustained up to 7 weeks post-boost. A potent B.1.1.529 pseudovirus-neutralizing activity was observed in mice receiving 5 μg of VFLIP-X (Fig. 4B). Although the binding IgG titers were detected at 2 weeks after the first immunization, the neutralizing titers were detectable after the second immunization. At 5 weeks post-boost, the levels of neutralizing antibodies were increased compared to one-week post-boost and sustained up to 10 weeks after the first immunization. These results demonstrate that the cross-neutralizing VFLIP-X spike expressed under the circRNA vaccine candidate is a potent immunogen.
Fig. 4.
Humoral and cellular immune responses against B.1.1.529 were raised by VFLIP-X. BALB/c mice (n = 3) were immunized at weeks 0 and 3 with 1 μg (pink) or 5 μg (red) of circRNA-LNP encoding VFLIP-X. Control mice were administered with PBS (white). Sera samples were collected at weeks 2, 5, 8, and 10. Splenocytes isolated from mice were collected at week 10. (A) SARS-CoV-2 Omicron (B.1.1.529) S-specific IgG levels of sera were assessed by ELISA. (B) Lentivirus-based pseudovirus expressing SARS-CoV-2 B.1.1.529 S 50% neutralization titers (pVNT50) of sera were determined. (C) IFN-γ (left) and IL-4 (right) ELISpot of seven weeks post-boost splenocytes restimulated with SARS-CoV-2 B.1.1.529 S peptide pool. (D) SARS-CoV-2 Omicron (B.1.1.529) S-specific IgG2a (left) and IgG1 (right) levels of seven weeks post-boost sera were determined by ELISA. In A, B, and D, data are presented as GMT ± geometric SD. Horizontal dotted lines represent assay limits of detection. In C, data are presented as mean ± SD.
Next, we characterized B.1.1.529 S specific cellular immune response in mice at 7 weeks post-boost. Enzyme-linked immunospot (ELISpot) assay was performed using isolated splenocytes from mice that received 1 or 5 μg of VFLIP-X restimulated with S peptide mixture to evaluate specific T cell responses. Following restimulation with B.1.1.529 S peptide pool, we found that splenocytes isolated from the immunization groups elicited strong IFN-γ, but low IL-4, responses (Fig. 4C). This result suggests that the vaccine candidate does not induce TH2-biased responses. We further elucidated the balance of TH1 and TH2 responses by comparing levels of IgG2a and IgG1, which are surrogate markers for TH1 and TH2 responses, respectively. Mice receiving 5 μg of VFLIP-X produced robust IgG2a and IgG1 levels, as shown by B.1.1.529 S-specific ELISA (Fig. 4D). Together, the IgG subclass and ELISpot profiles demonstrate that the immunization with VFLIP-X led to the induction of type 1 antiviral response rather than a TH2-skewed response.
3. Discussion
mRNA vaccines formulated with the first-generation wildtype spike antigen have proven to be highly effective against the ancestral SARS-CoV-2 variant. However, the emergence of variants of concern, especially the heavily mutated B.1.1.529 variant, poses the risk toward effectiveness and durability of protection conferred by the current vaccines. Indeed, several studies have recently reported that a booster dose with B.1.1529-specific vaccines in preclinical animal models does not provide greater immunity or protection than the current vaccines (Gagne et al., 2022; Hawman et al., 2022; Ying et al., 2022). Moreover, primary vaccination with B.1.1.529-targeted vaccines led to a limited cross-variant protective efficacy (Ying et al., 2022; Lee et al., 2022). This challenge has suggested the importance of development of structurally engineered vaccines with improved broader immunogenicity. Here we show the immunogenicity results of the SARS-CoV-2 circRNA vaccine VFLIP-X, rationally designed to provide broad immune responses against emerging SARS-CoV-2 VOCs and VOIs in mice. Our VFLIP-X spike comprises RBD mutations associated with emerging variants, including K417N, L452R, T478K, E484K, and N501Y, and the co-mutation at residue 614 (D614G). Topologically, T478 and E484 are positioned in the "peak" subsection of SARS-CoV-2 spike. K417 and L452 are positioned in the “valley” subsection. N501 is positioned in the "mesa" subsection (Hastie Kathryn et al., 2021). They have been shown to either enhance affinity between the spike protein and host ACE2 receptor or escape immunity derived from natural infection, vaccination, and monoclonal antibodies (Hastie Kathryn et al., 2021; Tatsi et al., 2021). The mutations K417N/L452R/T478K are found in Delta plus variant. Structurally, the aliphatic-to-positively charged mutation at T478K has been suggested to enhance binding between the Delta variant S and ACE2 via K478-Q24 interaction (McCallum et al., 2021a; Rehman et al., 2021). K417N, T478K, E484A and N501Y have been demonstrated to participate in Omicron's immune evasion (McCallum et al., 2022). Even though E484A caused more escape from D614G sera than E484K as determined by a deep mutational scanning study, the effect of single substitution at E484K in significantly poor neutralization is similar to that observed in human influenza viruses; a single amino acid substitution confers a large portion of the immunogenic difference (Wilks et al., 2022).
Following a prime-boost intramuscular immunization, VFLIP-X appeared to induce strong pseudovirus-neutralizing antibody response against B.1.1.529, whereas mice immunized with HexaPro-X generated little to no neutralizing antibody (Fig. 2). Thus, antigen expression and binding IgG titers were not predictive of neutralizing titers. Similar results have been observed by vaccination using an mRNA vaccine encoding full-length, HexaPro spike (Kalnin et al., 2021). By using VFLIP-X, potent neutralizing activity against emerging variants was elicited by a 5 μg dose (Fig. 3). We observed a robust B.1.1.529 pseudovirus-neutralizing response in this group, which is in line with a closer relationship between B.1.1.529 spike toward wildtype-X than toward other variants as determined by the phylogenetric tree analysis (Fig. 1B). We also noticed relatively low pseudovirus-neutralizing titers against C.37 group compared with other VOCs and VOIs. This may be due to a unique cluster of mutations in the C.37 which is not included in VFLIP-X. Previous studies have reported that the neutralizing activity tested against B.1.429, B.1.621, and C.37 VOIs was reduced in serum samples obtained from convalescence and vaccine recipients (Shen et al., 2021; McCallum et al., 2021b; Uriu et al., 2021; Messali et al., 2021; Á lvarez-D í az et al., 2022; Tada et al., 2021; Kimura et al., 2022).
Notably, neutralizing antibodies have been recently shown to mainly target prefusion conformation, suggesting that vaccines utilizing prefusion-stabilized spikes might elicit greater neutralizing titers (Bowen et al., 2021). In contrast to that notion, spike proteins kept in closed conformation, including VFLIP, have been demonstrated to elicit more potent neutralizing responses than the more opened conformation spikes S-2P and HexaPro (Olmedillas et al., 2021; Carnell George et al., 1128). In addition, the VFLIP spike displays more native-like glycosylation profiles than other prefusion-stabilized spikes, presumably better preserving the antigenicity of spike immunogen (Olmedillas et al., 2021; Chawla et al., 2022; Yao et al., 2020). Therefore, a balance in prefusion-stabilized and metastable states, opened and closed structures, as well as glycosylation profiles might be required when revising next-generation vaccines against SARS-CoV-2.
Our VFLIP-X candidate vaccine evaluated in this work elicited a balanced TH1-TH2 response, suggesting the protective immunity induction rather than enhanced disease (Fig. 4C and D). In preclinical evaluations of respiratory virus infection including SARS-CoV, MERS-CoV, and SARS-CoV-2 vaccines, T helper 2 cell (TH2)-biased immune responses have been associated with vaccine-associated enhanced respiratory disease. Indeed, whole-inactivated virus and protein subunit vaccines induce TH2-biased immune responses and antibodies with limited neutralization potency, resulting in severe lung pathogenesis (DiPiazza et al., 2021; Tseng et al., 2012; Kim et al., 1969; Ebenig et al., 2021). The IFN-γ and IL-4 ELISpot and IgG subclass titer results are consistent with other studies on mRNA vaccines against SARS-CoV-2 (Corbett et al., 2020; Vogel et al., 2021). our antigen candidate VFLIP-X confers a broad neutralizing activity over convalescence sera. Because immune protection observed by convalescence sera and vaccine protective efficacy are highly correlated with neutralizing antibody levels (Khoury et al., 2021), our work highlights the potential of a SARS-CoV-2 circRNA vaccine expressing a highly stable VFLIP-X spike immunogen as a next-generation COVID-19 vaccine protecting against current and emerging SARS-CoV-2 variants.
4. Methods
4.1. Ethics statement
Mouse experiments were performed under the Animal Ethics approved by Faculty of Science, Mahidol University (MUSC63-016-524). PCR-confirmed COVID-19 patients were hospitalized at Chakri Naruebodindra Medical Institute, Faculty of Medicine Ramathibodi Hospital, Mahidol University. Serum specimens were collected from patients 14–30 days post-infection. The study protocol and human ethics were approved by Faculty of Medicine Ramathibodi Hospital (COA. MURA2020/568).
4.2. Design of SARS-CoV-2 circRNA vaccine constructs
Antigens encoded by HexaPro-X and VFLIP-X vaccine candidates were designed on a background of the S sequence from SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank accession number MN908947.3). The codon-optimized nucleotide and amino acid sequences of VFLIP-X spike was deposited in GenBank with accession number ON806880. The sequence encoding SARS-CoV-2 spike with mutations in RBD (K417N, L452R, T478K, E484K, and N501Y), D614G, and structural changes within S1/S2 cleavage site and S2 subunit (HexaPro: 682-GSAS-685, F817P, A892P, A899P, A942P, K986P, and V987P; VFLIP: 675-GGSGGS-691, Y707C, F817P, T883C, A892P, A899P, A942P, and V987P) was codon optimized using GenSmart codon optimization algorithm (GenScript). The optimized DNA sequence was synthesized (Twist Bioscience), digested with AgeI and NotI, and cloned into a vector for producing circular RNA in vitro generating HexaPro-X and VFLIP-X constructs. To determine whether the rationally designed vaccines preserve the overall three-dimensional spike protein structure, molecular modeling of the HexaPro-X and VFLIP-X spikes was performed with the HexaPro prefusion SARS-CoV-2 spike template (PDB: 6xkl) using SWISS-MODEL's homology modeling. The modeled structures were visualized by UCSF ChimeraX software. An unrooted phylogenetic tree comparing amino acid sequences derived from wildtype spike, SARS-CoV-2 spike variants of concern (VOCs) and variants of interest (VOIs), and wildtype-X spike (wildtype spike with the six rationally substituted amino acids) was constructed by maximum likelihood using IQ-TREE software.
4.3. CircRNA synthesis
The circRNAs were produced by T7 RNA polymerase-based in vitro transcription as described previously (Wesselhoeft et al., 2018b). In brief, linearized plasmid DNA consisting of coding sequence of HexaPro-X and VFLIP-X spikes, CVB3 IRES, and permuted Anabaena pre-tRNA group I intron were used as a template. The in vitro transcription was done using HiScribe T7 High Yield RNA Synthesis Kit (NEB) according to the manufacturer's instructions. The reactions were then treated with DNase I to remove DNA template and purified by phenol-chloroform extraction. To circularize the RNA, GTP was added to a final concentration of 2 mM along with T4 RNA ligase buffer (NEB). RNA was then heated at 55 °C for 8 min. The circRNA products were further purified using Monarch RNA Cleanup Kit (NEB). The quality and quantity of RNA were evaluated by agarose gel electrophoresis and Qubit RNA HS Assay Kit (Invitrogen).
4.4. Lipid nanoparticle (LNP) formulation of circRNAs
LNPs were prepared by mixing ethanol and aqueous phase at a 1:3 volumetric ratio in the NanoAssemblr Benchtop microfluidic device (Precision Nanosystems, Vancouver, BC) using syringe pumps. In brief, ethanol phase was prepared by solubilizing a mixture of ionizable lipidoid SM-102 (Sinopeg), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, Sinopeg), cholesterol (Sinopeg), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG 2000, Sinopeg) at a molar ratio of 50:10:38.5:1.5. The aqueous phase was prepared in 25 mM acetate buffer (pH 4.0) with circRNA. LNPs were dialyzed against PBS in a Pur-A-Lyzer Maxi 3500 Dialysis Kit (Sigma-Aldrich) for 2 h at 4 °C. The concentration of circRNA encapsulated into LNPs was analyzed using Qubit RNA high sensitivity (Thermo Fisher) according to the manufacturer's protocol. The efficiency of circRNA encapsulation into LNPs was calculated by comparing measurements in the absence and presence of 0.5% (v/v) Triton X-100. Nanoparticle size and polydispersity index (PDI) were analyzed by dynamic light scattering (DLS).
4.5. In vivo bioluminescence imaging
For detection of in vivo expression of FLuc-encoding circRNA-LNP, 7-week-old female BALB/c mice were inoculated with 1 μg of the circRNA-LNP via intramuscular route. At indicated times post administration, animals were injected intraperitoneally with D-Luciferin (PerkinElmer) 10 min prior to imaging. Luminescence signals were collected by IVIS Spectrum (PerkinElmer).
4.6. Cell culture and transfection
HEK293T and HEK293T-hACE2 cells were cultured at 37 °C and 5% CO2 in Dulbecco's Modified Eagle's Medium (4.5 g/L glucose) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich). HEK293T-hACE2 cells were cultured with 1 μg/ml puromycin to maintain stable expression of hACE2. Cells were passaged every 3 days. For western blot analysis, HEK293T cells were seeded at 850,000 cells per well of a 6-well plate. After 16-h incubation, 2 μg of purified circRNA was transfected using Lipofectamine MessengerMax (Invitrogen) according to the manufacturer's instructions. For flow cytometry analysis, HEK293T cells were seeded at 170,000 cells per well of a 24-well plate. After 16-h incubation, cells were transfected with circRNA formulated as LNP. For immunofluorescence assay, a 24-well plate was coated with poly-L-lysine solution (P4707, Sigma-Aldrich) according to the manufacturer's instructions. HEK293T cells were seeded at 170,000 cells per well. After 16-h incubation, 2 μg of purified circRNA was transfected using Lipofectamine MessengerMax (Invitrogen).
4.7. Western blot analysis
Transfected HEK293T cells were harvested by scraping at 24-h post-transfection and lysed with NP-40 lysis buffer supplemented with protease inhibitor and PMSF. Proteins were collected from supernatant after centrifugation at 12,000 rpm for 20 min. The concentration of protein was determined by BCA protein assay kit (Millipore). The heated protein samples were then analyzed by SDS-PAGE using 10% acrylamide/bis-acrylamide and western blot. The proteins were transferred to a PVDF membrane using a wet transfer system. Blotted proteins were detected with a rabbit polyclonal antibody that recognizes SARS-CoV-2 spike RBD (40592-T62, SinoBiological) and a donkey anti-rabbit IgG-HRP antibody (sc-2077, Santa Cruz Biotechnology).
4.8. Flow cytometry
To detect surface protein expression, transfected HEK293T cells were stained with a rabbit polyclonal antibody that recognizes SARS-CoV-2 spike RBD (40592-T62, SinoBiological) and an Alexa Fluor 488-conjugated goat anti-rabbit IgG (A11034, Invitrogen). Cells were then acquired on a BD Accuri C6 plus and analyzed by FlowJo software version 10.6.2.
4.9. Immunofluorescence assay
Transfected HEK293T cells were fixed with 4% paraformaldehyde (PFA). Cells were then blocked with 20% FBS in PBS and incubated with a rabbit polyclonal antibody that recognizes SARS-CoV-2 spike RBD (40592-T62, SinoBiological) followed by an Alexa Fluor 594-conjugated goat anti-rabbit IgG (A11037, Invitrogen). DNA was stained with Hoechst. Images were acquired with a fluorescence microscope.
4.10. Mouse study designs
For immunogenicity studies, 7-week-old female BALB/c mice were used (n = 3 per group). The circRNA candidate vaccines (HexaPro-X and VFLIP-X) were administered via intramuscular injection in a 50 μL volume using insulin syringe. The placebo control group received 50 μL of PBS. For vaccine-tested group, circRNA formulations were diluted in 50 μL of PBS and administered to each mouse for 2 times at day 1 and day 21 (3-week interval as a single boost). Sera samples at 100 μL were collected at 2, 5, 8, and 10 weeks post-initial immunization by vena facialis blood collection without anesthesia, and were analyzed for SARS-CoV-2 specific IgG by ELISA, microneutralization of infectious SARS-CoV-2 variant isolates, and pseudovirus neutralization assays. The mice were sacrificed at 7 weeks following the second immunization to collect the spleen for measurement of SARS-CoV-2 specific T cell responses. Briefly, spleen single-cell suspensions were prepared in RPMI media supplemented with 10% FBS by mashing tissue against the surface of a 70-μm cell strainer.
4.11. Enzyme-linked immunosorbent assay (ELISA)
SARS-CoV-2 spike specific IgG titers were determined by ELISA. In brief, serial 2-fold dilutions of inactivated serum were added to blocked 96-well plates coated with 1 μg/ml recombinant SARS-CoV-2 B.1.1.529 spike antigen (40589-V08H26, SinoBiological) and plates were incubated for 1 h at 37 °C. After three washes with wash buffer, plates were added with goat anti-mouse IgG-HRP (G21040, Invitrogen), goat anti-mouse IgG1-HRP (PA174421, Invitrogen), or goat anti-mouse IgG2a-HRP (M32207, Invitrogen) and incubated for 45 min at 37 °C. Plates were then washed five times and added with TMB substrate solution (Abcam) followed by 15 min of incubation. The reaction was stopped by the addition of 1N HCl solution. The absorbance (450/630 nm) was read using a microplate reader. Endpoint titers were reported as the dilution that generated an optical density exceeding 3 times over the blank controls (secondary antibody alone).
4.12. Lentivirus-based pseudovirus neutralization assay
The production of lentivirus-based SARS-CoV-2 pseudovirus and neutralization assay were performed as described previously (Crawford et al., 2020). Briefly, the SARS-CoV-2 pseudovirus were produced by co-transfection of plasmids pHAGE-CMV-Luc2-IRES-ZsGreen-W, HDM-Hgpm2, HDM-tat1b, pRC-CMV-Rev1b, and SARS-CoV-2 spike expressing plasmid into HEK293T cells using jetPRIME transfection reagent.
The determination of 50% neutralization titer (pVNT50) of immunized mouse sera were performed in HEK293T-hACE2 cells. Cells were seeded in white wall 96-well plate (12,500 cells/well). After 24 h of incubation, heat-inactivated sera were serially 2-fold diluted starting at 1:40, incubated with 4 × 10 (Dejnirattisai et al., 2021a) relative light unit (RLU)/ml SARS-CoV-2 pseudovirus at 37 °C for 1 h, and added to each well. After 48 h, luciferase assay was performed using Bright-Glo Luciferase Assay (Promega) and RLU were determined by Cytation7 Cell Imaging Multi-Mode Reader (Bio Tek). Neutralization titers were defined as the reciprocal serum dilution at which RLU were reduced by 50% compared to the virus control wells after subtraction of background RLU in cell control wells.
4.13. Microneutralization assay of infectious SARS-CoV-2 variant isolates
The microneutralization assay was performed as described previously with some modifications (Seephetdee et al., 2021). Briefly, serially diluted heat-inactivated sera (starting with a dilution of 1:20) were pre-incubated with equal volumes of 100 TCID50 of SARS-CoV-2 for 1 h at 37 °C. 100 ml of the virus-serum mixtures were added to pre-seeded Vero E6/TMPRSS2 cell monolayers (1 × 10 (Bowen et al., 2021) cells/well) in duplicate on a 96-well microtiter plate. After 24 h (wildtype, B.1.1.7, B.1.351, and B.1.617.2) and 48 h (B.1.1.529) of incubation, viral protein in the virus-infected cells was detected by ELISA assay using anti-SARS-CoV-2 nucleocapsid mAb (40143-R001, SinoBiological) and HRP-conjugated goat anti-rabbit pAb (P0448, Dako). After 10 min incubation with TMB substrate, the reaction was stopped by the addition of 1N HCl. The absorbance was measured at 450 and 620 nm by a microplate reader (Tecan Sunrise). Neutralization titers were defined as the reciprocal of the highest serum dilution at which the optical density values were reduced by 50% relative to the virus control wells after subtraction of background optical density in cell control wells.
4.14. Enzyme-linked immunospot (ELISpot)
Mouse IFN-γ and IL-4 ELISpot assays were performed with ELISpot kits (Mabtech) according to the manufacturer's instructions. A total of 500,000 splenocytes were restimulated ex vivo with the full-length SARS-CoV-2 B.1.1.529 S 15-mer (overlapping by 11 amino acids) peptide pool (GenScript) in plates pre-coated with anti-IFN-γ or anti-IL-4 antibodies. Splenocytes were incubated with the peptide pool (1 μg/ml/peptide) at 37 °C for 18 h in a 5% CO2 incubator. Cells were removed and the plates were developed by a biotinylated detection antibody, followed by a streptavidin-ALP conjugate. After color development using BCIP/NBT-Plus substrate solution, spots were analyzed in an ELISpot reader. PHA, PMA/Ionomycin, and RPMI 1640/10% FBS served as assay controls.
Authorship contribution
CS, AT, SH and PW conceived and designed research. CS, KB, NB, PL1, PL2, SM, NP and PW conducted experiments. EO, EOS, SK, AT, SH and PW contributed reagents and analytical tools. CS, NB, PL1, PL2, SM, NP and PW analyzed data. CS and PW wrote the manuscript. All authors read and approved the manuscript.
Declaration of competing interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgements
This research project was supported by Mahidol University and Ramathibodi Foundation. C.S. was supported by the Science Achievement Scholarship of Thailand. P.W. was supported by Fundamental Fund, Mahidol University (FF65; BRF1-045-2565), Program Management Unit Competitiveness (PMU-C; C17F640219), Program Management Unit for Human Resources and Institutional Development, Research and Innovation (PMU-B; B05F640145), and National Research Council of Thailand and Mahidol University (N42A650359). AT was supported by PMU-C; C17F640221. IVIS Spectrum In Vivo Imaging System was supported by Mahidol University-Frontier Research Facility (MU-FRF) and Central Animal Facility, Faculty of Science, Mahidol University (MUSC-CAF). We thank Prof. Jetsumon Sattabongkot Prachumsri for NanoAssemblr Benchtop microfluidic device.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105370.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
In vivo expression of circular RNA in mice. BALB/c mice were inoculated with 1 μg of firefly luciferase-encoding circular mRNA-LNP via intramuscular injection and subjected to IVIS Spectrum imaging at the indicated times after administration.
Fig. S2.
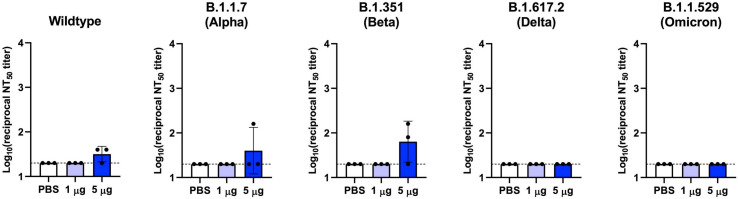
Neutralization of infectious SARS-CoV-2 variants by serum samples from HexaPro-X immunized mice. BALB/c mice (n = 3) were immunized at weeks 0 and 3 with 1 μg (light blue) or 5 μg (blue) of circRNA-LNP encoding HexaPro-X. Control mice were administered with PBS (white). Live virus 50% neutralization titers (NT50) of seven weeks post-boost sera were determined against the indicated infectious SARS-CoV-2 variants. Data are presented as GMT ± geometric SD. Horizontal dotted lines represent assay limits of detection.
Fig. S3.
Neutralization of SARS-CoV-2 variants in convalescent serum samples collected in 2020. (A) Infectious virus 50% neutralization titers (NT50) of convalescent sera were determined against the indicated infectious SARS-CoV-2 variant isolates. (B) Lentivirus-based pseudovirus 50% neutralization titers (pVNT50) of convalescent sera were determined against the indicated SARS-CoV-2 variants. Horizontal dotted lines represent assay limits of detection.
Table S1.
Physicochemical properties of LNP-RNAs (data are presented as mean ± SD, n = 3).
References
- Álvarez-Díaz D.A., et al. Low neutralizing antibody titers against the Mu variant of SARS-CoV-2 in 31 BNT162b2 vaccinated individuals in Colombia. Vaccines. 2022;10 doi: 10.3390/vaccines10020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.E., et al. SARS-CoV-2 spike conformation determines plasma neutralizing activity. bioRxiv. 2021 doi: 10.1101/2021.12.19.473391. 2012.2019, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell George, W. et al. SARS-CoV-2 spike protein stabilized in the closed state induces potent neutralizing responses. J. Virol. 95, e00203-00221, doi:10.1128/JVI.00203-21. [DOI] [PMC free article] [PubMed]
- Cerutti G., et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29:655–663. doi: 10.1016/j.str.2021.05.014. e654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla H., et al. Glycosylation and serological reactivity of an expression-enhanced SARS-CoV-2 viral spike Mimetic. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.-L., et al. Safety and immunogenicity of the SARS-CoV-2 ARCoV mRNA vaccine in Chinese adults: a randomised, double-blind, placebo-controlled, phase 1 trial. The Lancet Microbe. 2022;3:e193–e202. doi: 10.1016/S2666-5247(21)00280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12 doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200. doi: 10.1016/j.cell.2021.02.032. e2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954. doi: 10.1016/j.cell.2021.03.055. e2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPiazza A.T., et al. COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.06.018. 1869-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenig A., et al. In contrast to TH2-biased approaches, TH1 COVID-19 vaccines protect Syrian hamsters from severe disease in the absence of dexamethasone-treatable vaccine-associated enhanced respiratory pathology. bioRxiv. 2021 doi: 10.1101/2021.12.28.474359. 2012.2028, 2021. [DOI] [Google Scholar]
- Edwards A.M., Baric R.S., Saphire E.O., Ulmer J.B. Stopping pandemics before they start: lessons learned from SARS-CoV-2. Science (New York, N.Y.) 2022;375:1133–1139. doi: 10.1126/science.abn1900. [DOI] [PubMed] [Google Scholar]
- Gagne M., et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits comparable B cell expansion, neutralizing antibodies and protection against Omicron. bioRxiv. 2022 doi: 10.1101/2022.02.03.479037. 2002.2003, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S., Gilman M.S.A., McLellan J.S. Structure-based vaccine antigen design. Annu. Rev. Med. 2019;70:91–104. doi: 10.1146/annurev-med-121217-094234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., et al. A SARS-CoV-2 variant elicits an antibody response with a shifted immunodominance hierarchy. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.A.-O., et al. 2021. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie Kathryn M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman D.W., et al. Replicating RNA platform enables rapid response to the SARS-CoV-2 Omicron variant and elicits enhanced protection in naïve hamsters compared to ancestral vaccine. bioRxiv. 2022 doi: 10.1101/2022.01.31.478520. 2001.2031, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, N.Y.) 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen Y.F., et al. Phase I interim results of a phase I/II study of the IgG-Fc fusion COVID-19 subunit vaccine, AKS-452. Vaccine. 2022;40:1253–1260. doi: 10.1016/j.vaccine.2022.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnin K.V., et al. Immunogenicity and efficacy of mRNA COVID-19 vaccine MRT5500 in preclinical animal models. npj Vaccines. 2021;6:61. doi: 10.1038/s41541-021-00324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kim H.W., et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated VACCINE12. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- Kimura I., et al. The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.J., et al. Omicron-specific mRNA vaccine induced potent neutralizing antibody against Omicron but not other SARS-CoV-2 variants. bioRxiv. 2022 doi: 10.1101/2022.01.31.478406. 2001.2031, 2022. [DOI] [Google Scholar]
- McCallum M., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375:864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messali S., et al. A cluster of the new SARS-CoV-2 B.1.621 lineage in Italy and sensitivity of the viral isolate to the BNT162b2 vaccine. J. Med. Virol. 2021;93:6468–6470. doi: 10.1002/jmv.27247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Olmedillas E., et al. Structure-based design of a highly stable, covalently-linked SARS-CoV-2 spike trimer with improved structural properties and immunogenicity. bioRxiv. 2021 doi: 10.1101/2021.05.06.441046. 2005.2006, 2021. [DOI] [Google Scholar]
- Pallesen J., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman Z., et al. A computational dissection of spike protein of SARS-CoV-2 Omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.17.473260. 2012.2017, %J bioRxiv (2021) [DOI] [Google Scholar]
- Seephetdee C., et al. Mice immunized with the vaccine candidate HexaPro spike produce neutralizing antibodies against SARS-CoV-2. Vaccines. 2021;9 doi: 10.3390/vaccines9050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N. Engl. J. Med. 2021;384:2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., et al. SARS-CoV-2 lambda variant remains susceptible to neutralization by mRNA vaccine-elicited antibodies and convalescent serum. bioRxiv. 2021 doi: 10.1101/2021.07.02.450959. 2007.2002, 2021. [DOI] [Google Scholar]
- Tatsi E.-B., Filippatos F., Michos A. SARS-CoV-2 variants and effectiveness of vaccines: a review of current evidence. Epidemiol. Infect. 2021;149:e237. doi: 10.1017/S0950268821002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-T., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriu K., et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A.B., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks S.H., et al. Mapping SARS-CoV-2 antigenic relationships and serological responses. bioRxiv. 2022 doi: 10.1101/2022.01.28.477987. 2001.2028, %J bioRxiv (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B., et al. Boosting with Omicron-matched or historical mRNA vaccines increases neutralizing antibody responses and protection against B.1.1.529 infection in mice. bioRxiv. 2022 doi: 10.1101/2022.02.07.479419. 2002.2007, 2022. [DOI] [Google Scholar]