Abstract
Coronavirus Disease 2019 (COVID-19) still presents a pandemic trend globally. Detecting infected individuals and analyzing their status can provide patients with proper healthcare while protecting the normal population. Chest CT (computed tomography) is an effective tool for screening of COVID-19. It displays detailed pathology-related information. To achieve automated COVID-19 diagnosis and lung CT image segmentation, convolutional neural networks (CNNs) have become mainstream methods. However, most of the previous works consider automated diagnosis and image segmentation as two independent tasks, in which some focus on lung fields segmentation and the others focus on single-lesion segmentation. Moreover, lack of clinical explainability is a common problem for CNN-based methods. In such context, we develop a multi-task learning framework in which the diagnosis of COVID-19 and multi-lesion recognition (segmentation of CT images) are achieved simultaneously. The core of the proposed framework is an explainable multi-instance multi-task network. The network learns task-related features adaptively with learnable weights, and gives explicable diagnosis results by suggesting local CT images with lesions as additional evidence. Then, severity assessment of COVID-19 and lesion quantification are performed to analyze patient status. Extensive experimental results on real-world datasets show that the proposed framework outperforms all the compared approaches for COVID-19 diagnosis and multi-lesion segmentation.
Keywords: Automated diagnosis, Lesion segmentation, COVID-19, Adaptive multi-task learning, Explainable multi-instance learning
1. Introduction
Severe acute respiratory syndrome coronavirus (SARS-CoV) [1], middle east respiratory syndrome coronavirus (MERS-CoV) [2], and novel coronavirus (SARS-CoV-2) are highly pathogenic coronaviruses known to infect humans, whose infection can cause severe respiratory syndromes. At the beginning of 2020, SARS-CoV-2 quickly became a pandemic worldwide. SARS-CoV-2 infection can inflame the air sacs in the lungs and cause pleural effusion, which can lead to breathing difficulties, fever, cough, or other flu-like symptoms in patients. The diseases caused by this virus are collectively referred to as Coronavirus Disease 2019 (COVID-19) [3]. It has made a huge impact on the medical systems and economic activities around the globe. Molecular diagnostic testing and imaging testing are two main detection methods for COVID-19 infection. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) as a molecular diagnostic test performed in standard laboratories, it is a common standard for the detection of COVID-19. However, the current laboratory test has a long turnaround time, and RT-PCR testing for COVID-19 may be falsely negative due to specimen contamination or insufficient viral material in the specimen, which may not be sufficient to confirm infection or infection-free [4], [5].
Radiology imaging procedure has a faster turnaround time than RT-PCR, which facilitates the rapid screening of suspected COVID-19 patients in the severe and complicated novel coronavirus pneumonia epidemic. As one of the most common imaging tests, computed tomography (CT) is an effective tool for screening lung lesions and a means of diagnosing COVID-19 [6]. Chest CT images provide more detailed pathological information, which can quantitatively measure lesion size and the extent of lung involvement better [7]. Due to its convenience, accuracy, high positive rate, and good reproducibility, CT images play an indispensable role in screening COVID-19 and assessing the progression of patients. However, manual screening is a time-consuming and labor-intensive task. In addition, CT images of COVID-19 and some other common types of pneumonia have similar imaging characteristics. Sometimes even the most experienced physicians have difficulty analyzing them without relying on other testing methods. Therefore, reliable computer-aided diagnosis (CAD) methods based on CT images are needed to boost the efficiency of COVID-19 diagnosis and assessment. Developing such methods has been a research hotspot in the past year.
Because there are huge differences in the appearance, size, and location of the pneumonia lesions, it is difficult to design an appropriate CAD method to deal with the complex features of pneumonia lesions only using machine vision methods such as classic image processing techniques or traditional statistical learning methods.
Benefiting from the development of deep learning technology, the application of convolutional neural networks (CNNs) in automated COVID-19 diagnosis has become popular. For instance, there have published several recent works that focused on automated diagnosis [8], [9], [10] or lesion segmentation of COVID-19 [11], [12], [13]. However, most existing automated diagnosis methods are two-stage or depend heavily on expert knowledge and experience. And as a result, time cost and human factors will affect the efficiency and consistency of diagnosis. In another aspect, the diagnosis task of distinguishing COVID-19 from other common pneumonia with similar symptoms has received plenty of attention, but the severity of patients is rarely assessed which can be more challenging. In addition, the diagnosis results of deep learning methods also need to be explained with clinical significance. For lesion segmentation, some studies treat the segmentation task as an intermediate step in the diagnosis of COVID-19, and a detailed quantitative or qualitative analysis of segmentation results is not conducted. The segmentation of multiple lesion areas in the lung has been demanded due to clinical potential for disease progression analysis, while it is challenging as lesions are often similar and the sizes are quite small.
It is observed that most previous works consider automated diagnosis and lesion segmentation as two independent tasks or treat segmentation as a pre-step of disease diagnosis, thus ignoring the correlation between these two tasks. During the segmentation of lesion areas, rich spatial information and tissue type information can be obtained from CT images. Similarly, the diagnosis task also needs to pay attention to such information. Since diagnosis and segmentation are highly related tasks, the multi-task learning (MTL) scheme can utilize the potential representations between these two tasks to improve the performance on each task [14]. And this MTL method is also faster than the two-stage methods of first segmentation and then classification.
Based on the above analysis, this paper proposes a multi-task learning framework that combines automated diagnosis of COVID-19 and lung multi-lesion segmentation in CT images. Specifically, the regions related to pneumonia assessment are few and unevenly distributed in each CT image of the individual. To address this problem, we treat each CT image as a bag of CT slices through a weakly supervised multi-instance learning (MIL) strategy and use these instance bags as the inputs of the following model. A multi-task network is designed, including a shared encoder for feature extraction, a diagnosis branch and a segmentation branch. Here, the shared encoder learns common representations of both tasks and generates the information exchange, the diagnosis branch and the segmentation branch are used to solve the diagnosis and segmentation tasks, respectively.
In addition, based on the results of lesion segmentation, severity assessment of COVID-19 and lesion quantification are performed. And explainability of the diagnosis results can be implemented through a dedicatedly-designed Transformer MIL Pooling (TMP) layer. Extensive experiments are conducted on real-world COVID-19 datasets to verify the effectiveness of the proposed method. The contributions of this work can be summarized as follows:
-
1.
A multi-task learning framework is proposed to perform automated diagnosis of COVID-19 and lung multi-lesion segmentation in CT images simultaneously. Both tasks can adaptively exchange unique task-related information and learn common representations to improve the performance on each task.
-
2.
A novel explainable multi-instance learning strategy is designed, in which a TMP layer considers the expressive abilities of different instances, constructs the local to the global representations of CT images, and can endow the diagnosis results with a certain degree of explainability by suggesting which sets of instances exert similar effects on the diagnosis.
-
3.
The explainable multi-instance multi-task network (EMTN) in the framework is flexible, which has EM-Seg (EMTN with only the segmentation branch) and EM-Cls (EMTN with only the classification branch) two variants. Either variant of EMTN can be employed to perform diagnosis of COVID-19 or lesion segmentation in CT images.
The rest of the paper is organized as follows: Section 2 introduces the related works about the automated diagnosis and segmentation of COVID-19 based on CT images. In Section 3, we explain the proposed framework in detail. Then, the datasets and experimental setup are introduced in Section 4. The extensive experiments and discussions are presented in Section 5. Finally, Section 6 concludes this paper.
2. Related work
In this section, we briefly review the deep learning based COVID-19 classification and segmentation studies, including segmentation in CT images of COVID-19 and automated diagnosis of COVID-19 from CT images.
2.1. Segmentation of CT images infected with COVID-19
Segmentation is a key task in the assessment of COVID-19. Its main goal is to identify and mark the regions of interest (ROI) such as lung, lung lobes, or lesion areas in CT images. In terms of target ROI, the segmentation methods can be divided into two categories, namely, methods for lung fields and for lung lesions.
(1) Segmentation methods for lung fields: The methods aim to separate lung fields (i.e., the whole lung or lung lobes) from background areas. This is considered a basic task of segmentation in CT Images of COVID-19. For example, Chaganti et al. [11] proposed a two-stage method for segmenting lungs and lung lobes in CT images of COVID-19 patients. In the first stage, deep reinforcement learning was used to detect lung fields. Then, a depth image-to-image network was employed to segment the lungs and lung lobes from the detected lung fields. Xie et al. [12] introduced a non-local neural network module in the CNN to capture structural relationships of different tissues and perform the segmentation of lung lobes.
(2) Segmentation methods for lung lesions: The purpose is to segment lung lesions from other tissues in CT images. Since the lesions are of a variety of shapes and textures, and the sizes are usually small. A focus on lung lesions can improve the efficiency of follow-up in patients with COVID-19. Lung lesion segmentation is widely regarded as a challenging task. Some studies performed binary segmentation of lesions, that is, predicting the masks of all types of lesions without distinction [15], [16], [17]. For instance, Abdel-Basset et al. [15] adopted a dual-path network architecture, and designed a recombination and recalibration module that exchanges feature information to improve the segmentation of infected areas. Wu et al. [16] proposed an encoder–decoder CNN architecture with attentive feature fusion and deep supervision strategy, and obtained the locations of the infected areas. Chassagnon et al. [17] designed a CovidENet, which is an ensemble of 2D and 3D architectures based on AtlasNet [18]. CovidENet was employed to segment the lung lesions as a whole, and it can reach a segmentation level close to physicians in terms of dice coefficient and Hausdorff distance. Moreover, some scholars carried out researches on multi-lesion segmentation methods. Chaganti et al. [11] also paid attention to multi-lesion segmentation. They used a densely connected U-Net [19] to segment ground-glass opacities (GGO) and lung consolidation. Fan et al. [20] utilized a two-stage method with two CNNs for multi-lesion segmentation. The first CNN segmented the total lesions, and then based on the segmentation results of the first CNN, the second CNN was used to further segment the GGO and lung consolidation of total lesions. Similar to Fan et al. [20], Zhang et al. [13] also adopted a two-stage method to perform lesion segmentation. Total lesions and healthy lung tissues were first segmented. After that, different types of lesions were separated from total lesions, including GGO, lung fibrosis, lung consolidation, etc.
2.2. Automated diagnosis of COVID-19 from CT images
Automated diagnosis (classification) is one crucial task in COVID-19 detection. It aims to provide rapid and accurate judgments for the diagnosis of suspected COVID-19 patients. Considering the screening COVID-19 patients as the main target, the diagnosis of COVID-19 based on CT images mainly includes two categories: (1) binary classification (COVID-19 or non-COVID-19); (2) multi-class classification (COVID-19 and other types of pneumonia).
(1) Binary classification: In this category, many studies have been carried out to distinguish COVID-19 patients from non-COVID-19 patients. Li et al. [6] utilized a modified CheXNet [21] to diagnose COVID-19. The modified CheXNet was first pre-trained on a chest X-rays dataset, and then the pre-trained network was transferred to the target task by transfer learning. Wang et al. [22] utilized a 3D U-Net [23] based segmentation model to obtain lung segments in CT images, then coupled two 3D-ResNets into a classification model via a priority attention strategy, and finally predicted the type of patients through this classification model. A weakly-supervised automated diagnosis framework [24] was established. Specifically, a pre-trained U-Net was adopted to segment lung fields. Afterwards, the segmented lung fields and original CT images were as the inputs of a 3D deep neural network, which can determine whether patients are infected with COVID-19 or not. Shaban et al. [25] proposed an enhanced KNN classifier with hybrid feature selection, which selected significant features from CT images and detected COVID-19 patients based on these features. Ardakani et al. [8] extracted 1020 CT slices from CT images of 108 COVID-19 patients and 86 non-COVID-19 patients, and ten classical CNNs were employed to distinguish between COVID-19 patients and non-COVID-19 patients based on the extracted 2D slices. However, the classification results of Ardakani et al. were slice-level rather than individual-level. This may have the problem of data leakage. Bai et al. [9] segmented the lung regions in CT images and sliced them, then an EfficientNet B4 [26] was used to obtain the classification score of each 2D slice. After that, they integrated the predicted classification scores of several 2D CT slices to make final decisions at the individual level.
(2) Multi-class classification: COVID-19 has similar imaging features in CT images to other viral, bacterial, and community-acquired pneumonia, especially viral pneumonia. It is a challenging task for physicians to judge suspected patients if the pneumonia-related lesions are subtle. The classification of COVID-19 patients from other types of pneumonia patients can accelerate the screening of patients in clinical practice. Wang et al. [27] proposed a two-stage method to perform the classification of COVID-19 and other types of pneumonia. In the first stage, a 3D DenseNet121-FPN was adopted to segment lung fields in CT images, and then in the second stage, a COVID-19Net was designed to determine the label of each patient based on the patient’s clinical information and segmented lung fields. Han et al. [10] proposed a deep 3D multiple instance learning method based on the attention mechanism. With a certain number of 3D patches extracted from CT images as the inputs of a 3D CNN, the patch-level features were obtained through this CNN, and an attention-based pooling layer mapped the patch-level features into embedding space. Then, the features were transformed into the Bernoulli space, which can give the probabilities of patients with COVID-19. Song et al. [28] utilized OpenCV [29] to extract 15 complete lung slices from each CT image, and a diagnosis system was proposed to identify patients with COVID-19 from other types of pneumonia based on these slices, in which the system mainly consists of a ResNet50 fused with feature pyramid networks. Similarly, Wang et al. [30] manually selected slices with lesions according to the typical characteristics of pneumonia, and separated the CT slices into lung fields and other regions. Afterwards, they used these lung fields as the inputs of a pre-trained GoogLeNet [31] for the diagnosis of patients.
3. Methodology
3.1. Method overview
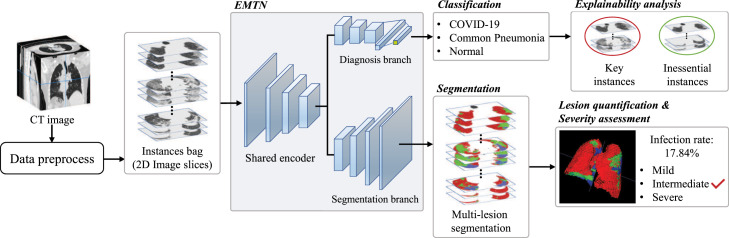
We construct a framework to perform automated diagnosis of COVID-19 and multi-lesion segmentation in CT images. The proposed framework is illustrated in Fig. 1. In order to weaken the influence of different CT images qualities, several preprocessing steps are adopted, and then a bag of instances is constructed by randomly choosing a set of 2D image slices. An explainable multi-instance multi-task network (EMTN) is designed to simultaneously perform classification and segmentation tasks based on the instance bags. As shown in Fig. 1, the architecture of EMTN consists of a shared encoder and two task branches (i.e., diagnosis and segmentation). The shared encoder plays a role in feature extraction of 2D image slices in each bag of instances, where the obtained features are fed into the following diagnosis and segmentation branches. In the diagnosis branch, these extracted features are used to construct the global representations of CT images, and the diagnosis of patients is determined through a classification layer learned from these global representations. In the segmentation branch, multi-lesion segmentation is performed via a decoding architecture symmetrical to the shared encoder. In the way of multi-task learning, task-related features such as textures and shapes can be properly utilized by both tasks to improve performance. In addition, EM-Seg (EMTN with only the segmentation branch) and EM-Cls (EMTN with only the classification branch) as two variants of EMTN, they can be employed to perform segmentation or diagnosis task individually.
Fig. 1.
Illustration of the pipeline of the proposed framework. For each CT image, a bag of instances (2D image slices) is constructed through the preprocess steps, which is fed into the explainable multi-instance multi-task network (EMTN) for the diagnosis of COVID-19 and multi-lesion segmentation. Following EMTN, explainability analysis endows the diagnosis results with explainability, lesion quantification and severity assessment provides the detailed information of patient status.
After classification and segmentation, lesion quantification and severity assessment of COVID-19 patients are carried out to further enrich the functionalities of the framework. Moreover, the diagnosis results can be more explainable by observing the key instances.
3.2. Network architecture
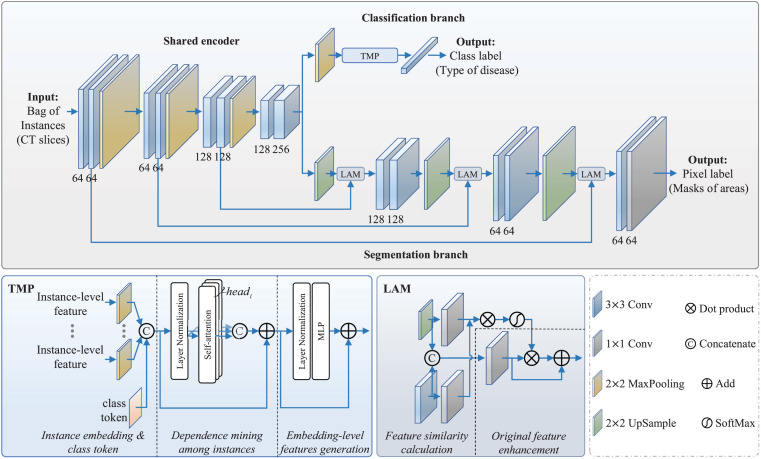
As above-emphasized, the proposed backbone network is an encoder-focused MTL model used to simultaneously perform classification of patients and segmentation of multi-lesion. As shown in Fig. 2, the forward-propagation direction of the bags or the features in the EMTN is from the shared encoder to the classification branch and the segmentation branch. Following this, the class label and the pixel label are determined by the corresponding branches.
Fig. 2.
The architecture of the proposed explainable multi-instance multi-task network (EMTN).
Specifically, the shared encoder contains four down-sampling blocks. The first three blocks include two convolutional (Conv) layers followed by a 2 × 2 max pooling layer. They are designed to down-sample the intermediate feature maps. The last down-sampling block only has two Conv layers, which is designed to ensure the size of the features extracted by the encoder. The number of channels for Conv layers in four blocks are [64, 64], [64, 64], [128, 128] and [128, 256], respectively. Each Conv layer has one convolutional layer with unit stride and zero padding, followed by batch normalization (BN) and rectified linear unit (ReLU) activation. The segmentation branch has three up-sampling blocks, which forms a symmetrical codec architecture with the shared encoder. The number of channels for Conv layers in three up-sampling blocks are [128, 128], [64, 64] and [64, 64], respectively. All up-sampling blocks have the same Conv layers as down-sampling blocks, and a lesion attention module (LAM) is adopted in each up-sampling block to concatenate the up-sampled feature maps and the outputs of the corresponding down-sampling block. The LAM mainly includes feature similarity calculation and original feature enhancement two core parts. In the feature similarity calculation part, linear transformation, dot product, and feature decoupling are performed on the up-sampled feature maps and corresponding down-sampled feature maps to obtain a similarity matrix. Then, in the feature enhancement part, this similarity matrix is multiplied by the dimensionality reduced features to get the enhanced features, where the dimensionality reduced features can be obtained by concatenating and transforming the up-sampled and down-sampled features. Finally, the enhanced features are added to the original feature maps as the output of LAM. The LAM is designed to better consider the global information in the original images and enhance some tiny features beneficial to segmentation. A successful application was presented in [32]. The segmentation branch outputs the masks for the four areas (i.e., lung areas, GGO, lung consolidation, and background areas) of image slices in the bag. The classification (and also diagnosis) branch contains a 2 × 2 max pooling layer, a TMP layer, and a classification layer. In this classification branch, local instance-level features are mapped to the embedding-level space through a novel explainable MIL strategy, and these embedding-level features are further processed by classification layer to obtain predicted diagnosis results.
Both tasks are simultaneously supervised by the MTL loss function which is usually a combination of single task loss functions. Since the loss functions of different tasks may have unbalanced contributions when optimizing the model parameters, a weight-adaptive multi-task learning loss function is designed in this paper. Specifically, let be a training set containing samples, where and denote the bag of instances for the th CT image and the corresponding class label, respectively; denotes the th instance in , and is the corresponding ground-truth segmentation mask for . For single task, the loss functions and for classification and segmentation tasks are as follows.
| (1) |
| (2) |
where is the cross-entropy loss, and is the aggregation of cross-entropy loss and dice loss. In terms of a given bag (e.g., ) being diagnosed as a specific class (e.g., ) or a given instance (e.g., ) being segmented as a predicted mask (e.g., ), function and denote the probability obtained by the classification branch and segmentation branch, respectively. In this work, three classification targets and one multi-categories semantic segmentation are performed, namely =3, =4.
Then, the single task loss function is weighted by the trade-off factors to form a joint loss function.
| (3) |
where and are the trade-off factors for the classification task and segmentation task, respectively. The factors are learnable parameters that can be adjusted in the process of model optimization. The regularization terms [33] are added to avoid the trade-off factor of each task reaching minimum, maximum, negative values, or zero solution.
3.3. Explainable multi-instance learning
COVID-19-related infection regions usually exist in some partial regions of the lung, while the regions in CT images are unlabeled, i.e., only the entire CT images have the corresponding labels. This situation is seen as a weakly supervised problem which can be solved with multi-instance learning (MIL) strategy. In this section, the details of the proposed explainable MIL strategy are introduced.
As mentioned in Section 3.1, a bag of image slices is constructed as the input of the network. Let denotes the bag which represents the th CT image, where represents the th slice of the th CT image. And then, the shared encoder performs feature extraction on these CT slices to obtain instance-level features , followed by a proposed Transformer MIL Pooling (TMP) layer to generate embedding-level features from instance-level features.
The proposed TMP layer is one of the few attempts that combine transformers [34] with MIL. Medical images usually have explicit sequences, such as modalities, slices, patches, etc. From these sequences, some important long-range dependence and semantic information can be mined. Transformers can effectively focus on long-range dependence in the data sequences. In the process of feature learning, MIL also needs to pay attention to the dependence among instances to generate embedding-level features. As a useful tool to process sequence relations, Transformers combine instance-level features with MIL, which can not only provide the degree of connection between instances and diagnosis results, but can also further improve the performance of the network. This is the motivation for us to propose TMP. Different from transformers which only analyze a single image, the TMP layer in this work is a core component of explainable MIL. It operates on features in the instance feature space and the embedding feature space, and generates embedding-level features, which can also provide the explainability of diagnosis results.
As shown in Fig. 2, the designed TMP layer contains the following three parts in series.
(1) Instance embedding and class token: For each extracted instance-level feature , is the number of channels, is the size of the feature map. In order to facilitate the propagation of features among different layers, we flatten the instance-level features and concatenate them into a sequence of embedded instance-level features , where is the reshaped instance-level feature map, is the number of instances, which also serves as the input sequence length for the TMP. Then, the embedded instance-level features are mapped to dimensions with a trainable linear projection. The above process is instance embedding.
Considering that it is unfair to choose one of the embedded instance-level features as the feature for subsequent classification, a learnable vector is prepended to the sequence of embedded instance-level features, which is referred to as class token . The class token plays the role of the bag representation, which will learn in the following steps and be used to predict the class label.
The resulting sequence of embedded instance-level features and the class token is as follows:
| (4) |
where and indicate the resulting sequence and the class token of th CT image, respectively. is the trainable linear projection.
(2) Dependence mining among instances: The core of this step is multi-head self-attention (MSA) [34] which mines the dependence in the resulting sequences based on the calculation of similarity among the elements of the sequence. MSA is an extension of self-attention (SA), which performs SA operation several times in parallel, and concatenates their outputs as the output of MSA.
For the th element in the resulting sequence , it is transformed into three vectors: query vector , key vector , and value vector , where and are used to compute an attention weight that indicates the dependence with the th element, represents the unique feature map of the element. Then, a weighted sum over the values of all elements in the sequence is computed (Eq. (5)).
| (5) |
| (6) |
| (7) |
where , and are the sets of , and , respectively. is a trainable linear projection which transforms the elements in the sequence to the corresponding , , and . is the attention weight which reflects the dependence between the class token and instances. In addition, the dependence among instances can also be calculated based on .
| (8) |
where represents performing h operations in parallel. is a trainable parameter matrix that controls the output size of feature maps as dimensions.
In this step, the MSA is with skip connections like residual networks and with a layer normalization (LN) to normalize each element in the sequence. Therefore, with the resulting sequence in the first step as input, the second step can be described by the following equation:
| (9) |
where is the output of the second step.
(3) Embedding-level feature generation: In this step, the embedding-level representations are generated with a multi-layer perceptron (MLP). The MLP is composed of two linear layers with dropout and a Gaussian error linear unit (GELU) activation which is inserted between these two layers. Like MSA in the second step, the MLP also has the residual connection and a layer normalization. This step can be written as follows.
| (10) |
where is the output embedding-level features. As mentioned in step 1, a class token is prepended to the sequence as the subsequently used bag representation. After three steps of learning, the learned class token is employed to predict the final classification.
The above three steps are the details of TMP. Furthermore, to provide the explainability of diagnosis results, the proposed explainable MIL strategy can not only process the weakly supervised problem but also suggest key instances which exert similar effects on the diagnosis. The acquisition process of the key instances is described as follows.
At each operation, we get an attention matrix that defines the attention maps from the output class token to the input instance-level space. To process the attention matrices of multiple operations, we take the average of these matrices across all operations and normalize them to get the averaged attention matrix . Then, the attention weight between the class token and each instance-level feature is obtained with . Specifically, the class token corresponds to bag representation, and the predicted result is obtained by mapping the bag representation to the label space. Thus, the class token can represent the bag of instances. Each instance-level feature corresponds to a unique instance in the bag (a 2D image slice). It is straightforward that the higher attention weight between the class token and one instance-level feature, the greater impact this instance-level feature has on the bag representation and the final diagnosis result. The key instances can be selected according to the values of the attention weights, and these instances usually contain more lungs or lesion areas than other instances, which provides the basis for the posterior test of the diagnosis results.
This explainable MIL is different from the standard multi-instance assumption. The standard multi-instance assumption states that a bag is positive if and only if it contains at least one positive instance, which relies heavily on the correctness of labels and is susceptible to false positive instances. The explainable MIL strategy considers the relationship between instances and bag representation, and assigns suitable weights for instance-level features to generate embedding-level features and bag representation. This allows the generated bag representation can better characterize the samples and reduce the interference of false positive instances.
3.4. Quantitative analysis of lesions and severity assessment of COVID-19
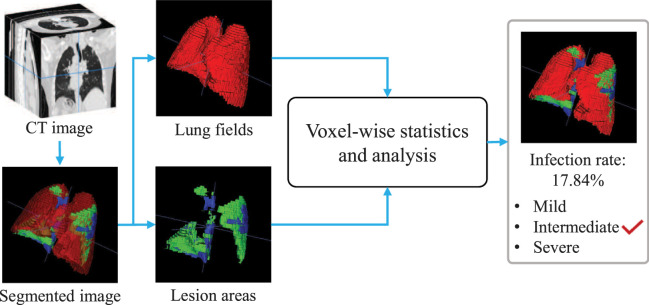
The quantitative analysis of lesions and severity assessment of COVID-19 can provide more detailed information for the evaluation of patient status, clinical treatment effect, or drug experiments. In this section, we use a voxel-wise analysis method to further analyze the segmentation results.
As shown in Fig. 3, at the segmentation branch in EMTN, the positions and sizes of the lung fields, as well as GGO and lung consolidation in CT images can be obtained. In order to facilitate the analysis, GGO and lung consolidation are collectively referred to as the lesion areas. Briefly, we count the number of voxels in lung fields and lesion areas. Let , and denote the number of voxels in the lung fields, GGO and lung consolidation, respectively. Then, the percentage of the number of voxels in the lesion areas to that in the lung fields is calculated as follows.
| (11) |
Fig. 3.
The pipeline of the lesion quantification and severity assessment via voxel-wise analysis.
where is the key indicator for quantitative analysis of lesions, i.e., infection rate. Furthermore, the severity of COVID-19 patients can be assessed with .
4. Experimental setup
4.1. Dataset and preprocess
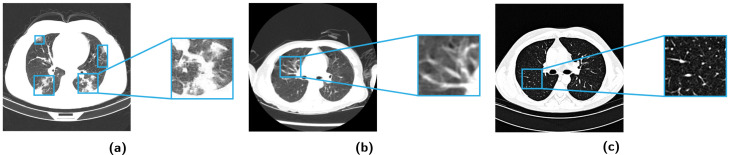
The real-world datasets of CT images are acquired from three sources: China Consortium of Chest CT Image Investigation (CC-CCII),1 COVID19-CT-dataset,2 and Radiopaedia.org.3 The combined dataset contains CT images of COVID-19 patients, common pneumonia patients, and normal control cases in three groups. Fig. 4 shows the axial surface of CT images in different groups.
Fig. 4.
Axial surface of CT images and variations in CT images of different groups. (a) One case of COVID-19 group. The COVID-19-specific patterns of lung infections are denoted by blue boxes, including ground-glass opacities and consolidation. (b) One case of common pneumonia group. The increased and/or disordered lung marking is (denoted by blue boxes) the CT manifestations of early common pneumonia patients. (c) One case of normal control group. The black area is normal lung tissue, and the white dots are normal physiological structures such as blood vessels, bronchi, etc.
The CC-CCII dataset consists of a total of 2778+ CT images from 2778 patients [13]. These patients and their CT images are collected from six hospitals, including 917 COVID-19 patients, 983 common pneumonia patients, and 878 normal control cases. All COVID-19 patients are tested positive by RT-PCR. The common pneumonia patients are confirmed based on standard clinical, radiological, and molecular test results. The segmentation of CT images of COVID-19 patients is manually annotated and reviewed by five experienced radiologists. The annotated range mainly contains lesion areas (i.e., GGO and lung consolidation) which are used to distinguish COVID-19 from other cases, and to distinguish lung fields from background areas in CT images. Since part of CT images masked other regions than the lungs, and this part images cannot provide complete information of patients, so these images are screened out. In addition, those CT images with a small number of slices are also screened out. After screening, the CC-CCII dataset includes 529 COVID-19 patients, 592 common pneumonia patients, and 520 normal control cases.
The COVID-19-CT-dataset is an open-access chest CT image repository, it consists of 3000+ CT images from 1013 patients with confirmed COVID-19 infections [35]. These patients and their CT images are collected from two general hospitals in Mashhad, Iran. All COVID-19 patients have positive RT-PCR tests and accompany by supporting clinical symptoms at the point of care in an inpatient setting. And CT images of patients are visually evaluated by two board-certified radiologists to confirm the presence of COVID-19 infection. For the CT images in which the first two radiologists are in disagreement, the final decision is rendered by a third more experienced radiologist.
Radiopaedia is a non-profit, international collaborative, open-edit radiology resource compiled by radiologists and other health professionals from across the globe. We collect 130 common pneumonia patients from Radiopaedia as a supplement. The collected common pneumonia patients are confirmed by their contributors based on standard clinical, radiological, or molecular test results.
The quality of CT images is inconsistent due to the use of different CT scanners or different scanning parameters during the image acquirement process, it is a challenging task to diagnose based on these images. To reduce the risk of data leakage and to ensure the quality of images in the datasets, we make sure that only one scan (CT image) per patient is selected and each CT image is checked. Then, the following processing procedures are performed.
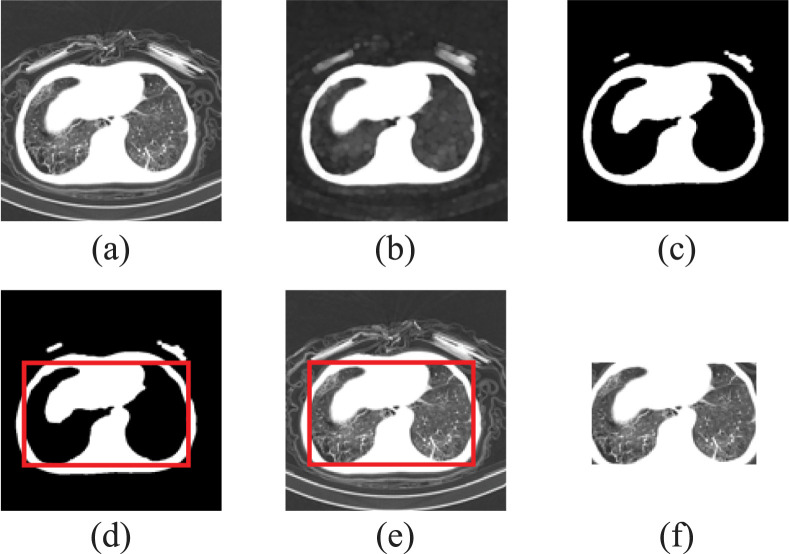
Specifically, the axial slices are first extracted from each CT image according to the physical spacing between slices [Fig. 5(a)]. To reduce the influence of noise in the images, each axial slice is processed with morphological operations, including structuring elements acquisition, erosion, and dilation [Fig. 5(b)]. Then, the images are binarized by Otsu algorithm [Fig. 5(c)]. With the above steps, the basic contours of human body and lung fields are identified. We detect the key points of the inner and the outer contours respectively, and identify the inner contour by the size of enclosed area [Fig. 5(d)]. Morphological operations, binarization, and contours detection are only used to determine the rectangle region of interest (ROI), therefore these operations will not lead to a loss of the image information. After that, according to the shape of the inner contours, the axial slices are cropped to obtain the corresponding ROIs [Fig. 5(e–f)]. Finally, with the MIL strategy, a set of preprocessed axial slices in each CT image are randomly selected to construct a bag of instances, and each bag can represent a corresponding individual and serve as the input of EMTN. In this way, we expect that the problems of uneven data quality and data discrepancies can be alleviated.
Fig. 5.
Procedures of preprocessing. (a) Extract the axial slices. (b) Morphological operation on the image. (c) Binarize image by thresholding. (d) Identify the basic contour and make mask. (e) Multiply the image with mask. (f) Crop the image to get the ROI.
4.2. Evaluation metrics
The proposed framework performs both classification of COVID-19 patients and segmentation of multi-lesion two tasks. In order to verify the effectiveness of the overall framework, 5-fold cross-validation is adopted, and different metrics are used in the performance evaluation.
For the classification task, five evaluation metrics are used to evaluate the classification performance, including Accuracy (ACC), Precision, Recall, F1 Score, and the area under the receiver operating characteristic (ROC) curve (AUC). The descriptions of these metrics are given below.
| (12) |
| (13) |
| (14) |
| (15) |
where TP, TN, FP, and FN are the number of true positive samples, true negative samples, false positive samples and false negative samples, respectively. It should be noted that ROC curve and AUC only partially summarize or explain the performance of diagnosis, especially when the data is unbalanced or the values of AUC of different classifiers are close, judging a better curve is difficult. The leftmost partial area of ROC curve is the region of interest for classifying fewer positives than negatives, and ROC curves of better classifiers usually go up quickly or stay to the left side. A concordant partial AUC () focuses on the region of interest in the ROC, and provides a good explanation for partial areas in ROC curves. The is a foundational partial measure, which has all the interpretations offered by the AUC [36]. For an ROC curve , the definition of is as follow:
| (16) |
where and are false positive rate (FPR) and true positive rate (TPR), respectively. denotes inverse function. In this work, area measures are performed for three parts of an ROC curve i = 1,2,3, including the leftmost partial curve (i=1, FPR=[0.00,0.33]), the middle partial curve (i=2, FPR=[0.33,0.66]), and the rightmost partial curve (i=3, FPR=[0.66,1.0]).
For the segmentation task, three commonly used metrics are adopted to evaluate the segmentation performance, including Dice Score (DC), Positive Predict Value (PPV), and Sensitivity (SEN). The evaluation metrics are defined as:
| (17) |
| (18) |
| (19) |
where and denote the predicted segmentation masks and the ground-truth, respectively.
4.3. Implementation details
For the CC-CCII dataset, 70% of the samples and 20% of the samples are respectively selected as the training set and validation set to supervise the training of EMTN, and 10% of the samples are as the testing set to evaluate the performance. Since there has no ground truth for segmentation contained in the COVID-19-CT dataset and common pneumonia patients collected from Radiopaedia, both of them are used as an additional test set to evaluate the performance of EMTN, which do not participate in the training stage of EMTN. Furthermore, the samples in these two datasets can be used to train and test a variant of EMTN, i.e., EMTN with only the classification branch, and the data split also follows a 7:2:1 ratio.
The proposed method is implemented with , in which EMTN is implemented based on the deep learning library. The training process is carried out on a workstation equipped with four NVIDIA GTX 1080Ti graphics cards. In both stages of training and testing, we randomly select a certain number of axial slices from each CT image to construct a bag of instances, where the bag size varies within , and the sizes of all bags constructed from different CT images are the same. During the training process, the EMTN is optimized by Stochastic Gradient Descent (SGD) algorithm with 100 epochs, and the weight decay is 1 . The initial learning rate is 0.0001, and it decays by 35% every 20 epochs.
5. Experiments and analysis
In this section, comparison study (Section 5.1) examines the superiority of the proposed method, ablation study (Section 5.2) evaluates the effectiveness of the MIL and MTL strategies. Both comparison study and ablation study are performed based on the CC-CCII dataset. Additional study (Section 5.3) evaluates the performance of the proposed method on the COVID-19-CT dataset and common pneumonia patients collected from Radiopaedia. Section 5.4 discusses the limitations of this work and future research direction.
5.1. Comparison study
We first test the performance of the proposed EMTN on both classification and segmentation tasks. Since the datasets in most of the methods listed in Section 2 are not accessible, the comparison of these methods is not possible. To show the superiority of the EMTN, it is compared with several state-of-the-art classification and segmentation methods. The competing classification methods in [37], [38] are the non-MIL method and the normal MIL method, respectively. ResNet18-Voting method [37] is one of the common ensemble methods and is often used in the field of patch-level or slice-level medical image analysis. It uses ResNet18 as the backbone network and performs individual-level classification through voting. Gated attention MIL method [38] combines attention mechanism with MIL to replace the pooling operators. Furthermore, two state-of-the-art segmentation methods U-Net [19] and U-Net++ [39] are compared for multi-lesion segmentation. U-Net [19] designs the commonly used symmetric U-shaped architecture, which is a classic image segmentation method. Based on U-Net, U-Net++ [39] adopts dense skip-connections to improve the fluidity of gradient, which connects the semantic gap between feature maps in the compression path and the expansion path. These methods are applied to the datasets used in this work, whose main architectures and parameter settings are consistent with those in their respective papers.
5.1.1. Classification of COVID-19
We compare and analyze the performance on three different classification targets of EMTN, EM-Cls, ResNet18-Voting, and Gated-Attention. EM-Cls represents a variant of EMTN, i.e., EMTN with only the classification branch. The classification targets include (1) COVID-19 patients vs. normal control cases (denoted as COVID-19 . NC), (2) COVID-19 patients vs. common pneumonia patients (denoted as COVID-19 . CP), and (3) common pneumonia patients vs. normal control cases (denoted as CP . NC). Table 1 shows the comparison on ACC and AUC.
Table 1.
Comparison of classification results of different methods on three targets (bag size=100).
| Targets | Method | ACC() | AUC() |
|---|---|---|---|
| COVID-19 NC | ResNet18-Voting | 88.97 |
90.08 |
| Gated-Attention | 94.48 |
94.63 |
|
| EM-Cls | 96.73 |
96.98 |
|
| EMTN | 98.62 |
98.90 |
|
| COVID-19 CP | ResNet18-Voting | 81.28 |
81.63 |
| Gated-Attention | 90.34 |
90.68 |
|
| EM-Cls | 93.10 |
92.53 |
|
| EMTN | 95.17 |
95.87 |
|
| CP NC | ResNet18-Voting | 84.93 |
85.72 |
| Gated-Attention | 91.03 |
91.00 |
|
| EM-Cls | 93.79 |
94.03 |
|
| EMTN | 95.86 |
96.46 |
|
The upper and lower bounds of 95% confidence interval are shown in [].
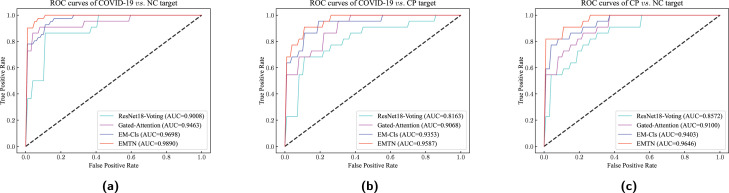
It can be observed that three MIL methods (i.e., EMTN, EM-Cls, and Gated-Attention) have satisfactory performance on different classification targets, which are better than the non-MIL method ResNet18-Voting. Taking the COVID-19 . CP target as an example, the ACC achieved by EMTN, EM-Cls and Gated-Attention are 17.09%, 14.54% and 11.15% higher than ResNet18-Voting, respectively, and AUC is 17.44%, 13.35% and 11.09% higher than ResNet18-Voting, respectively. In the MIL methods, the proposed EMTN achieves the best results on the three classification targets, which can reach 98.62% ACC and 98.90% AUC on COVID-19 . NC target, 95.17% ACC and 95.87% AUC on COVID-19 . CP target, 95.86% ACC and 96.46% AUC on CP . NC target. The ROC curves of these methods are illustrated in Fig. 6, and the of the corresponding methods are shown in Table 2 to explain the partial areas in ROC curves. It can be learned that the ROC curve of EMTN is better than others. And the EMTN achieves the best in the leftmost partial curve area, which means the ROC curves of EMTN rise faster and end with higher TPR than other curves. Table 1, Table 2, and Fig. 6 indicate that the proposed EMTN yields superior performance in these classification targets based on CT images.
Fig. 6.
ROC curves of different classification targets achieved by four methods.
Table 2.
Comparison of area measure of different methods on three targets (bag size=100).
| Targets | Method |
() |
||
|---|---|---|---|---|
| i = 1 | i = 2 | i = 3 | ||
| COVID-19 NC | ResNet18-Voting | 53.04 | 20.37 | 16.67 |
| Gated-Attention | 60.96 | 17.00 | 16.67 | |
| EM-Cls | 63.64 | 16.67 | 16.67 | |
| EMTN | 65.56 | 16.67 | 16.67 | |
| COVID-19 CP | ResNet18-Voting | 46.96 | 17.50 | 17.17 |
| Gated-Attention | 56.00 | 18.01 | 16.67 | |
| EM-Cls | 58.69 | 17.17 | 16.67 | |
| EMTN | 62.53 | 16.67 | 16.67 | |
| CP NC | ResNet18-Voting | 50.03 | 19.02 | 16.67 |
| Gated-Attention | 54.97 | 19.36 | 16.67 | |
| EM-Cls | 59.35 | 18.01 | 16.67 | |
| EMTN | 63.12 | 16.67 | 16.67 | |
5.1.2. Segmentation of multi-lesion
We then compare and analyze the performance of EMTN, EM-Seg, U-Net, and U-Net++ on multi-lesion segmentation. EM-Seg represents that EMTN with only the segmentation branch. Table 3 shows the segmentation results of different methods. The proposed EMTN achieves the best results, with the DC of 96.18%, the PPV of 96.26%, and the SEN of 96.09%. The other methods have similar results as EMTN in a part of evaluation metrics. For example, U-Net++ has the DC of 96.10%, and the SEN of 96.05%, which are only slightly lower than ours; U-Net yields the PPV of 96.00%, while the PPV in our method is 96.26%. Compared with other methods, the performance of EM-Seg in terms of evaluation metrics is decreased, but considering that the parameter number of EM-Seg is only 1.87M, the segmentation performance is acceptable. In addition, the segmentation branch in EMTN is based on EM-Seg, and with multi-task learning, EMTN can also have a better performance.
Table 3.
Comparison of multi-lesion segmentation results of different methods.
| Method | DC() | PPV() | SEN() |
|---|---|---|---|
| U-Net | 95.83 ± 3.1 | 96.00 ± 3.5 | 95.67 ± 2.7 |
| U-Net++ | 96.10 ± 2.3 | 96.16 ± 2.8 | 96.05 ± 1.9 |
| EM-Seg | 94.29 ± 3.4 | 94.49 ± 5.0 | 94.08 ± 3.9 |
| EMTN | 96.18 ± 3.7 | 96.26 ± 4.2 | 96.09 ± 3.8 |
The results are shown as mean standard deviation.
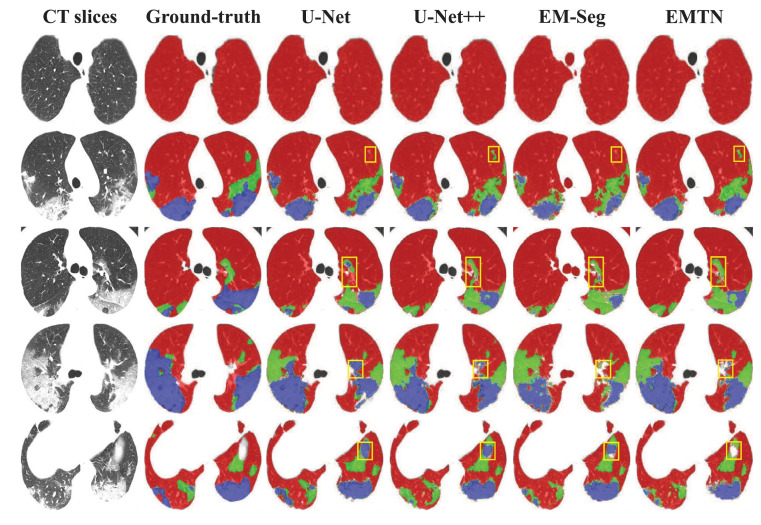
The visualization of segmentation results achieved by four different methods is shown in Fig. 7. It can be seen that the segmentation masks generated by EMTN are more complete and have fewer missing segmentation results than other methods. In addition, the GGO will transform to lung consolidation as the progression of the patient’s condition, it is difficult for segmentation methods and physicians to distinguish lesions in this transformation process. However, the EMTN ensures that the whole lesion areas in the lung are correctly segmented, which can be reliably used for subsequent severity assessment of COVID-19.
Fig. 7.
Visualization of multi-lesion segmentation results achieved by different methods on five COVID-19 cases. Red pixels represent normal lung areas, green pixels represent ground-glass opacities (GGO), and blue pixels represent lung consolidation. Some error-prone regions are denoted by yellow boxes.
5.2. Ablation study
In this section, we take the COVID-19 . NC classification target and multi-lesion segmentation as examples to evaluate the influence of MIL and MTL strategies on the performance of EMTN.
5.2.1. Influence of MIL strategy
To evaluate the influence of the explainable MIL strategy and its effectiveness, EM-Cls, the normal MIL method Gated-Attention, and the non-MIL method ResNet18-Voting are compared. Table 4 shows the comparison on evaluation metrics in terms of COVID-19 . NC classification target.
Table 4.
Evaluation of the explainable MIL strategy (bag size=100).
| Method | ACC() | Precision() | Recall() | F1 Score() | AUC() |
|---|---|---|---|---|---|
| ResNet18-Voting | 88.97 |
85.85 |
90.37 |
88.05 |
90.08 |
| Gated-Attention | 94.48 |
95.89 |
93.24 |
94.55 |
94.63 |
| EM-Cls | 96.73 |
97.22 |
94.59 |
95.89 |
96.98 |
The upper and lower bounds of 95% confidence interval are shown in [].
From Table 4, it can be learned that MIL methods (i.e., EM-Cls and Gated-Attention) yield better results in various metrics. One reason for these results is that non-MIL methods are susceptible to uneven distribution of lesions in CT images. This also proves that MIL methods can pay more attention to relationships between instances, which is helpful to improve the final expression abilities of instances when dealing with such weakly supervised problem. The proposed EM-Cls achieves the best classification performance in the MIL methods, with the ACC of 96.73%, the F1 Score of 95.89%, and the AUC of 96.98%, which is at least 1.39% higher than the metrics generated by Gated-Attention. These results reflect that the proposed explainable MIL strategy can further improve the classification performance and is more effective than normal MIL methods.
Furthermore, we analyze the performance of EMTN using different sizes of bags as inputs. Multi-lesion segmentation is a type of semantic segmentation task, where changing the bag size has little effect on the segmentation performance. Thus, Table 5 shows the classification performance corresponding to different bag sizes, and Table 7 shows the corresponding area measure . It can be observed that the performance of EMTN fluctuates with the changing bag sizes. The best classification performance is reached at the bag size of 100, and the of the leftmost partial curve is higher than others. When the bag size is smaller than 100, the performance gradually improves as the bag size increases; and the performance tends to be saturated when larger than 80, the of the leftmost partial curve under different bag sizes are higher than 63.84%. This indicates that the proposed EMTN needs at least 80 CT slices to obtain acceptable performance.
Table 5.
Classification performance of EMTN under different bag sizes.
| Bag size | ACC() | Precision() | Recall() | F1 Score() | AUC() |
|---|---|---|---|---|---|
| 30 | 95.17 |
93.99 |
94.59 |
94.29 |
95.11 |
| 50 | 96.55 |
95.86 |
95.80 |
95.83 |
96.78 |
| 80 | 97.41 |
96.05 |
97.13 |
96.59 |
97.23 |
| 100 | 98.62 |
97.33 |
98.65 |
97.99 |
98.90 |
| 120 | 98.27 |
98.63 |
97.30 |
97.96 |
98.21 |
| 150 | 97.93 |
96.05 |
98.65 |
97.33 |
97.72 |
The upper and lower bounds of 95% confidence interval are shown in [].
Table 7.
Area measure of EMTN under different bag sizes.
| Bag size |
() |
||
|---|---|---|---|
| i = 1 | i = 2 | i = 3 | |
| 30 | 61.77 | 16.67 | 16.67 |
| 50 | 63.44 | 16.67 | 16.67 |
| 80 | 63.89 | 16.67 | 16.67 |
| 100 | 65.56 | 16.67 | 16.67 |
| 120 | 64.87 | 16.67 | 16.67 |
| 150 | 63.84 | 17.21 | 16.67 |
As mentioned in Section 4.1, a random selection strategy is used to select instances and construct bags. And we further analyze the influence of different selection strategies on the performance of EMTN. Table 6 shows the classification performance using a random selection strategy and three other selection strategies to create the bags. The three selection strategies include selecting a set of slices in the front of CT sequence, in the back of CT sequence, and in the middle of CT sequence. For the convenience of description, they are referred to as front selection, back selection and middle selection, respectively. From Table 6, it can be learned that the results obtained by using other three selection strategies are close to that obtained by using random selection strategy, and creating the bags of instances by random selection can make the performance optimal. The distribution of lesions in CT images of different patients is diverse and not uniform, and the bags are unfavorable for the representations of corresponding individuals when the instances are selected on certain parts of CT images. For example, in the case of small bag size, there may be extreme situation that no positive instances in the bags of positive individuals. These results illustrate that using a random selection strategy to create the bags is more reasonable than the other three selection strategies. Furthermore, the performance achieved by using the other three selection strategies is close to the optimal and acceptable, which indicates that the multi-instance assumption in this work is effective and can reduce the interference caused by different instances selection strategies.
Table 6.
Classification performance of EMTN using different instances selection strategies.
| Bag size | Random selection |
Front selection |
Middle selection |
Back selection |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC() | AUC() | ACC() | AUC() | AUC() | AUC() | ACC() | AUC() | ||||
| 30 | 95.17 |
95.11 |
94.89 |
94.99 |
94.41 |
94.61 |
94.85 |
94.94 |
|||
| 50 | 96.55 |
96.78 |
95.17 |
95.37 |
95.00 |
94.94 |
96.46 |
96.54 |
|||
| 80 | 97.41 |
97.23 |
95.65 |
95.87 |
95.34 |
95.60 |
97.02 |
97.15 |
|||
| 100 | 98.62 |
98.90 |
96.34 |
96.44 |
95.80 |
95.89 |
97.57 |
97.81 |
|||
| 120 | 98.27 |
98.21 |
96.28 |
96.42 |
95.61 |
95.79 |
98.03 |
98.06 |
|||
| 150 | 97.93 |
97.72 |
95.86 |
96.16 |
95.55 |
95.70 |
97.79 |
97.94 |
|||
The upper and lower bounds of 95% confidence interval are shown in [].
5.2.2. Influence of MTL strategy
The proposed EMTN can simultaneously perform classification task and segmentation task in a weight-adaptive multi-task learning manner. We compare the performance of EMTN, EM-Cls, and EM-Seg to analyze the influence of MTL strategy. Table 8 summarizes the results of COVID-19 . NC classification and multi-lesion segmentation. As shown in Table 8, for classification task, MTL strategy can further improve the classification performance of EM-Cls. EMTN improves the ACC from 96.73% to 98.62%, the F1 Score from 95.89% to 97.99%, and the AUC from 96.98% to 98.90%. Meanwhile, for segmentation task, EMTN can achieve satisfactory segmentation results in terms of overall metrics with the influence of MTL strategy. In addition, Fig. 7 shows that EMTN generates more complete and detailed segmentation masks than EM-Seg. In general, the performance of EMTN based on the weight-adaptive MTL strategy is further improved on both classification of COVID-19 and multi-lesion segmentation tasks, which proves that the proposed MTL strategy can utilize task-related information to improve the performance on each task.
Table 8.
Evaluation of the weight-adaptive MTL strategy on classification task and segmentation task (bag size=100).
| Method | Classification task |
Segmentation task |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ACC() | Precision() | Recall() | F1 Score() | AUC() | DC() | PPV() | SEN() | ||
| EM-Cls | 96.73 |
97.22 |
94.59 |
95.89 |
96.98 |
– | – | – | |
| EM-Seg | – | – | – | – | – | 94.29 ± 3.4 | 94.49 ± 5.0 | 94.08 ± 3.9 | |
| EMTN | 98.62 |
97.33 |
98.65 |
97.99 |
98.90 |
96.18 ± 3.7 | 96.26 ± 4.2 | 96.09 ± 3.8 | |
Classification task results: the upper and lower bounds of 95% confidence interval are shown in [].
Segmentation task results: the results are shown as mean standard deviation.
The multi-task loss function with adaptive weights is designed to actively adjust the contribution ratio of different tasks to the training of the network parameters. We further analyze the influence of trade-off factors in Eq. (3). As mentioned in Section 3.2, and denote the trade-off factors for classification task and segmentation task, respectively, which are learnable parameters modified during the iteration. Considering that the segmentation task is a fine-grained pixel-level task, and the classification task is a coarse-grained individual-level task, the segmentation task makes a greater contribution to the network parameter optimization during the training process. Specifically, we vary the ratio of and within , and investigate the performance of EMTN when trade-off factors are different constants and learnable parameters. The comparison results are shown in Table 9, and area measure for classification task with different are shown in Table 10.
Table 9.
Results for classification task and segmentation task with different (bag size=100).
| Classification task |
Segmentation task |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ACC() | Precision() | Recall() | F1 Score() | AUC() | DC() | PPV() | SEN() | ||
| 1/1 | 97.44 |
96.16 |
97.53 |
96.84 |
97.52 |
95.13 ± 3.6 | 95.29 ± 4.5 | 94.97 ± 4.2 | |
| 0.6/1 | 98.37 |
98.61 |
97.25 |
97.93 |
98.48 |
95.94 ± 3.3 | 96.06 ± 4.4 | 95.82 ± 3.5 | |
| 0.2/1 | 96.86 |
95.95 |
96.37 |
96.16 |
97.03 |
95.42 ± 4.0 | 95.78 ± 4.7 | 95.07 ± 4.5 | |
| Learning | 98.62 |
97.33 |
98.65 |
97.99 |
98.90 |
96.18 ± 3.7 | 96.26 ± 4.2 | 96.09 ± 3.8 | |
Classification task results: the upper and lower bounds of 95% confidence interval are shown in [].
Segmentation task results: the results are shown as mean standard deviation.
Table 10.
Area measure for classification task with different (bag size=100).
|
() |
|||
|---|---|---|---|
| i = 1 | i = 2 | i = 3 | |
| 1/1 | 64.18 | 16.67 | 16.67 |
| 0.6/1 | 65.14 | 16.67 | 16.67 |
| 0.2/1 | 63.69 | 16.67 | 16.67 |
| Learning | 65.56 | 16.67 | 16.67 |
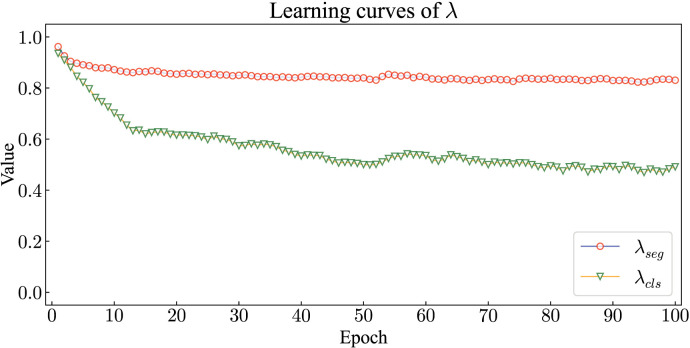
From Table 9, Table 10, it can be observed that the proposed EMTN can further improve the performance on both tasks when are learnable parameters. Fig. 8 shows the learning curves of the trade-off factors. The initial values of and are both set as 1, and the values of are recorded after each iteration. and stabilize after about 40 iterations. The experiments prove that EMTN generates the best performance after around 49 iterations. The final values of and are approximately 0.5 and 0.84, respectively. Through the analysis of learning process, the contribution ratio of different tasks to the training of network parameters can be determined, which can avoid the tedious process of manually adjusting the trade-off factors. In addition, the larger factor assigned to the segmentation task also indicates that a fine-grained task provides more support for network parameter optimization than a coarse-grained task.
Fig. 8.
Learning curves of trade-off factors . and respectively denote the trade-off factors for classification task and segmentation task.
5.2.3. Quantitative analysis of lesions and severity assessment of COVID-19
The quantitative analysis of lesions and severity assessment of COVID-19 are considered as the subsequent tasks of multi-lesion segmentation. Fig. 9 shows five assessment examples.
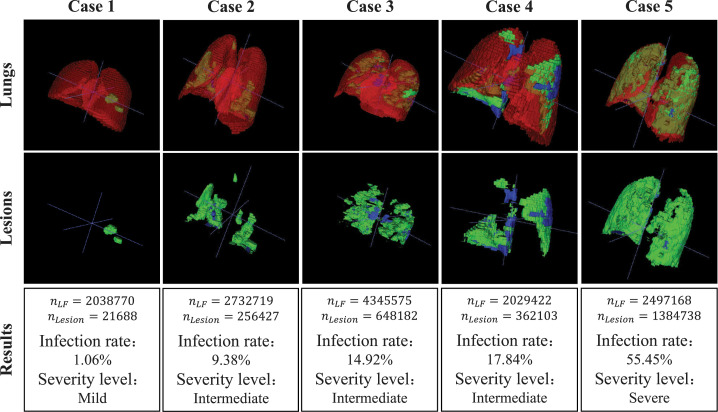
Fig. 9.
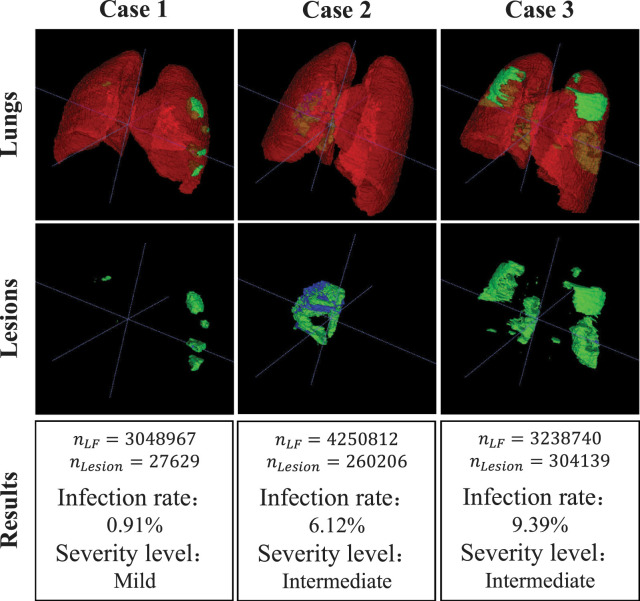
Assessment results on five COVID-19 cases from mild, intermediate, and severe groups. For each case, we show the 3D visualization results of the whole lung and its lesion areas. The severity level definitions are as follows: less than three GGO lesions and lesion areas less than 5% of the entire lung fields is defined as mild; lesion areas more than 5% of the entire lung fields is defined as intermediate; lesion areas more than 40% of the entire lung fields is defined as severe.
Specifically, the first, the second and the third rows of Fig. 9 present the segmentation results, the lesion areas and the assessment results, respectively. From the segmentation results and the separated lesion areas, it can be observed that the lesion areas of the entire lung fields from case 1 to case 5 are gradually increasing, while the infection rate of lung fields and the severity level of patients are unavailable. In the assessment results, the number of voxels in the lung fields and lesion areas, infection rate, and severity assessment of patients are shown. For example, the assessment results of case 3 suggest that the infection rate is 14.92% and the severity level of the patient is intermediate. Compared with direct observation, quantitative analysis and severity assessment can give the infection rate of lung fields and the severity level of patients, and these assessment results can serve as a reference for the diagnosis of COVID-19.
5.2.4. Explainability analysis of diagnosis results
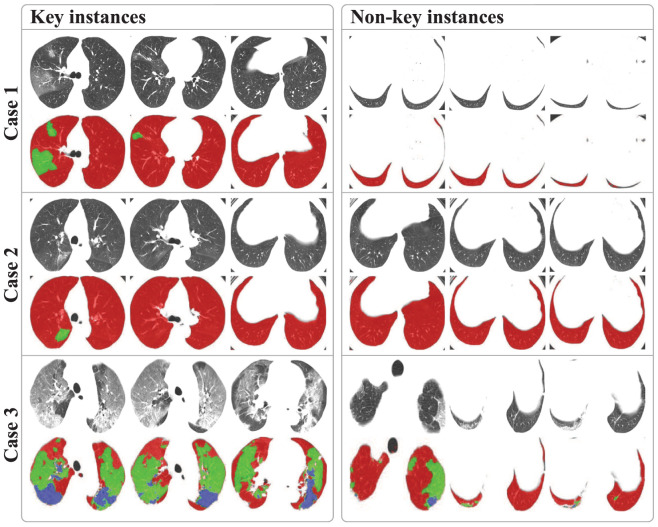
As mentioned in Section 3.3, the proposed MIL strategy can deal with the weakly supervised problem and make EMTN have explainability by suggesting the key instances. We further investigate the explainability of our method by deriving the attention weights between the class token and each instance-level feature. The comparison of some key instances and non-key instances is shown in Fig. 10.
Fig. 10.
Explainability analysis of diagnosis results of three cases by suggesting their key instances.
Specifically, we select the key instances based on the values of the attention weights, and the threshold is set to be 0.5. The instances with weights greater than the threshold are called key instances. Multiple experiments prove that these key instances usually account for about 20% or even less of the whole bag. As shown in Fig. 10, the first to third columns and the fourth to sixth columns are key instances and non-key instances, respectively; the second, the fourth and the last rows represent the segmentation masks of the first, the third and the fifth rows, respectively. For case 1, the weights of these six instances are 1.0, 0.5999, 0.5871, 0.0154, 0.0088, 0.0 from left to right; for case 2, the weights are 1.0, 0.6734, 0.6453, 0.0593, 0.0463, 0.0 from left to right; for case 3, the weights are 1.0, 0.8396, 0.7035, 0.1538, 0.0144, 0.0 from left to right. These weights are not normalized, and the larger weights mean that the corresponding instances have a greater influence on the diagnosis. It can be observed that the key instances of cases contain more lesion areas or complete lung fields than non-key instances, which can also roughly suggest the severity level of patients. Compared with key instances, the non-key instances contain many irrelevant areas, which have limited influence on the diagnosis of patients.
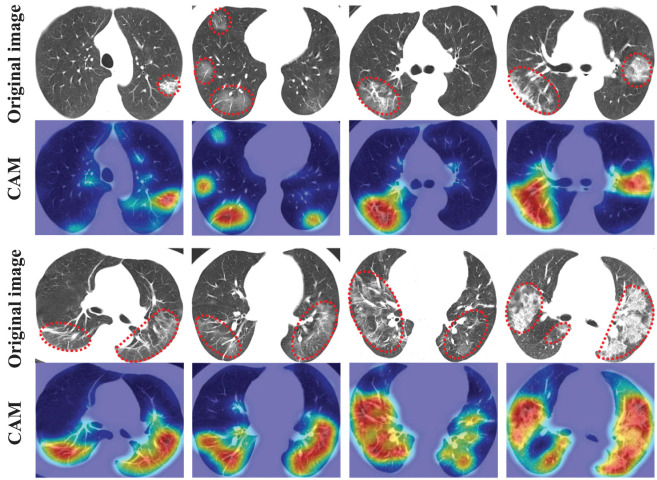
Furthermore, consistency is important for clinical diagnosis, and diagnostic decisions made by the same method are often influenced by similar images. To evaluate the consistency of the explainability part of the proposed framework, namely, to judge whether the suggested key instances can provide the posterior test of the diagnosis results, we give more explainable analysis from the aspect of semantic comprehension. We use Grad-CAM++ [40] to obtain class activation maps (CAMs) of several key instances from different patients. As shown in Fig. 11, the first and third rows are key instances (the lesion regions have been marked with dashed lines), the second and fourth rows are the corresponding class activation maps. It can be observed that the class activation maps of suggested key instances show the approximate locations of lesions, which indicates the key instances suggested by the explainability part of this framework are roughly consistent with the explainable analysis from the semantic perspective. These results imply that the proposed explainable MIL can provide the posterior test of the diagnosis results, and indicate the concern level of the network for different instances.
Fig. 11.
Class activation maps of suggested key instances obtained by Grad-CAM++.
5.3. Additional study
To further evaluate the proposed method, we analyze the performance of EMTN and EM-Cls on other open-source datasets referred to as COVID-19-CT & Radiopaedia (i.e., COVID-19-CT dataset and common pneumonia patients collected from Radiopaedia). Table 11, Table 12 show the classification results and area measure of EMTN and EM-Cls on the COVID-19-CT & Radiopaedia dataset, respectively. Fig. 12 shows three severity assessment examples based on multi-lesion segmentation masks generated by EMTN. It should be noted that due to the lack of ground truth for segmentation in the COVID-19-CT & Radiopaedia data, the dataset has not participated in the training stage of EMTN. The partial results in Table 11, Table 12, and Fig. 12 are obtained by directly using the EMTN which trained on the CC-CCII dataset for testing this dataset. For clarity, we illustrate the training and test data in the tables.
Table 11.
Results for classification task on COVID-19-CT & Radiopaedia dataset. (bag size=100).
| Method | Training data | Testing data | ACC() | Precision() | Recall() | F1 Score() | AUC() |
|---|---|---|---|---|---|---|---|
| EM-Cls | CC-CCII | COVID-19-CT & Radiopaedia |
88.37 |
87.93 |
87.29 |
87.61 |
89.17 |
| COVID-19-CT & Radiopaedia |
COVID-19-CT & Radiopaedia |
94.35 |
93.44 |
94.99 |
94.21 |
95.07 |
|
| EMTN | CC-CCII | COVID-19-CT & Radiopaedia |
89.92 |
90.29 |
89.05 |
89.67 |
90.12 |
The upper and lower bounds of 95% confidence interval are shown in [].
Table 12.
Area measure for classification task on COVID-19-CT & Radiopaedia dataset (bag size=100).
| Method | Training data | Testing data |
() |
||
|---|---|---|---|---|---|
| i = 1 | i = 2 | i = 3 | |||
| EM-Cls | CC-CCII | COVID-19-CT & Radiopaedia |
54.06 | 18.92 | 16.19 |
| COVID-19-CT & Radiopaedia |
COVID-19-CT & Radiopaedia |
61.03 | 17.37 | 16.67 | |
| EMTN | CC-CCII | COVID-19-CT & Radiopaedia |
56.15 | 17.48 | 16.49 |
Fig. 12.
Assessment results on three cases in COVID-19-CT & Radiopaedia dataset.
As shown in Table 11, without using COVID-19-CT & Radiopaedia dataset for training, the classification accuracy achieved by EMTN and EM-Cls on this dataset are 89.92% and 88.37%, respectively, and AUCs are about 90%. After training EM-Cls with COVID-19-CT & Radiopaedia dataset, the evaluation metrics are on average 7.20% higher than that of EM-Cls without using COVID-19-CT & Radiopaedia dataset for training. The ACC (94.35%) and AUC (95.07%) have 6.77% and 6.62% improvement, respectively. From Table 12, it can be observed that the ROC curve of EM-Cls trained with the COVID-19-CT & Radiopaedia dataset goes up faster while staying left, and it has a higher value of TPR than that trained with the CC-CCII dataset. In Fig. 12, the first, the second, and the third rows present the segmentation results, the lesion areas and the assessment results, respectively. The assessment results of these cases suggest the infection rate and the severity level of the patients. From case 1 to case 3, their assessment results are mild, intermediate, intermediate, and their infection rates are 0.91%, 6.12%, 9.39%.
Considering the influence of the differences between datasets on the model, though the classification results of EMTN and EM-Cls decline without using the COVID-19-CT & Radiopaedia as training data, these results are acceptable. In the situation of using the COVID-19-CT & Radiopaedia as training data, EM-Cls as a variant of EMTN can achieve satisfactory results. For the segmentation branch in EMTN, it accesses a large number of pixels and learns to correctly predict each pixel to the corresponding semantic category during the training process. Therefore, generating segmentation masks for COVID-19-CT & Radiopaedia dataset by EMTN trained on the CC-CCII dataset is less affected by the differences between datasets, and the generated masks are also relatively accurate. The above analysis indicates that the proposed method is applicable to the new datasets and can achieve satisfactory results.
5.4. Discussion on the limitation
This work is oriented towards an actual problem, that is, the auxiliary diagnosis of COVID-19. The effectiveness of the proposed framework has been verified by extensive experiments based on real-world datasets, yet there are still some limitations in our work. In this section, we discuss the limitations of this work and the gap with practical clinical applications.
As far as clinical data is concerned, data quality, data standards, and data amounts are all issues that need to be considered in the transition from algorithm to practical application. The data preprocessing procedures and multi-instance learning strategy adopted in this work can alleviate the problem of uneven data quality and diverse data standards, which is achieved by obtaining regions of interest and randomly selecting instances to construct a bag of instances and represent an individual. The real-world datasets of CT images used in this work have certain limitations, part of CT images have no ground truth for segmentation task, and the severity of COVID-19 has to be assessed through a robust voxel-wise analysis method. The proposed framework will be further perfected if more well-labeled datasets are available, such as those used for the severity assessment of COVID-19. In addition, considering the complexity of COVID-19, for some special cases, such as asymptomatic infections without obvious CT imaging features, decision-making cannot be performed only based on CT images. In this situation, the diagnosis of COVID-19 should be made under more comprehensive tests which may include RT-PCR or other clinical examinations. It also inspired that the information generated by these clinical examinations can be adopted as auxiliary diagnosis indexes to help construct a more robust computer-aided diagnosis system.
On the other hand, the interpretability of auxiliary diagnosis methods based on deep learning is weak, while the process of clinical diagnosis requires rigorous evidence. The black-box nature is one of the main reasons which limits the wide application of fully automated medical artificial intelligence (AI). In the European “Artificial Intelligence Act”, some clear guidance on the use of medical AI has been already provided, which constitutes a binding legal framework for the use of medical AI. This is the protection of human rights in the context of medical AI development. Stoeger et al. [41] point out that human oversight and explainability are required in medical AI, namely, one AI system must be explainable to be used in medicine. It not only coincides with the requirements of European fundamental rights, but also with the demands of computer science. Therefore, the importance of explainable medical AI is self-evident. Though the proposed explainable multi-instance learning can give the explainability analysis of diagnosis results by suggesting the key instances, it is not explainable in the mathematical sense but in the clinical sense. Furthermore, it also requires human oversight and a more complete explanation. Interactive machine learning with the ”human in the loop” could be a potential solution to this limitation of AI [42], [43]. It should be noted that physicians/radiologists have conceptual understanding and experience that no AI can fully learn. Combined with the conceptual understanding and experience of physicians/radiologists, the interactive machine learning with the ”human in the loop” can find the underlying explanatory factors for AI, which ensure that decisions made by AI can be human-controlled and clinically justified. This human-in-the-loop machine learning will be explored in our future work.
6. Conclusion
In this paper, we construct an integrated framework for segmenting lesion areas and diagnosing COVID-19 from CT images. It takes the explainable multi-instance multi-task network (EMTN) as the core, and the lesion quantification and severity assessment as important components. The EMTN can make proper use of task-related information to further improve the performance on diagnosis and segmentation, and it also has EM-Seg (EMTN with only the segmentation branch) and EM-Cls (EMTN with only the classification branch) two variants. These two variants can be respectively employed to perform diagnosis and segmentation, which improve the flexibility of EMTN. An explainable multi-instance learning strategy is proposed in EMTN for explainability analysis of diagnosis results, and a weight-adaptive multi-task learning strategy is proposed for the coordination between both tasks. Considering that the evaluation of patient status needs more detailed information, the lesion quantification and severity assessment are adopted as the subsequent tasks of multi-lesion segmentation. The experimental results show that the proposed EMTN has better performance over several mainstream methods on the diagnosis and multi-lesion segmentation of COVID-19, and can provide explicable diagnosis results that enhance the explainability of the network. Moreover, the proposed framework gives the assessment of COVID-19 patient status as an extra reference for the auxiliary diagnosis of COVID-19, including the lung infection rate and the severity level of patients.
CRediT authorship contribution statement
Minglei Li: Conceptualization, Methodology, Validation, Writing – original draft. Xiang Li: Conceptualization, Methodology, Writing – review & editing. Yuchen Jiang: Conceptualization, Methodology, Writing – review & editing. Jiusi Zhang: Conceptualization, Methodology. Hao Luo: Supervision, Writing – review & editing, Funding acquisition. Shen Yin: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
This work is supported by the Young Scientist Studio of Harbin Institute of Technology, China under Grant AUGA9803503221 and Interdisciplinary Research Foundation of Harbin Institute of Technology, China under Grant IR2021224.
References
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.De Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., et al. Commentary: Middle east respiratory syndrome coronavirus (mers-cov): announcement of the coronavirus study group. J. Virology. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: Relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhammad U., Hoque M.Z., Oussalah M., Keskinarkaus A., Seppänen T., Sarder P. SAM: Self-augmentation mechanism for COVID-19 detection using chest X-ray images. Knowl.-Based Syst. 2022 doi: 10.1016/j.knosys.2022.108207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C., Yang Y., Liang H., Wu B. Transfer learning for establishment of recognition of COVID-19 on CT imaging using small-sized training datasets. Knowl.-Based Syst. 2021;218 doi: 10.1016/j.knosys.2021.106849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in wuhan, China: A descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., Tran T.M.L., Choi J.W., Wang D.-C., Shi L.-B., et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., Wei H., Zhang W. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 11.Chaganti S., Grenier P., Balachandran A., Chabin G., Cohen S., Flohr T., Georgescu B., Grbic S., Liu S., Mellot F., et al. Automated quantification of CT patterns associated with COVID-19 from chest CT. Radiology: Artif. Intell. 2020;2(4) doi: 10.1148/ryai.2020200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W., Jacobs C., Charbonnier J.-P., Van Ginneken B. Relational modeling for robust and efficient pulmonary lobe segmentation in CT scans. IEEE Trans. Med. Imaging. 2020;39(8):2664–2675. doi: 10.1109/TMI.2020.2995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenhende S., Georgoulis S., Van Gansbeke W., Proesmans M., Dai D., Van Gool L. Multi-task learning for dense prediction tasks: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021 doi: 10.1109/TPAMI.2021.3054719. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Basset M., Chang V., Hawash H., Chakrabortty R.K., Ryan M. FSS-2019-nCov: A deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection. Knowl.-Based Syst. 2021;212 doi: 10.1016/j.knosys.2020.106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y.-H., Gao S.-H., Mei J., Xu J., Fan D.-P., Zhang R.-G., Cheng M.-M. Jcs: An explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 17.Chassagnon G., Vakalopoulou M., Battistella E., Christodoulidis S., Hoang-Thi T.-N., Dangeard S., Deutsch E., Andre F., Guillo E., Halm N., et al. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakalopoulou M., Chassagnon G., Bus N., Marini R., Zacharaki E.I., Revel M.-P., Paragios N. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2018. AtlasNet: Multi-atlas non-linear deep networks for medical image segmentation; pp. 658–666. [Google Scholar]
- 19.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 20.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: Automatic COVID-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 21.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Langlotz C., Shpanskaya K., et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 22.Wang J., Bao Y., Wen Y., Lu H., Luo H., Xiang Y., Li X., Liu C., Qian D. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020;39(8):2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- 23.Çiçek O., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- 24.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 25.Shaban W.M., Rabie A.H., Saleh A.I., Abo-Elsoud M. A new COVID-19 patients detection strategy (CPDS) based on hybrid feature selection and enhanced KNN classifier. Knowl.-Based Syst. 2020;205 doi: 10.1016/j.knosys.2020.106270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M., Le Q. International Conference on Machine Learning. PMLR; 2019. Efficientnet: Rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 27.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Wang R., Zhao H., Zha Y., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradski G., Kaehler A. ” O’Reilly Media, Inc.”; 2008. Learning OpenCV: Computer Vision with the OpenCV Library. [Google Scholar]
- 30.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 1–9.
- 32.Li X., Jiang Y., Li M., Yin S. Lightweight attention convolutional neural network for retinal vessel image segmentation. IEEE Trans. Ind. Inf. 2020;17(3):1958–1967. [Google Scholar]
- 33.Chen L., Jiang X., Liu X., Zhou Z. Logarithmic norm regularized low-rank factorization for matrix and tensor completion. IEEE Trans. Image Process. 2021;30:3434–3449. doi: 10.1109/TIP.2021.3061908. [DOI] [PubMed] [Google Scholar]
- 34.D. Alexey, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai, T. Unterthiner, M. Dehghani, An image is worth 16×16 words: Transformers for image recognition at scale, in: Proceedings of the International Conference on Learning Representations, Lisbon, Portuagal, 2021, pp. 7–8.
- 35.Shakouri S., Bakhshali M.A., Layegh P., Kiani B., Masoumi F., Ataei Nakhaei S., Mostafavi S.M. COVID19-CT-dataset: an open-access chest CT image repository of 1000+ patients with confirmed COVID-19 diagnosis. BMC Res. Notes. 2021;14(1):1–3. doi: 10.1186/s13104-021-05592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrington A.M., Fieguth P.W., Qazi H., Holzinger A., Chen H.H., Mayr F., Manuel D.G. A new concordant partial AUC and partial C statistic for imbalanced data in the evaluation of machine learning algorithms. BMC Med. Inform. Decis. Mak. 2020;20(1):1–12. doi: 10.1186/s12911-019-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietterich T.G. International Workshop on Multiple Classifier Systems. Springer; 2000. Ensemble methods in machine learning; pp. 1–15. [Google Scholar]
- 38.Ilse M., Tomczak J., Welling M. International Conference on Machine Learning. PMLR; 2018. Attention-based deep multiple instance learning; pp. 2127–2136. [Google Scholar]
- 39.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: A nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhay A., Sarkar A., Howlader P., Balasubramanian V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. 2018 IEEE Winter Conference on Applications of Computer Vision; WACV; IEEE; 2018. pp. 839–847. [Google Scholar]
- 41.Stöger K., Schneeberger D., Holzinger A. Medical artificial intelligence: The European legal perspective. Commun. ACM. 2021;64(11):34–36. [Google Scholar]
- 42.Sorantin E., Grasser M.G., Hemmelmayr A., Tschauner S., Hrzic F., Weiss V., Lacekova J., Holzinger A. The augmented radiologist: Artificial intelligence in the practice of radiology. Pediatr. Radiol. 2021:1–13. doi: 10.1007/s00247-021-05177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y., Li X., Luo H., Yin S., Kaynak O. Quo vadis artificial intelligence? Discov. Artif. Intell. 2022;2(1):1–19. [Google Scholar]