Abstract
Sexual crosses were used to determine the genetic basis of resistance to the sterol 14 α-demethylase inhibitor fungicide prochloraz in the cereal eyespot pathogen Tapesia yallundae. Three different crosses between sensitive parental strains (22-432 and 22-433 [the concentration required to inhibit growth by 50% {IG50} for each was ≤0.03 mg/liter]) and field isolates from France and New Zealand with differing levels of resistance (PR11 [IG50 = 0.5 mg/liter], PR1 [IG50 = 1.0 mg/liter], and 11-3-18 [IG50 = 2.4 mg/liter]) yielded progeny showing a bimodal distribution, with an even number of sensitive and resistant progeny. This indicated the segregation of a single major gene for resistance in each cross, which was confirmed by the use of backcrosses, crosses between F1 progeny, and control crosses between sensitive parents. However, there was also evidence of additional quantitative genetic components responsible for the increased IG50s of the more resistant isolates. A further cross was made between isolate PR11 and an F1 progeny arising from isolate 11-3-18, and this also yielded progeny which were entirely prochloraz resistant. This suggested that resistance genes were allelic in these two isolates, with resistance conferred by a gene at the same locus (or closely linked loci), despite the fact that the isolates (PR11 and 11-3-18) originated from different continents.
A major reason for the continued success of agricultural production in the developed world has been the availability of chemicals to control pests and diseases in crops. Of these, the sterol 14 α-demethylase inhibitor (DMI) fungicides represent the largest and most important group of modern antifungal compounds, possessing excellent protectant, curative, and eradicant properties against a wide range of fungal species (35). Most DMI fungicides are derivatives of imidazoles or triazoles and have remained highly effective in most field applications despite many years of intense agricultural use and their single-site mode of action. However, decreased sensitivity and field resistance to certain DMIs has been reported in at least 13 species of plant pathogen (10).
The imidazole prochloraz (1-{N-propyl-N-[2-(2,4,6-trichlorophenoxy)ethyl]carbamoyl}-imidazole; trade name, Sportak) was launched in 1977 and has since been registered for use on more than 30 different crops in 50 countries worldwide (37; Prochloraz—technical information, Hoechst Schering AgrEvo GmbH, Berlin, Germany, 1995). It has a broad spectrum of activity against several diseases of cereal, orchard, and horticultural crops (2; Prochloraz—technical information, Hoechst Schering AgrEvo GmbH, 1995). As a foliar treatment, prochloraz is active against a wide range of stem, leaf, and ear diseases of cereals and can therefore be used in situations where several diseases affecting different plant parts are present simultaneously (37). While it has been possible to produce mutants in vitro with increased resistance to prochloraz, resistance has rarely developed in field populations (17, 23, 33, 35). Indeed, prochloraz has remained effective in situations where a decline in sensitivity to related DMI fungicides has been reported (26). However, in recent years isolates of Tapesia yallundae and Tapesia acuformis (anamorph Pseudocercosporella herpotrichoides), causal agents of eyespot disease of cereals (14, 31), with significantly increased levels of resistance to prochloraz have been obtained from field locations in northwestern France and New Zealand (30; P. S. Dyer and R. E. Bradshaw, unpublished data), providing the first indication of a potential reduction in efficacy of control of eyespot disease by DMI fungicides. It also provides a possible model for development of prochloraz resistance, given that it is the properties of the chemical, rather than the target organism, which are thought to determine the nature of the resistance response (18).
A major factor determining the resistance risk of a particular fungicide is the genetic basis of resistance to the compound. An abrupt loss of effectiveness is more likely where resistance is conferred by mutation of a single major gene, as seen in the rapid development of resistance to benzimidazoles in populations of plant pathogenic fungi (29, 35). A synergistic interaction of two resistance genes may also lead to the appearance of field resistance (32). In contrast, a gradual directional shift in sensitivity in populations is likely when resistance is under polygenic control (18, 35). Various studies have been made to determine the genetic control of resistance to DMI fungicides. These studies have provided evidence of resistance conferred either by single major genes, the interaction of many additive genes, or a combination of both mechanisms, although the effect on fitness has not been determined in all cases (10, 34).
The objective of the present study was to determine the genetic basis of resistance to prochloraz in field isolates of T. yallundae, to assess whether resistance is primarily mono- or polygenic in nature. This analysis is now possible because techniques have been devised to allow in vitro sexual crossing of T. yallundae, and recombination of genetic markers using these techniques has demonstrated the presence of a two-allele heterothallic mating system (15).
MATERIALS AND METHODS
Fungal isolates and maintenance.
Field isolates of T. yallundae that had previously been identified as exhibiting different levels of fungicide resistance were selected (Table 1). Isolates were classified using symbols equivalent to those used for benomyl resistance to designate sensitivity or different levels of resistance to prochloraz (27). Isolates 22-432 and 22-433 (origin, United Kingdom) were sensitive to prochloraz (Prc-S) i.e., the concentration required to inhibit growth by 50% (IG50) was <0.05 mg/liter, while isolates PR11, PR1 (origin, North France), and 11-3-18 (origin, New Zealand) showed increasing levels of resistance to prochloraz. The latter isolates were respectively considered to have low resistance (IG50 range, 0.2 to 0.6 mg/liter), medium resistance (IG50 range, 0.7 to 1.5 mg/liter), and high resistance (IG50 range, 1.6 to 5.0 mg/liter) to prochloraz (Prc-LR, Prc-MR, and Prc-HR, respectively). Stock cultures were grown at 15°C on 1.5% tap water agar under white light for 8 weeks before storage at 4°C.
TABLE 1.
Characteristics of parental field isolates and F1 progeny used in fungicide assays
| Strain | Source (original code) | MATa | Phenotypeb |
|---|---|---|---|
| 22-432 | Wheat in Cambs., United Kingdom (C86/536/1) | MAT-2 | Prc-S |
| 22-433 | Wheat in Beds., United Kingdom (C85/484) | MAT-1 | Prc-S |
| PR1 | Wheat in northern France | MAT-2 | Prc-MR |
| PR11 | Wheat in northern France | MAT-2 | Prc-LR |
| 11-3-18 | Wheat stubble in Southland, New Zealand | MAT-1 | Prc-HR |
| A51 | F1 of cross 22-433 × PR11 | MAT-1 | Prc-LR |
| B32 | F1 of cross 22-433 × PR1 | MAT-1 | Prc-LR |
| B53 | F1 of cross 22-433 × PR1 | MAT-2 | Prc-LR |
| C39 | F1 of cross 22-432 × 11-3-18 | MAT-2 | Prc-LR |
| C69 | F1 of cross 22-432 × 11-3-18 | MAT-1 | Prc-LR |
| C99 | F1 of cross 22-432 × 11-3-18 | MAT-1 | Prc-LR |
MAT, mating type in accordance with the system described by Singh et al. (38).
Fungicide sensitivity or resistance. Prc-S, prochloraz sensitive (IG50, <0.05 mg/liter); Prc-LR, low resistance to prochloraz (IG50 range, 0.2 to 0.6 mg/liter); Prc-MR, medium resistance to prochloraz (IG50 range, 0.7 to 1.5 mg/liter); Prc-HR, high resistance to prochloraz (IG50 range, 1.6 to 5.0 mg/liter).
Experimental design and crossing of isolates of T. yallundae.
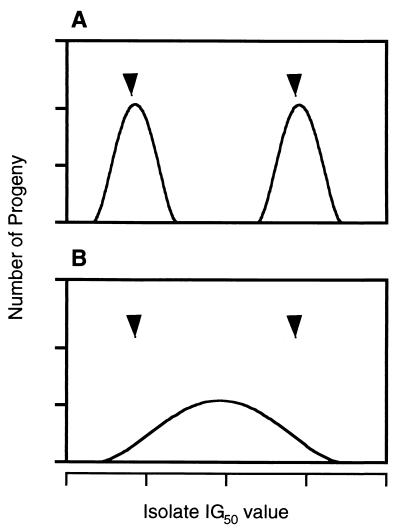
Two main patterns of inheritance of fungicide resistance in this haploid organism were envisaged, depending on the genetic nature of resistance (18, 34). If a single, major gene for resistance were present, then progeny from a cross between a resistant and a sensitive isolate would be predicted to segregate in equal numbers between resistance and susceptibility, with a bimodal distribution (Fig. 1A). In contrast, if resistance were polygenic in nature, arising from a series of individual genes at different loci, each with a small effect on resistance, then progeny from a cross between a resistant and a sensitive isolate would be predicted to show a continuous distribution between sensitivity and resistance, depending on the number of resistance genes present (Fig. 1B). Other patterns of inheritance of fungicide resistance are also possible, e.g., a 3:1 ratio arising from the synergistic action of two genes required for resistance or a skewed distribution as a result of the presence of a major gene with additional genetic loci (see Results). Three sexual crosses were therefore established between field isolates of opposite mating types exhibiting sensitivity or resistance to prochloraz (Table 2, crosses A, B, and C), and the fungicide sensitivities of approximately 100 progeny from these crosses were assayed in order to determine the pattern of inheritance of resistance. Crosses were made on winter barley straw, and progeny were recovered from apothecia by isolation of single ascospores on tap water agar plates as previously described (15). Recombination was confirmed in representative progeny by using randomly amplified polymorphic DNA molecular markers (15; H. M. Wood, unpublished data). Where relevant, backcrosses or crosses between sibling F1 progeny were performed to substantiate patterns of inheritance of fungicide resistance (see Results). Finally, a control cross was established between the sensitive parental strains 22-432 and 22-433 (both Prc-S) to ensure that no resistance to prochloraz arose as a result of the sexual cycle.
FIG. 1.
Distribution of progeny from a cross between fungicide-sensitive and -resistant parents in a haploid organism given monogenic resistance (A) or polygenic resistance (B). Arrowheads indicate IG50s for respective parents.
TABLE 2.
Summary of properties of offspring from crosses between isolates of T. yallundae differing in sensitivity to prochloraz
| Cross | Parental isolatesa | Ratio (S:R)b | χ2 | Probabilityc | Skew |
|---|---|---|---|---|---|
| A | 22-433 (Prc-S), PR11 (Prc-LR) | 43:57 | 1.96 | 0.16 | 0.01 |
| B | 22-433 (Prc-S), PR1 (Prc-MR) | 58:46 | 1.39 | 0.24 | 0.98 |
| C | 22-432 (Prc-S), 11-3-18 (Prc-HR) | 46:51 | 0.26 | 0.61 | 2.06 |
| D | PR11 (Prc-LR), A51 (Prc-LR) | 0:41 | —d | — | 1.18 |
| E | PR1 (Prc-MR), B32 (Prc-LR) | 0:41 | — | — | 2.84 |
| F | B32 (Prc-LR), B53 (Prc-LR) | 0:40 | — | — | 0.07 |
| G | 11-3-18 (Prc-HR), C39 (Prc-LR) | 0:44 | — | — | 1.65 |
| H | C39 (Prc-LR), C99 (Prc-LR) | 0:45 | — | — | 0.32 |
| I | PR11 (Prc-LR), C69 (Prc-LR) | 0:45 | — | — | 0.00 |
| J | 22-432 (Prc-S), 22-433 (Prc-S) | 15:0 | — | — | — |
Prc-S, prochloraz sensitive (IG50, <0.05 mg/liter); Prc-LR, low resistance to prochloraz (IG50 range, 0.2 to 0.6 mg/liter); Prc-MR, medium resistance to prochloraz (IG50 range, 0.7 to 1.5 mg/liter); Prc-HR, high resistance to prochloraz (IG50 range, 1.6 to 5.0 mg/liter).
For segregation purposes those isolates for which prochloraz IG50 was ≤0.05 mg/liter were considered sensitive (S) and those for which the IG50 was >0.05 mg/liter were considered resistant (R).
Probability of χ2 value (1 df) under the null hypothesis of 1:1 segregation.
—, not calculated or not applicable.
Fungicide testing.
Progeny and parents from crosses between strains sensitive to prochloraz and strains resistant to prochloraz were assayed on growth media amended with either 0.05, 0.15, 0.5, 1.0, 2.0 or 5.0 mg of prochloraz per liter, by inoculating plates with three replicate hyphal plugs (13). Values for the percent growth inhibition relative to that observed on unamended control plates were plotted graphically, and IG50s were derived from best-fit line equations. Variance testing of replicates (34) was not feasible due to the large numbers of plates required. Segregation data was then analyzed using χ2 tests, and where appropriate a measure of skew was determined for resistant progeny using the program Microsoft Excel 98 (8). Graphs of progeny distribution were plotted on axis scales appropriate to the IG50 range.
RESULTS
Inheritance of prochloraz resistance in crosses between sensitive and resistant field isolates.
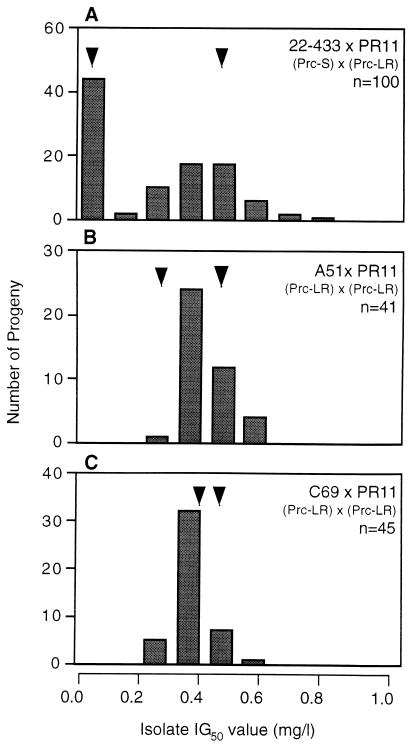
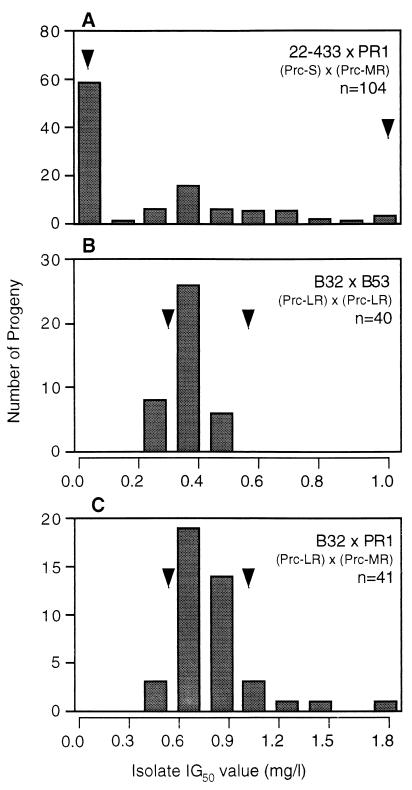
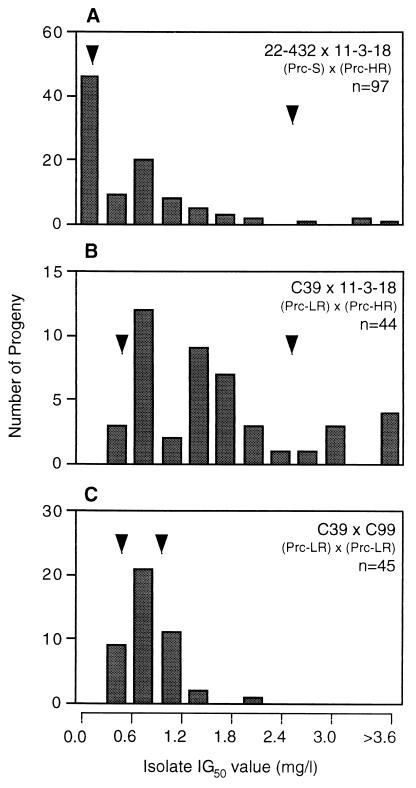
Three crosses between sensitive and resistant parental strains were initially set up. Analysis of individual progeny from a cross between 22-433 (Prc-S) and PR11 (Prc-LR) (designated cross A, with progeny identified by a subsequent serial number, e.g., F1 progeny A51) revealed a clear bimodal distribution (Fig. 2A; Table 2), indicative of a single major gene for resistance in PR11 (7, 18). Analysis of progeny arising from a cross between the same sensitive parent, 22-433, and PR1 (Prc-MR) (designated cross B) also revealed the presence of sensitive and resistant phenotypes in approximately equal proportions (Fig. 3A). However, the resistant progeny exhibited a positively skewed distribution (Table 2). Similar results were obtained in an analysis of progeny arising from a cross between 22-432 (Prc-S) and 11-3-18 (Prc-HR) (designated cross C). Sensitive and resistant phenotypes were present in approximately equal proportions (Fig. 4A), with the resistant progeny again exhibiting a positively skewed distribution (Table 2).
FIG. 2.
Distribution of prochloraz IG50s for progeny arising from crosses involving T. yallundae isolate PR11 (Prc-LR). Mating partners are shown on upper right of graph, with arrowheads indicating IG50s for respective parents, and n is the total number of progeny analyzed from each cross.
FIG. 3.
Distribution of prochloraz IG50s for progeny arising from crosses involving T. yallundae isolate PR1 (Prc-MR) or F1 offspring of PR1. Mating partners are shown on upper right of graph, with arrowheads indicating IG50s for respective parents, and n is the total number of progeny analyzed from each cross.
FIG. 4.
Distribution of prochloraz IG50s for progeny arising from crosses involving T. yallundae isolate 11-3-18 (Prc-HR) or F1 offspring of 11-3-18. Mating partners are shown on upper right of graph, with arrows indicating IG50s for respective parents, and n is the total number of progeny analyzed from each cross.
Experimental hypothesis.
The patterns of inheritance suggested the segregation of one major gene for resistance in each of the crosses, given the bimodal distribution with equal numbers of sensitive and resistant progeny, as seen particularly clearly with cross A. However, the skewed distribution of resistant progeny with crosses B and C suggested the presence of additional genetic components in parental isolates PR1 and 11-3-18, which were responsible for the long tails observed in the progeny distribution. These additional genetic components appeared to confer increased resistance to prochloraz but were only able to exert an effect if the single major gene for resistance was already present, i.e., the presence of these extra genetic components alone would not result in an increased IG50, but in the presence of the major resistance gene they appeared to confer a slight increase in resistance. The effect of the extra genetic components appeared to be mainly additive in nature, resulting in an increase in resistance level, as isolates for which the IG50s were notably below those of the most sensitive parent were not detected. Further crosses were then set up to test the hypothesis that resistance was conferred by a single, major gene in the resistant field isolates. These crosses consisted of backcrosses between parents and F1 progeny containing the putative single gene for resistance or between F1 progeny with the putative resistance gene. It was predicted that if the same single gene for resistance were present in all parental isolates then no sensitive strains would be generated in the progeny because the parental Prc-R genes would be allelic. This would be in contrast to the predicted results for a polygenic resistance model in which sensitive strains among the recombinant progeny would be expected because the parental mutations would occur at different loci (21). Furthermore, if additional genetic components for resistance were present in field isolates PR1 (Prc-MR) and 11-3-18 (Prc-HR) then backcrosses with these isolates might yield a progeny set for which the IG50s were in a wider range than those observed for progeny of crosses between F1 progeny with only the putative resistance gene. A final cross was set up between PR11 (Prc-LR) and C69 (Prc-LR; progeny from a cross involving 11-3-18) to determine whether resistance was conferred by a gene at the same locus, i.e., whether resistance genes were allelic in isolates from different sources.
Inheritance of prochloraz resistance in backcrosses and F1 progeny crosses.
Analyses were possible in those matings that produced fertile apothecia. A backcross between parental strain PR11 (Prc-LR) and F1 progeny A51 (Prc-LR) yielded a progeny set with a unimodal distribution with no sensitive progeny detected (Fig. 2B; Table 2, cross D). A backcross between parental strain PR1 (Prc-MR) and F1 progeny B32 (Prc-LR, i.e., with the putative single major gene but lacking any additional genetic component of PR1) yielded a unimodal distribution, with no sensitive progeny detected (Fig. 3C; Table 2, cross E). A similar distribution pattern was evident in the F2 progeny arising from a sibling cross between F1 progeny B32 and B53 (both Prc-LR), but IG50s for the progeny being below that for the parental strain PR1 (Prc-MR; IG50 = 1.0 mg/liter) (Fig. 3B; Table 2, cross F). A backcross between parental strain 11-3-18 (Prc-HR) and F1 progeny C39 (Prc-LR) yielded a complex distribution pattern, with a series of peaks and a relatively high value of skew, but, importantly, no sensitive progeny were detected (Fig. 4B; Table 2, cross G). In contrast, a sibling cross between F1 progeny C39 and C99 (both Prc-LR, lacking the putative additional genetic components of 11-3-18) yielded a near unimodal distribution pattern with a low value of skew, with no sensitive phenotypes detected, and IG50s for the progeny were below that observed for 11-3-18 (Fig. 4C; Table 2, cross H). In addition, a cross was set up between parental isolate PR11 (Prc-LR) and F1 progeny C69 (Prc-LR; containing the putative single major gene for resistance from 11-3-18 but no additional genetic component) to test whether the same genetic locus coded for resistance despite the fact that PR11 (Prc-LR) and 11-3-18 (Prc-HR) originated from different continents. The resulting progeny showed a unimodal pattern of distribution, with no sensitive progeny detected (Fig. 2C; Table 2, cross I). Finally, the IG50s for a progeny set from a control cross between the sensitive parental strains 22-432 and 22-433 (both Prc-S) (13) were determined to ensure that no resistance to prochloraz arose as a result of the sexual cycle. All progeny were found to be prochloraz sensitive (Table 2, cross J).
DISCUSSION
The question of how many genes are involved in the control of resistance to DMI fungicides has proved controversial (28). Early reports, based mainly on the use of laboratory mutants, suggested that resistance had a polygenic basis, with a complex heritable pattern involving additive genetic factors (10, 35). However, later studies involving isolates derived from field locations indicated that resistance was largely controlled by single major genes (4, 5, 34, 40). The present study used sexual crosses to investigate the genetic control of resistance to the DMI fungicide prochloraz in field isolates of the cereal eyespot pathogen T. yallundae. This represents the first such genetic analysis of resistance to prochloraz because other species in which resistance has been reported (Trichoderma harzianum, Rhizoctonia solani, and Colletotrichum coffeanum) lack amenable sexual stages (17, 25, 33).
Three crosses were made between prochloraz-sensitive parents and field isolates with differing levels of resistance to prochloraz (PR11 [Prc-LR], PR1 [Prc-MR], and 11-3-18 [Prc-HR]). The IG50s for the resulting progeny were determined, revealing an approximately even distribution of sensitive and resistant phenotypes. This bimodal pattern of inheritance provides clear evidence of a single major gene segregating for resistance in each of the different crosses (7, 18). This was substantiated by the use of three backcrosses and two crosses between sibling F1 progeny with the putative single resistance gene. The resulting progeny all exhibited a range of IG50s similar to that of the parents, with no evidence of recombinant sensitive progeny which might be expected if resistance were polygenically based (see reference 22). Analysis of a control cross between the two sensitive parental strains failed to detect any progeny with increased levels of resistance to prochloraz. Having established that resistance was primarily monogenic in nature, the related question of whether a single gene at the same locus conferred resistance in all three crosses, i.e., whether the resistance genes were allelic, arose. A further cross was therefore made between the French field isolate PR11 (Prc-LR) and an F1 progeny (C69 [Prc-LR]) of the New Zealand field isolate 11-3-18 (Prc-HR) selected to contain the putative single gene for resistance. This also yielded progeny which were entirely prochloraz resistant. This suggested that resistance was conferred by a gene at the same locus (or a closely linked locus) despite the geographic separation, since sensitive isolates would have been detected if the mutations were in different loci (39). It was not possible to cross PR11 and PR1 directly, as these were of the same mating type. Therefore, it is yet to be confirmed whether resistance is conferred by a gene with the same locus in PR1 (Prc-MR) as in the other two resistant field isolates. However, PR1 and PR11 were obtained from the same geographic area of northern France, so they may have been subject to similar selection pressures. Evidence for monogenic resistance to DMI fungicides has also been reported for resistance to triadimenol in Erysiphe graminis (4, 5), triadimenol in Nectria haematococca (9, 24), ketoconazole in Neurospora crassa (39), and fenarimol in Venturia inaequalis (40). This contrasts with reports of resistance to the fungicides imazalil in Aspergillus nidulans, fenarimol in N. haematococca (24), triadimenol in E. graminis (22), propiconazole in Pyrenophora teres (34), and triadimefon in Ustilago maydis (41), in which DMI resistance was polygenic in nature.
Although the segregation patterns for T. yallundae clearly indicated the presence of a single, major gene for resistance, there was also evidence for additional genetic components in field isolates PR1 (Prc-MR; IG50 = 1.0 mg/liter) and 11-3-18 (Prc-HR; IG50 = 2.4 mg/liter), which might explain the increased IG50s relative to that for PR11 (Prc-LR; IG50 = 0.5 mg/liter). Crosses between sensitive isolates and PR1 (Prc-HR) and 11-3-18 (Prc-HR) produced a peak IG50 for resistant progeny of approximately 0.5 mg/liter (Fig. 3A and 4A), corresponding to the single gene for low-level resistance of PR11 (Prc-LR). When crosses were made between F1 progeny for which the IG50 was in this range, the IG50s for the resulting F2 progeny also corresponded to the presence of the single low-level resistance gene, with a low value of skew in the progeny distribution (Fig. 3B and 4C; Table 2). However, when the same F1 progeny were backcrossed to PR1 (Prc-MR) or 11-3-18 (Prc-HR), a much longer tail was present in the progeny distribution, with a higher value of skew, indicating the presence of additional genetic components in the most resistant isolates (Fig. 3C and 4B; Table 2). These additional genetic components appeared to confer increased resistance to prochloraz, but an increase in IG50 only occurred if the single major gene for resistance was already present, i.e., the presence of these extra genetic components alone would not result in increased fungicide resistance. Similar results were obtained in an analysis of the inheritance of triadimenol resistance in six crosses of Pyrenophora teres, with evidence of one major segregating gene for resistance but with the presence of an extra 6 to 12 minor genes able to influence the resistance phenotype (34). Parasexual analysis has also indicated the presence of both major and minor genes for resistance to triadimenol in a Tapesia sp. (20), while varying levels of resistance to imazalil may be conferred by different genetic mechanisms in Penicillium italicum (19). This polygenic component may explain the directional, rather than disruptive, selection seen for DMI resistance in laboratory mutants of a Tapesia sp. (23). It is noted that for 4% of the progeny resulting from the 11-3-18 (Prc-HR) backcross, the IG50 was increased twofold over that observed for the resistant parent. A marked increase in resistance to DMI fungicides as a result of recombination during the sexual cycle has also been reported for P. teres and other crosses of T. yallundae (6, 31), emphasizing the possible importance of sexual reproduction in the field in the evolution of fungicide resistance.
A striking contrast was apparent in the levels of resistance conferred by the gene(s) for resistance to prochloraz and those conferred by the gene involved with resistance to the benzimidazole fungicide benomyl in T. yallundae. Resistance to benomyl is associated with single nucleotide mutations within the beta-tubulin gene (1), and resistance is inherited in a monogenic nature as reported for other pathogenic fungi (16, 36; P. Nicholson, unpublished data). Resistance to benomyl permits growth on media amended with very high levels of fungicide, whereas the growth of even the most prochloraz-resistant parent was inhibited at far lower concentrations of fungicide. This may be related to the mechanism of prochloraz resistance, which may involve decreased affinity of the target enzyme, detoxification, and/or increased efflux of fungicide through the action of ABC transporters (11, 12, 25, 28). The fact that even resistant isolates were affected by prochloraz may explain why there is no conclusive evidence that their presence leads to a lack of field performance by prochloraz (3). Studies are now in progress to clone genes conferring prochloraz resistance in T. yallundae in an attempt to determine the molecular genetic basis of resistance.
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom whom P.S.D. thanks for a David Phillips Research Fellowship and from whom IACR receives grant-aided support.
We thank Agrevo (UK) for providing isolates of T. yallundae. Overseas fungal isolates were imported into the United Kingdom under MAFF licence PHL 18/2846.
REFERENCES
- 1.Albernini C, Gredt M, Leroux P. Mutations of the β-tubulin gene associated with different phenotypes of benzimidazole resistance in the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis. Pestic Biochem Physiol. 1999;64:17–31. [Google Scholar]
- 2.Ballard D L, McLaughlan W T. Proceedings of the 35th New Zealand Weed and Pest Control Conference. Palmerston North, New Zealand: New Zealand Weed and Pest Control Society; 1982. Prochloraz—a new broad spectrum fungicide; pp. 269–273. [Google Scholar]
- 3.Birchmore R J, Russell P E, Buschhaus H. Sensitivity of eyespot to prochloraz. In: Heaney S, Slawson D, Hollomon D W, Smith M, Russell P E, Parry D W, editors. Fungicide resistance. Surrey, United Kingdom: British Crop Protection Council; 1994. pp. 27–34. [Google Scholar]
- 4.Blatter R H E, Brown J K, Wolfe M S. Genetic control of the resistance of Erysiphe graminis f. sp. hordei to five triazole fungicides. Plant Pathol. 1998;47:570–579. [Google Scholar]
- 5.Brown J K M, Jessop A C, Thomas S, Rezanoor H N. Genetic control of the response of Erysiphe graminis f. sp. hordei to ethirimol and triadimenol. Plant Pathol. 1992;41:126–135. [Google Scholar]
- 6.Campbell G F, Crous P W, Lucas J A. Genetic control of the response of Pyrenophora teres f. maculata, the cause of Pyrenophora leaf spot of barley in South Africa. Mycol Res. 1999;103:257–267. [Google Scholar]
- 7.Caten C E. Quantitative genetic variation in fungi. In: Thompson J N Jr, Thoday J M, editors. Quantitative genetic variation. New York, N.Y: Academic Press; 1979. pp. 33–59. [Google Scholar]
- 8.Clarke G M. Statistics and experimental design. London, United Kingdom: Edward Arnold; 1994. [Google Scholar]
- 9.Demopoulos V P, Ziogas B N. Studies on the mechanism of expression of a major gene mutation for resistance to triadimenol in the filamentous phytopathogenic ascomycete Nectria haematococca var. cucurbitae. Pestic Biochem Physiol. 1994;50:159–170. [Google Scholar]
- 10.De Waard M A. Resistance to fungicides which inhibit sterol 14 α-demethylation, an historical perspective. In: Heaney S, Slawson D, Hollomon D W, Smith M, Russell P E, Parry D W, editors. Fungicide resistance. Surrey, United Kingdom: British Crop Protection Council; 1994. pp. 3–10. [Google Scholar]
- 11.De Waard M A. Molecular genetics of resistance in fungi to azole fungicides. ACS (Am Chem Soc) Symp Ser. 1996;645:62–71. [Google Scholar]
- 12.De Waard M A. Significance of ABC transporters in fungicide sensitivity and resistance. Pestic Sci. 1997;51:271–275. [Google Scholar]
- 13.Dyer P S, Lucas J A. Incidence of apothecia of Tapesia yallundae at set-aside sites in England and sensitivity of the ascospore offspring to the fungicides benomyl and prochloraz. Plant Pathol. 1995;44:796–804. [Google Scholar]
- 14.Dyer P S, Nicholson P, Lucas J A, Peberdy J F. Tapesia acuformis as a causal agent of eyespot disease of cereals and evidence for a heterothallic mating system using molecular markers. Mycol Res. 1996;100:1219–1226. [Google Scholar]
- 15.Dyer P S, Nicholson P, Rezanoor H N, Lucas J A, Peberdy J F. Two-allele heterothallism in Tapesia yallundae, the teleomorph of the cereal eyespot pathogen Pseudocercosporella herpotrichoides. Physiol Mol Plant Pathol. 1993;43:403–414. [Google Scholar]
- 16.Faretra F, Pollastro S. Genetics of sexual compatibility and resistance to benzimidazole and dicarboxyimide fungicides in isolates of Botryotinia fuckeliana (Botrytis cinerea) from nine countries. Plant Pathol. 1993;42:48–57. [Google Scholar]
- 17.Figuerasroca M, Cristani C, Vannacci G. Sensitivity of Trichoderma isolates and selected resistant mutants to DMI fungicides. Crop Protect. 1996;15:615–620. [Google Scholar]
- 18.Georgopoulos S G, Skylakakis G. Genetic variation in the fungi and the problem of fungicide resistance. Crop Protect. 1986;5:299–305. [Google Scholar]
- 19.Guan J, Kapteyn J C, Kerkenaar A, De Waard M A. Characterisation of energy-dependent efflux of imazalil and fenarimol in isolates of Penicillium italicum with a low, medium and high degree of resistance to DMI-fungicides. Neth J Plant Pathol. 1992;98:313–324. [Google Scholar]
- 20.Hocart M J, McNaughton J E. Parasexual analysis of morpholine and triazole resistance in Pseudocercosporella. In: Heaney S, Slawson D, Hollomon D W, Smith M, Russell P E, Parry D W, editors. Fungicide resistance. Surrey, United Kingdom: British Crop Protection Council; 1994. pp. 325–329. [Google Scholar]
- 21.Hollomon D W. Genetic control of ethirimol resistance in a natural population of Erysiphe graminis f. sp. hordei. Phytopathology. 1981;71:536–540. [Google Scholar]
- 22.Hollomon D W, Butters J, Clark J. Proceedings of the British Crop Protection Conference 1984. Croydon, United Kingdom: British Crop Protection Council; 1984. Genetic control of triadimenol resistance in barley powdery mildew; pp. 477–482. [Google Scholar]
- 23.Julian A M, Hardy J E, Lucas J A. The induction and characterisation of isolates of Pseudocercosporella herpotrichoides with altered sensitivity to the EBI fungicide prochloraz. Pestic Sci. 1994;41:121–128. [Google Scholar]
- 24.Kalamarakis A E, Demopoulos V P, Ziogas B N, Georgopoulos S G. A highly mutable major gene for triadimenol resistance in Nectria haematococca var. cucurbitae. Neth J Plant Pathol. 1989;95(Suppl. 1):109–120. [Google Scholar]
- 25.Kapteyn J C, Pillmoor J B, De Waard M A. Biochemical mechanisms involved in selective fungitoxicity of two sterol 14 α-demethylation inhibitors, prochloraz and quinconazole: accumulation and metabolism studies. Pestic Sci. 1992;36:85–93. [Google Scholar]
- 26.Kendall S J, Hollomon D W, Cooke L R, Jones D R. Changes in sensitivity to DMI fungicides in Rhynchosporium secalis. Crop Protect. 1993;12:357–362. [Google Scholar]
- 27.Koenraadt H, Somerville S C, Jones A L. Characterisation of mutations in the beta-tubulin gene of benomyl resistant field strains of Venturia inaequalis and other plant pathogenic fungi. Phytopathology. 1992;82:1348–1354. [Google Scholar]
- 28.Köller W. Recent developments in DMI resistance. In: Lyr H, Russell P E, Sisler H D, editors. Modern fungicides and antifungal compounds. Andover, United Kingdom: Intercept; 1995. pp. 301–311. [Google Scholar]
- 29.Köller W, Scheinpflug H. Fungal resistance to sterol biosynthesis inhibitors: a new challenge. Plant Dis. 1987;71:1066–1074. [Google Scholar]
- 30.Leroux P, Gredt G. Evolution of fungicide resistance in the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis in France. Pestic Sci. 1997;51:321–327. [Google Scholar]
- 31.Lucas J A, Dyer P S, Murray T D. Pathogenicity, host-specificity, and population biology of Tapesia spp., causal agents of eyespot disease of cereals. Adv Bot Res. 2000;33:225–258. [Google Scholar]
- 32.Molnar A, Hornok L, Pesti M. The high level of benomyl tolerance in Fusarium oxysporum is determined by the synergistic interaction of two genes. Exp Mycol. 1985;9:326–333. [Google Scholar]
- 33.Mwang'ombe A W. Tolerance in isolates of Colletotrichum coffeanum Noack to prochloraz-manganese in Kenya. Pestic Sci. 1992;34:371–373. [Google Scholar]
- 34.Peever T L, Milgroom M G. Inheritance of tradimenol resistance in Pyrenophora teres. Phytopathology. 1992;82:821–828. [Google Scholar]
- 35.Russell P E. Fungicide resistance: occurrence and management. J Agric Sci. 1995;124:317–323. [Google Scholar]
- 36.Sanoamuang N, Gaunt R E, Fautrier A G. The segregation of resistance to carbendazim in sexual progeny of Monilinia fructicola. Mycol Res. 1995;99:677–680. [Google Scholar]
- 37.Schulz U, Scheinpflug H. Sterol biosynthesis inhibiting fungicides: antifungal properties and application in cereals. In: Berg D, Plempel M, editors. Sterol biosynthesis inhibitors: pharmaceutical and agrochemical aspects. Chichester, United Kingdom: Ellis Harwood; 1988. pp. 213–261. [Google Scholar]
- 38.Singh G, Dyer P S, Ashby A M. Intra-specific and inter-specific conservation of mating-type genes from the discomycete plant pathogenic fungi Pyrenopeziza brassicae and Tapesia yallundae. Curr Genet. 1999;36:290–300. doi: 10.1007/s002940050503. [DOI] [PubMed] [Google Scholar]
- 39.Staben C. Resistance to azole drugs in Neurospora crassa. Exp Mycol. 1995;19:163–165. doi: 10.1006/emyc.1995.1019. [DOI] [PubMed] [Google Scholar]
- 40.Stanis V F, Jones A L. Reduced sensitivity to sterol-inhibiting fungicides in field isolates of Venturia inaequalis. Phytopathology. 1985;75:1098–1101. [Google Scholar]
- 41.Wellmann H, Schauz Z. DMI-resistance in Ustilago maydis. I. Characterisation and genetic analysis of triadimefon-resistant laboratory mutants. Pestic Biochem Physiol. 1992;43:171–181. [Google Scholar]