Abstract
Few techniques are currently available for quantifying specific prokaryotic taxa in environmental samples. Quantification of specific genotypes has relied mainly on oligonucleotide hybridization to extracted rRNA or intact rRNA in whole cells. However, low abundance and cellular rRNA content limit the application of these techniques in aquatic environments. In this study, we applied a newly developed quantitative PCR assay (5′-nuclease assay, also known as TaqMan) to quantify specific small-subunit (SSU) rRNA genes (rDNAs) from uncultivated planktonic prokaryotes in Monterey Bay. Primer and probe combinations for quantification of SSU rDNAs at the domain and group levels were developed and tested for specificity and quantitative reliability. We examined the spatial and temporal variations of SSU rDNAs from Synechococcus plus Prochlorococcus and marine Archaea and compared the results of the quantitative PCR assays to those obtained by alternative methods. The 5′-nuclease assays reliably quantified rDNAs over at least 4 orders of magnitude and accurately measured the proportions of genes in artificial mixtures. The spatial and temporal distributions of planktonic microbial groups measured by the 5′-nuclease assays were similar to the distributions estimated by quantitative oligonucleotide probe hybridization, whole-cell hybridization assays, and flow cytometry.
Ribosomal RNA genes (rDNAs) are extensively used to study the diversity of microorganisms in environmental samples. To date, most surveys of microbial diversity in environmental samples have relied upon cloning and sequencing of rDNAs. These studies have shown that the diversity of microorganisms in natural ecosystems has been severely underestimated in culture collections and have led to the discovery of several new microbial lineages (5, 10, 20, 28). Despite the increased knowledge of the phylogenetic diversity of indigenous microbes, less is known about the abundance of particular groups or their spatial and temporal dynamics.
Few techniques are currently available for quantifying specific prokaryotic taxa in environmental samples. Recoveries of PCR-amplified rDNAs from clone libraries are subject to several quantitative biases (see reference 26 for a review). rRNA-based quantification therefore has relied mainly on radiolabeled oligonucleotide hybridization to extracted rRNA (11, 16, 21) or whole-cell hybridization using fluorescence-labeled oligonucleotides (1, 7). However, in many ecosystems the abundance of microorganisms is relatively low and many cells may have a low rRNA content, limiting the sensitivity of oligonucleotide hybridization techniques for quantification of specific microorganisms.
In individual cells, the number of copies of rDNAs may be several orders of magnitude lower than that of rRNAs; in DNA, rRNA coding sequences exist in a context of much higher genetic complexity. It is therefore difficult to quantify rDNAs by oligonucleotide hybridization. However, rDNA is suitable for amplification by the PCR. Provided that the amplification efficiencies of different rDNAs are comparable, the proportions of specific genes in PCR amplification products should reflect the proportions of the same rDNAs in the original samples.
Most described PCR-based quantification methods (e.g., dot blot hybridization [11], denaturing gradient gel electrophoresis [17], terminal restriction fragment length [TRFLP] analysis [14], and PCR amplicon length heterogeneity [LH-PCR] analysis [23]) rely on quantification after a number of replication cycles have been completed and reaction products are at concentrations sufficiently high to allow end-point detection. However, recent studies (23, 24) have shown that the accumulation of amplification products may bias the proportions of different amplicons in mixtures compared to the proportions in the original samples and have suggested that a minimum number of cycles be used. Furthermore, some of these techniques (TRFLP and LH-PCR) rely on a diagnostic fragment size, so microbial identification is presumptive, not determinative.
To circumvent the quantitative difficulties associated with other methods, we applied a recently developed PCR-based 5′-nuclease assay (also known as TaqMan [15]) to the quantification of SSU rDNAs in artificial mixtures and in environmental DNA samples. In contrast to other PCR-based techniques, this method detects amplified rDNAs in early cycles of the PCR, and quantification is performed during the exponential phase of the reactions. The method also allows the estimation of PCR amplification efficiency.
In this study, we developed general 5′-nuclease assays for the measurement of rDNAs belonging to the domains Archaea and Bacteria. We also designed specific assays for the quantification of SSU rDNAs of bacteria belonging to the genera Synechococcus and Prochlorococcus and two groups of uncultivated marine Archaea. We tested the assays using artificial DNA mixtures from cultivated strains as well as DNA extracted from bacterioplankton assemblages collected in California coastal waters. The results indicate that these assays can accurately estimate relative proportions of genes in complex mixtures. Our results also revealed some of the inherent limitations of the approach.
MATERIALS AND METHODS
Principles of 5′-nuclease assays.
The 5′-nuclease assays are based on the measurement of the fluorescence intensity of fluorochrome molecules that are produced during the extension step of PCR by the 5′-to-3′ exonuclease activity of Taq DNA polymerase. These fluorochromes are derived from an oligonucleotide probe with sequence complementary to a region between the amplification primers (TaqMan probe). In the intact probe, fluorescence emitted by the 5′ reporter dye is quenched by a dye attached to the 3′ end of the probe. During the PCR amplification cycle, the 5′ reporter dye is removed from the template-bound oligonucleotide by Taq DNA polymerase. The accumulation of cleaved reporter dye molecules is proportional to the initial copy numbers of the target genes in template DNA. Target gene copy numbers in an unknown sample are calculated from the number of cycles necessary for the fluorescence emission of reporter dyes to exceed a set threshold value (threshold cycle number [CT]) relative to standard controls with known numbers of copies. Slopes of standard curves (regression lines of CT versus log N, the log of initial gene copy numbers in standard templates) can be used to estimate PCR amplification efficiency in the assays. For a detailed description of the method, see reference 15.
Environmental DNA samples.
Water samples (4 liters) were collected at five depths at station M1, located 10.8 mi west of Moss Landing, Calif., on 3 December 1997 and 7 January 1998. Picoplankton collection and nucleic acid extraction were performed as previously described (16). DNA was further purified from inhibitory substances and RNA by CsCl buoyant equilibrium centrifugation with 300-μl tubes and an Optima TL ultracentrifuge (Beckman, Palo Alto, Calif.) as described previously (3), except that Centricon 100 centrifugal filter units (Millipore, Bedford, Mass.) were used to remove the CsCl from the DNA sample. All genomic DNA samples were stored at −20 to −80°C in pH 8.0 TE buffer (10 mM Tris-HCl, 1 mM EDTA) until analysis.
Cultivated strains.
Several cultivated organisms belonging to the domains Archaea and Bacteria were chosen to test primer and probe specificity and to serve as templates for the optimization of 5′-nuclease assays. Our choice of organisms was based on phylogenetic relationships and the availability of information on genome size and number of operons in the genome. All organisms used in this study are listed in Table 1. Nucleic acids were extracted from axenic cultures by the cetyltrimethylammonium bromide (CTAB) protocol (3), and DNA was separated from RNA by CsCl buoyant equilibrium centrifugation and stored as described above. Nucleic acids from Synechococcus strains WH7805 and WH8103 were kindly provided by D. Distel, Department of Biology, University of Maine. Genomic DNA concentrations were measured fluorometrically by PicoGreen (Molecular Probes, Eugene, Oreg.) staining with a FluorImager fluorescence imager (Molecular Dynamics, Sunnyvale, Calif.) according to the manufacturer's specifications.
TABLE 1.
Summary of primer specificity tests by three-step end-point PCR
| Domain | Groupa | Subgroupb | Organism or clonec | Amplificationd with the following primer set (annealing temp, °C):
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| BACT1 (59) | ARCH1 (59) | BACT2 (56) | ARCHGI (59) | ARCHGII (59) | PHPICO (60) | ||||
| Bacteria | Ver | MB11A01 | + | − | + | ||||
| Hgc | MB11A03 | + | − | + | − | ||||
| Cya | Syn | MB11A04 | + | − | + | + | |||
| Cya | Syn | MB11E08 | + | − | + | + | |||
| Cya | Syn | MB11E09 | + | − | + | + | |||
| Cya | Syn | MB11F02 | + | − | + | + | |||
| Cya | Syn | Synechococcus sp. strain WH7803 | + | − | + | + | |||
| Cya | Syn | Synechococcus sp. strain WH7805 | + | + | |||||
| Cya | Syn | Synechococcus sp. strain WH8103 | + | + | |||||
| Cya | Plastids | MB11B05 | + | − | + | − | |||
| Alp | Sar11 | MB11B07 | + | − | + | ||||
| Alp | Ros | Strain R2A57 | + | − | + | − | − | − | |
| Alp | Sar116 | MB11B03 | + | − | + | − | |||
| Gam | Escherichia coli DH10B | + | − | + | − | ||||
| Gam | Sar86 | MB11B08 | + | − | + | − | |||
| Gam | MB11B11 | + | − | + | − | ||||
| Lgc | Bacillus subtilis | + | − | + | − | − | − | ||
| Fbc | Strain R2A10 | + | − | + | |||||
| Archaea | Cre | Desulfurococcus ambivalens | − | + | − | − | − | ||
| Cre | GI | SBAR12 | ∗ | ∗ | ∗ | + | − | ||
| Cre | GI | SBAR5 | ∗ | ∗ | ∗ | + | ± | ||
| Cre | GI | 2i | ±e | + | ± | + | − | ||
| Eur | Haloferax mediterranei | − | + | − | − | − | − | ||
| Eur | Haloferax volcanii | − | + | − | − | − | |||
| Eur | Halococcus salinarum | − | + | − | − | − | |||
| Eur | Sulfolobus acidocaldarius | − | + | − | − | − | |||
| Eur | Methanobacterium thermoautotrophicum | − | + | − | − | − | |||
| Eur | Methanococcus jannaschii | − | + | − | − | − | |||
| Eur | Thermoplasma acidophilum | − | + | − | − | − | |||
| Eur | GII | SBAR16 | ∗ | ∗ | ∗ | − | + | ||
| Eur | GII | SBAR1A | ∗ | ∗ | ∗ | − | + | ||
| Eur | GII | WHARN | ∗ | ∗ | ∗ | − | + | ||
| Eur | GII | SB95-71 | ∗ | ∗ | ∗ | − | + | ||
| Eur | GII | SB95-72 | ∗ | ∗ | ∗ | − | + | ||
| Eur | GII | SB95-74 | ∗ | ∗ | ∗ | − | + | ||
Ver, Verrucomicrobiales; Hgc, high-G+C-content gram-positive bacteria; Cya, Cyanobacteria; Alp, α-Proteobacteria; Gam, γ-Proteobacteria; Lgc, low-G+C-content gram-positive bacteria; Fbc, Flexibacter, Bacteroides, and Cytophaga; Cre, Crenarchaeota; Eur, Euryarchaeota.
Syn, Synechococcus; Ros, Roseobacter; GI, group I marine Archaea; GII, group II marine Archaea. Sar11, Sar116, and Sar86 are groups according to reference 16b.
Boldfacing indicates organisms or clones used as standards in the 5′-nuclease assays.
+, strong amplification; ±, weak amplification; −, no amplification; ∗, clone does not include a priming site.
Likely due to contamination with E. coli genomic DNA.
Plasmid clones. (i) Environmental libraries.
Cloned rRNA genes were chosen as controls for all assays. This choice was based on the fact that the number of copies of the gene of interest can be more easily inferred from plasmid concentrations than from genomic DNA concentrations. Also, for many microorganisms, rDNA copy number, genome size, or both have not been determined. Finally, plasmid clones are also the only source of rDNA from uncultivated microorganisms.
Archaeal rDNA standards consisted of a cloned DNA fragment containing the rDNA from a planktonic crenarchaeote (Fosmid 4B7 [22]) and a small-subunit (SSU) rDNA clone from a marine euryarchaeote from the Santa Barbara Channel (SB95-72 [16]).
Bacterial rDNA clones were retrieved from a PCR library constructed with a surface seawater DNA sample collected on 5 June 1997 at station S297-67-90 (124°89/W, 35°44/N), 275 km from Moss Landing. Nucleic acids were extracted as previously described (16), and the DNA was purified by CsCl buoyant equilibrium centrifugation and resuspended in 200 μl of TE buffer, pH 8.0. DNA fragments containing nearly the entire SSU rDNA, the intergenic transcribed spacer (ITS), and about 1,900 bp of the SSU rDNA were amplified by PCR. In a final 50-μl volume, reaction mixtures contained 0.5 μl of TaqPlus Long 10× low-salt buffer (Stratagene, La Jolla, Calif.), 0.2 mM each deoxynucleoside triphosphate (dNTP) (Promega, Madison, Wis.), 0.5 μM each primer, 2 mM MgCl2, 4 μl of environmental DNA sample, and 2.5 U of TaqPlus Long polymerase mixture (Stratagene). The forward primer used was the SSU rDNA bacterial primer 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) (9), and the reverse primer used was the large-subunit (LSU) rDNA bacterial primer 1933eR (5′-ACC CGA CAA GGA ATT TCG C-3′) (2). A PE9700 thermal cycler (PE Biosystems Inc., Foster City, Calif.) was programmed to 5 min of precycling at 94°C and 30 cycles of 94°C denaturation for 30 s, 55°C annealing for 30 s, and 72°C extension for 3 min.
PCR products of triplicate reactions were combined and purified by phenol-chloroform extraction followed by ethanol precipitation (19), resuspended in 20 μl of deionized water, and blunt ended by treatment with Pfu DNA polymerase. In a final volume of 10 μl, the blunt-ending reaction mixtures contained 1 μl of recombinant Pfu DNA polymerase 10× buffer (Stratagene), 1 mM each dNTP, and 0.5 U of recombinant Pfu DNA polymerase (Stratagene). Reaction mixtures were incubated at 94°C for 20 min in a PE9700 thermal cycler.
Products of the blunt-ending reaction were purified by phenol-chloroform extraction followed by ethanol precipitation and resuspended in 10 μl of deionized water. Two microliters of the purified blunt-ended PCR product was ligated into the vector PCR Zero Blunt (Invitrogen, Carlsbad, Calif.) using the manufacturer's protocol, and the resulting ligation product was used to transform Escherichia coli One Shot TOP10 competent cells (Invitrogen). A total of 384 white colonies were picked, and plasmids were archived in 96-well microtiter dishes as previously described (3). A total of 190 clones were screened and grouped based on HaeIII restriction fragment length polymorphism analysis as previously described (25). Clones representing all HaeIII restriction fragment length polymorphism patterns were bidirectionally sequenced by the dideoxy termination reaction using an LC7200 automated DNA sequencer (LI-COR, Lincoln, Nebr.). These clones have the prefix MB1. Archaeal clones from additional libraries (5, 16) were used for the primer specificity tests (Table 1). These plasmids were purified using a Qiaprep Spin plasmid kit (Qiagen, Valencia, Calif.) or a Mini-Prep 24 machine (MacConnell Research, La Jolla, Calif.).
Plasmids used as standards for the 5′-nuclease assays were purified using a Qiagen Maxi plasmid kit according to the manufacturer's protocol and further purified by CsCl buoyant equilibrium centrifugation with 3.6-ml tubes and an Optima TL ultracentrifuge as described previously (3), except that Centricon 100 centrifugal filter units were used to remove the CsCl. Linearized plasmids were produced by digestion with the restriction endonuclease NotI (Promega) according to the manufacturer's protocol, purified by phenol-chloroform extraction, and subjected to three washes using Microcon 100 centrifugal filter units (Millipore) and a final desalting step using Micro Bio-spin columns (Bio-Rad, Hercules, Calif.) loaded with TE buffer. DNA concentrations of purified linearized plasmids were determined fluorometrically as described above. Supercoiled (not linearized) plasmid concentrations were determined spectrophotometrically.
(ii) Cultivated organisms.
The plasmid containing the SSU-ITS-LSU rDNA fragment of strain R2A57, a marine member of the α-Proteobacteria (25) (R57pCRbl), was prepared from extracted genomic DNA in the same manner as the environmental clones, except that the DNA polymerase used for amplification was TaqPlus Precision DNA polymerase mix (Stratagene) and the PCR products were directly cloned using a PCR ZeroBlunt kit (Invitrogen). The plasmid containing the SSU-ITS-LSU rDNA fragment of Haloferax volcanii (HvpCRbl) was produced in a similar fashion, except that the primers used were the degenerate archaeal SSU rDNA forward primer 21F (5′-TTC CGG TGG ATC CYG CCG GA-3′ [16]) and the degenerate prokaryotic LSU rDNA reverse primer 2445R (5′-CCC YGG GGT ARC TTT TCT ST-3′ [6]). These plasmids were purified using a Qiagen Maxi plasmid kit and isolated from genomic DNA via CsCl buoyant equilibrium centrifugation (3).
Primers and probes.
The TaqMan probe and reverse primers for the Bacteria and Archaea assays targeted the previously described universal regions homologous to Escherichia coli positions 1392 to 1406 (13) 1492 to 1510 (13), and 1518 to 1541 (9) (Table 2). Our PCR primers and probes were designed using ARB software graciously provided by O. Strunk and W. Ludwig, Technical University of Munich, Munich, Germany. A database of over 10,000 SSU rRNA sequences was used to check primer specificity and possible mismatches. Although all universal primers showed significant mismatches with some major bacterial and archaeal groups (Table 2), we opted to exclude the detection of genes belonging to these groups rather than to increase the numbers of degenerate positions in the primers, which would complicate the measurement of effective primer concentrations in the reactions. Group-specific primers and probes were designed using ARB software (Table 2). Since the target organisms were closely related, in the majority of cases it was possible to design primers with no degeneracies. All probes and primers were screened and optimized for the requirements of 5′-nuclease assays using Primer Express software (PE Biosystems).
TABLE 2.
PCR primers, 5′-nuclease assay probes, and optimal reaction conditions tested in this study
| Set | Target group | Annealing temp (°C) | MgCl2 (mM) | Type | Primer or probe name (optimized concn); 5′-to-3′ sequence; and target | Primer properties | General comments |
|---|---|---|---|---|---|---|---|
| BACT1 | Bacteria | 59 | 3 | Forward primer | BACT1369F (1,000 nM); CGGTGAATACGTTCYCGG; and Bacteria | Major mismatchesa with Chlorobium spp., Planctomyces phylum, Synechococcus, and Cytophagales; | Underestimates copy numbers of rDNA of Synechococcus; apparently underestimates total copy numbers of bacterial SSU rDNA in environmental samples |
| Reverse primer | PROK1541R (1,000 nM); AAGGAGGTGATCCRGCCGCA; and prokaryotes | Major mismatches with some Vibrio spp. Limited number of sequences for priming region | |||||
| Probe | TM1389F (500 nM); CTTGTACACACCGCCCGTC; and prokaryotes | ||||||
| BACT2 | Bacteria | 56 | 3 | Forward primer | BACT1369F (1,500 nM); CGGTGAATACGTTCYCGG; and Bacteria | Major mismatchesa with Chlorobium spp. and Planctomyces phylum Low melting temp | Best primer set for bacterial SSU rDNA; low annealing and extension temperatures seem to necessitate high-quality Taq DNA polymerase |
| Reverse primer | PROK1492R (1,000 nM); GGWTACCTTGTTACGACTT; and prokaryotes | ||||||
| Probe | TM1389F (500 nM); CTTGTACACACCGCCCGTC; and prokaryotes | ||||||
| ARCH1 | Archaea | 59 | 3 | Forward primer | ARCHMIX1369F (500 nM), 1:1 mixture of ARCH1-1369F, CGGTGAATACGTCCCTGC, and ARCH2-1369F, CGGTGAATATGCCCCTGC; and Archaea | Major mismatchesa with Methanococcales and Methanosarcinales | Apparently underestimates total copy numbers of group II Marine Archaea (Euryarchaeota) SSU rDNA in environmental samples |
| Reverse primer | PROK1541R (1,000 nM); AAGGAGGTGATCCRGCCGCA; and prokaryotes | ||||||
| Probe | TM1389F (500 nM); CTTGTACACACCGCCCGTC; and prokaryotes | ||||||
| PHPICO | Synechococcus and Prochlorococcus | 59 | 5 | Forward primer | PHPICO191F (500 nM); TGAAATGAATTTCGCCTGAG; and Synechococcus group | In Monterey Bay, appears to overestimate percentages of Prochlorococcus plus Synechococcus compared to flow cytometer cell counts | |
| Reverse primer | PHPICO420R (500 nM); AGAAAAGAGGTTTACAGCCCAG; and Synechococcus group | ||||||
| Probe | PHPICO283F (500 nM); CAGTAGCTGGTCTGAGAGGATGATC; and Synechococcus group | ||||||
| ARCHGI | Group I marine Archaea (Crenarcheota) | 59 | 5 | Forward primer | ARCHGI334F (1,000 nM); AGATGGGTACTGAGACACGGAC; and group I marine Archaea | ||
| Reverse primer | ARCHGI554R (500 nM); CTGTAGGCCCAATAATCATCCT; and group I marine Archaea | ||||||
| Probe | TM519AR (400 nM); TTACCGCGGCGGCTGGCAC; and Archaea | ||||||
| ARCHGII | Group II marine Archaea (Euryarchaeota) | 59 | 5 | Forward primer | ARCHGII333F (1,000 nM); GAGATGGATTCTGAGACACGAA; and group II marine Archaea | ||
| Reverse primer | ARCHGII554R (1,000 nM); TTAGGCCCAATAAAAKCGAC; and group II marine Archaea | ||||||
| Probe | TM519AR (400 nM); TTACCGCGGCGGCTGGCAC; and Archaea | ||||||
| pUC | pUC-derived plasmids | 59 | 5 | Forward primer | PUCF (500 nM); and CGCAACGCAATTAATGTGAGTTA | ||
| Reverse primer | PUCR (500 nM); and AATCATGGTCATAGCTGTTTCCTG | ||||||
| Probe | TMPUC (400 nM); and AAATTGTTATCCGCTCACAATTCCACAC |
Major mismatches are defined as two or more mismatches (excluding GT mismatches) in the last five 3′-end positions of the primer or four or more mismatches in the entire primer.
Performance of 5′-nuclease assays.
Although optimized 5′-nuclease assay parameters (i.e., primers, fluorogenic probe, and MgCl2 concentrations) between different primer and probe sets varied, the following conditions were identical for all. In a final volume of 25 μl, reaction mixtures contained the buffer recommended for the DNA polymerase used, 200 μM each dATP, dCTP, and dGTP, 400 μM dUTP, and 0.25 U of AmpErase uracyl N-Glycosylase (UNG; PE Biosystems). For all of the primer and probe sets, except for BACT2, 0.025 U of AmpliTaq Gold DNA polymerase (PE Biosystems) μl−1 was used. For the BACT2 set, 0.025 U of Platinum Taq DNA polymerase (Life Technologies, Rockville, Md.) μl−1 was used. All reactions were carried out with optical tubes or reaction trays (PE Biosystems), with 2.5 μl of a template being delivered into the tubes first using a Microman M10 positive-displacement pipette (Rainin, Emeryville, Calif.). A master mix (22.5 μl) was delivered using a Microman M100 positive-displacement pipette (Rainin), and tubes were sealed with optical caps. In experiments optimizing the effect of primer, probe, or MgCl2 concentrations, these reagents were excluded from the master mix and added to the tubes last. All reactions were performed with a model 7700 sequence detection system (PE Biosystems) programmed with a soak step of 2 min at 50°C, allowing AmpErase UNG to hydrolyze PCR amplicons possibly carried over from previous reactions. An enzyme activation soak step (15 min at 95°C for AmpliTaq Gold or 2 min at 94°C for Platinum Taq) followed the initial soak step. Finally, 40 cycles of 15 s of denaturation (95°C for AmpliTaq Gold or 94°C for Platinum Taq) and 1 min of annealing plus extension at the temperatures listed in Table 2 were performed. All results were analyzed with a PowerMac 4400 (Apple Computer Co., Cupertino, Calif.) computer using Sequence Detector v1.6.3 software (PE Biosystems).
Optimization of 5′-nuclease assays. (i) Primer melting temperature.
Since the BACT1 and ARCH1 primer and probe sets shared the same reverse primer and 5′-nuclease probe, we empirically tested the melting temperature of all of our primers via end-point PCR. In a final volume of 20 μl, reaction mixtures contained 1 μl of AmpliTaq Gold 10× buffer (PE Biosystems), 200 μM each dATP, dCTP, and dGTP, 400 μM dUTP, 1.5 mM MgCl2, 0.2 ng of template DNA, 0.05 U of AmpErase UNG, and 0.025 U of AmpliTaq Gold DNA polymerase μl−1. We used E. coli DH10B DNA to test the bacterial primer set and Methanobacterium thermoautotrophicum and Thermoplasma acidophilum to test the archaeal primer set. Reactions were run in a Robocycler Gradient96 thermal cycler (Stratagene) programmed to a 10-min enzyme activation soak step at 94°C and 25 cycles of 95°C denaturation for 52 s, a gradient of 56 to 65°C annealing for the bacterial primer set or 53 to 65°C annealing for the archaeal set for 30 s, and 72°C extension for 42 s. Mineral oil was overlaid on the samples to avoid evaporation, and 10 μl of the reaction products was run in 1% agarose minigels stained with ethidium bromide (50 μg/ml). Gels were scanned with an MD FSI fluorescence imager (Molecular Dynamics). We tested the melting temperature of the BACT2 set in a similar manner, except that the final reaction volume was 25 μl, the template used was strain R2A57 genomic DNA, and the gradient was 52 to 63°C.
(ii) Specificity.
Since most TaqMan probes were designed to target broad groups of organisms (i.e., prokaryotes or marine Archaea), we relied on the sequences of the primers for specificity. To examine possible cross-reactivity of the primers, we initially examined the formation of PCR products after 25 cycles in three-step PCRs. In a final volume of 10 μl, reaction mixtures contained 1 μl of AmpliTaq Gold 10× buffer, 0.2 mM each dNTP, 0.5 μM each primer, 1.5 mM MgCl2, and 0.05 U of AmpliTaq Gold DNA polymerase. Reactions were run in a PE9700 thermal cycler at the annealing temperatures listed in Table 1.
(iii) Primer concentration.
5′-Nuclease assay reactions were performed using a matrix of concentrations of forward and reverse primers to seek primer concentrations yielding minimal CT values and consequently the highest amplification efficiencies. Primer concentrations ranged from 100 to 1,500 nM. Annealing temperatures as well as MgCl2 concentrations were the same as those listed in Table 2. The remaining parameters were identical to those described above. For primer and probe sets using universal primers (i.e., PROK1541R and PROK1492R) or probes (i.e., ARCH519R and PROK1389F), we used specific target genomic or plasmid DNA alone as well as mixtures of target and nontarget genomic DNAs.
(iv) Probe concentration.
5′-Nuclease assay reactions were performed using optimized primer concentrations and a TaqMan probe concentration range. We determined the probe concentration yielding the minimal CT value for a specific template. Probe concentrations ranged from 200 to 800 nM, and the remaining reaction conditions were the same as used for the primer matrices. We used pure target genomic DNA as well as mixtures of target and nontarget genomic DNAs for primer and probe sets using universal probes (i.e., ARCH519R and PROK1389F), since the probes are complementary to both target and nontarget DNAs.
(v) MgCl2 concentration.
MgCl2 concentration has two main effects on PCRs. First, MgCl2 is required for Taq DNA polymerase activity. This was particularly important in 5′-nuclease assays, since the extension step was performed at suboptimal temperatures for the DNA polymerase. On the other hand, MgCl2 concentration affects the melting temperature of the primers and consequently their specificity. This was particularly critical for the BACT1, BACT2, and ARCH1 primer and probe sets, which relied only on the forward primers for specificity. For primer and probe sets BACT1 and ARCH1, we chose the minimal MgCl2 concentrations that allowed consistent amplification of templates at different concentrations and reasonably high amplification efficiencies, estimated from the slopes of standard curves empirically generated by 5′-nuclease assays. We used the same MgCl2 concentrations for primer and probe sets BACT2 and ARCH2, and 5 μM MgCl2 (final) was used for the remaining 5′-nuclease assay primer and probe sets.
Effect of template source and type.
To test whether genomic DNAs from different organisms might be differentially amplified by 5′-nuclease assays, standard curves for E. coli, Bacillus subtilis, and Deinococcus radiodurans genomic DNAs were compared. The 25-μl reaction mixtures contained 2.5 μl of AmpliTaq Gold 10× buffer, 1.5 mM MgCl2, 200 μM each dATP, dCTP, and dGTP, 400 μM dUTP, 500 nM each primers BACT1369F and PROK1541R, 200 nM probe TM1389F, 0.05 U of AmpErase UNG, 2.5 μl of templates with copy numbers ranging from 104 to 107 copies μl−1, and 0.025 U of AmpliTaq Gold DNA polymerase μl−1. Reactions were performed with a model 7700 sequence detection system and the cycling parameters were the same as those already described. Triplicate standard curves for each of the genomic DNA types were compared by analysis of variance (ANOVA).
Amplification efficiencies of the same SSU rDNA on chromosomal and plasmid DNAs were also compared using the optimized conditions for primer and probe sets BACT1 and ARCH1 (Table 2). H. volcanii was used for the ARCH1 set, and strain R2A57 was used for the BACT1 set. We compared the regression slopes and intercepts of triplicate standard curves for each of the templates as described previously (29).
Mixed-template experiments.
To test the applicability of 5′-nuclease assays for relative quantification, artificial mixtures of SSU rDNAs were prepared. In the first test, we used mixtures of strain R2A57 and H. volcanii genomic DNAs. R2A57 SSU rDNA copy numbers were estimated using the BACT1 set, and H. volcanii SSU rDNA copy numbers were estimated using the ARCH1 set. Plasmids R57pCRbl and HVpCRbl were used as standards. Reactions were carried out with the optimized conditions listed in Table 2. Three experiments were conducted with genomic DNA mixtures (106 SSU rDNA copies μl−1) adjusted to the following proportions of H. volcanii to R2A57: 0.1:0.9, 0.2:0.8, 0.4:0.6, 0.6:0.4, 0.8:0.2, and 0.9:0.1. In the second test, we used mixtures of strain R2A57 and Synechococcus sp. strain WH7803 genomic DNAs. Copy numbers of Synechococcus SSU rDNA were estimated using the PHPICO primer and probe set, the plasmid clone MB11A04 as a standard, and the optimized conditions listed in Table 2. Total bacterial SSU rDNAs were measured with both BACT1 and BACT2 primer sets using R57pCRbl and MB11A04 as standards. The same proportions of SSU rDNA and calculations were used.
Validation of 5′-nuclease assays for analyzing environmental samples.
To validate the use of 5′-nuclease assays as a quantitative tool for complex environmental samples, we compared results obtained with alternative methods. First, we estimated the proportion of SSU rDNA belonging to the Synechococcus and Prochlorococcus group relative to bacterial SSU rDNAs in Monterey Bay DNA extracts using the 5′-nuclease assays versus flow cytometry cell counts. Flow cytometry cell counts were determined at the Department of Oceanography, University of Hawaii, as previously described (4).
In a second set of experiments, the relative abundance of SSU rDNAs from group I and II marine Archaea in Monterey Bay DNA extracts was estimated by 5′-nuclease assays and compared to the results obtained using fluorescent in situ hybridization (FISH [6]) or RNA blot hybridization (16) for the same samples. SSU rDNA copies of group I marine Archaea in environmental samples from the Monterey Bay were estimated using the ARCHGI 5′-nuclease set and the optimized conditions listed in Table 2. SSU rDNA copies of group II marine Archaea were estimated using the ARCHGII primer set and clone SB9572 as a standard. FISH experiments and RNA blot hybridization experiments were performed as previously described (6, 16).
Analytical considerations.
The analytical precision of the model 7700 SDS instrument was relatively high. In general, triplicate measurements obtained simultaneously had a coefficient of variation of less than 0.15 (average, 0.092; n = 52) for all primer and probe sets tested (BACT1, BACT2, ARCH1, and PHPICO), although on rare occasions it was as high as 0.42. However, day-to-day variability in copy numbers estimated with the primer and probe sets BACT1, ARCH1, PHPICO, ARCHGI, and ARCHGII was much higher for replicate samples. In most cases, the coefficients of variation were higher than 0.30 (average, 0.74; n = 30).
RESULTS
Laboratory optimization of 5′-nuclease assays.
Initially we designed two broad primer and probe sets targeting Archaea and Bacteria (BACT1 and ARCH1). As discussed below, these primer and probe sets were shown to underestimate a number of groups that are present in a relatively high abundance in environmental samples; therefore, their utility for these samples appears limited. However, these primer and probe sets were very useful for optimizing and testing critical parameters of the 5′-nuclease assays.
(i) Specificity of and optimal conditions for 5′-nuclease assays.
The primer and probe sets BACT1 and ARCH1 utilized a common probe (PROK1389F) and a reverse primer (PROK1541R). This strategy was used to help standardize assays and to avoid the synthesis of multiple TaqMan probes. We relied on the specificity of forward primers to discriminate between rDNAs of bacterial and archaeal origins. Specificity tests showed that both primer and probe sets were specific for their intended groups. Optimal reaction conditions for all primer and probe sets are listed in Table 2.
(ii) Effect of template source and type.
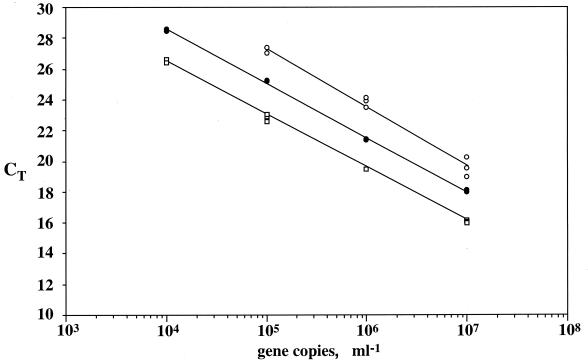
There were no significant differences in the slopes of standard curves for genomic DNAs from E. coli, B. subtilis, or D. radiodurans, as determined by ANOVA (Table 3; α = 0.05). The t-test comparison (29) between the slopes of standard curves obtained with supercoiled plasmids, linear plasmids, and genomic DNA containing SSU rDNA from strain R2A57 or H. volcanii also showed no significant differences (α = 0.05). Since gene copy numbers were more accurately calculated from plasmid DNA concentrations, we also compared the intercepts of standard curves for linear and supercoiled R57pCRbl and HvpCRbl. Comparison by t tests (29) revealed significant differences (α = 0.05) between the intercepts of supercoiled and linear HvpCRbl (Fig. 1) and R57pCRbl (data not shown) standard curves and underestimation of the copy numbers of supercoiled plasmids relative to linear plasmids by about 1 order of magnitude.
TABLE 3.
Effect of genomic DNA source on the efficiency of amplification of SSU rDNA by PCR
| Sources of genomic DNAs compared | Avg slope of curves for CT vs log no. of gene copies | Fa |
|---|---|---|
| E. coli | −3.530 | 0.037 |
| B. subtilis | −3.564 | |
| E. coli | −3.530 | 1.231 |
| D. radiodurans | −3.421 | |
| B. subtilis | −3.564 | 0.819 |
| D. radiodurans | −3.421 |
F values, determined by analysis of variance, are for comparisons of the sets of organisms listed at left. The critical F value (at an α of 0.05) for all comparisons was 7.708.
FIG. 1.
Effect of DNA quality on the amplification of SSU rDNA by PCR. Standard curves show plots of CT versus log number of gene copies for SSU rDNA from H. volcanii, as estimated by 5′-nuclease assays using the ARCH1 set. Open circles, SSU rDNA in supercoiled plasmids; closed circles, genomic DNA; open squares: SSU rDNA in linearized plasmids.
(iii) Detection limits.
BACT1 consistently produced a high background fluorescence signal in negative controls containing no template. This background signal was equivalent to the signal produced by approximately 2,500 SSU rDNA copies. Reactions carried out with both AmpliTaq Gold and Platinum Taq had equivalent backgrounds, and it is likely that the background originated from host DNA carryover in the cloned DNA polymerase preparations. Attempts to decrease the background by incubating the cocktail mixtures (without a template and primers) with exonucleases failed. Therefore, we assigned 25,000 copies of template per reaction tube as our detection limit for the bacterial domain primer and probe sets. The range for bacterial quantification was 2.5 × 104 to 2.5 × 108 copies per reaction tube, although higher values might still lie in the regression line of standard curves. In the majority of cases, the archaeal set ARCH1 produce no fluorescence signal in negative controls containing no template. The quantification range for archaeal and other group-specific assays was 2.5 × 102 to 2.5 × 108 copies (0.14 to 1.4 × 104 pg) per reaction tube, based on the genome size and rrn operon copy numbers in E. coli.
(iv) Mixed-template experiments.
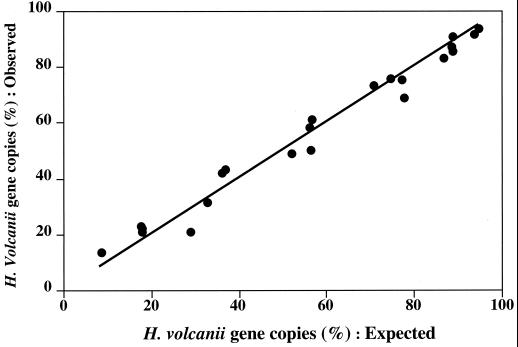
Primer and probe sets BACT1 and ARCH1 were successfully used to test the accuracy of estimating SSU rDNA percentages in genomic DNA mixtures. The percentage of H. volcanii genomic DNA in artificial mixtures with strain R2A57 genomic DNA estimated by the 5′-nuclease assays was not significantly different from the proportion in the templates (Fig. 2). We also used BACT1 to estimate total SSU rDNA copy numbers in artificial mixtures of genomic DNAs from Synechococcus strain WH7803 and strain R2A57. Surprisingly, the proportion of Synechococcus strain WH7803 estimated by the PHPICO set was overestimated twofold in mixtures when the percentage of Synechococcus strain WH7803 DNA in the template was high (over 80%). This result was clearly caused by an underestimation of total bacterial SSU rDNA by BACT1, likely due to mismatches with PROK1541R (3a; M. T. Suzuki et al., unpublished data). There are a limited number of public database sequences that include this SSU rDNA region, and some of the existing sequences are those of the PCR primers used to recover the SSU rDNA.
FIG. 2.
Estimation of the percentage of H. volcanii SSU rDNAs in mixtures with DNA of strain R2A57. The x axis shows expected values calculated using various mixture proportions and the numbers of SSU rDNA copies of H. volcanii and strain R2A57 per microliter prior to mixing. The y axis shows the percentage of H. volcanii SSU rDNA copies estimated by 5′-nuclease assays using set ARCH1. Total SSU rDNA copies were estimated by 5′-nuclease assays using primer and probe sets ARCH1 and BACT1. The line represents a theoretical line assuming a 100% match in the values.
Field application of 5′-nuclease assays.
We designed four additional primer and probe sets (BACT2, PHPICO, ARCHGI, and ARCHGII) to test the applicability of the 5′-nuclease assays to estimation of the proportions of different groups of prokaryotes in environmental DNA samples. Based on our initial tests and observations, we used the following procedure when applying 5′-nuclease assays to the quantification of prokaryotic rDNAs. (i) Quantification was performed relative to DNA mass or to rDNA copy numbers estimated by a domain-level primer and probe set. (ii) Linear plasmids were used as standards. (iii) Whenever possible, the same standards were used for group-specific and domain-specific primer and probe sets.
(i) Bacteria.
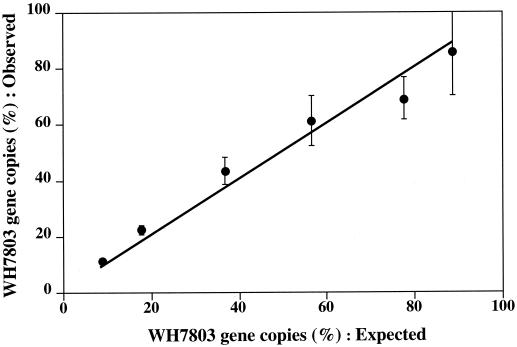
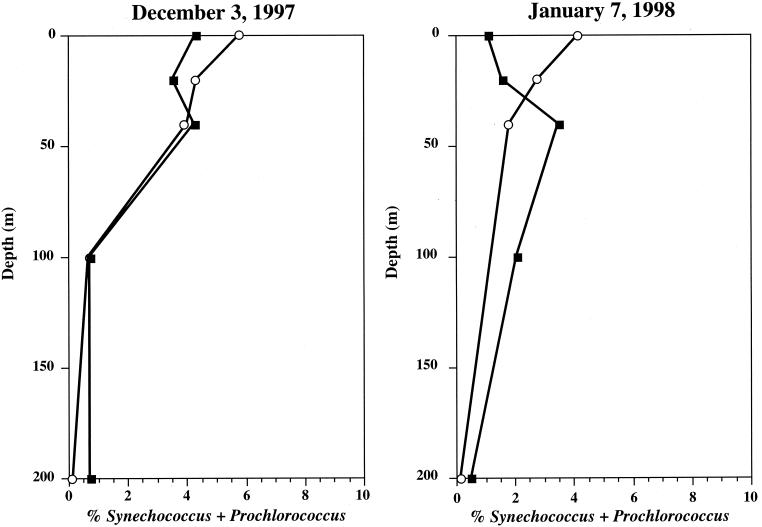
Since primer PROK1541R appears to miss several major groups of Bacteria, we used primer PROK1492R as the reverse primer for the bacterial assay for environmental samples. This primer has a melting temperature (56°C), which is lower than the optimal temperature for Taq DNA polymerase (72°C), more so than BACT1 (59°C). Also, we observed considerable variability in the activity of AmpliTaq Gold DNA polymerase, possibly caused by variability in enzyme activation (data not shown). Therefore, we used Platinum Taq DNA polymerase in assays with primer PROK1492R. The BACT2 set was tested for estimating the proportions of Synechococcus strain WH7803 genomic DNA in mixtures with strain R2A57. Results showed good agreement between the expected and observed proportions of Synechococcus strain WH7803 (Fig. 3). The BACT2 set was also used to estimate SSU rDNAs from Bacteria in samples from Monterey Bay. In this case, the proportions of SSU rDNAs of Synechococcus and Prochlorococcus estimated by the PHPICO set were similar to those estimated by cell counts, with the exception of surface and 20-m samples (Fig. 4). It is possible that this discrepancy between the proportions estimated by cell counts and 5′-nuclease assays was due to an underestimation of photoadapted cells by flow cytometry, which has been previously reported (18). As with BACT1, the range of quantification was 2.5 × 104 to 2.5 × 108 copies per reaction tube.
FIG. 3.
Estimation of the percentage of a Synechococcus sp. in mixtures with genomic DNA of strain R2A57. The x axis shows expected values calculated using various mixture proportions and the numbers of SSU rDNA copies of Synechococcus sp. strain WH7803 and strain R2A57 per microliter prior to mixing. The y axis shows the percentage of SSU rDNA from Synechococcus sp. strain WH7803 estimated by 5′-nuclease assays using set PHPICO. Total SSU rDNA was estimated by 5′-nuclease assays using set BACT2. Error bars indicate 1 standard deviation from the mean of triplicate reactions. The line represents a theoretical line assuming a 100% match in the values.
FIG. 4.
Depth profiles of Prochlorococcus and Synechococcus at station M1. Open circles, percentages of Prochlorococcus and Synechococcus SSU rDNAs measured by a 5′-nuclease assay using set PHPICO relative to bacterial SSU rDNA measured by a 5′-nuclease assay using set BACT2; closed squares, percentages of Prochlorococcus and Synechococcus cell numbers relative to total bacterioplankton cell numbers measured by flow cytometry.
(ii) Synechococcus and Prochlorococcus.
Set PHPICO was designed to target SSU rDNAs of Synechococcus and Prochlorococcus and to amplify a region homologous to positions 191 to 420 of E. coli SSU rDNA (Table 1). We used this probe to measure SSU rDNA of Synechococcus strain WH7803 in artificial mixtures with strain R2A57 and to estimate the proportions of Synechococcus and Prochlorococcus genes in DNA samples from Monterey Bay. The estimated proportions of SSU rDNAs correlated well with expected proportions. The tested range of quantification of the PHPICO primer and probe set was 2.5 × 103 to 2.5 × 107 SSU rDNA copies per reaction tube.
(iii) Marine Archaea.
Although we tested several different combinations of primer and probe sets targeting archaeal SSU rDNAs, currently we have no broadly encompassing set that is suitable for application to all environmental samples. Forward primer Arch1369F has several mismatches with SSU rDNAs of Methanosarcinales and Methanococcales, so that these groups likely are underestimated by the set containing this primer. Since these organisms were not likely to represent a large fraction of the archaeal population in planktonic samples, we used ARCH1 to estimate total archaeal SSU rDNAs in samples from Monterey Bay. The proportions of SSU rDNA of group I marine Archaea estimated by ARCHGI relative to total archaeal SSU rDNAs were similar to those estimated by rRNA blot hybridization or FISH. However, the proportions of SSU rDNA of group II marine Archaea estimated by ARCHGII were overestimated by 2 orders of magnitude, likely due to mismatches between group II marine Archaea SSU rDNA and the reverse primer PROK1541R (data not shown). Currently, there is no sequence information for the 3′ end of the SSU rDNA of group II marine Archaea, since the only sequences available are those from PCR clones with no sequence for this region of the SSU rDNA.
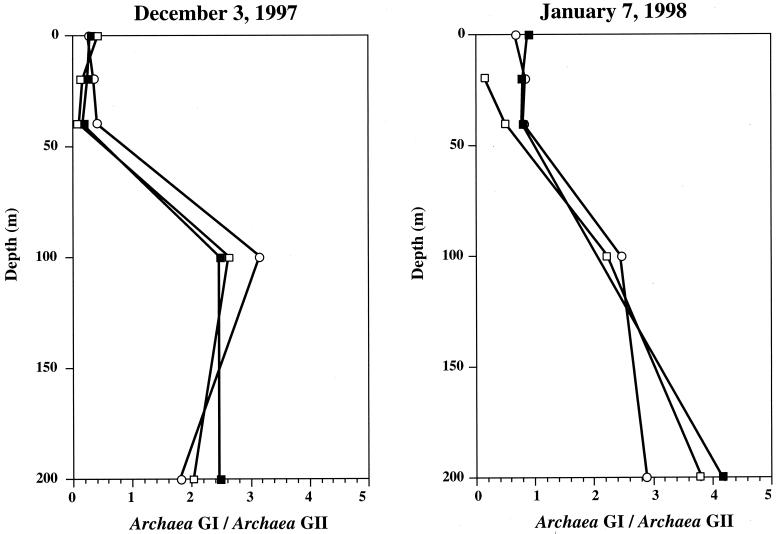
Although we encountered difficulties designing a primer and probe set suitable for all Archaea, we designed two primer and probe sets to target SSU rDNAs of two abundant groups of marine Archaea (Table 1). Set ARCHGI targeted genes of the marine Crenarchaeota (group I), and set ARCHGII targeted genes of the marine Euryarchaeota (group II). The ratios of SSU rDNAs from group I and II Archaea correlated well with the ratios of their rRNAs estimated in dot blot experiments using radiolabeled oligonucleotide probes (Fig. 5). These ratios also correlated well with those estimated from cell counts by polyribonucleotide FISH and agree with previous observations of the depth distribution of these groups (16). The range of quantification of both sets was 2.5 × 103 to 2.5 × 107 gene copies per reaction tube.
FIG. 5.
Depth profiles of ratios between group I (GI) marine Archaea and group II (GII) marine Archaea at station M1. Open circles, ratios of SSU rRNAs measured by radiolabeled oligonucleotide dot blot hybridization; open squares, ratios of cell numbers per milliliter estimated using fluorescence-labeled polyribonucleotide probes and FISH; closed squares, ratios of SSU rDNA copies per nanogram of DNA estimated by 5′-nuclease assays using primer and probe sets ARCHGI and ARCHGII for group I and group II, respectively.
DISCUSSION
We successfully developed and tested PCR-based fluorogenic 5′-nuclease assays for the relative quantification of SSU rDNAs of prokaryotes. It was possible to attain high sensitivity using small amounts of a sample, and the quantification range exceeded 4 orders of magnitude. These characteristics of 5′-nuclease assays have important implications for sampling strategies and experimental design in microbial ecology. The sensitivity and high throughput of the assays allow genes in DNA from small biomass samples to be detected and a large number of samples to be processed. Although we encountered difficulties in designing primer and probe sets that encompass entire domains, it was relatively easy to design primer and probe sets for rDNAs for narrower taxonomic groups. In the absence of domain-specific primer and probe sets for normalization, taxon-specific genes can be quantified relative to total DNA.
We attempted only relative quantification, for several reasons. (i) The quantification of SSU rDNA copy numbers in genomic DNA from DNA concentration requires a priori knowledge of rDNA copy number and genome size. Also, in the few instances when both values are known, a cell might contain more than one genome copy and consequently extra copies of the rrn operon. Therefore, gene copy numbers in stock solutions of genomic DNA were calibrated relative to gene copy numbers in plasmid DNA solutions using 5′-nuclease assays. (ii) We used linear plasmids as standards. Copy numbers were estimated from DNA concentrations in samples quantified by fluorometry. We did not attempt to improve the accuracy of copy number measurements by alternative methods (e.g., Poisson distribution [27]), since a further increase in the accuracy of copy number measurements was not justified when the next item was taken into consideration. (iii) Copy numbers of SSU rDNA were measured relative to the total amount of DNA extracted and purified from a mixed population. Due to methodological limitations associated with the study of uncultivated microorganisms, the accurate estimation of DNA extraction efficiency and, consequently, the amount of total DNA present in the original seawater sample was not possible. In view of these problems associated with absolute quantification, our strategy was to measure the proportions of rDNAs from different groups of Bacteria relative to all rDNAs detected from the domain Bacteria.
The amplification efficiency was independent of the template type (chromosomal versus plasmid clones); this is a critical consideration in the application of 5′-nuclease assays to environmental samples, since many naturally occurring but uncultured microbial species have not been cultivated, and for these organisms, only cloned rDNAs are available. Additionally, rDNAs from different bacterial phyla were amplified with similar efficiencies, further supporting the applicability of 5′-nuclease assays for mixed populations of bacteria. The significant difference between the y intercepts of standard curves for linear and supercoiled plasmids was likely due to differences in their concentration estimates and underscores the importance of normalization in relative quantification.
Variations in rrn operon copy numbers and genome sizes between different organisms are complex variables to be considered in the interpretation of relative proportions of SSU rDNAs in mixed populations. A recent report (8) reviewed the current information on prokaryotic genome sizes and operon copy numbers and concluded that genome size and operon copy number are relatively constant in certain phylogenetic groups. The authors suggested that estimation of organism numbers from SSU rDNA proportions—based on group averages of genome size and operon copy number—might be possible. However, no information is available on genome sizes and rrn operon copy numbers for many indigenous microorganisms, and in several instances, no cultivated species related to these organisms are available to allow inference of genome sizes and rrn operon copy numbers. Additionally, a recent report by Klappenbach et al. (12) suggested that rrn operon copy number may be related to growth strategy and not necessarily to phylogenetic affiliation. The approach suggested by Fogel et al. (8) therefore does not seem particularly applicable to complex, natural microbial assemblages.
By modifying the assays we describe here, it should also be possible to monitor proxies for metabolic activity. For instance, quantification of rRNA after an initial reverse transcription step and comparison of rRNA with rDNA levels could be performed using the same primer and probe sets. Furthermore, it is feasible to measure the relative expression of metabolic genes using reverse transcription of mRNA and PCR quantification by 5′-nuclease assays. Gene expression could then be quantified relative to specific phylogenetic markers (rDNAs or rRNAs), as described in this study. These approaches should provide much better estimates of the metabolic status of microorganisms in situ. Because of their high sensitivity and wide dynamic range, 5′-nuclease assays are particularly suited for samples containing a low level of biomass or slowly growing organisms.
ACKNOWLEDGMENTS
This work was supported by a grant from the David and Lucile Packard Foundation to MBARI.
We thank D. Distel for providing genomic DNA from Synechococcus strains WH8108 and WH7805 and B. Stevenson for providing clone pKK3535 used in preliminary experiments. We thank the crew of the RV Point Lobos and Tim Pennington for help during sampling. We thank Oded Béjà for editorial comments on the manuscript and Gianfranco de Feo for help during the early stages of development of our 5′-nuclease assays. We also thank two anonymous reviewers for their useful comments.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F A, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 3a.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorte, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. Environ. Microbiol., in press. [DOI] [PubMed]
- 4.Buck K R, Chavez F P, Campbell L. Basin-wide distributions of living carbon components and the inverted trophic pyramid of the central gyre of the North Atlantic Ocean, summer 1993. Aquat Microb Ecol. 1996;10:283–298. [Google Scholar]
- 5.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic Archaea and Bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 8.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 9.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 177–203. [Google Scholar]
- 10.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 11.Giovannoni S J, Rappe M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klappenbach J A, Dunbar J M, Schmidt T M. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 14.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 16.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Mullins T D, Britschgi T B, Kvest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:145–158. [Google Scholar]
- 17.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partensky F, Hess W R, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein J L, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki M, Rappe M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Wintzingoerode F, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Spadoro J. Determination of target copy number of quantitative standards used in PCR based diagnostic assays. In: Ferre F, editor. Gene quantification. Boston, Mass: Birkhauser; 1998. pp. 31–43. [Google Scholar]
- 28.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 29.Zar J. Biostatistical analysis. 4th ed. Upper Saddle Ridge, N.J: Prentice-Hall, Inc.; 1998. [Google Scholar]