Abstract
A DNA fragment encoding two enzymes leading to trehalose biosynthesis, maltooligosyltrehalose synthase (BvMTS) and maltooligosyltrehalose trehalohydrolase (BvMTH), was cloned from the nonpathogenic bacterium Brevibacterium helvolum. The open reading frames for the two proteins are 2,331 and 1,770 bp long, respectively, and overlap by four nucleotides. Recombinant BvMTS, BvMTH, and fusion gene BvMTSH, constructed by insertion of an adenylate in the overlapping region, were expressed in Escherichia coli. Purified BvMTS protein catalyzed conversion of maltopentaose to maltotriosyltrehalose, which was further hydrolyzed by BvMTH protein to produce trehalose and maltotriose. The enzymes shortened maltooligosaccharides by two glucose units per cycle of sequential reactions and released trehalose. Maltotriose and maltose were not catalyzed further and thus remained in the reaction mixtures depending on whether the substrates had an odd or even number of glucose units. The bifunctional in-frame fusion enzyme, BvMTSH, catalyzed the sequential reactions more efficiently than an equimolar mixture of the two individual enzymes did, presumably due to a proximity effect on the catalytic sites of the enzymes. The recombinant enzymes produced trehalose from soluble starch, an abundant natural source for trehalose production. Addition of α-amylase to the enzyme reaction mixture dramatically increased trehalose production by partial hydrolysis of the starch to provide more reducing ends accessible to the BvMTS catalytic sites.
Trehalose (α-d-glucopyranosyl-[1,1]-α-d-glucopyranose) is a nonreducing diglucoside found in various organisms, including bacteria, algae, fungi, yeasts, insects, and some plants (5). In nature, trehalose serves not only as a carbohydrate reserve but also as an agent that protects against a variety of physical and chemical stresses in various organisms (6, 21, 23, 24). Trehalose is known to have high water-holding activity and thus to preserve the integrity of biological membranes (3). As such, this sugar allows desert plants to tolerate naturally occurring stresses during cycles of dehydration and rehydration (4). In addition, the water-holding capability of trehalose has been applied to development of additives, stabilizers, and sweeteners that are quite useful in the food, cosmetic, and pharmaceutical industries (16). Investigations have focused on searching for efficient synthetic processes and abundant raw sources for production of trehalose.
Preparation of recombinant enzymes, especially multifunctional fusion, has great potential in enzyme technology. Fusion of structural genes encoding enzymes that catalyze sequential reactions has several advantages, such as simple expression of a single recombinant unit containing multiple genes and one-step purification of recombinant proteins. Physical proximity due to fusion of multiple enzymes might lead to faster rates of sequential enzyme reactions by facilitating transfer of reaction intermediates to the catalytic sites of the next enzymes.
In Escherichia coli and yeasts, biosynthesis of trehalose is accomplished through two enzymatic processes. Trehalose 6-phosphate (T6P) synthase converts UDP-glucose and glucose 6-phosphate to T6P, which is further dephosphorylated to trehalose by T6P phosphatase (7). Recently, we reported that a bifunctional fusion enzyme resulting from fusion between T6P synthase and T6P phosphatase catalyzes the sequential reactions more efficiently than a combination of the separate enzymes does (20).
In some bacteria, biosynthesis of trehalose is mediated by maltooligosyltrehalose synthase (MTS) and maltooligosyltrehalose trehalohydrolase (MTH). These two enzymes and their corresponding genes have been isolated from Arthrobacter sp. strain Q36 (14), Rhizobium sp. strain M-11 (15), Sulfolobus solfataricus KM1 (12), and Mycobacterium tuberculosis (18). MTS converts α-1,4-glycosidic linkages at reducing ends of maltooligosaccharides into α-1,1 linkages, producing maltooligosyltrehalose. MTH then hydrolyzes the second α-1,4-glycosidic linkage of the intermediate to release trehalose.
One nonpathogenic bacterium, Brevibacterium helvolum, is known to contain the enzymes that synthesize trehalose from maltooligosaccharides (16). In the present study, we cloned a DNA fragment that contains two new genes encoding MTS and MTH from B. helvolum (BvMTS and BvMTH, respectively). These genes were expressed in E. coli individually or in a fused form to produce recombinant enzymes. The recombinant enzymes were characterized to determine their in vitro activities during production of trehalose from soluble starch, an abundant source of maltodextrins in nature.
MATERIALS AND METHODS
Cloning of the BvMTS and BvMTH genes.
B. helvolum ATCC 11822 was grown in Luria-Bertani medium (19). Bacterial genomic DNA was isolated, digested by either EcoRI or SalI, separated on a 1.0% agarose gel, and hybridized on a blot (GenScreen Plus membrane; DuPont) at low stringency with the MTS-MTH gene probe from M. tuberculosis (18). Hybridized bands at 4.3 kb that resulted from EcoRI digestion and hybridized bands at 6.0 kb that resulted from SalI digestion were isolated from the gel, inserted into pUC18 at an EcoRI or SalI site, and transformed into E. coli MC1061. Overlapping MTS and MTH clones were screened by colony hybridization, and their nucleotide sequences were determined as described previously (19).
PCR.
A PCR was carried out in a 100-μl (total volume) mixture containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 100 ng of template DNA, 150 ng of each primer, and 2.5 U of Taq DNA polymerase (Boehringer Mannheim). DNA was amplified under the following conditions: 5 min at 94°C, followed by 30 cycles of 1 min 94°C, 1 min at 52°C, and 2 min at 72°C and a final extension at 72°C for 10 min.
Construction of recombinant expression plasmids.
Open reading frames (ORFs) of BvMTS and BvMTH were amplified by PCR. The BvMTS-specific primers were P1 (5′-GCGATATCATGAAGACTCCGGTCTCCAC-3′) and P2 (5′-GGGAATTCGTCCAACGTTGACCAAGGTCAT-3′), which contained translation initiation and termination codons (underlined), respectively. The PCR products were digested with EcoRV and EcoRI and cloned into the pRSET-C vector (Invitrogen Inc.) to produce pRBvMTS. The BvMTH-specific primers were P3 (5′-GGGGTACATGACCTTGGTCAACGTTG-3′) and P4 (5′-TTAAGCTTCAGGACTTGAGGACCG-3′). The PCR products were digested with KpnI and HindIII and cloned into the pRSET-B vector to produce pRBvMTH.
An expression recombinant for the fusion enzyme, pRBvMTSH, was constructed by using four primers. The ORF of BvMTS was produced by PCR by using primer P1 containing the translation initiation codon of BvMTS and primer P2m (5′-GAAAAGGCAATGACCTTG-3′) containing an additional adenine (underlined) inserted to modify the termination codon. The ORF of BvMTH was produced by PCR by using primers P3m (5′-CAAGGTCATTGCCTTTTC-3′) and P4 containing the termination codon of BvMTH. Primers P2m and P3m are complementary to each other. Employing the amplified ORFs for BvMTS and BvMTH as templates and mutual end primers, we amplified the BvMTSH fusion gene by using the P1 and P4 primers. The amplified BvMTSH fragments were introduced into pRSET-C at EcoRV and HindIII sites to produce pRBvMTSH.
Expression and purification of BvMTS, BvMTH, and the BvMTSH fusion enzyme.
pRBvMTS, pRBvMTH, and pRBvMTSH were transformed into E. coli BL21. Expression of the inserts was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to each culture and incubating it for 4 h at 37°C. Induced enzymes were purified with an Ni2+-nitrilotriacetic acid-agarose affinity column (Qiagen). Elution of the enzymes was performed by using a 10 to 300 mM imidazole gradient according to the manufacturer's instructions. Protein samples were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (19). Protein concentrations were measured by the Bradford method (1), using bovine serum albumin as a standard.
Enzyme assay.
Trehalose synthesis activity was measured by incubating 5 pmol of each purified enzyme in 100 μl of 50 mM sodium phosphate buffer (pH 7.0) containing 1 mM maltooligosaccharides (Sigma Chemical Co.). In some experiments, 1.0% (wt/vol) soluble starch or 45 mM maltopentaose was used as a substrate, as indicated below. The reaction was carried out at 37°C for up to 24 h and was terminated by heating the reaction mixture at 95°C for 5 min.
Products of the enzyme reactions were analyzed by thin-layer chromatography (TLC). Samples (5 μl) were spotted onto a TLC plate (Silica Gel F254; Merck), and the plate was developed twice with n-butanol–ethanol–water (5:3:2). To visualize the carbohydrate spots, the plate was sprayed with 20% sulfuric acid and charred. Each spot was quantified with a densitiometer.
To distinguish between trehalose and maltose, products and intermediates of the enzyme reactions were analyzed by high-pH ion chromatography (HPIC) at room temperature by using a Carbo-Pak PA1 column and a DX500 HPIC system (Dionex). The saccharides were eluted with a continuous 0 to 250 mM sodium acetate gradient (prepared in a 150 mM sodium hydroxide solution) for 30 min. The saccharides that eluted from the column were monitored with an ED40 potential amperometric detector. The amount of trehalose was determined from the following standard equation (R2 = 0.998); amount of trehalose (μg) = 0.2365 × (integrated peak area ×10−6).
Nucleotide sequence accession number.
The nucleotide sequences of B. helvolum genes and deduced amino acid sequences determined in this study have been deposited in the GenBank database under accession no. AF039919.
RESULTS
Molecular cloning of BvMTS and BvMTH genes.
Bacterial genomic DNA containing two genes encoding BvMTS and BvMTH was cloned from B. helvolum.
The translation initiation sites for the two enzymes were determined by comparison with MTS and MTH genes of other bacteria, such as Arthrobacter sp. strain Q36 (14), Rhizobium sp. strain M-11 (15), S. solfataricus KM1 (12), and M. tuberculosis (18). The translation initiation codon of the BvMTH gene overlaps the termination codon of the BvMTS gene by 4 nucleotides. The putative ribosome binding sites, 5′-AGACGCC-3′ for BvMTS and 5′-TGGAGAA-3′ for BvMTH, precede the ATG translation start codons of the two genes. The ORF of BvMTS is 2,331 bp long and encodes 776 amino acids, and the ORF of BvMTH is 1,770 bp long and encodes 589 amino acid residues. The estimated molecular weights of the two polypeptides are 85,800 and 64,200, respectively. The nucleotide sequences of BvMTS and BvMTH show 54 and 58% homology, respectively, to those of the M. tuberculosis genes.
Through GenBank database analysis, it was noticed that the deduced amino acid sequences encoded by the BvMTS and BvMTH genes contain the domains that are highly conserved in α-amylolytic enzymes. This family of enzymes includes α-amylases, pullulanases, cyclomaltodextrin glucanotransferases, and starch-branching enzymes (8, 25). The conserved domains are known to be the substrate-binding sites of the starch hydrolysis enzymes (9, 10).
Expression of BvMTS, BvMTH, and fusion gene BvMTSH.
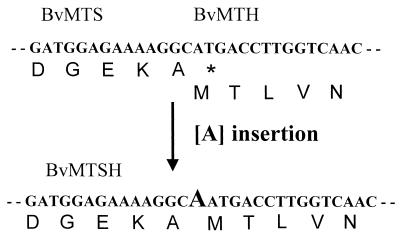
To overexpress each gene, either the BvMTS ORF or the BvMTH ORF was introduced into E. coli expression vector pRSET. In addition, genes encoding BvMTS and BvMTH were fused together in frame and expressed in E. coli as well. The ORF of the BvMTH gene overlaps that of the BvMTH gene by 4 nucleotides, as shown in Fig. 1. To construct an in-frame fusion gene, one additional adenine nucleotide was inserted next to the last amino acid codon (GCA) of BvMTS by PCR-mediated site-directed mutagenesis as described in Materials and Methods. The junction sequence between the two genes and the strategy used for construction of the BvMTSH fusion gene are illustrated in Fig. 1. As a result, a gene encoding fusion protein BvMTSH, in which the last amino acid (Ala) of BvMTS was fused directly to the first amino acid (Met) of BvMTH, was constructed. The construct was introduced into expression vector pRSET-C to produce pRBvMTSH.
FIG. 1.
DNA sequence of the fused region of BvMTS and BvMTH. The 3′ end of BvMTS overlaps the 5′ end of BvMTH by 4 nucleotides. One adenine nucleotide was inserted next to the last amino acid codon (GCA) of BvMTS by site-directed mutagenesis to produce the fused gene BvMTSH by using PCR as described in Materials and Methods.
BvMTS, BvMTH, and BvMTSH proteins produced in E. coli were purified by Ni2+-nitrilotriacetic acid affinity chromatography almost to homogeneity as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown). The estimated molecular weights of the recombinant BvMTS, BvMTH, and BvMTSH proteins were about 97,000, 66,000, and 160,000, respectively, including the hexahistidine domain derived from the pRSET vector. The sizes of the proteins observed on the gel were in accordance with those calculated from the deduced amino acids.
Production of trehalose from maltooligosaccharides.
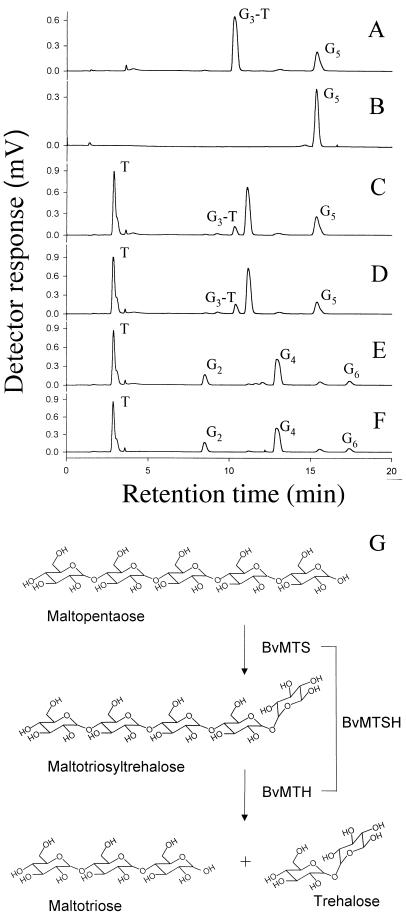
Enzymatic activities of the purified recombinant BvMTS and BvMTH proteins were analyzed by HPIC by using maltooligosaccharides as substrates. In the reaction with BvMTS, most of maltopentaose was converted to an intermediate (Fig. 2A). This intermediate has been identified as maltotriosyltrehalose, as determined by Maruta et al. (16) and Kato et al. (11). Maltopentaose did not react with BvMTH itself (Fig. 2B) but was converted into trehalose and maltotriose in the presence of both enzymes (Fig. 2C). These results suggest that BvMTS produces maltotriosyltrehalose from maltopentaose and then BvMTH hydrolyzes maltotriosyltrehalose to trehalose and maltotriose. The reactions catalyzed by the two enzymes are depicted in Fig. 2G.
FIG. 2.
HPIC analyses of reaction products obtained from maltooligosaccharides by enzymatic activity of recombinant BvMTS, BvMTH, and BvMTSH proteins. A 0.1-μmol portion of maltopentaose (A through D) or 0.1 μmol of maltohexaose (E and F) was reacted with 5 pmol of purified recombinant enzyme BvMTS alone (A), BvMTH alone (B), a mixture of BvMTS and BvMTH (C and E), or BvMTSH (D and F). Each reaction was terminated after 1 h. G6, maltohexaose; G5, maltopentaose; G4, maltotetraose; G3-T, maltotriosyltrehalose; G2, maltose; T, trehalose. (G) Trehalose synthesis from maltopentaose by BvMTS and BvMTH. BvMTS catalyzes the conversion of maltopentose to maltotriosyltrehalose, which is further hydrolyzed by BvMTH to produce trehalose and maltotriose. BvMTSH, the fusion protein consisting of the two individual enzymes, catalyzes the sequential reactions.
The recombinant BvMTSH fusion enzyme also converted maltopentaose to maltotriose and trehalose, as did the mixture of BvMTS and BvMTH (Fig. 2D). This result demonstrates that the BvMTSH fusion enzyme is functional and catalyzes two sequential reactions mediated by the two individual enzymes (Fig. 2G).
When maltohexaose was used as a substrate in the enzyme reactions, trehalose was also produced (Fig. 2E and F). In this reaction, maltotetraosyltrehalose might have been an intermediate (it was not detected) that was hydrolyzed to produce trehalose and maltotetraose. Maltotetraose then was converted to trehalose and maltose.
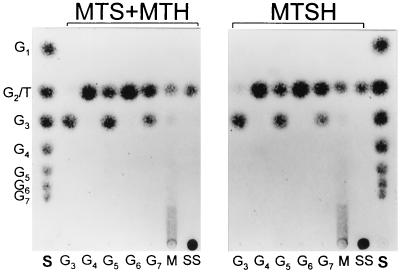
The enzyme activities of the recombinant BvMTS, BvMTH, and BvMTSH proteins with various sizes of maltooligosaccharides were further examined by identifying the reaction products on a TLC plate (Fig. 3). Both the enzyme mixture and the fusion enzyme were active on maltooligosaccharides longer than maltotriose. Less than 5% of the maltotriose reacted further with any of the enzymes, suggesting that maltotriose is not an efficient substrate for the enzymes. Most maltotetraose was converted to trehalose and maltose by the enzymes. Maltopentaose and maltoheptaose were converted into trehalose and maltotriose, while maltohexaose was converted to trehalose and maltose. Altogether, maltooligosaccharides were shorten by two glucose units per cycle of reactions by releasing a trehalose molecule. Maltooligosaccharides having an odd number of glucose units were converted to trehalose and maltotriose, and maltooligosaccharides with an even number of glucose units were converted to trehalose and maltose. There was no difference in substrate specificity between the BvMTS-BvMTH mixture and the BvMTSH fusion enzyme.
FIG. 3.
Reaction activities of BvMTS-BvMTH mixture and BvMTSH with various sizes of maltooligosaccharides. As indicated below the chromatograms, maltotriose (G3), maltotetraose (G4), maltopentaose (G5), maltohexaose (G6), maltoheptaose (G7), a mixture of maltooligosaccharides (M), or soluble starch (SS) was incubated with the BvMTS-BvMTH mixture or BvMTSH for 24 h, and the reaction products were analyzed on TLC plates. Lanes S contained a standard mixture of maltooligoglucosides (glucose through maltoheptaose) and trehalose. The positions of the reaction products, including glucose through maltoheptaose (G1 through G7) and trehalose (T), are indicated on the left.
The rate of conversion from maltotriose to trehalose by the BvMTSH fusion enzyme (50 nM) was higher by approximately 30% than the rate of conversion by an equimolar mixture of the individual enzymes (BvMTS and BvMTH) (Fig. 3). The optimal temperature for the enzyme reactions was 30 to 40°C (data not shown). At temperatures higher than 45°C, the enzyme activities were significantly reduced, resulting in little, if any, production of trehalose from maltooligosaccharides. The thermostablities of the individual recombinant enzymes and the recombinant fusion protein were identical.
Production of trehalose from soluble starch.
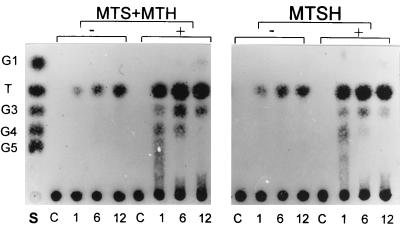
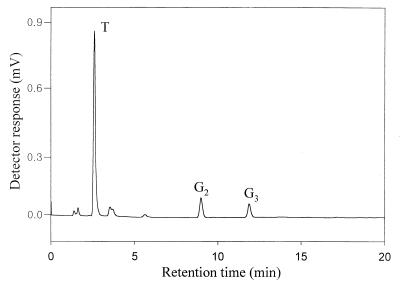
We examined if starch, the most abundant maltodextrin in nature, could be used as a substrate for the trehalose-producing enzymes. A mixture of the purified enzymes (BvMTS and BvMTH) or the purified fusion enzyme (BvMTSH) was incubated with 1.0% (wt/vol) soluble starch. Soluble starch was gradually converted to disaccharides by each enzyme preparation as the reaction time increased up to 12 h (Fig. 4). HPIC analysis revealed that the disaccharides were mainly trehalose (Fig. 5).
FIG. 4.
Effect of α-amylase on trehalose synthesis from soluble starch. In a 100-μl reaction mixture, 1% (wt/vol) soluble starch was incubated with 5 pmol of purified BvMTS-BvMTH mixture or 5 pmol of purified BvMTSH fusion enzyme. Reactions were carried out with (+) or without (−) 0.05 U of α-amylase for 1, 6, and 12 h. The reaction products were analyzed by TLC. Lane S contained a mixture of standard sugars, including glucose (G1), trehalose (T), maltose, maltotriose (G3), maltotetraose (G4), and maltopentaose (G5). Unreacted soluble starch was used as a control (lanes C).
FIG. 5.
HPIC analysis of reaction products obtained from soluble starch with the BvMTSH fusion enzyme. Soluble starch (1%, wt/vol) was incubated with 5 pmol of BvMTSH along with α-amylase (0.05 U per mg of starch) for 24 h. The reaction products were analyzed by HPIC. T, trehalose; G2, maltose; G3, maltotriose.
Addition of α-amylase to the reaction mixture dramatically increased trehalose production from soluble starch (Fig. 4). Without α-amylase, the mixture containing BvMTS and BvMTH had converted 22.1% (data not shown) of the soluble starch to trehalose after a 24-h reaction, and the fusion protein had converted 30.0% (Fig. 4). After addition of α-amylase (0.05 U/mg of soluble starch), 68.3% of the soluble starch was converted to trehalose after 24 h by the combined reactions of BvMTS and BvMTH (data not shown), and 70.4% was converted by the BvMTSH fusion enzyme (Fig. 4). The yield of trehalose was also affected by the concentration of α-amylase; the optimal concentration was 0.05 U per mg of soluble starch (data not shown). Concentrations of α-amylase higher than 0.05 U/mg slightly inhibited trehalose production.
DISCUSSION
In a group of bacteria, trehalose is synthesized by MTS and MTH (12, 14, 15, 18). It has been reported that the nonpathogenic bacterium B. helvolum contains enzymes similar to MTS and MTH (16). In the present study, a DNA clone encoding these two enzymes was isolated from B. helvolum.
Purified recombinant BvMTS proteins catalyzed conversion of maltopentose to maltotriosyltrehalose (Fig. 3). The intermediate was further hydrolyzed by BvMTH proteins to produce trehalose and maltotriose. We constructed a bifunctional fusion enzyme, BvMTSH, by in-frame fusion of structural genes for BvMTS and BvMTH (Fig. 1). The catalytic activity of the fusion protein for trehalose synthesis from maltopentaose was more efficient than that of an equimolar mixture of the two individual enzymes (Fig. 3). The improved catalytic activity of the fusion enzyme might be due to a proximity effect (17).
In many biological metabolic processes, proximity of enzymes provides highly attractive advantages for whole enzymatic processes. Such proximity allows a reaction intermediate to be directly transferred to the active site of the next enzyme in a sequential enzyme complex. By preventing serious diffusion of the intermediate, the reaction rates of whole enzymatic processes could be increased (2, 13, 22). Recently, we reported that a bifunctional fusion enzyme resulting from fusion between T6P synthase and trehalose 6-phosphatase of E. coli exhibited increased catalytic activity during trehalose synthesis (20).
We tested the possibility of using longer maltooligosaccharides as substrates in the enzyme reaction for trehalose production. The BvMTSH fusion protein or a mixture of the two individual enzymes shortened maltooligosaccharides by two glucose units per cycle of sequential reactions releasing trehalose, leaving either maltotriose or maltose depending on the length of the glucose units (Fig. 2).
The substrate specificity of the enzymes was extended to soluble starch (Fig. 3 and 4), the most abundant maltodextrin in nature. The yield of trehalose from soluble starch, however, was relatively low, 22.1 to 30.0% (Fig. 4). The low yield of trehalose might be because soluble starch could not provide many reducing ends available for BvMTS activity. Thus, we hypothesized that α-amylase could hydrolyze the soluble starch to maltooligosaccharides, generating more reducing ends. Addition of α-amylase to the enzyme reaction mixture along with soluble starch significantly increased the production of trehalose. In the presence of α-amylase, a mixture of the two individual enzymes converted 68.3% of the soluble starch into trehalose, and the fusion enzyme gave a 70.4% conversion yield.
Construction of a recombinant bifunctional fusion enzyme should provide a simple technique for preparation of purified enzymes that are essential in trehalose production, as demonstrated with BvMTSH in this study. In addition, the use of α-amylase in the enzyme reaction should provide a way to mass produce trehalose from an inexpensive raw material, such as soluble starch.
ACKNOWLEDGMENTS
This work was supported by a grant from the Agricultural Research & Processing Center of the Korean Ministry of Agriculture and Forestry and in part by a grant from the Genetic Engineering Program of the Korean Ministry of Education. Y.H.K. and J.J.C. acknowledge graduate and research fellowships provided by the Ministry of Education through the Brain Korea 21 Project.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Bülow L. Characterization of an artificial bifunctional enzyme, beta-galactosidase/galactokinase, prepared by gene fusion. Eur J Biochem. 1987;163:443–448. doi: 10.1111/j.1432-1033.1987.tb10889.x. [DOI] [PubMed] [Google Scholar]
- 3.Crowe J H, Crowe L M, Chapman D. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- 4.Drennan P M, Smith T M, Goldsworthy D, van Staden J. The occurrence of trehalose in the leaves of the desiccation-tolerant angiosperm Myrothamnus flabellifolius. Welw J Plant Physiol. 1993;142:493–496. [Google Scholar]
- 5.Elbein A. The metabolism of α,α-trehalose. Adv Carbohydr Chem Biochem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- 6.Eleutherio E C A, Araujo P S, Panek A D. Protective role of trehalose during heat stress in Saccharomyces cerevisiae. Cryobiology. 1993;30:591–596. doi: 10.1006/cryo.1993.1061. [DOI] [PubMed] [Google Scholar]
- 7.Giaever H M, Styrvoid O B, Kaasen I, Strom A R. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol. 1988;170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itkor P, Tsukagoshi N, Udaka S. Nucleotide sequence of the raw-starch-digesting amylase gene from Bacillus sp. B1018 and its strong homology to the cytodextrin glucanotransferase genes. Biochem Biophys Res Commun. 1990;166:630–636. doi: 10.1016/0006-291x(90)90855-h. [DOI] [PubMed] [Google Scholar]
- 9.Jespersen H M, MacGregor E A, Henrissat B, Sierks M R, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 10.Jespersen H M, MacGregor E A, Sierks M R, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato M, Miura Y, Kettoku M, Shindo K, Iwamatsu A, Kobayashi K. Purification and characterization of new trehalose-producing enzymes isolated from the hyperthermophilic archae, Sulfolobus solfataricus KM1. Biosci Biotechnol Biochem. 1996;60:546–550. doi: 10.1271/bbb.60.546. [DOI] [PubMed] [Google Scholar]
- 12.Kobayash K, Kato M, Miura Y, Kettoku M, Komeda T, Iwamatsu A. Gene cloning and expression of new trehalose-producing enzyme from the hyperthermophilic archaeum Sulfolobus solfataricus KM1. Biosci Biotechnol Biochem. 1996;60:1882–1885. doi: 10.1271/bbb.60.1882. [DOI] [PubMed] [Google Scholar]
- 13.Ljungerantz P, Carlsson H, Mansson M O, Buckel P, Mosbach K, Bülow L. Construction of an artificial bifunctional enzyme, beta-galactosidase/galactose dehydrogenase, exhibiting efficient galactose channeling. Biochemistry. 1989;28:8786–8792. doi: 10.1021/bi00448a016. [DOI] [PubMed] [Google Scholar]
- 14.Maruta K, Hattori K, Nakada T, Kubota M, Sugimoto T, Kurimoto M. Cloning and sequencing of trehalose biosynthesis genes from Arthrobacter sp. Q36. Biochim Biophys Acta. 1996;1289:10–13. doi: 10.1016/0304-4165(95)00139-5. [DOI] [PubMed] [Google Scholar]
- 15.Maruta K, Hattori K, Nakada T, Kubota M, Sugimoto T, Kurimoto M. Cloning and sequencing of trehalose biosynthesis genes from Rhizobium sp. M-11. Biosci Biotechnol Biochem. 1996;60:717–720. doi: 10.1271/bbb.60.717. [DOI] [PubMed] [Google Scholar]
- 16.Maruta K, Nakada T, Kubota M, Chaen H, Sugimoto T, Kurimoto M. Formation of trehalose from maltooligosaccharides by a novel enzymatic system. Biosci Biotechnol Biochem. 1995;59:1829–1834. doi: 10.1271/bbb.59.1829. [DOI] [PubMed] [Google Scholar]
- 17.Meek T D, Garvey E P, Santi D V. Purification and characterization of the bifunctional thymidylate synthetase-dihydrofolate reductase from methotrexate-resistant Leishmania tropica. Biochemistry. 1985;24:678–686. doi: 10.1021/bi00324a021. [DOI] [PubMed] [Google Scholar]
- 18.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Seo H S, Koo Y J, Lim J Y, Song J T, Kim C H, Kim J K, Lee J S, Choi Y D. Characterization of a bifunctional fusion enzyme between trehalose 6-phosphate synthase and trehalose 6-phosphate phosphatase of Escherichia coli. Appl Environ Microbiol. 2000;66:2484–2490. doi: 10.1128/aem.66.6.2484-2490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strøm A R, Kassen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 22.Tamada Y, Swanson B A, Arabshahi A, Frey P A. Preparation and characterization of a bifunctional fusion enzyme composed of UDP-galactose 4-epimerase and galactose-1-puridyltransferase. Bioconjugate Chem. 1994;5:660–665. doi: 10.1021/bc00030a023. [DOI] [PubMed] [Google Scholar]
- 23.Van Laere A. Trehalose, reserve and/or stress metabolite? FEMS Microbiol Rev. 1989;63:201–210. [Google Scholar]
- 24.Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. J Gen Microbiol. 1990;58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- 25.Yong J, Choi J N, Park S S, Park C S, Park K H, Choi Y D. Secretion of heterologous cyclodextrin glycosyltransferase of Bacillus sp. E1 from Escherichia coli. Biotechnol Lett. 1996;18:1223–1228. [Google Scholar]