Abstract
Enrichment experiments with North Sea bacterioplankton were performed to test if rapid incubation-induced changes in community structure explain the frequent isolation of members of a few particular bacterial lineages or if readily culturable bacteria are common in the plankton but in a state of dormancy. A metabolic inhibitor of cell division (nalidixic acid [NA]) was added to substrate-amended (S+) and unamended (S−) grazer-free seawater samples, and shifts in community composition and per cell DNA and protein content were compared with untreated controls. In addition, starvation survival experiments were performed on selected isolates. Incubations resulted in rapid community shifts towards typical culturable genera rather than in the activation of either dormant cells or the original DNA-rich bacterial fraction. Vibrio spp. and members of the Alteromonas/Colwellia cluster (A/C) were selectively enriched in S+ and S−, respectively, and this trend was even magnified by the addition of NA. These increases corresponded with the rise of cell populations with distinctively different but generally higher protein and DNA content in the various treatments. Uncultured dominant γ-proteobacteria affiliating with the SAR86 cluster and members of the culturable genus Oceanospirillum were not enriched or activated, but there was no indication of substrate-induced cell death, either. Strains of Vibrio and A/C maintained high ribosome levels in pure cultures during extended periods of starvation, whereas Oceanospirillum spp. did not. The life strategy of rapidly enriched culturable γ-proteobacteria could thus be described as a “feast and famine” existence involving different activation levels of substrate concentration.
Our knowledge about the phylogenetic lineages that contribute to the marine bacterioplankton is presently obtained from three sources: isolation of various bacterial strains (33, 43), clone libraries of 16S ribosomal DNA (rDNA) (31, 37, 43), and hybridizations to whole cells or isolated nucleic acids (22, 33, 38). The results of isolation and of clone libraries often disagree. During the last decade the discrepancy between isolation and cloning has commonly been regarded as an indication of cultivation-induced shifts (4). Yet, since cloning does not reveal community structure either, this view is actually based on little experimental evidence. On the contrary, by using quantitative genome probe hybridizations against community DNA, some isolates (Sphingomonas and Caulobacter spp.) have been shown to represent a significant amount of the total bacterioplankton in brackish Baltic Sea waters (33). A marine isolate related to Vibrio was described to exhibit remarkable annual variation in population density, ranging from undetectably low to ≥100 of total community DNA (38). Is this high relative abundance of typical culturable bacteria the exception or the rule? In a recent study on North Sea bacterioplankton (14), we found that the most readily culturable bacteria on media low in organic carbon, such as Vibrio, Alteromonas, and Pseudoalteromonas, did not significantly contribute to the bacterioplankton community during different seasons, as determined by fluorescence in situ hybridization (FISH) with specific oligonucleotide probes. In contrast, a FISH probe targeted to 16S rDNA clones affiliating with a cosmopolitan γ-proteobacterial lineage, SAR86 (1, 14, 18, 31), detected a prominent fraction (up to 10%) of the microbial community in situ. However, no corresponding isolates were obtained in spite of extensive cultivation efforts.
It has, however, been claimed that a supposedly typical marine isolate was undetectable in situ by FISH because of its low per cell ribosome content (40). This raises the question of whether the readily culturable bacteria of our previous study were really rare in situ, or whether they were simply not detectable by fluorescent probes. If FISH sensitivity limits are interfering with the in situ quantification of such cells, their “activation” should be observable during enrichment on substrates successfully used for their cultivation. If frequently isolated bacteria are, however, found to be rare in situ, they should then be able to take advantage of cultivation-associated changes in their environment more rapidly than their competitors. Monitoring dilutions of North Sea bacterioplankton with seawater that is free of bacteria by flow cytometry and subsequent FISH of sorted cells have provided first evidence that members of the γ-subclass of the Proteobacteria may indeed be selectively enriched (17), but it is unknown if those γ-proteobacteria were affiliated with typical marine isolates. In this context, the other side of the observed phylogenetic differences between marine isolates and rDNA clones needs to be addressed, too: how do so-called “unculturable” bacteria develop during the early phases of cultivation attempts or during typical cultivation-associated procedures, such as filtration, confinement, substrate addition, temperature variation, etc.?
We set up enrichments with substrate-amended (S+) and unamended (S−) North Sea water and subsequently analyzed community composition and changes in bacterial per cell DNA and protein content by FISH and flow cytometry. The antibiotic nalidixic acid (NA) (27) was added to half of the treatments. It inhibits prokaryotic DNA replication, yet allows cells to increase in volume. In our study, NA was not applied for the quantification of active bacteria. We rather wanted to test if readily culturable bacteria are frequent but inactive or dormant, and if consequently their low per cell ribosome content could be the reason why we found low in situ abundances of such genera by FISH in a previous study (14). In addition, the FISH detectability of different γ-proteobacterial isolates during starvation was monitored.
MATERIALS AND METHODS
Sampling site and fixation.
In August 1998, surface water was collected at a 1-m depth in acid-washed, seawater-prerinsed 50-liter polyethylene containers at station Helgoland Roads (54.09 N, 7.52 E) near the island of Helgoland, which is situated approximately 50 km offshore in the German Bay of the North Sea. Water was stored at 4°C and further processed within approximately 1 h. Samples for flow cytometry were fixed with formaldehyde (final concentration, 2% [wt/vol]) and stored frozen. For FISH, portions of 10 to 100 ml of unfiltered seawater were fixed with formaldehyde (final concentration, 2% [wt/vol]) for several hours, collected on white polycarbonate filters (diameter, 47 mm; pore size, 0.2 μm; type GTTP; Millipore, Eschborn, Germany), and rinsed with distilled water. Filters were stored at −20°C until further processing.
Total cell counts and protein and DNA content per cell.
Determination of total cell numbers and relative DNA and protein content of bacteria after double staining with Hoechst 33342 and SYPRO (Molecular Probes, Eugene, Oreg.) was performed by flow cytometry on a FACStar Plus flow cytometer as described (Becton Dickinson, Mountain View, Calif.) (48). At least 2,000 Hoechst 33342-positive cells were counted per sample.
Growth experiments.
For the experimental enrichments, seawater was gently filtered through cellulose nitrate filters (diameter, 47 mm; pore size, 1.2 μm; Sartorius AG, Göttingen, Germany). Half of the prefiltered samples were supplemented with NA (30 mg/liter) (27). Triplicate 150-ml aliquots were incubated at the in situ temperature (16°C) on a rotation shaker (100 rpm) either unamended (S−) or amended (S+) with a mix of monomers (alanine, l-aspartate, dl-leucine, l-glutamate, l-ornithine, and dl-serine [1 μM]; glucose, fructose, galactose, glycolate, succinate, and mannitol [10 μM]; acetate, lactate, ethanol, and glycerol [15 μM]). At the beginning of the experiment and after 20 and 43 h, 10-ml aliquots were fixed for FISH, and 2-ml aliquots were fixed for flow cytometry (see above).
Batch cultures.
For starvation experiments 150-ml triplicate samples inoculated with either Alteromonas sp. isolate KT1113 (GenBank accession number AF173965), Oceanospirillum sp. isolate KT0923 (AF173967), or Vibrio sp. isolate KT0901 (AF172840) (14) were incubated at the in situ temperature (16°C) on a rotation shaker (100 rpm) in synthetic seawater (14) to which trace elements, vitamins, and the mix of monomers used for the field incubation were added. At four time points within a period of 50 days, 1.5-ml aliquots were fixed for FISH, immobilized on polycarbonate filters (diameter, 47 mm; pore size, 0.22 μm; type GTTP; Millipore, Eschborn, Germany).
FISH.
Cells on filter sections were hybridized with group-specific oligonucleotide probes EUB338 (3), ALF968 (20% formamide) (32), GAM42a (30), and CF319a (29). In addition, probes for subgroups of α- and γ-proteobacteria (Table 1) were used. Counterstaining with 4,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) and mounting for microscopic evaluation were performed as described previously (3, 21).
TABLE 1.
Oligonucleotide probes for in situ hybridization
| Probe | Specificity | Probe sequence (5′→3′) | Target sitesa (16S rRNA positions) | % Formamide in buffer | Reference |
|---|---|---|---|---|---|
| ALT1413 | A/C | TTTGCATCCCACTCCCAT | 1413–1430 | 40 | 14 |
| SAR86-1249 | SAR86 cluster | GGCTTAGCGTCCGTCTG | 1249–1265 | 50 | 14 |
| G V | Vibrio | AGGCCACAACCTCCAAGTAG | 841–822 | 30 | 20 |
| OCE232 | Oceanospirillum | AGCTAATCTCACGCAGGC | 232–249 | 40 | 14 |
| G Rb | Rhodobacter/Roseobacter | GTCAGTATCGAGCCAGTGAG | 645–626 | 30 | 20 |
Escherichia coli numbering (9).
RESULTS
Changes in community composition.
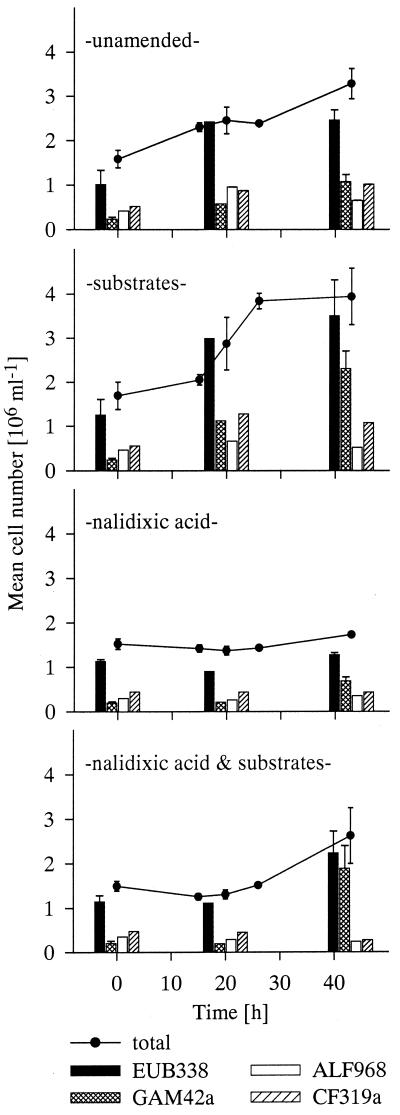
During a 43-h enrichment, total cell number increased from 1.6(±0.2) × 106 (mean ± standard deviation, n = 12) cells per ml by a factor of about 2 in the unamended (S−) and about 2.5-fold in the substrate-amended (S+) samples (Fig. 1). This difference between S− and S+ was not statistically significant (Student's t test, P > 0.05). After NA addition, no significant changes in cell numbers occurred during the first 20 h in both amended and unamended treatments. Total cell number increased slightly in S−NA+ thereafter. In S+NA+, total cell number almost doubled during the second half of the incubation in spite of the antibiotic. Detection rates of probe EUB338 ranged around 75% ± 12% (n = 6) in both S−NA− and S−NA+ throughout the experiment. FISH detection in the substrate-amended treatments increased from 75% ± 1.5% to 87% ± 1.5% of total cells at the end of the incubations.
FIG. 1.
Mean cell numbers of the total bacterial assemblage (lines) during the enrichment experiments and of cells hybridized with group-specific fluorescent probes (bars). Solid bars, cells stained with probe EUB338; dotted bars, γ-subclass of the Proteobacteria; open bars, α-subclass of the Proteobacteria; hatched bars, C/F cluster. Error bars indicate standard deviations (n = 3).
The amount of cells hybridizing with the group-specific probe for the γ-subclass of the Proteobacteria, GAM42a, increased from 2.1(±0.4) × 105 (n = 12) cells per ml 4- to 5-fold and 9- to 10-fold in S− and S+, respectively. This effect was enhanced by incubation with NA. Thirteen percent ± 3% (n = 6) of total cells hybridized with probe GAM42a in the beginning and 40% (34 to 44%, S−NA+) and 72% (67 to 76%, S+NA+) after 43 h of incubation.
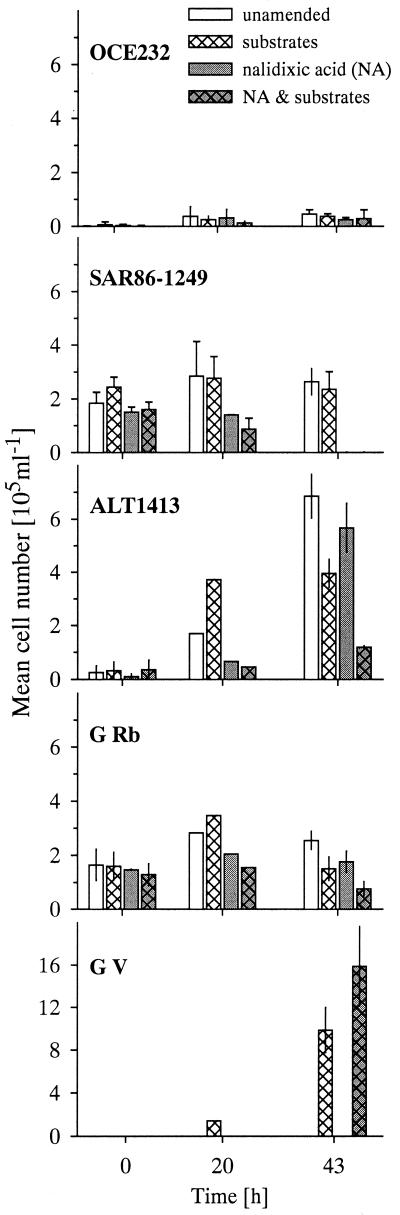
Members of the SAR86 cluster, which are small rods (approximately 0.5 μm in width and 1 μm in length), were detected by FISH with probe SAR86-1249. They showed only weak FISH signals and could not be enriched during different treatments (Fig. 2). Their absolute cell numbers remained constant in both S−NA− and S+NA− treatments (estimated generation time, 83 h). Incubation with NA resulted in a continuous decrease in SAR86 cell numbers within 43 h of incubation. The relative abundances of this phylogenetic group dropped from 11.3% (9.0 to 14.1%) to 6.7% (5.2 to 9.1%) or below the detection level (<1% DAPI) in the various treatments.
FIG. 2.
Mean cell numbers of cells hybridized with probes for various lineages within the γ- and α-proteobacteria in the different treatments. Error bars without caps indicate ranges of replicates; error bars with caps indicate standard deviations of triplicates. Note the different y scale in the bottom panel.
Bacteria targeted by the oligonucleotide probe OCE232, specific for the genus Oceanospirillum (Table 1), showed only weak fluorescence and were initially present in small numbers. During incubation, Oceanospirillum spp. were not enriched significantly in any of the treatments, and their relative abundances hardly exceeded the lower limit of FISH detection rates. The Alteromonas/Colwellia cluster (A/C), as identified by oligonucleotide probe ALT1413, showed a different response (Fig. 2). These bacteria, usually large cells compared to other marine bacteria, showed bright FISH signals. In the beginning of the experiments, they constituted approximately 1.5% of total bacteria, but increased significantly in the S−, S+, and, during the second half of the incubation period, in both NA+ treatments. Concomitant with a rise in numbers, the cell volume of these large cells increased even more (Fig. 3). A/C constituted 6% and 20 ± 2% of total bacteria in S− after 20 and 43 h, respectively. From abundance changes, we estimated a generation time (g) of 9 h. This enrichment was enhanced by the presence of NA. The relative abundance of A/C was 33% ± 5% in the S−NA+ treatment at the end of the experiment. After a strong initial increase in absolute numbers in S+NA−, numbers stagnated after 20 h of incubation, resulting in 10% ± 0.2% relative abundance (g = 11.8 h). S+NA+ treatments resulted in little increase in A/C in absolute numbers.
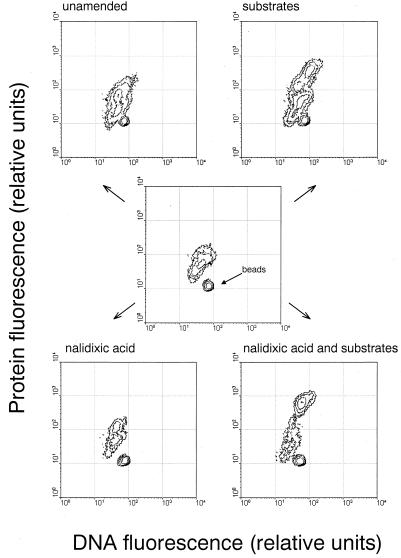
FIG. 3.
Relative per cell DNA and protein content (arbitrary units) of the bacterial assemblages at the beginning and end of the various treatments.
Vibrio spp. were enriched more drastically than any other group during incubation, but only in the S+ treatments (Fig. 2). The increase in cell numbers was even stronger in the S+NA+ treatment. At the end of the experiments Vibrio spp. constituted 25% ± 1% (g = 6.3 h) and 65% ± 1% of total bacteria in the S+NA− and S+NA+ treatments, respectively.
The two other studied groups, α-proteobacteria and Cytophaga/Flavobacterium (C/F), which constituted 23% (17 to 27%) and 31% (26 to 34%) of total bacteria in the beginning of the experiment, respectively, exhibited much lower growth during the enrichments. In absolute numbers, members of the C/F cluster almost doubled from 4.9(±0.9) × 105 (n = 8) to 10.4(±1.5) × 105 (n = 4) cells per ml in both S− and S+, whereas α-proteobacteria only grew in the S− treatments. Both groups decreased little in their relative abundances during incubations without NA. In contrast, α-proteobacteria constituted less than half and members of the C/F cluster about one third of their original relative abundances in the S+NA+ treatment.
The morphologically diverse Rhodobacter/Roseobacter subgroup of the α-subclass of the Proteobacteria constituted 9% ± 3% of total bacteria and a significant fraction (40%) of α-proteobacteria (Table 1). Mean cell numbers of Rhodobacter/Roseobacter increased from 1.5(±0.5) × 105 (n = 8) by 1.7-fold and by 2.2-fold in the first 20 h of incubation in the S− and S+ treatments, respectively, whereas the increase in NA+ treatments was smaller. Within the second half of incubation, numbers of cells targeted by probe G Rb changed little in all treatments and dropped below the original value in the substrate-amended treatments (Fig. 2). Their relative abundances in all but the NA+ treatments decreased during 43 h of incubation. Rhodobacter/Roseobacter constituted only about 4% ± 1% in S+ but up to 7% ± 1% of bacteria in the S− treatments.
Changes in per cell DNA and protein content.
The flow cytometric signature of double-stained bacterioplankton cells revealed treatment-specific changes during the incubations (Fig. 3). At the end of the experiment, the cytograms from the unamended treatments with and without NA showed pronounced differences from those of the original community. In both, a second cell population with higher protein content was discernible after 43 h. Incubation with substrates either with or without NA resulted in the appearance of cells with significantly higher DNA and protein content than in the unamended sample. In S+NA+ treatments, the fraction of these large cells was much higher (56.0%) than in S+NA− (19.8%).
Starvation experiment.
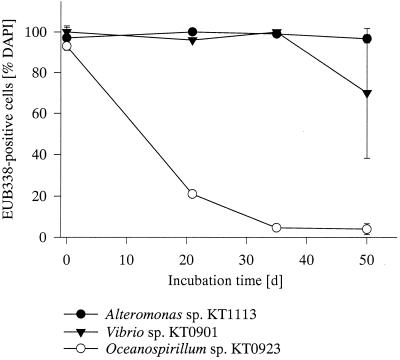
Representative isolates obtained from the North Sea (14) hybridizing with probe ALT1413, OCE232, or G V were starved for more than 50 days (Fig. 4). The percentage of intact bacteria was determined as the fraction of ribosome-containing cells, i.e., by their EUB338 signal. A/C and Vibrio spp., which were readily enriched during the field growth experiment, showed no significant loss of EUB338 detection rate versus DAPI during 50 days. In contrast, the detection rates of Oceanospirillum spp. decreased rapidly within the first 20 days, and cells from this strain were almost not detectable in the last half of the starvation experiment. The addition of fresh medium did not result in an increase in FISH-detectable cell numbers of Oceanospirillum within 10 h of incubation.
FIG. 4.
FISH detection rates of different genera of the gamma subclass of the Proteobacteria in stationary-phase batch cultures (probe EUB338, DAPI counterstaining). Oceanospirillum sp. KT0923, ○; Vibrio sp. KT0901, ▾; Alteromonas sp. KT1113, ●.
DISCUSSION
Shifts in bacterioplankton community composition during enrichment.
Enrichment cultures have a long tradition in microbiology (8). This experimental strategy, arranged intentionally or not, eventually resulted in the isolation of the presently known variety of marine bacteria. The classic ZoBell approach of 1946 (33, 47) has been repeatedly improved or modified, e.g., by differential filtration (15), dilution (10), and the use of specific substrates (24). Knowledge about the spatiotemporal occurrence or physiological features of particular phylogenetic groups may allow the design of more directed experiments (12, 23, 36). However, enrichment attempts always represent substantial interferences with microbial life and their environment, even in the absence of additional substrates. For example, prefiltration may influence bacterioplankton composition by removal of large filamentous and most of the particle-attached cells (1, 13). Cellulose ester filters of 1.2-μm pore size were found to retain up to almost 50% of unfiltered bacterial abundances in coastal waters (19). We could, however, not verify such a reduction in our samples (means ± 1 standard deviation: unfiltered, 1.52(±0.04) × 106 cells ml−1, n = 3; prefiltered, 1.57(±0.19) × 106 cells ml−1, n = 9). Even gentle filtration may increase substrate concentrations (e.g., of dissolved free amino acids) due to damage of phytoplankton cells (15) and disrupt the link between dissolved and particulate organic matter (34). The absence of protistan grazers will relieve bacteria from selective mortality (41), and confinement will put an end to the dynamic equilibrium between the formation and decomposition of organic matter (45).
Already in 1984, Ammerman et al. (5) and Ferguson et al. (15) had shown an increase in population size and average cell volume during undirected bacterioplankton growth in unamended seawater. More recently, FISH in combination with flow cytometry revealed changes in the taxonomic community composition of North Sea bacterioplankton in dilution culture (17). In our experiments, changes in community structure occurred more rapidly than reported by Suzuki, who did not detect taxonomic shifts in filtered seawater samples for a period of 24 h (42). γ-Proteobacteria had increased overproportionally already after 20 h of incubation in S−, whereas α-proteobacteria and C/F members did not (Fig. 1). The addition of organic substrates in micromolar concentrations (5.7 mg of C per liter) did not result in significantly higher total cell numbers after 48 h compared to S− (Fig. 1), but in an even more pronounced change in community structure. γ-Proteobacteria increased from about 15 to 60% and the fraction of C/F again remained constant, but α-proteobacteria decreased by half in relative abundance. Concomitantly, the development of cell populations with higher protein and DNA content was observed in S+ and S− (Fig. 3), and these large cells thus mainly belonged to the rapidly growing fraction within the γ-proteobacteria.
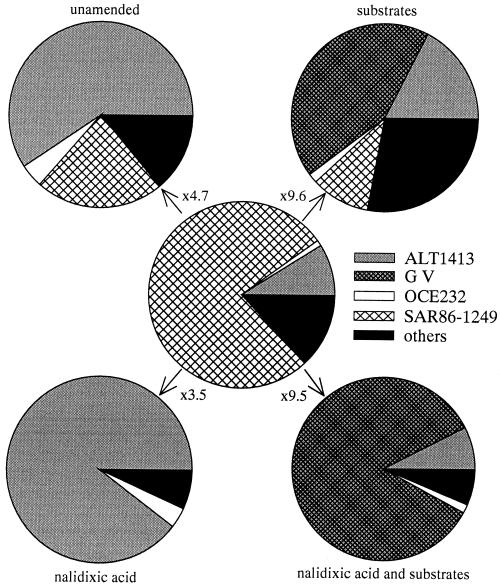
Our study extends previous findings in several respects. We present the response of several individual groups within the marine γ-proteobacteria to different treatments. Evidence is provided that the dominant members of this lineage in situ were rapidly outcompeted during enrichment culture. In the prefiltered seawater, about 14% of total cell numbers, corresponding to approximately 2 × 105 cells ml−1, belonged to the γ-subclass of Proteobacteria. Members of a single phylogenetic lineage, the uncultured SAR86 cluster, formed 90% of all γ-proteobacteria in the beginning of the experiments (Fig. 5). SAR86 belongs to the free-living fraction of the pelagic bacterioplankton, as determined by clone libraries of prefiltered seawater (1, 14) or visualization by FISH (14). Several typical culturable γ-proteobacterial genera were detected in very low numbers, either attached (Vibrio and Alteromonas) or freeliving (Oceanospirillum) in the original North Sea pelagic community (14). In contrast to Vibrio and A/C, SAR86 and Oceanospirillum were not enriched in any of the treatments; the absolute abundances of these groups remained constant, and we did not observe a significant increase in either cell size or FISH signal intensity. Members of the SAR86 cluster and Oceanospirillum were therefore neither visibly subjected to substrate-accelerated death (35) nor activated in either S− or S+. Enhanced mortality of SAR86 was, however, observed as a consequence of the antibiotic treatment, and already within the first 20 h of incubation, the abundance of SAR86 had decreased significantly in the NA+ incubations (Fig. 2). NA might, therefore, have acted as a cell toxin for members of this lineage (2) or represented a stress factor that caused the lysis of virus-infected cells (46).
FIG. 5.
Percentage of different lineages within the gamma subclass of the Proteobacteria at the beginning and at the end of enrichment experiments. Probes: A/C group, ALT1413; Oceanospirillum, OCE232; Vibrio, G V; SAR86 cluster, SAR86-1249. Factors, increase in γ-proteobacteria compared to the original sample.
On the other hand, Vibrio and A/C responded rapidly to our simulated culturing conditions, with lag phases ranging from 5 to 10 h and generation times of between 7 and 12 h, as estimated from the abundance changes during the 43-h experimental period. This corresponds well with the high numbers of Vibrio-related sequences found in clone libraries of stationary-phase dilution cultures from Mediterranean Sea samples (20), and with the selective enrichment of bacteria affiliating with Alteromonas macleodii during enclosure incubations in the same system (39). Presently we cannot distinguish between the different potential causes for the observed community shifts, such as shorter response times to substrate upshifts, but also antibacterial or autocrine growth factors released by the rapidly growing groups (44).
No activation of dormant culturable bacteria.
The high relative contribution of microbes affiliated with Vibrio, Alteromonas, or Colwellia to the colony-forming bacteria (14, 28) might be attributed to rapid cell multiplication and short lag times. Alternatively, it may be the consequence of a high fraction of dormant cells from these genera in the original community that are activated by culturing effects like substrate addition or the presence of solid surfaces (26). The combined incubation with NA and substrates causes an abnormal increase in cell size, and consequently ribosome content (11), by delaying cell division until the eventual appearance of NA-resistant strains (Fig. 3). Therefore, dormant bacteria that respond to substrate addition should become FISH detectable in such a treatment. In our experiments, the offered substrate mix was appropriate to activate, e.g., Vibrio, Alteromonas, and Colwellia, as it has been successfully utilized for their isolation previously (14). However, no increase in the relative abundances of these typical culturable bacteria was observed during the initial period of incubation with NA (0 to 20 h) (Fig. 1 and 2). In contrast, there was a clear rise of the two groups during the same incubation period in the treatments lacking NA. This is evidence that no or few dormant, FISH-undetectable bacteria affiliated with Vibrio or A/C were present in the water column. In fact, no initial increase in total FISH detection rates with the bacterial probe EUB338 was observed during incubations with NA. This suggests either that in general there was no activation of dormant cells by our incubation conditions or else that no cells escaped FISH detectability due to their low ribosome content.
Interestingly, the addition of the cell division-inhibiting agent NA did not result in the dominance of one particular resistant bacterial group irrespective of substrate levels, but rather amplified the success of the most competitive lineage within the respective treatment. In the substrate-unamended enrichments, resistant strains of the A/C cluster increased to similar absolute numbers in NA+ as in NA− after 43 h of incubation. However, in the presence of the antibiotic, they constituted a much larger fraction, about one third of the total community. The addition of substrate always specifically favored Vibrio. This group constituted 65% of total bacteria in NA+, which was almost three times as much as in the NA− treatment (Fig. 5). On the other hand, A/C, in spite of being potentially NA resistant (Fig. 5), was almost completely suppressed in substrate-amended NA− treatments, and the antibiotic shifted the competition between the two groups towards Vibrio. It would be premature to draw general conclusions from an unplanned observation in a single sample. However, the study of the combined effects of growth-promoting and growth-inhibiting factors on microbial competition might be a fruitful field for future investigations.
Enrichable culturable genera: “feast-or-famine” strategists.
Our data do not support the hypothesis that readily culturable pelagic bacteria are in general rapidly enriched in filtered or substrate-amended seawater. During extensive cultivation at Hegoland Roads (14), 33 of 145 different bacterioplankton isolates affiliated with genera which also dominated our enrichments. However, another nine isolates were related to Oceanospirillum, which did not grow during the incubations. Strains related to Vibrio and A/C maintained large amounts of cellular ribosomes during starvation in pure culture, whereas the FISH detectability of Oceanospirillum declined rapidly (Fig. 4). A high total per cell rRNA content of nongrowing cells apparently provides the potential for a more rapid response to changes in growth conditions (16). We conclude that rapidly enriched culturable bacteria like Vibrio and A/C are able to maintain a high potential to react to changes in growth conditions even during extended periods of nongrowth. This life strategy goes beyond the simplified dichotomy of r versus K selection (6), and the growth of the two r strategists A/C and Vibrio was apparently triggered at different ambient substrate concentrations (Fig. 5). Members of both lineages have been found associated with marine metazoans (7, 25), which would agree with a concept of a feast-or-famine existence.
This bacterial life strategy will confront microbiologists trying to culture as yet uncultured bacteria with fundamental problems. Some representatives (e.g., A/C) grow on unamended seawater and media with a relatively low carbon content (14). On the other hand, they maintain a high potential for growth during starvation and show immediate response to the environmental changes caused by sampling. Moreover, members of several readily culturable genera survived and rapidly resisted the stress factor NA. In summary, new strategies are required to enrich and eventually isolate yet uncultured bacteria in plankton samples, and for this purpose molecular methods that monitor the changes in community composition will be essential.
ACKNOWLEDGMENTS
We acknowledge J. Trotter for providing the freeware program WinMDI. We thank Christian Schütt (BiologischeAnstalt Helgoland, Dept. of Microbiology) for sampling and use of the laboratory facility. We thank Gunnar Gerdts and Antje Wichels for inspiring discussions.
This work was supported by the Max Planck Society (Germany).
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertini S, Chetelat A A, Miller B, Muster W, Pujadas E, Strobel R, Gocke E. Genotoxicity of 17 gyrase and four mammalian topoisomerase II poisons in prokaryotic and eukaryotic test systems. Mutagenesis. 1995;10:343–351. doi: 10.1093/mutage/10.4.343. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ammerman J W, Fuhrman J A, Hagstöm A, Azam F. Bacterioplankton growth in seawater. I. Growth kinetics and cellular characteristics in seawater cultures. Mar Ecol Prog Ser. 1984;18:31–39. [Google Scholar]
- 6.Andrews J H, Harris R F. r- and K-selection and microbial ecology. Adv Microb Ecol. 1986;9:99–147. [Google Scholar]
- 7.Baumann P, Schubert R H W. Family II, Vibrionaceae. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Williams; 1984. pp. 516–550. [Google Scholar]
- 8.Beijerinck M W. Anhäufungsversuche mit Ureumbakterien. Ureumspaltung durch Urease und durch Katabolismus. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt II. 1901;7:33–61. [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Button D K, Schut F, Quang P, Martin R, Robertson B. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper S. Bacterial growth and division. London, U.K: Academic Press, Inc.; 1991. [Google Scholar]
- 12.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 14.Eilers H, Pernthaler J, Glöckner F O, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson R L, Buckley E N, Palumbo A V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984;47:49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flärdh K, Cohen P S, Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992;174:6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs B M, Zubkov M V, Sahm K, Burkill P H, Amann R. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ Microbiol. 2000;2:191–201. doi: 10.1046/j.1462-2920.2000.00092.x. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasol J M, Morán X A G. Effects of filtration on bacterial activity and picoplankton community structure as assessed by flow cytometry. Aquat Microb Ecol. 1999;16:251–264. [Google Scholar]
- 20.Giuliano L, De Domenico E, Höfle M G, Yakimov M M. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb Ecol. 1999;37:77–85. doi: 10.1007/s002489900132. [DOI] [PubMed] [Google Scholar]
- 21.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 22.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González J M, Mayer F, Moran M A, Hodson R E, Withman W B. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 24.González J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmström C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 26.Kjelleberg S, Humphrey B A, Marshall K C. Effect of interfaces on small, starved marine bacteria. Appl Environ Microbiol. 1982;43:1166–1172. doi: 10.1128/aem.43.5.1166-1172.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1978;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 28.Lebaron P, Servais P, Troussellier M, Courties C, Vives-Rego J, Muyzer G, Bernard L, Guindulain T, Schäfer H, Stackebrandt E. Changes in bacterial community structure in seawater mesocosms differing in their nutrient status. Aquat Microb Ecol. 1999;19:225–267. [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 30.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 31.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 32.Neef A. Ph.D. thesis. Munich, Germany: Techical University; 1997. [Google Scholar]
- 33.Pinhassi J, Zweifel U L, Hagström A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ploug H, Grossart H P, Azam F, Jorgensen B B. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar Ecol Prog Ser. 1999;179:1–11. [Google Scholar]
- 35.Postgate J R, Hunter J R. Accelerated death of Aerobacter aerogenes starved in the presence of growth limiting substrates. J Gen Microbiol. 1964;34:459–473. doi: 10.1099/00221287-34-3-459. [DOI] [PubMed] [Google Scholar]
- 36.Prokic I, Brümmer F, Brigge T, Gortz H D, Gerdts G, Schütt C, Elbrächter M, Müller M E G. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist. 1998;149:347–357. doi: 10.1016/S1434-4610(98)70041-0. [DOI] [PubMed] [Google Scholar]
- 37.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 38.Rehnstam A S, Backman S, Smith D C, Azam F, Hagström A. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol. 1993;102:161–166. [Google Scholar]
- 39.Schäfer H, Servais P, Muyzer G. Successional changes in the genetic diversity of a marine assemblage during confinement. Arch Microbiol. 2000;173:138–145. doi: 10.1007/s002039900121. [DOI] [PubMed] [Google Scholar]
- 40.Schut F. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1994. [Google Scholar]
- 41.Simek K, Kojecka P, Nedoma J, Hartman P, Vrba J, Dolan D R. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol Oceanogr. 1999;44:1634–1644. [Google Scholar]
- 42.Suzuki M T. Effect of protistan bacterivory on costal bacterioplankton diversity. Aquat Microb Ecol. 1999;20:261–272. [Google Scholar]
- 43.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takamoto S, Yamada K, Ezura Y. Producing of bacteriolytic enzymes during the growth of a marine bacterium Alteromonas sp. no. 8-R. J Gen Appl Microbiol. 1994;40:499–508. [Google Scholar]
- 45.Waksman S A, Carey C L. Decomposition of organic matter in sea water by bacteria. I. Bacterial multiplication in stored sea water. J Bacteriol. 1935;29:531–543. doi: 10.1128/jb.29.5.531-543.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinbauer M G, Suttle C A. Lysogeny and prophage induction in coastal offshore bacterial communities. Aquat Microb Ecol. 1999;18:217–225. [Google Scholar]
- 47.ZoBell C E. Marine microbiology: a monograph on hydrobacteriology. Waltham, Mass: Chronica Botanica Company; 1946. [Google Scholar]
- 48.Zubkov M V, Fuchs B M, Eilers H, Burkill P H, Amann R. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl Environ Microbiol. 1999;65:3251–3257. doi: 10.1128/aem.65.7.3251-3257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]