Abstract
Pfiesteria complex species are heterotrophic and mixotrophic dinoflagellates that have been recognized as harmful algal bloom species associated with adverse fish and human health effects along the East Coast of North America, particularly in its largest (Chesapeake Bay in Maryland) and second largest (Albermarle-Pamlico Sound in North Carolina) estuaries. In response to impacts on human health and the economy, monitoring programs to detect the organism have been implemented in affected areas. However, until recently, specific identification of the two toxic species known thus far, Pfiesteria piscicida and P. shumwayae (sp. nov.), required scanning electron microscopy (SEM). SEM is a labor-intensive process in which a small number of cells can be analyzed, posing limitations when the method is applied to environmental estuarine water samples. To overcome these problems, we developed a real-time PCR-based assay that permits rapid and specific identification of these organisms in culture and heterogeneous environmental water samples. Various factors likely to be encountered when assessing environmental samples were addressed, and assay specificity was validated through screening of a comprehensive panel of cultures, including the two recognized Pfiesteria species, morphologically similar species, and a wide range of other estuarine dinoflagellates. Assay sensitivity and sample stability were established for both unpreserved and fixative (acidic Lugol's solution)-preserved samples. The effects of background DNA on organism detection and enumeration were also explored, and based on these results, we conclude that the assay may be utilized to derive quantitative data. This real-time PCR-based method will be useful for many other applications, including adaptation for field-based technology.
Pfiesteria complex species are heterotrophic and mixotrophic dinoflagellates that have been recognized as harmful algal bloom (HAB) species. Many HAB species are believed to be increasing in frequency and worldwide distribution, with negative effects on the economy, human health, and the environment (12, 13, 20). Of the approximately 5,000 recognized species of marine phytoplankton (21), about 300 can occur in sufficient concentration to discolor the water while at least 90 of these are classified as HAB species because they can produce potent toxins that have adverse effects on fish and human health (2, 3, 13). Other species, although harmless to humans, may have direct effects on fish through damage to their gills (13) or by leading to low dissolved-oxygen concentrations (2).
Toxicity-associated Pfiesteria species have been identified in both the Chesapeake Bay (Maryland) and Albermarle-Pamlico Sound (North Carolina) estuaries, where adverse fish and human health effects attributed to these organisms have been reported (1, 5, 7, 10, 22). In 1997, detection of Pfiesteria piscicida was correlated with three major fish kills affecting the Pocomoke, Chicamacomico, and Manokin Rivers in Maryland. In that same year, five major fish kill-disease events occurred in the Neuse and Pamlico estuaries in North Carolina.
Watermen and other individuals exposed to those affected river systems at these times complained of symptoms including gastrointestinal disturbance, headache, respiratory difficulties, burning skin, eye irritation and, for some, confusion and memory difficulty (8–10). In addition to complaints of these symptoms, reversible deficits in learning efficiency and concentration were observed among individuals who were clinically evaluated in Maryland shortly after exposure to Pfiesteria-related fish kills (5, 10, 11, 17). Laboratory staff who worked with toxic, fish-killing P. piscicida cultures previously had been reported to have similar symptoms (8). Thus, a tentative linkage between human health effects and exposure to partially characterized toxins present during environmental, as well as laboratory, exposure to Pfiesteria-associated fish kill-disease events was established. Although no correlation was or has been made between seafood consumption and illness, public concern led to significant impacts on the seafood industry along the eastern seaboard and consequently affected the livelihoods of many watermen (16).
In consideration of the association of toxic Pfiesteria species (P. piscicida Steidinger and Burkholder and a second species, P. shumwayae sp. nov.; 7, 22) with human health and the adverse economic impact of the 1997 events, comprehensive monitoring programs were developed and implemented by several Atlantic coast states (19). In Maryland, Virginia, and North Carolina, monitoring programs are now in place, with weekly to bimonthly collection of biophysical parameter data, including efforts to identify and enumerate Pfiesteria spp. Assessment of algal communities and fish health monitoring programs have also been implemented. Furthermore, programs have been established to rapidly assess these same parameters in response to reports of fish health disturbance or of human illness in association with estuarine exposure to toxic Pfiesteria outbreaks.
However, detection and quantification of Pfiesteria spp. have been problematic. The two known organisms (P. piscicida and P. shumwayae sp. nov.) are relatively nondescript heterotrophic-mixotrophic dinoflagellates (5, 15). Their life cycles are complex and may include multiple flagellated, amoeboid, and cyst forms with a considerable size range (major cell axis, 5 to 750 μm; 4, 5). These forms or stages cannot be positively identified by light microscopy (LM) alone because they closely resemble various other flagellates and amoebae. Moreover, specific antibodies or lectins for organism labeling are not yet available. Pfiesteria spp. (flagellated zoospores) can be identified by scanning electron microscopy (SEM) of membrane-stripped or suture-swollen cells (7, 23); however, this painstaking process requires considerable time and expertise, thus limiting the number of specimens that can be analyzed. Until recently, no genetic sequence data were available to permit development of sequence-based detection methods. This bottleneck was recently overcome (18), permitting development of new assays for these organisms.
We developed and implemented real-time PCR-based assays utilizing the 5′-to-3′ exonuclease activity of Taq polymerase (Taqman; 14, 26) for detection of P. piscicida and P. shumwayae sp. nov. in both fixative-preserved and unpreserved environmental estuarine water samples and cultures. In these assays, detection of amplified target DNA requires annealing of fluorescently labeled oligonucleotide probes, resulting in an added level of specificity compared with assays based on traditional PCR methodology. As the reaction proceeds, the 5′-to-3′ exonuclease activity of Taq polymerase cleaves the probe. This cleavage frees the quencher dye from the emitter dye, which is then able to fluoresce. Amplification was observed via real-time fluorescence monitoring on the Lightcycler.
The specificity of both Pfiesteria sp. assays was tested against a panel of dinoflagellate cultures characterized by SEM or LM. After specificity was determined, it was imperative to test the sensitivity of the assays on both fixative (acidic Lugol's solution)-preserved (24) and unpreserved (fresh) culture and environmental samples to aid in designing the optimal protocol for sample collection and storage until the time of processing. In addition, given the availability of archived samples and an interest in investigating prior algal blooms and fish kill events, it was essential to determine the long-term stability of preserved samples. Given the anticipated use of the assay in environmental screening and the marked heterogeneity (species composition and relative abundance) of estuarine water samples, the effect of variable background DNA concentrations on assay performance was investigated.
MATERIALS AND METHODS
Cultures.
For dilution experiments, two P. piscicida zoospore cultures were utilized: strain 113-3 (Aquatic Botany Laboratory, North Carolina State University [NCSU], Raleigh) and a strain (MDFDEPMR23, characterized by K. Steidinger, Florida Department of Environmental Protection [FL DEP], St. Petersburg) maintained by Horn Point Environmental Laboratories (University of Maryland Center for Environmental Studies, Cambridge) using previously described methods (5). P. piscicida zoospores were quantified from acidic Lugol's solution-preserved samples (24) using a Palmer-Maloney counting chamber (25) and an Olympus IMT-2 inverted microscope (magnification, ×600, phase contrast). Four additional P. piscicida cultures were utilized for assay specificity experiments (NCSU cultures 102-1 and 97-1, Provasoli-Guillard National Center for Culture of Marine Phytoplankton [CCMP] culture 1831, and FL DEP culture MMRCC981020BR01C5). P. shumwayae sp. nov. cultures (B-Vandemere, 7-28-T, and BP) were provided by NCSU.
Additional cultures were received from the Horn Point Environmental Laboratory, including Gymnodinium galatheanum, three Ciliophora cultures, and Rhodomonas sp. P. piscicida and Pfiesteria-like (morphologically similar to Pfiesteria complex species) cultures were provided by CCMP (R. Anderson, West Boothbay Harbor, Maine), and additional Pfiesteria-like dinoflagellate cultures were supplied by Old Dominion University (H. Marshall, Norfolk, Va.). Culture material characterization was confirmed by at least two methods and in at least two laboratories in all cases. Table 1 lists the cultures and isolates used in this study.
TABLE 1.
Specificity of species-selective primers
| Organism or collection site and date (mo/yr) | Source; strain(s) | P. piscicida PCR | P. shumwayae sp. nov. PCR |
|---|---|---|---|
| Prorocentrum minimum | CCMP; 699 | Negb | Neg |
| Heterocapsa triquetra | CCMP; 449 | Neg | Neg |
| Amphidinium carterae | CCMP; 1314 | Neg | Neg |
| Katodinium rotundatum | CCMP; 1542 | Neg | Neg |
| Pyrocystis lunula | CCMP; 731 | Neg | Neg |
| Ceratium longipes | CCMP; 1770 | Neg | Neg |
| Lingoludinium polyedrum | CCMP; 407 | Neg | Neg |
| Gymnodinium breve | CCMP; 718 | Neg | Neg |
| Chattonella subsalsa | CCMP; 217 | Neg | Neg |
| (Heterokontophyta) | |||
| Gymnodinium galatheanum | CCMP; 415, 416 | Neg | Neg |
| Prorocentrum hoffmannianum | CCMP; 683 | Neg | Neg |
| Prorocentrum triestinum | CCMP; 700 | Neg | Neg |
| Scrippsiella faroensae | CCMP; 771 | Neg | Neg |
| Scrippsiella sp. | CCMP; 772, 735 | Neg | Neg |
| Gambierdiscus toxicus | CCMP; 1600 | Neg | Neg |
| Protoceratium reticulatum | CCMP; 1889 | Neg | Neg |
| Adenoides eludens | CCMP; 1891 | Neg | Neg |
| Coolia monotis | CCMP; 305 | Neg | Neg |
| Gymnodinium varians | CCMP; 421 | Neg | Neg |
| Gymnodinium mikimotoi | CCMP; 429, 430 | Neg | Neg |
| Heterocapsa niei | CCMP; 447 | Neg | Neg |
| Amphidinium operculatum | CCMP; 1342 | Neg | Neg |
| Prorocentrum sp. | CCMP; 1541 | Neg | Neg |
| Prorocentrum compressum | CCMP; 1786 | Neg | Neg |
| Thecadinium inclinatum | CCMP; 1890 | Neg | Neg |
| Alexandrium tamarense | CCMP; 116 | Neg | Neg |
| Oxyrrhis marina (Alveolata) | CCMP; 604, 605, 1739 | Neg | Neg |
| Peridinium foliaceum | CCMP; 1326 | Neg | Neg |
| Gonyaulax cochlea | CCMP; 1592 | Neg | Neg |
| Gymnodinium sanguineum | CCMP; 417 | Neg | Neg |
| Prorocentrum balticum | CCMP; 1260 | Neg | Neg |
| Prorocentrum sp. | CCMP; 703 | Neg | Neg |
| Gyrodinium impudicum | CCMP; 1678 | Neg | Neg |
| Gyrodinium instriatum | CCMP; 431 | Neg | Neg |
| Gymnodinium simplex | CCMP; 419 | Neg | Neg |
| Gyrodinium uncatenum | CCMP; 1310 | Neg | Neg |
| Gymnodinium catenatum | CCMP; 414 | Neg | Neg |
| Prorocentrum mexicanum | CCMP; 1370 | Neg | Neg |
| Rhodomonas sp. | CCMP; 767 | Neg | Neg |
| Rhodomonas sp. | CCMP; 768 | Neg | Neg |
| Pfiesteria piscicida | NCSU; 102-1 | Posc | Neg |
| Pfiesteria piscicida | FL DEP; MDFDEPMR23 | Pos | Neg |
| Pfiesteria piscicida | NCSU; 97-1 | Pos | Neg |
| Pfiesteria piscicida | CCMP; 1831 | Pos | Neg |
| Pfiesteria piscicida | FL DEP; MMRCC981020BR01C5 | Pos | Neg |
| Pfiesteria shumwayae sp. nov. | Species ‘B’ (GenBank AF218805) | Neg | Pos |
| Pfiesteria shumwayae sp. nov. | NCSU; B-Vandemere | Neg | Pos |
| Pfiesteria shumwayae sp. nov. | NCSU; 7-28-T | Neg | Pos |
| Pfiesteria shumwayae sp. nov. | NCSU; BP | Neg | Pos |
| Cryptoperidiniopsis sp. (gen. nov.) | CCMP; 1827a | Neg | Neg |
| Cryptoperidiniopsis sp. (gen. nov.) | CCMP; 1827b | Neg | Neg |
| Gymnodinium galatheanum | HPELa; GE | Neg | Neg |
| Pfiesteria-like isolate sites | |||
| Neuse River (N.C.) 10/98 | CCMP; 1872 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1873 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1874 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1875 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1876 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1877 | Neg | Neg |
| Neuse River Isolate (N.C.) 12/98 | CCMP; 1878 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1879 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1880 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1881 | Neg | Neg |
| Wilmington River (Ga.) 11/98 | CCMP; 1882 | Neg | Neg |
| Pocomoke River (Md.) 1/98 | CCMP; A8925 | Pos | Neg |
| Chicamacomico River (Md.) 1/98 | CCMP; A8932 | Pos | Neg |
| Pocomoke Sound (Md.) 1/98 | CCMP; A8942 | Neg | Neg |
| Pocomoke Sound (Md.) 1/98 | CCMP; A8941 | Neg | Neg |
| Kings Creek (Md.) 9/97 | CCMP; 1827 | Neg | Neg |
| Kings Creek (Md.) 9/97 | CCMP; 1828 | Neg | Neg |
| Rhode River (Md.) 9/97 | CCMP; 1829 | Neg | Neg |
| Chicamacomico River (Md.) 1/98 | CCMP; 1830 | Pos | Neg |
| Chicamacomico River (Md.) 1/98 | CCMP; 1832 | Neg | Neg |
| Chicamacomico River (Md.) 1/98 | CCMP; 1833 | Neg | Neg |
| Pocomoke River (Md.) 1/98 | CCMP; 1834 | Pos | Neg |
| Pocomoke Sound (Md.) 1/98 | CCMP; 1835 | Neg | Neg |
| Pocomoke Sound (Md.) 1/98 | CCMP; 1836 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1838 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1839 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1840 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1841 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1842 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1843 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1844 | Neg | Neg |
| Neuse River (N.C.) 2/98 | CCMP; 1845 | Neg | Neg |
| Ciliophora spp. | |||
| Mesodinium pulex | HPEL | Neg | Neg |
| Strombium sp. | HPEL | Neg | Neg |
| Tontonia sp. | HPEL | Neg | Neg |
HPEL, Horn Point Environmental Laboratories (University of Maryland Center for Environmental Studies).
Neg, negative.
Pos, positive.
Acidic Lugol's solution fixation.
For fixation of cultures and environmental estuarine water samples, acidic Lugol's solution (hydrated iodine-potassium iodide, acetic acid solution; 24) was used at a final concentration of 1% (Sigma, St. Louis, Mo.).
DNA extraction.
For all experiments, sample aliquots were filtered through a 5-μm-pore-size hydrophilic Durapore filter (Millipore, Bedford, Mass.). The filter was then placed into an Eppendorf tube, and DNA extraction was performed by following the protocol supplied with the DNeasy Plant Kit (Qiagen, Valencia, Calif.). DNA was eluted with 100 μl of elution buffer and stored at −20°C.
PCR.
The primers and probes were designed utilizing the Primer Express software (Test Version; Perkin-Elmer) and an alignment of >100 dinoflagellate small-subunit ribosomal DNA sequences. The alignment was constructed using the Pileup software (Genetics Computer Group) and sequences downloaded from GenBank (in addition to multiple unpublished dinoflagellate sequences [T. Tengs, University of Maryland, unpublished data]). The alignment included P. piscicida (GenBank accession no. AF077055) and P. shumwayae sp. nov. (GenBank accession no. AF218805), and primers and probes were designed to target signature sequences unique to these species. PCR assays with these assays were performed on the Lightcycler (Idaho Technology, Idaho Falls, Idaho). The following reagents were added for a 10-μl P. piscicida-specific reaction: primers 107 (5′-CAGTTAGATTGTCTTTGGTGGTCAA-3′) and 320 (5′-TACCATATCACTTTCTGACCTATCA-3′), each at a final concentration of 0.2 μM (Operon, Alameda, Calif.); a P. pisc. probe labeled with FAM (carboxyfluorescein) and TAMRA (carboxytetramethylrhodamine) (5′-FAM-CATGCACCAAAGCCCGACTTCTCG-TAMRA-3′) at a final concentration of 0.15 μM (Operon); Taq polymerase at a final concentration of 0.1 U μl−1 (Life Technologies, Rockville, Md.); MgCl2 at a final concentration of 4 mM (Life Technologies); a deoxynucleoside triphosphate mixture with each deoxynucleoside triphosphate at a final concentration of 0.2 mM (Bioline, Reno, Nev.); bovine serum albumin at a final concentration of 0.25 mg ml−1 (Idaho Technologies); PCR buffer at a final concentration of 1× (Life Technologies); approximately 10 ng of template DNA; and PCR grade water to a final volume of 10 μl (Sigma). For a 10-μl P. shumwayae-specific reaction, primers Pshumfor (5′-TGCATGTCTCAGTTTAAGTCA-3′) and Pshumrev (5′-TCGATCATCAAATACACTAAAACTGTTTT-3′) each at a final concentration of 0.2 μM (Operon), were used. The probe used in this assay, at a final concentration of 0.30 μM, was P. shum (5′-FAM-TACGGCGAAACTGCGAATGGCTCAT-TAMRA-3′). The same reagents and concentrations were used as described above to obtain a 10-μl reaction mixture. Seven microliters of the reaction mixture was added to a cuvette (Idaho Technologies) and pulse spun on a tabletop centrifuge (Sorvall). Cuvettes were loaded into the Lightcycler, and the following quantification cycling protocol was used: 50 cycles at 94°C for 0 s and 60°C for 20 s, with a temperature transition time of 20°C s−1. Fluorescence acquisition was 100 ms after each incubation at 60°C, and the display mode was CH1 1−1 with the gain set at 1.
RESULTS
Assay specificity.
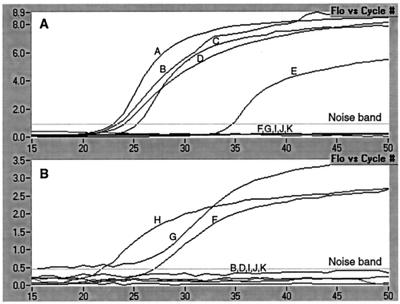
DNA extraction and PCR were performed utilizing SEM-verified P. piscicida and P. shumwayae sp. nov. culture DNA and panels of control organism DNA. Extensive specificity testing was performed with a panel of 36 well-characterized dinoflagellate cultures, 2 cryptophyte prey cultures, other protist representatives (Heterokontophyta and Alveolata), three Ciliophora representatives, and a panel of 32 dinoflagellate cultures characterized as Pfiesteria-like by the reference laboratory from which they were obtained (CCMP). Of these 32 cultures, 4 were positive by the PCR assay (Table 1) and have been confirmed via SEM and/or 18S rDNA sequencing to be P. piscicida. The remaining 28 cultures, all heterotrophic estuarine dinoflagellates, have been demonstrated through either 18S rDNA sequencing or heteroduplex mobility assay (18) to be distinct from P. piscicida (data available upon request). Figure 1A and B and Table 2 depict the specificity of the P. piscicida and P. shumwayae sp. nov. PCR assays against a representative panel of dinoflagellates, including SEM- and small-subunit ribosomal DNA sequence-validated P. piscicida (five cultures), P. shumwayae sp. nov. (three cultures), and the morphologically similar (Pfiesteria-like) dinoflagellates G. galatheanum and Cryptoperidiniopsis sp. Controls containing no template DNA were negative.
FIG. 1.
Specificity of P. piscicida (A) and P. shumwayae sp. nov. (B) real-time PCR assays. DNA was extracted from five cultures (A, B, C, D, and E) determined to be P. piscicida by either SEM or LM (coupled with 18S rDNA sequence analysis) and analyzed with the real-time PCR assay specific for P. piscicida. DNA was extracted from three cultures (F, G, and H) determined to be P. shumwayae sp. nov. by SEM and analyzed with the real-time PCR assay specific for P. shumwayae sp. nov. Negative results in both graphs (below the noise band) represent morphologically close relatives. The negative (no-DNA) controls were negative. The corresponding results obtained are presented in Table 2.
TABLE 2.
Specificity of P. piscicida and P. shumwayae sp. nov. real-time PCR assays (see Fig. 1)
| Samplea | Species | Method | P. piscicida PCR | P. shumwayae sp. nov. PCR |
|---|---|---|---|---|
| A; NCSU; 102-1 | P. piscicida | SEM | + | − |
| B; FL DEP; MDFDEPMR23 | P. piscicida | SEM | + | − |
| C; NCSU; 97-1 | P. piscicida | SEM | + | − |
| D; CCMP; 1831 | P. piscicida | LM-18S rDNA sequencing | + | − |
| E; FL DEP; MMRCC981020BR01C5 | P. piscicida | SEM | + | − |
| F; NCSU; B-Vandemere | P. shumwayae sp. nov. | SEM | − | + |
| G; NCSU; 7-28-T | P. shumwayae sp. nov. | SEM | − | + |
| H; NCSU; BP | P. shumwayae sp. nov. | SEM | − | + |
| I; CCMP; 1827a | Cryptoperidiniopsis sp. (gen. nov.) | SEM | − | − |
| J; CCMP; 1827b | Cryptoperidiniopsis sp. (gen. nov.) | SEM | − | − |
| K; Horn Point; GE | G. galatheanum | LM-18S rDNA sequencing | − | − |
The first letter corresponds to a designation in Fig. 1, and the source and the strain designation follow.
Sensitivity.
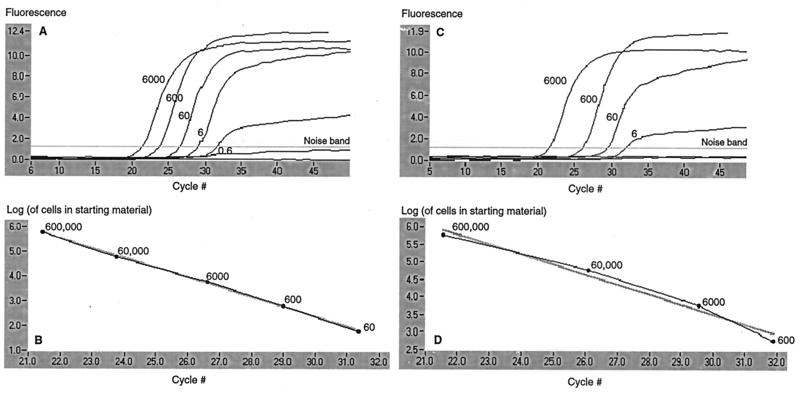
The sensitivity of the P. piscicida assay was assessed by performing PCR on fixative (acidic Lugol's solution)-preserved and unpreserved 10-fold serial dilutions of a pure P. piscicida culture (NCSU strain 113-3). Figure 2A reflects the sensitivity limits of the P. piscicida-specific assay on an unpreserved culture, with a detection limit of approximately 0.6 cell in a reaction. This value corresponds to DNA extracted from a total of 60 cells, assuming 100% extraction efficiency with the protocol used (under our experimental conditions, 1 μl of extracted DNA from 100 μl of total eluate was used as a template). Sensitivity decreased by 1 log with a fixative-preserved culture (Fig. 2B).
FIG. 2.
Real-time P. piscicida PCR assay on the Lightcycler to detect the organism in 10-fold serial dilutions of unpreserved and fixative (acidic Lugol's solution)-preserved culture material. A 10-ml volume of each dilution was filtered through a 5-μm-pore-size filter, and DNA was extracted from the retained organism. In graphs A and C (unpreserved and fixative preserved, respectively), fluorescence acquired from dilutions detected with the probe is plotted against the cycle number. The numbers indicate the equivalent numbers of cells (genomes) aliquoted into the PCR (i.e., extracted DNA was eluted in 100 μl, and 1 μl 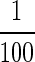 was assayed). In graphs B and D (unpreserved and fixative preserved, respectively), the log of the number of cells in the starting material is plotted against the cycle number at which the signal exceeded the threshold (set at 10% of the total fluorescence for the data set). In the unpreserved dilution, fewer than one cell per reaction could be detected, while in the fixative-preserved sample, the lower limit of detection was six cells per reaction.
was assayed). In graphs B and D (unpreserved and fixative preserved, respectively), the log of the number of cells in the starting material is plotted against the cycle number at which the signal exceeded the threshold (set at 10% of the total fluorescence for the data set). In the unpreserved dilution, fewer than one cell per reaction could be detected, while in the fixative-preserved sample, the lower limit of detection was six cells per reaction.
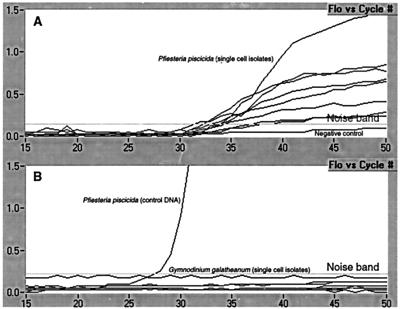
Sensitivity was further assessed by performing a single-cell PCR assay. Single P. piscicida strain MDFDEPMR23 cells were isolated with a capillary tube and placed directly into reaction cuvettes, and a PCR assay was performed immediately. Amplification was evident in all eight single-cell trials (Fig. 3).
FIG. 3.
Single-cell specificity and sensitivity of P. piscicida real-time PCR-based assay. (A) Results of PCR performed on eight replicates of single P. piscicida cells (all detectable). (B) Results of PCR performed on G. galatheanum (seven replicates), a close morphological relative, to test assay specificity. The positive control was total DNA isolated from a P. piscicida culture. In both graphs, the values for the negative control are below the noise band.
Stability.
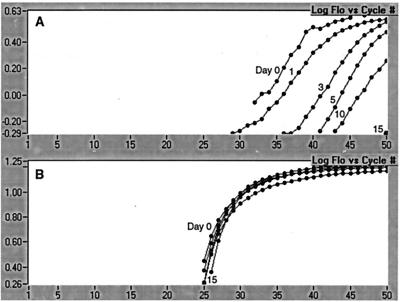
The ability to recover and detect P. piscicida DNA over time from fixative (acidic Lugol's solution)-preserved and unpreserved environmental water samples spiked with a known number of organisms was assessed. Environmental water samples collected from the Choptank River (Maryland) tested negative for the presence of P. piscicida with our PCR-based assay. Two 950-ml aliquots of this Choptank River water were spiked with 50 ml of a P. piscicida culture of 60,000 cells ml−1 (NCSU strain 113-3) for a final concentration of 3,000 cells ml−1. One sample was preserved with 1% acidic Lugol's solution, and both samples were maintained at room temperature on the benchtop. DNA was extracted from 40-ml aliquots on days 0, 1, 3, 5, 10, and 15. PCR was performed on all of the samples in the same run.
Detection of P. piscicida in the unpreserved sample was dramatically reduced over time, with undetectable levels by day 15 (Fig. 4A). In contrast, the fixative-preserved sample was markedly more stable, with P. piscicida at detectable levels throughout the experimental period and fluorescence detection consistent for all time points (Fig. 4B).
FIG. 4.
Detection of P. piscicida over time in unpreserved (A) and fixative (acidic Lugol's solution)-preserved (B) environmental water spiked with a known number of organisms. Spiked samples were stored on the benchtop, and DNA was extracted from 40-ml aliquots on the days indicated.
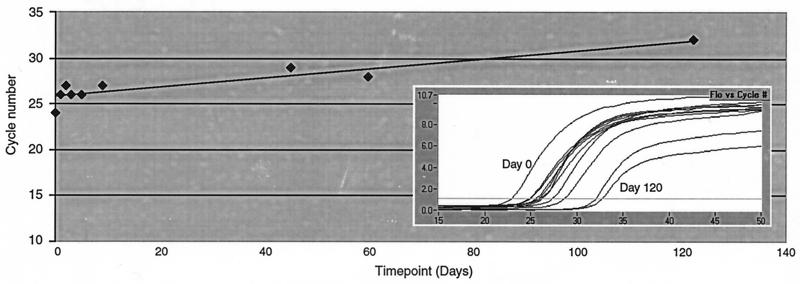
A further experiment was designed to assess the long-term stability of a fixative-preserved sample. A 22-ml aliquot of a P. piscicida culture (NCSU strain 113-3; concentration, 60,000 cells ml−1) was preserved with 1% acidic Lugol's solution and stored at room temperature on the benchtop. DNA was extracted from 2-ml aliquots on days 0, 1, 2, 3, 5, 9, 45, 60, and 120. A PCR assay was performed on all of the samples in the same run, and the cycle number at which fluorescence was detected at each time point was recorded (Fig. 5). Although there was an approximate shift of five cycles over the course of 4 months, long-term stability was apparent.
FIG. 5.
Detection of P. piscicida to 120 days in a fixative (acidic Lugol's solution)-preserved culture. At time point indicated, DNA was extracted from a 2-ml aliquot of the culture. DNA from all time points was assayed with the P. piscicida probe assay in the same Lighcycler run. The inset is a graph depicting fluorescence versus cycle number for each time point.
Effects of background DNA.
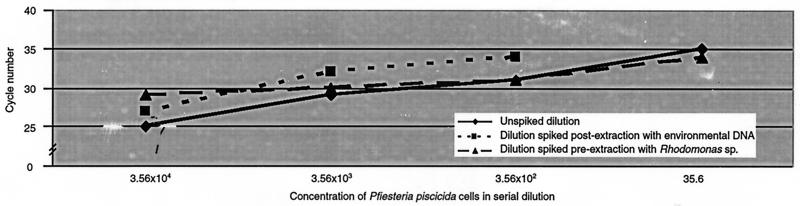
The performance of the P. piscicida assay was assessed in the presence of various background DNA concentrations either present prefiltration as prey organisms in the culture or introduced postfiltration through addition of extraneous organism DNA derived from environmental water. Three 10-fold serial dilution sets were prepared from a pure culture (strain MDFDEPMR23; concentration, 35,000 cells ml−1). One set was filtered, and DNA was extracted. The second set was filtered, DNA was extracted, and aliquots were then spiked with 640 ng of background environmental DNA (for a total of 12.8 ng in the PCR) to represent postfiltration spiking. In the third serial dilution set prepared from the same strain, a total of 1,860,000 Rhodomonas sp. cells were spiked into each dilution prior to filtration and DNA extraction.
PCR was performed on all three sets of serial dilutions in the same run. A 1-log decrease in the sensitivity of P. piscicida detection was observed when high extraneous background DNA concentrations were added to samples postextraction (Fig. 6). However, assay sensitivity was not affected by high background DNA concentrations when they were present as high extraneous organism loads in samples to be filtered, a condition more closely approximating screening of environmental samples. Regardless of the presence or absence of exogenous DNA, correlation of cell cycle number at detection versus concentration of target cells was highly significant (R values for the unspiked, spiked postextraction, and spiked preextraction conditions were 0.98, 0.94, and 0.91, respectively).
FIG. 6.
Effects of background DNA on detection of P. piscicida. Three 10-fold serial dilutions were prepared from a pure P. piscicida strain MDFDEPMR23 culture. Aliquots from one dilution set were spiked postfiltration with 12.8 ng of organism DNA extracted from a heterogeneous environmental water sample (Choptank River in Maryland). The third dilution set was spiked with 1,860,000 cells of Rhodomonas sp. prefiltration.
DISCUSSION
Based on the testing of available characterized cultures of P. piscicida and P. shumwayae sp. nov., a wide array of cultures representing morphological and genetically closely related organisms, and representatives of other photosynthetic protist groups, the real-time PCR-based assays described here have proven to be highly specific and sensitive for the detection of P. piscicida and P. shumwayae sp. nov. In our experience, the use of fluorescein-labeled species-specific probes in conjunction with species-specific primers added additional assay specificity in comparison to detection with SyBr Green or other double-stranded DNA intercalating dyes (data not shown), probably due to the conserved nature of the ribosomal gene targets assayed.
The demonstration of PCR assay sensitivity utilizing fixed (acidic Lugol's solution) samples over time will prove valuable for ongoing investigations of Pfiesteria biology. As demonstrated, the confounding effects of variable time intervals between sample collection and laboratory analysis, an often unavoidable consequence of oceanographic field work, can be addressed with a standard fixation methodology that has minimal (and consistent) impacts on downstream molecular analysis. The fixation method is simple to use, and it provides the means to assay archived samples. Further experiments will include assessment of assay stability over longer time periods (i.e., greater than 1 year) and efficiency of DNA extraction from samples preserved with other fixatives (glutaraldehyde, formalin).
In addition to a high level of specificity and stability of detection over time, the P. piscicida PCR assay demonstrated high sensitivity, with a detection limit of 0.6 cell. Further results showing detection of single P. piscicida cells in a PCR support the assay's sensitivity. Future efforts will include comparison of single-cell PCR assays of various described life stages (zoospores, cysts, and amoebae). The assay cannot yet be used in an absolutely quantitative manner due to (i) the fact that the number of 18S gene copies per cell is unknown and (ii) the possible variance of 18S gene copy number during the growth cycle. However, it can and currently is being used to determine relative concentrations of P. piscicida in environmental field samples, permitting statistical assessment of parameters believed to be associated with Pfiesteria blooms.
SEM methods are regarded by dinoflagellate systematists as the “gold standard” for identification of Pfiesteria spp. (e.g., see references 7 and 23). However, these procedures require membrane stripping or suture swelling techniques which are tedious and limit SEM's utility for environmental monitoring (7). Limitations also arise in utilizing SEM methods for detection of Pfiesteria spp. in estuarine water samples because these organisms are often minor components of the species composition (101 to 103 cells ml−1 versus 105 or more total phytoplankton cells ml−1; 5). In contrast, our real-time PCR assays developed for these organisms may be run rapidly with large sample sets and thus have proven to be useful tools for the detection of these species in both culture and environmental samples.
Molecular methods are rapid and allow phylogenetic analyses based on genetic data, but they also have limitations. For example, molecular techniques are subject to uncertainty in species specificity because various Pfiesteria-like estuarine dinoflagellates have not yet been formally described (22). In addition, the assay, which detects nuclear encoded DNA sequences, does not differentiate between Pfiesteria cultures in a toxic versus a nontoxic state as assayed in laboratory settings by estimation of toxin detectable in a reporter gene assay (6) or by ichthyotoxicity (4). This limitation can be addressed when the genetics of Pfiesteria toxicity are determined, permitting development of assays targeting toxicity-associated mRNA transcripts.
In summary, we have developed a highly sensitive and specific assay for detection of toxicity-associated dinoflagellates (P. piscicida and P. shumwayae sp. nov.) that can be used to explore Pfiesteria biology and the epidemiology of human health impacts of the organisms. The methods developed can be applied to a variety of critically important environmental monitoring initiatives (for instance, water quality screening for the presence of fecal coliforms or cryptosporidia). Fundamental questions about Pfiesteria biology, such as characterization of toxins and of mechanisms of toxin production, determinants of population blooms, and the full range of impacts on human health, must be resolved. The assays described here can be used as tools to address these important questions.
ACKNOWLEDGMENTS
We thank Diane Stoecker and Dan Gustafson from Horn Point Environmental Laboratories (University of Maryland Center for Environmental Studies, Cambridge) for providing P. piscicida MDFDEPMR23 culture material and Karen Steidinger (FL DEP, St. Petersburg) for SEM characterization of this culture. We also thank Robert Anderson (CCMP, West Boothbay Harbor, Maine), Harold Marshall, and David Seaborn (Old Dominion University, Norfolk, Va.) for additional P. piscicida, Pfiesteria-like, and various dinoflagellate cultures.
This work was supported by EPA grant R-827084 under the ECOHAB (Ecology and Oceanography of Harmful Algal Blooms) program.
Footnotes
ECOHAB publication number 008.
REFERENCES
- 1.Burkholder J M, Hobbs C W, Glasgow H B., Jr Distribution and environmental conditions for fish kills linked to a toxic ambush-predator dinoflagellate. Mar Ecol Prog Ser. 1995;124:43–61. [Google Scholar]
- 2.Burkholder J M. Implications of harmful marine microalgae and heterotrophic dinoflagellates in management of sustainable marine fisheries. Ecol Applications. 1998;8(Suppl.):S37–S62. [Google Scholar]
- 3.Burkholder, J. M. Critical needs in harmful algal bloom research, p. 232–269. In S. Vaupel (ed.), Proceedings of the Workshop on Opportunities for Environmental Application of Marine Biotechnology, in press. National Academy of Sciences, Washington, D.C.
- 4.Burkholder J M, Glasgow H B., Jr Interactions of a toxic estuarine dinoflagellate with microbial predators and prey. Arch Protistenkd. 1995;145:177–188. [Google Scholar]
- 5.Burkholder J M, Glasgow H B., Jr Pfiesteria piscicida and other toxic Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls. Limnol Oceanogr. 1997;42:1052–1075. [Google Scholar]
- 6.Fairey E R, Edmunds J S G, Deamer-Melia N J, Glasgow H B, Jr, Johnson F M, Moeller P R, Burkholder J M, Ramsdell J S. Reporter gene assay for fish-killing activity produced by Pfiesteria piscicida. Environ Health Perspect. 1999;107:711–714. doi: 10.1289/ehp.99107711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasgow H B., Jr . The biology and impacts of toxic Pfiesteria complex species. Ph.D. dissertation. Raleigh: Department of Marine, Earth & Atmospheric Sciences, North Carolina State University; 2000. [Google Scholar]
- 8.Glasgow H B, Jr, Burkholder J M, Schmechel D E, Tester P A, Rublee P A. Insidious effects of a toxic dinoflagellate on fish survival and human health. J Toxicol Environ Health. 1995;46:501–522. doi: 10.1080/15287399509532051. [DOI] [PubMed] [Google Scholar]
- 9.Golub J E, Haselow D T, Hageman J C, Lopez A S, Oldach D W, Grattan L M, Perl T M. Pfiesteria in Maryland: preliminary epidemiological findings. Md Med J. 1998;47:137–43. [PubMed] [Google Scholar]
- 10.Grattan L M, Oldach D, Perl T M, Lowitt M H, Matuszak D L, Dickson C, Parrott C, Shoemaker R C, Wasserman M P, Hebel J R, Charache P, Morris J G., Jr Problems in learning and memory occur in persons with environmental exposure to waterways containing toxin-producing Pfiesteria or Pfiesteria-like dinoflagellates. Lancet. 1998;352:532–539. doi: 10.1016/S0140-6736(98)02132-1. [DOI] [PubMed] [Google Scholar]
- 11.Grattan L M. Current status and future directions for the investigation and management of the human health effects of exposure to Pfiesteria piscicida or Pfiesteria-like dinoflagellates. Md Med J. 1998;47:148–51. [PubMed] [Google Scholar]
- 12.Hallegraeff G M. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32:79–99. [Google Scholar]
- 13.Hallegraeff G M. Harmful algal blooms, a global overview. In: Hallegraeff G M, Anderson D M, Cembella A D, editors. Manual on harmful marine microalgae. Intergovernmental Oceanographic Commission manuals and guides no. 33. Paris, France: United Nations Educational, Scientific, and Cultural Organization; 1995. pp. 1–22. [Google Scholar]
- 14.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewitus A J, Glasgow H B, Jr, Burkholder J M. Kleptoplastidy in the toxic dinoflagellate, Pfiesteria piscicida. J Phycol. 1999;35:303–312. [Google Scholar]
- 16.Lipton D W. Pfiesteria's economic impact on seafood industry sales and recreational fishing. In: Gardner B L, Koch L, editors. Proceedings of the Conference, Economics of Policy Options for Nutrient Management and Pfiesteria. College Park: Center for Agricultural and Natural Resource Policy, University of Maryland; 1999. pp. 35–38. [Google Scholar]
- 17.Oldach D W, Grattan L M, Morris J G. Pfiesteria piscicida and human health. In: Scheld W M, Craig W A, Hughes J M, editors. Emerging infections 3. Washington, D.C.: American Society for Microbiology; 1999. pp. 135–151. [Google Scholar]
- 18.Oldach D W, Delwiche C F, Jakobsen K S, Tengs T, Brown E G, Kempton J W, Schaefer E F, Bowers H A, Glasgow H B, Jr, Burkholder J M, Steidinger K A, Rublee P A. Heteroduplex mobility assay-guided sequence discovery: elucidation of the small subunit (18S) rDNA sequences of Pfiesteria piscicida and related dinoflagellates from complex algal culture and environmental sample DNA pools. Proc Natl Acad Sci USA. 2000;97:4303–8. doi: 10.1073/pnas.97.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rublee P A, Kempton J, Schaefer E, Burkholder J M, Glasgow H B, Jr, Oldach D. PCR and FISH detection extends the range of Pfiesteria piscicida in estuarine waters. Va J Sci. 1999;50:325–336. [Google Scholar]
- 20.Smayda T J. Primary production and the global epidemic of phytoplankton blooms in the sea: a linkage? In: Cosper E M, Bricelj V M, Carpenter E J, editors. Novel phytoplankton blooms. Coastal and estuarine studies, no. 35. New York, N.Y: Springer-Verlag; 1989. pp. 449–483. [Google Scholar]
- 21.Sournia A, Chretiennot-Dinet M J, Ricard M. Marine phytoplankton: how many species in the world ocean? J Plankton Res. 1991;13:1093–1099. [Google Scholar]
- 22.Steidinger K A, Burkholder J M, Glasgow H B, Jr, Truby E, Garrett J, Noga E J, Smith S A. Pfeisteria piscicida (Pfiesteriaceae, fam. nov.), a new toxic dinoflagellate with a complex life cycle and behavior. J Phycol. 1996;32:157–164. [Google Scholar]
- 23.Steidinger K A, Landsberg J H, Truby E W, Blakesley B A. The use of scanning electron microscopy in identifying small “gymnodinioid” dinoflagellates. Nova Hedwigia. 1996;112:415–422. [Google Scholar]
- 24.Vollenweider R A, editor. A manual on methods for measuring primary production in aquatic environments, 2nd ed. International biological programmes handbook no. 12. Oxford, England: Blackwell Scientific Publications; 1974. [Google Scholar]
- 25.Wetzel R G, Likens G E. Limnological analyses. 2nd ed. Philadelphia, Pa: W. B. Saunders; 1991. [Google Scholar]
- 26.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]