Abstract
Positioning organelles at the right place and time is critical for their function and inheritance. In budding yeast, mitochondrial and nuclear positioning require the anchoring of mitochondria and dynein to the cell cortex by clusters of Num1. We have previously shown that mitochondria drive the assembly of cortical Num1 clusters, which then serve as anchoring sites for mitochondria and dynein. When mitochondrial inheritance is inhibited, mitochondrial-driven assembly of Num1 in buds is disrupted and defects in dynein-mediated spindle positioning are observed. Using a structure-function approach to dissect the mechanism of mitochondria-dependent dynein anchoring, we found that the EF hand–like motif (EFLM) of Num1 and its ability to bind calcium are required to bias dynein anchoring on mitochondria-associated Num1 clusters. Consistently, when the EFLM is disrupted, we no longer observe defects in dynein activity following inhibition of mitochondrial inheritance. Thus, the Num1 EFLM functions to bias dynein anchoring and activity in nuclear inheritance subsequent to mitochondrial inheritance. We hypothesize that this hierarchical integration of organelle positioning pathways by the Num1 EFLM contributes to the regulated order of organelle inheritance during the cell cycle.
INTRODUCTION
Organelle inheritance is a highly regulated process that ensures that every new cell receives a full complement of organelles during cell division. In asymmetrically dividing cells, organelle inheritance depends on active positioning mechanisms, which ensure that essential organelles that cannot be generated de novo, such as the nucleus and mitochondria, are faithfully inherited. It is clear that these organelle positioning mechanisms are under tight spatiotemporal regulation to ensure that organelles are placed at the right location in the cell at the right time (Fagarasanu and Rachubinski, 2007; Eves et al., 2012; Knoblach and Rachubinski, 2015). In the budding yeast Saccharomyces cerevisiae, organelle inheritance occurs in a specific order; cortical endoplasmic reticulum (ER) and peroxisomes are inherited during bud emergence, followed by vacuoles and mitochondria at the small bud stage, and finally nuclear and perinuclear ER inheritance during the large bud stage (Segal and Bloom, 2001; Fagarasanu and Rachubinski, 2007; Eves et al., 2012; Li et al., 2021). Recent evidence indicates that the preferred order of organelle inheritance is not exclusively regulated by cell cycle signaling, suggesting that additional modes of regulation exist (Li et al., 2021). While it has been thought that the inheritance of one organelle is largely independent of the inheritance of others, recent studies suggest that some organelle inheritance pathways impact others, with some being interdependent on one another (Eves et al., 2012; Kraft and Lackner, 2017). These findings raise the possibility that yet-to-be-described surveillance mechanisms may help regulate the timing and order of organelle inheritance by monitoring the inheritance of one organelle and coupling it to the inheritance of another.
In budding yeast, the protein Num1 functions in both mitochondrial and nuclear positioning (Kormanec et al., 1991; Farkasovsky and Küntzel, 1995, 2001; Heil-Chapdelaine et al., 2000; Cerveny et al., 2007; Tang et al., 2012; Klecker et al., 2013; Lackner et al., 2013). For its role in mitochondrial positioning, Num1 tethers mitochondria to the plasma membrane (PM) and cortical ER, forming a tripartite membrane contact site called the mitochondria-ER-cortex-anchor (MECA; Lackner et al., 2013). The interaction between Num1 and mitochondria drives the assembly of Num1 into clusters, and once assembled, Num1 clusters stably and persistently anchor the organelle to the cell cortex (Figure 1A; Kraft and Lackner, 2017). The only other known protein component of MECA, Mdm36, functions as an accessory component that bridges Num1–Num1 interaction to enhance Num1 clustering (Hammermeister et al., 2010; Lackner et al., 2013; Omer et al., 2020).
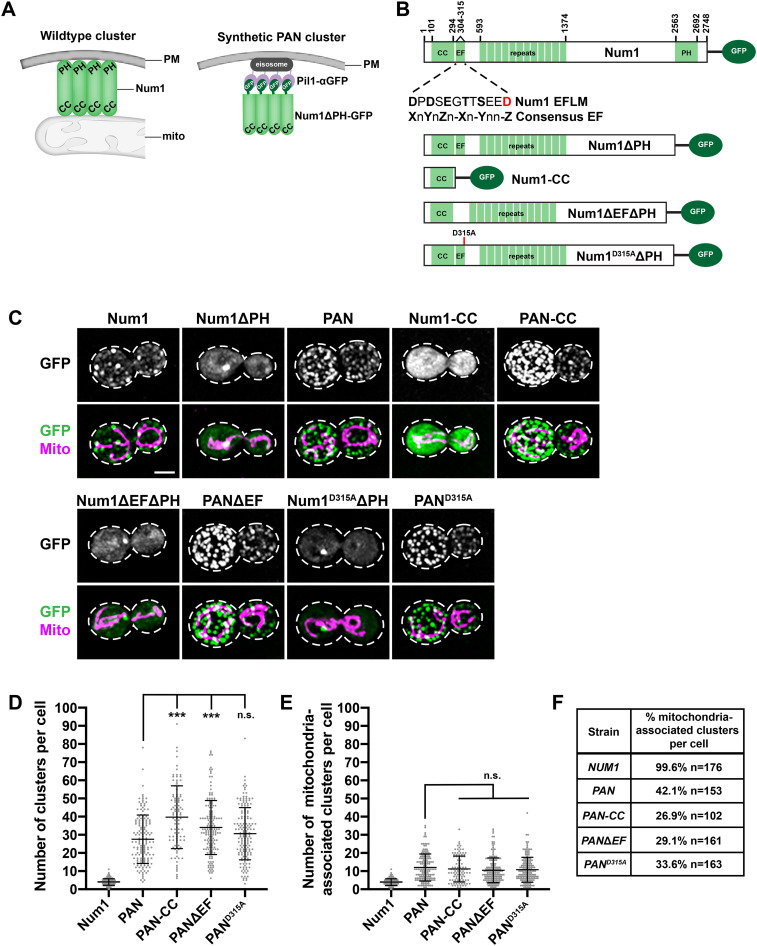
FIGURE 1:
Synthetically clustered Num1 truncations can tether mitochondria. (A) Schematic depicting the αGFP-GFP nanobody targeting system used to synthetically cluster Num1 on the PM. The left schematic represents a WT Num1 cluster, which requires the presence of mitochondria for assembly. The schematic on the right represents a synthetic Num1 cluster formed via the αGFP-GFP nanobody targeting system, independent of mitochondria. Mito, mitochondria; PAN, Pil1-associated Num1; PM, plasma membrane. (B) Schematics of all endogenously expressed Num1 constructs used in this study. The inset below the full-length Num1 schematic displays the protein sequence for the Num1 EFLM and the consensus EF hand sequence. X, Y, Z, –X, –Y, –Z = any calcium coordinating residue (D, E, N, Q, S, T, Y); n = any residue. The red residue corresponds to the D315A mutation. CC, coiled-coil domain; EF, EF-like hand motif; PH, pleckstrin homology domain. (C) Cells expressing Num1, Num1ΔPH, PAN, Num1-CC, PAN-CC, Num1ΔEFΔPH, PANΔEF, Num1D315AΔPH, and PAND315A as GFP fusions and mito-Red were analyzed by fluorescence microscopy. Whole-cell max projections are shown. The top panel shows the GFP signal in grayscale, and the bottom panel shows a merge of the GFP and mito-Red signals. The cell cortex is outlined with a dashed white line. Bar, 2 µm. (D) Quantification of the total number of clusters per cell for Num1, PAN, PAN-CC, PANΔEF, and PAND315A is shown as the mean ± SD. Each dot represents a single cell. p values are in comparison to PAN cells. *** p ≤ 0.0001; n.s., not significant. Num1 (n = 176), PAN (n = 153), PAN-CC (n = 102), PANΔEF (n = 161), and PAND315A (n = 163). (E) Quantification of mitochondria-associated clusters per cell for Num1, PAN, PAN-CC, PANΔEF, and PAND315A is shown as the mean ± SD (right). To be scored as a mitochondria-associated cluster, mitochondria had to remain associated with the cluster for ≥1.5 min. Each dot represents a single cell. p values are in comparison to PAN cells. n.s., not significant. Num1 (n = 176), PAN (n = 153), PAN-CC (n = 102), PANΔEF (n = 161), and PAND315A (n = 163). (F) Quantification of the percentage of Num1-yEGFP, PAN, PAN-CC, PANΔEF, and PAND315A clusters that are mitochondria-associated per cell. To be scored as a mitochondria-associated cluster, mitochondria had to remain associated with the cluster for ≥1.5 min. n = number of cells.
A subset of Num1 clusters also serve as cortical attachment sites for dynein. Specifically, dynein, in an inhibited conformation, is trafficked to the plus ends of astral microtubules (aMTs) emanating from the mitotic spindle. Dynein is then offloaded onto Num1 clusters at the cortex and subsequently activated. This allows dynein to capture and walk along the aMT, creating the force required for spindle migration and nuclear inheritance (Eshel et al., 1993; Carminati and Stearns, 1997; Adames and Cooper, 2000; Heil-Chapdelaine et al., 2000; Farkasovsky and Küntzel, 2001; Lee et al., 2005; Lammers and Markus, 2015).
Num1 is a 313 kDa multidomain protein consisting of an N-terminal coiled-coil (CC) domain, an adjacent EF hand–like motif (EFLM), a highly repetitive region of 12 64-amino-acid repeats, and a C-terminal pleckstrin homology (PH) domain (Figure 1B; Kormanec et al., 1991). The CC domain facilitates Num1 self-interaction, directly interacts with the mitochondrial outer membrane and Mdm36, and is required for the association of Num1 with dynein (Tang et al., 2012; Lackner et al., 2013; Ping et al., 2016; Won et al., 2021). The PH domain binds with high specificity to PI(4,5)P2, localizing Num1 to the PM (Yu et al., 2004; Tang et al., 2009). With the exception of the CC and PH domains, which are required for Num1 to tether mitochondria and anchor dynein, a majority of Num1’s domains have uncharacterized functions.
We have previously shown that mitochondria-assembled Num1 clusters that are actively tethering mitochondria can function as cortical attachment sites for dynein. Consistently, when mitochondrial-driven assembly of Num1 clusters is disrupted, defects in dynein-mediated spindle positioning are observed (Kraft and Lackner, 2017). Using a synthetic system that bypasses the requirement for mitochondria in Num1 cluster formation (Figure 1A), we have been able to create two populations of Num1 clusters—those that are associated with mitochondria and those that are not. We find that dynein is preferentially anchored to clusters that are mitochondria-associated, further highlighting a role for mitochondria in dynein anchoring and function (Schmit et al., 2018). Contrasting studies suggest that the Num1 clusters that anchor mitochondria are distinct from those that anchor dynein (Omer et al., 2018, 2020). In these studies, Num1 clustering is either enhanced by overexpression of Mdm36 or diminished by the deletion of Scs2, an integral ER protein thought to interact with Num1 (Hammermeister et al., 2010; Lackner et al., 2013; Chao et al., 2014; Ping et al., 2016; Omer et al., 2018). In more wild-type (WT) situations, however, our work demonstrates that mitochondrial inheritance positively impacts Num1 cluster formation in buds and dynein function in spindle positioning (Kraft and Lackner, 2017; Schmit et al., 2018).
To better understand the interplay between Num1’s role in mitochondria tethering and dynein anchoring, we used our synthetic clustering system in combination with a structure-function approach to identify the region of Num1 responsible for mitochondria-dependent dynein anchoring. The synthetic clustering system provides a way to examine the functions of Num1 downstream of cluster formation to determine whether these functions are dependent on an interaction with mitochondria. We found that the EFLM of Num1, which we determined is a bona fide Ca2+-binding EF hand, is important for the mitochondria-dependent association of dynein with Num1. When the Ca2+ binding by the Num1 EFLM is disrupted in our synthetic clustering system, we find a dramatic increase in non–mitochondria associated cortical dynein. Furthermore, in an otherwise WT background, when the EFLM is disrupted, cells no longer exhibit defects in dynein-mediated spindle positioning when mitochondrial inheritance is inhibited. Together, our in vivo and biochemical data indicate that the Ca2+-bound EFLM of Num1 favors dynein anchoring in the presence of mitochondria, consequently biasing dynein-dependent nuclear inheritance to occur subsequent to mitochondrial inheritance. We hypothesize that this integration of mitochondrial and nuclear positioning pathways provides a spatiotemporal mechanism to bias mitochondrial inheritance before inheritance of the nucleus and the subsequent completion of the cell cycle. Unlike nuclear inheritance, in which clear inheritance checkpoint mechanisms exist (Amon, 1999; Matellán and Monje-Casas, 2020), very little is understood about the monitoring of mitochondrial inheritance, which is surprising given the importance of mitochondria for cell viability. By coupling dynein-mediated nuclear inheritance to the presence of mitochondria in the bud, mitochondria-dependent dynein anchoring may function as a mitochondrial inheritance surveillance system.
RESULTS
Synthetically clustering the Num1 CC domain uncouples mitochondria tethering and dynein anchoring
To identify regions of Num1 that are important for the protein’s mitochondria tethering and dynein anchoring activities following cluster formation, we examined truncated versions of Num1 in our previously described synthetic clustering system (Figure 1, A and B; Schmit et al., 2018). For the synthetic clustering approach, Pil1 is expressed from its endogenous locus as an α-GFP nanobody fusion (Fridy et al., 2014). Pil1 is a major component of stable, multiprotein, PM-associated structures known as eisosomes (Walther et al., 2006). Truncated versions of Num1 are then expressed as yEGFP fusions from the endogenous NUM1 locus in the cells expressing Pil1-αGFP, resulting in recruitment of Num1 to eisosomes and its clustering on the PM (Figure 1, A and C; Schmit et al., 2018). The synthetic clustering system bypasses the need for mitochondrial association in cluster formation, allowing us to create populations of Num1 that are and are not associated with mitochondria. Thus, we can use this system as a tool to uncouple Num1 cluster formation at the PM from mitochondrial association. Cells expressing the components of the synthetic clustering system are referred to as PAN (for Pil1-associated Num1) cells, and the synthetic protein clusters are referred to as PAN clusters.
The CC domain of Num1 is required for both mitochondria tethering and dynein anchoring (Tang et al., 2012; Lackner et al., 2013). Thus, we reasoned that this minimal domain would be functional in our synthetic system. To test this, we expressed Num1(aa1-303)-yEGFP (Num1-CC) in cells expressing Pil1-αGFP (Num1-CC-yEGFP Pil1-αGFP, referred to as PAN-CC; Figure 1B). Importantly, we found that while PAN-CC clusters can tether mitochondria to the PM, most PAN-CC clusters are not associated with mitochondria (Figure 1, C–F). This is similar to PAN clusters, in which Num1ΔPH-yEGFP is synthetically clustered, but distinct from WT Num1 clusters where the majority of clusters are associated with mitochondria (Figure 1, C–F). There is also an overall increase in the number of mitochondria-associated clusters per cell for PAN and PAN-CC compared with WT Num1. This increase is expected due to the increased number of total PAN and PAN-CC clusters per cell, a result of targeting the Num1 truncations to eisosomes, which are highly abundant on the PM (Figure 1, D and E; Walther et al., 2006; Schmit et al., 2018).
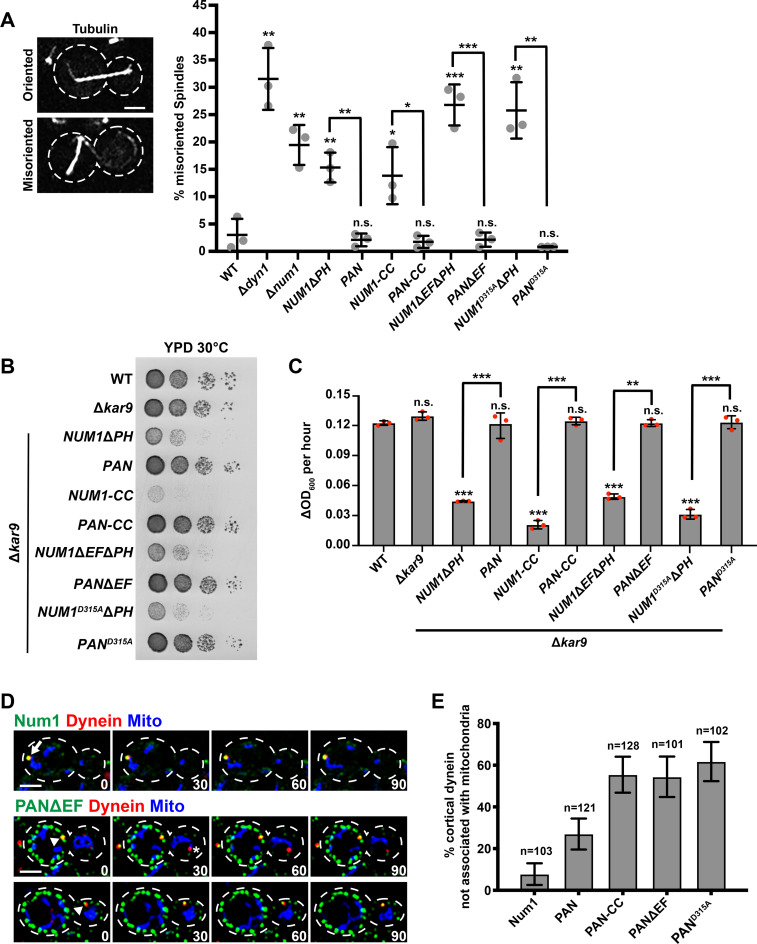
We next asked whether this minimal domain of Num1 could function in the dynein-mediated nuclear inheritance pathway. To test for dynein function, we used a mitotic spindle orientation assay to quantify the percentage of cells with misoriented spindles. Cells in which the dynein pathway is disrupted, such as Δdyn1, Δnum1, and NUM1ΔPH cells, have defects in spindle orientation (Figure 2A; Eshel et al., 1993; Li et al., 1993; Farkasovsky and Küntzel, 1995; Schmit et al., 2018). We found that NUM1-CC cells in the absence of Pil1-αGFP have spindle orientation defects, similar to Δnum1 and NUM1ΔPH cells (Figure 2A). In contrast, the percentage of PAN-CC cells with misoriented spindles was similar to that of WT cells, indicating that dynein is functional. We also examined dynein function by assessing cell growth in the absence of Kar9, a protein required for a partially redundant spindle positioning pathway. In the absence of both the dynein- and Kar9-dependent spindle positioning pathways, cells exhibit a severe growth defect or are inviable (Miller and Rose, 1998). While cells expressing Num1-CC without Pil1-αGFP grew normally in the presence of Kar9 (Supplemental Figure S1D), a severe growth defect was observed in the absence of Kar9, both visually on plates and quantitatively in a microplate reader growth assay (Figure 2, B and C). In contrast, deletion of KAR9 in PAN-CC cells did not affect cell growth (Figure 2, B and C), supporting the conclusion that dynein is functional in PAN-CC cells.
FIGURE 2:
Synthetically clustered Num1 truncations can function in the dynein pathway, but mitochondria and dynein anchoring are uncoupled. (A) WT W303, Δdyn1, Δnum1, NUM1ΔPH, PAN, NUM1-CC, PAN-CC, NUM1ΔEFΔPH, PANΔEF, NUM1D315AΔPH, and PAND315A cells expressing mRuby-Tub1 were visualized by fluorescence microscopy. Representative whole-cell max projections of cells with spindles scored as correctly oriented and misoriented are shown. Large buds with spindles ≥1.75 μm were scored. n = 3 independent experiments of ≥102 spindles for each replicate. Quantification of the percent of cells with misoriented spindles is shown as the mean ± SD with each dot representing an individual replicate for each strain. The cell cortex is outlined with a dashed line. Bars, 2 μm. p values are in comparison to WT W303 except when denoted by brackets. *** p < 0.001, ** p < 0.01, * p < 0.04; n.s., not significant. (B) Tenfold serial dilutions of WT W303, Δkar9, Δkar9 NUM1ΔPH, Δkar9 PAN, Δkar9 NUM1-CC, Δkar9 PAN-CC, Δkar9 NUM1ΔEFΔPH, Δkar9 PANΔEF, Δkar9 NUM1D315AΔPH, and Δkar9 PAND315A were plated on YPD medium and grown at 30°C for 24 h. This is a representative example of four replicates. (C) The change in OD600 per hour for the linear portion of growth curves obtained using a microplate reader for WT, Δkar9, Δkar9 NUM1ΔPH, Δkar9 PAN, Δkar9 NUM1-CC, Δkar9 PAN-CC, Δkar9 NUM1ΔEFΔPH, Δkar9 PANΔEF, Δkar9 NUM1D315AΔPH, and Δkar9 PAND315A. Three individual biological replicates are shown, each represented by a dot. Each biological replicate is the average of three to four technical replicates; mean ± SD is shown. p values are in comparison to WT W303 unless otherwise denoted by brackets. *** p < 0.0001, ** p < 0.0003; n.s., not significant. (D) Representative images of cortical dynein foci associated with mitochondria (top) and not associated with mitochondria (bottom). NUM1 or PANΔEF cells expressing Dyn1-mKate and mito-TagBFP were visualized by fluorescence microscopy over time. A max projection of three consecutive 0.4 μm slices are shown. Time is in seconds. Arrow indicates a colocalization event with mitochondria. Arrowhead indicates a colocalization event without mitochondria. Asterisk indicates a noncortical dynein focus. The cell cortex is outlined with a dashed line. Bar, 2 μm. (E) Quantification of the percent of cortical dynein foci that are not associated with mitochondria for Num1-yEGFP, PAN, PAN-CC, PANΔEF, and PAND315A. n = number of cortical dynein foci. Error bars represent the SE of proportion with a 95% confidence interval. To be considered cortically anchored, a Dyn1-mKate focus had to remain stably associated with a cluster at the cell cortex for ≥1.5 min.
We next examined the ability of PAN-CC clusters to anchor dynein at the cell cortex. We observed a cortical dynein focus in 42.1% of PAN-CC cells (n = 152 cells), which is a dramatic increase in comparison to WT and PAN cells in which cortically anchored dynein was observed in 2.5% and 16.5% of cells, respectively (n = 320 and 284 cells, respectively). PAN cells do exhibit a small increase in the percentage of cells with a cortical dynein, but this is likely due to an increase in total PAN clusters compared with WT Num1 clusters per cell (Figure 1D). Interestingly, in PAN-CC cells, more than 50% of cortical dynein foci were anchored at PAN-CC clusters that are not associated with mitochondria (Figure 2, D and E). This differed from WT and PAN cells, in which the majority of cortical dynein foci are anchored at Num1 and PAN clusters that are associated with mitochondria (Figure 2, D and E; Kraft and Lackner, 2017; Schmit et al., 2018). The difference in cortical dynein anchoring at clusters not associated with mitochondria between PAN and PAN-CC cells was not due to differences in the number of mitochondria-associated clusters per cell (Figure 1, E and F). Thus, in comparison to PAN clusters, the association of dynein with PAN-CC clusters is less biased by the presence of mitochondria.
Synthetically clustering Num1 EFLM mutants reveals a role for the EFLM in the regulation of dynein anchoring
On the basis of the above results, we hypothesized that Num1-CC is missing a regulatory region that biases dynein anchoring at mitochondria-associated Num1 clusters. Directly downstream of the CC domain of Num1 is a predicted EF hand–like motif (EFLM) (Figure 1B). The EFLM of Num1 has a 12-residue loop nearly identical to canonical Ca2+-binding domains. However, the regions flanking the 12-residue loop differ from canonical EF hand domains with helix-loop-helix structures (Kormanec et al., 1991; Grabarek, 2006; Gifford et al., 2007). Given that EF hands often serve regulatory functions, we hypothesized that the EFLM plays a role in regulating the association of dynein with Num1 clusters. To test this idea, we deleted as well as mutated the EFLM in our synthetic clustering system. The final residue of canonical calcium coordinating EF hand loops is invariably a glutamate or aspartate (Grabarek, 2006; Gifford et al., 2007). Consistently, the Num1 EFLM harbors an aspartic acid at the final loop position (Figure 1B), so we mutated this highly conserved residue to alanine (D315A) to disrupt the putative small metal binding pocket of the EFLM. Thus, we created PANΔEF (Num1ΔEFΔPH-yEGFP Pil1-αGFP) and PAND315A (Num1D315AΔPH-yEGFP Pil1-αGFP) cells (Figure 1B).
We examined how PANΔEF and PAND315A affected the activities of Num1 in mitochondria tethering as well as dynein anchoring and function. We observed that PANΔEF and PAND315A clusters are able to tether mitochondria to the cell cortex (Figure 1C) and that the numbers of total and mitochondria-associated Num1 clusters in PANΔEF and PAND315A cells are similar to those of PAN and PAN-CC cells (Figure 1, D–F). We also find that PANΔEF and PAND315A cells do not have spindle orientation defects or a growth defect in the absence of KAR9, indicating that dynein is functional in the synthetically clustered EFLM mutant cells (Figure 2, A–C).
Surprisingly, in comparison to PAN clusters, we observed that >50% of cortical dynein foci were anchored at PANΔEF and PAND315A clusters that were not associated with mitochondria, similar to PAN-CC (Figure 2, D and E). We observed cortical dynein anchoring events in 15.8% and 20.3% of PANΔEF and PAND315A cells, respectively (n = 558 and 295, respectively), similar to PAN cells (16.5%, n = 284). While Num1 protein levels for Num1-CC in PAN-CC cells was approximately five times greater than the Num1 protein levels in PAN cells, the Num1 protein levels in PANΔEF and PAND315A cells were comparable to the Num1 protein levels in PAN cells (Supplemental Figure S1, A and B). Thus, the increase in cortical dynein anchored at clusters without mitochondria in PAN-CC, PANΔEF, and PAND315A cells cannot be attributed to changes in Num1 protein levels alone. Together, the data indicate that, by disrupting the EFLM in our synthetic clustering system, the bias of anchoring dynein on mitochondria-associated clusters is reduced. These results support the hypothesis that the EFLM is important for the regulation of mitochondria-biased dynein anchoring.
The Num1 EFLM is a bona fide calcium-binding EF hand
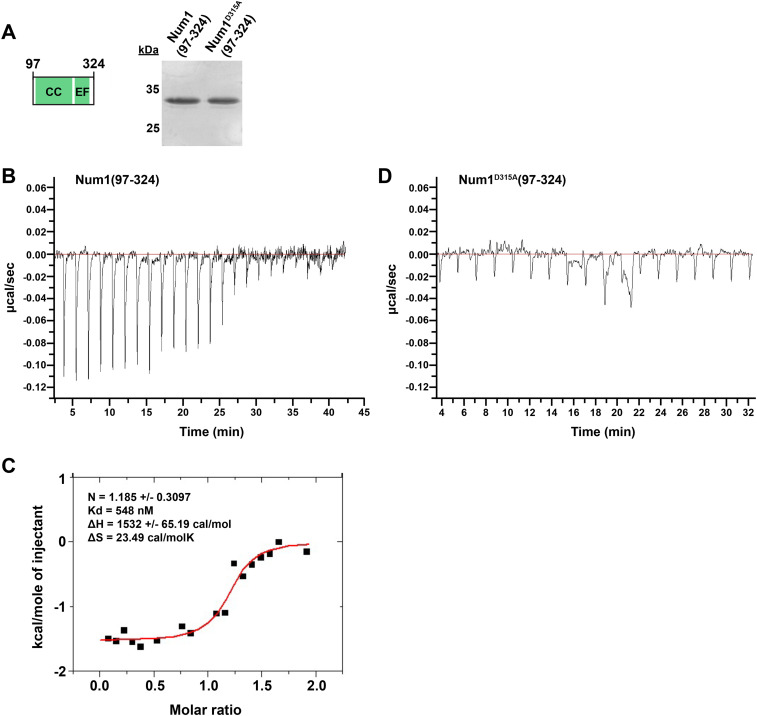
Canonical EF hand motifs are known to modulate regulatory functions often through calcium binding; however, it is currently unknown whether the Num1 EFLM can bind calcium. To test whether the Num1 EFLM can bind calcium, we purified Num1(97-324) and Num1D315A(97-324), which harbors a mutation in the highly conserved terminal EFLM residue (D315A), to disrupt the putative small metal binding pocket of the EFLM (Figures 1B and 3A and Supplemental Figure S2A; Grabarek, 2006; Gifford et al., 2007). We examined their ability to bind Ca2+ using isothermal titration calorimetry (ITC). ITC experiments with Num1(97-324) indicated that the protein binds Ca2+ with an affinity of 548 nM (Figure 3, B and C), consistent with the reported binding affinities for EF hands (Yap et al., 1999). To obtain reasonable enthalpy measurements, Num1(97-324) required treatment with the Ca2+ chelator ethylene glycol tetraacetic acid (EGTA) before ITC, suggesting that the protein is bound to Ca2+ during purification (Supplemental Figure S2C). ITC experiments with Num1D315A(97-324) under buffer conditions identical to those used for Num1(97-324) did not show any detectable Ca2+ binding by the mutant protein (Figure 3D). Together, these data indicate that the Num1 EFLM is a bona fide Ca2+-binding EF hand and that Ca2+ binding is disrupted by the D315A mutation.
FIGURE 3:
The Num1 EFLM is a bona fide Ca2+-binding domain. (A) Schematic representation of the protein construct Num1(97-324) used for ITC. Silver-stained 15% SDS–PAGE gel showing purified Num1(97-324) and Num1D315A(97-324). The 25- and 35-kD markers are indicated on the left; the predicted molecular mass of both proteins is 29 kD. (B–D) Raw ITC data for titration of 50 µM Num1(97-324) (B) or Num1D315A(97-324) (D) with 750 µM CaCl2 at 25°C in buffer containing 20 mM tricine, pH 8.7, 50 mM NaCl. Proteins were pretreated with EGTA, and the EGTA was removed before ITC. The ITC data for Num1(97-324) were also integrated, and the heat of reaction plotted vs. the molar ratio of the injectant at each injection point is shown (C). N = reaction stoichiometry, Kd = binding constant, ΔH = enthalpy, ΔS = entropy. See Supplemental Figure S2B for buffer control ITC experiment.
Synthetically clustered Num1 EFLM mutants rescue dynein function in the absence of mitochondrial inheritance
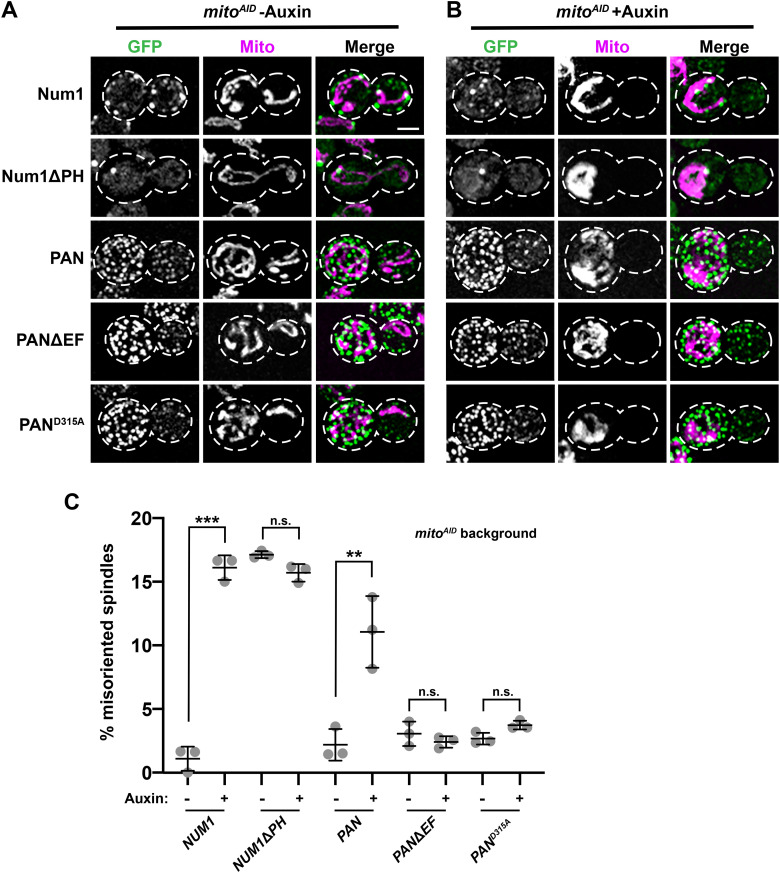
To further explore the role that the EFLM of Num1 and its ability to bind Ca2+ plays in the integration of the mitochondria tethering and dynein anchoring, we examined how the inhibition of mitochondrial inheritance affects the ability of PANΔEF and PAND315A clusters to support dynein function. We have previously described a system to conditionally inhibit mitochondrial inheritance called mitoAID (Kraft and Lackner, 2017). This strain lacks Ypt11, one of the two adaptors required for Myo2-driven transport of mitochondria to the bud, and expresses the other adaptor, Mmr1, as a fusion with an auxin-inducible degron tag (AID; Itoh et al., 2002, 2004; Boldogh et al., 2004; Frederick et al., 2008; Nishimura et al., 2009; Eves et al., 2012; Chernyakov et al., 2013; Lewandowska et al., 2013). Thus, in the presence of auxin, mitochondrial transport into buds is inhibited in mitoAID cells (compare Figure 4, A and B). This allows us to examine the impact mitochondrial inheritance has on dynein function by examining spindle orientation in mitoAID cells grown in the absence and presence of auxin (Figure 4).
FIGURE 4:
Synthetically clustered Num1 EFLM mutants rescue defects in dynein function observed in the absence of mitochondrial inheritance. (A, B) Representative fluorescence microscopy images of mitoAID cells expressing Num1-yEGFP, Num1ΔPH-yEGFP, PAN, PAN-CC, PANΔEF, and PAND315A and mito-TagBFP (magenta) grown in the absence (A) and presence (B) of 1 mM auxin. Whole-cell max projections are shown. The cell cortex is outlined with a dashed white line. Bar, 2 µm. (C) Quantification of misoriented spindles for mitoAID cells expressing Num1, PAN, PANΔEF, or PAND315A (as described in Figure 2A) in the absence and presence of auxin, as indicated under the graph. Large buds with spindles >1.25 μm were scored. n ≥ 292 spindles from three independent experiments of ≥78 spindles per replicate. Quantification of the percent of cells with misoriented spindles is shown as the mean ± SD with each dot representing an individual replicate. p value comparisons are designated by brackets. *** p < 0.0001, ** p < 0.001; n.s., not significant.
Using mitoAID, we have previously shown that when mitochondrial inheritance is disrupted following the addition of auxin, WT Num1 clusters do not form in buds and dynein function in spindle orientation is disrupted (Figure 4, A–C; Kraft and Lackner, 2017). As the formation of PAN clusters does not depend on mitochondria, PAN clusters are still present in buds when mitochondrial inheritance is inhibited. However, dynein function in spindle orientation is compromised in these cells, as previously described (Figure 4, A–C; Schmit et al., 2018). Given that we observed an increase in dynein anchoring at clusters not associated with mitochondria in cells expressing the synthetically clustered EFLM mutants (Figure 2E), we examined spindle orientation in these cells when mitochondrial inheritance is inhibited. We observed that PANΔEF and PAND315A clusters are able to form in the absence of mitochondrial inheritance, similar to PAN clusters (Figure 4, A and B). Interestingly, in contrast to PAN cells, in which spindle misorientation increases following treatment with auxin, the addition of auxin had no effect on spindle orientation in PANΔEF and PAND315A cells (Figure 4C). These data indicate that the defects in dynein function observed in the absence of mitochondrial inheritance are rescued by the synthetically clustered EFLM mutants.
Disrupting the EFLM in full-length Num1 does not affect mitochondria tethering, dynein anchoring, or ER association
Previous structure-function studies have not been able to clearly define the function of the EFLM in full-length Num1. While the overexpression of Num1 lacking the EFLM and all but one repeat does not rescue the nuclear segregation defect observed in NUM1 deficient cells (Farkasovsky and Küntzel, 1995), dynein function does not appear to be affected in cells expressing a Num1 mutant that lacks only the EFLM in an otherwise WT background (Tang et al., 2012). Interestingly, results obtained using our synthetic clustering system reveal a potential role for the EFLM in the regulation of dynein function. To better understand the role of Num1’s EFLM in a more native state, we disrupted the EFLM by either deleting the EFLM from or introducing the D315A mutation into full-length Num1-yEGFP in an otherwise WT background (NUM1ΔEF and NUM1D315A). The steady-state protein levels of the EFLM mutants were similar to that of WT Num1 (Supplemental Figure S1, A and C).
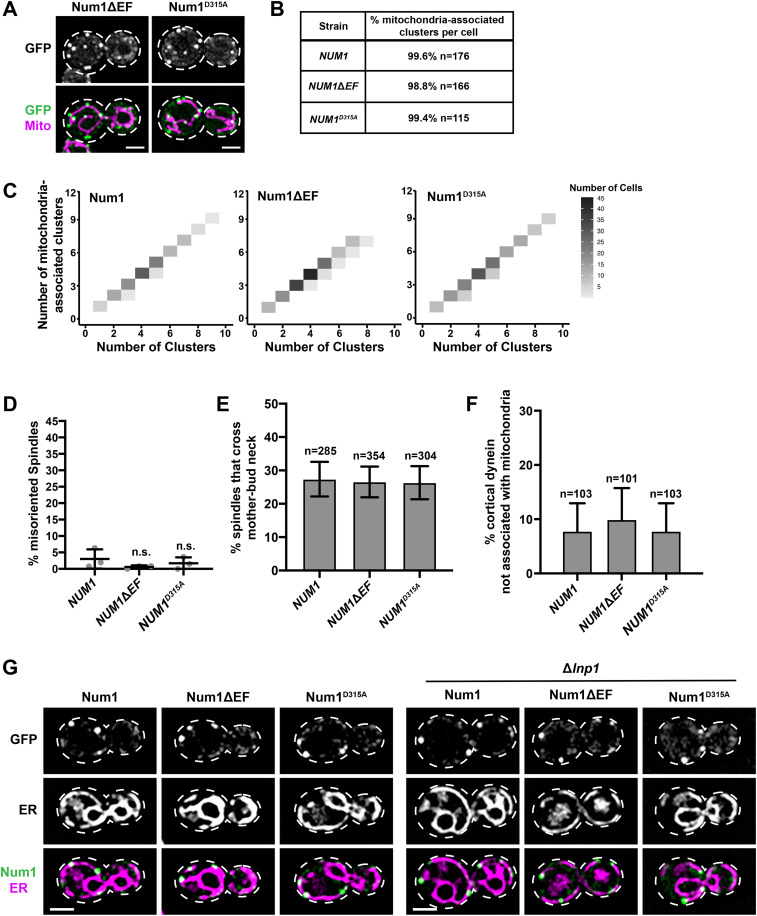
To characterize the EFLM mutants, we first examined the relationship between Num1 clusters and mitochondria. We observed no obvious phenotypic differences between Num1ΔEF and Num1D315A clusters when compared with WT clusters (Figures 1C and 5A). We found that the percentage of mitochondria-associated clusters per cell in cells expressing Num1ΔEF and Num1D315A was similar to that of cells expressing WT Num1 (Figure 5, A and B). When the number of total clusters versus mitochondria-associated clusters was plotted, an almost exclusively 1-to-1 relationship was observed for all three strains (Figure 5C). Thus, similar to what has been observed for WT Num1 (Kraft and Lackner, 2017), the vast majority of Num1ΔEF and Num1D315A clusters are mitochondria associated.
FIGURE 5:
Characterization of clusters, tethers, dynein function, and ER association for full-length Num1 EFLM mutants. (A) Representative images of cells expressing Num1ΔEF-yEGFP and Num1D315A-yEGFP with mito-Red. Whole-cell max projections are shown. The cell cortex is outlined with a dashed white line. Bar, 2 µm. (B) The table describes the average percentage of Num1ΔEF-yEGFP and Num1D315A-yEGFP clusters that are mitochondria-associated per cell. To be scored as a mitochondria-associated cluster, mitochondria had to remain associated with the cluster for ≥1.5 min. n = number of cells. (C) The number of mitochondria-associated clusters per cell was plotted against the total number of clusters per cell for NUM1 (n = 176), NUM1ΔEF (n = 166), and NUM1D315A (n = 115) cells. The color of each rectangle represents the number of cells, as indicated by the key (right). (D) Quantification of misoriented spindles (as described in Figure 2A) for NUM1 (replicated from Figure 2A), NUM1ΔEF, and NUM1D315A cells. Large buds with spindles ≥1.75 μm were scored. n = 3 independent experiments of ≥102 spindles per replicate. Quantification of the percent of cells with misoriented spindles is shown as the mean ± SD with each dot representing an individual replicate for each strain. n.s. = not significant in comparison to NUM1. (E) Quantification of the percentage of HU-arrested spindles that cross the M-B neck in a 10-min-time-lapse movie for NUM1, NUM1ΔEF, and NUM1D315A cells. n = number of cells. SE of the proportion with 95% confidence interval is shown. (F) Quantification of the percent of cortical dynein foci that do not colocalize with mitochondria at Num1, Num1ΔEF, and Num1D315A clusters. n = number of cortical dynein foci. Error bars represent the SE of proportion with a 95% confidence interval. To be considered cortically anchored, a Dyn1-mKate focus had to remain stably associated with a cluster at the cell cortex for ≥1.5 min. (G) Representative images of cells expressing Num1-yEGFP, Num1ΔEF-yEGFP, and Num1D315A-yEGFP with Pho88-mCherry (ER marker) in the presence (left) or absence (right) of Lnp1. Single slices are shown. The cell cortex is outlined with a dashed white line. Bar, 2 µm.
Next, we examined the ability of Num1ΔEF and Num1D315A to support dynein function. We found that NUM1ΔEF and NUM1D315A cells have no growth defects in the absence of KAR9 (Supplemental Figure S3, A and B). In addition, no defects in spindle orientation in NUM1ΔEF and NUM1D315A cells were observed (Figure 5D). We also examined dynein function using a spindle oscillation assay, which more directly examines the pulling force that dynein exerts on the mitotic spindle. For this assay, cells are arrested in a large budded preanaphase state using hydroxyurea (HU; Moore et al., 2009; Tang et al., 2012). Spindles are visualized using fluorescent tubulin, and time-lapse movies are taken to track the spindles over time. We found that the percentages of spindles that cross the mother-bud (M-B) neck were similar in WT, NUM1ΔEF, and NUM1D315A celIs (Figure 5E). Together, these results indicate that Num1ΔEF and Num1D315A are able to support dynein function, consistent with a previous result demonstrating that NUM1ΔEF cells do not exhibit defects in nuclear segregation (Tang et al., 2012). We also found that only a very small percentage of cortical dynein anchoring events observed in WT, NUM1ΔEF, and NUM1D315A cells occurred at Num1 clusters that are not associated with mitochondria (Figure 5F), which is expected given that the vast majority (>98%) of Num1 clusters in all three strains are associated with mitochondria (Figure 5, A–C).
We also examined the relationship between full-length Num1 EFLM mutant clusters and the ER, which is a component of MECA. Interestingly, the ER can locally concentrate calcium levels (Cunningham, 2011). Thus, it is possible that calcium binding by the EFLM could be regulated by association with the ER. Additionally, the EFLM region of Num1 contains a putative FFAT motif, composed of amino acids 306–330 (Figure 1B), which has been suggested to mediate the interaction between Num1 and the integral ER protein Scs2 (Chao et al., 2014). However, our previous work indicates that versions of Num1 that lack the EFLM are still able to associate with the ER (Lackner et al., 2013), and there is debate about whether amino acids 306–330 compose a bona fide FFAT motif (Chao et al., 2014; Murphy and Levine, 2016). To determine whether NUM1ΔEF and NUM1D315A cells have a defect in the association of Num1 with the ER resulting from disruption of the putative FFAT motif, we examined the localization of Num1ΔEF and Num1D315A relative to the ER (Figure 5G). Similar to Num1 clusters, Num1ΔEF and Num1D315A clusters localized to regions of the cell cortex that are occupied by cortical ER (Figure 5G, left). Because the cortical ER covers a significant portion of the cell cortex, we also examined Num1-ER localization in Δlnp1 cells, in which cortical ER localization is disrupted and large regions of the cortex devoid of cortical ER are produced (Chen et al., 2012). In this background, Num1 association with the ER should be more apparent. Indeed, the ER association for WT Num1, Num1ΔEF, and Num1D315A clusters was highly evident in the Δlnp1 background (Figure 5G, right). It has also been shown in previous studies that when the Num1-ER association is disrupted through deletion of Scs2, Num1 clustering is affected (Chao et al., 2014; Omer et al., 2018). However, we observe no obvious clustering defects for Num1ΔEF and Num1D315A (Figure 5A), further suggesting that the EFLM likely does not interact with the ER through Scs2. These results indicate that the deletion of the EFLM or introduction of the D315A mutation does not interfere with the association of Num1 clusters with the ER.
Disrupting the EFLM in full-length Num1 can rescue dynein function in the absence of mitochondrial inheritance
Given that the synthetically clustered EFLM mutants were able to rescue defects in dynein function observed in the absence of mitochondrial inheritance, we next examined how the inhibition of mitochondrial inheritance affected the ability of full-length Num1ΔEF and Num1D315A to support dynein-mediated spindle orientation. In contrast to NUM1 mitoAID cells, in which defects in spindle orientation are observed in the absence of mitochondrial inheritance (+auxin), the addition of auxin and consequent inhibition of mitochondrial inheritance had no effect on spindle orientation in NUM1ΔEF mitoAID and NUM1D315A mitoAID cells (Figure 6A). This suggests that dynein function is not compromised in the absence of mitochondrial inheritance when the EFLM is disrupted in an otherwise WT NUM1.
FIGURE 6:
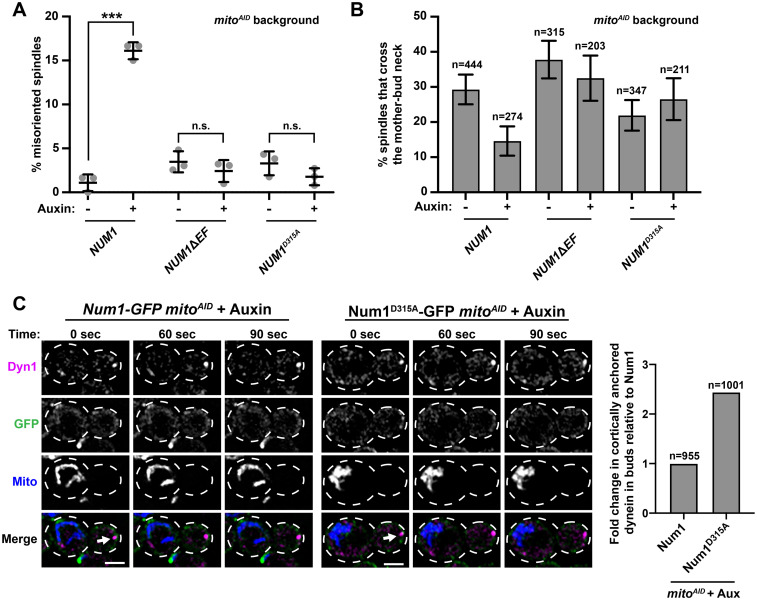
Num1 EFLM mutants rescue defects in dynein function observed in the absence of mitochondrial inheritance. (A) Quantification of misoriented spindles (as described in Figure 2A) for NUM1 (replicated from Figure 4C), NUM1ΔEF, and NUM1D315A cells in the presence and absence of auxin as indicated under the graph. Large buds with spindles ≥1.25 μm were scored. n = 3 independent experiments of ≥78 spindles per replicate. Quantification of the percent of cells with misoriented spindles is shown as the mean ± SD with each dot representing individual replicates for each strain. p value comparisons are designated by brackets. *** p < 0.0001; n.s., not significant. (B) Quantification of the percentage of HU-arrested spindles that crossed the M-B neck in the absence and presence of auxin, as indicate under the graph, during a 10 min-time-lapse movie for NUM1, NUM1ΔEF, and NUM1D315A cells. n = number of spindles. SE of the proportion with 95% confidence interval is shown. (C) Examples of bud localized cortical dynein in the absence of mitochondrial inheritance for Num1 mitoAID (left) and Num1D315A mitoAID (right) cells expressing Dyn1-yoHalo and MitoBlue (+auxin). A max projection of three consecutive 0.4 μm slices is shown. Time is in seconds. Arrow indicates cortical dynein in the absence of mitochondria. The cell cortex is outlined with a dashed line. Bar, 2 μm. The graph displays the fold change in cortically anchored dynein in buds that lack mitochondria compared with Num1 for Num1 mitoAID and Num1D315A mitoAID cells in the presence of auxin. n = number of cells.
To further study dynein function in the EFLM mutants, we also examined spindle oscillation in NUM1 mitoAID, NUM1ΔEF mitoAID, and NUM1D315A mitoAID cells. In NUM1 mitoAID cells in the absence of auxin, approximately 29.3% of spindles crossed the M-B neck (Figure 6B), which is comparable to what is observed in WT cells (Figure 5E, 27.3%). When mitochondrial inheritance was inhibited by the addition of auxin in NUM1 mitoAID cells, the percentage of spindles that crossed the M-B neck was dramatically reduced to 14.6% (Figure 6B), consistent with previous studies (Kraft and Lackner, 2017). Spindle oscillation for NUM1ΔEF and NUM1D315A mitoAID cells in the absence of auxin was similar to that for NUM1 mitoAID, despite subtle deviations (Figure 6B). However, in contrast to NUM1 mitoAID cells, a significant reduction in spindle oscillation was not observed in NUM1ΔEF mitoAID and NUM1D315A mitoAID cells when mitochondrial inheritance was inhibited by the addition of auxin (Figure 6B). To confirm that the spindle oscillations we observed in all conditions were dynein-dependent, we quantified spindle oscillations in NUM1 mitoAID, NUM1ΔEF mitoAID, and NUM1D315A mitoAID cells in the absence of Dyn1. In the Δdyn1 background, the percentage of cells in which spindle oscillation was observed was extremely low for all strains examined (Supplemental Figure S3C). Together, these results indicate that when the EFLM of Num1 is disrupted, dynein function is no longer impacted by the absence of mitochondria.
The observation that dynein is functional for spindle orientation and oscillation in the EFLM mutant strains when mitochondrial inheritance is disrupted suggests that dynein is anchored in the bud in the absence of mitochondria-assembled Num1 clusters. To address this hypothesis, we simultaneously imaged dynein, Num1, and mitochondria in both WT Num1 and Num1D315A mitoAID cells. While the vast majority of cortically anchored dynein foci are found in the mother cell and are rarely observed in buds (Lee et al. 2005), we were able to capture 10 examples of cortical dynein anchoring in the bud of Num1D315A mitoAID cells treated with auxin to inhibit mitochondrial inheritance (Figure 6C, ∼1% of budded cells, n = 1001). When we imaged a similar number of cells for WT Num1 mitoAID cells, we observed only four examples (Figure 6C, ∼0.42% of budded cells, n = 955). This is equivalent to a 2.5-fold increase in bud cortical dynein anchoring in the absence of mitochondrial inheritance for Num1D315A compared with WT Num1 (Figure 6C). The Num1 assemblies present at the sites of cortical dynein anchoring were smaller and more diffuse than mitochondria-assembled Num1 clusters, consistent with previous descriptions of dynein anchoring by Num1 in the absence of mitochondria (Omer et al., 2018, 2020). The fact that we were able to capture more examples of cortically bud anchored dynein for Num1D315A cells than WT cells is consistent with our findings that we observe properly oriented spindles as well as WT-level spindle oscillation in cells expressing Num1D315A in the absence of mitochondria (Figure 6, A and B). These data indicate that in certain mutant conditions, such as disruption of the Num1 EFLM (as shown here), overexpression of Mdm36 (Omer et al., 2020), or deletion of Scs2 (Omer et al., 2018), the non–mitochondria associated pool of the Num1 can anchor dynein and support dynein function. However, in a WT system, it is clear that dynein anchoring by Num1 is biased by the presence of mitochondria and this bias is dependent on the ability of the EFLM of Num1 to bind Ca2+.
DISCUSSION
By leveraging two molecular tools that we previously developed—our synthetic Num1 clustering system and conditional mitochondrial inheritance strain—we were able to reveal a novel role for the EFLM of Num1 in the regulation of dynein function, which was masked in previous structure-function studies (Tang et al., 2012; Kraft and Lackner, 2017; Schmit et al., 2018). We have previously shown that the vast majority of Num1 clusters in WT cells are mitochondria-associated and these clusters serve as cortical anchor sites for dynein (Figure 5, B and C; Kraft and Lackner, 2017). Based on these findings, there are two questions we wanted to address: 1) whether mitochondria influence dynein activity downstream of anchoring at Num1, and 2) why mitochondria tethering is functionally linked to a nuclear inheritance pathway. However, it was not possible to address these outstanding questions without being able to uncouple Num1 clustering from an interaction with mitochondria as well as dynein anchoring from mitochondria tethering. Using our synthetic clustering system to bypass the requirement of mitochondria for Num1 clustering, we found that the EFLM is required to bias dynein anchoring on Num1 clusters that are mitochondria associated. We then used EFLM mutants, which uncouple dynein anchoring from mitochondria tethering, in combination with our conditional mitochondrial inheritance strain to address whether mitochondria impact the activity of dynein after it is anchored. Importantly, we found that mitochondria do not appear to affect dynein activity downstream of dynein anchoring. Thus, mitochondria do not directly activate dynein activity or processivity per se, but instead help to arrange Num1 in clusters at the cell cortex that are competent to anchor dynein, and subsequently dynein is activated. As shown here in this study with the Num1 EFLM mutants and as is suggested by other studies that alter Num1 assembly (Omer et al., 2018, 2020), it is possible to bypass the role of mitochondria in facilitating dynein anchoring in certain mutant conditions. However, in WT conditions, it is clear that the presence of mitochondria in the bud positively impacts dynein function in nuclear inheritance and this is dependent on a functional EFLM.
Our findings suggest a direct role for the Num1 EFLM in mitochondrial-dependent dynein anchoring. Specifically, we find that, in the presence of a functional EFLM, Num1-dependent anchoring of dynein is heavily weighted toward Num1 clusters that are associated with mitochondria and this bias is reduced when the EFLM is disrupted. Consistently, when the EFLM is disrupted, we do not observe defects in dynein-dependent spindle orientation and oscillation in the absence of mitochondrial inheritance. These results suggest that the EFLM negatively impacts the association of dynein with Num1 before the association of Num1 with mitochondria and that the Num1-mitochondria association relieves the negative effect of the EFLM. At this point, we cannot rule out an alternative explanation in which disrupting the EFLM results in a gain of function phenotype that promotes dynein anchoring by Num1 proteins that are not associated with mitochondria. However, in either scenario, the presence of a functional EFLM serves to couple dynein-mediated nuclear inheritance to mitochondrial inheritance.
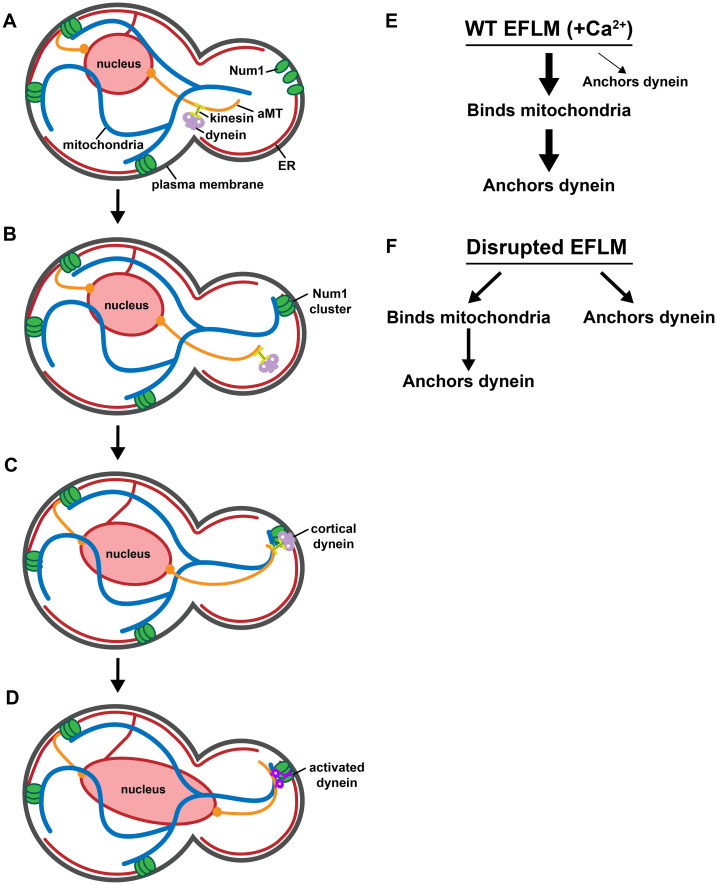
We hypothesize that the coupling of inheritance pathways provides a spatiotemporal mechanism of organelle inheritance that favors mitochondrial inheritance before that of the nucleus. In our working model, Num1 exists in an unassembled state on the PM in small buds before mitochondrial inheritance (Figure 7A). As the cell cycle progresses, mitochondria are transported to the bud and interact with unassembled Num1. This interaction drives the assembly of Num1 into stable, ER-associated, cortical clusters that persistently tether mitochondria to the PM (Figure 7B). Mitochondrial-driven assembly of Num1 is likely enhanced by Mdm36 (not shown in the figure for simplicity) and additional factors that have yet to be characterized. The negative impact that the EFLM has on dynein anchoring by Num1 is relieved in the mitochondria-assembled and -associated clusters, and the clusters are competent to anchor dynein. Dynein, which is trafficked by the kinesin Kip2 in an inhibited state to the minus end of an aMT (Carvalho et al., 2004; Markus and Lee, 2011), is then offloaded onto a Num1 cluster (Figure 7C). Once anchored, dynein is activated and subsequently captures and walks along the aMT to facilitate the orientation of the mitotic spindle and its migration into the bud (Figure 7D). This sequential coupling of mitochondrial and nuclear positioning pathways is dependent on the EFLM of Num1 and its ability to bind Ca2+ (Figure 7E). Disrupting the EFLM uncouples the mitochondria tethering and dynein anchoring activities of Num1 (Figure 7F), which we speculate will result in an altered timing of organelle inheritance and a fitness cost to the cells. We are working to explore this exciting possibility.
FIGURE 7:
The Num1 EFLM coordinates the integration of the mitochondrial and nuclear positioning pathways. (A) Mitochondria are inherited early in the cell cycle at the small bud stage. (B) The assembly of Num1 clusters is triggered by interaction with mitochondria, and the assembled Num1 clusters then serve to tether mitochondria to the cell cortex. (C) Dynein, in an inhibited state, bound to kinesin is localized to the plus ends of aMTs by kinesin and is offloaded onto mitochondria-assembled Num1 clusters. (D) Dynein is activated and subsequently captures and walks on aMTs to facilitate the orientation and migration of the mitotic spindle. (E) In WT cells, in which Num1 has an intact EFLM that can bind calcium, Num1 binds mitochondria, which drives the formation of Num1 clusters that persistently tether mitochondria. Dynein can then be offloaded on and anchored by these Num1 clusters. While it is possible for dynein to be anchored by Num1 clusters not associated with mitochondria, this is rarely observed. Thicker arrows indicate bias toward a pathway. (F) When the EFLM is disrupted, dynein anchoring is no longer biased toward mitochondria-assembled and -associated Num1 clusters. Dynein anchoring can still occur at mitochondria-assembled Num1 clusters but also at smaller Num1 assemblies that are not associated with mitochondria.
The question remains of why the cell would want to use Num1 and MECA, the mitochondrial membrane contact site it forms, to couple nuclear inheritance with mitochondrial inheritance. While nuclear inheritance checkpoint mechanisms are well characterized at the molecular level (Amon, 1999; Matellán and Monje-Casas, 2020), the existence of a mitochondrial inheritance checkpoint is a matter of debate. Interestingly, cells lacking Mdm10 or Mmm1, components of the ER-mitochondria tether ERMES (Kornmann et al., 2009), have defects in mitochondrial inheritance that have been linked to the inhibition of cytokinesis (García-Rodríguez et al., 2009). These data suggest that cell cycle progression could be linked to mitochondrial inheritance; however, the mechanism by which the absence of mitochondria in the bud is sensed is not known. In contrast, when Myo2-dependent inheritance of mitochondria into buds is inhibited, cytokinesis is able to progress, producing daughters that lack mitochondria and eventually die (Chernyakov et al., 2013). Our work raises the interesting possibility that mitochondria-dependent dynein anchoring may function as a mitochondrial inheritance monitoring/surveillance system by coupling the spatiotemporal regulation of dynein anchoring and, consequently, function with the inheritance of mitochondria (Figure 7). In this proposed surveillance system, Num1 senses mitochondria in the bud via a direct interaction with the organelle. This interaction then triggers Num1 assembly into clusters that are competent for dynein anchoring, which serves as the functional readout of mitochondrial inheritance.
Our biochemical data indicate that the Num1 EFLM is a bona fide Ca2+-binding EF hand. Ca2+ binding by EF hands often serves a regulatory role, allowing a protein’s function to be integrated with cellular physiology. Past studies have suggested that Num1 could act as an activator of dynein (Lammers and Markus, 2015). Interestingly, human dynein requires adaptor proteins for activity (McKenney et al., 2014; Schlager et al., 2014), and the dynein adaptors CRACR2a (calcium release–activated calcium channel regulator 2A), Rab45 (Ras-related protein Rab-45), FIP3 (Rab11 family–interacting protein 3), and NIN (Ninein) contain EF hand motifs (Reck-Peterson et al., 2018; Lee et al., 2020). CRACR2a, which contains both EF hand motifs and a CC domain, has been shown to activate dynein in vitro in a calcium-dependent manner (Wang et al., 2019; Lee et al., 2020). Our data suggest that in mitotically growing cells, the EFLM of Num1 is likely in a Ca2+-bound state, as we observed a bias in dynein anchoring on Num1 clusters that are mitochondria associated in WT and PAN cells that is lost in PAND315A cells (Figure 2E). Therefore, the EFLM and Ca2+ may play more of a structural role within a Num1 cluster in mitotically growing cells (Zhou et al., 2006). In many EF hand–containing proteins, the EF hand motifs occur in pairs. However, proteins that contain a single EF hand often oligomerize, with oligomerization being important for function (Zhou et al., 2006; Gifford et al., 2007). This is the case for Num1, which likely exists as a dimer in the unassembled state and a cluster of ∼31 molecules in the assembled state (Ping et al., 2016; Omer et al., 2020; Won et al., 2021). Whether all or just a subset of the EFLMs in a Num1 cluster need to be Ca2+-bound to weight dynein anchoring toward mitochondria-associated clusters is unclear at this point. There are likely conditions in which the EFLM is not occupied by Ca2+ and therefore mitochondria anchoring and dynein tethering by Num1 clusters would be uncoupled. However, what these conditions may be and what function uncoupling these Num1 activities may serve are at this point unclear.
In addition to positioning mitochondria and the nucleus, Num1 is associated with a third organelle, the ER (Figure 5G; Lackner et al., 2013; Chao et al., 2014; Omer et al., 2018). Thus, the tripartite membrane contact site formed by Num1 could be used to functionally link the inheritance of three essential organelles, the ER, mitochondria, and the nucleus. In this scenario, Num1 would act as a sensor for both ER and mitochondrial inheritance, forming clusters capable of anchoring dynein only in the presence of both organelles. While mitochondria do not serve as calcium stores in yeast, the ER concentrates calcium ∼100-fold relative to the cytoplasm (Cunningham, 2011), which ranges from 50 to 200 nM (Cui et al., 2009). Given that Num1 associates with the cortical ER, it is possible that Ca2+ release from the ER results in a higher ER-proximal concentration of Ca2+ that may help maintain the EFLM in a Ca2+-bound state. Interestingly, the Schizosacharomyces pombe homologue of Num1 lacks an EFLM and does not associate with the ER, raising the possibility that the presence of the EFLM and the association of Num1 with the ER are functionally linked (Saito et al., 2006; Yamashita and Yamamoto, 2006; Kraft and Lackner, 2019). The role for the ER in the formation and function of Num1 clusters is an area of active investigation.
Interestingly, in both yeast and higher eukaryotes the inheritance of mitochondria has been linked to the functional state of the organelle. In stem cells, older mitochondria are partitioned to the cell destined to differentiate, while the cell that maintains stem cell properties retains younger and more functional mitochondria (Katajisto et al., 2015; Wu et al., 2019). Similarly in budding yeast, the more functional, or fit, mitochondria are inherited by the daughter cell, while the mother cell retains less-fit mitochondria (Klinger et al., 2010; McFaline-Figueroa et al., 2011; Higuchi et al., 2013; Pernice et al., 2016). Interestingly, spindle pole body inheritance has been linked to the inheritance of the fittest mitochondria, suggesting that mitochondrial and nuclear inheritance pathways may impact one another in multiple ways (Manzano-López et al., 2019). While Num1 is not conserved at the protein level, similar mechanisms of interdependent organelle positioning are likely present in other cell types, especially those that divide asymmetrically. Given the physical and functional associations between Num1 and mitochondria, the ER, and the nucleus, we speculate that Num1 functions as a cortical hub, integrating temporal, spatial, and functional cues to promote and regulate organization within cells.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Strains and plasmids
Supplemental Tables 1 and 2 list all primers and strains, respectively, used in this study. W303 Num1∆EF∆PH-GFP-yEGFP::HIS was obtained by PCR-based targeted homologous recombination with two PCR products (pLL#500 template with primers LL#513/940 and pLL#54 with primers LL#259/178) into W303 Δnum1::KAN cells. W303 Num1D315A∆PH-GFP-yEGFP::KAN was obtained by PCR-based targeted homologous recombination with two PCR products (pLL#501 with primers LL#513/940 and pLL#55 with primers LL#259/178) into W303 Δnum1::HIS cells. W303 Num1∆EF-yEGFP::HIS was obtained by PCR-based targeted homologous recombination with two PCR products (pLL#500 with primers LL#513/514 and pLL#54 with primers LL#177/178) into W303 Δnum1::KAN cells. W303 Num1D315A-yEGFP::HIS was obtained by PCR-based targeted homologous recombination with two PCR products (pLL#501 template with primers LL#513/514 and pLL#54 with primers LL#177/178) into W303 Δnum1::KAN cells. W303 Δlnp1::KAN was obtained by replacing the complete open reading frame of the gene by the indicated cassette using PCR-based targeted homologous recombination. All other strains were obtained through crossing, sporulation, and tetrad analysis or through transformations with plasmids.
The following plasmids were previously described: pXY142-mitodsRED (mito-RED::LEU; Friedman et al., 2011); p414-GalL::yEGFP-NUM1 (pLL#336; Lackner et al., 2013); pET22b modT7prom::His6-T7 (Ping et al., 2016); pFA6a-link-yEGFP::SpHIS5 (pKT128 pLL#54) and pFA6a-link-yEGFP-KAN (pKT127 pLL#55; Sheff and Thorn, 2004); pFA6-link-LaG16-CaURA3 (αGFP; Schmit et al., 2018); and pFA6-link-mKate-spHIS5 and pHIS3p:mRuby2-Tub1+3′UTR::LEU (mRuby-Tub1::LEU; Kraft and Lackner, 2017). mRuby-Tub1::LEU was digested with XhoI before transformation into yeast. pHIS3p:mRuby2-Tub1 + 3′UTR::HPH (mRuby-Tub1::HYG; Addgene plasmids #50,633) was a gift from W. Lee, University of Massachusetts Amherst (Markus et al., 2015). Ruby-Tub1::HYG was digested with XbaI before transformation into yeast. pNHK53 (YIp, ADH1::OsTIR1-9Myc1; TIR1) was obtained from the National BioResource Project, Japan (depositor: M. Kanemaki, Osaka University). pRS305-Pho88-mCherry::LEU was a gift from the Brickner Lab (Northwestern University, Evanston, IL). Pho88-mCherry::LEU was digested with ClaI before transformation into yeast. pFA6a-yoHalo::CaUra3 (pLL#743) was a gift from J. Nunnari (University of California, Davis; Subramanian et al., 2019).
pLL#189 pET22b mod-T7prom::His6-T7-Num1(97-324) was constructed by PCR amplification of Num1(97-324) from genomic DNA of LLY#92 using primers LL#112/115 to add BamHI and XhoI sites. Digested PCR product was ligated into pET22b modT7prom::His6-T7 using BamHI and XhoI cut sites.
pLL#779 pRS304-TEF::mito-TagBFP::TRP was constructed by cloning mito-TagBFP from p416-GPD::mito-TagBFP into pRS304-TEF1::TRP using BcuI and XhoI sites. TEF::mito-TagBFP::TRP was digested with Eco32I before transformation into yeast.
pLL#500 p416-Met25::yEGFP-Num1ΔEF::URA was constructed using three PCR products from pLL#336 with primers #226/434, #433/457, and #456/437, which were inserted into p416-MET25::yEGFP-Num1 (Lackner et al., 2013) cut with BamHI and HindIII using gap repair in W303 Δnum1.
pLL#501 p416-Met25::yEGFP-Num1(D315A)::URA was constructed using three PCR products from pLL#336 with primers #226/434, #433/459, and #458/437, which were inserted into p416-MET25::yEGFP-Num1 (Lackner et al., 2013) cut with BamHI and HindIII using gap repair in W303 Δnum1.
pLL#849 pET22b mod-T7prom::His6-T7-Num1D315A(97-324) was constructed by PCR amplification of Num1D315A(97-324) from genomic DNA of LLY#3603 using primers LL#112/523 to add BamHI and XhoI sites. Digested PCR product was ligated into pET22b modT7prom::His6-T7 using BamHI and XhoI cut sites.
pLL#866 pFa6-yoHalo::HIS and pLL#867 pFa6-yoHalo::KAN were constructed by cloning yoHalo from pFA6a-yoHalo::CaUra3 (pLL#743) into pFA6a-link-yEGFP-SpHIS5 (pLL#54) or pFA6a-link-yEGFP-KAN (pLL#55), respectively, using BglII and SacI cut sites.
Imaging
For Figure 1C, all strains harboring mito-RED::LEU (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) LEU + 2% (wt/vol) dextrose media with 2× adenine. Single-time-point images were captured.
For Figure 1D, all strains (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine. Three-minute-time-lapse movies were captured.
For Figure 2A, all strains harboring mRuby-Tub1::HYG (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine. Single- time-point images were captured.
For Figure 2C, all strains (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine at 24°C. Three-minute-time-lapse movies were captured.
For Figure 4, A and B, all strains (as indicated in the figure legend) were grown at 30°C for 2 h in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine, pH 6.4. Auxin (α-naphthalene acetic acid; HiMedia), to a final concentration of 1 mM, or dimethyl sulfoxide (DMSO) was added, and cells were grown for an additional 3 h at 30°C. Single-time-point images were captured.
For Figure 5A, all strains harboring mito-RED::LEU (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) LEU + 2% (wt/vol) dextrose media with 2× adenine. Single-time-point images were captured.
For Figure 5D, all strains harboring mRuby-Tub1::HYG (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine. Single- time-point images were captured.
For Figure 5E, all strains (as indicated in the figure legend) were grown 30°C for 4 h in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine. HU (USBiological) was added to a final concentration of 200 mM, and the cells were grown for an additional 2.5 h at 30°C. Ten-minute-time-lapse movies were captured.
For Figure 5F, all strains (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine at 24°C. Three-minute-time-lapse movies were captured.
For Figure 5G, all strains (as indicated in the figure legend) were grown to mid–log phase in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine at 24°C. Single-time-point images were captured.
For Figure 6A, all strains (as indicated in the figure legend) were grown at 30°C for 2 h in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine, pH 6.4. Auxin, to a final concentration of 1 mM, or DMSO was added, and cells were grown for an additional 3 h at 30°C. Single-time-point images were captured.
For Figure 6B, all strains (as indicated in the figure legend) were grown at 30°C for 2 h in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine, pH 6.4. Auxin, to 1 mM, or DMSO was added, and cells were grown for an additional 3 h at 30°C. HU was added to a final concentration of 200 mM, and the cells were grown for an additional 2 h at 30°C. Ten-minute-time-lapse movies were captured.
For Figure 6C, all strains (as indicated in the figure legend) were grown at 30°C for 2 h in synthetic complete (SC) + 2% (wt/vol) dextrose media with 2× adenine, pH 6.4. Auxin, to a final concentration of 1 mM, or DMSO was added, and cells were grown for an additional 3 h at 30°C. JFX650 HaloTag ligand (Grimm et al., 2021) to a final concentration of 1 μM for 30 min at 30°C with shaking. The auxin-containing media was removed by filter washing, and the cells were resuspended by pipetting on the filter before HaloTag ligand addition. Excess dye was removed by a second round of filter washing, and cells were resuspended in SC + 2% (wt/vol) dextrose media, pH 6.4, containing 1 mM auxin or DMSO. Three-minute-time-lapse movies were captured.
For all imaging, cells were grown to mid–log phase, concentrated by centrifugation, and mounted on a 4% wt/vol agarose pad. All imaging was performed at 24°C. Z-series of cells were imaged at a single time point or over time using a Leica Spinning Disk Confocal System (Leica) fitted with a CSU-X1 spinning-disk head (Yokogawa). A PLAN APO 63X (1.32 NA objective; Leica), and an Evolve 512 Delta EMCCD camera (Photometrics) were used for image capture. A step size of 0.2 μm was used for Figures 1C and 5G and 0.4 μm for all other figures. Images were captured with Metamorph (Molecular Devices) and deconvolved using AutoQuant X3’s (Media Cybernetics) iterative, constrained three-dimensional deconvolution method. Linear adjustments to brightness and contrast were performed using FIJI and Photoshop (Adobe). Deconvolved images are shown in the figures.
For the quantification of spindle orientation, large budded cells with spindles >1.25 μm or >1.75 μm were analyzed, as indicated in the figure legends. To include only anaphase spindles in our assay, we quantified preanaphase spindles for WT and mitoAID cells (±auxin) using a HU arrest (Supplemental Figure S4). For all non-mitoAID cells, we used a cutoff of >1.75 μm spindles, which excludes >95% of preanaphase spindles (see top red dashed line in Supplemental Figure S4). For all mitoAID cells we used a cutoff of >1.25 μm spindles, which excludes >95% of preanaphase spindles (see bottom red dashed line in Supplemental Figure S4). To be correctly oriented, the spindle must cross the M-B neck or be within 45° of a line perpendicular to the neck. Misoriented spindles did not cross the neck and were greater than 45° off of the M-B axis (Kraft and Lackner, 2017). Measurements of bud size, spindle length, and orientation were all done in FIJI. Statistical analyses were performed in GraphPad using a two-tailed unpaired t test with a 95% confidence interval (the Shapiro–Wilk test was used to test for normality), with the exception of Figure 1, D and E, for which the Mann–Whitney test with a 95% confidence interval was used. Designations of significance are indicated in the figure legends.
A cluster is defined as an accumulation of Num1 above background fluorescence that persists for at least three frames of time-lapse imaging (>1.5 min). The dimmer accumulations of Num1 are dynamic over time and do not result in a productive cluster or tether point. For the quantification of mitochondria tethers, mitochondria and a Num1 cluster had to remain associated for ≥1.5 min. For the quantification of cortical dynein foci, a Dyn1-mKate focus had to stably remain at the cell cortex for ≥1.5 min.
For the spindle oscillation assay, cells were diluted from an overnight culture to 0.1 OD600 in SC + 2% (wt/vol) dextrose media with 2× adenine and grown at 30°C for 4 h. HU was added to a final concentration of 200 mM, and the culture was grown for another 2.5 h before imaging. For the mitoAID spindle oscillation assay, cells were diluted from an overnight culture to 0.1 OD600 in SC + 2% (wt/vol) dextrose media with 2× adenine (pH 6.4) and grown at 30°C for 2 h. Auxin was added to a final concentration of 1 mM and grown at 30°C for 2 h. DMSO was used in place of auxin for control experiments (–auxin). HU was then added to a final concentration of 200 mM, and the culture was grown for another 2 h at 30°C before imaging. For both spindle oscillation assays, 10-min-time-lapse movies with 30 s intervals were taken. All image analysis was done using FIJI. Bright-field images taken before and after the 10 min were overlaid to ensure that there was no drifting of the cells over time. The mRuby-TUB1 channel was overlaid with a bright-field image to quantify the preanaphase spindles that cross the M-B neck. Any cells that were not large-budded were excluded from analysis. For the mitoAID spindle orientation and oscillation assays, the small percentage of cells that inherited mitochondria in auxin conditions were excluded from analysis.
For the HaloTag labeling and imaging, cells starting at an OD600 of 0.1 were grown in SC + 2% (wt/vol) dextrose media, pH 6.4, for 2 h at 30°C, treated with 1 mM auxin, and grown for a further 3 h at 30°C. Owing to the presence of auxin in the media interfering with HaloTag labeling, the auxin-containing media was removed by filter washing the cells by pushing 6 ml of fresh medium through a 0.22 μm syringe filter. Washed cells were resuspended by pipetting on the filter. JFX650 HaloTag ligand (Grimm et al., 2021) was added from a 1 mM stock in DMSO to a final concentration of 1 μM, and labeling was performed for 30 min at 30°C with shaking. Excess dye was removed by a second round of filter washing, and cells were resuspended in SC + 2% (wt/vol) dextrose media, pH 6.4, containing 1 mM auxin, mounted on a 4% wt/vol agarose pad, and imaged on a Leica spinning-disk confocal microscope.
Plate reader growth assay
Growth assays were performed in 96-well clear bottom plates (Nunclon Delta Surface;Thermo Scientific) with OD600 measurements taken every 10 min for 24 h at 30°C with no shaking on a SpectraMax iD3 (Molecular Devices) plate reader. Each plate was prepared with four technical replicates of each strain in yeast extract, peptone + 2% (wt/vol) dextrose (YPD) media, and the plate was sealed with a clear breathable membrane to prevent evaporation. The change in OD600 per hour (slope) was calculated from the linear portion of the growth curve between 320 and 720 min (determined from WT for an R2 value of 0.99). Each strain has three biological replicates with three to four technical replicates each. Outliers were determined using the following criteria: the end point OD600 was outside the upper and lower limits of quartiles 1 and 3 (Q1 and Q3, respectively). The lower limit was calculated using Q1–[1.5*(Q3–Q1)]. The upper limit was calculated using Q3+[1.5*(Q3–Q1)].
Spot test assay
For analysis of growth by serial dilution, cells were grown overnight in YPD media, pelleted, and resuspended in water at a concentration of 0.2 OD600/ml. Tenfold serial dilutions were performed, and cells were spotted onto YPD plates and grown at 30°C for 24 h.
Purification of Num1(97-324) and Num1 D315A(97-324)
Num1(97-324) and Num1D315A(97-324) were purified from Escherichia coli as follows: Starter cultures of BL21(λDE3)/RIPL cells harboring plasmid pET22b modT7-His6-T7-Num1(97-324) (pLL#189) or pET22b modT7-His6-T7-Num1D315A(97-324) (pLL#849) were grown overnight in Luria–Bertani (LB) medium with 150 µg/ml ampicillin, 25 µg/ml chloramphenicol, and 0.04% glucose. Five milliliters of an overnight starter culture was used to inoculate 4 l of LB medium with 150 µg/ml ampicillin, 25 µg/ml chloramphenicol, and 0.04% glucose. The cells were grown at 37°C until an OD600 of ∼0.5 was reached. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to 250 µM to induce protein expression, and growth was continued for an additional 16 h at 18°C. The cells were then harvested by centrifugation at 3500 × g for 15 min. The resulting pellet was resuspended in a 1/200 volume of resuspension buffer (RB; 20 mM Tris, pH 8.0, 500 mM NaCl, and 1.89 mM 2-mercaptoethanol), quickly frozen in liquid N2, and stored at −80°C. The cell suspension was quickly thawed in a room-temperature water bath, phenylmethylsulfonyl fluoride (PMSF) was added to 0.5 mM, and the thawed cell suspension was subjected to two more freeze–thaw cycles. The homogenate was sonicated briefly with pulses to further lyse cells and clarified by centrifugation at 17,000 × g for 45 min at 4°C. The protein was purified from the supernatant using HisPur Ni-NTA resin (Thermo Fisher Scientific). The supernatant was incubated with resin at a 1/7 volume for 1 h at 4°C, and the resin was then pelleted at 3000 × g for 3 min. The resin was washed three times with RB and three times with wash buffer (RB + 30 mM imidazole) and loaded into a chromatography column. Protein was eluted from the column using a step gradient of RB + 60–300 mM imidazole. Four 500 µl elutions were collected for each imidazole step: 60, 90, 120, and 150; 16 500 µl elutions of 300 mM imidazole were collected. Then, 5 µl of each elution was mixed with sample buffer (SB; 62.5 mM Tris-HCl, pH 6.8, 2.5% SDS, 0.002% bromophenol blue, 0.7135 M β-mercaptoethanol, 10% glycerol), run on a 12.5% SDS–PAGE gel, and Coomassie stained. The elutions containing Num1(97-324) or Num1D315A(97-324) were pooled and dialyzed overnight in 20 mM tricine, pH 8.7, 50 mM NaCl. For the chelation treatment before ITC experiments, EGTA was added to a final concentration of 1 mM for two rounds of 2 h dialysis followed by two rounds without EGTA (2 h and overnight) of dialysis to remove the EGTA. The concentration of the purified proteins was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific).
Silver stain
Purified Num1(97-324) and Num1D315A(97-324) were run on a 15% SDS–PAGE gel in denaturing condition with SB. The gel was washed for 10 min with 50% methanol followed by 10 min with 10% methanol and then washed for 10 min with 32 µM dithiothreitol. The gel was rinsed briefly with water, then incubated with 0.1% (wt/vol) silver nitrate. The gel was then incubated with 3% (wt/vol) sodium bicarbonate and 0.0185% (vol/vol) formaldehyde for ∼15 min until bands were developed. Acetic acid (10%) was used to stop the reaction, and the gel was imaged on an Azure c600.
Western blot
The indicated strains in Supplemental Figure S1A were grown to mid–log phase in YPD media. Cells (1.0 OD600) were harvested, and whole-cell protein extracts were prepared using a NaOH lysis and trichloroacetic acid (TCA) precipitation procedure. Each TCA pellet was resuspended in 50 µl MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea). Whole-cell extracts were run on 6% SDS–PAGE gels in denaturing conditions followed by Western analysis using anti-GFP (Abclonal rabbit anti–GFP-Tag pAb [AE011]; https://abclonal.com/catalog-antibodies/RabbitantiGFPTagpAb/AE011) as the primary antibody and goat anti-rabbit immunoglobulin G (IgG) DyLight 800 (ThermoFisher; https://www.thermofisher.com/antibody/product/Goat-anti-Rabbit-IgG-H-L-Secondary-Antibody-Polyclonal/SA5-35571) as the secondary antibody. Licor total protein stain () was used for normalization of protein levels. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences).
ITC
All experiments were performed on a MicroCal ITC200 instrument (Malvern Panalytical) in the Keck Biophysics Facility at Northwestern University. Before each experiment, the reference cell and syringe were filled with deionized filtered water and then washed extensively with the reaction buffer (20 mM tricine, pH 8.7, 50 mM NaCl). The solutions were degassed for 10 min, 40 μl of the CaCl2 solution was loaded into the titrating syringe, and 280 μl of the protein solution was placed in the ITC cell. After the instrument was equilibrated at 298 K and 750 rpm syringe rotational speed, a first injection of 0.1 μl was performed, followed by a series of 1.8 μl injections. The first injection was discarded from the analysis of the integrated data in order to avoid artifacts related to diffusion through the injection port during the equilibration period. To measure residual heats, a separate control experiment was performed with the same reaction parameters, except that only the dialysis buffer (20 mM tricine, pH 8.7, 50 mM NaCl) was placed in the ITC cell (Supplemental Figure S3B). Titration data were processed with the MicroCal Origin 7.0 software package as described (Biltonen and Langerman, 1979). Individual injection heats (qi)—obtained by integrating the corresponding injection peaks—were normalized for ligand concentration and corrected for dilution heats. A nonlinear regression fit to a single set of sites model provided the stoichiometry of binding N, binding constant Kd, enthalpy change ΔH, and entropy change ΔS (Privalov and Dragan, 2007).
Supplementary Material
Acknowledgments
We thank members of the Lackner lab and Jennifer Brace for suggestions and critical scientific discussions. We also thank Northwestern’s Cell Biology Supergroup and the Wignall-Lackner Cell Biology Group for constructive feedback on the project. We are extremely grateful to Jessica Hornick and Arabela Grigorescu for their help and advice on imaging and ITC, respectively. All microscopy was performed at the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), supported by the Chemistry for Life Processes Institute, the Northwestern University Office for Research, the Department of Molecular Biosciences, and the Rice Foundation. The ITC was performed by the Northwestern University Keck Biophysics Facility, supported by a Cancer Center Support Grant (NCI CA060553). We also thank Luke Lavis from Janelia for providing the JFX650 dye for the HaloTag imaging experiments. H.L.A. was supported by American Heart Association Predoctoral Fellowship 19PRE34381053. L.L.L. is supported by National Institutes of Health, National Institute of General Medical Sciences grant R01GM120303.
Abbreviations used:
- AID
auxin-inducible degron
- aMT
astral microtubule
- CC
coiled-coil
- cER
cortical ER
- DMSO
dimethylsulfoxide
- EFLM
EF hand-like motif
- EGTA
ethylene glycol tetraacetic acid
- ER
endoplasmic reticulum
- ERMES
ER-mitochondria encounter structure
- IPTG
Isopropyl β- d-1-thiogalactopyranoside
- MECA
mitochondria-ER-cortex anchor
- PAN
Pil1-associated Num1
- PH
pleckstrin homology
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PM
plasma membrane
- SC
synthetic complete
- TCA
trichloroacetic acid
- yEGFP
yeast enhanced GFP
- YPD
yeast extract peptone dextrose
- WT
wild-type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E21-12-0610-T) on January 5, 2022.
REFERENCES
- Adames NR, Cooper JA (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol 149, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A (1999). The spindle checkpoint. Curr Opin Genet Dev 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Biltonen RL, Langerman N (1979). Microcalorimetry for biological chemistry: experimental design, data analysis, and interpretation. Methods Enzymol 61, 287–318. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Ramcharan SL, Yang H-C, Pon LA (2004). A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol Biol Cell 15, 3994–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol 138, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML Jr, Hoyt MA, Pellman D (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell 6, 815–829. [DOI] [PubMed] [Google Scholar]
- Cerveny KL, Studer SL, Jensen RE, Sesaki H (2007). Yeast mitochondrial division and distribution require the cortical Num1 protein. Dev Cell 12, 363–375. [DOI] [PubMed] [Google Scholar]
- Chao JT, Wong AKO, Tavassoli S, Young BP, Chruscicki A, Fang NN, Howe LJ, Mayor T, Foster LJ, Loewen CJR (2014). Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158, 620–632. [DOI] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S (2012). ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol 14, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Santiago-Tirado F, Bretscher A (2013). Active segregation of yeast mitochondria by Myo2 is essential and mediated by Mmr1 and Ypt11. Curr Biol 23, 1818–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Kaandorp JA, Ositelu OO, Beaudry V, Knight A, Nanfack YF, Cunningham KW (2009). Simulating calcium influx and free calcium concentrations in yeast. Cell Calcium 45, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW (2011). Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium 50, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR (1993). Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA 90, 11172–11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves PT, Jin Y, Brunner M, Weisman LS (2012). Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J Cell Biol 198, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Rachubinski RA (2007). Orchestrating organelle inheritance in Saccharomyces cerevisiae. Curr Opin Microbiol 10, 528–538. [DOI] [PubMed] [Google Scholar]
- Farkasovsky M, Küntzel H (1995). Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol 131, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Küntzel H (2001). Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol 152, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, Okamoto K, Shaw JM (2008). Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics 178, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, et al. (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods 11, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011). ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez LJ, Crider DG, Gay AC, Salanueva IJ, Boldogh IR, Pon LA (2009). Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Curr Biol 19, 1730–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem J 405, 199–221. [DOI] [PubMed] [Google Scholar]
- Grabarek Z (2006). Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol 359, 509–525. [DOI] [PubMed] [Google Scholar]
- Grimm JB, Xie L, Cassler JC, Patel R, Tkachuk AN, Falco N, Choi H, Lippincott-Schwartz J, Brown TA, Glick BS, et al. (2021). A general method to improve fluorophores using deuterated auxochromes. JACS Au 1, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammermeister M, Schödel K, Westermann B (2010). Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol Biol Cell 21, 2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA (2000). The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA (2013). Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol 23, 2417–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Toh-E A, Matsui Y (2004). Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J 23, 2520–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Watabe A, Toh-E A, Matsui Y (2002). Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol 22, 7744–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto P, Döhla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM (2015). Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker T, Scholz D, Förtsch J, Westermann B (2013). The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J Cell Sci 126, 2924–2930. [DOI] [PubMed] [Google Scholar]
- Klinger H, Rinnerthaler M, Lam YT, Laun P, Heeren G, Klocker A, Simon-Nobbe B, Dickinson JR, Dawes IW, Breitenbach M (2010). Quantitation of (a)symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol 45, 533–542. [DOI] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA (2015). Sharing the cell’s bounty—organelle inheritance in yeast. J Cell Sci 128, 621–630. [DOI] [PubMed] [Google Scholar]
- Kormanec J, Schaaff-Gerstenschläger I, Zimmermann FK, Perecko D, Küntzel H (1991). Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol Gen Genet 230, 277–287. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft LM, Lackner LL (2017). Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J Cell Biol 216, 3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft LM, Lackner LL (2019). A conserved mechanism for mitochondria-dependent dynein anchoring. Mol Biol Cell 30, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Ping H, Graef M, Murley A, Nunnari J (2013). Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci USA 110, E458–E467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers LG, Markus SM (2015). The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J Cell Biol 211, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-G, Cason SE, Alqassim SS, Holzbaur ELF, Dominguez R (2020). A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat Commun 11, 5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-L, Kaiser MA, Cooper JA (2005). The offloading model for dynein function: differential function of motor subunits. J Cell Biol 168, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska A, Macfarlane J, Shaw JM (2013). Mitochondrial association, protein phosphorylation, and degradation regulate the availability of the active Rab GTPase Ypt11 for mitochondrial inheritance. Mol Biol Cell 24, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW, Lu MS, Iwamoto Y, Drubin DG, Pedersen RTA (2021). A preferred sequence for organelle inheritance during polarized cell growth. J Cell Sci 134, jcs258856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K (1993). Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA 90, 10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-López J, Matellán L, Álvarez-Llamas A, Blanco-Mira JC, Monje-Casas F (2019). Asymmetric inheritance of spindle microtubule-organizing centres preserves replicative lifespan. Nat Cell Biol 21, 952–965. [DOI] [PubMed] [Google Scholar]
- Markus SM, Lee W-L (2011). Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell 20, 639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Omer S, Baranowski K, Lee W-L (2015). Improved plasmids for fluorescent protein tagging of microtubules in Saccharomyces cerevisiae. Traffic 16, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matellán L, Monje-Casas F (2020). Regulation of mitotic exit by cell cycle checkpoints: lessons from Saccharomyces cerevisiae. Genes 11, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA (2011). Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD (2014). Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 140, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Sept D, Cooper JA (2009). Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci USA 106, 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta 1861, 952–961. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6, 917–922. [DOI] [PubMed] [Google Scholar]
- Omer S, Brock K, Beckford J, Lee W-L (2020). Overexpression of Mdm36 reveals Num1 foci that mediate dynein-dependent microtubule sliding in budding yeast. J Cell Sci 133, jcs246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer S, Greenberg SR, Lee W-L (2018). Cortical dynein pulling mechanism is regulated by differentially targeted attachment molecule Num1. eLife 7, e36745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernice WM, Vevea JD, Pon LA (2016). A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat Commun 7, 10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping HA, Kraft LM, Chen W, Nilles AE, Lackner LL (2016). Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J Cell Biol 213, 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov PL, Dragan AI (2007). Microcalorimetry of biological macromolecules. Biophys Chem 126, 16–24. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, Carter AP (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol 19, 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Okuzaki D, Nojima H (2006). Mcp5, a meiotic cell cortex protein, is required for nuclear movement mediated by dynein and microtubules in fission yeast. J Cell Biol 173, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP (2014). In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33, 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit HL, Kraft LM, Lee-Smith CF, Lackner LL (2018). The role of mitochondria in anchoring dynein to the cell cortex extends beyond clustering the anchor protein. Cell Cycle 17, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Bloom K (2001). Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol 11, 160–166. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jochem A, Le Vasseur M, Lewis S, Paulson BR, Reddy TR, Russell JD, Coon JJ, Pagliarini DJ, Nunnari J (2019). Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J Cell Biol 218, 1353–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Germain BS, Lee W-L (2012). A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J Cell Biol 196, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Punch JJ, Lee W-L (2009). A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle 8, 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P (2006). Eisosomes mark static sites of endocytosis. Nature 439, 998–1003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Huynh W, Skokan TD, Lu W, Weiss A, Vale RD (2019). CRACR2a is a calcium-activated dynein adaptor protein that regulates endocytic traffic. J Cell Biol 218, 1619–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J, Choi Y, Yun Y, Lee HH (2021). Biochemical characterization of the Num1-Mdm36 complex at the mitochondria-plasma membrane contact site. Mol Cells 44, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-J, Chen Y-S, Kim MR, Chang C-C, Gampala S, Zhang Y, Wang Y, Chang C-Y, Yang J-Y, Chang C-J (2019). Epithelial-mesenchymal transition directs stem cell polarity via regulation of mitofusin. Cell Metab 29, 993–1002.e6. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Yamamoto M (2006). Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase. Genetics 173, 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Ames JB, Swindells MB, Ikura M (1999). Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins 37, 499–507. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13, 677–688. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang W, Kirberger M, Lee H-W, Ayalasomayajula G, Yang JJ (2006). Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins 65, 643–655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.