Abstract
Actin nucleation is achieved by collaborative teamwork of actin nucleator factors (NFs) and nucleation-promoting factors (NPFs) into functional protein complexes. Selective inter- and intramolecular interactions between the nucleation complex constituents enable diverse modes of complex assembly in initiating actin polymerization on demand. Budding yeast has two formins, Bni1 and Bnr1, which are teamed up with different NPFs. However, the selective pairing between formin NFs and NPFs into the nucleation core for actin polymerization is not completely understood. By examining the functions and interactions of NPFs and NFs via biochemistry, genetics, and mathematical modeling approaches, we found that two NPFs, Aip5 and Bud6, showed joint teamwork effort with Bni1 and Bnr1, respectively, by interacting with the C-terminal intrinsically disordered region (IDR) of formin, in which two NPFs work together to promote formin-mediated actin nucleation. Although the C-terminal IDRs of Bni1 and Bnr1 are distinct in length, each formin IDR orchestrates the recruitment of Bud6 and Aip5 cooperatively by different positioning strategies to form a functional complex. Our study demonstrated the dynamic assembly of the actin nucleation complex by recruiting multiple partners in budding yeast, which may be a general feature for effective actin nucleation by formins.
INTRODUCTION
Dynamic actin assembly requires collaborative recruitment of multiple actin monomers by paired actin nucleation factors (NFs) and nucleation promoting factors (NPFs) that spatiotemporally regulate actin polymerization for a multitude of cellular activities (Pollard, 2007; Quinlan et al., 2007; Chesarone and Goode, 2009; Firat-Karalar and Welch, 2011; Zoncu et al., 2011; Breitsprecher et al., 2012; Graziano et al., 2013; Xie et al., 2019). Several NPFs have been known to promote NF-mediated actin nucleation via tunable protein interactions and composition stoichiometry of the formed nucleation complex, such as Wiskott–Aldrich syndrome protein (WASP) and WAVE (WASP family veroprolin homolog) regulatory complex (WRC) for Arp2/3 complex in diverse eukaryotes, as well as Bud6 and Aip5 for yeast and fungal formins (Quinlan et al., 2007; Graziano et al., 2011; Tu et al., 2012; Xie et al., 2020; Zheng et al., 2020). In addition, the hierarchical assembly of multiple participating partners was shown to result in macromolecular condensation of WASP that exhibited enhanced NPF activity (Campellone et al., 2008; Padrick et al., 2008; Sallee et al., 2008; Su et al., 2016; Case et al., 2019). Such synergistic nucleation of F-actin was also demonstrated by the association of two NFs or the interactions of two NPFs on the same NF, such as Spire-Fmn2/Capu, adenomatous polyposis coli (APC)-mDia1, and Aip5-Bud6-Bni1 (Machesky et al., 1999; Moseley and Goode, 2005; Quinlan et al., 2007; Okada et al., 2010; Graziano et al., 2011; Bieling et al., 2019; Case et al., 2019; Glomb et al., 2019; Xie et al., 2019). In Drosophila melanogaster, the WASP homology 2 class-NF Spire synergizes with the actin nucleation activity of formin class-NF Cappuccino (Quinlan et al., 2007; Vizcarra et al., 2011). In mammalian cells, APC protein has applied the rocket launcher mechanism to boost the nucleation activity of formin mDia1 (Okada et al., 2010; Breitsprecher et al., 2012). Recently, Aip5 was identified as another fungal NPF for Bni1 in both Saccharomyces cerevisiae and Candida albicans by binding to actin monomer and Bni1 through its C-terminus (1110–1234 a.a for ScAip5C and 786–896 a.a for CaAip5C) (Xie et al., 2019, 2020); aip5∆ exhibited synthetic sickness in the background of bni1∆, suggesting a potential function of Aip5 in collaborating with the remaining formin Bnr1 in bni1∆. Interestingly, while the C-terminal tail of Bni1 (1767–1953 a.a) hosts both NPFs, Aip5 and Bud6, for complex formation, Bnr1 likely works with NPFs differently because the C-terminal intrinsically disordered region (IDR) of Bnr1 is much shorter (1288–1375 a.a) than Bni1-C (1767–1953 a.a) and does not seem to interact with Bud6 on its own (Graziano et al., 2013). The active region of NPF Bud6, Bud6C (residues 489–788), is located close to the FH2 domain of Bnr1, which thereby impairs NPF function if not recruiting the alleviation factor Bil1 (Graziano et al., 2013). Due to the intrinsically disordered nature of both formin’s C-terminal tails, it has also been challenging to determine the structural position of Bud6C and Aip5C in the Bni1-Bud6-Aip5 complex in coordinating their interactions and activities. Thus, how actin cables were nucleated by collaborative pairing for two distinct nucleators, Bni1 and Bnr1, and two NPFs, Aip5 and Bud6, into NPF-NF complexes has remained enigmatic.
Here we unraveled a common paradigm of orchestrated actin cable nucleation from the partnership of a formin, Bni1 or Bnr1, with two NPFs, Bud6 and Aip5, in budding yeast. In vitro biochemical assays and in vivo genetics have validated the collaborative functions of two NPFs, Aip5 and Bud6, on distinct formins. Through molecular dynamics (MD) simulation, we obtained interactive modes for two sets of NF-NPF complexes in which each component could adopt an appropriate conformation to avoid steric hindrance for efficient nucleation of actin cables. We also found that unlike Bni1-C, a helix of the Bnr1 FH2 region is required to stabilize its tri-protein complex. Our results shed light on the orchestrated complex formation of yeast formin NFs and NPFs, which might be a general mechanism underlying actin assembly in the fungal kingdom.
RESULTS
Aip5 is a nucleation-promoting factor for both Bni1 and Bnr1
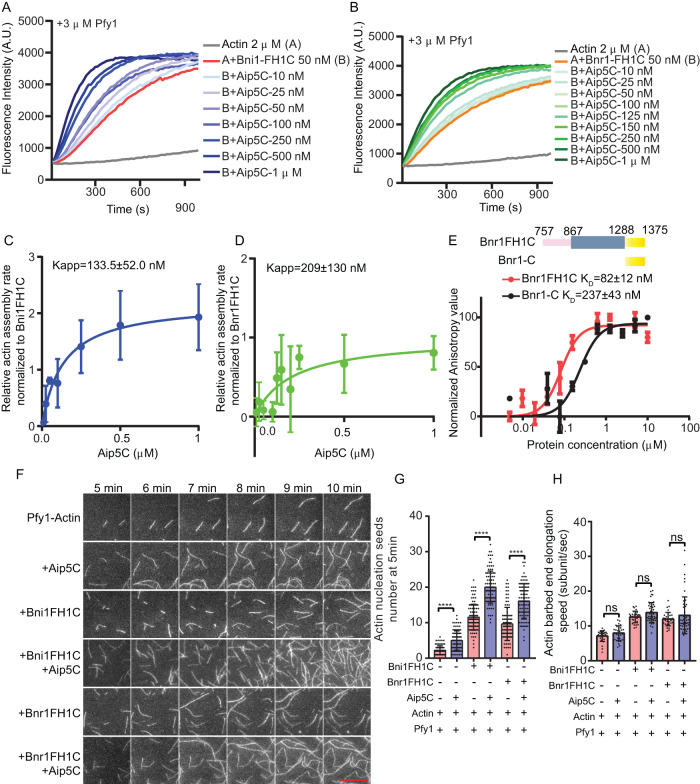
To investigate whether Aip5 has conversed NPF activities for both yeast formins, Bni1 and Bnr1, we examined actin nucleation biochemically using the recombinant protein domain of Aip5 (Aip5C, residues 1110–1234 a.a.), Bni1 (Bni1FH1C, residues 1227–1953 a.a.), and Bnr1 (Bnr1FH1C, residues 757–1375 a.a.), in the presence of Pfy1 (Supplemental Figure S1, A–C). Consistent with previously reported results, Bnr1FH1C stimulated actin polymerization in a bulk actin assembly assay in a dose-dependent manner in the presence of profilin (Figure 1, A and B and Supplemental Figure S1D) (Moseley and Goode, 2005). Interestingly, we observed that Aip5C is capable of promoting the actin nucleation of both Bni1FH1C and Bnr1FH1C in an Aip5 dose-dependent manner (Figure 1, A and B). By analyzing the half polymerization of actin polymerization, Aip5C exhibited a slightly higher potency in stimulating Bni1 than Bnr1 for actin nucleation (Figure 1, C and D).
FIGURE 1:
Aip5 binds to both Bni1 and Bnr1 and stimulates their nucleation activities. (A, B) Pyrene actin polymerization of 2 µM G-actin with 3 µM yeast Pfy1. An increasing amount of Aip5C was added to 50 nM of Bni1FH1C and Bnr1FH1C, respectively, at the indicated concentrations. (C, D) The relative actin assembly rates of Aip5C were normalized to Bni1FH1C and Bnr1FH1C, respectively, where the Kapp was deduced from three independent biological replicates. (E) Fluorescence anisotropy binding measurements of Aip5C (Alexa 488 labeled, 30 nM) that were titrated by a serial concentration of Bnr1FH1C and Bnr1-C, respectively, as indicated. Data are represented by circles using the average value of three biological replicates and fitted by the Hill equation to determine the KD. (A.U., arbitrary unit.). (F) The representative TIRF images of actin nucleation seeds formed at 5 and 10 min, respectively. The control actin filament was assembled from 1 µM actin (10% Oregon green 488-labeled actin and 0.5% biotin-actin) with 3 µM yeast Pfy1, and the used proteins were indicated as follows: 10 nM Bni1FH1C; 2 nM Bnr1FH1C and 20 nM Aip5C. The scale bar represents 10 µm. (G) Quantification of actin nucleation by measuring the seeds number at 5 min with the indicated combinations of proteins. (n = 86, 98, 114, 100, 94, 99 for each sample from ROI = 894 µm2) (H) Quantification of actin filament barbed end elongation speed of indicated protein combinations as shown in A. (n = 31, 30, 31, 45, 31, 49 for each sample) P values was determined by the one-way ANOVA, ns = not significant, **p < 0.01, ****p < 0.0001. Error bar, SD.
Because Aip5C directly interacts with Bni1-C (residues 1767–1953 a.a.) (Xie et al., 2019), we next examined whether Aip5C similarly interacts with Bnr1. Using a fluorescence anisotropy binding assay, we found that Bnr1-C directly interacts with Aip5C at a KD of 237 ± 43 nM (Figure 1E), although Bnr1-C (residues 1288–1375 a.a.) is much shorter than the corresponding IDR of Bni1-C (Supplemental Figure S1, A, B, and E). In addition, Bnr1FH1C displayed a stronger affinity and positive cooperativity to Aip5 than Bnr1-C with a KD of 82±12 nM (Figure 1E), reminiscent of the similar interactions between Aip5C and Bni1FH1C (Xie et al., 2019). To further confirm the different promotion effects of Aip5C on the two formins, we next utilized a total internal reflection fluorescence (TIRF) actin polymerization assay to determine the Aip5C functions in formin-mediated nucleation in the presence of profilin (Pfy1). First, 10 nM Bni1FH1C and 2 nM Bnr1FH1C displayed similar nucleation activity, both of which generated ∼5-fold more actin seeds than the control of profilin-actin at the initial period of polymerization (Figure 1, F and G). In the presence of 20 nM Aip5C, Bni1FH1C- and Bnr1FH1C-initiated actin nucleation was significantly enhanced by ∼1.73- and ∼1.69-fold, respectively (Figure 1, F and G). This observation agreed with the bulk actin assembly result, where Aip5C promoted both Bni1- and Bnr1-mediated actin polymerization (Figure 1, A–D). By examining the actin filament elongation rate over 5 min, Aip5C did not alter the barbed end elongation for Bni1FH1C- or Bnr1FH1C in the presence of Pfy1 (Figure 1, F and H), indicating an NPF function of Aip5C for both yeast Bni1 and Bnr1.
Teamwork efforts of Aip5 and Bud6 on formin nucleation activity
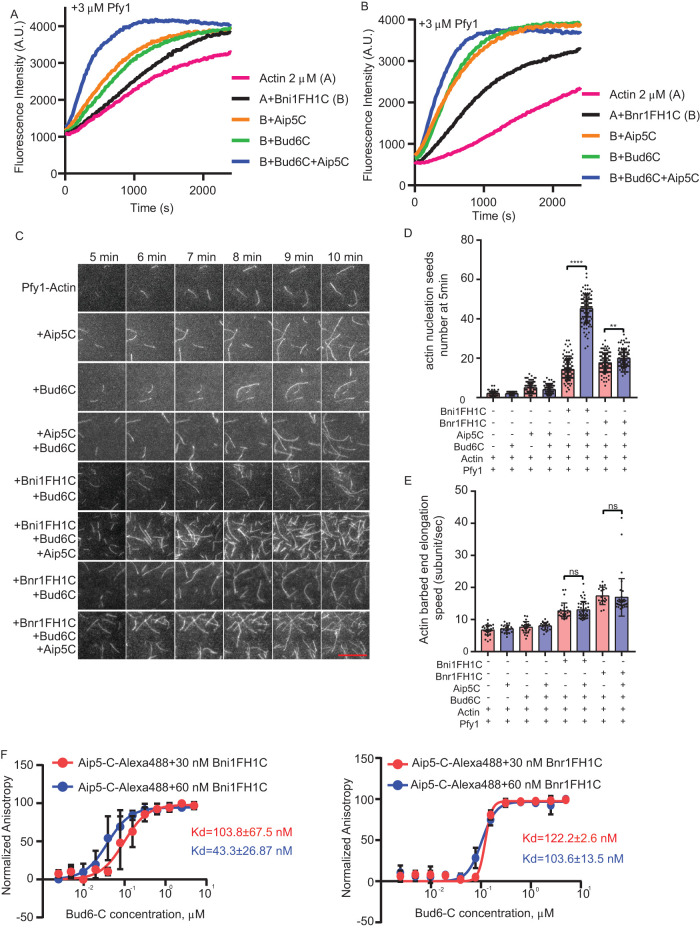
Bud6 is a well-studied NPF for budding yeast Bni1 and Bnr1 (Moseley and Goode, 2005; Graziano et al., 2011; Tu et al., 2012; Graziano et al., 2013). We next sought to investigate whether Aip5 cooperates with Bud6 to boost formin-mediated actin nucleation. We first used a pyrene actin assembly assay to examine actin polymerization under different combinations of NFs and NPFs using the short C-terminus of Bud6 (Bud6C, residues 550–789 a.a.), which is sufficient to activate Bnr1 without hindering the FH2 domain and masking its activity in actin nucleation (Graziano et al., 2013). In the presence of Pfy1, Bud6C enhanced actin polymerization triggered by Bni1 and Bnr1 in the pyrene actin assay (Figure 2, A and B). Interestingly, an additional supplement of Aip5C to the protein mixtures of formins and Bud6C further activated actin polymerization (Figure 2, A and B). To distinguish the roles of Aip5 and Bud6 in formin-mediated nucleation and elongation, we performed a TIRF actin polymerization assay. We found that Aip5C promoted the nucleation activity of both Bni1-Bud6 without altering the barbed end elongation in the presence of Pfy1 (Figure 2, C–E and Supplemental Movie S1). Compared with Bni1FH1C-mediated actin nucleation in the presence of Bud6C, additional Aip5C resulted in an ∼3.4-fold increase in the generation of actin seeds, whereas Aip5C enhanced Bnr1FH1C-Bud6C-mediated actin nucleation by ∼1.13-fold (Figure 2, C and D). The presence of two NPFs showed higher promotion rates on Bni1 (∼3.9-fold)- and Bnr1 (∼2.1-fold)-mediated nucleation than one NPF, Bud6C, or Aip5C. The NPF-NF pairs showed a 1.3-2-fold increase in their promotion of NF activity (Figures 1, F and G and 2, C and D). The above results indicate the teamwork of two NPFs on Bni1 by working simultaneously. However, Bni1 and Bnr1 showed slightly different modes of partnership with the two NFPs. Aip5C and Bud6C showed a synergistic effect on Bni1FH1C by having a combined promotion rate (∼3.9-fold) that was greater than the multiplied rates from two single NFPs, Aip5C (∼1.73-fold) and Bud6C (∼1.23-fold), on their own. (Figure 1, F and G, and Figure 2, C and D). However, the overall promotion rate of the two NFPs on Bnr1FH1C (∼2.1-fold) was slightly lower than the multiplied rates (∼ 3.5) of Bud6C (∼1.83) and Aip5C (∼1.69), indicating an additive effect instead of a synergy of the two NPFs on Bnr1 (Figure 1, F and G and Figure 2, C and D). To further understand the teamwork modes of the two NPFs on one NF, we performed an anisotropic assay to characterize the change in interactions between Aip5 and formin by supplementation with Bud6.
FIGURE 2:
Aip5 and Bud6 synergizes formin-mediated actin nucleation. (A, B) Pyrene actin polymerization with different combinations of proteins as indicated, 2 µM monomeric actin, 3 µM yeast profilin, 200 nM Aip5C, 40 nM Bud6C, 20 nM Bni1FH1C, and 20 nM Bnr1FH1C. (C) The representative TIRF images of actin nucleation seeds formed from 5 to 10 min using the indicated combination of proteins. The actin filament was assembled by mixing 1 µM actin (10% Oregon green 488-labeled actin and 0.5% biotin-actin) with 3 µM yeast profilin, 10 nM Bni1FH1C, 2 nM Bnr1FH1C, 5 nM Bud6C, or 20 nM Aip5C. The scale bar represents 10 µm. (D) Quantification of actin nucleation seeds number at 5 min with the indicated combinations of proteins (n = 80, 60, 98, 94, 114, 99, 100, 100 for each sample from ROI = 894 µm2). (E) Quantification of actin filament barbed end elongation speed of indicated protein combinations as shown in A (n = 28, 21, 24, 23, 27, 43, 19, 37 for each sample). (F) Fluorescence anisotropy binding measurements of 60 nM Alexa 488-labeled Aip5C were preincubated with different concentrations of Bni1FH1C and Bni1FH1C, respectively, before being titrated by a serial concentration of Bud6C. P values was determined by the one-way ANOVA, ns = not significant, ****p < 0.0001. Error bar, SD.
Supporting Movie S1.
We previously characterized NFs and NPFs in vivo to understand their physiologically relevant range of concentrations and stoichiometry at their concentrated sites. In vivo Aip5 and Bni1 have a concentration of 120 nM and 80 nM, respectively, at the bud tip (Xie et al., 2019). To inform the design of biochemical assays comparing the nucleation activities of two formin-NPF complexes, Bni1-Aip5-Bud6 and Bnr1-Aip-Bud6, and facilitate the interpretation of their physiological relevance, we determined a Bud6 concentration of ∼100 nM at the bud tip and a Bnr1 concentration of ∼200 nM at the bud neck (Supplemental Figure S1F), as previously reported for the determination of in vivo Bni1 and Aip5 (Xie et al., 2019). Bni1 and Bnr1 have differential localizations during early polar growth. Aip5 and Bud6 could switch their localization from the bud tip to the neck along with cell cycle progression. We measured the concentrations of the two formins at both the tip and the neck. In contrast, we only measured Aip5 and Bud6 concentrations at one location to simplify the stoichiometry estimation of NF-NPFs at a snapshot moment along the cell cycle without considering the migration of Aip5 and Bud6. The C-terminal fragment of Aip5 (1000–1234) has been shown to interact with the N-terminal fragment of Bud6 (1–141) (Glomb et al., 2019), indicating the intermolecular interaction between Bni1, Aip5, and Bud6 as a tri-protein nucleation complex. It is technically challenging to express and purify all these full-length formin and NPF proteins using our current prokaryotic expression system. It is challenging to examine complex formations with full lengths with all possible interactions. Hence we used a minimum-component reconstitution approach to dissect the molecular mechanism by which two NPFs work on one formin simultaneously. We reconstituted the tri-protein complex interaction using fragments of Aip5C and Bud6C, which contain the formin-binding NPF domains of each protein but lack the domains known to mediate interactions between Aip5 and Bud6 (Glomb et al., 2019). It is possible that these domains could even further elevate the collaborative NPF effects of Aip5 and Bud6 on formins. Here we added increasing concentrations of Bud6C to the premixed sample of Alexa 488-labeled Aip5C and Bni1FH1C in which the changes in anisotropic value would likely reflect the interaction changes between Aip5 and Bni1FH1C-Bud6C. Notably, all three proteins used here are in their dimeric state (Moseley et al., 2004; Xu et al., 2004; Tu et al., 2012; Garabedian et al., 2018; Xie et al., 2019). We found that Bud6C started to obviously enhance the anisotropy signal of Aip5 (60 nM) and Bni1 (30 nM) from a lower concentration and stoichiometry than for the pair of Aip5 (60 nM) and Bnr1 (30 nM). A likely explanation is that the interactions between Aip5 and Bni1FH1C are stimulated by the presence of Bud6C, starting from the concentrations of ∼30 nM Bud6C, which has a Bni1FH1C:Aip5C:Bud6C stoichiometry of 1:2:1 (Figure 2F). However, we could not exclude the possibility that the tumbling rate of Aip5C was changed simply by the increasing partition of Bud6C within the macromolecular complex. Interestingly, a higher Bni1FH1C:Aip5C stoichiometry at 1:1 (60 nM each) demonstrated a further enhanced interaction in response to increasing concentrations of Bud6C, such as 60 nM Bud6C:Bni1FH1C:Aip5C at a stoichiometry of 1:1:1. The above results suggest that Bni1C has sufficient binding sites to host both Aip5C and Bud6C, creating a joint effort of two NPFs toward a formin protein. In contrast, Bud6C exhibited slower promotion effects on the mix of Aip5C and Bnr1FH1C, which only started to show noticeable enhancement in interactions after adding more than 200 nM of Bud6C even under conditions with twice the amount of Bnr1 at 60 nM (Figure 2F). The different binding kinetics of Bni1-Aip5 and Bnr1-Aip5 in response to Bud6 also indicated distinct interaction modes within the two tri-protein nucleation complexes, similar to their different activities in nucleating actin polymerization (Figure 2, C and D).
MD simulation of the complex conformation of formin-Bud6-Aip5
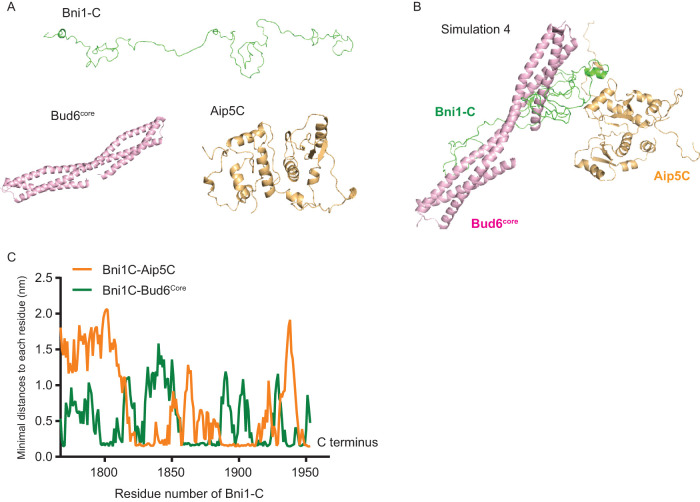
Since Aip5 and Bud6 collaboratively activate both formins, we next sought to understand the conformation and the orchestral functions of two different sets of tri-protein complexes. Because it is challenging to obtain structural information on the tri-protein complex by protein structural studies, due to the flexible nature of formin C-terminal IDR, we used an all-atom MD simulation. We utilized Bni1-C and the functional and structural regions of the NPFs Aip5C (PDB: 6ABR) (Xie et al., 2019) and Bud6core (residues 550–688 a.a., PDB: 3ONX) (Tu et al., 2012) (Figure 3A) for MD simulation. We assigned the initial structure of Bni1-C as a random coil (Figure 3A) due to its IDR nature (Xie et al., 2019). We placed Bud6core and Aip5C at eight different initial positions relative to Bni1-C in MD simulations to avoid bias by using specific relative positioning. All MD simulations converged after 200 ns, as shown by root-mean-square deviation (RMSD) plots (Supplemental Figure S2A), suggesting a relatively stable complex conformation. The representative conformations of the tri-protein complex were determined by choosing the lowest free energy conformation from the two-dimensional free energy surface made by principal component analysis (PCA) (Supplemental Figure S2B). To further compare the relative stabilities of the eight low free energy conformations, we calculated the scores of the conformations by the Rosetta program (Alford et al., 2017) (Supplemental Figure S2C) where the most stable Bni1-Bud6-Aip5 tri-protein complex with the lowest Rosetta scores is shown in Figure 3B. Interestingly, Aip5C preferentially associates with the C-terminus part of Bni1-C, whereas Bud6core interacts more toward the N-terminus of the C-terminal tails of Bni1-C (Figure 3C). We did not observe close contact between Bud6core and Aip5C, which is consistent with the result tested by fluorescence polarization (Supplemental Figure S2D). Furthermore, Aip5 C-terminus (residues 1000–1234 a.a.) was reported to associate with the N-terminus of Bud6 (residues 1–141 a.a.) (Glomb et al., 2019).
FIGURE 3:
Computer simulation of the complex assembly of Bni1-Bud6-Aip5. (A) Initial structures of Bni1-C, Aip5C, and Bud6core for setting up the MD simulations. (B) The most stable complex conformation (Simulation 4 in Supplemental Figure S2) from MD simulations among the eight randomly positioned nucleation complexes (Bni1-C, green; Aip5C orange; Bud6C, pink). (C) The minimal distance plot of each residue of Bni1-C from Aip5C (orange) and Bud6core(green), respectively, in the complex conformation in B.
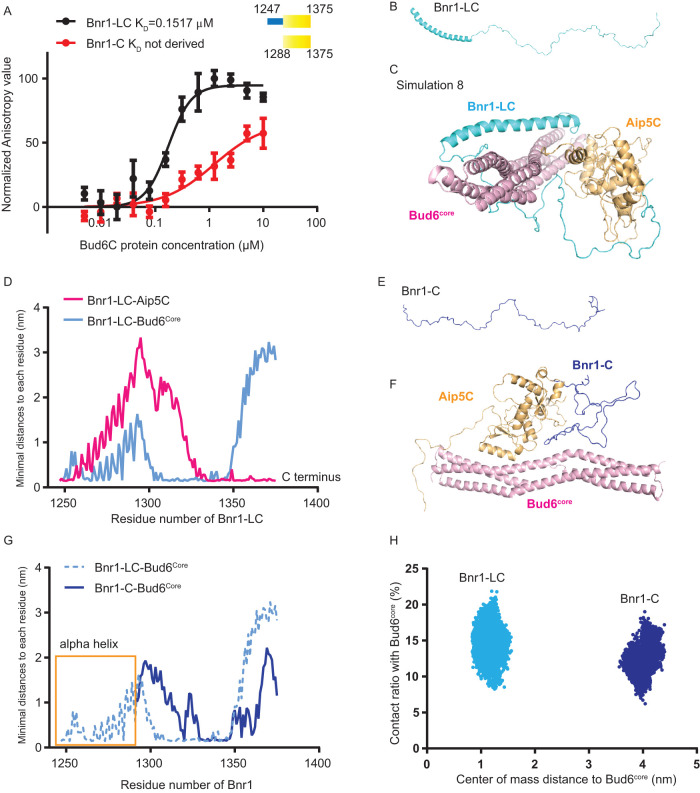
A similar strategy of MD simulation was applied to study the Bnr1-Bud6-Aip5 tri-protein complex conformation. Although the C-terminal regions of both formins are essential for direct binding with Aip5C, Bnr1-C (1288–1375 a.a.) alone is insufficient to interact with Bud6C (Figure 4A) (Graziano et al., 2013). This is consistent with the previously identified Bnr1 FH2 domain as the interactive domain in association with Bud6C. Nevertheless, the NPF activity of Bud6 still requires the C-terminus tail of Bnr1 (Graziano et al., 2013). By observing the resolved crystal structure of the Bni1 FH2 domain (PDB: 1UX5), there is a long αT helix extending out from the “doughnut”-shaped structure, and such an αT helix is conserved in Bnr1 (residues 1247–1288 a.a.) (Supplemental Figure S3, A and B) (Xu et al., 2004). Hence we hypothesized that the αT helix of Bnr1 that is adjacent to the C-terminus might be crucial to interact with Bud6C directly. Thus we constructed another Bnr1 C-terminus variant, which includes the αT helix (Bnr1-LC, residues 1247–1375 a.a.). Indeed, we found that Bnr1-LC displayed a high affinity and positive cooperativity in interacting with Bud6C (Figure 4A). A circular dichroism (CD) analysis also showed an ∼40% increase in the alpha-helix conformation of Bnr1-LC compared with Bnr1-C (Supplemental Figure S3, C and D). Based on the above experimental results, we ran MD simulations for Bnr1-LC by assigning an α-helix followed by a random coil as the initial structure (Figure 4B). Similar to the MD simulation setup for the Bni1-Bud6-Aip5 tri-protein set, we placed Bud6core and Aip5C at eight different initial positions relative to Bnr1-LC. The representative stable complexes, which were evaluated by RMSD, were chosen from all simulation trajectories using free energy analysis by PCA (Supplemental Figure S4, A and B). Furthermore, Rosetta scoring identified the most stable Bnr1-Bud6-Aip5 tri-protein complex from the eight simulated conformations (Figure 4C and Supplemental Figure S4C). Interestingly, the Aip5C protein preferentially associates with the C-terminus part of Bnr1-LC, similar to what we observed in the Bni1-Bud6-Aip5 tri-protein complex, whereas Bud6core associates more toward the N-terminus region adjacent to the αT helix in the Bnr1-LC (Figure 4D), suggesting a potential role of the αT helix in stabilizing the interaction between Bud6core and Bnr1-C, consistent with the experimentally validated interaction between Bnr1-LC and Bud6C (Figure 4A). To further test this hypothesis, we performed MD simulations of the interactions between the αT helix-lacking Bnr1-C, Bud6core, and Aip5C starting from the initial positioning the same as the most stable Bnr1-LC complex in Figure 4, B and C (Figure 4E and Supplemental Figure S4, D and E). In the results, the simulation showed that by lacking the αT helix, Bnr1-C is located further away from the Bud6core with reduced direct contacts compared with Bnr1-LC (Figure 4, F–H). In general, for each simulated NF-NPF pair, electrostatic interactions and hydrogen bonds contribute to the majority of interactions (Supplemental Figure S5).
FIGURE 4:
Computer simulation of the complex assembly of Bnr1-Bud6-Aip5. (A) Fluorescence anisotropy binding measurements of Alexa 488-labeled Bnr1-LC (1245-1375 a.a., 30 nM) and Bnr1-C (1288-1375 a.a., 30 nM) by a serial concentration of Bud6C, respectively. The average values from three biological replicates are fitted by the Hill equation to determine the KD. (A.U., arbitrary unit.). (B) The initial structure of Bnr1-LC. (C) The most stable complex conformation (Simulation 8 in Supplemental Figure S4) among the eight sets of Bnr1-Bud6-Aip5 MD simulations (Bnr1-LC, cyan; Aip5C, orange; Bud6core, pink). (D) The minimal distance plot of each residue between Bnr1-LC and Aip5C(pink)/Bud6core (blue) in the final stable complex conformation, as shown in Figure 3C. (E) The initial structure of Bnr1-C. (F) The final Bnr1-C- Bud6core -Aip5C complex conformation after MD simulation (Bnr1-C, blue; Aip5C, orange; Bud6core, pink). (G) The minimal distance plot of each residue between Bnr1-LC (light blue, dashed curve) and Bnr1-C (dark blue, solid curve) with Bud6core in the final stable complex conformation. (H) The conformational distribution of Bnr1-LC/Bnr1-C as a function of the contact ratio (percentage of residues contacting with Bud6core) and the center of mass distance to Bud6core.
Aip5 regulates actin cable formation in vivo with formins and Bud6
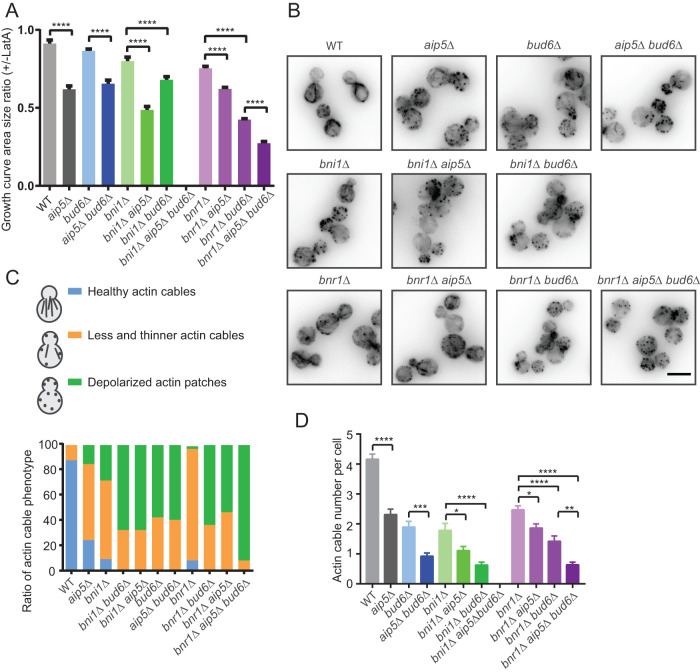
To investigate the in vivo teamwork effort of Bud6 and Aip5 in promoting formin-mediated actin nucleation, we generated multiple single/double/triple mutants that knocked out different NFs and NPFs. Notably, the triple mutant bni1∆ aip5∆ bud6∆, which deleted three polarisome components, was lethal, suggesting an essential function of three NF-NPF proteins for cell physiology (Supplemental Figure S6A). Subsequently, we tested the cell growth of different mutants in the presence of a low dose of 1 µM latrunculin A (LatA), which slightly disrupts the actin cytoskeleton (Supplemental Figure S6, B–D) (Miao et al., 2013, 2016). To compare the sensitivity of different mutants to actin perturbation, we measured the area size under each growth curve, which was used to calculate the ratio change between growth conditions with or without LatA (Figure 5A and Supplemental Figure S6, E and F). Under such a mild perturbation in actin polymerization, wild-type yeast reduces cell growth by ∼10%. However, aip5∆ exhibited high sensitivity to LatA treatment in which we observed an ∼40% growth reduction, which was the highest growth retardation among all the single mutants (Figure 5A); bni1∆ and bnr1∆ led to only an approximately 20–30% reduction in growth on F-actin perturbation (Figure 5A). Further removal of AIP5 in any NF or NPF mutant attributed to an additional sensitivity to LatA by a further decrease in growth rate (Figure 5A), supporting the crucial function of Aip5 in F-actin turnover by itself and cooperation with other nucleation regulatory factors. The above observations are consistent with the dual roles of Aip5 in budding yeast as an NPF for formins and a weak NF (Xie et al., 2019). When comparing different double mutants, the removal of BUD6 or AIP5 seems to provide different sensitivity to LatA treatment in the background that deletes different formins. The triple mutants bnr1∆ aip5∆ bud6∆ demonstrated the greatest extent of genetic sickness in the presence of LatA. Because the triple mutant lacks functional assembly of both formin complexes, Bni1-NPFs and Bnr1-NPFs, our results suggest the teamwork function of the two NPFs in regulating Bnr1-mediated actin turnover for cell growth. To better characterize the protein functions in regulating actin assembly, we examined different mutants expressing the actin cable marker Abp140-3GFP. Compared with the wild type, aip5∆ exhibited a reduction in the population of healthy and intact actin cables by ∼60%, whereas single mutants of formin lost ∼70% (Figure 5, B–D). Consistent with previous findings (Graziano et al., 2011), bud6∆ displays severe disruption of actin cables (Figure 5, B–D). Removal of AIP5 in the mutant background of BNR1 or BNI1 resulted in more cells carrying depolarized actin cables by 30–50% and reduced actin cable numbers, suggesting Aip5 roles in coordinating the function of both formins in vivo (Figure 5, B–D). Together with the lethality of the bni1∆ aip5∆ bud6∆ triple mutant and the severe impairment of actin cable in the bnr1∆ bud6∆ aip5∆ mutant (Figure 5, B–D), our results suggest indispensable tricomponent complex assembly for forming the nucleation core of NF-NPFs underlying actin cable polymerization and cell growth for both Bni1 and Bnr1.
FIGURE 5:
Aip5 regulates actin assembly together with formins in vivo. (A) Quantification of the area ratio of the growth curve of indicated yeast cells. The ratio is calculated by normalizing the untreated cells by 1 µM LatA-treated cells. (B) Representative fluorescence images of actin cable marker Abp140-3GFP from indicated yeast strains. The scale bar is 5 µm. (C) The overall actin cable phenotypes are characterized by normal actin cables, less and thinner actin cables, and depolarized actin cables. The cell number quantified for each strain: wild type (WT), n = 55; aip5∆, n = 58; bni1∆, n = 32; bni1∆ bud6∆, n = 40; bni1∆ aip5∆, n = 49; bud6∆, n = 33; aip5∆ bud6∆, n = 47; bnr1∆, n = 35; bnr1∆ bud6∆, n = 49; bnr1∆ aip5∆, n = 52; bnr1∆ bud6∆ aip5∆, n = 45. (D) Actin cable number analysis in different yeast strains in (B). Error bar, SE; From left to right: wild type (WT), n = 63; aip5∆, n = 70; bni1∆, n = 58; bni1∆ bud6∆, n = 79; bni1∆ aip5∆, n = 60; bud6∆, n = 115; aip5∆ bud6∆, n = 105; bnr1∆, n = 79; bnr1∆ bud6∆, n = 133; bnr1∆ aip5∆, n = 65; bnr1∆ bud6∆ aip5∆, n = 118 cells.
DISCUSSION
Common machinery of cooperative actin nucleation
Actin nucleation is a complex and tunable process regulated by the assembly of NFs and their binding partners. Increasing evidence has demonstrated a common mechanistic framework in which collaborative actin nucleation can be achieved by the multimerization of NPFs or by multicomponent participation. Such teamwork in actin nucleation by different combinations of NFs and NPFs requires the generation of biomolecular complexes that are assembled at lower or higher orders depending on the interaction modes of the complex components. There are many reports in which protein oligomerization creates local connectivity and cooperativity of NF or NPFs, thereby increasing nucleation activities. Dynamic interplays could occur between different NFs in which a joint effort of NFs was implemented for actin nucleation and elongation, such as Formin Cappuccino-Spir and mDia1-APC (Quinlan et al., 2007; Okada et al., 2010; Pfender et al., 2011; Breitsprecher et al., 2012). Several types of multimerization of NPF have also been shown to enhance nucleation activity, such as dimerization of WASP by GST tagging (Higgs and Pollard, 2000) and WASP clustering by the pathogen effector EspFU (Sallee et al., 2008) or SNX9 (Yarar et al., 2007). Furthermore, processive actin polymerase Ena/VASP family proteins were found to enhance their activities in processive actin elongation once clustered by IRSp53 (insulin receptor substrate of 53 kDa) (Disanza et al., 2013; Brühmann et al., 2017; Cheng and Mullins, 2020). Fascin-mediated F-actin bundling also increases the processivity of the nucleator Ena/VASP (Harker et al., 2019), likely through a similar mechanism that increases the location concentration of contact sites and elongates local residence times of VASP proteins. Recently, activation of immune receptor signaling triggered high-order macromolecular assembly, and condensation of WASP increased WASP dwell time locally and thereby enhanced actin nucleation, undergoing liquid-liquid phase separation (Su et al., 2016; Case et al., 2019).
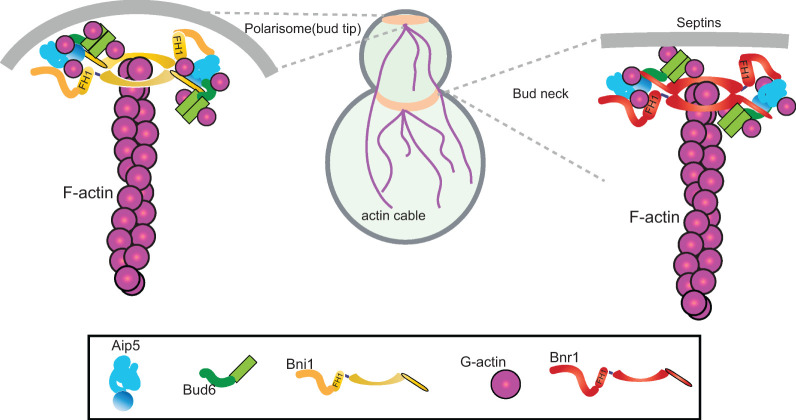
Here we describe an additional cooperative mode in two budding yeast NPFs, Bud6 and Aip5, that jointly promote actin nucleation activities of the same formin, Bni1 or Bnr1, with different modes of interplay. Budding yeast Bud6 was identified as an effective NPF for both Bni1 and Bnr1 using its formin-binding region Bud6core (residues 550–688) and G–actin-binding domain Bud6flank (residues 699–788) (Tu et al., 2012; Graziano et al., 2013). Recently, Aip5 was found to serve as an NPF of Bni1 for nucleating actin cables in budding yeast via the C-terminal Bni1-binding region (Aip5C) (Xie et al., 2019). The C. albicans CaAip5C (residues 786–896 a.a.) and CaBud6C (residues 411–701 a.a.) exhibited cooperative association with CaBni1-C (residues 1545–1733 a.a.) in promoting Bni1-mediated actin nucleation (Xie et al., 2020). Here we found a similar orchestrated pairing for the tri-protein core of the NF-NPF complex for budding yeast Bni1 and Bnr1 and propose a consensus mode between budding yeast Bni1 and filamentous fungal Bni1 as well as between differently localized Bni1 and Bnr1 in assembling the protein core for delivering G-actin into a ring-shaped FH2 structure. One of the mechanisms we have identified is the enhanced interprotein interactions between Aip5 and formin by having Bud6 (Figure 6). However, Bud6 enhanced the affinity of Aip5 to Bni1 and Bnr1 at different kinetics. The Bni1-Aip5 pair seems to be more responsive to engaging Bud6 in changing their interactions than Bnr1-Aip5, which raised an interesting hypothesis that IDR-mediated intermolecular interactions seem to create tunable interactions, packing modes, and functions by modulating the compositional stoichiometry and changing the participating sequences.
FIGURE 6:
NF-NPFs complex for Bni1 and Bnr1 in budding yeast. Bud tip and bud neck localized formin Bni1 and Bnr1, respectively, form tricomponents complex with Aip5 and Bud6 dimers for barbed end nucleation in budding yeast.
The IDRs of Aip5 and Bud6 could drive macromolecular assembly in heterogeneous states under different cellular conditions (Miao et al., 2018; Xie et al., 2019; Xie and Miao, 2021). Although Aip5C does not interact with Bud6C, Aip5C interacts with the N-terminal fragment of Bud6 (1–141) (Glomb et al., 2019); whether full-length Aip5 and Bud6 collaborate to promote formin function needs to be further studied to achieve a more comprehensive understanding of tunable regulation of formin activities by having higher-order assembly of the complex.
Hierarchical assembly of nucleation complexes for tunable activities
Unlike the self-clustering of NPF (WASP) for Arp2/3 complex-mediated nucleation, the cooperative assembly of two formin NPFs might create more diverse assemblies of NF-NPFs for actin polymerization on various signaling events. By having a long IDR, Aip5 proteins localize to the bud tip through inter- and intramolecular interactions with Spa2, as well as cytoplasmic condensates, which contribute to stress adaptation under ATP-depleted conditions (Xie et al., 2019). Such IDR participation introduces dynamic inter- and intracomponent interactions and tunable viscoelastic properties of the macromolecular assemblies. IDRs have highly variable binding affinities for diverse modes of molecular interactions and exhibit a broad range of binding modes with biomolecules depending on the length and amino acid composition, which results in nonlinearities in inter- and intramolecular interactions (Li et al., 2020; Clemens et al., 2021). Our formin-NPF-based interactions all showed positive cooperativity. We still do not understand how different association states of formin-NPFs correspond to actin assembly activities. It is worth future studies to have a comprehensive understanding of how cooperative NPF activities of Aip5 and Bud6 on formins are regulated by their local concentrations, structural flexibility, functional conformation, and molecular equilibrium between the dilute phase in the cytoplasm and the dense phase around the bud neck or bud tip. Nevertheless, the functional core of both sets of the formin-Aip5-Bud6 complex in budding yeast likely requires the direct tri-protein complex formation we have described here, which defines the minimum complex core for potential higher-order assemblies. During cell signal transduction under physiological and pathological conditions, IDR creates diverse states of macromolecular assembly in spatiotemporally regulated manners, which could provide tunable activities of functional complexes, such as NF-NFPs. It has been well documented that phase separation dynamically regulates actin nucleator activities, such as the WASP and Arp2/3 complex during immune signaling (Su et al., 2016; Case et al., 2019) or plant formin during bacterial infection (Sun et al., 2018; Qiao et al., 2019; Ma and Miao, 2020; Ma et al., 2021a,b; Sun et al., 2021). In both of the above scenarios, spatiotemporally regulated compositional stoichiometry plays a critical role in fine-tuning actin remodeling. While Bnr1 has a relatively stable bud neck localization, Bni1 seems to have more exchange between the dense phase (tip and bud neck) and the dilute phase (cytoplasm) (Xie and Miao, 2021). Compared with Bnr1, the dynamic molecular behavior of Bni1 might offer it a more flexible spatiotemporal association with Bud6 and Aip5 than the Bnr1 set. Therefore, Bni1 is likely more sensitive than Bnr1 to environmental perturbations, such as chemical or physical cues, which influence the diffusion, condensation, and activities of Bni1 and its NPFs at emerging bud sites or bud necks (Segal et al., 2000; Ozaki-Kuroda et al., 2001; Buttery et al., 2007; Glomb et al., 2019; Xie et al., 2019). In vivo and in vitro reconstitution systems using the full-length recombinant proteins Aip5, Bud6, and formins might address such complex regulations of actin nucleation in budding yeast. Here our results of multimember teamwork modes for both Bni1 and Bnr1 lay a foundation for future studies of complex and tunable actin remodeling during cell signaling by changing their inter- and intramolecular interactions.
METHODS
Protein expression and purification
The Escherichia coli expression vectors for Bni1FH1C, Bnr1FH1C, Bnr1-C, Aip5C, and Pfy1 contain N-terminal His6-TEV, and Bud6C is fused with N-terminal GST-Prescission-His6-TEV. All the proteins are expressed and purified from E. coli (BL21(DE3) Rosetta T1R) as previously described (Xie et al., 2019). Cells were cultured in 5 ml of TB medium (24 g/l yeast extract, 20 g/l tryptone, 4 ml/l glycerol, phosphate buffer, pH 7.4) for overnight and transferred to 1 liter of TB medium for 4–5 h culture at 37°C. Afterward, 0.5 mM isopropyl-thio-b-D-galactoside was added for protein induction at 18°C for overnight culture. The cells were harvested the next morning and resuspended into the 50 ml lysis buffer (20 mM HEPES, pH 7.4, 500 mM NaCl, 20 mM imidazole) with 1 mM phenylmethylsulfonyl fluoride and one tablet of Pierce protease inhibitor (ThermoFisher). The cells were sonicated for 5 min (10 s on, 30 s off), and the resulting lysate was further clarified by centrifugation at 40,000 × g, 1 h at 4°C. The supernatant was filtered by 0.22 µm Minisart Syringe Filter before loading into a 5 ml HisTrap HF column (GE Healthcare Life sciences) connected to the FPLC system (GE ÄKTA FPLC). The protein was eluted over a gradient injection of 500 mM imidazole. The collected protein peak fractions were further purified by size-exclusion chromatography using a HiLoad 16/600 Superdex200 column (GE Healthcare) in 20 mM HEPES, pH 7.4, 500 mM NaCl. The protein elution peak fractions were examined by the SDS–PAGE and followed by GelCode Blue Stain (ThermoFisher Scientific). The collected target proteins were concentrated and aliquoted to 5–10 µl each tube and frozen in liquid N2.
To prepare monomeric G-actin, the purification procedure is described as before (Xie et al., 2019). In brief, 2 g of rabbit muscle acetone powder was dissolved in 60 ml of ice-cold G-buffer (5 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM dithiothreitol [DTT]) for 30 min, and the solution was filtered by cheesecloth to collect the actin-rich extracts. Such a procedure was repeated two more times on the dissolved rabbit muscle powder to collect a total of 180 ml actin-rich extracts. Furthermore, the actin-rich extract was subjected to centrifugation at 18,000 × g, and the supernatant was collected. To induce actin polymerization, 50 mM KCl and 2 mM MgCl2 were added to the solution for 1 h. Subsequently, 0.8 M of KCl was added and stirred slowly for 30 min to remove actin-binding proteins. To collect polymerized actin filaments, the solution was subjected to centrifugation at 4°C, 95,800 × g for 3 h. Afterward, the F-actin pellets were transferred to the 10 ml homogenizer with a 7 ml G-buffer to be homogenized with the grinder. Furthermore, the F-actin was depolymerized by sonication with 3 s on and 10 s off, and such cycle was repeated four times, followed by dialysis against G-buffer for 1–2 d. We then performed centrifugation at 167,000 × g for 2.5 h to collect the top 2/3 supernatant and further purified through column HiPrep 16/60 SephacrylTM S-300 HR. The collected monomeric actin was then stored at 4 °C with 1 mM sodium azide added and used within 1 mo.
Actin assembly assay
The assembly reaction contains 2 µM G-actin with 5% pyrene actin (Cytoskeleton) where 3 µM yeast profilin was added. First, 10 µM G-actin was converted to Mg2+-ATP-actin for 5 min on ice. Once the desired proteins were mixed with G-actin, the actin polymerization was initiated by adding 10× KME buffer mix (10 mM MgCl2, 10 mM EGTA, and 500 mM KCl), at a total reaction volume of 120 µl. The pyrene-actin fluorescence signal was monitored in a plate reader Cytation 5 (BioTek, USA) at excitation and emission wavelengths of 365 and 407 nm, respectively. To determine the percentage increase in promoting formin activity (P), we first fitted line at the half polymerization of each reaction to reflect the actin assembly rate of each condition, and we calculated P based on function (Sa-Sf)/Sf, where Sa is the slope value of each NPF+formin condition and Sf is the slope value of formin. Afterward, hyperbola fitting function P = Bmax*c/(Kapp + c) was used to determine the Kapp, where c is the concentration of NPF, and Kapp = 0.5 * Bmax, which corresponds to the concentration of NPF that is required for half-maximal stimulation of formin-mediated actin assembly. The values shown in the graph were the average data from three times of independent biological replicates experiments.
Fluorescence anisotropy
The fluorescence anisotropy assay was performed as previously described (Xie et al., 2019). In brief, 60 nM Aip5C-Alexa Fluor 488 was mixed with Bnr1FH1C or Bnr1-C in an equal volume. The reaction buffer was 20 mM HEPES, pH 7.4, 150 mM NaCl, and proteins were incubated in low-volume nonbinding black 384-well plates at room temperature for 1 h. The plate reader Cytation 5 (BioTek, USA) with fluorescence polarization mode (excitation filter: 485 nm, 20 nm bandpass; emission filter: 510 nm, 20 nm bandpass) was used to record the parallel reading I‖ and perpendicular reading I⊥. The fluorescence anisotropy was then calculated by the following equation:
Furthermore, the fluorescence anisotropy value was normalized by blank reading and plotted against the corresponding ligand concentration in Prism Graphpad 8. The binding affinity KD can be determined from the binding curves fitted with the Hill equation. The final graph was shown with means from three independent experiments.
TIRF microscopy
The TIRF microscopy experiment was performed as described before (Xie et al., 2019). In brief, 25 × 50-mm coverslips (Marienfeld Superior) were soaked in 20% sulfuric acid overnight and rinsed thoroughly with sterile water. Afterward, the coverslips were coated with 2 mg/ml methoxy-PEG-silane (Mw: 2000; Laysan Bio) and 2 µg/ml biotin-PEG-silane (Mw: 3400; Laysan Bio) in 80% ethanol (pH 2.0, adjusted by HCl) at 70 °C for overnight. The next day, coverslips were rinsed thoroughly with sterile water and dried in N2 stream, which can be kept at −80 °C for long-term storage. Before each experiment, the functionalized coverslip was attached to a plastic flow cell chamber (Ibidi, sticky-Slide VI 0.4). The flow cell was first incubated for 30 s with buffer HBSA (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, and 1% bovine serum albumin), incubated for 60 s in 0.1 mg/ml streptavidin in buffer HEKG10 (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, 10% [vol/vol] glycerol). Finally, the flow cell chamber was washed with 1× TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.3 mM ATP, 50 mM DTT, 15 mM glucose, 100 µg/ml glucose oxidase, and 0.5% methylcellulose [4000 cP], pH 7.4). Proteins were mixed in TIRF buffer with 1 µM G-actin (10% Oregon Green 488 labeled, 0.5% biotin labeled) with 3 µM yeast profilin and then mixed with an equal volume of 2× TIRF buffer before flowing into the chamber. The still images were acquired at 4–5 min for quantifying actin nucleation seeds number; 5-s interval movies were acquired for 10 min using Apochromat TIRF 100× NA 1.49 (Nikon Instruments) on Nikon ECLIPSE Ti-S inverted microscope with iLAS2 motorized TIRF illuminator (Roper Scientific, Evry Cedex, France). The illumination source, microscope stage, and a Prime 95B sCMOS camera (Photometrics) were all under the control of MetaMorph 7.8 software (Molecular Device, Sunnyvale, CA). The focus was maintained using the Perfect Focus System. To measure actin filament elongation rate, the individual filament in each sample was traced manually for at least 2 min each. To quantify the formin-mediated elongation speed, we selected the fast-growing barbed end population, as profilin-actin has been well characterized to accelerate the formin-mediated elongation speed. The measured actin filament length was then divided by the corresponding time to determine the elongation rate. We used the conversion factor of 370 subunits per micrometer of F-actin to estimate the barbed end elongation rate.
All-atom MD simulation
The MD simulations were performed using the GROMACS 5.1.2 software (Van Der Spoel et al., 2005). The protein structure information was derived from our previous structural studies and approaches (Tanaka et al., 2008; Chen et al., 2012; Deng et al., 2012; Xie et al., 2019, 2020). The Charmm36m force field (Huang et al., 2017) was used to describe proteins with the TIP3P water model (Mackerell Jr et al., 2004) for solvent molecules. The temperature of water and proteins was kept constant, coupled independently for each group of molecules at 300 K with a V-rescale thermostat (Bussi et al., 2007). The pressure was coupled with a Parrinello-Rahman (Parrinello and Rahman, 1981) barostat at 1 atm separately in each of the three dimensions. The temperature and pressure time constants of the coupling were 0.1 and 2 ps, respectively. Integration of the equations of motion was performed by using a leapfrog algorithm with a time step of 2 fs. Periodic boundary conditions were implemented in all systems. A cutoff of 1 nm was implemented for the Lennard–Jones interactions and for the direct space part of the Ewald sum for Coulombic interactions. The Fourier space part of the Ewald splitting was computed by using the particle-mesh-Ewald method (Darden et al., 1993) with a grid length of 0.16 nm and a cubic spline interpolation.
CD
Three hundred microliters of 0.15 mg/ml purified Bnr1-C and 0.075 mg/ml purified Bnr1-LC proteins were prepared in a buffer (50 mM phosphate-buffered saline, pH 7.4) and then loaded in the water-jacketed, 1-mm path length cylindrical quartz cuvette (Hellma). The CD spectra were collected using a Jasco J-710 spectropolarimeter at 1 nm resolution and a scan rate of 200 nm /min at room temperature. Reported molar ellipticities were calculated by subtracting the background spectrum in the Chriascan CD Spectrometer. The CD spectrum was then subjected to DichroWeb (http://dichroweb.cryst .bbk.ac.uk/html/home.shtml) with K2D program for secondary structure analysis (Whitmore and Wallace, 2004, 2008).
Yeast cell growth assay
The overnight culture of yeast cells was reinoculated into fresh YPD (10 g/l yeast extract, 20 g/l peptone, 20 g/l glucose) medium starting from O.D. 600 = 0.2 and allowed to grow to O.D. 600 around 0.8 to 1. Subsequently, 130 µl of cultures were added into a transparent 96-well plate with or without 1 µM LatA. Each condition was replicated four times and monitored at O.D. 600 every 15 min at 25°C. A constant shaking was maintained between each measurement to prevent cell precipitation. The O.D.600 was recorded by the plate reader Cytation 5 (BioTek, USA) for 20 h. To quantify the area size below each growth curve, O.D. 600 = 0.1 was used as the baseline; the area size was calculated by Prism Graphpad 8.
Live-cell fluorescence imaging
All yeast strains used in this study are listed in Supplemental Table S1. Yeast strains were cultured overnight at 25°C in the synthetic complete media + 2% glucose without tryptophan (to minimize auto-fluorescence) and reinoculated into fresh medium to culture until O.D. 600 = 0.6–0.8. Cells were then immobilized onto Concanavalin A (1 mg/ml)-coated coverslips and imaged by wide-field microscope Leica DMi8 (Leica Microsystems) equipped with an ORCA-Flash4.0 LT scientific CMOS camera (Hamamatsu Photonics, Japan) and a Leica× 100 oil immersion objective lens (NA 1.4). For whole-cell imaging, images were acquired continuously at a 0.25-µm interval for a total range of 7.5 µm in the z direction using an exposure time of 200 ms and 1× binning. To characterize actin cable phenotype, the maximum Z-projection images were created from live-cell imaging data by ImageJ.
In vivo concentration measurements for polarisome proteins
We have quantified in vivo protein concentration of Bud6 and Bnr1 using three reference strains expressing Syp1-GFP, Crn1-GFP, and Sec3-GFP as reference. The cytosolic protein concentrations of Syp1, Crn1, and Sec3 were previously determined by fluorescence cross-correlation spectroscopy (Boeke et al., 2014; Picco et al., 2017). Sum slices-Z projection images were used for intensity analysis. As previously reported, we measured the total cytosol signal intensity of reference and polarisome proteins from the ROIs of 10 × 10 pixels from 20 cells (Rowlinson and Widom, 2013). The same methods were used to measure the background and auto-fluorescence signal from the cytosol. A signal intensity standard curve and equation were derived from the reference cytosolic concentration and examined cytosolic signal of Syp1, Crn1, and Sec3. The cytosol concentrations of Bud6 and Bnr1 were calculated based on the above equation in Supplemental Figure S5, whereas tip concentration was derived from the signal intensity ratio.
Protein sequence analysis
Sequence alignment of formins (Bni1 and Bnr1) was carried out through the online server Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the figure was generated by software Jalview.
Statistical analysis
All statistical analyses were performed in GraphPad Prism 8. P values were determined by one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 and ns = no significant). Error bars indicate the SD.
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank Jessica Henty-Ridilla (SUNY Upstate Medical University) for sharing the actin preparation protocol. We thank the National Supercomputing Center Singapore (https://www.nscc.sg/) for assisting the computational work. This study was supported by a NTU startup grant (M4081533), Skin Research Institute of Singapore (SRIS, SIG18002), and MOE Tier 2 (MOE2016-T2-1-005), MOE Tier 3 (MOE2019-T3-1-012) to Y.M., and MOE Tier 1 (2018-T1-001-096) to L.L. in Singapore.
Abbreviations used:
- APC
adenomatous polyposis coli
- CD
circular dichroism
- IDR
intrinsically disordered region
- LatA
latrunculin A
- MD
molecular dynamics
- NF
nucleation factor
- NPF
nucleation promoting factor
- PCA
principal component analysis
- RMSD
root-mean-square deviation
- WASP
WASP family veroprolin homolog
- WAVE
Wiskott–Aldrich syndrome protein
- WRC
WAVE regulatory complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-06-0285) on November 24, 2021.
REFERENCES
- Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K (2017). The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput 13, 3031–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Mullins D, Fletcher D (2019). Force Regulation of Capping and Arp2/3 Nucleation of Branched Actin Networks. Biophys J 116, 252a. [Google Scholar]
- Boeke D, Trautmann S, Meurer M, Wachsmuth M, Godlee C, Knop M, Kaksonen M (2014). Quantification of cytosolic interactions identifies Ede1 oligomers as key organizers of endocytosis. Mol Syst Biol 10, 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL (2012). Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brühmann S, Ushakov DS, Winterhoff M, Dickinson RB, Curth U, Faix J (2017). Distinct VASP tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc Natl Acad Sci USA 114, E5815–E5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M (2007). Canonical sampling through velocity rescaling. J Chem Phys 126, 014101. [DOI] [PubMed] [Google Scholar]
- Buttery SM, Yoshida S, Pellman D (2007). Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell 18, 1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Cheng H-C, Robbins D, Siripala AD, McGhie EJ, Hayward RD, Welch MD, Rosen MK, Koronakis V, Leong JM (2008). Repetitive N-WASP—binding elements of the enterohemorrhagic Escherichia coli effector EspF U synergistically activate actin assembly. PLoS Pathog 4, e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK (2019). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kuo CC, Kang H, Howell AS, Zyla TR, Jin M, Lew DJ (2012). Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol Biol Cell 23, 3814–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KW, Mullins RD (2020). Initiation and disassembly of filopodia tip complexes containing VASP and lamellipodin. Mol Biol Cell 31, 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL (2009). Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol 21, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens L, Dushek O, Allard J (2021). Intrinsic disorder in the T cell receptor creates cooperativity and controls ZAP70 binding. Biophys J 120, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L (1993). Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J Chem Physics 98, 10089–10092. [Google Scholar]
- Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, Lee J, Aguilar C, Ahrens CH, Chang C, et al. (2012). Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci USA 109, 15479–15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Bisi S, Winterhoff M, Milanesi F, Ushakov DS, Kast D, Marighetti P, Romet-Lemonne G, Müller HM, Nickel W (2013). CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J 32, 2735–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD (2011). New mechanisms and functions of actin nucleation. Curr Opin Cell Biol 23, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian MV, Stanishneva-Konovalova T, Lou C, Rands TJ, Pollard LW, Sokolova OS, Goode BL (2018). Integrated control of formin-mediated actin assembly by a stationary inhibitor and a mobile activator. J Cell Biol 217, 3512–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glomb O, Bareis L, Johnsson N. (2019). YFR016c/Aip5 is part of an actin nucleation complex in yeast. Biol Open 8, bio044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL (2011). Mechanism and cellular function of Bud6 as an actin nucleation–promoting factor. Mol Biol Cell 22, 4016–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL (2013). Ligand-induced activation of a formin—NPF pair leads to collaborative actin nucleation. J Cell Biol 201, 595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker AJ, Katkar HH, Bidone TC, Aydin F, Voth GA, Applewhite DA, Kovar DR (2019). Ena/VASP processive elongation is modulated by avidity on actin filaments bundled by the filopodia cross-linker fascin. Mol Biol Cell 30, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2000). Activation by Cdc42 and PIP2 of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol 150, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, MacKerell AD (2017). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Xie M, Bruschweiler R (2020). Quantitative Cooperative Binding Model for Intrinsically Disordered Proteins Interacting with Nanomaterials. J Am Chem Soc 142, 10730–10738. [DOI] [PubMed] [Google Scholar]
- Ma Z, Liu X, Nath S, Sun H, Tran TM, Yang L, Mayor S, Miao Y (2021a). Formin nanoclustering-mediated actin assembly during plant flagellin and DSF signaling. Cell Rep 34, 108884. [DOI] [PubMed] [Google Scholar]
- Ma Z, Miao Y (2020). Review: F-Actin remodelling during plant signal transduction via biomolecular assembly. Plant Sci 301, 110663. [DOI] [PubMed] [Google Scholar]
- Ma Z, Sun Y, Zhu X, Yang L, Chen X, Miao Y (2021b). Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses. Plant Cell, koab261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD (1999). Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA 96, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD Jr, Feig M, Brooks CL III (2004). Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25, 1400–1415. [DOI] [PubMed] [Google Scholar]
- Miao Y, Han X, Zheng L, Xie Y, Mu Y, Yates JR III, Drubin DG (2016). Fimbrin phosphorylation by metaphase Cdk1 regulates actin cable dynamics in budding yeast. Nat Commun 7, 11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Tipakornsaowapak T, Zheng L, Mu Y, Lewellyn E (2018). Phospho-regulation of intrinsically disordered proteins for actin assembly and endocytosis. FEBS J 285, 2762–2784. [DOI] [PubMed] [Google Scholar]
- Miao Y, Wong CC, Mennella V, Michelot A, Agard DA, Holt LJ, Yates JR, Drubin DG (2013). Cell-cycle regulation of formin-mediated actin cable assembly. Proc Natl Acad Sci USA 110, E4446–E4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL (2005). Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem 280, 28023–28033. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL (2004). A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell 15, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL (2010). Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol 189, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y (2001). Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol 21, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Cheng H-C, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM (2008). Hierarchical regulation of WASP/WAVE proteins. Mol Cell 32, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, Rahman A (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys 52, 7182–7190. [Google Scholar]
- Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M (2011). Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol 21, 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco A, Irastorza-Azcarate I, Specht T, Boke D, Pazos I, Rivier-Cordey AS, Devos DP, Kaksonen M, Gallego O (2017). The In Vivo Architecture of the Exocyst Provides Structural Basis for Exocytosis. Cell 168, 400–412.e418. [DOI] [PubMed] [Google Scholar]
- Pollard TD (2007). Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct 36, 451–477. [DOI] [PubMed] [Google Scholar]
- Qiao Z, Sun H, Ng JTY, Ma Q, Koh SH, Mu Y, Miao Y, Gao YG (2019). Structural and computational examination of the Arabidopsis profilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly. J Biol Chem 294, 18650–18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E (2007). Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol 179, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlinson JS, Widom B (2013). Molecular Theory of Capillarity, Mineola, NY: Dover Publications. [Google Scholar]
- Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, Mayer BJ, Lim WA (2008). The pathogen protein EspF u hijacks actin polymerization using mimicry and multivalency. Nature 454, 1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Bloom K, Reed SI (2000). Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell 11, 3689–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Qiao Z, Chua KP, Tursic A, Liu X, Gao YG, Mu Y, Hou X, Miao Y (2018). Profilin negatively regulates formin-mediated actin assembly to modulate PAMP-triggered plant immunity. Curr Biol 28, 1882–1895.e1887. [DOI] [PubMed] [Google Scholar]
- Sun H, Zhu X, Li C, Ma Z, Han X, Luo Y, Yang L, Yu J, Miao Y (2021). Xanthomonas effector XopR hijacks host actin cytoskeleton via complex coacervation. Nat Commun 12, 4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sakamoto S, Kuroda M, Goda S, Gao YG, Tsumoto K, Hiragi Y, Yao M, Watanabe N, Ohta T, Tanaka I (2008). A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus. Structure 16, 488–496. [DOI] [PubMed] [Google Scholar]
- Tu D, Graziano BR, Park E, Zheng W, Li Y, Goode BL, Eck MJ (2012). Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc Natl Acad Sci USA 109, E3424–E3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005). GROMACS: fast, flexible, and free. J Comput Chem 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ (2011). Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci USA 108, 11884–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace B (2004). DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32, W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA (2008). Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400. [DOI] [PubMed] [Google Scholar]
- Xie Y, Loh ZY, Xue J, Zhou F, Sun J, Qiao Z, Jin S, Deng Y, Li H, Wang Y, et al. (2020). Orchestrated actin nucleation by the Candida albicans polarisome complex enables filamentous growth. J Biol Chem 295, 14840–14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Miao Y (2021). Polarisome assembly mediates actin remodeling during polarized yeast and fungal growth. J Cell Sci 134, jcs247916. [DOI] [PubMed] [Google Scholar]
- Xie Y, Sun J, Han X, Turšic´-Wunder A, Toh JD, Hong W, Gao Y-G, Miao Y (2019). Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization. Nat Commun 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723. [DOI] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2007). SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell 13, 43–56. [DOI] [PubMed] [Google Scholar]
- Zheng P, Nguyen TA, Wong JY, Lee M, Nguyen T-A, Fan J-S, Yang D, Jedd G (2020). Spitzenkörper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds. Nat Commun 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM (2011). mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.