Abstract
Prediction of hepatocellular carcinoma (HCC) risk is an urgent unmet need in patients with non-alcoholic fatty liver disease (NAFLD). In cohorts of 409 patients with NAFLD from multiple global regions, we defined and validated hepatic-transcriptome and serum-secretome signatures predictive of long-term HCC risk in patients with NAFLD. A 133-gene signature, Prognostic Liver Signature [PLS]-NAFLD, predicted incident HCC over up to 15 years of longitudinal observation. High-risk PLS-NAFLD was associated with IDO1+ dendritic cells and dysfunctional CD8+ T cells in fibrotic portal tracts along with impaired metabolic regulators such as fibroblast growth factor (FGF)19, FGF21, and farnesoid X receptor signaling. PLS-NAFLD was validated in independent cohorts of patients with NAFLD who were HCC-naïve (HCC incidence rates at 15 years were 22.7% and 0% in high- and low-risk patients, respectively) or HCC-experienced (de novo HCC recurrence rates at 5 years were 71.8% and 42.9% in high- and low-risk patients, respectively). PLS-NAFLD was bioinformatically translated into a 4-protein secretome signature, PLSec-NAFLD, which was validated in an independent cohort of HCC-naïve patients with NAFLD and cirrhosis (HCC incidence rates at 15 years were 37.6% and 0% in high- and low-risk patients, respectively). Combination of PLSec-NAFLD with our previously defined etiology-agnostic PLSec-AFP yielded improved HCC risk stratification. PLS-NAFLD was modified by bariatric surgery, lipophilic statin, and IDO1 inhibitor, suggesting that the signature can be used for drug discovery and as a surrogate endpoint in HCC chemoprevention clinical trials. Collectively, PLS/PLSec-NAFLD may enable NAFLD-specific HCC risk prediction and facilitate clinical translation of NAFLD-directed HCC chemoprevention.
One Sentence Summary:
Hepatic transcriptome and serum secretome-derived signatures predict long-term HCC risk and preventive intervention efficacy in patients with NAFLD.
Introduction
Non-alcoholic fatty liver disease (NAFLD), alongside the global obesity epidemic, is rapidly emerging as a dominant liver disease etiology that leads to progressive liver fibrosis and its terminal stage, cirrhosis (1). Cirrhosis is a well-established high-risk condition for developing hepatocellular carcinoma (HCC), the fastest rising cancer-related mortality in the U.S. (2). Given the survival benefit of diagnosing early-stage HCC, practice guidelines recommend semi-annual HCC screening in patients with cirrhosis (3). However, in real-world clinical practice, regular screening is applied in <25% of at-risk patients, resulting in frequent late-stage diagnoses and poor survival. These gaps in care highlight an issue of insufficient medical resources to conduct recommended ultrasound-based screening among the large at-risk population (4). Furthermore, HCC screening in NAFLD is more challenging compared to other etiologies such as viral hepatitis due to the vast size of the patient population and associated low HCC incidence rate; the estimated global prevalence of NAFLD is 25%, whereas annual HCC incidence rate is only 0.1–0.6% in biopsy-proven NAFLD cirrhosis, which is below the cut-off of 1.5% to justify HCC screening as cost-effective (5–7). Thus, HCC risk prediction is particularly critical in NAFLD to identify a subset of patients with elevated likelihood of developing HCC among the larger, mostly indolent NAFLD population.
To date, several clinical risk scores have been proposed to identify patients with elevated HCC risk, although refined risk prediction is needed to further reduce the number of patients with NAFLD who should be closely monitored by HCC screening. We previously developed biomarkers predictive of long-term HCC risk using liver tissue (Prognostic Liver Signature [PLS]) (8–11) and serum (Prognostic Liver Secretome signature [PLSec]) (12) in patients with chronic liver disease from viral and metabolic etiologies, including NAFLD. However, there is still room for improvement, especially in identifying patients with negligible risk, who may not require HCC screening as well as identify those at highest risk to whom intensive screening efforts could be targeted (13). To address the challenge, we identified a NAFLD-specific HCC risk signature as a “plug-in” module to refine the etiology-agnostic risk prediction by utilizing prospective-specimen-collection, retrospective-blinded-evaluation (PRoBE) design for biomarker validation (14).
Results
Study design
Study design and patient demographics are summarized in Fig. 1 and Table 1. Because the HCC incidence rate is low in HCC-naïve patients with NAFLD (<1% per year) (6, 7), it is challenging to identify a molecular signature reliably associated with time to HCC development with sufficient statistical power (Fig. S1A). We previously addressed the challenge when we developed the etiology-agnostic PLS by defining the biomarker in patients with a history of prior HCC based on the two following assumptions. First, their livers are already primed as soil for metachronous and multicentric carcinogenesis with approximately three-time higher incidence rate compared to HCC-naïve patients. Second, HCC incidences (clinically recognized as HCC recurrences) in these patients are likely de novo HCC recurrence, that is, metachronous recurrence clonally independent from previously treated tumor, as shown in our previous loss-of-heterozygosity analysis (15). Subsequently, PLS was successfully validated in independent cohorts of HCC-naïve as well as HCC-experienced patients with chronic liver disease (9–11). Herein, we followed the same strategy to identify and validate a NAFLD-specific HCC risk signature (PLS-NAFLD). We first defined the signature in a cohort of 48 patients with prior history of HCC (derivation set) for emergence of de novo HCC recurrence (Fig. S1B). The signature was subsequently validated in independent external cohorts of 106 HCC-naïve patients with NAFLD (tissue validation set 1) and 59 HCC-experienced patients with NAFLD who previously underwent curative surgical tumor resection (tissue validation set 2). Further, we translated PLS-NAFLD to a serum-protein panel, PLSec-NAFLD, using our computational algorithm, TexSEC (12), and externally validated it in an independent cohort of 59 HCC-naïve patients with NAFLD and cirrhosis (serum validation set). Our objectives were to confirm that NAFLD-specific HCC risk biomarker achieves risk stratification with HR > 2 that would enable cost-effective risk-stratified HCC screening, and to confirm improved prognostic prediction by our etiology-agnostic HCC risk biomarker, PLS/PLSec. Prognostic association of the etiology-agnostic PLS in the derivation and tissue validation sets was limited, supporting exploration of NAFLD-specific biomarker to refine the risk prediction (Fig. S1C–E).
Fig. 1. Study design.
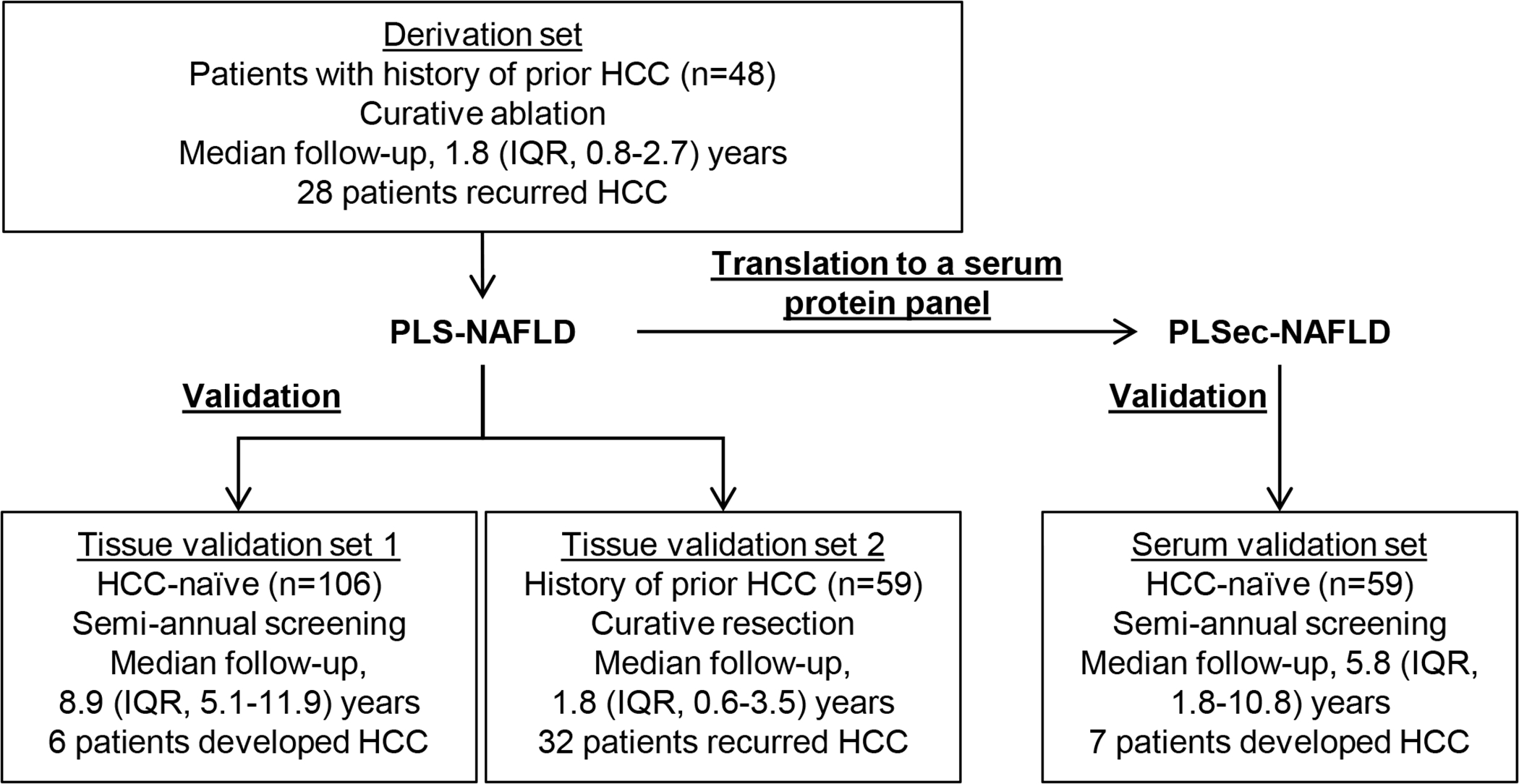
PLS-NAFLD was derived in a cohort of patients with history of prior NAFLD-associated HCC who underwent curative ablation therapies and validated in HCC-naïve and HCC-experienced patients with NAFLD. PLS-NAFLD was translated to serum protein-based PLSec-NAFLD and validated in an independent cohort of HCC-naïve patients with cirrhosis from NAFLD.
Table 1.
Clinical demographics of derivation and validation sets.
| Variables | PLS-NAFLD | PLSec-NAFLD | ||
|---|---|---|---|---|
| Derivation set (n=48) HCC-experienced |
Tissue validation set 1 (n=106) HCC-naïve |
Tissue validation set 2 (n=59) HCC-experienced |
Serum validation set (n=59) HCC-naïve |
|
| Age (y) | 73 (67–75) | 53 (42–62) | 72 (65–77) | 55 (50–62) |
| Male sex | 16 (33%) | 64 (60%) | 44 (75%) | 23 (39%) |
| BMI (kg/m2) | 27.0 (25.4–29.2) | 26.5 (24.7–29.7) | 23.4 (22.0–24.9) | 33.7 (28.6–38.1) |
| Obesity | 37 (79%) | 77 (73%) | 10 (24%) | 29 (49%) |
| Diabetes | 31 (65%) | 59 (57%) | 18 (43%) | 42 (71%) |
| AST (IU/L) | 44 (36–60) | 39 (29–64) | 24 (19–32) | 50 (36–72) |
| ALT (IU/L) | 36 (22–47) | 73 (50–105) | 22 (16–36) | 41 (33–67) |
| Albumin (g/dL) | 3.7 (3.4–4.1) | 4.6 (4.3–4.8) | 3.9 (3.7–4.2) | 3.4 (3.1–3.7) |
| Total bilirubin (mg/dL) | 0.9 (0.7–1.2) | 0.8 (0.6–1.0) | 0.7 (0.5–0.9) | 1.2 (0.8–1.8) |
| Platelet count (×103/μL) | 123 (75–165) | 221 (192–261) | 175 (122–254) | 97 (73–137) |
| Child-Pugh class A | 40 (83%) | 106 (100%) | 58 (98%) | 23 (39%) |
| Fibrosis stage (0–2/3–4) | 5/43 (10%/90%) |
80/26 (75%/25%) |
35/24 (59%/41%) |
- |
| NAFLD activity score | 3.0 (2.0–4.3) | 4.0 (3.0–5.0) | 1.0 (0–2.0) | - |
| NAFLD fibrosis score | 3.79 (3.04–4.38) | 0.78 (−0.27–1.87) | 2.30 (0.93–3.03) | 1.65 (0.56–3.09) |
| FIB-4 index | 4.56 (2.80–8.03) | 1.05 (0.74–1.69) | 2.01 (1.36–2.95) | 4.87 (3.34–7.24) |
| AJCC stage I | 33 (69%) | - | 47 (80%) | - |
| Single nucleotide polymorphism (SNP)* | ||||
| PNPLA3 rs738409 (CC/CG/GG) | 2/19/27 (4%/40%/56%) | 16/48/42 (15%/45%/40%) | 8/32/19 (14%/54%/32%) | - |
| TM6SF2 rs58542926 (CC/CT/TT) | 41/6/1 (85%/13%/2%) | 83/21/2 (78%/20%/2%) | 53/6/0 (90%/10%/0%) | - |
| MBOAT7 rs641738 (CC/CT/TT) | 48/0/0 (100%/0%/0%) | 106/0/0 (100%/0%/0%) | 58/1/0 (98%/2%/0%) | - |
| GCKR rs1260326 (TT/TC/CC) | 41/6/1 (85%/13%/2%) | 80/18/8 (76%/17%/8%) | 53/3/3 (90%/5%/5%) | - |
| HSD17B13 rs72613567 (TT/TTA/TATA) | 38/5/5 (79%/10%/10%) | 61/24/21 (58%/23%/20%) | 47/4/8 (80%/7%/14%) | - |
| MTARC1 rs2642438 (GG/GA/AA) | 41/3/4 (85%/6%/8%) | 45/14/47 (43%/13%/44%) | 45/0/14 (76%/0%/24%) | - |
| Genetic Risk Score† | 3 (3–4) | 3 (2–4) | 3 (2.5–4) | - |
Categorical variables are shown as n (%). Continuous variables are shown as median (IQR).
Most right shows risk genotype for PNPLA3, TM6SF2, MBOAT7, and GCKR and protective genotype for HSD17B13 and MTARC1.
A combined score of PNPLA3, TM6SF2, and HSD17B13 (57).
PLS, Prognostic Liver Signature; NAFLD, non-alcoholic fatty liver disease; PLSec, Prognostic Liver Secretome Signature; BMI, body mass index; AST, aspartate transaminase; ALT, alanine transaminase; AJCC, American Joint Committee of Cancer; IQR, interquartile range.
Derivation of tissue-based NAFLD HCC risk signature, PLS-NAFLD
In the derivation set we defined a 133-gene PLS-NAFLD consisting of 80 high- and 53 low-risk genes (Fig S2A, Table S1), which classified 19 (40%), 9 (18%), and 20 (42%) patients into high-, intermediate-, and low-risk groups, respectively. Of note, high- and intermediate-risk groups showed similar HCC incidence rates (Fig. S2B), suggesting that the main utility of this signature would be to distinguish patients with low risk of HCC development. Indeed, annual rate of (assumedly metachronous) HCC recurrence was substantially less frequent in the low-risk group (7.2%) compared to the rest (60.4%). Therefore, for subsequent validation, we merged the high- and intermediate-risk groups as high-risk to be compared with the low-risk group (Fig. 2A, B). PLS-NAFLD was independent of any clinical, histological, or NAFLD-related genetic features, including fibrosis stage and single nucleotide polymorphisms (Fig. S2C–E), supporting that PLS-NAFLD provides complementary prognostic information beyond these clinically available variables. High-risk PLS-NAFLD showed a robust prognostic association in various multivariable models adjusted for potential confounding variables (Table S2, 3). None of previously reported gene expression signatures for NAFLD was predictive of long-term HCC risk (Table S2). There was no significant prognostic association of PLS-NAFLD in patients affected with other etiologies such as hepatitis C, hepatitis B, or alcohol-related liver disease (Fig. S2F, G), supporting NAFLD-specificity of the signature (8, 16, 17).
Fig. 2. Derivation of PLS-NAFLD.
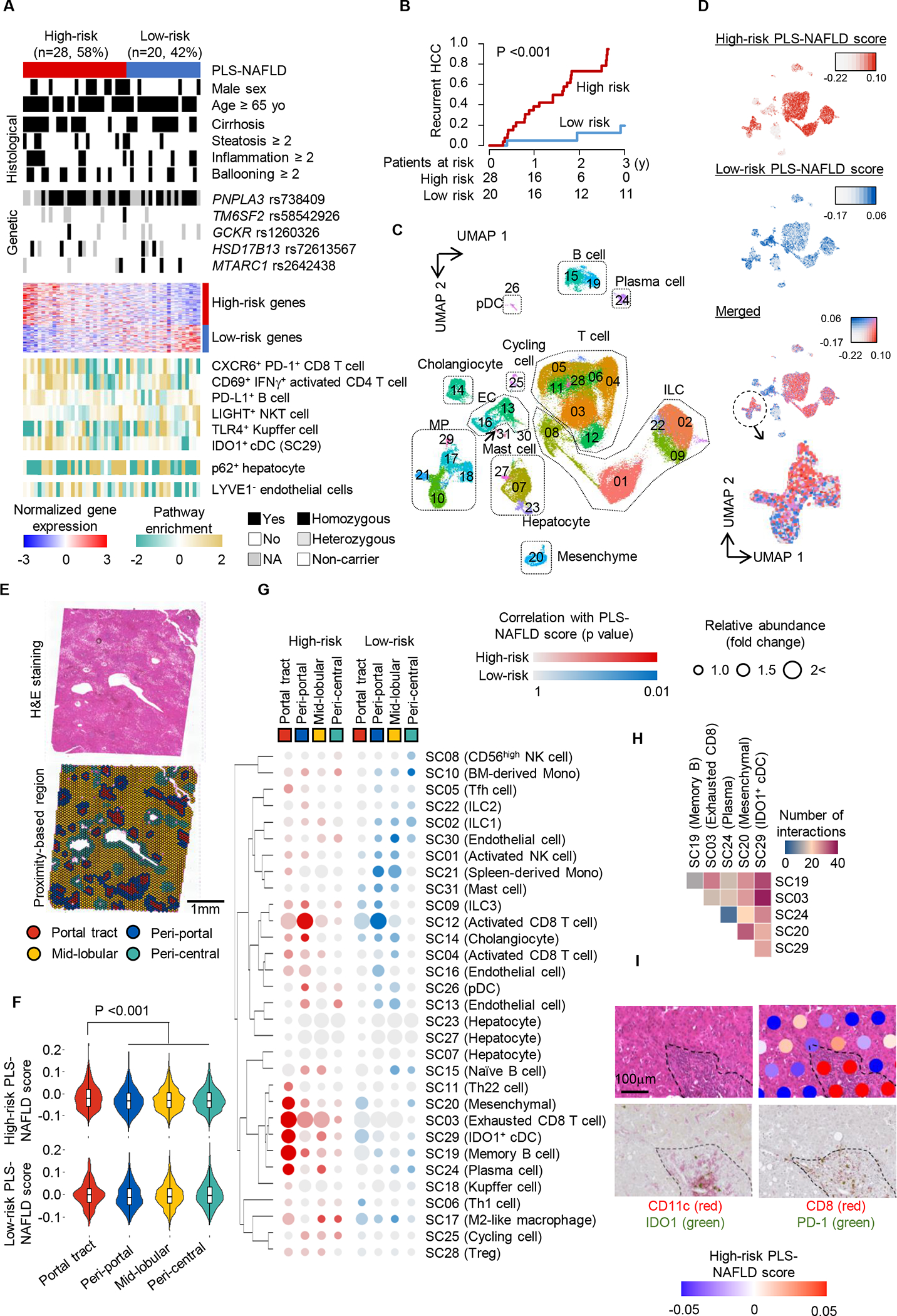
(A) Expression of PLS-NAFLD genes and clinico-histological and genetic features in the derivation set. (B) Prognostic association of PLS-NAFLD. (C) Clusters of human liver-derived single cells from a meta-analysis of four scRNA-seq datasets representing healthy to NAFLD-affected livers. (D) Induction of high- and low-risk PLS-NAFLD genes measured by the average relative expression (“score”) across human hepatic single-cell clusters. (E) H&E staining (upper panel) and histological architecture determination over grid-like “spots” for spatial transcriptome profiling (lower panel) of liver tissue from a patient with NAFLD. (F) Induction of high- and low-risk PLS-NAFLD genes measured by the “score” across the four histological architectures. (G) Correlation of high- and low-risk PLS-NAFLD scores with relative abundance of the hepatic single cell clusters across the four histological architectures. (H) Number of inferred cell-cell interactions between the five hepatic single cell clusters co-presenting in the portal tracts and contributing to the high-risk PLS-NAFLD induction. (I) Representative portal tract (outlined by dotted line) with the high-risk PLS-NAFLD induction determined by the spatial transcriptome profiling (right upper panel), where IDO1+ cDCs (left lower panel) and PD-1+ CD8+ T cells (right lower panel) were co-localized in close proximity.
Molecular dysregulations encoded in PLS-NAFLD in hepatic cells
Molecular pathway analyses revealed that the high-risk pattern of PLS-NAFLD was associated with T cell-mediated immune reaction, B cell activation, and macrophage migration, as well as T cell exhaustion (Fig. S2H, Table S4). High-risk PLS-NAFLD was also associated with previously reported pathways involved in NAFLD-driven disease progression and hepatocarcinogenesis, including the tumor necrosis factor (TNF) pathway (18), autophagy and YAP signaling (19), and disrupted circadian rhythm (20). Activation of STAT3, but not STAT1, was observed in patients with high-risk PLS-NAFLD, consistent with previous observations (21). In contrast, metabolic pathway regulators involved in homeostatic maintenance of normal liver function, particularly bile acid metabolism as indicated by fibroblast growth factor (FGF)19 (18) and FGF21 (22) and nuclear receptor signaling, for example the farnesoid X receptor (FXR) pathway, were downregulated in association with high-risk PLS-NAFLD. A transcriptional target gene signature of an FXR agonist, obeticholic acid (23), was also suppressed, suggesting that patients with high-risk PLS-NAFLD prediction may be candidates for obeticholic acid treatment. Despite no prognostic association with histological inflammation grade (Table S2), PLS-NAFLD encoded diverse inflammatory pathways. This highlights limitation of gross histological evaluation of inflammation (semi-quantitative scoring of lymphocyte infiltration) and the importance of characterizing immune cell subsets and their functional status in predicting long-term HCC risk over up to 11 years.
We next determined cellular sources of the high-risk PLS-NAFLD based on a meta-analysis of four human hepatic single-cell RNA sequencing (scRNA-seq) datasets (108,855 cells in total) representing all major cell types anticipated in healthy to NAFLD-affected cirrhotic livers (Table S5) (24–28). We identified 31 single-cell clusters (SC01 to SC31) based on similarity of transcriptome pattern (Fig. 2C, Fig. S3A, Table S6). As expected, a uniform manifold approximation and projection (UMAP) plot showed clustering of the cells by cell type, not by dataset (Fig. 2C, Fig. S3B). Among the liver cell clusters, we confirmed the presence of NAFLD-associated cell types such as CXCR6+PD-1+ CD8 T cells (29) and TLR4+ Kupffer cells (Fig. S3C) (30), some of which were associated with the high-risk pattern of PLS-NAFLD.
The high-risk PLS-NAFLD score defined by average relative expressions of signature genes (31) was mainly associated with immune cell clusters, whereas the low-risk score was mostly associated with the endothelial cell clusters (Fig. 2D). Within the mononuclear phagocyte (MP) cluster, high- and low-risk scores were enriched in distinct subclusters (for example, high- and low-risk genes in clusters SC29 and SC10, respectively) (Fig. 2D, Fig. S3D). We found that the high-risk-related SC29 cluster contained XCR1+ type 1 conventional DCs implicated in NAFLD progression, but not yet in HCC risk (Fig. S3E, F) (32). In contrast, the low-risk-related SC10 cluster contained bone-marrow-derived monocytes (BMDCs), which are located at the bifurcation of differentiation to either matured macrophages or DCs in hepatic cirrhosis (25). These results suggest that mutually exclusive presence of these cell types contributes to the generation of an HCC-prone hepatic microenvironment in NAFLD.
Across all cell type clusters, we found that cells in SC29 almost exclusively expressed IDO1, encoding indoleamine 2,3-dioxygenase 1 (IDO1) (Fig. S3G). IDO1 is a rate-limiting enzyme that converts L-tryptophan to L-kynurenine, which suppresses anti-tumor immunity via induction of tolerogenic cDCs and T cell anergy (33). Indeed, enrichment of SC29 signature significantly correlated with PLS-NAFLD-based risk prediction (rho=0.44; p=0.002) (Fig. 2A), suggesting that IDO1 may be a therapeutic target to modulate high-risk PLS-NAFLD and reduce HCC incidence in NAFLD.
We next spatially mapped PLS-NAFLD onto histological architecture in human NAFLD-affected liver (F-stage, 2; NAFLD activity score, 5) using genome-wide spatial transcriptome profiling (Fig. S4A). First, we determined portal tract, peri-portal, mid-lobular, and peri-central areas (based on grid-like regions called “spots” in the assay) according to histologically-determined portal area and central vein and computationally-defined proximity to these two histological architectures (Fig. 2E, Fig. S4B, C). The spots with elevated high-risk PLS-NAFLD score were mainly observed in portal tracts, whereas elevated low-risk scores distributed across the four types of the area/spot (Fig. 2F).
Elevated high-risk score in portal tracts was associated with presence of SC29 (IDO1+ cDCs), SC03 (exhausted CD8+ T cells), SC19 (memory B cells), SC20 (mesenchymal cells), and SC24 (plasma cells), whereas low-risk score in peri-central and mid-lobular areas was associated with SC30 (ECs) and SC10 (BMDMs) based on a non-negative matrix factorization-based cell type deconvolution (Fig. 2G) (34). The five high-risk-PLS-NAFLD-related cell types as well as their co-localization were most frequently observed in portal tracts (Fig. S4D, E). Among the five cell types, cell-cell interactions between SC29 (IDO+ cDCs) and SC03 (exhausted CD8+ T cells) stood out (Fig. 2H, Fig. S4F) (35). IDO1+ cDCs and PD-1+ CD8+ T cells were indeed in close proximity in portal tracts with elevated high-risk PLS-NAFLD scores (Fig. 2I).
Independent validation of PLS-NAFLD in HCC-naïve patients with NAFLD
We externally tested PLS-NAFLD in an independent cohort of 106 HCC-naïve patients with NAFLD (tissue validation set 1) (Fig. 3A, Table 1). During a median follow-up of 8.9 (interquartile range [IQR], 5.1–11.9) years, HCC developed in 6 patients (annual incidence rate, 0.6%). The signature classified 71 (67%) and 35 (33%) patients into high- and low-risk groups, respectively (Fig. 3B). There was no association between age and high-risk PLS-NAFLD (p=0.14). Annual HCC incidence rates were 0.9% and 0% and 15-year probabilities were 22.7% (95% confidence interval [CI], 3.6%−38.0%) and 0% (95% CI, 0%−0%) in the high- and low-risk groups, respectively (Fig. 3C). High-risk PLS-NAFLD was independently associated with incident HCC (adjusted hazard ratio [aHR] with Firth’s correction (36), 260.0; 95% confidence interval [CI], 1.02-Inf) (Table S3). Of note, three HCC cases developed in patients with minimal fibrosis (F-stage, 1), a patient population typically excluded from the recommended regular HCC screening, who were identified as high-risk by PLS-NAFLD.
Fig. 3. Independent validation of PLS-NAFLD in HCC-naïve and HCC-experienced patients with NAFLD.
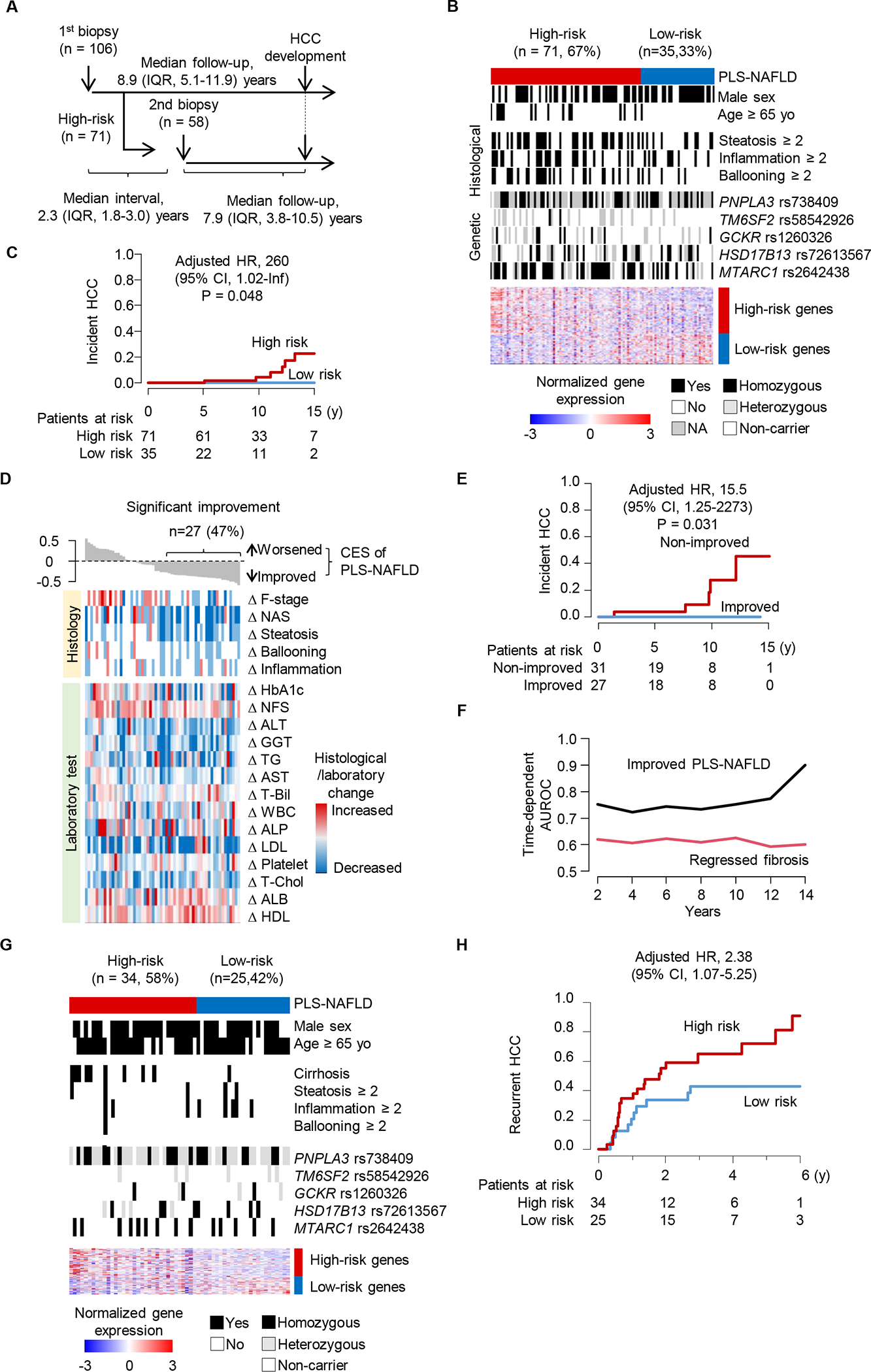
(A) Overview of tissue sampling and clinical follow-up in the tissue validation set 1. (B) Expression of PLS-NAFLD genes and clinico-histological and genetic features in tissue validation set 1. (C) Prognostic association of PLS-NAFLD with incident HCC in tissue validation set 1. (D) Longitudinal changes in PLS-NAFLD-based HCC risk measured by combined enrichment score (CES) and clinico-histological features and laboratory tests between the serial biopsies. (E) Prognostic association of improved PLS-NAFLD. (F) Time-dependent AUROC of improved PLS-NAFLD and regressed fibrosis (decreased F-stage). (G) Expression of PLS-NAFLD genes with clinico-histological and genetic features in tissue validation set 2. (H) Prognostic association of PLS-NAFLD in tissue validation set 2.
Refined HCC risk prediction by repeated PLS-NAFLD measurement
Clinical biomarkers such as alpha-fetoprotein (AFP) often fluctuate over time, and their change over time-series measurements better inform prognosis compared to a single cross-sectional measurement (37). We hypothesized that repeated measurement of PLS-NAFLD might improve HCC risk prediction, which we tested in tissue validation set 1 using follow-up liver biopsies. Given that there was no incident HCC in the low-risk group, we profiled 58 out of 71 high-risk patients who had a second biopsy with a median interval of 2.3 (IQR, 1.8–3.0) years. We used the combined enrichment score (CES) (11) to quantify modulated PLS-NAFLD-based prognostic risk between the serial biopsies. There were 27 (47%) patients with decreased HCC risk, 22 (38%), with stable risk, and 9 (16%) with worse PLS-NAFLD-based risk of developing HCC (Fig. 3D). All HCCs developed in patients without improvement in PLS-NAFLD-based risk. Change in PLS-NAFLD risk was significantly associated with HCC development (aHR with Firth’s correction (36), 15.5; 95% CI, 1.25–2273) and showed stably high discrimination ability (Fig. 3E, F). Annual incidence rates were 2.2% and 0% in PLS-NAFLD-non-improved and improved patients, respectively. The incidence rate among non-improved patients is higher than the traditionally used cut-off of 1.5% to justify cost-effectiveness of the HCC screening (3). CES was significantly correlated with change in fibrosis stage (rho=0.41; false discovery rate [FDR]=0.028) (Fig. 3D). The CES also showed better prognostic capability than change in fibrosis stage (Fig. 3F, Table S7). Collectively, repeated assessment of PLS-NAFLD would refine HCC risk estimation in NAFLD.
Independent validation of PLS-NAFLD in clinical NAFLD with history of prior HCC
We further externally validated PLS-NAFLD in 59 HCC-experienced patients with NAFLD who underwent complete tumor resection with no radiological/pathological residual tumor (tissue validation set 2). PLS-NAFLD classified 34 (58%) and 25 (42%) patients into high- and low-risk groups, respectively. The high-risk prediction was associated with HCC recurrence after curative resection (aHR, 2.38; 95% CI, 1.07–5.25) (Fig. 3G, H, Table S3). HCC incidence rates were 34.0% and 13.8% at 1 year and 71.8% and 42.9% at 5 years in high- and low-risk patients, respectively.
Serum protein-based surrogate marker of PLS-NAFLD (PLSec-NAFLD)
Requirement of liver tissue would be a major bottleneck for widespread use of tissue-based PLS-NAFLD, particularly in HCC-naïve patients who do not undergo liver surgery. To address this limitation, we converted tissue-transcriptome-based PLS-NAFLD into a blood-based four-protein secretome panel (PLSec-NAFLD) using our previously developed computational pipeline, TexSEC (www.texsec-app.org) (12) (Fig. S5A). PLSec-NAFLD consists of 2 high-risk proteins (lymphotactin and progranulin, encoded by XCL1 and GRN, respectively) and 2 low-risk proteins (angiopoietin 2 and hepatocyte growth factor receptor, encoded by ANGPT2 and MET, respectively). Lymphotactin is a ligand of XCR1, suggesting that the panel monitors status of the XCR1+ cDCs in the liver (38). PLSec-NAFLD was implemented in a US Food and Drug Administration (FDA)-approved multiplex clinical diagnostic platform, xMAP assay (Luminex), and a cut-off of >1 was defined for prediction of high risk in two independent optimization sets (n=73 and n=72) covering broad stages of NAFLD (Fig. S5B).
PLSec-NAFLD was subsequently externally validated in an independent cohort of 59 HCC-naïve patients with NAFLD cirrhosis (serum validation set). PLSec-NAFLD identified 42 (71%) high-risk and 17 (29%) low-risk patients (Fig. 4A). Similar to tissue-based PLS-NAFLD, PLSec-NAFLD-based prediction was independent of known HCC-risk-associated clinical variables. Annual HCC incidence rates were 2.7% and 0% and 15-year probabilities were 37.6% and 0% in high- and low-risk patients, respectively (Fig. 4B). High-risk predictions were significantly associated with HCC development (aHR with Firth’s correction (36), 32.4; 95% CI, 1.13–1.96×1010), and PLSec-NALFD was well-calibrated over time (Fig. 4C, Table S3). Of note, all HCC developed in high-risk patients, supporting that the biomarker might discriminate low-risk patients who are spared from the regular HCC screening because of negligible HCC risk.
Fig. 4. Independent validation of serum-based PLSec-NAFLD in HCC-naïve patients with NAFLD cirrhosis.
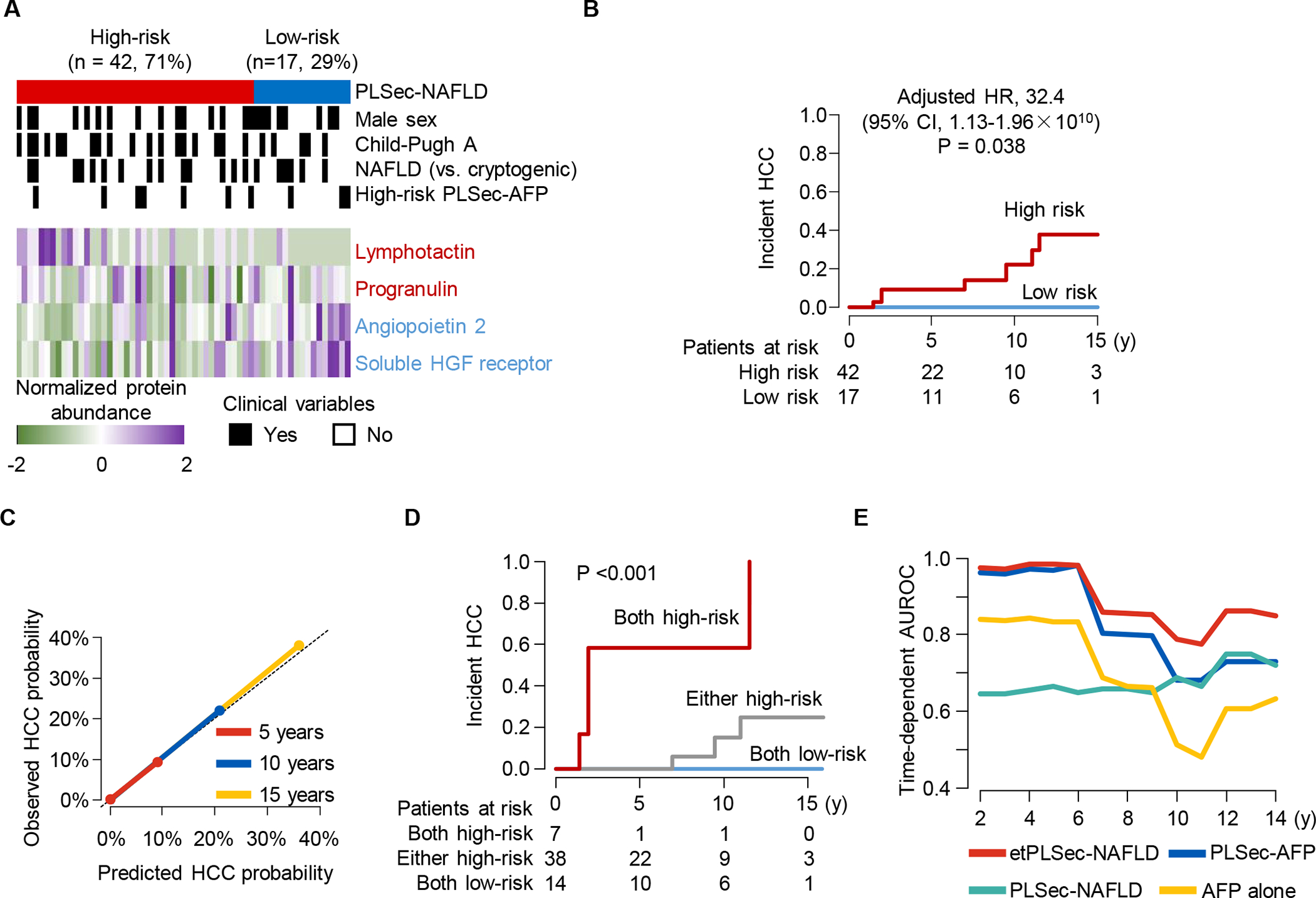
(A-B) PLSec-NAFLD protein abundance (A) and prognostic association of high-risk PLSec-NAFLD (B) in the serum validation set. (C) Calibration plot of PLSec-NAFLD at 5, 10, and 15 years after blood collection for PLSec-NAFLD assessment. The diagonal dotted line indicates ideal calibration. (D) Prognostic association of etPLSec-NAFLD. (E) Time-dependent AUROC of etiology-agnostic PLSec-AFP, etiology-specific PLSec-NAFLD, and their combination named etPLSec-NAFLD.
Last, to test whether the etiology-specific PLSec-NAFLD can serve as a “plug-in” module to improve prognostic prediction by the etiology-agnostic PLSec-AFP (12), we tested a combination of these HCC risk predictive signatures as an integrative etiology-specific method of predicting NAFLD-related HCC risk (etPLSec-NAFLD) in the serum validation set. First, we confirmed that high-risk PLSec-AFP (n=10, 17%) was associated with HCC development (Fig. S5C). High-risk predictions with PLSec-AFP and PLSec-NAFLD were independent, suggesting their complementary prognostic utility (Fig. S5D). Integration of two signatures into etPLSec-NAFLD substantially improved HCC risk stratification. Annual HCC incidence rates were 13.3%, 1.2%, and 0% in patients with high-risk prediction by both (etPLSec-NAFLD high-risk group), either (intermediate-risk group), and neither (low-risk group) of the signatures, respectively (Fig. 4D). High-risk etPLSec-NAFLD scores were significantly associated with HCC development (aHR with Firth’s correction (36), 100.7; 95% CI, 2.95-Inf for high-risk compared to low-risk patients) (Table S3). In addition, this integration achieved consistently high prognostic performance compared to its individual components over time (Fig. 4E).
Therapeutic modulation of PLS-NAFLD as surrogate biomarker of future HCC incidence
Various NAFLD therapeutics have been actively explored, targeting specific pathologies such as steatosis, inflammation, and fibrosis (39). However, it is difficult to estimate the impact of their short-term therapeutic modulation on long-term prognosis within the typical timeframe of clinical trial (15, 40). To address this challenge, we evaluated the utility of PLS-NAFLD as a surrogate biomarker of HCC development. As proof of concept, we assessed the magnitude of PLS-NAFLD modulation and its association with future HCC risk in patients with NAFLD in the following three distinct types of therapeutic interventions.
The first example is bariatric surgery, which is associated with reduced HCC incidence in NAFLD patients (41). We analyzed hepatic transcriptome profiles of paired liver biopsies obtained before and after therapies in 79 patients with NAFLD (Fig. 5A) (42, 43). Based on pre-treatment hepatic transcriptomes, 68 (86%) and 11 (14%) patients were classified to high- and low-risk groups, respectively. Among the high-risk patients, bariatric surgery showed more frequent significant reduction of PLS-NAFLD-based HCC risk compared to lifestyle-only intervention (odds ratio [OR], 0.21; 95% CI, 0.06–0.73), which is comparable with the magnitude of reduction in HCC incidence by bariatric surgery in the previous epidemiological study (HR, 0.32) (41), whereas no low-risk patients showed improved PLS-NAFLD status (Fig. 5B). The second example is use of lipophilic statins such as atorvastatin and simvastatin, which is associated with reduced HCC incidence in NAFLD patient cohorts (HR 0.36) (44). Prevalence of high-risk PLS-NAFLD in lipophilic statin users was three-fold lower compared to non-statin users (conditional OR, 0.33; 95% CI, 0.16–0.68) in severely obese patients, which is again comparable to the magnitude of reduction in HCC incidence (Fig. 5C, D) (45). The third example is use of an IDO1 inhibitor, which could potentially target the IDO1+ cDCs we identified in the single-cell and spatial transcriptome profiling. In our PLS-inducible cell culture model (cPLS system) (46), we confirmed that high-risk pattern of PLS-NAFLD induced by free fatty acid treatment was reversed by an IDO1 inhibitor under clinical development for malignancies, epacadostat, in a dose-dependent manner (Fig. 5E, F).
Fig. 5. Therapeutic modulation of PLS-NAFLD.
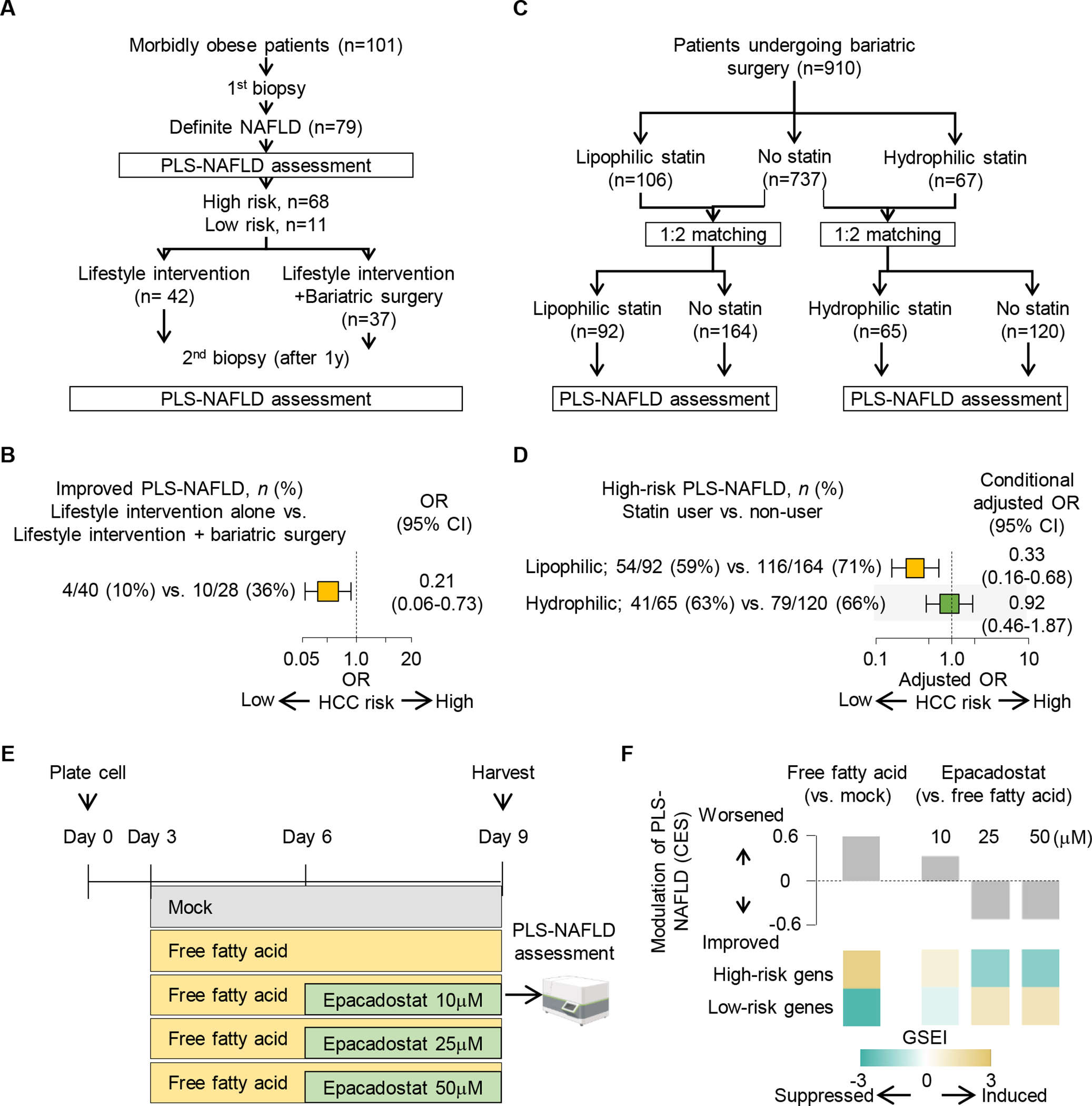
(A) Study design to assess PLS-NAFLD modulation by bariatric surgery in patients with NAFLD under lifestyle intervention. (B) Proportion of patients with significantly improved PLS-NAFLD with bariatric surgery in patients with NAFLD under lifestyle intervention. (C) Study design to assess association of PLS-NAFLD status with statin use in patients who underwent bariatric surgery. (D) Association of PLS-NAFLD-based HCC risk prediction with lipophilic or hydrophilic statin use. (E) Experimental design to assess the effect of an IDO1 inhibitor, epacadostat, in a PLS-inducible cell culture model (cPLS system). (F) Induction of high-risk pattern of PLS-NAFLD by free fatty acid and its reversal with epacadostat in the cPLS system.
These examples collectively support the utility of PLS-NAFLD to monitor therapeutic modulation of HCC risk to gauge anticipated HCC-preventive effect as a surrogate endpoint in clinical trials of anti-NAFLD therapies. Additionally, presence of high-risk PLS-NAFLD may justify enrollment to HCC chemoprevention clinical trials because of elevated HCC risk. PLS-NAFLD might also be used for cell-based high-throughput screening of candidate chemoprevention agents for NAFLD-related HCC.
DISCUSSION
The global shift of the etiology of liver cirrhosis and HCC from viral hepatitis to NAFLD highlights an urgent need for developing a risk-stratification markers to allocate limited medical resources such as regular HCC screening to those who would benefit most. We previously developed etiology-agnostic biomarkers, tissue-based PLS and serum-based PLSec-AFP, to identify patients with chronic liver disease at high risk of developing HCC (8–10, 12). Although these biomarkers showed promising prognostic capability, there was room to improve the identification of low-risk patients who do not require HCC screening. Here we found that PLS/PLSec-NAFLD identified low-risk patients with NAFLD who had negligible HCC risk consistently in derivation and validation sets. This is a substantial step toward refining HCC screening because over-screening of low-risk patients can cause unnecessary physical, economical, and psychological harms, and under-screening of high-risk patients likely results in late HCC diagnosis and misses the chance of curative treatment (47). Although serum-based PLSec-NAFLD might be preferred for HCC risk stratification given the invasiveness of liver biopsy, tissue-based PLS-NAFLD could still be useful to gauge HCC-preventive effect of anti-NAFLD agents in clinical trials, in which liver biopsy is typically performed, along with other tissue-based endpoints. Furthermore, we show proof of principle that combining an etiology-specific biomarker with an etiology-agnostic biomarker can improve prognostication as a “plug-in” module, which can be extended to HBV-infected, HCV-cured, and patients with alcoholic liver disease. PLS-NAFLD may also guide application of screening biomarkers such as circulating cell-free methylated DNA according to predicted HCC risk.
Several hepatic gene expression-based signatures for NAFLD have been proposed (48–50). However, all of them were derived from cross-sectionally obtained samples; therefore, it was challenging to know whether these signatures reflect cause or consequence of disease progression as evidenced by the lack of their prognostic association. PLS-NAFLD is the first gene expression signature to predict future HCC risk, reflecting status of molecular drivers of hepatocarcinogenesis in HCC-naive NAFLD livers. Indeed, we found IDO1+ cDCs associated with high-risk signatures, and suggesting they may be possible targets for HCC chemoprevention in NAFLD.
There is increased interest in HCC chemoprevention using generic agents such as aspirin and statins as well as other molecular-targeted agents (15). However, it is practically infeasible to follow patients in a clinical trial until significant reduction of HCC incidence is observed because of the low incidence rate (<1% per year). Our study showed that PLS-NAFLD can be modulated in a short time period (in several days to weeks) by potential medical interventions reducing HCC risk, suggesting that PLS-NAFLD might serve as a surrogate endpoint to estimate their long-term prognostic benefit in clinical trials. This concept is already incorporated in our ongoing clinical trials using the etiology-agnostic PLS/PLSec as companion biomarkers (NCT02273362, NCT04172779).
Although PLS/PLSec-NAFLD showed promising prognostic capability, there are several limitations. First, the number of events was small in the HCC-naïve cohorts (the tissue validation set 1 and serum validation set) as typically observed in clinical studies, which may obscure accurate magnitude of risk difference between the high- and low-risk groups. Nevertheless, the absence of HCC incidence in low-risk patients supports the capability of PLS/PLSec-NAFLD to identify patients with negligible HCC risk. Second, given the heterogeneous clinical demographics between the cohorts, PLS/PLSec-NAFLD should be prospectively validated in future studies to eliminate potential influence of confounding factors before its clinical translation. Third, association of therapeutic modulation of the biomarkers and future HCC incidence should be prospectively confirmed. Fourth, although we employed a selective gene signature development method based on the leave-one-out procedure, further shaving of the gene list may lower the bar for its clinical application.
In summary, we developed and validated tissue-transcriptome- and serum-secretome-based signatures, PLS/PLSec-NAFLD, for predicting long-term HCC risk and estimating effects of therapeutic interventions in patients with NAFLD. These signatures may lead to improvement of the poor prognosis of NAFLD-related HCC.
Materials and Methods
Study design
We first defined the signature in a cohort of 48 patients with prior history of HCC (derivation set) for emergence of de novo HCC recurrence (Fig. S1B). The signature was subsequently validated in independent external cohorts of 106 HCC-naïve patients with NAFLD (tissue validation set 1) and 59 HCC-experienced patients with NAFLD who previously underwent curative surgical tumor resection (tissue validation set 2). Further, we translated PLS-NAFLD to a serum-protein panel, PLSec-NAFLD, using our computational algorithm, TexSEC (12), and externally validated it in an independent cohort of 59 HCC-naïve patients with NAFLD and cirrhosis (serum validation set).
Patient cohorts
Diagnoses of NAFLD and HCC were made based on the clinical practice guidelines from the American Association for the Study of Liver Diseases (3, 51) for patients in the PLS/PLSec-NAFLD derivation and validation sets detailed below. Histological grading and staging of NAFLD were performed in a centralized manner by an experienced liver pathologist blinded to clinical information according to the established criteria (52). In these prospective-retrospective cohorts (53), biospecimens were collected and analyzed by utilizing the PRoBE design for biomarker validation (14). The study was approved by the institutional review board at respective institutions with written informed consent or exemption for use of archived de-identified samples (protocol numbers: STU062018-058, STU072018-071, 2010P000220/PHS, and HS13-00159)
The PLS-NAFLD derivation set includes 48 patients who underwent curative ultrasound-guided percutaneous radiofrequency ablation (RFA) for early-stage NAFLD-related HCC (AJCC T1/2 tumor without extrahepatic lesion) at the University of Tokyo between May 2006 and December 2018 (54). Absence of residual tumor was radiologically confirmed with contrast-enhanced multiphase CT/MRI. Non-cancerous liver biopsy (16-gauge needle) tissues were collected at the time of treatment and immediately fixed in formalin. Post treatment follow-up was performed with multiphase CT/MRI and tumor markers every 3–4 months. Time to HCC development was defined as the interval between RFA and HCC diagnosis or the last follow-up including death as a censored observation. During a median follow-up of 1.8 (IQR, 0.8–2.7) years, 28 patients developed HCC recurrence. The recurrence hazard curve suggested that the observed recurrences were assumed to be dominantly de novo HCC recurrence (Fig. S1B) (15).
The tissue validation set 1 included 106 non-cirrhotic HCC-naïve patients with NAFLD who underwent diagnostic liver biopsy at Hiroshima University between May 2003 and February 2015 and regularly followed up using ultrasound every 6 months for a median of 8.9 (IQR, 5.1–11.9) years, during which 6 patients developed HCC (55). Time to HCC development was defined as the interval between biopsy and HCC diagnosis or the last follow-up. The tissue validation set 2 included 59 patients with early-stage NAFLD-related HCC who underwent curative surgical resection for HCC at Toranomon Hospital or Kumamoto University between January 2003 and November 2011 (11). Post-treatment follow-up, diagnosis of HCC, and determination of time to HCC development were similarly performed as in the derivation set. During a median follow-up of 1.8 (IQR, 0.6–3.5) years, 32 patients developed HCC recurrence. The serum validation set included 59 patients with NAFLD cirrhosis enrolled at University of Michigan between January 2004 and September 2006, and regularly followed up using ultrasound and AFP every 6 months for a median follow-up of 5.8 (1.8–10.8) years, during which 7 patients developed HCC. Time to HCC development was defined as the interval between the dates of blood sampling and HCC diagnosis or the last follow-up as a censored observation.
Derivation of PLS-NAFLD in hepatic tissue transcriptome profiles
To define a transcriptome signature correlating with time to HCC development, we employed the leave-one-out procedure as previously described (8). Briefly, in the derivation set including 48 patients, genes that were less variably expressed across patients were excluded based on the cut-offs of CV <0.1 and a difference between maximum and minimum expression <100. This gene filtering did not rely on any prior knowledge related to the HCC status during follow-up. Subsequently, one patient was excluded, and genes associated with time to HCC development were selected in the remaining patients by using Cox score (56) calculated with the following equation:
, where i is indices of samples, xi is gene expression for sample i, ti is time for sample i, k ∈1, …, K is indices of unique death times z1, z2, …, zK, dk is number of deaths at time zk, mk is number of samples in Rk = i: ti ≥ zk, , and . Genes with a random permutation test p-value less than 0.05 were selected for the HCC risk signature in the leave-one-out step. This procedure was repeated for each of the 48 patients, and 48 HCC risk signatures were derived. Genes consistently selected throughout the 48 HCC risk signatures were regarded as the PLS-NAFLD member genes (80 high-risk and 53 low-risk genes) to be subsequently evaluated for prognostic performance in the independent validation sets. Cox scores calculated in 48 patients for the final signature genes were used as a weight vector for prognostic prediction in independent cohorts as described in the Supplementary Materials and Methods.
Statistical analysis
Categorical and continuous variables were tested by Fisher’s exact test and Wilcoxon rank-sum test, respectively. Time-to-event analyses were performed using the Kaplan-Meier method and uni/multivariable Cox regression modeling. Proportional-hazards assumption was confirmed by using cox.zph function in survival R package (Table S8). The matched case-control series was analyzed by conditional logistic regression model (Fig. 5D). Estimated hazard ratios were adjusted by the Firth’s correction using coxphf R package (36) when no event occurred in a patient group and therefore hazard ratios could not be reliably calculated. To evaluate the robustness of our signature’s association with prognosis, we built multiple multivariable models with clinically known confounding variables (Table S3). Correction for multiple hypothesis testing was applied using FDR as needed. A two-tailed p-value <0.05 was regarded as statistically significant. All data analyses were performed using R statistical language otherwise specified (www.r-project.org).
Supplementary Material
Materials and Methods
Fig. S1. Sample size calculation, HCC recurrence hazard, and performance of etiology-agnostic PLS in the derivation and validation sets.
Fig. S2. Clinical and molecular correlates of PLS-NAFLD.
Fig. S3. Cell type-level deconvolution of PLS-NAFLD based on human hepatic single-cell transcriptome profiles.
Fig. S4. Spatial deconvolution of PLS-NAFLD on histological architectures in NAFLD-affected human liver.
Fig. S5. Derivation, optimization, and independent validation of PLSec-NAFLD.
Table S1. PLS-NAFLD signature genes.
Table S2. Prognostic association of PLS-NAFLD and clinical/molecular variables with HCC risk in the derivation set using univariable Cox regression.
Table S3. Prognostic association of PLS-/PLSec-NAFLD and etPLSec-NAFLD with HCC risk in the serum validation set using multivariable Cox regression.
Table S4. Molecular pathways associated with PLS-NAFLD in the derivation set.
Table S5. Hepatic single-cell RNA-seq datasets for meta-analysis of human NAFLD and healthy livers,
Table S6. Hepatic single-cell cluster annotation based on meta-analyses of single-cell transcriptome profiles of human NAFLD and healthy livers.
Table S7. Comparisons of prognostic capability between change of PLS-NAFLD and fibrosis stage over time in tissue validation set 1.
Table S8. Test for proportional hazard assumption of the molecular biomarkers.
Table S9. Signatures extracted from public datasets and database.
Table S10. Antibodies for immunohistochemical staining.
Acknowledgments
Funding:
This work was supported by National Institutes of Health grant R01DK099558 (YH), R01CA233794 (YH), U01CA226052 (AGS, YH), U01CA230694 (AGS), and R01CA222900 (AGS); Cancer Prevention and Research Institute of Texas RR180016 (YH), RR180014 (ZZ), and RP200197 (SL); American Association for the Study of Liver Disease AASLDF 50028 (SL); Uehara Memorial Foundation Postdoctoral award (NF); AMED JP21fk0210090 (KC), JP22fk0210115 (NH), and JP21fk0210059 (NH); KAKENHI 21H02892 (NH); European Commission ERC-2014-AdG-671231 (TFB, YH) and ERC-2020-ADG-101021417 (TFB, YH); and Inserm Plan Cancer and TheraHCC 20 (TFB).
Footnotes
Competing interests: YH serves as an advisory board member for Helio Health and founding share holder for Alentis Therapeutics. YH received a research funding from Morphic Therapeutics. TFB is founder, shareholder and advisor of Alentis Therapeutics. AGS has served as a consultant or on advisory boards for Genentech, AstraZeneca, Bayer, Eisai, Exelixis, BMS, Eli Lilly, FujiFilm Medical Sciences, Exact Sciences, Roche, Glycotest, GRAIL, and TARGET RWE. The other authors declare that they have no competing interests.
Data and materials availability:
All data associated with this study are in the paper and supplementary materials. All data are publicly available at the NCBI GEO (accession number, GSE193084). Raw data from figures is in data file S1. The R codes for Gene Set Enrichment Index based on single sample-based signature enrichment analysis (eseach algorithm) (11) and TexSEC are available at 10.5281/zenodo.6519385. Of note, data with patient names and other identifiers cannot be shared.
References and notes
- 1.Loomba R, Friedman SL, Shulman GI, Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokdad AH, Dwyer-Lindgren L, Fitzmaurice C, Stubbs RW, Bertozzi-Villa A, Morozoff C, Charara R, Allen C, Naghavi M, Murray CJ, Trends and Patterns of Disparities in Cancer Mortality Among US Counties, 1980–2014. JAMA 317, 388–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK, Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 68, 723–750 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wolf E, Rich NE, Marrero JA, Parikh N, Singal AG, Utilization of hepatocellular carcinoma surveillance in patients with cirrhosis: A systematic review and meta-analysis. Hepatology, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, Wai-Sun Wong V, Yilmaz Y, George J, Fan J, Vos MB, Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology 69, 2672–2682 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Simon TG, Roelstraete B, Sharma R, Khalili H, Hagstrom H, Ludvigsson JF, Cancer Risk in Patients With Biopsy-Confirmed Nonalcoholic Fatty Liver Disease: A Population-Based Cohort Study. Hepatology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanyal AJ, Van Natta ML, Clark J, Neuschwander-Tetri BA, Diehl A, Dasarathy S, Loomba R, Chalasani N, Kowdley K, Hameed B, Wilson LA, Yates KP, Belt P, Lazo M, Kleiner DE, Behling C, Tonascia J, Network NCR, Prospective Study of Outcomes in Adults with Nonalcoholic Fatty Liver Disease. N Engl J Med 385, 1559–1569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, Reich M, Chan JA, Glickman JN, Ikeda K, Hashimoto M, Watanabe G, Daidone MG, Roayaie S, Schwartz M, Thung S, Salvesen HB, Gabriel S, Mazzaferro V, Bruix J, Friedman SL, Kumada H, Llovet JM, Golub TR, Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 359, 1995–2004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshida Y, Villanueva A, Sangiovanni A, Sole M, Hur C, Andersson KL, Chung RT, Gould J, Kojima K, Gupta S, Taylor B, Crenshaw A, Gabriel S, Minguez B, Iavarone M, Friedman SL, Colombo M, Llovet JM, Golub TR, Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 144, 1024–1030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King LY, Canasto-Chibuque C, Johnson KB, Yip S, Chen X, Kojima K, Deshmukh M, Venkatesh A, Tan PS, Sun X, Villanueva A, Sangiovanni A, Nair V, Mahajan M, Kobayashi M, Kumada H, Iavarone M, Colombo M, Fiel MI, Friedman SL, Llovet JM, Chung RT, Hoshida Y, A genomic and clinical prognostic index for hepatitis C-related early-stage cirrhosis that predicts clinical deterioration. Gut 64, 1296–1302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa S, Wei L, Song WM, Higashi T, Ghoshal S, Kim RS, Bian CB, Yamada S, Sun X, Venkatesh A, Goossens N, Bain G, Lauwers GY, Koh AP, El-Abtah M, Ahmad NB, Hoshida H, Erstad DJ, Gunasekaran G, Lee Y, Yu ML, Chuang WL, Dai CY, Kobayashi M, Kumada H, Beppu T, Baba H, Mahajan M, Nair VD, Lanuti M, Villanueva A, Sangiovanni A, Iavarone M, Colombo M, Llovet JM, Subramanian A, Tager AM, Friedman SL, Baumert TF, Schwarz ME, Chung RT, Tanabe KK, Zhang B, Fuchs BC, Hoshida Y, Precision C Liver Cancer Prevention, Molecular Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell 30, 879–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara N, Kobayashi M, Fobar AJ, Hoshida A, Marquez CA, Koneru B, Panda G, Taguri M, Qian T, Raman I, Li QZ, Hoshida H, Sezaki H, Kumada H, Tateishi R, Yokoo T, Yopp AC, Chung RT, Fuchs BC, Baumert TF, Marrero JA, Parikh ND, Zhu S, Singal AG, Hoshida Y, A blood-based prognostic liver secretome signature and long-term hepatocellular carcinoma risk in advanced liver fibrosis. Med (N Y) 2, 836–850 e810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goossens N, Singal AG, King LY, Andersson KL, Fuchs BC, Besa C, Taouli B, Chung RT, Hoshida Y, Cost-Effectiveness of Risk Score-Stratified Hepatocellular Carcinoma Screening in Patients with Cirrhosis. Clin Transl Gastroenterol 8, e101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD, Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 100, 1432–1438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara N, Friedman SL, Goossens N, Hoshida Y, Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 68, 526–549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW, A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 70, 10202–10212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trepo E, Goossens N, Fujiwara N, Song WM, Colaprico A, Marot A, Spahr L, Demetter P, Sempoux C, Im GY, Saldarriaga J, Gustot T, Deviere J, Thung SN, Minsart C, Serste T, Bontempi G, Abdelrahman K, Henrion J, Degre D, Lucidi V, Rubbia-Brandt L, Nair VD, Moreno C, Deltenre P, Hoshida Y, Franchimont D, Combination of Gene Expression Signature and Model for End-Stage Liver Disease Score Predicts Survival of Patients With Severe Alcoholic Hepatitis. Gastroenterology 154, 965–975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa H, Umemura A, Taniguchi K, Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek MA, Seki E, Hidalgo J, Koike K, Kaufman RJ, Karin M, ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 26, 331–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YA, Noon LA, Akat KM, Ybanez MD, Lee TF, Berres ML, Fujiwara N, Goossens N, Chou HI, Parvin-Nejad FP, Khambu B, Kramer EGM, Gordon R, Pfleger C, Germain D, John GR, Campbell KN, Yue Z, Yin XM, Cuervo AM, Czaja MJ, Fiel MI, Hoshida Y, Friedman SL, Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat Commun 9, 4962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, Katchy CA, Lee C, Moore DD, Fu L, Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grohmann M, Wiede F, Dodd GT, Gurzov EN, Ooi GJ, Butt T, Rasmiena AA, Kaur S, Gulati T, Goh PK, Treloar AE, Archer S, Brown WA, Muller M, Watt MJ, Ohara O, McLean CA, Tiganis T, Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell 175, 1289–1306 e1220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barb D, Bril F, Kalavalapalli S, Cusi K, Plasma Fibroblast Growth Factor 21 Is Associated With Severity of Nonalcoholic Steatohepatitis in Patients With Obesity and Type 2 Diabetes. J Clin Endocrinol Metab 104, 3327–3336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attia YM, Tawfiq RA, Gibriel AA, Ali AA, Kassem DH, Hammam OA, Elmazar MM, Activation of FXR modulates SOCS3/Jak2/STAT3 signaling axis in a NASH-dependent hepatocellular carcinoma animal model. Biochem Pharmacol 186, 114497 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grun D, A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, Taylor RS, Efremova M, Vento-Tormo R, Carragher NO, Kendall TJ, Fallowfield JA, Harrison EM, Mole DJ, Wigmore SJ, Newsome PN, Weston CJ, Iredale JP, Tacke F, Pollard JW, Ponting CP, Marioni JC, Teichmann SA, Henderson NC, Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payen VL, Lavergne A, Alevra Sarika N, Colonval M, Karim L, Deckers M, Najimi M, Coppieters W, Charloteaux B, Sokal EM, El Taghdouini A, Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep 3, 100278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfister D, Nunez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, Gupta R, Qiu M, Deczkowska A, Weiner A, Muller F, Sinha A, Friebel E, Engleitner T, Lenggenhager D, Moncsek A, Heide D, Stirm K, Kosla J, Kotsiliti E, Leone V, Dudek M, Yousuf S, Inverso D, Singh I, Teijeiro A, Castet F, Montironi C, Haber PK, Tiniakos D, Bedossa P, Cockell S, Younes R, Vacca M, Marra F, Schattenberg JM, Allison M, Bugianesi E, Ratziu V, Pressiani T, D’Alessio A, Personeni N, Rimassa L, Daly AK, Scheiner B, Pomej K, Kirstein MM, Vogel A, Peck-Radosavljevic M, Hucke F, Finkelmeier F, Waidmann O, Trojan J, Schulze K, Wege H, Koch S, Weinmann A, Bueter M, Rossler F, Siebenhuner A, De Dosso S, Mallm JP, Umansky V, Jugold M, Luedde T, Schietinger A, Schirmacher P, Emu B, Augustin HG, Billeter A, Muller-Stich B, Kikuchi H, Duda DG, Kutting F, Waldschmidt DT, Ebert MP, Rahbari N, Mei HE, Schulz AR, Ringelhan M, Malek N, Spahn S, Bitzer M, Ruiz de Galarreta M, Lujambio A, Dufour JF, Marron TU, Kaseb A, Kudo M, Huang YH, Djouder N, Wolter K, Zender L, Marche PN, Decaens T, Pinato DJ, Rad R, Mertens JC, Weber A, Unger K, Meissner F, Roth S, Jilkova ZM, Claassen M, Anstee QM, Amit I, Knolle P, Becher B, Llovet JM, Heikenwalder M, NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 592, 450–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudek M, Pfister D, Donakonda S, Filpe P, Schneider A, Laschinger M, Hartmann D, Huser N, Meiser P, Bayerl F, Inverso D, Wigger J, Sebode M, Ollinger R, Rad R, Hegenbarth S, Anton M, Guillot A, Bowman A, Heide D, Muller F, Ramadori P, Leone V, Garcia-Caceres C, Gruber T, Seifert G, Kabat AM, Mallm JP, Reider S, Effenberger M, Roth S, Billeter AT, Muller-Stich B, Pearce EJ, Koch-Nolte F, Kaser R, Tilg H, Thimme R, Boettler T, Tacke F, Dufour JF, Haller D, Murray PJ, Heeren R, Zehn D, Bottcher JP, Heikenwalder M, Knolle PA, Auto-aggressive CXCR6(+) CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Carpino G, Del Ben M, Pastori D, Carnevale R, Baratta F, Overi D, Francis H, Cardinale V, Onori P, Safarikia S, Cammisotto V, Alvaro D, Svegliati-Baroni G, Angelico F, Gaudio E, Violi F, Increased Liver Localization of Lipopolysaccharides in Human and Experimental NAFLD. Hepatology 72, 470–485 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA, Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deczkowska A, David E, Ramadori P, Pfister D, Safran M, At The B, Giladi A, Jaitin DA, Barboy O, Cohen M, Yofe I, Gur C, Shlomi-Loubaton S, Henri S, Suhail Y, Qiu M, Kam S, Hermon H, Lahat E, Ben Yakov G, Cohen-Ezra O, Davidov Y, Likhter M, Goitein D, Roth S, Weber A, Malissen B, Weiner A, Ben-Ari Z, Heikenwalder M, Elinav E, Amit I, XCR1(+) type 1 conventional dendritic cells drive liver pathology in non-alcoholic steatohepatitis. Nat Med 27, 1043–1054 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Munn DH, Mellor AL, IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol 37, 193–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elosua-Bayes M, Nieto P, Mereu E, Gut I, Heyn H, SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res 49, e50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R, CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Heinze G, Schemper M, A solution to the problem of monotone likelihood in Cox regression. Biometrics 57, 114–119 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Singal AG, Nabihah T, Mehta A, Marrero JA, El-Serag H, Jin Q, Saenz de Viteri C, Fobar A, Parikh ND, GALAD Demonstrates High Sensitivity for HCC Surveillance in a Cohort of Patients with Cirrhosis. Hepatology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroczek RA, Henn V, The Role of XCR1 and its Ligand XCL1 in Antigen Cross-Presentation by Murine and Human Dendritic Cells. Front Immunol 3, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuppalanchi R, Noureddin M, Alkhouri N, Sanyal AJ, Therapeutic pipeline in nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol, (2021). [DOI] [PubMed] [Google Scholar]
- 40.Qian T, Fujiwara N, Koneru B, Ono A, Kubota N, Jajoriya AK, Tung MG, Crouchet E, Song WM, Marquez CA, Panda G, Hoshida A, Raman I, Li QZ, Lewis C, Yopp A, Rich NE, Singal AG, Nakagawa S, Goossens N, Higashi T, Koh AP, Bian CB, Hoshida H, Tabrizian P, Gunasekaran G, Florman S, Schwarz ME, Hiotis SP, Nakahara T, Aikata H, Murakami E, Beppu T, Baba H, Warren A, Bhatia S, Kobayashi M, Kumada H, Fobar AJ, Parikh ND, Marrero JA, Rwema SH, Nair V, Patel M, Kim-Schulze S, Corey K, O’Leary JG, Klintmalm GB, Thomas DL, Dibas M, Rodriguez G, Zhang B, Friedman SL, Baumert TF, Fuchs B, Chayama K, Zhu S, Chung RT, Hoshida Y, Molecular signature predictive of long-term liver fibrosis progression to inform anti-fibrotic drug development. Gastroenterology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rustgi VK, Li Y, Gupta K, Minacapelli CD, Bhurwal A, Catalano C, Elsaid MI, Bariatric Surgery Reduces Cancer Risk in Adults With Nonalcoholic Fatty Liver Disease and Severe Obesity. Gastroenterology 161, 171–184 e110 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Haas JT, Vonghia L, Mogilenko DA, Verrijken A, Molendi-Coste O, Fleury S, Deprince A, Nikitin A, Woitrain E, Ducrocq-Geoffroy L, Pic S, Derudas B, Dehondt H, Gheeraert C, Van Gaal L, Driessen A, Lefebvre P, Staels B, Francque S, Dombrowicz D, Transcriptional Network Analysis Implicates Altered Hepatic Immune Function in NASH development and resolution. Nat Metab 1, 604–614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefebvre P, Lalloyer F, Bauge E, Pawlak M, Gheeraert C, Dehondt H, Vanhoutte J, Woitrain E, Hennuyer N, Mazuy C, Bobowski-Gerard M, Zummo FP, Derudas B, Driessen A, Hubens G, Vonghia L, Kwanten WJ, Michielsen P, Vanwolleghem T, Eeckhoute J, Verrijken A, Van Gaal L, Francque S, Staels B, Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARalpha-regulated dermatopontin. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinyopornpanish K, Al-Yaman W, Butler RS, Carey W, McCullough A, Romero-Marrero C, Chemopreventive Effect of Statin on Hepatocellular Carcinoma in Patients With Nonalcoholic Steatohepatitis Cirrhosis. Am J Gastroenterol, (2021). [DOI] [PubMed] [Google Scholar]
- 45.Margerie D, Lefebvre P, Raverdy V, Schwahn U, Ruetten H, Larsen P, Duhamel A, Labreuche J, Thuillier D, Derudas B, Gheeraert C, Dehondt H, Dhalluin Q, Alexandre J, Caiazzo R, Nesslany P, Verkindt H, Pattou F, Staels B, Hepatic transcriptomic signatures of statin treatment are associated with impaired glucose homeostasis in severely obese patients. BMC Med Genomics 12, 80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crouchet E, Bandiera S, Fujiwara N, Li S, El Saghire H, Fernandez-Vaquero M, Riedl T, Sun X, Hirschfield H, Juhling F, Zhu S, Roehlen N, Ponsolles C, Heydmann L, Saviano A, Qian T, Venkatesh A, Lupberger J, Verrier ER, Sojoodi M, Oudot MA, Duong FHT, Masia R, Wei L, Thumann C, Durand SC, Gonzalez-Motos V, Heide D, Hetzer J, Nakagawa S, Ono A, Song WM, Higashi T, Sanchez R, Kim RS, Bian CB, Kiani K, Croonenborghs T, Subramanian A, Chung RT, Straub BK, Schuppan D, Ankavay M, Cocquerel L, Schaeffer E, Goossens N, Koh AP, Mahajan M, Nair VD, Gunasekaran G, Schwartz ME, Bardeesy N, Shalek AK, Rozenblatt-Rosen O, Regev A, Felli E, Pessaux P, Tanabe KK, Heikenwalder M, Schuster C, Pochet N, Zeisel MB, Fuchs BC, Hoshida Y, Baumert TF, A human liver cell-based system modeling a clinical prognostic liver signature for therapeutic discovery. Nat Commun 12, 5525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singal AG, Patibandla S, Obi J, Fullington H, Parikh ND, Yopp AC, Marrero JA, Benefits and Harms of Hepatocellular Carcinoma Surveillance in a Prospective Cohort of Patients With Cirrhosis. Clin Gastroenterol Hepatol 19, 1925–1932 e1921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govaere O, Cockell S, Tiniakos D, Queen R, Younes R, Vacca M, Alexander L, Ravaioli F, Palmer J, Petta S, Boursier J, Rosso C, Johnson K, Wonders K, Day CP, Ekstedt M, Oresic M, Darlay R, Cordell HJ, Marra F, Vidal-Puig A, Bedossa P, Schattenberg JM, Clement K, Allison M, Bugianesi E, Ratziu V, Daly AK, Anstee QM, Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci Transl Med 12, (2020). [DOI] [PubMed] [Google Scholar]
- 49.Hoang SA, Oseini A, Feaver RE, Cole BK, Asgharpour A, Vincent R, Siddiqui M, Lawson MJ, Day NC, Taylor JM, Wamhoff BR, Mirshahi F, Contos MJ, Idowu M, Sanyal AJ, Gene Expression Predicts Histological Severity and Reveals Distinct Molecular Profiles of Nonalcoholic Fatty Liver Disease. Sci Rep 9, 12541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerhard GS, Legendre C, Still CD, Chu X, Petrick A, DiStefano JK, Transcriptomic Profiling of Obesity-Related Nonalcoholic Steatohepatitis Reveals a Core Set of Fibrosis-Specific Genes. J Endocr Soc 2, 710–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ, The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 67, 328–357 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ, Nonalcoholic N Steatohepatitis Clinical Research, Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Simon RM, Paik S, Hayes DF, Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 101, 1446–1452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiina S, Tateishi R, Arano T, Uchino K, Enooku K, Nakagawa H, Asaoka Y, Sato T, Masuzaki R, Kondo Y, Goto T, Yoshida H, Omata M, Koike K, Radiofrequency ablation for hepatocellular carcinoma: 10-year outcome and prognostic factors. Am J Gastroenterol 107, 569–577; quiz 578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daijo K, Nakahara T, Inagaki Y, Nanba M, Nishida Y, Uchikawa S, Kodama K, Oya K, Morio K, Fujino H, Ono A, Murakami E, Yamauchi M, Kawaoka T, Miki D, Tsuge M, Hiramatsu A, Hayes CN, Imamura M, Aikata H, Ochi H, Chayama K, Risk factors for histological progression of non-alcoholic steatohepatitis analyzed from repeated biopsy cases. J Gastroenterol Hepatol 35, 1412–1419 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Bair E, Tibshirani R, Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2, E108 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gellert-Kristensen H, Richardson TG, Davey Smith G, Nordestgaard BG, Tybjaerg-Hansen A, Stender S, Combined Effect of PNPLA3, TM6SF2, and HSD17B13 Variants on Risk of Cirrhosis and Hepatocellular Carcinoma in the General Population. Hepatology 72, 845–856 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP, GenePattern 2.0. Nat Genet 38, 500–501 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Hoshida Y, Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One 5, e15543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goossens N, Hoshida Y, Song WM, Jung M, Morel P, Nakagawa S, Zhang B, Frossard JL, Spahr L, Friedman SL, Negro F, Rubbia-Brandt L, Giostra E, Nonalcoholic Steatohepatitis Is Associated With Increased Mortality in Obese Patients Undergoing Bariatric Surgery. Clin Gastroenterol Hepatol 14, 1619–1628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ono A, Goossens N, Finn RS, Schmidt WN, Thung SN, Im GY, Hoshida Y, Precision C Liver Cancer Prevention, Persisting risk of hepatocellular carcinoma after hepatitis C virus cure monitored by a liver transcriptome signature. Hepatology 66, 1344–1346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, Xiao G, Li Y, Allen J, Zhong R, Chen B, Kim M, Wang T, Heiser LM, Realubit R, Mattioli M, Alvarez MJ, Shen Y, Community N-D, Gallahan D, Singer D, Saez-Rodriguez J, Xie Y, Stolovitzky G, Califano A, Community N-D, A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 32, 1213–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Candia J, Bayarsaikhan E, Tandon M, Budhu A, Forgues M, Tovuu LO, Tudev U, Lack J, Chao A, Chinburen J, Wang XW, The genomic landscape of Mongolian hepatocellular carcinoma. Nat Commun 11, 4383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juhling F, Hamdane N, Crouchet E, Li S, El Saghire H, Mukherji A, Fujiwara N, Oudot MA, Thumann C, Saviano A, Roca Suarez AA, Goto K, Masia R, Sojoodi M, Arora G, Aikata H, Ono A, Tabrizian P, Schwartz M, Polyak SJ, Davidson I, Schmidl C, Bock C, Schuster C, Chayama K, Pessaux P, Tanabe KK, Hoshida Y, Zeisel MB, Duong FH, Fuchs BC, Baumert TF, Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut 70, 157–169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deltenre P, Trepo E, Fujiwara N, Goossens N, Marot A, Dubois M, Spahr L, Henrion J, Moreno C, Hoshida Y, Gene signature-MELD score and alcohol relapse determine long-term prognosis of patients with severe alcoholic hepatitis. Liver Int 40, 565–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Auwera GA, O’Connor BD, Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. (O’Reilly Media, 2020). [Google Scholar]
- 72.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K, dbSNP: the NCBI database of genetic variation. Nucleic acids research 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv, 201178 (2017). [Google Scholar]
- 74.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, Cheng ML, Liu LY, Camat D, Chung SW, Seliga RK, Shao Z, Lee E, Ogawa S, Ogawa M, Wilson MD, Fish JE, Selzner M, Ghanekar A, Grant D, Greig P, Sapisochin G, Selzner N, Winegarden N, Adeyi O, Keller G, Bader GD, McGilvray ID, Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9, 4383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fisher RA, Statistical Methods for Research Workers. (Oliver and Boyd, London, 1932). [Google Scholar]
- 76.Ahrens M, Ammerpohl O, von Schonfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, Erhart W, Egberts J, Sipos B, Schreiber S, Hasler R, Stickel F, Becker T, Krawczak M, Rocken C, Siebert R, Schafmayer C, Hampe J, DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab 18, 296–302 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM, Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 145, 1076–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fujiwara N, Fobar AJ, Raman I, Li QZ, Marrero JA, Parikh ND, Singal AG, Hoshida Y, A Blood-Based Prognostic Liver Secretome Signature Predicts Long-term Risk of Hepatic Decompensation in Cirrhosis. Clin Gastroenterol Hepatol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujiwara N, Trepo E, Raman I, Li QZ, Degre D, Gustot T, Moreno C, Hoshida Y, Plasma-Signature-Model for End-Stage Liver Disease Score to Predict Survival in Severe Alcoholic Hepatitis. Clin Gastroenterol Hepatol 20, 651–657 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas JT, Vonghia L, Mogilenko DA, Verrijken A, Molendi-Coste O, Fleury S, Deprince A, Nikitin A, Woitrain E, Ducrocq-Geoffroy L, Pic S, Derudas B, Dehondt H, Gheeraert C, Van Gaal L, Driessen A, Lefebvre P, Staels B, Francque S, Dombrowicz D, Author Correction: Transcriptional network analysis implicates altered hepatic immune function in NASH development and resolution. Nat Metab 1, 744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. Sample size calculation, HCC recurrence hazard, and performance of etiology-agnostic PLS in the derivation and validation sets.
Fig. S2. Clinical and molecular correlates of PLS-NAFLD.
Fig. S3. Cell type-level deconvolution of PLS-NAFLD based on human hepatic single-cell transcriptome profiles.
Fig. S4. Spatial deconvolution of PLS-NAFLD on histological architectures in NAFLD-affected human liver.
Fig. S5. Derivation, optimization, and independent validation of PLSec-NAFLD.
Table S1. PLS-NAFLD signature genes.
Table S2. Prognostic association of PLS-NAFLD and clinical/molecular variables with HCC risk in the derivation set using univariable Cox regression.
Table S3. Prognostic association of PLS-/PLSec-NAFLD and etPLSec-NAFLD with HCC risk in the serum validation set using multivariable Cox regression.
Table S4. Molecular pathways associated with PLS-NAFLD in the derivation set.
Table S5. Hepatic single-cell RNA-seq datasets for meta-analysis of human NAFLD and healthy livers,
Table S6. Hepatic single-cell cluster annotation based on meta-analyses of single-cell transcriptome profiles of human NAFLD and healthy livers.
Table S7. Comparisons of prognostic capability between change of PLS-NAFLD and fibrosis stage over time in tissue validation set 1.
Table S8. Test for proportional hazard assumption of the molecular biomarkers.
Table S9. Signatures extracted from public datasets and database.
Table S10. Antibodies for immunohistochemical staining.
Data Availability Statement
All data associated with this study are in the paper and supplementary materials. All data are publicly available at the NCBI GEO (accession number, GSE193084). Raw data from figures is in data file S1. The R codes for Gene Set Enrichment Index based on single sample-based signature enrichment analysis (eseach algorithm) (11) and TexSEC are available at 10.5281/zenodo.6519385. Of note, data with patient names and other identifiers cannot be shared.


