Summary:
Lysosomes require an acidic lumen between pH 4.5 and 5.0 for effective digestion of macromolecules. This pH optimum is maintained by proton influx produced by the V-ATPase and efflux through an unidentified “H+ leak” pathway. Here we show that TMEM175, a genetic risk factor for Parkinson’s disease (PD), mediates the lysosomal “H+ leak” by acting as a proton-activated, proton-selective channel on the lysosomal membrane (LyPAP). Acidification beyond the normal range potently activated LyPAP to terminate further acidification of lysosomes. An endogenous polyunsaturated fatty acid and synthetic agonists also activated TMEM175 to trigger lysosomal proton release. TMEM175 deficiency caused lysosomal over-acidification, impaired proteolytic activity, and facilitated α-synuclein aggregation in vivo. Mutational and pH normalization analyses indicated that the channel’s H+ conductance is essential for normal lysosome function. Hence, modulation of LyPAP by cellular cues may dynamically tune the pH optima of endosomes and lysosomes to regulate lysosomal degradation and PD pathology.
Keywords: Proton channel, lysosome, pH optimum, acidification, degradation
Graphical Abstract
eTOC:
TMEM175 is a proton-activated proton channel under lysosomal physiological condition, and mediates the lysosomal proton efflux to maintain a steady-state pH.
Introduction:
Intracellular endosomes and lysosomes require a vacuolar-type H+ ATPase (V-ATPase) to establish a 50- to 5,000-fold proton concentration gradient across their membranes (Mindell, 2012; Ohkuma et al., 1982). This proton gradient can be utilized to drive the active transport of ions and metabolites between the vesicular lumen and the cytosol, and an acidic lumen (pH 4.5-5.0) is required for the optimal activities of most hydrolytic enzymes in the lysosome (Kalatzis et al., 2001; Kolter and Sandhoff, 2005; Li et al., 2019; Mindell, 2012; Xiong and Zhu, 2016; Xu and Ren, 2015). Abnormal lysosomal pH impairs lysosomal degradation, cargo loading, catabolite export, vesicle movement, and nutrient sensing (Ballabio and Bonifacino, 2019; Xu and Ren, 2015), contributing to pathologies of Alzheimer’s disease (AD), Parkinson’s disease (PD), and lysosomal storage diseases (LSDs) (Holopainen et al., 2001; Lee et al., 2010; Xu et al., 2014). Because lysosomal pH normalization (optimization) was shown to effectively remove toxic protein aggregates in these neurodegenerative diseases, lysosomal pH homeostasis likely plays a pivotal role in lysosome function, cellular health, and neuropathology (Bonam et al., 2019; Bourdenx et al., 2016; Nicoli et al., 2019).
Lysosomal pH is heterogeneous, and the “steady-state” set-point pH for individual lysosomes is determined by the dynamic equilibrium between proton influx and efflux across the lysosomal membranes (Ishida et al., 2013; Johnson et al., 2016). Inhibition of the V-ATPase quickly leads to organelle alkalization, suggesting the presence of unidentified “H+ leak” channel-mediated conductance(s) on the membranes of intracellular acidic compartments (Christensen et al., 2002; Li et al., 2019; Xu and Ren, 2015). In order to maintain a “steady-state” lysosomal pH at the set-point pH, a small “H+ leak” (roughly estimated to be 104 - 105 ions/sec or 0.1-1 pA across lysosomal membranes) is required to offset the proton influx produced by the V-ATPase (Johnson et al., 2016; Sakai et al., 2006). Two families of mammalian proton-selective channels have been identified: Otopetrins (OTOP1-3) and the voltage-gated proton channel (Hv1), but their normal location is the plasma membrane of specific cell types (Ramsey et al., 2006; Sasaki et al., 2006; Tu et al., 2018). Here we report that TMEM175, a ubiquitously-expressed lysosomal membrane protein (Cang et al., 2015), is highly and selectively permeable to protons when its luminal face is exposed to the acidic pH found within lysosomes, mediating the lysosomal “H+ leak” that balances V-ATPase activity to maintain lysosome pH homeostasis. TMEM175 was previously reported to be a “K+ leak” channel in lysosomes (Brunner et al., 2020; Cang et al., 2015; Lee et al., 2017; Oh et al., 2020). However, those studies were conducted with luminal-side pH above 7.0, which is non-physiological for lysosomal channels (Li et al., 2019; Xu and Ren, 2015). Under the normal lysosomal pH (4.5-5.0), we found that TMEM175 is approximately 105 times more permeable to protons than to potassium or sodium, and that in native lysosomes under conditions normally found in lysosomes, >90% of the ion flow through TMEM175 is mediated by protons. Furthermore, lysosomal TMEM175 could be activated by an endogenous lipid and two synthetic chemicals. Thus, rather than being a constitutively-active potassium-selective channel, lysosomal TMEM175 is a proton- or lipid-gated, proton-selective channel under lysosomal physiological condition.
Results:
A proton “leak” channel conductance exists in endosomes and lysosomes.
To study lysosomal proton flux mechanisms, we performed patch-clamp recordings (Fig. 1A) on late endosomes and lysosomes (LELs) chemically enlarged by vacuolin-1 and manually isolated from various types of mammalian cells (Chen et al., 2017b; Dong et al., 2008). To facilitate the detection of proton currents, the large ions N-methyl-D-glucamine (NMDG+) and methanesulfonic acid (MSA−) were used to replace the normal physiological cations and anions (Na+, K+, Ca2+, Mg2+, and Cl−) in both pipette/luminal and bath/cytosolic solutions, and a proton gradient between the cytosolic and luminal sides was established using appropriate pH buffers. When luminal pH (pHL) was set to 4.6 with a cytosolic pH (pHC) of 7.2, detectable (> 5 pA at −120 mV, Vm= VCytosol - VLumen) inward “leak-like” currents were found in a small portion (31 out of 128) of the enlarged LELs and the currents reversed at above + 80 mV (Supplemental Fig. S1A–B). When pHL was lowered to 3.5, all LELs (7 out of 7) showed large (> 100 pA at −120 mV) inward currents (Fig. 1B–C). When both sides were equally acidic (pHL/pHC of 4.6/4.6 or 3.5/3.5), significant currents were seen in both the inward and outward directions, and the reversal potential (Erev) shifted to near 0 mV (Fig. 1B and Supplemental Fig. S1A), as would be expected for a proton-selective channel. No measurable current was seen at all potentials when pHL and pHC were both set to 7.2 (Supplemental Fig. S1C and Fig. 1C). Similar pH-dependent currents were seen regardless of the major cation (NMDG+, TMA+, Choline+, K+, Na+, Ca2+, or Mg2+) or anion (MSA− or Cl−) present in the luminal and cytosolic sides of recording solutions (see Supplemental Fig. S1D for a current trace with Na+ as the major cation). Hereafter, we refer to the channel carrying this H+-dependent “leak” current as the lysosomal, proton-activated, proton-permeant channel (LyPAP). LyPAP currents (ILyPAP) were present in all cell types that we investigated: COS1, HAP1, HEK293T, HeLa, mouse embryonic fibroblasts, and mouse hippocampal neurons (Supplemental Fig. S1E). Similar albeit smaller currents were also observed in early endosomes (Supplemental Fig. S1F). Hence, LyPAP is a ubiquitously-expressed lysosomal proton channel with its activity beginning to be detectable at a luminal pH of 4.6, the typical lower edge of the optimal pH range for lysosomes (i.e., set-point pH) (Forgac, 2007; Mindell, 2012), but increasing dramatically when luminal pH is further lowered.
Figure 1. TMEM175 is necessary and sufficient for lysosomal proton-activated proton-permeant (LyPAP) currents.
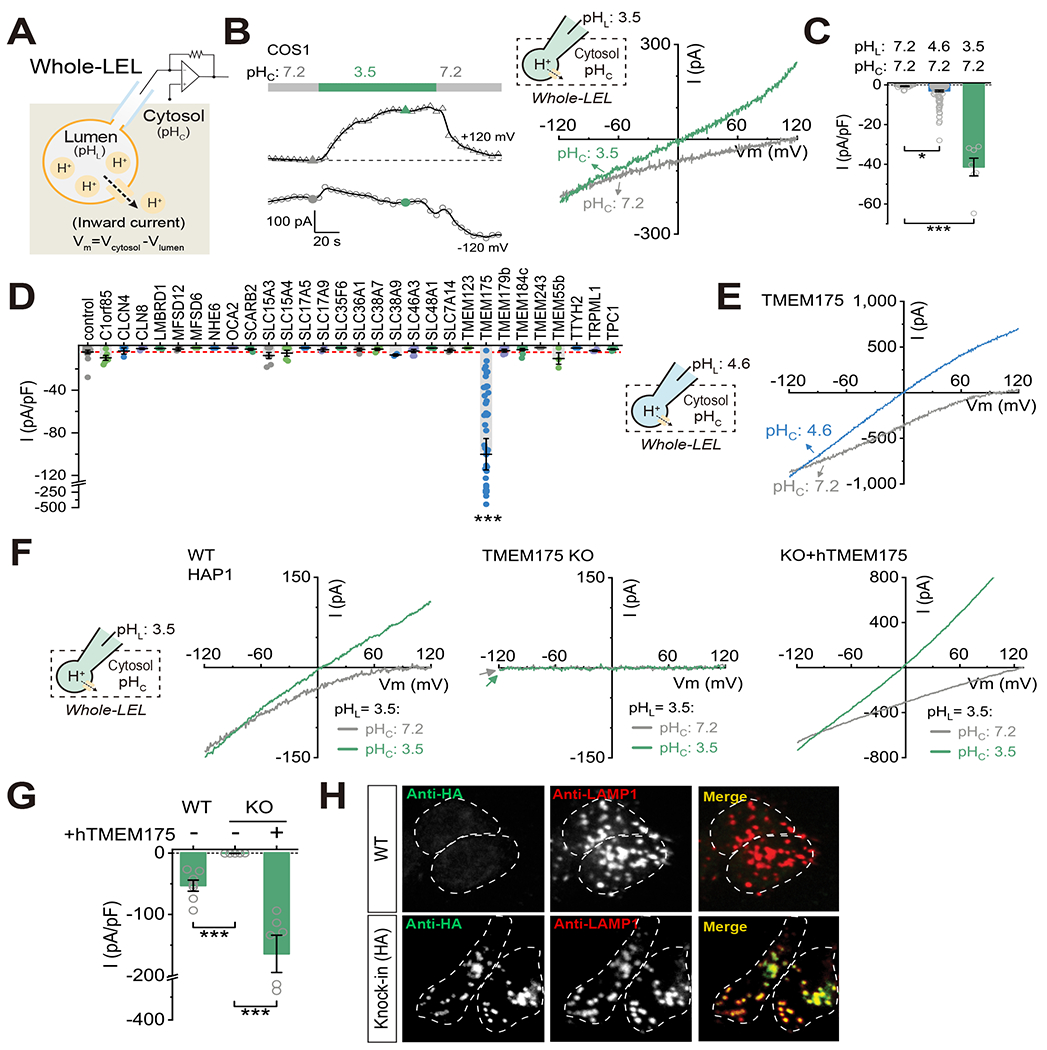
(A) Diagram of whole-LEL (late endosome and lysosome) recording method. Cation flow out of the LEL is defined as inward current. (B) Whole-LEL H+ “leak” currents from a COS1 cell. Luminal pH (pHL) was set to 3.5 and cytosolic pH (pHC) varied as indicated. Unless otherwise indicated, all recording solutions contained NMDG-MSA as the major ions. The I-V relations at the time of the colored points are illustrated. (C) Summary of whole-LEL H+ “leak” current density (pA/pF) at −120 mV (Vm= VCytosol – VLumen) from COS1 cells. Representative recordings when pHL was set to 4.6 or 7.2 are shown in Supplemental Fig. S1A and S1C (pHL 7.2/4.6/3.5: n = 18/128/7 LELs, mean ± SEM, * p <0.05, *** p <0.001). (D) Candidate expression screening revealed that only human TMEM175 overexpression resulted in a dramatic increase of whole-LEL H+ “leak” currents from COS1 cells (pHL = 4.6 and pHC = 7.2). The red dashed line represents the mean level from non-transfected control cells (control/TMEM175/others: n = 16/39/2-9 LELs, mean ± SEM, *** p <0.001). (E) Typical whole-LEL H+ “leak” currents from a COS1 cell overexpressing TMEM175. (F) Examples of whole-LEL H+ “leak” currents from WT HAP1 cells, TMEM175 KO cells, and KO cells re-expressing human TMEM175 (hTMEM175, pHL = 3.5). (G) Summary of current density at −120 mV from experiments as in (F) (WT/KO/KO+hTMEM175: n = 7/5/6, mean ± SEM, *** p <0.001). (H) Assessment of co-localization of TMEM175 with LAMP1 in TMEM175-HA knock-in and WT control HAP1 cells that were immuno-stained with anti-HA (green) and anti-LAMP1 (red) antibodies. Dashed lines indicate cell perimeters. Scale bar = 10 μm.
Lysosomal membrane protein TMEM175 is required for the LyPAP current.
To search for the molecular identity of LyPAP, we performed an expression screen with the candidate library consisting of a list of putative channel/transporter-like lysosomal membrane proteins based on proteomic studies (Schroder et al., 2010; Wyant et al., 2018). To make recognizing a positive hit easier, the screening was conducted in COS1 cells with pHL set to 4.6, which only allows very small endogenous ILyPAP (Fig. 1C). The only positive hit was TMEM175, for which the average current amplitude at −120 mV was more than 20 times larger than those from control (un-transfected) cells, cells transfected with the other candidates, or cells transfected with known lysosomal channels (e.g., TRPML1 or TPC1) that were previously suggested to be proton-permeable (Pitt et al., 2014; Soyombo et al., 2006) (Fig. 1D–E and Supplemental Fig. S1G–H). Like endogenous ILyPAP, Erev of the whole-LEL currents from TMEM175-transfected cells shifted from > +80 mV to 0 mV when pHC was changed from 7.2 to 4.6 (equaling pHL; Fig. 1E).
In TMEM175 knockout (KO) HAP1, HeLa, and HEK293T cells (Supplemental Fig. S2A) that were generated using Crispr-Cas9 (Ran et al., 2013), whole-LEL ILyPAP was absent even with a pHL of 3.5, but was reliably observed in unmodified wild-type (WT) cells or from KO cells re-expressing human TMEM175 (hTMEM175, Fig. 1F–G). In contrast, ILyPAP was intact in TPC1/TPC2 double knockout (DKO) HAP1 cells or TRPML1 KO human fibroblasts (Shen et al., 2012; Zhang et al., 2019b) (Supplemental Fig. S2B–C). Thus, TMEM175 is both necessary and sufficient for LyPAP currents in endolysosomes.
Consistent with previous reports (Cang et al., 2015; Jinn et al., 2019), fluorescently-tagged EGFP-TMEM175 puncta exhibited extensive co-localization with a marker for LELs (LAMP1), partial co-localization with a marker for early endosomes (EEA1), but no co-localization with the mitochondrial marker MitoTracker™ green (Supplemental Fig. S2D). A suitable antibody for immunodetection of endogenous TMEM175 proteins was unavailable, so we generated HA-tagged TMEM175 knock-in HAP1 cells (Supplemental Fig. S2E); in these cells, most HA-positive puncta were observed to overlap with LAMP1 (Fig. 1H).
TMEM175 is highly selective for protons.
We found that in TMEM175-transfected HEK293T cells, large whole-cell currents (ITMEMI175) were evoked by acidic extracellular pH; such currents were never observed in the vector-transfected (sham) cells (Fig. 2A–B). Hence, overexpressed, but not endogenous TMEM175, can traffic to the plasma membrane, as reported for some other lysosomal channels in heterologous overexpression systems (Li et al., 2019; Xu and Ren, 2015). The extracellular face of TMEM175 present on the plasma membrane is topologically equivalent to its luminal face in LEL membranes (Lee et al., 2017). We therefore studied the pH-dependence of ITMEM175 using whole-cell recordings, in which the extracellular pH (referred to as pHE/L) can be easily switched with a perfusion system to mimic changes in the luminal pH of endolysosomes. When pHC was set to 7.2 with NMDG+ as the only major cation in the recording solutions, the current amplitude at −120 mV of whole-cell ITMEM175 increased dramatically upon reducing pHE/L (Fig. 2B–C). Similar pHL-dependence was observed in lumen-side-out lysosomal recordings (Supplemental Fig. S3B–C), validating the characterization of heterologously-overexpressed TMEM175 using whole-cell recordings. The estimated macroscopic conductance of both human and mouse TMEM175 increased drastically as the extracellular/luminal side [H+] became greater (Supplemental Fig. S3A, S3C–D), suggestive of an activation effect of pHE/L.
Figure 2. The TMEM175 channel is highly proton selective.
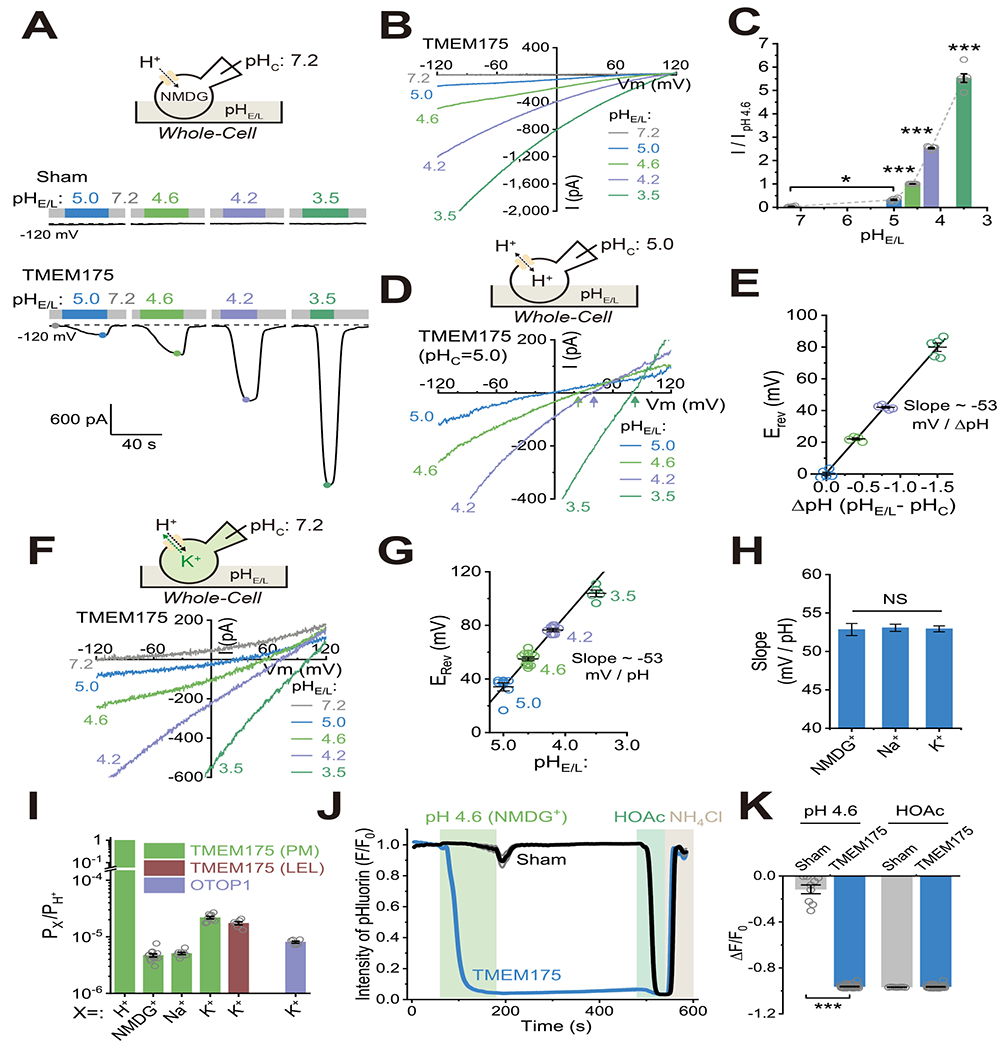
(A-B) Whole-cell currents recorded in an HEK293T cell overexpressing TMEM175 or a vector transfected (sham) cell. pHC was set to 7.2 and extracellular pH (pHE/L) varied as indicated. Unless otherwise indicated, all recording solutions (bath/extracellular and pipette/cytosolic) contained NMDG-MSA as the major ions. (C) Summary of whole-cell TMEM175 currents (ITMEM175) at −120 mV normalized to current at pHE/L = 4.6 (n = 6 cells, mean ± SEM, * p <0.05, *** p <0.001). Summary of whole-cell macroscopic conductance is shown in Supplemental Fig. S3A. (D) Erev measured in an HEK293T cell overexpressing TMEM175 with pHc set to 5.0 and pHE/L varied as indicated. (E) Erev as a function of ΔpH (= pHE/L- pHC) from experiments as in (D). The line fit to the data had a slope of −53 mV/ΔpH (n = 4 - 5, mean ± SEM). (F) Erev of ITMEM175 under “bi-ionic” conditions. The cytosolic solution contained K-MSA instead of NMDG-MSA as the major ions (pHc = 7.2). (G) Analysis of pH-dependence of Erev from experiments as in (F). The line is a least square linear regression fit to the averages of Erev measured at pHE/L 5.0 to 4,2 (n = 5 - 11, slope = −53 mV/ΔpH, mean ± SEM). (H) Summary of slope values in the pH-Erev experiments when NMDG+ (as in D-E), Na+, and K+ (as in F-G) were used as the major ions (n = 4 - 6, mean ± SEM, NS, p > 0.05). (I) Analysis of relative H+ permeability over NMDG+, Na+, or K+ based on Erev measurement (n = 6 - 13). The estimated PK/PH for the OTOP1 H+ channel, measured using the same recording conditions, is shown for comparison. Note that an incomplete block (by TEA+) of background K+ channels present in the HEK293T plasma membrane (PM) might make a contaminating contribution to PK, hence leading to an underestimated PH/PK value for TMEM175 on the PM compared with on LELs. Additionally, the outward currents may also contain a contaminating component of H+, currents due to experimentally-induced cytosolic H+ accumulation (see Methods). (J) Average pHlourin fluorescence in TMEM175-transfected HEK293T cells (n > 30 cells per coverslip) in response to stimuli as indicated. Imaging solutions contained NMDG-MSA as the major ions. HOAc (pH 5.0) and NH4Cl (pH 7.2), which freely enter the cytosol, served as positive and negative controls to acidify or alkalinize the cytosol, respectively. (K) Summary of relative pHluorin fluorescence intensity in response to pHE/L 4.6 and HOAc (sham/TMEM175: n = 9/35, mean ± SEM, *** p <0.001). See Supplemental Fig. S3K–O for similar experiments using other imaging solutions. N represents the number of cells randomly selected from at least three independent biological replicates.
To assess the selectivity of TMEM175 channels for protons, we set pHC to 5.0 (to make the outward current much larger than that at pHC 7.2) and measured Erev as a function of the proton gradient (ΔpH = pHE/L - pHC). With either NMDG+, Na+, or K+ as the major cation, the change in Erev closely followed the Nernst equation for a proton-selective channel with a linear fit of −53 mV/ΔpH unit (Fig. 2D–H). TMEM175 was previously reported to be a K+-selective channel (Cang et al., 2015), but those experiments were conducted under neutral pH conditions, which are physiological for plasma membrane channels but non-physiological for endolysosomal channels (Li et al., 2019; Xu and Ren, 2015). Under normal physiological conditions for endolysosomes, where the luminal side is highly acidic and the cytosolic side is neutral with a high concentration of potassium (Li et al., 2019; Xu and Ren, 2015), protons were much more permeable than potassium. For example, at pHE/L = 5.0, even though the cytosolic side [K+] was 14,000 times higher than the extracellular/luminal side [H+], the Erev was quite positive (Fig. 2F), indicating that H+ is much more permeable than K+ through TMEM175. Based on Erev measurement under “bi-ionic” conditions, as well as calculations based on the ΔpH-dependent Erev shifts, we estimated the relative permeabilities to be PH/PK = 48,000 ± 2,000, PH/PNa > 200,000, and PH/PNMDG > 200,000 (Fig. 2I), which are comparable to those of canonical proton-selective channels (Chen et al., 2019; Decoursey, 2003; Ramsey et al., 2006; Sasaki et al., 2006; Tu et al., 2018) (also see Supplemental Fig. S3E). Consistent with a linear fit to the whole Erev-pH plot (see Fig. 2E and 2G), the proton selectivity was independent of pHE/L (Supplemental Fig. S3F).
The basal whole-cell outward K+ currents in the TMEM175-transfected cells were typically too small to be reliably differentiated from the background K+ currents (see Methods). However, a subset of transfected cells had outward currents at +120 mV far larger than ever observed in untransfected cells, so we could attribute most of these currents to K+ flow through TMEM175. In these cells, the currents at all potentials were decreased ~ 50% when pHE/L was changed from 7.2 to either 6.0 or 4.6 (see Supplemental Fig. S3G), as long as precautions to avoid pH buffer saturation were followed (see Methods), suggesting that K+ flux through TMEM175 is suppressed at physiological pH in endolysosomes (also see (Pergel et al., 2021). Two approaches, one based on separate measurements of IH alone in K+-free solution and IK alone in K+-containing solution at neutral pH and then taking into consideration the IK inhibition by acidic pH (Supplemental Fig. S3H–I; also see Fig. 2B, F & Supplemental Fig. S4E, G), and the other by deriving the individual IH and IK currents from the total current (IH + IK) in “bi-ionic” conditions at pH 4.6 or 3.5 (see Fig. 2F), both led to the conclusion that in native lysosomes at their normal pH (4.6) and membrane potential (−110 to 0 mV) (Li et al., 2019; Saminathan et al., 2021), the majority (> 90%) of ion flow through TMEM175 is mediated by H+ (Supplemental Fig. S3J).
H+ flow through overexpressed TMEM175 on the plasma membrane would rapidly alter cytosolic pH if a large inward H+ gradient was present. We tested this prediction using both genetically-encoded (pHluorin; see (Miesenbock et al., 1998; Wang et al., 2017b)) and chemical (pHrodo™ Red) fluorescent indicators for intracellular pH (pHi) (Fig. 2J–K). Regardless of the major cation (NMDG+, TMA+, K+, Na+, Ca2+, or Mg2+) or anion (MSA− or Cl−) present in the extracellular solution, exposure to pHE/L 4.6 (with membrane-impermeable pH buffers) elicited rapid and substantial reductions in pHi in TMEM175-transfected, but not in sham HEK293T cells (Supplemental Fig. S3K–P & Supplemental Video S1).
Arachidonic acid and synthetic chemicals activate TMEM175 and endogenous LyPAP currents.
Arachidonic acid (ArA), a polyunsaturated fatty acid involved in numerous cellular signaling events, reportedly increases permeability to K+ and H+ in isolated lysosomes (Zhang et al., 2006). At pHL of 4.6 or 6.0 (in symmetric NMDG+), ArA application dramatically increased the endogenous whole-LEL H+ currents ILyPAP of COS1 cells Fig. 3A–B) and whole-cell H+ currents of TMEM175-transfected HEK293T cells (Fig. 3C–D & Supplemental Fig. S4A). Two small-molecule synthetic chemicals, DCPIB, previously known as an inhibitor of VRAC channels (Decher et al., 2001), and ML 67-33, previously known as an activator of K2P channels (Bagriantsev et al., 2013), were also found to be potent activators of LyPAP and TMEM175-mediated H+ and K+ currents (Fig. 3E–H & Supplemental Fig. S4A–H). Fluorescent pHluorin assays showed that, with a pHE/L 6.0 extracellular solution, ArA or DCPIB induced substantial H+ entry in TMEM175-transfected, but not sham HEK293T cells (Supplemental Fig. S4I–L).
Figure 3. Arachidonic Acid and synthetic chemicals activate TMEM175.
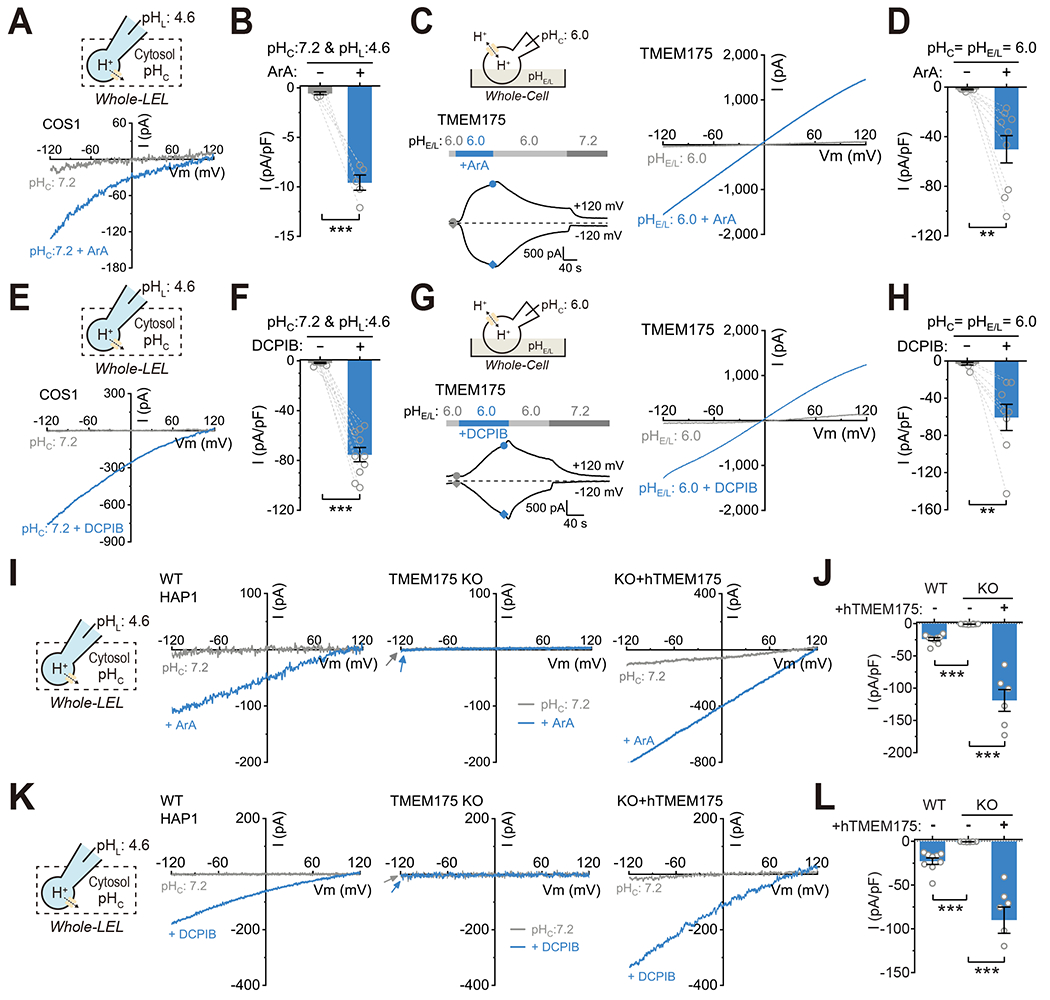
(A) Effects of Arachidonic Acid (ArA, 200 μM) on whole-LEL H+ currents in COS1 cells. The luminal solution contained NMDG-MSA with pHL set to 4.6. (B) Summary of current density at −120 mV from isolated enlarged LELs as in (A) (n = 5 LELs, mean ± SEM, *** p <0.001). (C-D) Effects of ArA (100 μM) on whole-cell H+ currents in TMEM175-transfected HEK293T cells. Current density (−120 mV) before and after ArA application is summarized in (D) (n = 9 cells, mean ± SEM, ** p <0.01), A representative recording from a non-transfected control cell is shown in Supplemental Fig. S4A. (E-F) Effects of DCPIB (100 μM) on whole-LEL H+ currents in COS1 cells. Current density (−120 mV) is summarized in (F) (n = 10 LELs, mean ± SEM, *** p <0.001). (G-H) Effects of DCPIB (50 μM) on whole-cell H+ currents in TMEM175-transfected HEK293T cells. Current density (−120 mV) before and after DCPIB application is summarized in (H) (n = 8 cells, mean ± SEM, ** p <0.01). (I) Representative traces of whole-LEL H+ currents from WT HAP1 cells, TMEM175 KO cells, and KO cells re-expressing human TMEM175 (KO+hTMEM175) in response to ArA (100 μM) with pHL = 4.6 and pHC = 7.2. (J) Summary of whole-LEL H+ currents (−120 mV) from experiments as in (I) (WT/KO/KO+hTMEM175, n = 9/10/6, mean ± SEM, *** p <0.001). (K) Representative traces of whole-LEL H+ currents from WT HAP1 cells, TMEM175 KO cells, and KO cells re-expressing human TMEM175 (KO+hTMEM175) in response to DCPIB (100 μM) with pEL = 4.6 and pHC = 7.2. (L) Summary of whole-LEL H+ currents (−120 mV) from experiments as in (K) (WT/KO/KO+hTMEM175, n = 9/8/7, mean ± SEM, *** p <0.001).
At the pHL of 4.6, large whole-LEL H+ currents elicited by ArA or DCPIB were reliably observed in WT HAP1 cells, but absent in TMEM175 KO HAP1 cells (Fig. 3I–L). When pHC and PHE/L were both set to 7.2 to suppress H+ permeation, K+ or Cs+ was able to generate large TMEM175-dependent whole-LEL currents, but only in the presence of ArA or DCPIB (Supplemental Fig. S4E–H); in most native lysosomes, lysosomal K+/Cs+ leak was undetectable (Supplemental Fig. S4E and S4G; also see Supplemental Fig. S3I). As the activation effect was more than 100-fold relative to the basal current, which was rarely observed in our hands (Li et al., 2019), lysosomal TMEM175 is likely a “gated” channel, not a constitutively-active “leak” channel as previously reported (Cang et al., 2015).
It was recently reported that lysosomal TMEM175 currents were stimulated by SC-79, a small-molecule AKT activator (Wie et al., 2021). However, in conditions that would facilitate the detection of either K+ or H+ currents through TMEM175 (symmetric K+ and pH 6.0), no detectable increase of whole-LEL or whole-cell currents was observed in TMEM175-transfected cells even with prolonged (10 minutes) bath application of SC-79 (10-100 μM; Supplemental Fig. S5A and S5C). In contrast, DCPIB readily evoked large H+ and K+ currents in the same recordings, in both non-treated cells and cells that were pretreated with AKT inhibitors (MK-2206 and ARQ-092; Supplemental Fig. S5A–C). The efficacies of these small-molecule AKT modulators were confirmed using specific biochemical assays (Supplemental Fig. S5D). Hence, in our hands, AKT activation is neither necessary nor sufficient for TMEM175 channel activity.
Luminal side protons activate TMEM175.
The pHL-dependence of the macroscopic conductance (see Supplemental Fig. S3A and S3C) of TMEM175 could arise from at least two separate biophysical mechanisms: pHL-dependent activation (gating) or increase of single channel conductance (permeation). To distinguish between these two possibilities, we set pHC to 4.6 instead of the normal physiological value of 7.2 (Fig. 4A–B), because this is expected to reveal large outward currents through open (activated), but not closed proton channels. Under the whole-cell configuration, with pHE/L initially set to 7.2 to establish a proton gradient of the same amplitude, but in the opposite direction from the normal situation in lysosomes (shown in Fig. 2A–B), i.e., ΔpH = pHE/L- pHC = +2.6, there was no significant current at any potential from −120 to +120 mV (Fig. 4A–B). Changing pHE/L to 4.6 (matching pHC, ΔpH = 0) elicited large H+ currents, in both the inward and outward directions, that reversed near 0 mV (Fig. 4A–B). Whereas changing pHE/L back to 7.2 slowly, but completely, de-activated the currents, stepping pHE/L from 4.6 to an intermediate pH (e.g., 5.6) resulted in current decreases with a concurrent left-ward shift in Erev (Supplemental Fig. S5E). Similar results were obtained with lumen-side-out lysosomal recordings (Supplemental Fig. S5F). Thus, H+ can only permeate the channel when there is an acidic environment facing the luminal side of TMEM175. In other words, TMEM175 is gated by luminal protons. With the reversed proton gradient (ΔpH = +2.6; pHE/L=7.2 and pHC = 4.6), DCPIB also activated substantial TEMEM175-mediated outward currents (Supplemental Fig. S5G–H) with a very negative Erev (~−120 mV) expected for a proton-selective channel. Hence, both luminal protons and non-proton agonists can activate the proton permeation pathway through TMEM175.
Figure 4. The TMEM175 channel is activated by luminal protons.
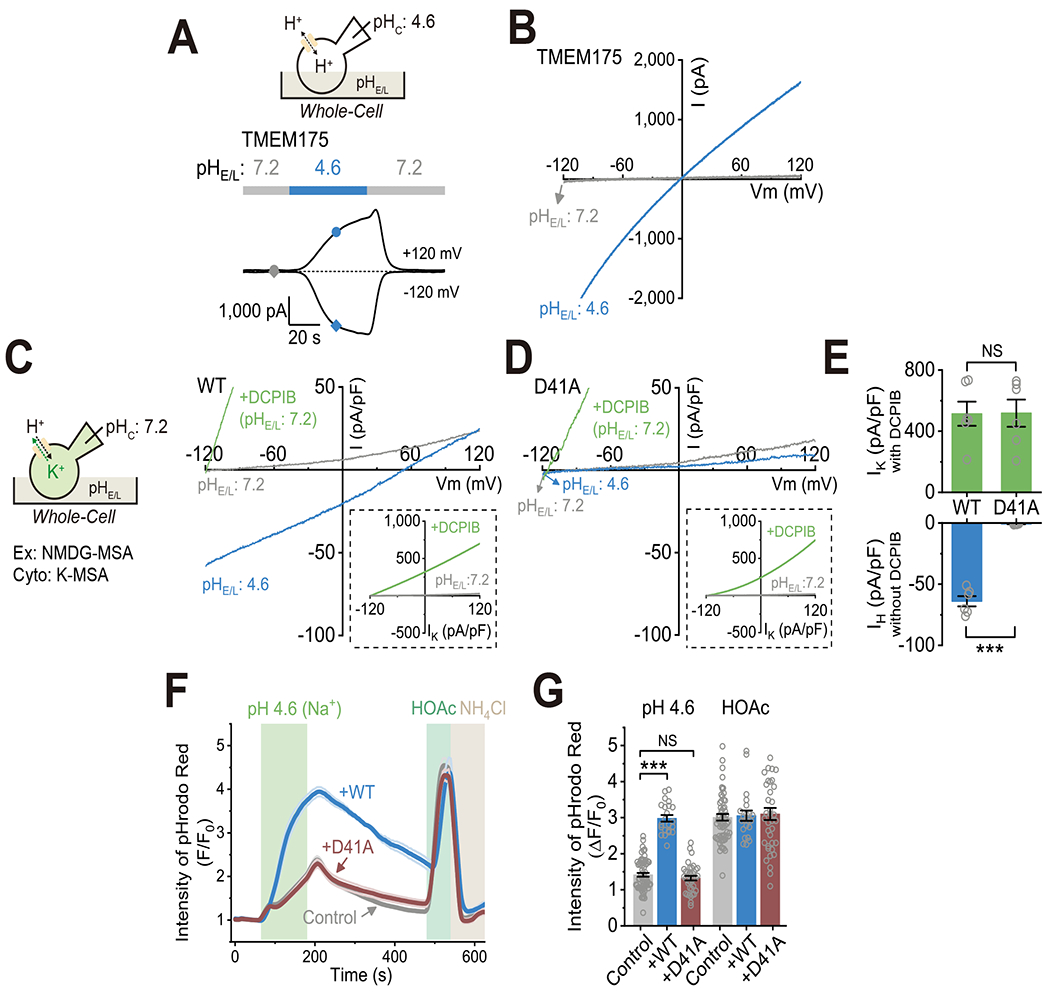
(A-B) Whole-cell currents recorded in aTMEM175-overexpressing HEK293T cell with the proton gradient reversed from what is normally found across lysosomal membranes. Cytosolic pHC was set to 4.6 and the extracellular pHE/L varied from 7.2 to 4.6 and back. See Supplemental Fig. S5F for a lumen-side-out lysosomal recording. (C-D) Whole-cell currents recorded in an HEK293T cell overexpressing WT TMEM175 or the D41A mutant. The cytosolic solution contained K-MSA with pHC set to 7.2. The large outward K+ currents in the presence of DCPIB are shown on a different scale in the boxed inset. (E) Summary of H+ (IH) and K+ (IK) currents recorded in cells from experiments as in (C-D). IH was recorded at pHE/L 4.6 at −120 mV without any activator, and IK was recorded in the presence of 50 μM DCPIB at pHE/L 7.2 at +120 mV (WT/D41A: n = 6/6 cells, mean ± SEM, *** p <0.001). (F) Average fluorescence changes of pHrodo™ Red in HEK293T cells overexpressing WT EGFP-TMEM175 or EGFP-TMEM175-D41A in response to the stimuli as indicated. (G) Summary of the relative fluorescent intensity of pHrodo™ Red in response to pH 4.6 and HOAc (control/WT/D41A, n = 65/25/38 cells; mean ± SEM, *** p <0.001, NS, p > 0.05). N represents the number of cells randomly selected from at least three independent biological replicates.
A pore mutation selectively eliminates TMEM175’s H+ permeability without altering its K+ permeability.
To identify the molecular determinants involved with pHL activation and/or H+ permeability of TMEM175, we systematically mutated amino acid residues with side chains capable of H+ binding (Aspartate, glutamate or Histidine) in the pore region and luminal loops (see Supplemental Fig. S6A), as revealed in the atomic-resolution structures of TMEM175 (Brunner et al., 2020; Lee et al., 2017; Oh et al., 2020). Mutant channels were tested in the whole-cell recording configuration using a high-K+ cytosolic solution with pHC = 7.2 to allow us to isolate either inward H+ currents IH or outward K+ currents IK In contrast to the WT and most tested mutant channels (Supplemental Fig. S6B–C), the D41A mutant TMEM175 channel showed no indication of inward IH at pHE/L of 4.6 (ΔpH = −2.6), but retained normal outward IK that was activated by DCPIB at pHE/L of 7.2 (ΔpH = 0, Fig. 4C–E) and inhibited by acidic pHE/L of 4.6 or 6.0 (Fig. 4D and Supplemental Fig. S6D). Thus, D41A is a selective H+-conductance-deficient mutant of TMEM175. Conversely, compared with the WT and most tested mutant channels, the S45A mutant channel had normal H+ permeation, but a partial (~ 2-fold) loss of K+ permeability relative to H+ or Na+ (Supplemental Fig. S6F–I). Intracellular pH imaging confirmed the incapability of D41A to mediate significant H+ flux (Fig. 4F–G), although the mutant proteins were correctly localized to lysosomes (Supplemental Fig. S6E), generating large DCPIB-activated whole-LEL IK under neutral pHL.
Genome Wide Associated Studies (GWAS) have identified several TMEM175 variants, especially p.M393T, as Parkinson Disease-risk factors (Blauwendraat et al., 2019; Chang et al., 2017; Krohn et al., 2020). In the heterologous overexpression experiments, the M393T mutation did not cause any change in the channel properties of TMEM175, e.g., H+ and K+ permeability (Supplemental Fig. S6F–G). However, in M393T knock-in HEK293T cells, where the expression level was controlled by the endogenous promoter, we observed a ~50% reduction in whole-LEL/ILyPAP (Supplemental Fig. S6J–K), suggesting that consistent with previous observations (Jinn et al., 2019; Wie et al., 2021), M393T is likely a trafficking mutant.
Activation of TMEM175 induces proton release from the lysosome lumen.
When pHluorin-expressing HEK293T cells were imaged to monitor the intracellular pH, with a pHE 7.2 or pH 7.6 (ΔpH = 0 or + 0.4) bathing solution, application of DCPIB or ArA induced substantial, TMEM175-dependent decreases in pHi (Fig. 5A–B and Supplemental Fig. S7A–B). Nullifying the membrane potential of the cells using a high K+ extracellular solution yielded similar results. Hence, in the absence of an electrochemical gradient of protons across the plasma membrane, DCPIB or ArA likely activated lysosome-localized overexpressed TMEM175 channels to release luminal H+ into the cytosol. Indeed, pre-treating cells with GPN (glycyl-L-phenylalanine 2-naphthylamide) to dissipate the lysosomal H+ gradient (Jadot et al., 1984), abolished the effects of DCPIB or ArA on cytosolic pH (Fig. 5A–B and Supplemental Fig. S7A–B). In addition to activating TMEM175, DCPIB is also known to inhibit LRRC8A-mediated Lyso-VRAC currents (Li et al., 2020b). Smaller DCPIB-induced, GPN-pre-treatment-sensitive pHi decreases were seen in WT, but not in TMEM175 KO HEK293T cells (Fig. 5C–D), suggesting that chemical activation of endogenous TMEM175 proteins is sufficient to induce lysosomal proton release, and that the effect of DCPIB on lysosomal proton release is mediated by TMEM175.
Figure 5. Activation of lysosomal TMEM175 induces proton efflux from lysosomes.
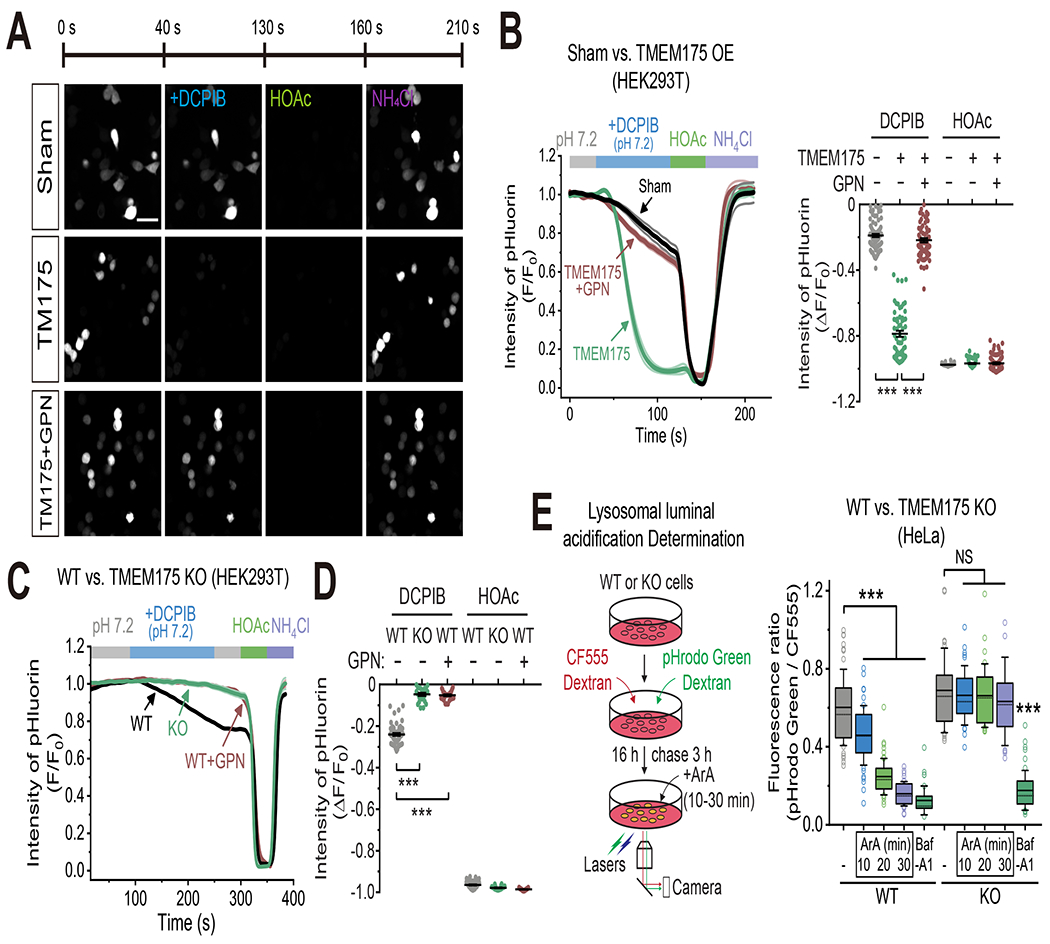
(A) The fluorescence intensity of pHluorin in HEK293T cells dually transfected with pHluorin and mCherry-TMEM175 or mCherry (sham) in response to stimuli as indicated (scale bar = 50 μm). Imaging solution was set to pHE/L 7.2 to minimize H+ influx across the plasma membrane (ΔpH = 0). DCPIB (50 μM) was bath applied to activate TMEM175. In the GPN experiment, cells were pre-treated with GPN (200 μM) one hour before imaging to dissipate the lysosomal H+ gradient. (B) Average time series from experiments as in (A) (mean ± SEM). The right panel shows the summary of relative pHluorin fluorescence intensity in response to DCPIB and HOAc (sham/TM175/TM175+GPN, n = 88/63/73 randomly selected cells from at least three independent biological repeats; mean ± SEM, *** p <0.001). See Supplemental Fig. S7A–B for the effect of ArA. (C) Average pHlourin fluorescence in pHlourin-expressing WT or TMEM175 KO HEK293T cells in response to stimuli as indicated (mean ± SEM). DCPIB (20 μM) was bath-applied to activate endogenous lysosomal TMEM175. In the GPN experiment, cells were pretreated with GPN (200 μM) one hour before imaging. Imaging solutions contained Na-MSA as the major ions. HOAc and NH4Cl served as controls to acidify or alkalinize the cytosol. (D) Summary of relative pHluorin fluorescence intensity in response to pHE/L 4.6 and HOAc from experiments as in (C) (WT/KO/WT+GPN: n = 57/26/31 cells, mean ± SEM, *** p <0.001). (E) Time-dependent effects of ArA application (400 μM) on lysosomal acidity in WT and KO HeLa cells, which was determined using a ratiometric pH dye combination (pHrodo Green dextran and CF555 dextran). Note that the slow de-acidification effect of ArA on lysosomes might be due to a combination of slow membrane diffusion, delayed lysosomal delivery, and the existence of counteracting acidifying force. Baf-A1 (1 μM, 1h) served as a positive control. Plots show the fluorescence ratios of pHrodo Green vs. CF555 (n = 26-52 randomly selected cells from three biological replicates; *** p <0.001, NS, p > 0.05) (see Methods for the details about the box plots).
We also investigated the effect of TMEM175 activation on luminal pH using a ratiometric pH-sensitive dye combination (pH-sensitive pHrodo Green Dextran + pH-insensitive CF555 Dextran), which can be loaded to the lysosome lumen via endocytosis. ArA caused a dramatic, time-dependent lysosomal de-acidification (alkalization) in WT, but not TMEM175 KO HeLa cells (Fig. 5E); the V-ATPase inhibitor Bafilomycin-A1 (Baf-A1) (Bowman et al., 1988) produced luminal de-acidification in both WT and KO cells. DCPIB induced lysosomal de-acidification in both WT and LRRC8A KO HAP1 cells, but not in TMEM175 KO HAP1 cells (Supplemental Fig. S7C–D), suggesting that DCPIB’s effect on lysosomal pH is through its activation effect on TMEM175, not through its inhibition effect on Lyso-VRAC.
Proton conductance through TMEM175 is required for lysosomal pH homeostasis.
Steady-state lysosomal pH likely reflects a balance between proton import mediated by the V-ATPase and proton efflux through TMEM175. In multiple lines of TMEM175 KO cells (HAP1, HEK293T, and HeLa), LysoTracker (LysoTracker™ Red DND-99, a pH-sensitive dye) staining was more intense than in WT cells, yet LAMP1 staining was comparable (Fig. 6A–B & Supplemental Fig. S7E), suggesting that the lysosomes of TMEM175 KO cells are hyper-acidified (also see (Cang et al., 2015; Jinn et al., 2017)). Similar results were also seen when lysosomal pH was more accurately determined by using three separate sets of ratiometric dyes: pHrodo Green Dextran (Fig. 5E), LysoSensor Yellow/Blue DND-160 (Supplemental Fig. S7F), and Oregon Green™ 488 Dextran (Fig. 6C & Supplemental Fig. S7G). When the fluorescence ratios were calibrated to pH values, we found that TMEM175 KO HeLa cells displayed significant lysosomal hyperacidity (~ 0.3 pH unit), which was brought back toward normal by partially inhibiting the V-ATPase with a low concentration (1 nM) of Baf-A1 (Fig. 6C). Conversely, HEK293T cell lines stably overexpressing TMEM175 showed significant lysosomal hypoacidity (Supplemental Fig. S7H–I). The lysosomal hyperacidity of KO cells was corrected by re-expressing WT TMEM175 or the S45A mutant, but not the D41A mutant (Fig. 6D–E & Supplemental Fig. S7J–K). Together with the ArA/DCPIB effects on lysosomal acidity, these results suggest that the H+ conductance, but not K+ conductance of TMEM175, is required for the regulation of lysosomal pH homeostasis. Whereas increasing TMEM175 expression/activity raises steady-state lysosomal pH (alkaline shift in the set-point pH; hypo-acidification), decreasing TMEM175 expression/activity reduces steady-state lysosomal pH (acidic shift in the set-point pH; hyper-acidification).
Figure 6. Proton permeation through TMEM175 is required for lysosomal pH homeostasis and effective proteolytic degradation.
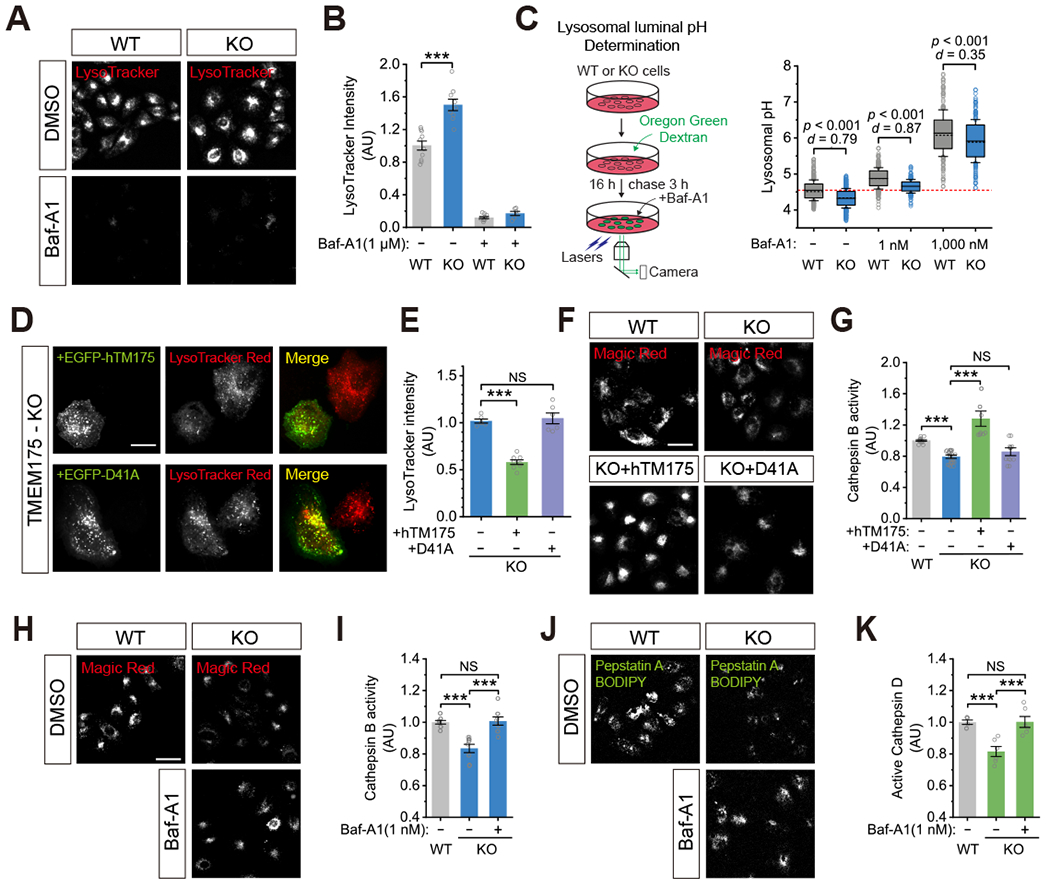
(A-B) Lysosomal acidity assessed by LysoTracker (red) staining in WT and TMEM175 KO HeLa cells (scale bar = 50 μm). Plots show the overall LysoTracker intensity per cell (n = 6-10 randomly selected images from at least three independent biological repeats, *** p <0.001). Cells incubated with 1 μM Baf-A1 for 1 hour served as a positive control. (C) Lysosomal pH in WT and KO HeLa cells determined using Oregon Green Dextran. Baf-A1 was applied to mildly (1 nM) or maximally (1,000 nM) block the V-ATPase to alkalize the lysosomes. The red dashed line represents the mean level from WT cells (n = 177-487 cells per group; Cohen’s d > 0.75 indicates large effects and d = 0.25-0.50 indicates small effects. (D-E) Lysosomal acidity assessed by LysoTracker staining in TMEM175 KO HeLa cells re-expressing WT EGFP-TMEM175 or EGFP-TMEM175-D41A (scale bar = 20 μm, n = 6-8, *** p < 0.001, NS, p > 0.05). (F-G) Lysosomal Cathepsin B activity assayed by Magic Red staining in WT, KO, and KO HeLa cells stably expressing WT TMEM175 or D41A (scale bar = 50 μm, n = 6-13, NS, p > 0.05, *** p < 0.001). (H-I) Lysosomal Cathepsin B activity in WT, KO, and KO cells incubated with Baf-A1 (1 nM) for 30 min (scale bar = 50 μm; n = 6-8, NS, p > 0.05, *** p < 0.001). (J-K) Lysosomal active amount of Cathepsin D assayed by Pepstatin-A-BODIPY-FL staining in WT, KO, and KO cells incubated with Baf-A1 (1 nM) for 30 min (scale bar = 50 μm; n = 6, NS, p > 0.05, *** p < 0.001). All plots show the results from at least three independent biological replicates for each experimental condition.
Proton permeation through lysosomal TMEM175 is required for the optimal activity of lysosomal hydrolases.
Many lysosomal hydrolases, such as Cathepsin B and Cathepsin D, require a narrow optimal pH range for their enzymatic activities (Briozzo et al., 1988; Kirschke et al., 1977; Yoshida et al., 2015). The overall lysosomal hydrolytic activity, assayed by DQ™-BSA-red dye staining (Ashcom and Jacobson, 1989) , was decreased in TMEM175 KO HeLa cells compared with WT cells, but was fully restored in KO cells stably re-expressing WT TMEM175, but not the D41A mutant (Supplemental Fig. S7L–M). Using Magic Red, a fluorogenic substrate for Cathepsin B (Creasy et al., 2007), we found that Cathepsin B activity was impaired in KO cells compared with WT cells, but was fully restored in KO cells stably re-expressing WT TMEM175, but not the D41A mutant (Fig. 6F–G). Moreover, 1 nM Baf-A1, the concentration that normalized the lysosomal pH of KO cells (pH optimization; see Fig. 6C), brought Cathepsin B activity back to the normal level (Fig. 6H–I). Similarly, the amount of active Cathepsin D, assayed with Pepstatin A BODIPY FL (Chen et al., 2000; Lee et al., 2010; Rahman et al., 2016), was also reduced in the TMEM175 KO cells, but fully restored by pH optimization with 1 nM Baf-A1 (Fig. 6J–K). The protein levels of Cathepsins B and D were not decreased in KO cells compared with WT cells (Supplemental Fig. S7N). Taken together, these results suggest that the H+ conductance of TMEM175 plays an essential role in the pH-dependent regulation of optimal activities of lysosomal hydrolases.
TMEM175 deficiency causes pathological α-synuclein aggregation in the mouse brain.
To investigate the in vivo role of TMEM175, we generated Tmem175 KO (Tmem175−/− mice using Cripsr-Cas9 (Ran et al., 2013) (Fig. 7A). Motor skills are compromised in a similar line of Tmem175 KO mice (Wie et al., 2021). In cultured hippocampal neurons isolated from Tmem175−/− mice, we confirmed the essential roles of TMEM175 in mediating LyPAP (Fig. 7B–C), regulation of lysosomal steady-state pH (Fig. 7D), agonist-induced lysosomal proton release (Fig. 7E), and Cathepsin B activity (Fig. 7F). TMEM175 has been recently identified as a genetic risk factor for PD (Blauwendraat et al., 2019; Chang et al., 2017; Hopfner et al., 2020; Iwaki et al., 2019; Krohn et al., 2020; Li et al., 2020a; Rudakou et al., 2020). A hallmark pathology of PD is α-synuclein aggregation in the brain (Spillantini et al., 1997), and α-synuclein is a known substrate of Cathepsin B/D (Cullen et al., 2009; McGlinchey and Lee, 2015; Qiao et al., 2008). We used an established assay to facilitate α-synuclein aggregation in non-aged animals via a single striatal injection of preformed α-synuclein fibrils (pff) (Luk et al., 2012). Thirty days after pff injection, we observed much greater aggregation of phosphorylated α-synuclein, the pathogenic form of α-synuclein (Luk et al., 2012) which is known to be accumulated in the Lewy body inclusions containing excessive lysosomal membranes and contents (Shahmoradian et al., 2019), in the striatum of Tmem175−/− mice compared with WT littermates (Fig. 7G–H).
Figure 7. TMEM175 deficiency in mouse neurons causes lysosomal over-acidification, impaired lysosomal hydrolytic activity, and α-synuclein aggregation in the brain.
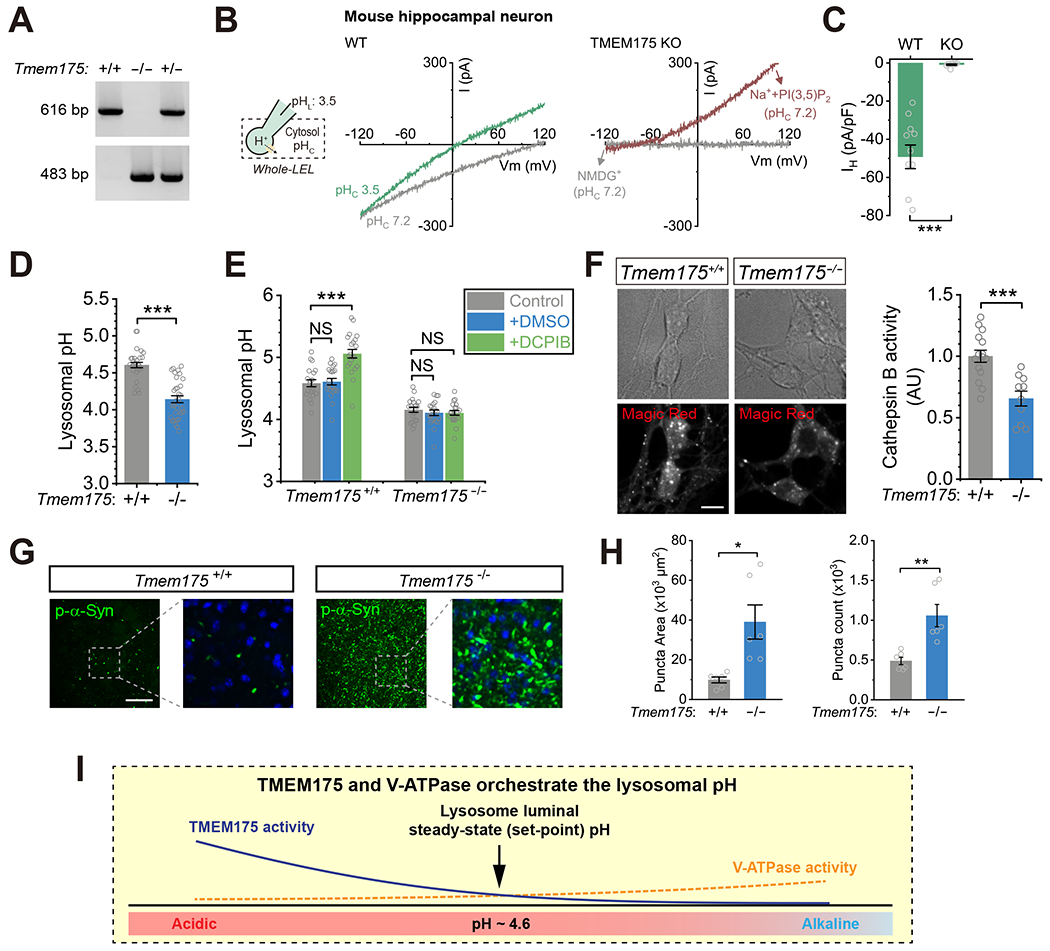
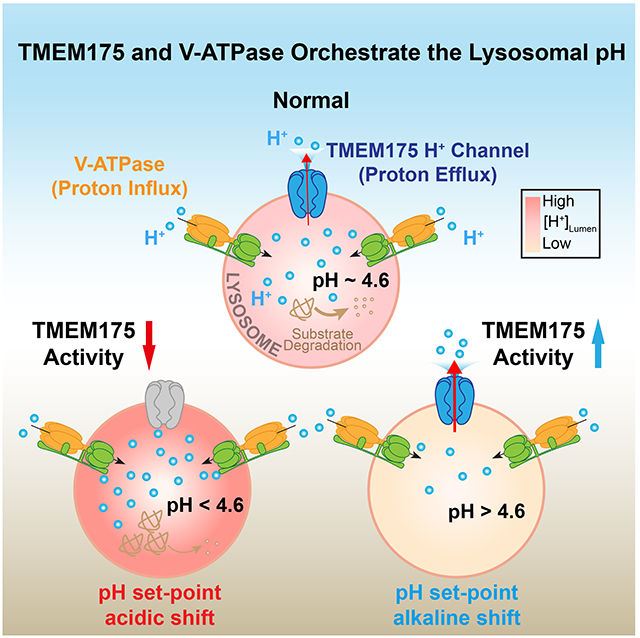
(A) PCR genotype analysis of Tmem175+/+ (WT), Tmem175+/− (Heterozygous), and Tmem175−/− (KO) mice. (B) Whole-LEL ILyPAP in the cultured hippocampal neurons isolated from WT and Tmem175 KO mice (pHL = 3.5 and pHC = 7.2). The TPC-mediated outward Na+ current evoked by PI(3,5)P2 (Wang et al., 2012) served as a positive control that the recording was in the whole-LEL mode. (C) Average current density at −120 mV from experiments as in (B) (n = 9 - 11 LELs, mean ± SEM, *** p <0.001). (D) Lysosomal pH in WT and Tmem175 KO hippocampal neurons determined using Oregon Green Dextran (n = 30 - 32 randomly selected images from at least three independent repeats, mean ± SEM, *** p <0.001). (E) Effects of DCPIB (50 μM) on lysosomal pH in Tmem175+/+ and Tmem175−/− hippocampal neurons determined using Oregon Green Dextran (n = 20-21 images, mean ± SEM, NS p > 0.05, *** p <0.001). (F) Lysosomal Cathepsin B activity assayed with Magic Red staining in Tmem175+/+ and Tmem175−/− hippocampal neurons (scale bar = 10 μm, n = 10 - 15, *** p < 0.001). (G) Representative images of phosphorylated α-synuclein puncta in striatal slices collected from Tmem175+/+ and Tmem175−/− mice (scale bar = 100 μm). The boxed regions are shown at higher magnification on the right together with DAPI staining. (H) Summary of puncta area and count of phosphorylated α-synuclein in striatum (n = 6 slices from 3 mice for each group, mean ± SEM, * p < 0.05, ** p < 0.01). (I) A working model for how TMEM175, a lysosomal luminal proton-activated proton release channel, regulates lysosome pH set-point, optimum, and hydrolytic activity. Lysosomal pH is heterogeneous within the range of pH 4.5-5.0 that is required for the optimal hydrolytic activity of lysosomal enzymes. For individual lysosomes, the steady-state luminal pH is determined by the relative rates of V-ATPase-mediated proton influx (which decreases with luminal acidification (Sakai et al., 2006)) vs. TMEM175-mediated proton efflux. For a typical lysosome with a steady-state pH of 4.6 (the set-point), only a small lysosomal proton efflux through TMEM175 is needed to balance out the proton influx produced by the V-ATPase. Because TMEM175 is far from maximally activated at pH 4.6, any stimulus that acidifies the lysosome to below 4.6 causes TMEM175 currents to rapidly increase as if a pH “threshold” has been crossed. Increasing the expression/activity of TMEM175 causes an alkaline shift in the pH set-point and steady-state pH. When the activity of TMEM175 is compromised, as in the TMEM175 knockout cells, lysosomes are over-acidified due to an unopposed proton influx produced by V-ATPase, causing impaired lysosomal hydrolysis, a-synuclein aggregation in the central neurons, and PD-like pathology.
Discussion:
To maintain the luminal pH optimal for lysosomal degradation (pH 4.5- 5.0), the proton influx and efflux pathways need to function coordinately on the lysosomal membrane. While the V-type H+ ATPase actively imports H+ into the lysosome lumen, proton efflux pathways are required to passively release H+ into the cytosol (see Fig. 7I). There are several H+-coupled transporters on the lysosomal membrane that may mediate slow H+ leak during their active transport cycle (Casey et al., 2010; Kalatzis et al., 2001; Mindell, 2012). For example, CLCN7, by acting as a lysosomal Cl−/H+ exchanger, may consume the H+ gradient while transporting Cl− (Graves et al., 2008). However, lysosomes lacking CLCN7 appear to have a normal steady-state pH (Kasper et al., 2005; Steinberg et al., 2010). Hence, the slow, transporter-mediated proton “leak” pathways are unlikely to be the primary proton efflux mechanism that maintains the lysosomal pH optimum. Instead, a fast, channel-mediated “H+ leak” conductance can effectively prevent lysosomal over-acidification by responding dedicatedly to the trigger: a change in the luminal status of pH (pHL). At normal lysosomal pH (pH 4.5-5.0), this “H+ leak” is slightly active; during hyper-acidification, e.g., < pH 4.5, the activity of “H+ leak” is dramatically elevated; during hypo-acidification, e.g., > pH 5.0, the activity of “H+ leak” is suppressed (see Fig. 7I). We identified TMEM175 as a pHL-activated H+-selective channel that is required for this dedicated “H+ leak”. In contrast, TRPML1 and TPC1, which were previously suggested to encode the “H+ leak” channel in lysosomes (Pitt et al., 2014; Soyombo et al., 2006), displayed no measurable proton permeability in lysosomal patch-clamp studies (Dong et al., 2008; Wang et al., 2012; Xu and Ren, 2015) and had no role in lysosomal “H+ leak”. The slow, non-dedicated “H+ leak” pathways, e.g., those mediated by CLCN7 or other H+-coupled transporters, might make supplementary or compensatory contributions to lysosomal pH homeostasis, especially outside of normal pH range, as was observed in V-ATPase-inhibited cells or TMEM175 KO cells.
TMEM175 was previously reported to be a lysosomal K+-permeable/selective channel under neutral pH (Cang et al., 2015), which is non-physiological for healthy lysosomes. We demonstrated in the current study that with the normal physiological H+ and K+ gradients across lysosomes (Li et al., 2019), TMEM175-mediated K+ flux is suppressed but H+ flux is stimulated, so ion flow through TMEM175 is almost exclusively carried by protons (> 90%) at physiological lysosomal membrane potential (Koivusalo et al., 2011; Saminathan et al., 2021). Consistent with our observations, a recent study published during the final review process of our current work demonstrated that at acidic pH, TMEM175 is not only permeant to proton, but also undergoes pH-dependent structural changes that reduce K+ permeation (Zheng et al., 2022). Supporting the dominance of proton flux through endogenous TMEM175 in native lysosomes, the basal K+/Cs+ current in the lysosome is barely detectable, while H+ current is sizable. Hence, just like Ca2+-selective channels that allow Na+ or K+ permeation in the absence of Ca2+ (Sather and McCleskey, 2003), as an H+-selective channel, TMEM175 may permit K+ permeation in the nominal “absence” of H+ under certain experimental conditions or if localized in non-lysosomal compartments.
Loss of TMEM175 was previously proposed to affect lysosomal pH homeostasis through an indirect regulation via TMEM175-mediated K+ flux (Cang et al., 2015), e.g., as a counter ion mechanism (Mindell, 2012; Steinberg et al., 2010). In this hypothesis, K+ efflux from lysosomes would provide the counter ion required for continuous V-ATPase-mediated pumping H+ into lysosomes (Mindell, 2012; Steinberg et al., 2010). However, under physiological conditions, the electrochemical gradient of K+ favors K+ influx into lysosomes (Li et al., 2019). In addition, the inhibitory effect of H+ on TMEM175-mediated K+ flux is also contradictory to a role in providing the counter ion. Furthermore, if TMEM175’s K+ conductance indeed plays a role in this counter ion regulation of lysosomal pH, one would expect that lysosomes are hypo-acidified in TMEM175 KO cells, and hyper-acidified in TMEM175-overexpressing cells. However, opposite results were observed: lysosomes are hypo-acidified in TMEM175-overexpressing cells, and hyper-acidified in the KO cells. Hence, the H+ conductance, but not K+ conductance of TMEM175 seems to be primarily responsible for all the pH-dependent lysosomal functions of TMEM175 investigated in the current study: pHL-regulated lysosomal H+ release, setting the lysosomal pH optimum, agonist-induced regulation of lysosomal pH, and pH-dependent Cathepsin B/D activity. Supporting this conclusion are the results from the mutational analyses (e.g., D41A and S45A) and pH normalization experiments.
The proton permeation pathway of TMEM175 can be activated by at least two distinct mechanisms. First, TMEM175 is activated by luminal protons. At the normal lysosomal pH (i.e. the set-point), a small LyPAP current is present to offset basal proton influx produced by V-ATPase, but when the lysosome lumen is over-acidified by the V-ATPase to below the set-point, i.e., pHL < 4.5, both the H+ driving force and channel macroscopic conductance increase dramatically. Because the key pH-sensitive site (D41) may be located in the conduction pathway, proton activation and permeation may initially reinforce each other via a positive-feedback mechanism, resulting in a rapid surge in proton efflux once lysosomes are hyper-acidified below the set-point pH. Subsequently, the substantial proton release will effectively prevent further luminal acidification (see Fig. 7I). Thus, in native lysosomes, a negative feedback loop is operative to “clamp” lysosomal pH close to the set-point. What is unique about this negative feedback loop is that the sensor and the response element are integrated into one protein: a proton-activated proton-permeant channel. Second, polyunsaturated fatty acids such as arachidonic acid can exert a non-pH control over TMEM175 activity. Therefore, cellular cues that modulate TMEM175 may effectively change the pH-dependent activation of TMEM175, resulting in a new set-point and steady-state pH. The various compartments of the endocytic pathway (early endosomes, late endosomes, lysosomes, and secretory vesicles) have luminal pHs between 4.5 and 6.5 (Casey et al., 2010). The observations that TMEM175 expression is highly enriched in lysosomes, and that the activation pH for endogenous TMEM175 is close to pH 4.6, are consistent with the idea that these channels may play a particularly important role in regulating lysosomal pH homoeostasis. However, in the presence of endogenous agonists such as arachidonic acid or a related lipid, proton flow through TMEM175 could occur in less acidic organelles such as early endosomes. We hypothesize that both the V-ATPase and TMEM175 are regulated in their activity and lysosomal abundance to achieve the dynamicity and variety of endolysosomal pH in cells.
Compromised lysosome function due to abnormal lysosomal pH may play a causative role in the pathologies of a range of diseases, including LSDs, AD, and PD (Bourdenx et al., 2016; Holopainen et al., 2001; Lee et al., 2010; Xu et al., 2014). TMEM175 has been identified by several recent GWAS studies as a genetic risk factor for PD (Blauwendraat et al., 2019; Chang et al., 2017; Krohn et al., 2020). Our findings that TMEM175, as a proton-activated, proton-selective channel required for the maintenance of lysosomal pH optimum, regulates Cathepsins B/D activity at the cellular level and the clearance of α-synuclein aggregation in vivo may provide a new opportunity for understanding the pathologies of PD and LSDs. Importantly, pH optimization or restoring TMEM175’s H+ permeability results in normal lysosomal proteolytic activity in TMEM175 KO cells, provide a proof of concept that modulation of TMEM175 activity in the lysosome could help develop therapies for neurodegenerative diseases.
Limitations of the study.
Although our collective evidence suggests that the H+ conductance of TMEM175 is likely more important than its K+ conductance, without a mutation that selectively and completely abolishes K+ permeability (S45A is only a partial K+-conductance-deficient mutant), one cannot completely rule out the roles of TMEM175’s K+ permeability in regulating lysosomal functions. Specifically, at lysosomal pH, the K+ permeability of TMEM175, albeit very small, could still contribute to K+ flux in native lysosomes under certain untested cellular conditions. Moreover, even though an acidic environment favors H+ over K+ flux, the presence of endogenous non-proton agonists (e.g., ArA), which activate both H+ and K+ conductance, may increase the relative contribution of K+ flux. Hence, the K+ permeability, an evolutionarily-preserved biophysical feature of TMEM175, which is well documented and confirmed in the current study, might play a role in mammalian cells when TMEM175 is located in less acidic compartments, e.g., early endosomes and newly formed (un-acidified) autolysosomes, or under neutral pH conditions, e.g., at the plasma membranes when cells undergo lysosomal exocytosis (Li et al., 2019). Future development of synthetic agonists/inhibitors selective for the K+ or H+ permeation pathway and analyses of TMEM175 mutant variant (e.g., D41A) knock-in mice may reveal the roles of H+ vs. K+ flux in lysosome function, cell physiology, and PD pathology.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Requests for resources, reagents, and further information regarding this manuscript should be addressed to and will be fulfilled by the lead contact, Meiqin Hu (meiqinhu@umich.edu).
Materials availability
Plasmids and cell lines newly generated in this work are available upon request to the lead contact.
Data and code availability
Accession numbers are listed in the key resources table. Original traces of electrophysiological recordings, western blots, and microscopical images will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse anti-human LAMP1 (H4A3) antibody | DSHB | Cat# h4a3, RRID: AB_2296838 |
| Rat anti-HA (3F10) antibody | Roche | Cat# 11867423001, RRID: AB_390918 |
| Rabbit Anti-Human alpha Synuclein, phospho (Ser129) antibody | Abcam | Cat# ab51253, RRID: AB_869973 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Texas Red-X | Thermo Fisher Scientific | Cat# T-862, RRID: AB_2556781 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006, RRID: AB_2534074 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_253579 |
| Mouse anti-GFP (clone 7.1 and 13.1) antibody | Roche | Cat# 11814460001, RRID: AB_390913 |
| Rabbit anti-Phospho-Akt(Thr308) (C31E5E) antibody | Cell Signaling Technology | Cat #2965, RRID: AB_2255933 |
| Rabbit anti-Akt (pan) (C67E7) antibody | Cell Signaling Technology | Cat# 4691, RRID: AB_915783 |
| Rabbit anti-Cathepsin B antibody [EPR4323] | Abcam | Cat# ab125067, RRID: AB_10972167 |
| Rabbit anti-Cathepsin D antibody [EPR3057Y] | Abcam | Cat# ab75852, RRID: AB_1523267 |
| Mouse Anti-beta-Actin Antibody (AC-74) | Sigma-Aldrich | Cat# A5316, RRID: AB_476743 |
| IRDye® 680RD Goat anti-mouse IgG (H+L) secondary antibody | Li-cor | Cat# 926-68070, RRID: AB_10956588 |
| IRDye® 800CW Goat anti-Rabbit IgG (H+L) secondary antibody | Li-cor | Cat# 926-32211, RRID: AB_621843 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N-methyl-D-glucamine (NMDG) | Sigma-Aldrich | M2004; CAS: 6284-40-8 |
| Methanesulfonic acid (MSA) | Sigma-Aldrich | 471356; CAS: 75-75-2 |
| HCl | Fisher Scientific | A144C-212; CAS: 7647-01-0 |
| Tetramethylammonium (TMA) | Sigma-Aldrich | T-7505; CAS: 10424-65-4 |
| K-Gluconate | Sigma-Aldrich | P1847; CAS: 299-27-4 |
| KOH | Fisher Scientific | P250-500; CAS: 1310-58-3 |
| NaOH | Fisher Scientific | S318-500; CAS: 1310-73-2 |
| KCl | Sigma-Aldrich | P9541; CAS: 7447-40-7 |
| NaCl | Sigma-Aldrich | S9888; CAS: 7647-14-5 |
| CaCl2 | Sigma-Aldrich | C8106; CAS: 10035-04-8 |
| MgCl2 | Sigma-Aldrich | 208337; CAS: 7786-30-3 |
| NH4Cl | Sigma-Aldrich | A4514; CAS: 12125-02-9 |
| NaH2PO4 | Sigma-Aldrich | S3139; CAS: 7558-80-7 |
| Sucrose | Sigma-Aldrich | S0389; CAS: 57-50-1 |
| Tetraethylammonium chloride (TEA-Cl) | Sigma-Aldrich | T2265; CAS: 56-34-8 |
| Cs-Methanesulfonate (Cs-MSA) | Sigma-Aldrich | C1426; CAS: 2550-61-0 |
| HEPES | Sigma-Aldrich | H3375; CAS: 7365-45-9 |
| MES | Sigma-Aldrich | M8250; CAS: 1266615-59-1 |
| Citric acid | Sigma-Aldrich | 27109; CAS: 77-92-9 |
| Acetic acid | Fisher Scientific | A38-212; CAS: 64-19-7 |
| Ethanol | Decon laboratories inc. | 2701; CAS: 64-17-5 |
| Methanol | Fisher Scientific | A52-4; CAS: 67-56-1 |
| Isopropanol | Fisher Scientific | A416-4; CAS: 67-63-0 |
| Glucose | Sigma-Aldrich | G6152; CAS: 50-99-7 |
| Triton-X 100 | Sigma-Aldrich | T8787; CAS: 9002-93-1 |
| NaF | Sigma-Aldrich | S7920; CAS: 7681-49-4 |
| Na-orthovanadate | Sigma-Aldrich | S6508; CAS: 13721-39-6 |
| EDTA | Sigma-Aldrich | 03609; CAS: 60-00-4 |
| EGTA | Sigma-Aldrich | E4378; CAS: 67-42-5 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D2650; CAS: 67-68-5 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A3059; CAS: 9048-46-8 |
| Paraformaldehyde (PFA) 4% in PBS | Thermo Fisher Scientific | J19943-K2 |
| Phosphate-buffered saline (PBS) | Gibco | 10010 |
| RIPA | Boston Bioproducts | BP-115 |
| Protease inhibitor cocktail | Sigma Aldrich | P8340 |
| Phosphatase inhibitor cocktail I | Sigma Aldrich | P0044 |
| LDS sample buffer (4x) | Invitrogen | NP0007 |
| Pierce™ BCA™ Protein Assay | Invitrogen | 23227 |
| PVDF membrane | Millipore | IPFL00010 |
| Fluoromount-G | Southern Biotech | 0100-01 |
| Magic Red Cathepsin B assay kit | ImmunoChemistry Technologies | 937 |
| Pepstatin A BODIPY™ FL | Invitrogen | P12271 |
| DQ™-BSA red dye | Invitrogen | D12051 |
| CF®488a conjugated dextran-10 kd | Biotium | 80110 |
| CF®555 conjugated dextran-10 kd | Biotium | 80112 |
| LysoSensor™ Yellow/Blue DND-160 | Invitrogen | L7545 |
| LysoTracker™ Red DND-99 | Invitrogen | L7528 |
| pHrodo™ Green conjugated dextran-10 kd | Invitrogen | P35368 |
| Oregon Green™ 488 conjugated dextran-10 kd | Invitrogen | D7170 |
| pHrodo™ Red AM | Invitrogen | P35372 |
| MitoTracker™ Green FM | Invitrogen | M7514 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma Aldrich | D9542; CAS: 28718-90-3 |
| Lipofectamine 2000 | Invitrogen | 11668-027 |
| DMEM medium without phenol red | Gibco | 21063029 |
| DMEM medium high glucose | Gibco | 11965 |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | Gibco | 12440 |
| Neurobasal™ Medium | Gibco | 21103049 |
| Fetal bovine serum (FBS) | Gemini Bio-Products | 100-106 |
| B-27™ Supplement (50X), serum free | Gibco | 17504044 |
| Trypsin-EDTA (0.05%), phenol red | Gibco | 25300054 |
| Puromycin Dihydrochloride | Gibco | A1113803 |
| Antibiotic-Antimycotic | Gibco | 15240062 |
| MycoAlertTM Mycoplasma Detection Kit | Lonza | LT07-218 |
| Poly-D-lysine solution | Millipore | A-003-E |
| Poly-L-lysine hydrobromide | Sigma Aldrich | P1399; CAS: 25988-63-0 |
| Vacuolin-1 | Sigma Aldrich | 673000; CAS: 351986-85-1 |
| 4-[(2-Butyl-6,7-dichloro-2-cyclopentyl-2,3-dihydro-1-oxo-1H-inden-5-yl)oxy]butanoic acid (DCPIB) | Tocris Bioscience | 1540; CAS: 82749-70-0 |
| 2,7-Dichloro-9,10-dihydro-9,9-dimethyl-10-[2-(2H-tetrazol-5-yl)ethyl]acridine (ML 67-33) | Tocris Bioscience | 6886; CAS: 1443290-89-8 |
| Bafilomycin-A1 | Cayman Chemical | 11038; CAS: 88899-55-2 |
| Arachidonic acid-sodium | Cayman Chemical | 10006607; CAS: 6610-25-9 |
| Arachidonic acid | Cayman Chemical | 90010; CAS: 506-32-1 |
| SC-79 | Cayman Chemical | 14972; CAS: 305834-79-1 |
| MK-2206 | Cayman Chemical | 11593; CAS: 1032350-13-2 |
| ARQ-092 | Cayman Chemical | 21388; CAS: 1313881-70-7 |
| Wortmannin | Cayman Chemical | 10010591; CAS: 19545-26-7 |
| Latrunculin B from Latruncula magnifica | Sigma Aldrich | L5288; CAS: 76343-94-7 |
| PI(3,5)P2 diC8 | Echelon Biosciences | P-3508 |
| ML-SA5 | This paper | N. A. |
| isoflurane (Fluriso™) | VetOne | 501017 |
| Tissue-Tek® O.C.T. (Optimal Cutting Temperature) compound | Sakura Finetek | 4583 |
| Recombinant mouse WT α-synuclein preformed fibril (5 mg/mL) | a gift from Dr. Kelvin C. Luk, the University of Pennsylvania Perelman School of Medicine | N. A. |
| Critical Commercial Assays | ||
| QuikChange II Site-Directed Mutagenesis Kit | Agilent | 200523 |
| Gibson Assembly Cloning Kit | New England Biolabs | E5510S |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | M0491S |
| GoTaq™ master mixes | Promega | M7122 |
| Experimental Models: Cell Lines | ||
| Monkey: COS1 | ATCC | CRL-1650, RRID: CVCL_0223 |
| Human: HEK293T cells | ATCC | CRL-3216; RRID: CVCL_0063 |
| Human: HeLa cells | ATCC | CCL-2; RRID: CVCL_0030 |
| Human: HAP1 cells | Horizon Discovery Ltd. | C631; RRID: CVCL_Y019 |
| Human: TMEM175 KO HAP1 cells | This paper | N. A. |
| Human: TMEM175-HA Knock-in HAP1 cells | This paper | N. A. |
| Human: TPC1&TPC2 DKO HAP1 cells | This paper | N. A. |
| Human: LRRC8A KO HAP1 cells | (Li et al., 2020b) | N. A. |
| Human: TMEM175 KO HEK293T cells | This paper | N. A. |
| Human: HEK293T cells stably overexpressing human TMEM175 (L1&L2) | This paper | N. A. |
| Human: TMEM175 KO HeLa cells (KO-1&KO-2) | This paper | N. A. |
| Human: TMEM175 KO HeLa cells stably re-expressing human TMEM175 | This paper | N. A. |
| Human: TMEM175 KO HeLa cells stably re-expressing human TMEM175-D41A | This paper | N. A. |
| Human: TRPML1-KO fibroblasts | Coriell Institute for Medical Research | GM02048; RRID: CVCL_M927 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tmem175 −/− / C57BL/6N | This paper | N. A. |
| Oligonucleotides | ||
| sgRNA targeting sequence: hTMEM175 (#1): 5’- CGCGTGGAAGCCTTCAGCGA -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: hTMEM175 (#2): 5’- TGTCCTCCCTGGTCCATCTC -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: hTMEM175 (#3): 5’- CGGCCGCCAAACCACACCGA -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: mTmem175 (#1): 5’- GCACTAATACTCTTAGCCTG -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: mTmem175 (#2): 5’- GGACCCCTTACAATATGCCT -3’ | http://crispr.mit.edu | N. A. |
| Genotyping primer: 5’- CATTCCTGTTGCTCCGTCTCCTTGT -3’ and 5’- CGCCAGGGAAGCTCTGTACTTTTGA -3’ | This paper | N. A. |
| for Tmem175−/− mice | ||
| Genotyping primer: 5’- TGCTATGCTGAGCTTATAGCCTTG -3’ and 5’- AACGTTCCAAGAAGGAGGATCGCC -3’ for Tmem175−/− mice | This paper | N. A. |
| Recombinant DNA | ||
| pEZ-M55-mCherry-hTMEM175 | GeneCopoeia | EX-V0772-M55 |
| pEZ-M29-EGFP-hTMEM175 | GeneCopoeia | EX-V0772-M29 |
| pEZ-M97-hOTOP1-EGFP | GeneCopoeia | EX-E1341-M97 |
| pSpCas9 (BB)-2A-puro vector | Addgene | 48139 |
| pHluorin (in modified pEYFP-N1) | This paper | N. A. |
| Software and Algorithms | ||
| MetaMorph 7.10 | Molecular Devices | https://www.moleculardevices.com/ |
| pClamp 10.7 | Molecular Devices | https://www.moleculardevices.com/ |
| OriginPro 2018 | Originlab | https://www.originlab.com/ |
| GraphPad Prism 8 | Graphpad | https://www.graphpad.com/ |
| Fiji/ImageJ | Fiji contributors | https://imagej.net/Fiji |
| Image Studio lite | Li-cor | https://www.licor.com/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mammalian cell lines
COS1, HEK293T, and HeLa cells were purchased from American Type Culture Collection (ATCC) and cultured in a high glucose DMEM (Gibco) medium with 10% fetal bovine serum (FBS, 100-106, Gemini Bio-Products). HAP1 cells (Horizon Discovery Ltd.) were maintained in IMDM (Gibco) with 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen). All cells were used at low passages with occasional authentication for mycoplasma contamination using MycoAlert™ Mycoplasma Detection Kit (LT07-218, Lonza).
Generation of TMEM175 knockout and HA-tagged knock-in cells
TMEM175 knockout (KO) HAP1, HeLa, HEK293T, TMEM175-HA knock-in (KI) HAP1 cell lines, and M393T knock-in HEK293T cells were created using a Crispr-Cas9 system (Ran et al., 2013). The guide RNA (gRNA) targeting exon 10 (5’-CGCGTGGAAGCCTTCAGCGA-3’) was designed to generate KO cell lines that resulted in a frameshift near the IIS1 helix (see Supplemental Fig. S2A). For HA-tagged KI cells, a 2.2 kb human genomic DNA region spanning exon 11 with ~1.4 kb intron sequence was PCR amplified and inserted into a backbone vector. HA tag (YPYDVPDYA) linked by five (5) glycine was then inserted at the end of the Open Reading Frame (ORF) using site-directed mutagenesis to generate the template plasmid. The gRNA targeting the intron downstream of exon 11 (5’-TGTCCTCCCTGGTCCATCTC-3’) was designed to trigger homologous recombination (see Supplemental Fig. S2E). The gRNA was cloned into pSpCas9 (BB)-2A-puro vector (Addgene) and transfected into target cells alone (for KO) or together with the template plasmid (for KI) using Lipofectamine 2000 (Invitrogen). After puromycin (Gibco) selection for 48 hours, the remaining cells were trypsin-digested, and plated into 96-well plates in order to select single monoclonal cell populations. The frameshift in KO cells and the in-place insertion of HA-tag in KI cells were confirmed with PCR and DNA sequencing. M393T knock-in HEK293T cells were generated using the sgRNA (5’-CGGCCGCCAAACCACACCGA-3’) and site-directed mutagenesis.
Targeted deletion of Tmem175 in mice
Tmem175 knockout (Tmem175−/−) mice were generated using Crispr-Cas9 technology (Ran et al., 2013) with two single guide RNAs: sgRNA1: 5’-GCACTAATACTCTTAGCCTG-3’; sgRNA2: 5’-GGACCCCTTACAATATGCCT-3’, which were designed to create a ~8.2 kb chromosomal deletion (exons 2-11) at the Tmem175 loci in the mouse genome. After the transcription of sgRNA in vitro, zygote microinjection was performed on the C57BL/6N mouse strain. Heterozygous intercrossing generated Tmem175−/− mice with the expected Mendelian ratio. The genotype of newborn mice was confirmed by PCR electrophoresis with primers: WT-1: 5’-CATTCCTGTTGCTCCGTCTCCTTGT-3’; WT-2: 5’-CGCCAGGGAAGCTCTGTACTTTTGA-3’; KO-1: 5’-TGCTATGCTGAGCTTATAGCCTTG-3’; KO-2: 5’-AACGTTCCAAGAAGGAGGATCGCC-3’. All animals were used according to approved animal protocols and institutional animal care guidelines at the University of Michigan and were maintained in a standard 12-hour light and dark cycle with ad libitum access to food and water. For the neuronal culture experiments, Tmem175+/+ and Tmem175−/− mice in the C57BL/6N background (both sex) at postnatal day 1 were used. For the α-Syn aggregation experiments, Tmem175+/+ and Tmem175−/− mice in the C57BL/6N background (male only) between 3 to 4 months of age were used.
METHOD DETAILS
Generation of TMEM175 site-directed mutants
Site-directed mutagenesis was performed on the template of EGFP-TMEM175 (pEZ-M29, GeneCopoeia) or mCherry-TMEM175 (pEZ-M55, GeneCopoeia) using either a QuikChange II Site-Directed Mutagenesis Kit (200523, Agilent) or a Gibson Assembly Cloning Kit (E5510S, New England Biolabs). All constructs were confirmed by DNA sequencing.
Lysosome membrane protein (LMP) library
All cDNA plasmid constructs in the LMP library are from the human genes and the proteins were tagged with either EGFP or mCherry. Among them, C1orf85, CLCN4, SCARB2, and SLC36A1 were purchased from Origene; CLN8, LMBRD1, MFSD12, MFSD6, OCA2, SLC15A3, SLC15A4, SLC17A5, SLC17A9, SLC35F6, SLC38A7, SLC38A9, SLC46A3, SLC48A1, SLC7A14, TMEM123, TMEM175, TMEM179b, TMEM184c, TMEM243, TMEM55b, and TTYH2 were purchased from GeneCopoeia; the NHE6 (pEGFP-N3) plasmid was a gift from Dr. Rajini Rao (Johns Hopkins University School of Medicine).
Whole-cell patch-clamp electrophysiology
Ionic currents were recorded in the whole-cell configuration under voltage-clamp using an Axonpatch 200B amplifier equipped with a Digidata 1440A digitizer (Molecular Devices). Unless otherwise stated, both pipette (cytosolic) and bath (extracellular) solutions contained NMDG-MSA as the major ions. The pipette (cytosolic) solution usually contained (in mM): 150 NMDG and 20 HEPES (pH 7.2 adjusted with MSA). The pipette solution was supplemented with essential Cl− (2 mM) needed to stably conduct electricity through Ag/AgCl electrode. To vary the pH of the bath (extracellular) solution from 7.2-3.5, an appropriate pH buffer (20 mM HEPES, 20 mM MES, or 10 mM citrate acid) with its pKa near the target pH was used, and the solution was supplemented with sufficient NMDG (120-150 mM) to bring the final osmolarity to ~ 300 mOsm (pH adjusted with MSA). All bath solutions were applied via a perfusion system that allowed us to achieve complete solution exchange within a few seconds. The Cl− free solution used extracellularly was essential to prevent activation of H+-activated Cl− currents on the plasma membrane (Yang et al., 2019). An agar bridge filled with 3M KCl was used for the reference electrode. Glass electrodes were pulled with a resistance of 2-4 MΩ. A ramp protocol (−120 mV to +120 mV, 400 ms, holding at 0 mV) was used to record the currents at 2 second intervals. In some experiments, the holding potential was set to Erev in order to minimize proton flux between recordings. Data were collected using a Clampex 10.7 software (Molecular Devices). All experiments were performed at room temperature (22-25°C), and all data were analyzed with Clampfit 10.7 (Molecular Devices), Origin 2018 (OriginLab) or Prism 8.4 (Graphpad).
Whole-LEL patch-clamp electrophysiology
Endolysosomal electrophysiology was performed in isolated enlarged endolysosomes using a modified patch-clamp method (Dong et al., 2008). Briefly, cells were treated with 1 μM vacuolin-1 (673000, Millipore Sigma), a lipid-soluble polycyclic triazine that can selectively increase the size of late endosomes and lysosomes (LELs) (Huynh and Andrews, 2005), for at least 1h and up to 12h. Whole-LEL recordings were performed on enlarged endolysosomes manually isolated from various cells. In brief, a glass pipette was pressed against a cell and quickly pulled away to break the cell membrane. Enlarged LELs were released out of the hole in the plasma membrane and visualized under an inverted microscope (IX71, Olympus). If applicable, the fluorescence emission was confirmed on the membrane of isolated endolysosomes expressing fluorescent proteins. After formation of a giga-seal between the glass pipette and the isolated endolysosome, capacitance transients were compensated. Voltage steps of several hundred millivolt (mV) with a 1-10 millisecond (ms) duration were then applied to break the vacuolar membrane. The whole-LEL configuration was verified by the re-appearance of capacitance transients after break-in.
Unless otherwise stated, both bath (cytosolic) and pipette (luminal) solutions contained NMDG-MSA as the major ions. The bath (cytosolic) solution contained (in mM): 150 NMDG and 20 HEPES (pH 7.2 adjusted with MSA); the pipette (luminal) solution at pH 4.6 contained (in mM): 150 NMDG and 20 MES (pH adjusted with MSA). The pipette (luminal) solution at pH 3.5 contained (in mM): 110 NMDG and 25 citrate acid (pH adjusted with MSA). The pipette solution was supplemented with essential Cl− (2 mM) needed to stably conduct electricity through the Ag/AgCl reference electrode. An agar bridge filled with 3M KCl was used for the reference electrode. Lysosomal transmembrane potential (Vm) is defined as VCytosol - VLumen (Li et al., 2019) for consistency with whole-cell recordings (Vm = Vcytosol - VExtracellular). A ramp protocol (−120 mV to +120 mV, 400 ms, holding at 0 mV) was used to record the currents at 2 seconds intervals. In some experiments, the holding potential was set to Erev in order to minimize proton flux between recordings. Data were collected using an Axonpatch 200B amplifier, Digidata 1440A Digitizer, and Clampex 10.7 software (Molecular Devices). All experiments were performed at room temperature (22-25°C), and all data were analyzed with Clampfit 10.7 (Molecular Devices), Origin 2018 (OriginLab) or Prism 8.4 (Graphpad).
Endolysosomal excised patch-clamp electrophysiology
Endolysosomal lumen-side-out recordings were performed on giant excised patches as previously described (Wang et al., 2017a). In brief, after formation of a giga-seal between the glass pipette and isolated endolysosome, the glass pipette was pulled away to obtain a giant excised patch, in which the luminal side faced outside. Both pipette (cytosolic) and bath (luminal) solutions were the same as those used in the whole-cell recordings. Glass electrodes were pulled with the resistance 1-2 MΩ.
Determination of relative permeability (PH/PX)
The permeability of TMEM175 for H+ relative to other monovalent cations (K+, Na+, or NMDG+) was estimated based on the Goldman-Hodgkin-Katz current equations (Lewis, 1979) and the measured Erev using “bi-ionic” conditions:
where R, T, F and V are the gas constant, absolute temperature, Faraday’s constant and the reversal potential, respectively. Note that because [H+] was several orders of magnitude less than [Na+] or [K+], the term “bi-ionic” is loosely adopted. Assumed ion activity coefficients are 1 for monovalent ions. The liquid junction potentials were measured and corrected as described (Neher, 1992). The pipette (cytosolic) solution, buffered with 20 mM HEPES (adjusted to pH 7.2), contained either 150 mM NMDG-MSA, Na-MSA, or 140 mM K-MSA. The pipette solution containing NMDG+ or Na+ was supplemented with Cl− (2 mM) in order to stably conduct electricity through the Ag/AgCl electrode. In the whole-cell recording, the pipette solution containing K-MSA also included 10 mM tetraethylammonium (TEA+) chloride that was used to block the endogenous basal outward K+ currents in HEK293T cells. An agar bridge filled with 3 M KCl was used for the reference electrode. Erev was determined using a ramp protocol (−120 mV to +120 mV, 400 ms, holding at 0 mV) in response to bath (extracellular) solutions at varied pH (see whole-cell patch clamp electrophysiology section). Note that in the whole-cell recordings, after prolonged exposures to acidic pHE/L, the pipette/cytosolic solution can become acidified due to repeated ramp currents and sustained inward currents at the holding potential that may saturate the pH buffers. Subsequently, the outward H+ currents will grow, and the H+ reversal potential will gradually shift leftward. When such experimentally-induced H+ accumulation in the cytosolic side occurred, the current traces were excluded from the Erev analysis.
Cytosolic pH imaging
Cytosolic pH dynamics were determined by time-lapse imaging using pHluorin, a genetically-encoded pH indicator (Miesenbock et al., 1998), or the chemical pH sensor pHrodo™ Red. When binding H+ intracellularly, the brightly fluorescent pHluorin is quenched to a lower level, while the dim fluorescence of pHrodo™ Red gets brighter.
The pHluorin plasmid was sub-cloned from NPY-pHluorin (a gift from Dr. Zhuan Zhou, Peking University, China) (Wang et al., 2017b). Cells seeded on glass coverslips were transfected with pHluorin together with mCherry-TMEM175 or mCherry sham vector (both in pEZ-M55) using Lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours after transfection, cells were washed with the imaging solution at pH 7.2 and imaged with a spinning-disk confocal microscope (IX81, Olympus; CSU-X1, Yokogawa) equipped with a 20 × objective lens and a temperature controller that kept the cells at 37 °C. The imaging solution at 7.2 or 4.6 usually contained 100 mM pH buffer (HEPES or MES) supplemented with the either N-methyl-D-glucamine methanesulfonate (NMDG-MSA), tetramethylammonium methanesulfonate (TMA-MSA), potassium methanesulfonate (K-MSA), or sodium methanesulfonate (Na-MSA). The final osmolarity was ~ 300 mOsm for all imaging solutions. In a subset of experiments, Tyrode’s solution at pH 7.4 (145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose) and a modified Tyrode’s-like solution at pH 4.6 (replacing HEPES with MES) were used as imaging solutions. Prior to imaging, a single image was acquired using excitation at 561 nm to identify the cells expressing mCherry-TMEM175 or mCherry. The fluorescence emissions excited by at 488 nm were then acquired every 5 seconds with an EMCCD camera (Andor, iXon ultra 897U) controlled by MetaMorph software. Un-dissociated acetic acid (HOAc; 150 mMNaCl, 10 mM HEPES, and 10 mM HOAc, pH 5.0 adjusted by NaOH) freely entering the cytosol, where it dissociates into a H+ and an acetic ion, served as a positive control to acidify the cytosol (Ylitervo et al., 2014). Free NH3 (NH4Cl; 110mM NaCl, 50 mM NH4Cl, 20 mM HEPES, pH 7.2 adjusted by NaOH) may enter cytosol, where it associates with H+ to generate NH4+, served as an alkalizing agent to confirm the availability of pHluorin (Decoursey, 2003).
The chemical-based intracellular pH indicator, pHrodo™ Red AM (P35372, Invitrogen), was also used to determine intracellular pH. Cells were transfected with EGFP-TMEM175 (pEZ-M29, GeneCopoeia) or EGFP sham vector using Lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours after transfection, cells were loaded with the pHrodo™ Red AM according to the manufacturer’s instructions and washed with the imaging solution at pH 7.2. In these experiments, a single image was acquired using excitation at 488 nm to identify the cells expressing EGFP-TMEM175. The fluorescence emissions excited by at 561 nm were then acquired every 5 seconds. The fluorescence intensity in cells were quantified using the Fiji/ImageJ software.
Lysosomal pH determination
Lysosomal pH measurement with a ratiometric pH-sensitive dye Oregon Green™ 488 conjugated dextran-10 kd (D7170, Molecular Probes) was performed as described previously (Christensen et al., 2002; Davis et al., 2012). In brief, cells seeded on glass coverslips were loaded with 150 μg/mL Oregon Green™ 488 Dextran overnight and chased in medium without dye for three hours before imaging. Cells were washed with Ringer’s buffer (in mM): 155 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 2 NaH2PO4, 10 HEPES, and 10 glucose, pH 7.4) and imaged using a Nikon TE300 inverted microscope equipped with a 60× objective lens (N.A. = 1.4) and a temperature-controlled stage. The fluorescence emission (530 nm) excited at 440 nm and 490 nm wavelengths were acquired with a cooled digital CCD camera (Quantix Photometries) controlled by MetaMorph software. pH calibration was performed to generate the pH standard curve. pH standard buffers (in mM: 130 KCl, 1 MgCl2, 15 HEPES, and 15 MES) were prepared at varied pH ranging from 3.0 to 8.0 supplemented with 10 μM valinomycin and 10 μM nigericin. The ratio of fluorescence intensities excited at 440 nm and 490 nm in response to varied pH was fitted to sigmoidal equation to generate the pH standard curve. The lysosomal pH values were then calculated with the resulting intensity ratio (440 nm / 490 nm) images based on the standard curve.
Determination of luminal acidity in lysosomes.
Lysosomal luminal acidity was estimated with the fluorescence ratio between a pH-sensitive dye pHrodo™ Green conjugated dextran-10 kd (P35368, Invitrogen) and a pH-insensitive dye CF555 conjugated dextran-10 kd (80112, Biotium). In brief, cells seeded on glass coverslips were loaded with 20 μg/mL of each dextran overnight and chased in medium without dye for 3 hours before imaging. The HEPES buffered DMEM medium without phenol red (21063029, Gibco) was used as the imaging solution to eliminate the short time starvation side effects during imaging process. Cells were washed with the imaging solution and imaged using an inverted microscope (IX81 Olympus) equipped with a 60× objective lens and a temperature-controlled stage. The fluorescence emission excited at 488 nm and 561 nm wavelengths were acquired with an EM-CCD camera (Andor, iXon ultra 897U) controlled by MetaMorph software. The fluorescence intensity of pHrodo™ Green and CF555 were quantified by Fiji/ImageJ.
LysoTracker and LysoSensor staining
For a rough determination of lysosomal acidity, cells were cultured in a 24-well dish one day before the experiment. To visualize the acidic organelles, 50 nM LysoTracker Red DND-99 (L7528, Invitrogen) was added into the cell culture medium and incubated at 37°C for 30 min. Cells were then washed twice with Tyrode’s solution and kept in Tyrode’s for imaging. Images were acquired using an inverted fluorescence microscope (IX81, Olympus), and the fluorescence intensity of LysoTracker was quantified by Fiji/ImageJ software.
For LysoSensor staining, LysoSensor Yellow/Blue DND-160 (L7545, Invitrogen) was added into the cell culture medium and incubated at 37°C for 30 min. The fluorescence emission (530 nm) excited at 340 nm and 380 nm wavelengths were acquired with a cooled digital CCD camera. The ratio of fluorescence intensities excited at 340 nm and 380 nm in response to varied pH was fitted to sigmoidal equation to generate the pH standard curve. The lysosomal pH values were calculated with the resulting intensity ratio (340 nm / 380 nm) images based on the standard curve.
Lysosomal hydrolytic activity determination
Cathepsin B and Cathepsin D activities in the lysosomes were determined using Magic Red Cathepsin B assay kit (937, ImmunoChemistry Technologies) and Pepstatin A BODIPY™ FL (P12271, Invitrogen), respectively. Overall lysosomal hydrolytic activity was determined with DQ™-BSA red dye (D12051, Invitrogen). Magic Red, Pepstatin A BODIPY™ FL, and DQ™-BSA red stock solutions were prepared according to the manufacturer’s instructions. Cells were incubated with Magic Red (1:1,000 dilution) or Pepstatin A, BODIPY FL (2 μM at 37°C for 1 hour and washed twice with Tyrode’s solution before imaging. For overall lysosomal hydrolytic activity detection, cells were loaded with DQ-BSA red dye (10 μg/mL) for 16 hours. Images were taken using an inverted fluorescence microscope (IX81, Olympus) and the fluorescence intensities of Magic Red/BODIPY FL/DQ-BSA were quantified with Fiji (ImageJ).
Immunocytochemistry
Cells seeded on glass coverslips were washed with Phosphate-buffered saline (PBS) and fixed with 4% Paraformaldehyde (PFA) or cold anhydrous methanol. Afterwards, 0.3 % Triton-X 100 in block solution (PBS supplemented with 2% bovine serum albumin) was used to permeate the phospholipid membrane (only for PFA fixation). Cells were incubated with a primary antibody overnight at 4 °C and with a secondary antibody for one hour at room temperature. Primary antibodies (anti-human LAMP1 (H4A3), DSHB, 1:200; anti-HA (3F10), 11867423001, Roche, 1:200) and second antibodies (anti-mouse conjugated to Texas Red, T862, Life Technologies, 1:500; anti-rat conjugated to Alexa Fluor 488, A11006, Life Technologies, 1:500) were diluted into the block solution proportionally as indicated. Coverslips were mounted on glass slides with Fluoromount-G (0100-01, Southern Biotech). Images were acquired with an inverted confocal microscope (IX81, Olympus) equipped with an oil 60 × objective lens.
Immunoblotting
To analyze the level of specific proteins in cells, cells grown to 70-80% confluence in a 35 mm dish were lysed with 100 μl of ice-cold lysis buffer (RIPA, cat. No. BP-115, Boston Bioproducts) supplemented with protease inhibitor cocktail (P8340, Sigma Aldrich). For detection of phosphorylated proteins, the lysis buffer was additionally supplemented with 20 mM NaF, 1 mM Na-orthovanadate, 2 mM EDTA, and Phosphatase inhibitor cocktail I (P0044, Sigma). The lysates were then centrifuged at 15,000 g for 15 min at 4 °C. The supernatant was collected, mixed with 4X LDS sample buffer (NP0007, Invitrogen), heated at 75 °C for 15 min, and subjected to immunoblotting. Pierce™ BCA™ Protein Assay (23227, Invitrogen) was used to quantify the protein concentration. Primary antibodies [anti-GFP (clone 7.1 and 13.1), 11814460001, Roche, 1:1000; anti-p-Akt(Thr308), 2965, CST, 1:1000; anti-Akt(pan) (C67E7), 4691, CST, 1:1000; Anti-Cathepsin B (EPR4323), ab125067, Abcam, 1:1000; Anti-Cathepsin D (EPR3057Y), ab75852, Abcam, 1:1000; anti-β-Actin (AC-74), A5316, Sigma, 1:1000] were diluted into the block solution (PBS supplemented with 0.1% Tween-20 and 2% BSA) proportionally as indicated. Odyssey CLx imaging system (Li-cor) was used to scan PVDF membrane (IPFL00010, Millipore) after incubation with a secondary antibody [IRDye® 680RD Goat anti-mouse IgG (H+L), 926-68070, Li-cor, 1:5000; IRDye® 800CW Goat anti-Rabbit IgG (H+L), 926-32211, Li-cor, 1:5000] at room temperature for 1 hour. Band intensities were quantified using Image Studio software (Li-cor).
Isolation and culture of primary neurons
For the isolation of mouse hippocampal neurons, hippocampi were dissected from mice at postnatal day 1 and treated with 0.25% trypsin at 37°C for 30 min. Neurons were plated on Poly-D-lysine (A-003-E, Millipore) coated glass coverslips in 12-well plates and settled down in DMEM (Gibco) supplemented with 10% FBS for 3 hours. Afterwards the culture medium was replaced with Neurobasal (Gibco) supplemented with 0.5 mM Glutamine and 2% B-27 (Gibco) for longtime culture. Cultured neurons were fed every 2 days by replacing half of the old medium with fresh Neurobasal medium. Cultured neurons at 7 - 14 DIV were used for all the imaging and electrophysiological experiments.
Intra-striatal inoculation of preformed α-synuclein fibrils (pff)
Recombinant mouse WT α-Syn fibril stock (α-Syn pff, 5 mg/mL) (a gift from Dr. Kelvin C. Luk, the University of Pennsylvania Perelman School of Medicine) was diluted with sterile Dulbecco’s phosphate-buffered saline (D-PBS, without Ca2+ and Mg2+) to 2 mg/mL in a tube (520216, Covaris) for sonication for 30 min (30 seconds ON, 30 seconds OFF, repeating for 30 cycles at a constant temperature of 10 °C) with a ultra-sonicator (M220, Covaris).
α-Syn pff was injected into mouse brain following an established procedure as described (Zhang et al., 2019a). Mice between 3 to 4 months of age were anesthetized with isoflurane (Fluriso™, 501017, VetOne). Sonicated pff (2.5 μL) or vehicle control (D-PBS) was stereotaxically injected in the dorsal striatum (+0.2 mm from Bregma, 2 mm from midline, −2.6 mm beneath the brain surface) of one hemisphere. Injections were performed using a 10 μL syringe (Hamilton, NV) at a rate of 0.1 μL/min with the needle in place for > 5 min at each injection site. Animals were monitored regularly following recovery from surgery. Thirty days after injection, animals were sacrificed by overdose of isoflurane. After perfusion with phosphate-buffered saline (PBS) and 70% ethanol (in 150 mM NaCl, pH 7.4), the brain was taken out and put in 70% ethanol (in 150 mM NaCl, pH 7.4) overnight at 4 °C for post fixation. The fixed brain was then transferred to a PBS solution with 30% sucrose at 4 °C until the tissue sank to the bottom, before being processed and embedded in O.C.T. (Optimal Cutting Temperature) compound. Coronal brain sections of 30 μm thickness were collected onto glass slides by cryo-sectioning before being utilized for immunohistochemical experiments.
For immunohistochemical study of α-Syn aggregation, the brain slices were blocked in 8% goat serum supplemented with 0.3% Triton-X 100 for one hour at room temperature before being incubated at 4 °C overnight with primary antibodies [α-Syn phospho S129 (EP1536Y), ab51253, Abcam, 1:3,000], The brain slices were then washed with PBS and incubated with secondary antibodies and 4’,6-diamidino-2-phenylindole (DAPI). The puncta number and area of α-Syn aggregations in brain slices were quantified by the Fiji/ImageJ software. The images were evaluated by the Analyze Particle function of Fiji/ImageJ to get a readout of puncta counts and area. Puncta sizes between 6.4-256 μm2 were included.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM (standard error). When box plots were used, the box represents the 25-75% range, with the solid line within them indicating the mean level and the dashed line indicating the median level. The whiskers represent the standard deviation and all points outside the standard deviation are shown as individual dots.
For experiments with only two groups, Student’s t-test was used to check the significance, while for the experiments with more than two groups and multi-testing was required, one-way ANOVA with Sidak’s test was adopted to reduce the type 1 error and obtain adjusted p value. Significance is indicated as NS (not significant), * p <0.05, ** p <0.01, and ***p < 0.001.
For experiments with large sample size, effect size between groups was calculated (Cohen’s d). The effects are defined as no effect (d < 0.25), small effect (0.25 < d < 0.5), medium effect (0.5 < d < 0.75), and large effect (d > 0.75).
Supplementary Material
Supplemental Figure S1. TMEM175 mediates small inward H+ “leak” currents in endolysosomes at luminal pH 4.6. Related to Figure 1.
(A) Whole-LEL H+ “leak” currents from a COS1 cell with luminal pH (pHL) set to 4.6 and cytosolic pH (pHC) set to either 7.2 or 4.6 as indicated. (B) Whole-LEL current amplitude distribution with pHL set to 4.6 and pHC to 7.2 from experiments as in (A) (n = 128 LELs, bin size = 5 pA). (C) Whole-LEL H+ “leak” currents are undetectable (grey trace) in a COS1 cell with both pHL and pHC set to pH 7.2. The outward TPC-mediated Na+ currents (Wang et al., 2012) evoked by PI(3,5)P2 (red trace) served as a control that the recording was in the whole-LEL mode. (D) Whole-LEL H+ “leak” currents from a COS1 cell with pHL set to 4.6 and pHC set to 7.2. Both luminal and cytosolic solutions contained Na-MSA as major ions. (E) Average whole-LEL H+ “leak” currents from COS1, HAP1, HEK293T, HeLa, mouse embryonic fibroblast (MEF) cells, and mouse primary hippocampal neurons (mHippo) with pHL set to 3.5 and pHC to 7.2 (n = 4-11 LELs). (F) Whole-EE (early endosome) H+ “leak” currents in a chemically (Wortmannin + Latrunculin B)-enlarged early endosome (Chen et al., 2017a) from a COS1 cell. (G-H) Whole-LEL H+ “leak” currents were tiny in COS1 cells overexpressing TRPML1 or TPC1 with pHL set to 4.6. Luminal and cytosolic (Cyto) solutions contained indicated major ions required for the ML-SA5-activated TRPML1 or PI(3,5)P2-activated TPC1 positive-control currents.
Supplemental Figure S2. Lysosome-localized TMEM175 is required for inward H+ “leak” currents in endolysosomes. Related to Figure 1.
(A) Crispr-Cas9 strategy used to create TMEM175 knock-out (KO) human cell lines. The top of the panel shows the amino acid sequence for the IIS1 region of TMEM175 and the genomic sequence with the guide RNA (gRNA) sequence and protospacer adjacent motif (PAM) motif highlighted in red and blue, respectively. The bottom of the panel shows the Sanger sequencing confirming that frameshift was produced in HAP1 (haploid), HeLa, and HEK293T cells. (B-C) Whole-LEL H+ “leak” currents from a Mucolipidosis IV patient-derived (TRPML1 KO) fibroblast or a TPC1&TPC2 double knockout (TPC1&2 DKO) HAP1 cell with pHL set to 3.5 and recording solutions as indicated. (D) The subcellular localization of overexpressed TMEM175 in HEK293T cells was determined by labeling markers for three different organelles: LAMP1-mCherry (LELs), EEA1-mCherry (early endosomes), and MitoTracker™ Green (mitochondria). Scale bar = 20 μm. Magnified versions of the boxed regions are displayed on the right side. (E) Scheme used to produce HA-tagged TMEM175 knock-in (HA-KI) HAP1 cells using a Crispr-Cas9 strategy. A 2.2 kb human genomic DNA region spanning exon 11 with ~1.4 kb intron sequence was PCR amplified (arrows indicate the primers) and inserted into a backbone vector. HA tag was subsequently flanked downstream to the Open Reading Frame (ORF) using site-directed mutagenesis. The gRNA targeting the intron downstream of exon 11 was designed to trigger homologous recombination.
Supplemental Figure S3. TMEM175 is a pH-dependent, proton-permeant channel. Related to Figure 2.
(A) pHE/L-dependent effects on TMEM175 macroscopic chord conductance (−120 mV) for a series of cells studied as in Fig. 2B. For each cell, the conductance (at −120 mV) at each pHL was normalized to the conductance at pHL 3.5 (n = 5, mean ± SEM, * p <0.05, *** p <0.001). The chord conductance was calculated using the Ohm’s law-based equation: G= Im/(Em-Eion), where G is the chord conductance, Im is the recorded current, Em is the membrane holding potential, and Eion is the reversal potential by a specific ion calculated using the Nernst equation. (B) Representative H+ currents from a lumen-side-out recording on an enlarged LEL from a TMEM175-transfected COS1 cell when pHC was set to 7.2 and pHL varied from 7.2 to 3.5. In this configuration, an inward (negative) current indicates cation efflux from the luminal side to the cytosolic side. (C) Summary of TMEM175 macroscopic conductance (−120 mV) from experiments as in (B). For each patch, the conductance (−120 mV) at each pHL was normalized to the conductance at pHL 3.5 (n = 5, mean ± SEM, * p <0.05, *** p <0.001). (D) Whole-cell currents recorded in an HEK293T cell overexpressing mouse TMEM175. pHC was set to 7.2 and extracellular pH (pHE/L) varied as indicated. (E) Representative whole-cell current traces from a human OTOP1-transfected HEK293T cell using the same recording conditions as in Fig. 2F. (F) Relative permeability of H+ vs. K+ of TMEM175 under different pHE/L based on Erev measurement from Fig. 2G (n = 6 - 13, Mean ± SEM, NS, p > 0.05). (G) The inhibitory effect of pHE/L of 4.6 and 6.0 on basal outward K+ currents in TMEM175-transfected HEK293T cells. Note that significant basal outward K+ currents were only seen in a sub-set of TMEM175-transfected cells, which were chosen to study pH effect on K+ currents. The recording conditions were the same as those in Fig. 2F. Right panel shows the changes of outward K+ currents at +120 mV when pHE/L was varied from 7.2 to 6.0 or 4.6 (n = 9/6, Mean ± SEM, NS, p > 0.05). (H) Representative whole-cell recordings of IH and IK in HEK293T cells overexpressing TMEM175. IK (gray trace) was recorded using a K-MSA cytosolic solution with both pHE/L and pHC set to 7.2; IH (Red trace) was recorded using K+ free solutions (NMDG-MSA) with pHE/L set to 4.6 and pHC to 7.2. The estimated IK at pHE/L 4.6 (dashed green trace) was calculated from the measured IK at pHE/L 7.2 (gray trace) by rescaling to account for the 50% inhibition of K+ current at this pH seen in panel G. (I) Representative endogenous whole-LEL IH (pHL 4.6 or 3.5) and IK in WT HAP1 cells under recording conditions similar as (H). (J) The calculated ratios of IH/(IH + IK) from −120 to 0 mV based on the data in panels H and I (pHE/L 4.6, [H+]/[K+] = 0.00018). At physiological lysosomal membrane potential, the estimated fractional currents of IH increased further to > 99% at pH 3.5 ([H+]/[K+]=0.0023), but decreased to <75% at pH 5.0 ([H+]/[K+]=0.00007), and ~30% at pH 6.0 ([H+]/[K+]=0.000007). (K-O) The fluorescence intensity of pHluorin in mCherry-TMEM175-transfected HEK293T cells (n = 31 – 60, mean ± SEM) and vector-transfected (sham) cells (n = 9 – 31, *** p <0.001) in response to the stimuli as indicated (see Methods for details of the imaging solutions). The average changes of pHlourin fluorescence and the summary of relative pHluorin fluorescence intensity in response to pH 4.6 and HOAc are shown on the right side of each panel. (P) The fluorescence intensity of the pHrodo™ Red dye in EGFP-TMEM175-transfected cells (n = 35, mean ± SEM) or vector transfected (sham) cells (n = 25, *** p <0.001) in response to the stimuli as indicated.
Supplemental Figure S4. Synthetic chemicals activate TMEM175 H+ currents. Related to Figure 3.
(A) Lack of effect of ArA (100 μM), DCPIB (50 μM) or ML 67-33 (50 μM) on whole-cell currents of un-transfected HEK293T cells. Both pHE/L and pHC were set to 6.0. (B) Effects of ML 67-33 (50 μM) on whole-cell H+ currents from TMEM175-transfected HEK293T cells (n = 5, mean ± SEM, ** p <0.01). (C) ArA, DCPIB, and ML 67-33 evoked whole-cell H+ and K+ currents in TMEM175-transfected HEK293T cells. Cytosolic solutions contained 10 mM K-MSA (supplemented with NMDG-MSA to maintain osmolality) with pHC set to 7.2. (D) Summary of Erev from experiments as in (C) (n = 5, NS, p >0.05 between groups). (E) Representative traces of whole-LEL Cs+ currents from WT, TMEM175 KO, and KO HAP1 cells re-expressing human TMEM175 (KO+hTMEM175) in response to ArA (100 μM) with both pHL and pHC set to 7.2. The major ions were Cs-MSA in the luminal solution and NMDG-MSA in the cytosolic solution, respectively. (F) Summary of whole-LEL Cs+ currents (−120 mV) from experiments as in (E) (WT/KO/KO+hTMEM175, n = 7/6/7 LELs, mean ± SEM, *** p <0.001). (G) Examples of whole-LEL Cs+ currents from WT, TMEM175 KO, and KO HAP1 cells re-expressing human TMEM175 (KO+hTMEM175) in response to DCPIB (100 μM) with both pHL and pHC set to 7.2. (H) Summary of whole-LEL Cs+ currents (−120 mV) from experiments as in (G) (WT/KO/KO+hTMEM175, n = 7/12/6 LELs, mean ± SEM, *** p <0.001). (I-L) The fluorescence intensity of pHluorin in HEK293T cells dually transfected with pHluorin and mCherry-TMEM175 in response to stimuli as indicated (scale bar = 20 μm). The control imaging solution was set to pH 7.2. ArA (50 μM) or DCPIB (20 μM) dissolved in the pH 6.0 solution was bath applied to activate TMEM175. Panels (J) and (L) show the average time series (mean ± SEM) from experiments as in (I) and (K). The right panels show the summary of relative pHluorin fluorescence intensity in response to vehicle (Veh), ArA, or DCPIB application at pH 6.0 (n = 9/10; mean ± SEM, *** p <0.001).
Supplemental Figure S5. Luminal protons, but not AKT, activate TMEM175. Related to Figure 4
(A) Representative whole-LEL currents recorded in a TMEM175-overexpressing COS1 cell in response to SC-79 (an AKT activator, 100 μM) or DCPIB (50 μM) with both luminal and cytosolic solution set to pH 6.0 and K-MSA as the major ions. NMDG-MSA cytosolic solution of pH 7.2 served as a control. Current densities at +120 mV in response to SC-79 or DCPIB are summarized in the right panel (n = 6 LELs, mean ± SEM, NS, p > 0.05, *** p <0.001). (B) Representative DCPIB (50 μM)-activated whole-LEL currents in a TMEM175-expressing COS1 cell that was pre-treated with MK-2206 (20 μM) and ARQ-092 (20 μM) for 3 hours. The recording conditions were the same as for (A). (C) Whole-cell currents recorded in TMEM175-overexpressing HEK293T cells in response to SC-79 (40 μM) or DCPIB (50 μM). Both cytosolic and extracellular solutions contained K-MSA with both pHE/L and pHC set to 7.2. NMDG-MSA extracellular solution (pHE/L=7.2) served as a control. (D) Protein expression levels of phosphorylated AKT (Thr-308) and total AKT in HeLa cells that were pre-treated with SC-79 (20 μM, 3 h), MK-2206 (20 μM, 3 h), or ARQ-092 (20 μM, 3 h). β-actin served as the loading control. (E) Whole-cell currents recorded in a TMEM175-overexpressing HEK293T cell with pHC set to 4.6 and pHE/L varied as indicated. The red arrow indicates the Erev recorded under pHE/L 5.6. (F) Representative H+ currents from a lumen-side-out recording in a TMEM175-overexprewssing COS1 cell with pHC set to 4.6 and pHL varied from 7.2 to 4.6 and back. (G-H) Whole-cell H+ currents from a TMEM175-transfected HEK293T cell in response to DCPIB (50 μM) with pHC set to 4.6 and pHE/L to 7.2.
Supplemental Figure S6. Luminal protons activate the proton permeation pathway of TMEM175 through a negatively-charged amino acid residue in the pore region. Related to Figure 4
(A) The membrane topology of one subunit of human TMEM175 based on the reported Cryo-EM structure (PDB: 6wc9). The human TMEM175 channel is a dimer composed of two identical subunits, each of which has 12 transmembrane segments containing two similar repeats (IS1-IS6 and IIS1-IIS6). The structure shows a putative K+ pathway formed by IS1 and IIS1 of both subunits. The locations of aspartate (D), glutamate (E), and histidine (H) sites in IS1, the IS1-IS2 loop, IIS1, and the IIS1-IIS2 loop, as well as S45, are indicated. (B) Representative whole-cell H+ currents in HEK293T cells overexpressing each TMEM175 mutant. Multiple mutations were made simultaneously in two constructs: 4D4A (D67A, D283A, D287A, and D290A) and 4E4A (E59A, E53A, E282A, and E293A). (C) Summary of whole-cell H+ currents at −120 mV from experiments as in (B) (n = 3-14, mean ± SEM). (D) The inhibitory effect of pHE/L of 4.6 and 6.0 on basal outward K+ currents in D41A-transfected HEK293T cells. The recording conditions were the same as in (B). The inhibition (%) of outward K+ currents through WT and D41A is summarized in the right panel (n = 5 - 7, Mean ± SEM, NS p > 0.05). (E) Predominant lysosomal localization of EGFP-TMEM175 and EGFP-TMEM175-D41A, as determined by co-localization with LAMP1-mCherry. Scale bars = 20 μm. (F) Whole-cell currents in HEK293T cells overexpressing WT, M393T, and S45A mutant TMEM175 channels. The cytosolic solution contained K-MSA with pHC set to 7.2. Large outward K+ currents were activated by DCPIB (50 μM) in the extracellular solutions (pHE/L 7.2) with NMDG-MSA or Na-MSA as the major ions. (G) Summary of Erev from experiments as in (F) (n = 4-12, mean ± SEM, *** p < 0.001, NS p > 0.05). (H-I) Analysis of relative K+ permeability over H+ or Na+ of WT, M393T, and S45A mutant channels based on Erev measurement from (G) (n = 5-9, mean ± SEM, *** p < 0.001, NS p > 0.05). Note that the PK/PH value for OTOP1 was replotted from Fig. 2I for comparison. (J) Representative whole-LEL H+ currents from WT and TMEM175-M393T KI HEK293T cells. The luminal pHL was set to 3.5. (K) Summary of current densities at −120 mV from experiments as in (J) (WT/M393T-KI: n = 8/7, mean ± SEM, *** p <0.001).
Supplemental Figure S7. Proton permeation through TMEM175 regulates lysosomal pH homeostasis and hydrolytic activity. Related to Figure 5 and Figure 6.
(A) pHluorin imaging of HEK293T cells dually transfected with pHluorin and mCherry-TMEM175 in response to stimuli as indicated (scale bar = 50 μm). Imaging solution was set to pHE/L 7.2 to minimize H+ influx across the plasma membrane. ArA (400 μM) was bath applied to activate TMEM175. In the GPN experiment, cells were pre-treated with GPN (200 μM) one hour before imaging to abolish the lysosomal H+ gradient. (B) Summary of relative pHluorin fluorescence intensity in response to ArA and HOAc (sham/TMEM175/TMEM175+GPN, n = 26/30/23 cells; mean ± SEM, *** p <0.001). (C) Representative Lyso-VRAC currents activated by a low ionic strength (Γ) solution (40 mM NaCl supplemented with mannitol to reach isotonicity (see (Li et al., 2020b)) from WT and TMEM175 KO HAP1 cells. Right panel shows the summary of current densities at +120 mV (WT/TMEM175-KO: n = 7/7, mean ± SEM, NS, p > 0.05). (D) The effects of DCPIB application (20 μM, 30 min) on lysosomal acidity in WT, LRRC8A-KO, and TMEM175-KO HAP1 cells, determined using a ratiometric pH dye combination (pHrodo Green dextran and CF555 dextran). Baf-A1 (1 μM, 30 min) served as a positive control (n = 12-23 images, *** p <0.001, NS, p > 0.05). (E) LAMP1 immunostaining (scale bar = 20 μm) in WT and KO HeLa cells (n = 14-18 images, NS, p >0.05). (F) Lysosomal pH of WT HeLa cells and two KO cell lines (KO-1 and KO-2) determined by LysoSensor Yellow/Blue dye (n = 10-12 images, *** p <0.001). (G) Lysosomal pH distribution of WT and KO HeLa cells determined by Oregon Green dextran from experiments as in Fig. 6C. Bin size = 0.2 pH. (H) LysoTracker imaging of WT cells and two HEK293T cell lines stably expressing TMEM175 (L1 and L2). Scale bar = 50 μm. (I) Quantification of LysoTracker fluorescence intensity of all cells from three biological replicate experiments as in (H) (n = 8-9 images, *** p <.001). Cells were incubated with 1 μM Baf-A1 for 10 minutes immediately before imaging. In the right panel, the relative LysoTracker intensity was calculated from the ratio between Baf-A1 and DMSO treated groups. (J-K) Lysosomal acidity assessed by LysoTracker staining in TMEM175 KO cells re-expressing WT EGFP-TMEM175 or EGFP-TMEM175-S45A (scale bar = 20 μm, n = 6-7 images, *** p < 0.001). (L-M) Lysosomal overall hydrolytic activity estimated by staining with DQ™-BSA red in WT, TMEM175 KO, and KO HeLa cells stably expressing TMEM175 or D41A (scale bar = 50 μm, n = 5-8 images, NS, p > 0.05, *** p <0.001). (N) Protein expression levels of Cathepsin B and Cathepsin D in WT, TMEM175 KO, and KO HeLa cells re-expressing TMEM175 or D41A. Anti-EGFP antibody was used to indicate the expression of TMEM175 or D41A. β-actin served as the loading control.
Supplemental Video S1: pHluorin imaging of TMEM175-transfected HEK293T cells. Related to Figure 2.
Basal pHluorin fluorescence was acquired in an imaging solution at pH 7.2 followed by an acidic imaging solution at pH 4.6. The major ions in both solutions were TMA-MSA. The mcherry-TMEM175-positive cells and sham cells are shown side by side. Acetic acid (HOAc, pH 5.0) and NH4Cl (pH 7.2), freely entering the cytosol, served as positive and negative controls to acidify or alkalinize the cytosol, respectively.
Highlights:
TMEM175 is the proton “leak” channel of lysosomes and endosomes.
TMEM175 is a highly proton-selective channel that is gated by luminal protons.
An endogenous lipid can also activate TMEM175 to trigger lysosomal proton release.
TMEM175 sets the lysosomal pH optimum via a classic negative feedback mechanism.
Acknowledgements:
Research reported in this publication was supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK115474 (HX & RIH), the National Institute of General Medical Sciences under award number R35GM131720 (JAS), University of Michigan Mcubed and PFD disease initiative grants (HX), and funds from the Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals (MH, PL, and XF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We appreciate the encouragement and helpful comments provided by our Xu laboratory colleagues.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
H.X. is the scientific co-founder and a partial owner of Lysoway Therapeutics Inc (Boston, MA). Other authors declare no competing interests.
References:
- Ashcom JD, and Jacobson LA (1989). Self-quenched fluorogenic protein substrates for the detection of cathepsin D and other protease activities. Analytical biochemistry 176, 261–264. [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Ang KH, Gallardo-Godoy A, Clark KA, Arkin MR, Renslo AR, and Minor DL Jr. (2013). A high-throughput functional screen identifies small molecule regulators of temperature- and mechano-sensitive K2P channels. ACS chemical biology 8, 1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A, and Bonifacino JS (2019). Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. [DOI] [PubMed] [Google Scholar]
- Blauwendraat C, Heilbron K, Vallerga CL, Bandres-Ciga S, von Coelln R, Pihlstrom L, Simon-Sanchez J, Schulte C, Sharma M, Krohn L, et al. (2019). Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and alpha-synuclein mechanisms. Movement disorders : official journal of the Movement Disorder Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam SR, Wang F, and Muller S (2019). Lysosomes as a therapeutic target. Nat Rev Drug Discov 18, 923–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx M, Daniel J, Genin E, Soria FN, Blanchard-Desce M, Bezard E, and Dehay B (2016). Nanoparticles restore lysosomal acidification defects: Implications for Parkinson and other lysosomal-related diseases. Autophagy 12, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, and Altendorf K (1988). Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proceedings of the National Academy of Sciences of the United States of America 85, 7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briozzo P, Morisset M, Capony F, Rougeot C, and Rochefort H (1988). In vitro degradation of extracellular matrix with Mr 52,000 cathepsin D secreted by breast cancer cells. Cancer research 48, 3688–3692. [PubMed] [Google Scholar]
- Brunner JD, Jakob RP, Schulze T, Neldner Y, Moroni A, Thiel G, Maier T, and Schenck S (2020). Structural basis for ion selectivity in TMEM175 K(+) channels. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang C, Aranda K, Seo Y.-j., Gasnier B, and Ren D (2015). TMEM175 Is an Organelle K+ Channel Regulating Lysosomal Function. Cell 162, 1101–1112. [DOI] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, and Orlowski J (2010). Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11, 50–61. [DOI] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, International Parkinson’s Disease Genomics, C., and Me Research T, Kerchner GA, Ayalon G, et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Butz ES, Chao YK, Grishchuk Y, Becker L, Heller S, Slaugenhaupt SA, Biel M, Wahl-Schott C, and Grimm C (2017a). Small Molecules for Early Endosome-Specific Patch Clamping. Cell chemical biology 24, 907–916.e904. [DOI] [PubMed] [Google Scholar]
- Chen CC, Cang C, Fenske S, Butz E, Chao YK, Biel M, Ren D, Wahl-Schott C, and Grimm C (2017b). Patch-clamp technique to characterize ion channels in enlarged individual endolysosomes. Nat Protoc 12, 1639–1658. [DOI] [PubMed] [Google Scholar]
- Chen CS, Chen WN, Zhou M, Arttamangkul S, and Haugland RP (2000). Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. Journal of biochemical and biophysical methods 42, 137–151. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zeng W, She J, Bai XC, and Jiang Y (2019). Structural and functional characterization of an otopetrin family proton channel. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KA, Myers JT, and Swanson JA (2002). pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci 115, 599–607. [DOI] [PubMed] [Google Scholar]
- Creasy BM, Hartmann CB, White FK, and McCoy KL (2007). New assay using fluorogenic substrates and immunofluorescence staining to measure cysteine cathepsin activity in live cell subpopulations. Cytometry Part A : the journal of the International Society for Analytical Cytology 71, 114–123. [DOI] [PubMed] [Google Scholar]
- Cullen V, Lindfors M, Ng J, Paetau A, Swinton E, Kolodziej P, Boston H, Saftig P, Woulfe J, Feany MB, et al. (2009). Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Molecular brain 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MJ, Gregorka B, Gestwicki JE, and Swanson JA (2012). Inducible renitence limits Listeria monocytogenes escape from vacuoles in macrophages. Journal of immunology (Baltimore, Md : 1950) 189, 4488–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decher N, Lang HJ, Nilius B, Bruggemann A, Busch AE, and Steinmeyer K (2001). DCPIB is a novel selective blocker of I(Cl,swell) and prevents swelling-induced shortening of guinea-pig atrial action potential duration. British journal of pharmacology 134, 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoursey TE (2003). Voltage-gated proton channels and other proton transfer pathways. Physiological reviews 83, 475–579. [DOI] [PubMed] [Google Scholar]
- Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, and Xu H (2008). The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M (2007). Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8, 917–929. [DOI] [PubMed] [Google Scholar]
- Graves AR, Curran PK, Smith CL, and Mindell JA (2008). The Cl-/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature 453, 788. [DOI] [PubMed] [Google Scholar]
- Holopainen JM, Saarikoski J, Kinnunen PK, and Jarvela I (2001). Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs). Eur J Biochem 268, 5851–5856. [DOI] [PubMed] [Google Scholar]
- Hopfner F, Mueller SH, Szymczak S, Junge O, Tittmann L, May S, Lohmann K, Grallert H, Lieb W, Strauch K, et al. (2020). Rare Variants in Specific Lysosomal Genes Are Associated with Parkinson’s Disease. Movement disorders : official journal of the Movement Disorder Society. [DOI] [PubMed] [Google Scholar]
- Huynh C, and Andrews NW (2005). The small chemical vacuolin-1 alters the morphology of lysosomes without inhibiting Ca2+-regulated exocytosis. EMBO reports 6, 843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Nayak S, Mindell JA, and Grabe M (2013). A model of lysosomal pH regulation. J Gen Physiol 141, 705–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki H, Blauwendraat C, Leonard HL, Liu G, Maple-Grodem J, Corvol JC, Pihlstrom L, van Nimwegen M, Hutten SJ, Nguyen KH, et al. (2019). Genetic risk of Parkinson disease and progression:: An analysis of 13 longitudinal cohorts. Neurol Genet 5, e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadot M, Colmant C, Wattiaux-De Coninck S, and Wattiaux R (1984). Intralysosomal hydrolysis of glycyl-L-phenylalanine 2-naphthylamide. The Biochemical journal 219, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn S, Blauwendraat C, Toolan D, Gretzula CA, Drolet RE, Smith S, Nalls MA, Marcus J, Singleton AB, and Stone DJ (2019). Functionalization of the TMEM175 p.M393T variant as a risk factor for Parkinson disease. Human molecular genetics 28, 3244–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn S, Drolet RE, Cramer PE, Wong AH, Toolan DM, Gretzula CA, Voleti B, Vassileva G, Disa J, Tadin-Strapps M, et al. (2017). TMEM175 deficiency impairs lysosomal and mitochondrial function and increases alpha-synuclein aggregation. Proceedings of the National Academy of Sciences of the United States of America 114, 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Ostrowski P, Jaumouille V, and Grinstein S (2016). The position of lysosomes within the cell determines their luminal pH. The Journal of cell biology 212, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatzis V, Cherqui S, Antignac C, and Gasnier B (2001). Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. The EMBO journal 20, 5940–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper D, Planells-Cases R, Fuhrmann JC, Scheel O, Zeitz O, Ruether K, Schmitt A, Poet M, Steinfeld R, Schweizer M, et al. (2005). Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. The EMBO journal 24, 1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke H, Langner J, Wiederanders B, Ansorge S, and Bohley P (1977). Cathepsin L. A new proteinase from rat-liver lysosomes. Eur J Biochem 74, 293–301. [DOI] [PubMed] [Google Scholar]
- Koivusalo M, Steinberg BE, Mason D, and Grinstein S (2011). In situ measurement of the electrical potential across the lysosomal membrane using FRET. Traffic (Copenhagen, Denmark) 12, 972–982. [DOI] [PubMed] [Google Scholar]
- Kolter T, and Sandhoff K (2005). Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol 21, 81–103. [DOI] [PubMed] [Google Scholar]
- Krohn L, Ozturk TN, Vanderperre B, Ouled Amar Bencheikh B, Ruskey JA, Laurent SB, Spiegelman D, Postuma RB, Arnulf I, Hu MTM, et al. (2020). Genetic, Structural, and Functional Evidence Link TMEM175 to Synucleinopathies. Annals of neurology 87, 139–153. [DOI] [PubMed] [Google Scholar]
- Lee C, Guo J, Zeng W, Kim S, She J, Cang C, Ren D, and Jiang Y (2017). The lysosomal potassium channel TMEM175 adopts a novel tetrameric architecture. Nature 547, 472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. (2010). Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA (1979). Ion-concentration dependence of the reversal potential and the single channel conductance of ion channels at the frog neuromuscular junction. The Journal of physiology 286, 417–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ou R, Chen Y, Gu X, Wei Q, Cao B, Zhang L, Hou Y, Liu K, Chen X, et al. (2020a). Mutation analysis of TMEM family members for early-onset Parkinson’s disease in Chinese population. Neurobiol Aging. [DOI] [PubMed] [Google Scholar]
- Li P, Gu M, and Xu H (2019). Lysosomal Ion Channels as Decoders of Cellular Signals. Trends in biochemical sciences 44, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Hu M, Wang C, Feng X, Zhao Z, Yang Y, Sahoo N, Gu M, Yang Y, Xiao S, et al. (2020b). LRRC8 family proteins within lysosomes regulate cellular osmoregulation and enhance cell survival to multiple physiological stresses. Proceedings of the National Academy of Sciences of the United States of America 117, 29155–29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, and Lee VM (2012). Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, and Lee JC (2015). Cysteine cathepsins are essential in lysosomal degradation of alpha-synuclein. Proceedings of the National Academy of Sciences of the United States of America 112, 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, and Rothman JE (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394. [DOI] [PubMed] [Google Scholar]
- Mindell JA (2012). Lysosomal acidification mechanisms. Annu Rev Physiol 74, 69–86. [DOI] [PubMed] [Google Scholar]
- Neher E (1992). Correction for liquid junction potentials in patch clamp experiments. Methods in enzymology 207, 123–131. [DOI] [PubMed] [Google Scholar]
- Nicoli ER, Weston MR, Hackbarth M, Becerril A, Larson A, Zein WM, Baker PR 2nd, Burke JD, Dorward H, Davids M, et al. (2019). Lysosomal Storage and Albinism Due to Effects of a De Novo CLCN7 Variant on Lysosomal Acidification. American journal of human genetics 104, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Paknejad N, and Hite RK (2020). Gating and selectivity mechanisms for the lysosomal K(+) channel TMEM175. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma S, Moriyama Y, and Takano T (1982). Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proceedings of the National Academy of Sciences of the United States of America 79, 2758–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergel E, Veres I, Csigi GI, and Czirják G (2021). Translocation of TMEM175 Lysosomal Potassium Channel to the Plasma Membrane by Dynasore Compounds. International Journal of Molecular Sciences 22, 10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt SJ, Lam AK, Rietdorf K, Galione A, and Sitsapesan R (2014). Reconstituted human TPC1 is a proton-permeable ion channel and is activated by NAADP or Ca2+. Science signaling 7, ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, Xie ZL, Speake LD, Parks R, Crabtree D, et al. (2008). Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Molecular brain 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, Ramos-Espiritu L, Milner TA, Buck J, and Levin LR (2016). Soluble adenylyl cyclase is essential for proper lysosomal acidification. J Gen Physiol 148, 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, and Clapham DE (2006). A voltage-gated proton-selective channel lacking the pore domain. Nature 440, 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudakou U, Yu E, Krohn L, Ruskey JA, Asayesh F, Dauvilliers Y, Spiegelman D, Greenbaum L, Fahn S, Waters CH, et al. (2020). Targeted sequencing of Parkinson’s disease loci genes highlights SYT11, FGF20 and other associations. Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Kawawaki J, Moriura Y, Mori H, Morihata H, and Kuno M (2006). pH dependence and inhibition by extracellular calcium of proton currents via plasmalemmal vacuolar-type H+-ATPase in murine osteoclasts. J Physiol 576, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan A, Devany J, Veetil AT, Suresh B, Pillai KS, Schwake M, and Krishnan Y (2021). A DNA-based voltmeter for organelles. Nature nanotechnology 16, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Takagi M, and Okamura Y (2006). A voltage sensor-domain protein is a voltage-gated proton channel. Science 312, 589–592. [DOI] [PubMed] [Google Scholar]
- Sather WA, and McCleskey EW (2003). Permeation and selectivity in calcium channels. Annual review of physiology 65, 133–159. [DOI] [PubMed] [Google Scholar]
- Schroder BA, Wrocklage C, Hasilik A, and Saftig P (2010). The proteome of lysosomes. Proteomics 10, 4053–4076. [DOI] [PubMed] [Google Scholar]
- Shahmoradian SH, Lewis AJ, Genoud C, Hench J, Moors TE, Navarro PP, Castaño-Díez D, Schweighauser G, Graff-Meyer A, Goldie KN, et al. (2019). Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nature Neuroscience 22, 1099–1109. [DOI] [PubMed] [Google Scholar]
- Shen D, Wang X, Li X, Zhang X, Yao Z, Dibble S, Dong X.-p., Yu T, Lieberman AP, Showalter HD, et al. (2012). Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nature Communications 3, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, and Kiselyov K (2006). TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. The Journal of biological chemistry 281, 7294–7301. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, and Goedert M (1997). Alpha-synuclein in Lewy bodies. Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- Steinberg BE, Huynh KK, Brodovitch A, Jabs S, Stauber T, Jentsch TJ, and Grinstein S (2010). A cation counterflux supports lysosomal acidification. The Journal of cell biology 189, 1171–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y-H, Cooper AJ, Teng B, Chang BR, Artiga DJ, Turner HN, Mulhall EM, Ye W, Smith AD, and Liman ER (2018). An evolutionarily conserved gene family encodes proton-selective ion channels. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang X, Gao Q, Lawas M, Yu L, Cheng X, Gu M, Sahoo N, Li X, Li P, et al. (2017a). A voltage-dependent K+ channel in the lysosome is required for refilling lysosomal Ca2+ stores. The Journal of cell biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang X, Dong XP, Samie M, Li X, Cheng X, Goschka A, Shen D, Zhou Y, Harlow J, et al. (2012). TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu Q, Hu M, Liu B, Chai Z, Huang R, Wang Y, Xu H, Zhou L, Zheng L, et al. (2017b). Ligand- and voltage-gated Ca2+ channels differentially regulate the mode of vesicular neuropeptide release in mammalian sensory neurons. Science signaling 10. [DOI] [PubMed] [Google Scholar]
- Wie J, Liu Z, Song H, Tropea TF, Yang L, Wang H, Liang Y, Cang C, Aranda K, Lohmann J, et al. (2021). A growth-factor-activated lysosomal K(+) channel regulates Parkinson’s pathology. Nature 591, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, and Zhu MX (2016). Regulation of lysosomal ion homeostasis by channels and transporters. Science China Life sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, and Ren D (2015). Lysosomal physiology. Annu Rev Physiol 77, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Liu K, Swaroop M, Sun W, Dehdashti SJ, McKew JC, and Zheng W (2014). A phenotypic compound screening assay for lysosomal storage diseases. J Biomol Screen 19, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen J, Del Carmen Vitery M, Osei-Owusu J, Chu J, Yu H, Sun S, and Qiu Z (2019). PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylitervo P, Franzen CJ, and Taherzadeh MJ (2014). Continuous ethanol production with a membrane bioreactor at high acetic Acid concentrations. Membranes 4, 372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Ohta M, Kuwahara K, Cao MJ, Hara K, and Osatomi K (2015). Purification and Characterization of Cathepsin B from the Muscle of Horse Mackerel Trachurus japonicus. Mar Drugs 13, 6550–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kehm V, Gathagan R, Leight SN, Trojanowski JQ, Lee VM, and Luk KC (2019a). Stereotaxic Targeting of Alpha-Synuclein Pathology in Mouse Brain Using Preformed Fibrils. Methods in molecular biology (Clifton, NJ) 1948, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Yi YP, and Zhang GJ (2006). Effects of arachidonic acid on the lysosomal ion permeability and osmotic stability. Journal of bioenergetics and biomembranes 38, 75–82. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen W, Li P, Calvo R, Southall N, Hu X, Bryant-Genevier M, Feng X, Geng Q, Gao C, et al. (2019b). Agonist-specific voltage-dependent gating of lysosomal two-pore Na(+) channels. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Shen C, Wang L, Rawson S, Xie WJ, Nist-Lund C, Wu J, Shen Z, Xia S, Holt JR, et al. (2022). pH regulates potassium conductance and drives a constitutive proton current in human TMEM175. Science advances 8, eabm1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. TMEM175 mediates small inward H+ “leak” currents in endolysosomes at luminal pH 4.6. Related to Figure 1.
(A) Whole-LEL H+ “leak” currents from a COS1 cell with luminal pH (pHL) set to 4.6 and cytosolic pH (pHC) set to either 7.2 or 4.6 as indicated. (B) Whole-LEL current amplitude distribution with pHL set to 4.6 and pHC to 7.2 from experiments as in (A) (n = 128 LELs, bin size = 5 pA). (C) Whole-LEL H+ “leak” currents are undetectable (grey trace) in a COS1 cell with both pHL and pHC set to pH 7.2. The outward TPC-mediated Na+ currents (Wang et al., 2012) evoked by PI(3,5)P2 (red trace) served as a control that the recording was in the whole-LEL mode. (D) Whole-LEL H+ “leak” currents from a COS1 cell with pHL set to 4.6 and pHC set to 7.2. Both luminal and cytosolic solutions contained Na-MSA as major ions. (E) Average whole-LEL H+ “leak” currents from COS1, HAP1, HEK293T, HeLa, mouse embryonic fibroblast (MEF) cells, and mouse primary hippocampal neurons (mHippo) with pHL set to 3.5 and pHC to 7.2 (n = 4-11 LELs). (F) Whole-EE (early endosome) H+ “leak” currents in a chemically (Wortmannin + Latrunculin B)-enlarged early endosome (Chen et al., 2017a) from a COS1 cell. (G-H) Whole-LEL H+ “leak” currents were tiny in COS1 cells overexpressing TRPML1 or TPC1 with pHL set to 4.6. Luminal and cytosolic (Cyto) solutions contained indicated major ions required for the ML-SA5-activated TRPML1 or PI(3,5)P2-activated TPC1 positive-control currents.
Supplemental Figure S2. Lysosome-localized TMEM175 is required for inward H+ “leak” currents in endolysosomes. Related to Figure 1.
(A) Crispr-Cas9 strategy used to create TMEM175 knock-out (KO) human cell lines. The top of the panel shows the amino acid sequence for the IIS1 region of TMEM175 and the genomic sequence with the guide RNA (gRNA) sequence and protospacer adjacent motif (PAM) motif highlighted in red and blue, respectively. The bottom of the panel shows the Sanger sequencing confirming that frameshift was produced in HAP1 (haploid), HeLa, and HEK293T cells. (B-C) Whole-LEL H+ “leak” currents from a Mucolipidosis IV patient-derived (TRPML1 KO) fibroblast or a TPC1&TPC2 double knockout (TPC1&2 DKO) HAP1 cell with pHL set to 3.5 and recording solutions as indicated. (D) The subcellular localization of overexpressed TMEM175 in HEK293T cells was determined by labeling markers for three different organelles: LAMP1-mCherry (LELs), EEA1-mCherry (early endosomes), and MitoTracker™ Green (mitochondria). Scale bar = 20 μm. Magnified versions of the boxed regions are displayed on the right side. (E) Scheme used to produce HA-tagged TMEM175 knock-in (HA-KI) HAP1 cells using a Crispr-Cas9 strategy. A 2.2 kb human genomic DNA region spanning exon 11 with ~1.4 kb intron sequence was PCR amplified (arrows indicate the primers) and inserted into a backbone vector. HA tag was subsequently flanked downstream to the Open Reading Frame (ORF) using site-directed mutagenesis. The gRNA targeting the intron downstream of exon 11 was designed to trigger homologous recombination.
Supplemental Figure S3. TMEM175 is a pH-dependent, proton-permeant channel. Related to Figure 2.
(A) pHE/L-dependent effects on TMEM175 macroscopic chord conductance (−120 mV) for a series of cells studied as in Fig. 2B. For each cell, the conductance (at −120 mV) at each pHL was normalized to the conductance at pHL 3.5 (n = 5, mean ± SEM, * p <0.05, *** p <0.001). The chord conductance was calculated using the Ohm’s law-based equation: G= Im/(Em-Eion), where G is the chord conductance, Im is the recorded current, Em is the membrane holding potential, and Eion is the reversal potential by a specific ion calculated using the Nernst equation. (B) Representative H+ currents from a lumen-side-out recording on an enlarged LEL from a TMEM175-transfected COS1 cell when pHC was set to 7.2 and pHL varied from 7.2 to 3.5. In this configuration, an inward (negative) current indicates cation efflux from the luminal side to the cytosolic side. (C) Summary of TMEM175 macroscopic conductance (−120 mV) from experiments as in (B). For each patch, the conductance (−120 mV) at each pHL was normalized to the conductance at pHL 3.5 (n = 5, mean ± SEM, * p <0.05, *** p <0.001). (D) Whole-cell currents recorded in an HEK293T cell overexpressing mouse TMEM175. pHC was set to 7.2 and extracellular pH (pHE/L) varied as indicated. (E) Representative whole-cell current traces from a human OTOP1-transfected HEK293T cell using the same recording conditions as in Fig. 2F. (F) Relative permeability of H+ vs. K+ of TMEM175 under different pHE/L based on Erev measurement from Fig. 2G (n = 6 - 13, Mean ± SEM, NS, p > 0.05). (G) The inhibitory effect of pHE/L of 4.6 and 6.0 on basal outward K+ currents in TMEM175-transfected HEK293T cells. Note that significant basal outward K+ currents were only seen in a sub-set of TMEM175-transfected cells, which were chosen to study pH effect on K+ currents. The recording conditions were the same as those in Fig. 2F. Right panel shows the changes of outward K+ currents at +120 mV when pHE/L was varied from 7.2 to 6.0 or 4.6 (n = 9/6, Mean ± SEM, NS, p > 0.05). (H) Representative whole-cell recordings of IH and IK in HEK293T cells overexpressing TMEM175. IK (gray trace) was recorded using a K-MSA cytosolic solution with both pHE/L and pHC set to 7.2; IH (Red trace) was recorded using K+ free solutions (NMDG-MSA) with pHE/L set to 4.6 and pHC to 7.2. The estimated IK at pHE/L 4.6 (dashed green trace) was calculated from the measured IK at pHE/L 7.2 (gray trace) by rescaling to account for the 50% inhibition of K+ current at this pH seen in panel G. (I) Representative endogenous whole-LEL IH (pHL 4.6 or 3.5) and IK in WT HAP1 cells under recording conditions similar as (H). (J) The calculated ratios of IH/(IH + IK) from −120 to 0 mV based on the data in panels H and I (pHE/L 4.6, [H+]/[K+] = 0.00018). At physiological lysosomal membrane potential, the estimated fractional currents of IH increased further to > 99% at pH 3.5 ([H+]/[K+]=0.0023), but decreased to <75% at pH 5.0 ([H+]/[K+]=0.00007), and ~30% at pH 6.0 ([H+]/[K+]=0.000007). (K-O) The fluorescence intensity of pHluorin in mCherry-TMEM175-transfected HEK293T cells (n = 31 – 60, mean ± SEM) and vector-transfected (sham) cells (n = 9 – 31, *** p <0.001) in response to the stimuli as indicated (see Methods for details of the imaging solutions). The average changes of pHlourin fluorescence and the summary of relative pHluorin fluorescence intensity in response to pH 4.6 and HOAc are shown on the right side of each panel. (P) The fluorescence intensity of the pHrodo™ Red dye in EGFP-TMEM175-transfected cells (n = 35, mean ± SEM) or vector transfected (sham) cells (n = 25, *** p <0.001) in response to the stimuli as indicated.
Supplemental Figure S4. Synthetic chemicals activate TMEM175 H+ currents. Related to Figure 3.
(A) Lack of effect of ArA (100 μM), DCPIB (50 μM) or ML 67-33 (50 μM) on whole-cell currents of un-transfected HEK293T cells. Both pHE/L and pHC were set to 6.0. (B) Effects of ML 67-33 (50 μM) on whole-cell H+ currents from TMEM175-transfected HEK293T cells (n = 5, mean ± SEM, ** p <0.01). (C) ArA, DCPIB, and ML 67-33 evoked whole-cell H+ and K+ currents in TMEM175-transfected HEK293T cells. Cytosolic solutions contained 10 mM K-MSA (supplemented with NMDG-MSA to maintain osmolality) with pHC set to 7.2. (D) Summary of Erev from experiments as in (C) (n = 5, NS, p >0.05 between groups). (E) Representative traces of whole-LEL Cs+ currents from WT, TMEM175 KO, and KO HAP1 cells re-expressing human TMEM175 (KO+hTMEM175) in response to ArA (100 μM) with both pHL and pHC set to 7.2. The major ions were Cs-MSA in the luminal solution and NMDG-MSA in the cytosolic solution, respectively. (F) Summary of whole-LEL Cs+ currents (−120 mV) from experiments as in (E) (WT/KO/KO+hTMEM175, n = 7/6/7 LELs, mean ± SEM, *** p <0.001). (G) Examples of whole-LEL Cs+ currents from WT, TMEM175 KO, and KO HAP1 cells re-expressing human TMEM175 (KO+hTMEM175) in response to DCPIB (100 μM) with both pHL and pHC set to 7.2. (H) Summary of whole-LEL Cs+ currents (−120 mV) from experiments as in (G) (WT/KO/KO+hTMEM175, n = 7/12/6 LELs, mean ± SEM, *** p <0.001). (I-L) The fluorescence intensity of pHluorin in HEK293T cells dually transfected with pHluorin and mCherry-TMEM175 in response to stimuli as indicated (scale bar = 20 μm). The control imaging solution was set to pH 7.2. ArA (50 μM) or DCPIB (20 μM) dissolved in the pH 6.0 solution was bath applied to activate TMEM175. Panels (J) and (L) show the average time series (mean ± SEM) from experiments as in (I) and (K). The right panels show the summary of relative pHluorin fluorescence intensity in response to vehicle (Veh), ArA, or DCPIB application at pH 6.0 (n = 9/10; mean ± SEM, *** p <0.001).
Supplemental Figure S5. Luminal protons, but not AKT, activate TMEM175. Related to Figure 4
(A) Representative whole-LEL currents recorded in a TMEM175-overexpressing COS1 cell in response to SC-79 (an AKT activator, 100 μM) or DCPIB (50 μM) with both luminal and cytosolic solution set to pH 6.0 and K-MSA as the major ions. NMDG-MSA cytosolic solution of pH 7.2 served as a control. Current densities at +120 mV in response to SC-79 or DCPIB are summarized in the right panel (n = 6 LELs, mean ± SEM, NS, p > 0.05, *** p <0.001). (B) Representative DCPIB (50 μM)-activated whole-LEL currents in a TMEM175-expressing COS1 cell that was pre-treated with MK-2206 (20 μM) and ARQ-092 (20 μM) for 3 hours. The recording conditions were the same as for (A). (C) Whole-cell currents recorded in TMEM175-overexpressing HEK293T cells in response to SC-79 (40 μM) or DCPIB (50 μM). Both cytosolic and extracellular solutions contained K-MSA with both pHE/L and pHC set to 7.2. NMDG-MSA extracellular solution (pHE/L=7.2) served as a control. (D) Protein expression levels of phosphorylated AKT (Thr-308) and total AKT in HeLa cells that were pre-treated with SC-79 (20 μM, 3 h), MK-2206 (20 μM, 3 h), or ARQ-092 (20 μM, 3 h). β-actin served as the loading control. (E) Whole-cell currents recorded in a TMEM175-overexpressing HEK293T cell with pHC set to 4.6 and pHE/L varied as indicated. The red arrow indicates the Erev recorded under pHE/L 5.6. (F) Representative H+ currents from a lumen-side-out recording in a TMEM175-overexprewssing COS1 cell with pHC set to 4.6 and pHL varied from 7.2 to 4.6 and back. (G-H) Whole-cell H+ currents from a TMEM175-transfected HEK293T cell in response to DCPIB (50 μM) with pHC set to 4.6 and pHE/L to 7.2.
Supplemental Figure S6. Luminal protons activate the proton permeation pathway of TMEM175 through a negatively-charged amino acid residue in the pore region. Related to Figure 4
(A) The membrane topology of one subunit of human TMEM175 based on the reported Cryo-EM structure (PDB: 6wc9). The human TMEM175 channel is a dimer composed of two identical subunits, each of which has 12 transmembrane segments containing two similar repeats (IS1-IS6 and IIS1-IIS6). The structure shows a putative K+ pathway formed by IS1 and IIS1 of both subunits. The locations of aspartate (D), glutamate (E), and histidine (H) sites in IS1, the IS1-IS2 loop, IIS1, and the IIS1-IIS2 loop, as well as S45, are indicated. (B) Representative whole-cell H+ currents in HEK293T cells overexpressing each TMEM175 mutant. Multiple mutations were made simultaneously in two constructs: 4D4A (D67A, D283A, D287A, and D290A) and 4E4A (E59A, E53A, E282A, and E293A). (C) Summary of whole-cell H+ currents at −120 mV from experiments as in (B) (n = 3-14, mean ± SEM). (D) The inhibitory effect of pHE/L of 4.6 and 6.0 on basal outward K+ currents in D41A-transfected HEK293T cells. The recording conditions were the same as in (B). The inhibition (%) of outward K+ currents through WT and D41A is summarized in the right panel (n = 5 - 7, Mean ± SEM, NS p > 0.05). (E) Predominant lysosomal localization of EGFP-TMEM175 and EGFP-TMEM175-D41A, as determined by co-localization with LAMP1-mCherry. Scale bars = 20 μm. (F) Whole-cell currents in HEK293T cells overexpressing WT, M393T, and S45A mutant TMEM175 channels. The cytosolic solution contained K-MSA with pHC set to 7.2. Large outward K+ currents were activated by DCPIB (50 μM) in the extracellular solutions (pHE/L 7.2) with NMDG-MSA or Na-MSA as the major ions. (G) Summary of Erev from experiments as in (F) (n = 4-12, mean ± SEM, *** p < 0.001, NS p > 0.05). (H-I) Analysis of relative K+ permeability over H+ or Na+ of WT, M393T, and S45A mutant channels based on Erev measurement from (G) (n = 5-9, mean ± SEM, *** p < 0.001, NS p > 0.05). Note that the PK/PH value for OTOP1 was replotted from Fig. 2I for comparison. (J) Representative whole-LEL H+ currents from WT and TMEM175-M393T KI HEK293T cells. The luminal pHL was set to 3.5. (K) Summary of current densities at −120 mV from experiments as in (J) (WT/M393T-KI: n = 8/7, mean ± SEM, *** p <0.001).
Supplemental Figure S7. Proton permeation through TMEM175 regulates lysosomal pH homeostasis and hydrolytic activity. Related to Figure 5 and Figure 6.
(A) pHluorin imaging of HEK293T cells dually transfected with pHluorin and mCherry-TMEM175 in response to stimuli as indicated (scale bar = 50 μm). Imaging solution was set to pHE/L 7.2 to minimize H+ influx across the plasma membrane. ArA (400 μM) was bath applied to activate TMEM175. In the GPN experiment, cells were pre-treated with GPN (200 μM) one hour before imaging to abolish the lysosomal H+ gradient. (B) Summary of relative pHluorin fluorescence intensity in response to ArA and HOAc (sham/TMEM175/TMEM175+GPN, n = 26/30/23 cells; mean ± SEM, *** p <0.001). (C) Representative Lyso-VRAC currents activated by a low ionic strength (Γ) solution (40 mM NaCl supplemented with mannitol to reach isotonicity (see (Li et al., 2020b)) from WT and TMEM175 KO HAP1 cells. Right panel shows the summary of current densities at +120 mV (WT/TMEM175-KO: n = 7/7, mean ± SEM, NS, p > 0.05). (D) The effects of DCPIB application (20 μM, 30 min) on lysosomal acidity in WT, LRRC8A-KO, and TMEM175-KO HAP1 cells, determined using a ratiometric pH dye combination (pHrodo Green dextran and CF555 dextran). Baf-A1 (1 μM, 30 min) served as a positive control (n = 12-23 images, *** p <0.001, NS, p > 0.05). (E) LAMP1 immunostaining (scale bar = 20 μm) in WT and KO HeLa cells (n = 14-18 images, NS, p >0.05). (F) Lysosomal pH of WT HeLa cells and two KO cell lines (KO-1 and KO-2) determined by LysoSensor Yellow/Blue dye (n = 10-12 images, *** p <0.001). (G) Lysosomal pH distribution of WT and KO HeLa cells determined by Oregon Green dextran from experiments as in Fig. 6C. Bin size = 0.2 pH. (H) LysoTracker imaging of WT cells and two HEK293T cell lines stably expressing TMEM175 (L1 and L2). Scale bar = 50 μm. (I) Quantification of LysoTracker fluorescence intensity of all cells from three biological replicate experiments as in (H) (n = 8-9 images, *** p <.001). Cells were incubated with 1 μM Baf-A1 for 10 minutes immediately before imaging. In the right panel, the relative LysoTracker intensity was calculated from the ratio between Baf-A1 and DMSO treated groups. (J-K) Lysosomal acidity assessed by LysoTracker staining in TMEM175 KO cells re-expressing WT EGFP-TMEM175 or EGFP-TMEM175-S45A (scale bar = 20 μm, n = 6-7 images, *** p < 0.001). (L-M) Lysosomal overall hydrolytic activity estimated by staining with DQ™-BSA red in WT, TMEM175 KO, and KO HeLa cells stably expressing TMEM175 or D41A (scale bar = 50 μm, n = 5-8 images, NS, p > 0.05, *** p <0.001). (N) Protein expression levels of Cathepsin B and Cathepsin D in WT, TMEM175 KO, and KO HeLa cells re-expressing TMEM175 or D41A. Anti-EGFP antibody was used to indicate the expression of TMEM175 or D41A. β-actin served as the loading control.
Supplemental Video S1: pHluorin imaging of TMEM175-transfected HEK293T cells. Related to Figure 2.
Basal pHluorin fluorescence was acquired in an imaging solution at pH 7.2 followed by an acidic imaging solution at pH 4.6. The major ions in both solutions were TMA-MSA. The mcherry-TMEM175-positive cells and sham cells are shown side by side. Acetic acid (HOAc, pH 5.0) and NH4Cl (pH 7.2), freely entering the cytosol, served as positive and negative controls to acidify or alkalinize the cytosol, respectively.
Data Availability Statement
Accession numbers are listed in the key resources table. Original traces of electrophysiological recordings, western blots, and microscopical images will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse anti-human LAMP1 (H4A3) antibody | DSHB | Cat# h4a3, RRID: AB_2296838 |
| Rat anti-HA (3F10) antibody | Roche | Cat# 11867423001, RRID: AB_390918 |
| Rabbit Anti-Human alpha Synuclein, phospho (Ser129) antibody | Abcam | Cat# ab51253, RRID: AB_869973 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Texas Red-X | Thermo Fisher Scientific | Cat# T-862, RRID: AB_2556781 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006, RRID: AB_2534074 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_253579 |
| Mouse anti-GFP (clone 7.1 and 13.1) antibody | Roche | Cat# 11814460001, RRID: AB_390913 |
| Rabbit anti-Phospho-Akt(Thr308) (C31E5E) antibody | Cell Signaling Technology | Cat #2965, RRID: AB_2255933 |
| Rabbit anti-Akt (pan) (C67E7) antibody | Cell Signaling Technology | Cat# 4691, RRID: AB_915783 |
| Rabbit anti-Cathepsin B antibody [EPR4323] | Abcam | Cat# ab125067, RRID: AB_10972167 |
| Rabbit anti-Cathepsin D antibody [EPR3057Y] | Abcam | Cat# ab75852, RRID: AB_1523267 |
| Mouse Anti-beta-Actin Antibody (AC-74) | Sigma-Aldrich | Cat# A5316, RRID: AB_476743 |
| IRDye® 680RD Goat anti-mouse IgG (H+L) secondary antibody | Li-cor | Cat# 926-68070, RRID: AB_10956588 |
| IRDye® 800CW Goat anti-Rabbit IgG (H+L) secondary antibody | Li-cor | Cat# 926-32211, RRID: AB_621843 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N-methyl-D-glucamine (NMDG) | Sigma-Aldrich | M2004; CAS: 6284-40-8 |
| Methanesulfonic acid (MSA) | Sigma-Aldrich | 471356; CAS: 75-75-2 |
| HCl | Fisher Scientific | A144C-212; CAS: 7647-01-0 |
| Tetramethylammonium (TMA) | Sigma-Aldrich | T-7505; CAS: 10424-65-4 |
| K-Gluconate | Sigma-Aldrich | P1847; CAS: 299-27-4 |
| KOH | Fisher Scientific | P250-500; CAS: 1310-58-3 |
| NaOH | Fisher Scientific | S318-500; CAS: 1310-73-2 |
| KCl | Sigma-Aldrich | P9541; CAS: 7447-40-7 |
| NaCl | Sigma-Aldrich | S9888; CAS: 7647-14-5 |
| CaCl2 | Sigma-Aldrich | C8106; CAS: 10035-04-8 |
| MgCl2 | Sigma-Aldrich | 208337; CAS: 7786-30-3 |
| NH4Cl | Sigma-Aldrich | A4514; CAS: 12125-02-9 |
| NaH2PO4 | Sigma-Aldrich | S3139; CAS: 7558-80-7 |
| Sucrose | Sigma-Aldrich | S0389; CAS: 57-50-1 |
| Tetraethylammonium chloride (TEA-Cl) | Sigma-Aldrich | T2265; CAS: 56-34-8 |
| Cs-Methanesulfonate (Cs-MSA) | Sigma-Aldrich | C1426; CAS: 2550-61-0 |
| HEPES | Sigma-Aldrich | H3375; CAS: 7365-45-9 |
| MES | Sigma-Aldrich | M8250; CAS: 1266615-59-1 |
| Citric acid | Sigma-Aldrich | 27109; CAS: 77-92-9 |
| Acetic acid | Fisher Scientific | A38-212; CAS: 64-19-7 |
| Ethanol | Decon laboratories inc. | 2701; CAS: 64-17-5 |
| Methanol | Fisher Scientific | A52-4; CAS: 67-56-1 |
| Isopropanol | Fisher Scientific | A416-4; CAS: 67-63-0 |
| Glucose | Sigma-Aldrich | G6152; CAS: 50-99-7 |
| Triton-X 100 | Sigma-Aldrich | T8787; CAS: 9002-93-1 |
| NaF | Sigma-Aldrich | S7920; CAS: 7681-49-4 |
| Na-orthovanadate | Sigma-Aldrich | S6508; CAS: 13721-39-6 |
| EDTA | Sigma-Aldrich | 03609; CAS: 60-00-4 |
| EGTA | Sigma-Aldrich | E4378; CAS: 67-42-5 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D2650; CAS: 67-68-5 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A3059; CAS: 9048-46-8 |
| Paraformaldehyde (PFA) 4% in PBS | Thermo Fisher Scientific | J19943-K2 |
| Phosphate-buffered saline (PBS) | Gibco | 10010 |
| RIPA | Boston Bioproducts | BP-115 |
| Protease inhibitor cocktail | Sigma Aldrich | P8340 |
| Phosphatase inhibitor cocktail I | Sigma Aldrich | P0044 |
| LDS sample buffer (4x) | Invitrogen | NP0007 |
| Pierce™ BCA™ Protein Assay | Invitrogen | 23227 |
| PVDF membrane | Millipore | IPFL00010 |
| Fluoromount-G | Southern Biotech | 0100-01 |
| Magic Red Cathepsin B assay kit | ImmunoChemistry Technologies | 937 |
| Pepstatin A BODIPY™ FL | Invitrogen | P12271 |
| DQ™-BSA red dye | Invitrogen | D12051 |
| CF®488a conjugated dextran-10 kd | Biotium | 80110 |
| CF®555 conjugated dextran-10 kd | Biotium | 80112 |
| LysoSensor™ Yellow/Blue DND-160 | Invitrogen | L7545 |
| LysoTracker™ Red DND-99 | Invitrogen | L7528 |
| pHrodo™ Green conjugated dextran-10 kd | Invitrogen | P35368 |
| Oregon Green™ 488 conjugated dextran-10 kd | Invitrogen | D7170 |
| pHrodo™ Red AM | Invitrogen | P35372 |
| MitoTracker™ Green FM | Invitrogen | M7514 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma Aldrich | D9542; CAS: 28718-90-3 |
| Lipofectamine 2000 | Invitrogen | 11668-027 |
| DMEM medium without phenol red | Gibco | 21063029 |
| DMEM medium high glucose | Gibco | 11965 |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | Gibco | 12440 |
| Neurobasal™ Medium | Gibco | 21103049 |
| Fetal bovine serum (FBS) | Gemini Bio-Products | 100-106 |
| B-27™ Supplement (50X), serum free | Gibco | 17504044 |
| Trypsin-EDTA (0.05%), phenol red | Gibco | 25300054 |
| Puromycin Dihydrochloride | Gibco | A1113803 |
| Antibiotic-Antimycotic | Gibco | 15240062 |
| MycoAlertTM Mycoplasma Detection Kit | Lonza | LT07-218 |
| Poly-D-lysine solution | Millipore | A-003-E |
| Poly-L-lysine hydrobromide | Sigma Aldrich | P1399; CAS: 25988-63-0 |
| Vacuolin-1 | Sigma Aldrich | 673000; CAS: 351986-85-1 |
| 4-[(2-Butyl-6,7-dichloro-2-cyclopentyl-2,3-dihydro-1-oxo-1H-inden-5-yl)oxy]butanoic acid (DCPIB) | Tocris Bioscience | 1540; CAS: 82749-70-0 |
| 2,7-Dichloro-9,10-dihydro-9,9-dimethyl-10-[2-(2H-tetrazol-5-yl)ethyl]acridine (ML 67-33) | Tocris Bioscience | 6886; CAS: 1443290-89-8 |
| Bafilomycin-A1 | Cayman Chemical | 11038; CAS: 88899-55-2 |
| Arachidonic acid-sodium | Cayman Chemical | 10006607; CAS: 6610-25-9 |
| Arachidonic acid | Cayman Chemical | 90010; CAS: 506-32-1 |
| SC-79 | Cayman Chemical | 14972; CAS: 305834-79-1 |
| MK-2206 | Cayman Chemical | 11593; CAS: 1032350-13-2 |
| ARQ-092 | Cayman Chemical | 21388; CAS: 1313881-70-7 |
| Wortmannin | Cayman Chemical | 10010591; CAS: 19545-26-7 |
| Latrunculin B from Latruncula magnifica | Sigma Aldrich | L5288; CAS: 76343-94-7 |
| PI(3,5)P2 diC8 | Echelon Biosciences | P-3508 |
| ML-SA5 | This paper | N. A. |
| isoflurane (Fluriso™) | VetOne | 501017 |
| Tissue-Tek® O.C.T. (Optimal Cutting Temperature) compound | Sakura Finetek | 4583 |
| Recombinant mouse WT α-synuclein preformed fibril (5 mg/mL) | a gift from Dr. Kelvin C. Luk, the University of Pennsylvania Perelman School of Medicine | N. A. |
| Critical Commercial Assays | ||
| QuikChange II Site-Directed Mutagenesis Kit | Agilent | 200523 |
| Gibson Assembly Cloning Kit | New England Biolabs | E5510S |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | M0491S |
| GoTaq™ master mixes | Promega | M7122 |
| Experimental Models: Cell Lines | ||
| Monkey: COS1 | ATCC | CRL-1650, RRID: CVCL_0223 |
| Human: HEK293T cells | ATCC | CRL-3216; RRID: CVCL_0063 |
| Human: HeLa cells | ATCC | CCL-2; RRID: CVCL_0030 |
| Human: HAP1 cells | Horizon Discovery Ltd. | C631; RRID: CVCL_Y019 |
| Human: TMEM175 KO HAP1 cells | This paper | N. A. |
| Human: TMEM175-HA Knock-in HAP1 cells | This paper | N. A. |
| Human: TPC1&TPC2 DKO HAP1 cells | This paper | N. A. |
| Human: LRRC8A KO HAP1 cells | (Li et al., 2020b) | N. A. |
| Human: TMEM175 KO HEK293T cells | This paper | N. A. |
| Human: HEK293T cells stably overexpressing human TMEM175 (L1&L2) | This paper | N. A. |
| Human: TMEM175 KO HeLa cells (KO-1&KO-2) | This paper | N. A. |
| Human: TMEM175 KO HeLa cells stably re-expressing human TMEM175 | This paper | N. A. |
| Human: TMEM175 KO HeLa cells stably re-expressing human TMEM175-D41A | This paper | N. A. |
| Human: TRPML1-KO fibroblasts | Coriell Institute for Medical Research | GM02048; RRID: CVCL_M927 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tmem175 −/− / C57BL/6N | This paper | N. A. |
| Oligonucleotides | ||
| sgRNA targeting sequence: hTMEM175 (#1): 5’- CGCGTGGAAGCCTTCAGCGA -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: hTMEM175 (#2): 5’- TGTCCTCCCTGGTCCATCTC -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: hTMEM175 (#3): 5’- CGGCCGCCAAACCACACCGA -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: mTmem175 (#1): 5’- GCACTAATACTCTTAGCCTG -3’ | http://crispr.mit.edu | N. A. |
| sgRNA targeting sequence: mTmem175 (#2): 5’- GGACCCCTTACAATATGCCT -3’ | http://crispr.mit.edu | N. A. |
| Genotyping primer: 5’- CATTCCTGTTGCTCCGTCTCCTTGT -3’ and 5’- CGCCAGGGAAGCTCTGTACTTTTGA -3’ | This paper | N. A. |
| for Tmem175−/− mice | ||
| Genotyping primer: 5’- TGCTATGCTGAGCTTATAGCCTTG -3’ and 5’- AACGTTCCAAGAAGGAGGATCGCC -3’ for Tmem175−/− mice | This paper | N. A. |
| Recombinant DNA | ||
| pEZ-M55-mCherry-hTMEM175 | GeneCopoeia | EX-V0772-M55 |
| pEZ-M29-EGFP-hTMEM175 | GeneCopoeia | EX-V0772-M29 |
| pEZ-M97-hOTOP1-EGFP | GeneCopoeia | EX-E1341-M97 |
| pSpCas9 (BB)-2A-puro vector | Addgene | 48139 |
| pHluorin (in modified pEYFP-N1) | This paper | N. A. |
| Software and Algorithms | ||
| MetaMorph 7.10 | Molecular Devices | https://www.moleculardevices.com/ |
| pClamp 10.7 | Molecular Devices | https://www.moleculardevices.com/ |
| OriginPro 2018 | Originlab | https://www.originlab.com/ |
| GraphPad Prism 8 | Graphpad | https://www.graphpad.com/ |
| Fiji/ImageJ | Fiji contributors | https://imagej.net/Fiji |
| Image Studio lite | Li-cor | https://www.licor.com/ |


