Abstract
Background:
Perfluorooctane sulfonate (PFOS) is a persistent organic pollutant that can cause hepatotoxicity. The underlying toxicological mechanism remains to be investigated. Given the critical role of fecal microbiota in liver function, it is possible that fecal microbiota may contribute to the liver toxicity induced by PFOS.
Objectives:
We aimed to investigate the role of liver-fecal microbiota axis in modulating PFOS-induced liver injury in mice.
Methods:
Male and female mice were exposed to PFOS or vehicle for 14 d. In this investigation, 16S rDNA sequencing and metabolomic profiling were performed to identify the perturbed fecal microbiota and altered metabolites with PFOS exposure. In addition, antibiotic treatment, fecal microbiota transplantation, and bacterial administration were conducted to validate the causal role of fecal microbiota in mediating PFOS-induced liver injury and explore the potential underlying mechanisms.
Results:
Both male and female mice exposed to PFOS exhibited liver inflammation and steatosis, which were accompanied by fecal microbiota dysbiosis and the disturbance of amino acid metabolism in comparison with control groups. The hepatic lesions were fecal microbiota-dependent, as supported by antibiotic treatment and fecal microbiota transplantation. Mice with altered fecal microbiota in antibiotic treatment or fecal microbiota transplantation experiments exhibited altered arginine concentrations in the liver and feces. Notably, we observed sex-specific lower levels of key microbiota, including Lactobacillus, Enterococcus, and Akkermansia. Mice treated with specific bacteria showed lower arginine levels and lower expression of the phosphorylated mTOR and P70S6K, suggesting lower activity of the related pathway and mitigation of the pathological differences observed in PFOS-exposed mice.
Conclusions:
Our study demonstrated the critical role of the fecal microbiota in PFOS-induced liver injury in mice. We also identified several critical bacteria that could protect against liver injury induced by PFOS in male and female mice. Our present research provided novel insights into the mechanism of PFOS-induced liver injury in mice. https://doi.org/10.1289/EHP10281
Introduction
Perfluorooctane sulfonate (PFOS) is a ubiquitous environmental pollutant, which has accumulated excessively among the higher trophic level of the food chain through biological concentration and magnification. It has a long elimination half-life1 and distributes ubiquitously in wildlife2,3 and humans.4,5 It is important to note that PFOS has been commonly detected in cord blood6 and human breast milk,7 indicating the potential adverse effect of maternal PFOS exposure in offspring.8,9 In 2009, PFOS was listed as one of the nine new persistent organic pollutants in the Stockholm Convention.10
The widespread contamination and the persistence of PFOS have raised concerns about its possible side effects. Experimental studies have revealed numerous toxicities of PFOS in mice11–14 and in rats.14–16 The liver is the major target of PFOS toxicity in mice.11,17 PFOS was reported to disturb the expressions of hepatic genes involved in fatty acid metabolism, as well as hepatic fatty acid and cholesterol contents in mice17 and in fetal rats.18 A cross-sectional study demonstrated that concurrent exposure to PFOS was associated with increased risk for nonalcoholic steatohepatitis and higher plasma levels of metabolites in children diagnosed with nonalcoholic fatty liver disease (NAFLD).19 Stratakis et al. also found that prenatal exposure to PFOS could contribute to liver injury in childhood.20 Furthermore, previous research indicated that males showed more severe histological hepatic changes than females in PFOS-exposed zebrafish, which was likely related to the higher excretion rate of females.21 Over the last few decades, numerous studies have revealed gut microbiota as critical regulators in liver functions.22–24 Research found that restored gut homeostasis might alleviate hepatic steatosis and inflammation in mice.25 Moreover, it was highlighted that gut microbiota had an impact on host metabolic disorders related to energy metabolism and bile acid diversity in mice,26,27 indicating the key role of gut microbiota metabolites in host physiology. Recently, there have been reports indicating the alterations of gut microbiota and host metabolism by PFOS exposure in mice.28,29 However, the causal relationship has not been directly investigated, and the detailed contributions of gut microbiota to the liver injury induced by PFOS remain elusive.
In this study, we aimed to investigate the potential mechanisms of hepatotoxicity induced by PFOS in mice from the perspective of fecal microbiota. In this investigation, 16S rDNA-based microbiota analysis, metabolomic profiling, antibiotic (ABX) treatment, fecal microbiota transplantation (FMT) experiments, and bacterial administration were conducted to reveal a potential mechanistic link among the sex-specific microbiota signature, metabolic homeostasis, and mammalian target of rapamycin (mTOR)-P70S6K pathway in the modulation of PFOS-induced liver injury.
Methods
Chemicals and Reagents
The alanine aminotransferase (ALT; C009-2-1, microplate test), aspartate aminotransferase (AST; C010-2-1, microplate test), and alkaline phosphatase (ALP; a059-2-2, microplate test) assay kits were purchased from the Nanjing Jiancheng Bioengineering Institute. Vancomycin was purchased from MedChemExpress. Neomycin was purchased from Alfa Aesar, and 4-Chloro-phenylalanine (4-Cl-Phe), metronidazole, ampicillin, and PFOS (Product No. 77282, potassium salt) were purchased from Sigma Aldrich. L. reuteri (BNCC192190), E. faecalis (BNCC186075), and Akk. muciniphila (DSM22959) were purchased from BeNa Culture Collection. De Man, Rogosa and Sharpe (MRS) broth, and brain heart infusion (BHI) broth were obtained from Qingdao Hope Bio-Technology Co., Ltd. Porcine mucin was purchased from Yuanye Bio-Technology. Cysteine was purchased from Aladdin. Antibody for p-mTOR (Ser2448, AF3308) was purchased from Affinity. Antibody for p-P70S6K (Thr389, Thr412, PA5-104,842) was purchased from Thermo Fisher Scientific. Antibodies for mTOR (A11354) and P70S6K (A16658) were purchased from ABclonal. HRP-Polymer anti-Rabbit antibody (MaxVision, KIT5005) was purchased from MXB Biotechnologies.
Hepatotoxicity Experiment
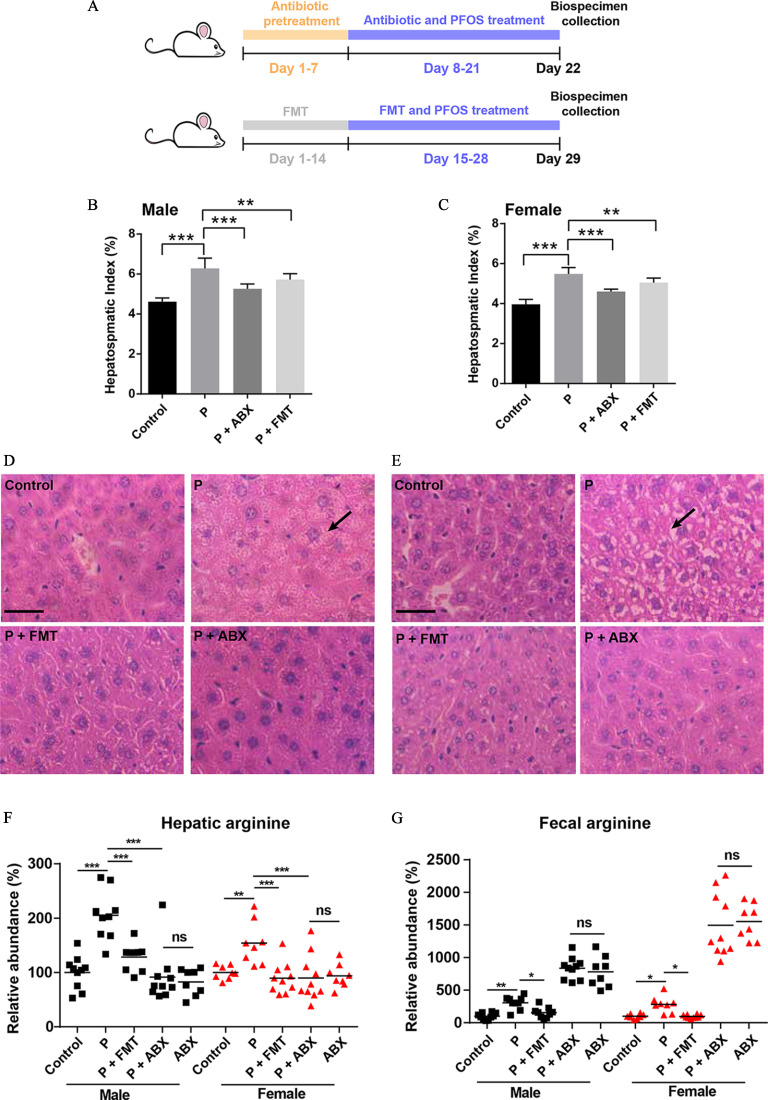
Male and female specific pathogen-free (SPF) ICR mice, 4–5 wk of age (Chinese University of Hong Kong) were used. All experiments were conducted according to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health, and the procedures were approved by the Hong Kong Department of Health. Mice were treated humanely and were housed in the Hong Kong Baptist University facility for a week before the start of experimentation, where they were allowed to consume tap water ad libitum. At the start of experimentation, mice were randomly assigned to either a control or PFOS-treated group ( for each group of male and female mice). PFOS was dissolved in 2% Tween 80 and given via daily oral gavage to mice once daily for 14 d at doses of , , and . Control mice received 2% Tween 80 without PFOS. The administered doses were selected to induce liver injury without causing death, based on a previous study, with minor modification.17 Twenty-four hours after the last dose, mice were anesthetized by isoflurane, and biological samples were then collected. The mice were euthanized by cervical dislocation. Blood was collected via eyeball enucleation. Serum samples were collected by centrifugation at for 15 min at 4°C. The livers were then collected, washed with phosphate buffered saline (PBS) and blotted with filter paper. The hepatosomatic index was calculated as liver weight (g) per gram of body weight, multiplied by 100. Fresh feces were collected during necropsy and rapidly quenched in liquid nitrogen and then stored at until further analysis. The experimental design for this study is shown in Figure 1A.
Figure 1.
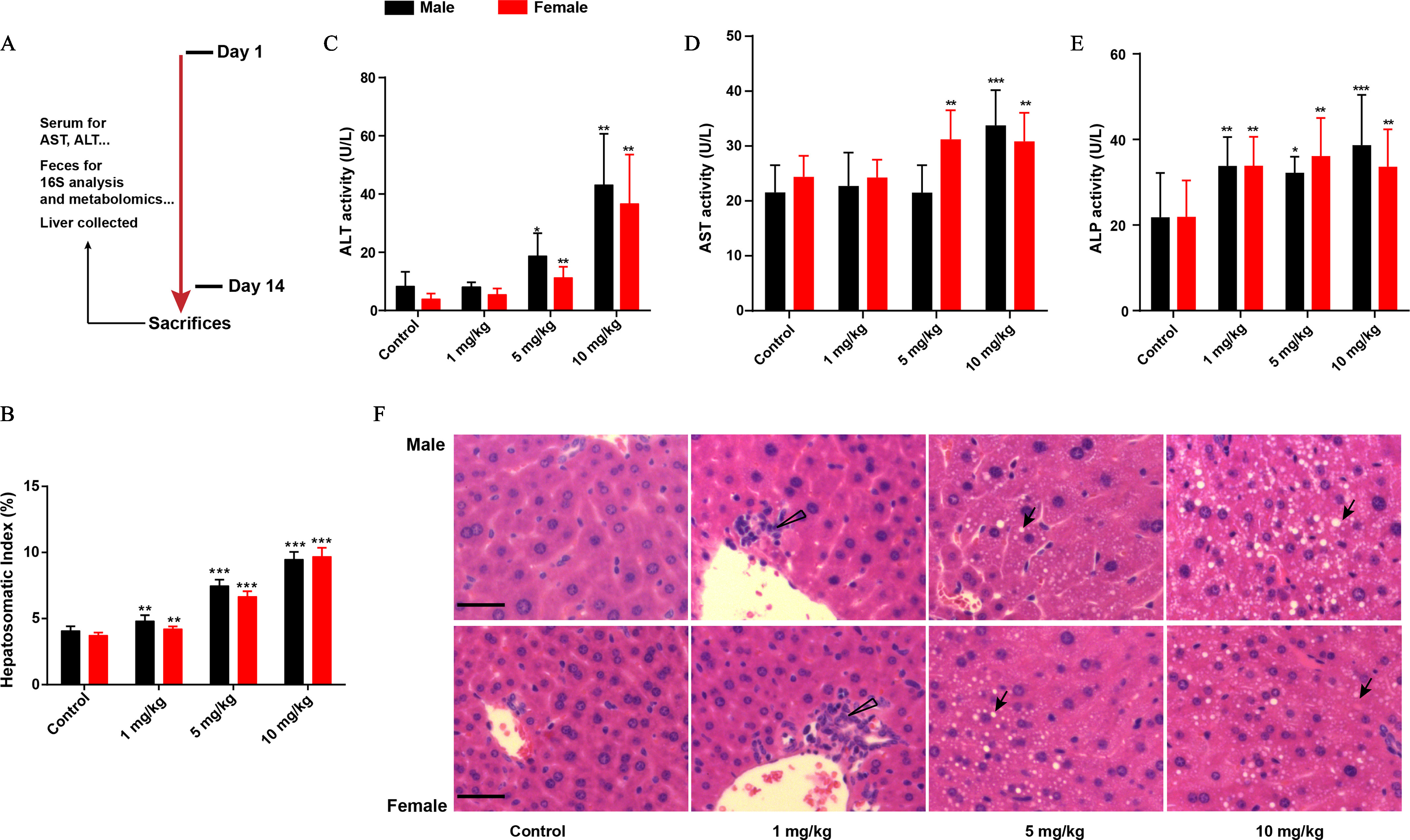
Evaluation of liver function in male and female mice with PFOS exposure. (A) Mice were administered 0, , , or PFOS for 2 wk. The mice were finally sacrificed and subjected to various analyses as indicated below. (B) Hepatosomatic index of mice treated with or without PFOS. Liver enzyme activities (U/L) of (C) ALT, (D) AST, and (E) ALP. Histopathology of liver tissues (F, upper: male mice, lower: female mice, from left to right are control, , , and PFOS). Hepatosomatic index was calculated as liver weight (g) divided by body weight (g) multiplied by 100. Results were presented as the , . . Summary data can be found in Table S2. Statistical significance was analyzed by one-way ANOVA among multiple groups. Note: ALP, alkaline phosphatase; ALT, alanine aminotransferase; ANOVA, analysis of variance; arrow, steatosis; AST, aspartate aminotransferase; PFOS, perfluorooctane sulfonate; SD, standard deviation; triangle, inflammatory cells. *. **. *** in comparison with the control group.
Biochemical Analysis and Histopathologic Examination
The serum ALT activity, AST activity, and ALP activity were determined using a microplate reader (PerkinElmer 2030), according to the instructions of the manufacturers. Briefly, for ALT activity analysis, ALT reacted with alanine and and produced pyruvic acid. Pyruvic acid was then reacted with 2,4-dinitrophenylhydrazine (DNPH) to form phenylhydrazone, which could be dissolved in sodium hydroxide and then detected at . For AST activity analysis, AST catalyzed aspartate and to form oxalacetate and glutamate. Oxalacetate could decarboxylate automatically to pyruvic acid, which could react with DNPH and then be detected at . For ALP activity analysis, ALP catalyzed disodium phenyl phosphate to form phenol, which could react with 4-aminoantipyrine and potassium ferricyanide and could be detected at . For pathological analysis, the mice liver tissues were fixed in 10% neutral buffered formaldehyde solution for 24 h, paraffin-processed (Excelsior AS and HistoStar; Thermo Scientific) and sectioned at (Leica RM2125RTS). The sections were stained with hematoxylin-eosin (HE) and examined for histopathological changes under the microscope (Olympus DX45). The images were taken by the built-in software installed in the microscope.
16S rDNA Amplicon Sequencing
Total DNA was extracted from fecal pellets using E.Z.N.A. Bacterial DNA Kit (Omega Bio-Tek) according to the manufacturer’s instructions. The DNA was quantified by Qubit 2.0 (Invitrogen, Thermo Scientific) and stored at until analysis. DNA was amplified using primers of 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) (Invitrogen, Thermo Scientific) to target the V4 regions of 16S rDNA of bacteria. Samples were sequenced on the Illumina Hiseq2500 platform producing 250-bp paired end reads (Illumina). The reads with quality scores of , the contaminated reads, the reads with ambiguous bases and low complexity (Reads with 10 consecutive same base) were removed. The operational taxonomic unit (OTU) was clustered using USEARCH at a threshold of 97% similarity. Representative sequence for each OTU was assigned to Greengenes Database (V201305, gg_13_5) using RDP Classifier (v2.2). Quantitative Insights into Microbial Ecology (QIIME) and Mothur were used for analysis including alpha and beta diversities. The taxonomic assignment was achieved at different levels, including phylum, class, order, family, genus, and species. Sequencing and quality control was done by BGI Co., Ltd.
Metabolomic Profiling of Liver and Fecal Samples
A total of of liver tissue was homogenized with of 80% methanol by using Bullet Blender (Next Advance). For feces, of fecal sample was homogenized with of 80% methanol, then ultrasonicated in ice water for 30 min and vortex-mixed for 2 min. All the extracts were centrifuged at for 10 min at 4°C, and the supernatant was then dried using an IR concentrator (N-BIOTEK). The dried supernatant was redissolved with of 50% methanol for metabolomic profiling; 4-Cl-Phe () was used as internal standard (IS).
Analysis was performed using an Ultimate 3000 rapid separation liquid chromatography (LC) system, coupled with a Q Exactive Focus Orbitrap MS (mass spectrometry) (Thermo Scientific). Full scan mode was applied in positive and negative ion modes with ESI source. Chromatographic separation was achieved on an ultra-performance liquid chromatography (UPLC) HSS T3 column (, ; Waters). The mobile phase consisted of 0.1% formic acid-water (v/v; A) and 0.1% formic acid-acetonitrile (v/v; B) at the flow rate of . The liquid chromatographic gradient was programmed as follows: 0 min, 2% B; 1 min, 2% B; 19 min, 100% B; 21 min, 100% B; 21.1–25 min, 2% B. The MS parameters were set as reported by our previous publication.30
Targeted Amino Acid Analysis
Amino acids in the liver and feces were analyzed using Ultimate 3000 rapid separation LC coupled with TSQ Quantiva triplequadrupole MS (QqQ, Thermo Scientific). Chromatographic separation was performed on an Xbridge BEH Amide column (, ; Waters). Liver and fecal samples were processed using the methods as mentioned in metabolic profiling. An aliquot () of the final supernatant was subjected for liquid chromatography–mass spectrometry (LC-MS) analysis, with 4-Cl-Phe () to be the IS. The mobile phase consisted of 0.1% formic acid–water (v/v) containing ammonium acetate (A) and acetonitrile (B) at the flow rate of . The liquid chromatographic gradient was programmed as follows: 0 min, 85% B; 12 min, 55% B; 13 min, 30% B; 14 min, 30% B; 14.5–18 min, 85% B. The MS parameters were set as previously published.31
ABX and FMT Experiments
To ablate the fecal microbiota, mice were gavaged with a solution of vancomycin (), neomycin (), and metronidazole () twice daily for 7 d in advance ( for each group in male and female mice). The dosages of the antibiotics were selected based on the literature.32 Ampicillin () was provided ad libitum in drinking water. With regard to the selection of PFOS dosage for fecal microbiota transplantation and antibiotic treatment studies, preexperiments were conducted to establish a dose that resulted in moderate liver injury in mice. One to three mice each were exposed to either 2 or PFOS via daily oral gavage once daily for 14 d. Twenty-four hours after the last dose, the liver was collected, and the hepatosomatic index was calculated to evaluate the hepatotoxicity. Although PFOS dosage induced severe liver injury in mice (Figure 1B and Table S2; hepatosomatic index 6.7%–7.5%), dosage exposure (14 d) induced moderate liver injury (Table S1; hepatosomatic index 4.9%–5.6%) and was selected for further study. The mice were treated with PFOS in the presence of antibiotics for 2 wk from the eighth day. In fecal transfer experiments, 5–6 fecal pellets were collected from healthy mice (consumed water and feed ad libitum without any treatment) and resuspended in PBS. The mixtures were vortexed and then centrifuged at for 1 min at 4°C. A supernatant was used for orogastric gavage 2 wk in advance. The mice were then exposed to PFOS with fecal supernatant for 2 wk from the 15th day. The experimental design for the antibiotic and fecal transfer studies is shown in Figure 4A.
Figure 4.
The effects of ABX treatment and FMT on PFOS-induced liver injury. (A) Experimental design: Mice were orally administered solvent with ABX for 7 d, followed by the addition of PFOS and ABX for days 8–21. The FMT group was fed fresh feces from an untreated mouse once daily for 14 d, followed by the addition of PFOS and fresh feces for days 15–28. (B) and (C) Hepatosomatic indices of various groups in male (B) and female (C) mice. (D) Histopathology of liver tissues in male mice. (E) Histopathology of liver tissues in female mice. (F) and (G) The relative abundance of arginine in livers (F) and feces (G) by LC/MS analysis in the indicated groups. Results were presented as the . Hepatosomatic index was calculated as liver weight (g) divided by body weight (g) multiplied by 100. The relative abundance was calculated by the ratio between the level of metabolite in exposure group and the average level of control group. . Summary data can be found in Table S5. Statistical significance was analyzed by one-way ANOVA among multiple groups. . Note: ABX, antibiotic treatment; ANOVA, analysis of variance; arrow, steatosis; FMT, fecal microbiota transplantation; LC/MS, liquid chromatography–mass spectrometry; ns, not significant; P and PFOS, perfluorooctane sulfonate; SD, standard deviation. *. **. *** in comparison with the indicated group.
Mouse Intervention Study with Specific Species
L. reuteri was cultured anaerobically in MRS broth. E. faecalis were cultured aerobically in MRS broth. Akk. muciniphila was cultured anaerobically in BHI broth supplemented with 0.05% porcine mucin and 0.05% cysteine. L. reuteri, E. faecalis, and Akk. muciniphila were cultured for 2, 4, and 7 d, respectively. For bacteria identification, the bacterial suspension was amplified by Phanta Max Super-Fidelity DNA Polymerase (Product No. P505;Vazyme Biotech Co. Ltd.) with the universal primers (27F: AGAGTTTGATCMTGGCTCAG and 1492R: GGTTACCTTGTTACGACTT; Sangon Biotech), according to the instruction. The reaction was programmed using a 2720 Thermal Cycler as follows: 95°C for 3 min; 35 cycles of 95°C for 15 s; 55°C for 15 s, followed by 72°C for 60 s; and 72°C for 5 min. The product was then sequenced by 3730XL [Applied Biosystems, BGI Health (HK) Co. Ltd., or Sangon Biotech], and the result sequences were identified with the BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, 2.13.0). The bacteria were quantified by plate count method. The bacteria were diluted , , and times with PBS after cultivation. A total of of suspension was then cultured on MRS or BHI agar plates (L. reuteri and Akk. muciniphila: anaerobic environment, 37°C, 5% CO2; E. faecalis: aerobic environment, 37°C). The number of bacteria were counted when the obvious colonies appeared. The experiments were repeated three times. For bacterial administration, at least colony-forming units (CFUs) of bacteria were administered to mice via oral gavage ( for each group in male and female mice). Mice were treated with bacteria once daily for 21 d in advance; then the mice were exposed to PFOS with bacteria for 2 wk from the 22nd day. In sham-controlled trials, bacteria were heat-killed at 121°C under high pressure (0.1 MPa) for 20 min (Hirayama autoclave, HV-50). The experimental design for this study can be found in Figure 5B.
Figure 5.
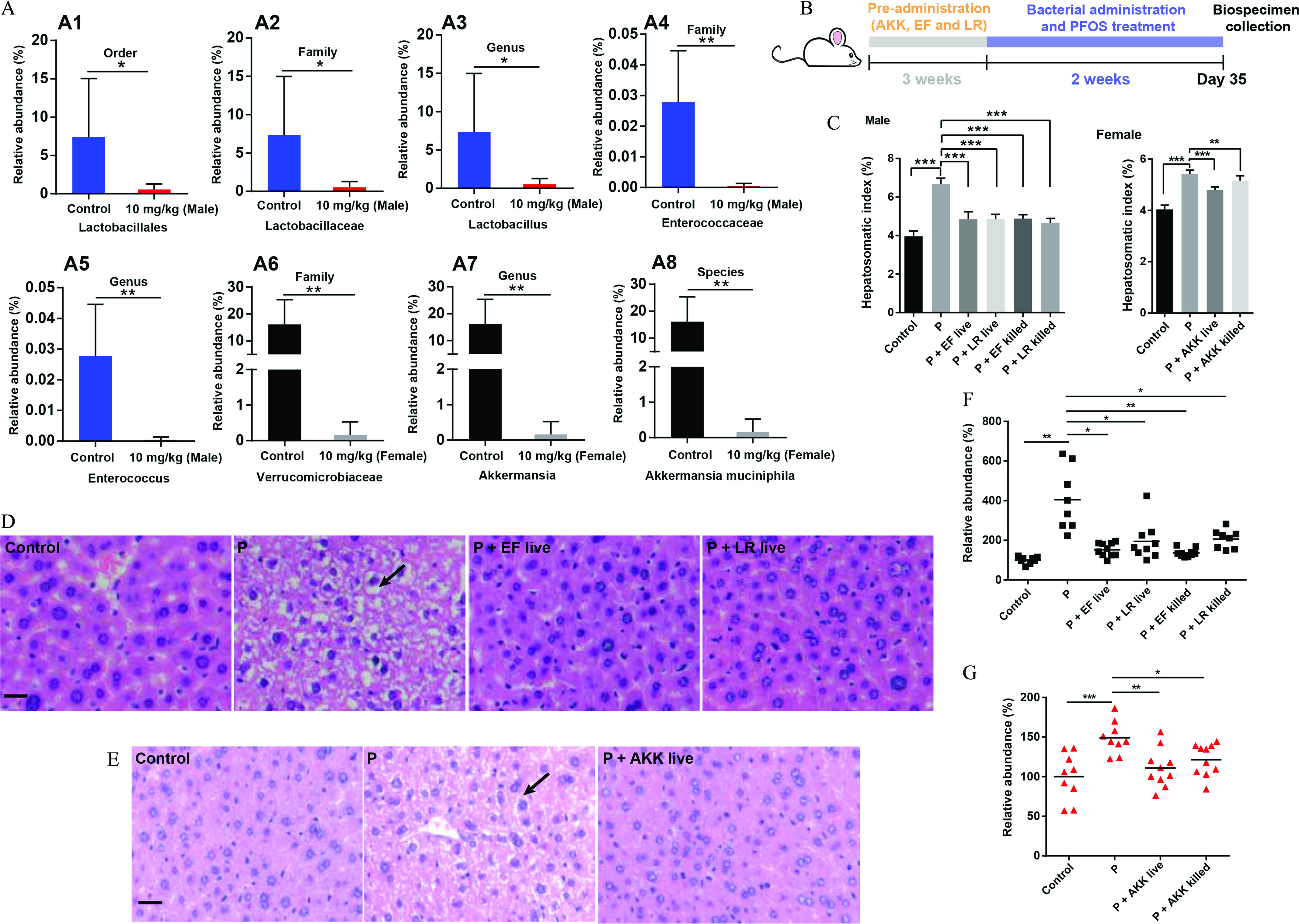
The effects of bacterial administration on PFOS-induced liver injury. (A) Comparison proportion of order, family, genus, and species levels of specific bacteria in feces detected by sequencing analysis. (B) Experimental design: Mice were colonized specific bacteria for 3 wk and subsequently orally administered with PFOS and specific bacteria for an additional 2 wk. (C) Hepatosomatic indices of various groups in male (left) and female (right) mice. (D) Histopathology of liver tissues in male mice. (E) Histopathology of liver tissues in female mice. (F) The relative abundance of arginine in livers of male mice by LC/MS analysis. (G) The relative abundance of arginine in livers of female mice by LC/MS analysis. Hepatosomatic index was calculated as liver weight (g) divided by body weight (g) multiplied by 100. The relative abundance was calculated by the ratio between the level of metabolite in exposure group and the average level of control group. . Summary data can be found in Table S6. Statistical significance was analyzed by one-way ANOVA among multiple groups. . Results were presented as the . Note: ANOVA, analysis of variance; AKK, Akk. muciniphila; arrow, steatosis; EF, E. faecalis; F, female; F1, control group; F4, . LR, L. reuteri; M, male, M1, control group; M4, ; P and PFOS, perfluorooctane sulfonate; SD, standard deviation. *. **. *** in comparison with the indicated group.
Quantification of the Abundance of Bacteria in Feces
The fecal samples were collected during necropsy at the end of the experiment and frozen at until analysis. Fecal DNA was extracted using the TIANamp Stool DNA Kit [DP 328, TIANGEN Biotech (Beijing) Co. Ltd.] according to the manufacturer’s instructions. The DNA concentration was then measured by a Nano-100 micro-spectrophotometer (Hangzhou AllSheng Instruments Co. Ltd.). Quantitative real-time polymerase chain reaction (qPCR) reactions were performed using the PIKOREAL 96 Real-Time PCR System (Thermo Fisher Scientific) with the TB Green Premix Ex Taq (Takara Biotechnology), to determine the amounts of bacteria. The primers sequences used to amplify the bacteria are described as follows () [BGI Tech Solutions (Beijing Liuhe) Co., Ltd.]:
Akk. muciniphila: Forward: CAGCACGTGAAGGTGGGGAC
Reverse: CCTTGCGGTTGGCTTCAGAT;
E. faecalis: Forward: AGAAATTCCAAACGAACTTG
Reverse: CAGTGCTCTACCTCCATCATT;
L. reuteri: Forward: GCGTTGATGTTGTTGAAGGAATGAGCTTTG
Reverse: CATCAGCAATGATTAAGAGAGCACGGCC;
Universal: Forward: AAACTCAAAKGAATTGACGG
Reverse: CTCACRRCACGAGCTGAC.
A previously described method33 was used to quantify the total bacterial DNA in fecal samples using 16S primers (338F: ACTCCTACGGGAGGCAGCAG, 518R: ATTACCGCGGCTGCTGG) [BGI Tech Solutions (Beijing Liuhe) Co., Ltd.], with minor modification. The quantification was also conducted using the PIKOREAL 96 Real-Time PCR System with the TB Green Premix Ex Taq. The standard curve was plotted with E. coli DNA [Sangon Biotech (Shanghai) Co., Ltd.]. The concentration of total bacterial DNA was presented as lg (copy number) per milligram feces. The reaction was programmed as follows34: 95°C for 30 s; 50 cycles of 95°C for 5 s; 60°C for 35 s, followed by 95°C for 15 s; 60°C for 60 s; and 95°C for 15 s.
Immunohistochemistry
Paraffin-embedded liver sections were deparaffinized and rehydrated, and antigen retrieval was performed using antigen retrieved buffer (KGIHC001, KeyGEN BioTECH). Then the sections were blocked with 1% bovine serum albumin (BSA) for 20 min, incubated with p-mTOR (1:50), p-P70S6K (1:100), mTOR (1:200), and P70S6K (1:200) at 4°C overnight, followed by incubation with HRP-Polymer anti-rabbit antibody (1:200) for 20 min at room temperature. After washing, samples were incubated in diaminobenzidine and counterstained in hematoxylin. The images were visualized by a digital pathological section scanner (NanoZoomer 2.0 RS, Hamamatsu) and were quantified using Image J.
Statistical Analysis
The results of biological assays are presented as . The differences among the groups were analyzed using Student’s t-test (two groups) or one-way analysis of variance (ANOVA) (more than two groups). Statistical analysis was performed using SPSS (version 20; IBM) software. All tests were two-sided and were considered statistically significant at . Partial least squares discrimination analysis (PLS-DA) was performed to identify the discrimination of variables in metabolomic analysis. Differential metabolites were defined as those with variable importance in the projection , fold change , and .
Results
Evaluation of Liver Function in Male and Female Mice with PFOS Exposure
We first addressed the interaction between PFOS treatment and liver injury (Figure 1A). In comparison with the control group, PFOS-exposed male and female mice had significantly higher hepatosomatic indices in a dose-dependent manner (Figure 1B). The impacts of PFOS on ALT, AST, and ALP activities were further assessed using the corresponding kits. As can be seen in Figure 1C–E, male and female mice treated with PFOS had higher serum ALT activities in a dose-dependent manner, with significant alterations at the dosages of and in comparison with the control groups (Figure 1C). In contrast to the control group, male mice treated with had significantly higher AST activities. Similarly, in female mice, PFOS exposure at the dosages of 5 and resulted in higher activities of AST in serum (Figure 1D). Additionally, significantly higher ALP activities were observed in PFOS-treated male and female mice at 1, 5, and when compared with the control groups (Figure 1E). The histopathologic results (Figure 1F) suggested that male and female mice exposed to PFOS exhibited inflammatory cell infiltration at the dose of , and a dose-dependent steatosis necrosis could be found in livers of mice with 5 and dosages.
Exploration of Fecal Microbiota Composition in PFOS-Treated Mice
To test whether the fecal microbiota played a role in PFOS-mediated liver injury, we explored the composition of fecal microbiota by performing a 16S rDNA amplicon sequencing analysis of bacteria in feces. Principal component analysis (PCA) revealed a distinct clustering of microbiota composition between control and PFOS treatment groups in male mice (Figure 2A). Nevertheless, there was no significant difference in the observed OTUs between control and PFOS-treated male mice (Figure 2B). Similarly, in female mice, the fecal microbiota community structures of the PFOS-treated group were readily differentiated from those of the control group (Figure 2C). Higher alpha diversity was observed in PFOS-exposed female mice (Figure 2D), suggesting the enrichment of particular bacterial taxa with PFOS exposure. Using the linear discriminant analysis (LDA) effect size (LEfSe) pipeline, marked alterations in PFOS-exposed mice were observed in several taxa, such as Lactobacillaceae (g), Enterococcaceae (e), and Allobaculum (m) in male mice (Figure 2G; Figure S1A), Akkermansia (a1), Bifidobacterium (a), and Turicibacter (l) in female mice (Figure 2H; Figure S1B).
Figure 2.
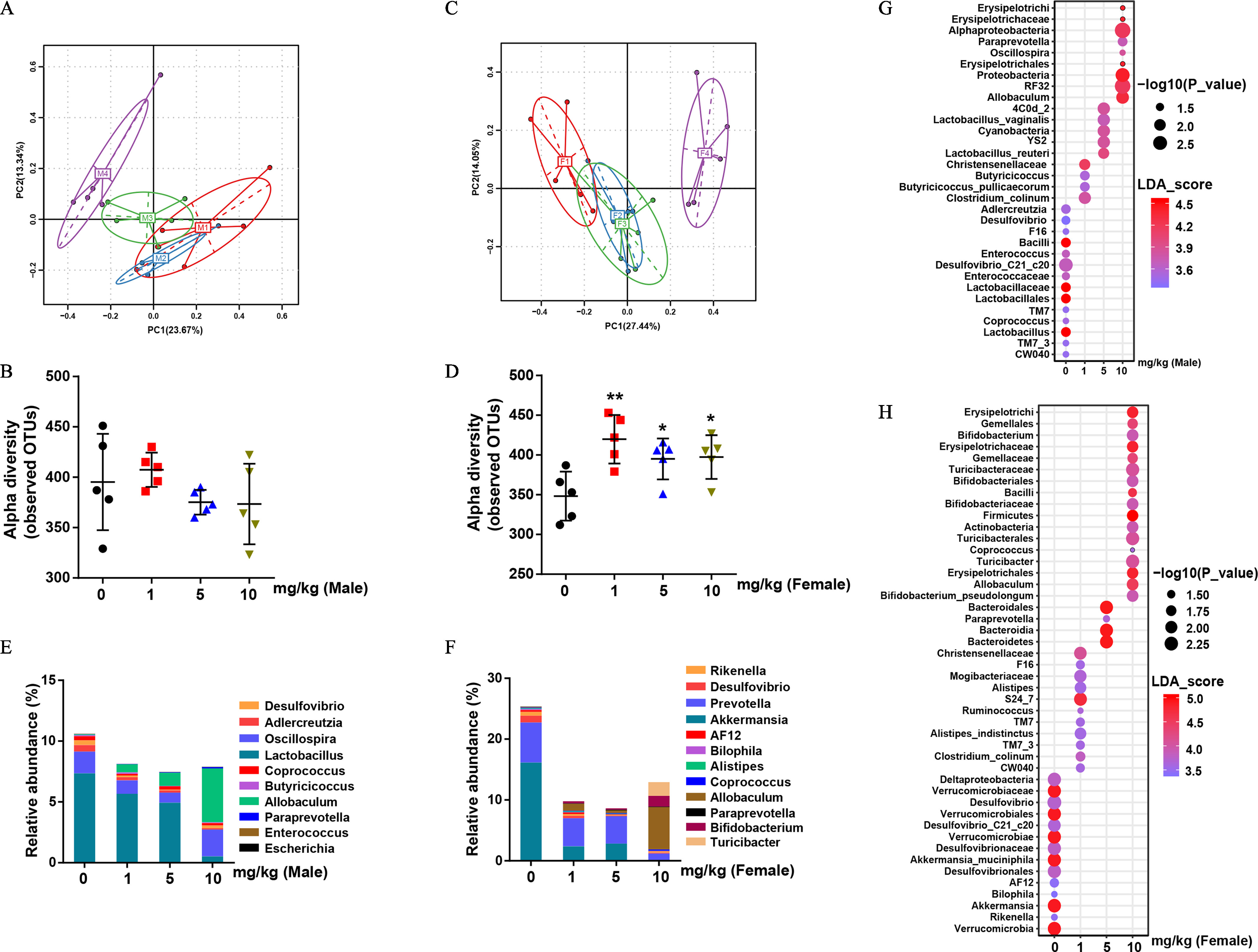
Bacterial 16S rDNA-based analysis of fecal microbiota of PFOS-exposed mice. (A) PCA plot based on bacterial 16S rDNA gene sequence abundance in the feces of male mice. (B) Alpha diversity of fecal 16S rDNA sequencing data from male mice. (C) PCA plot based on bacterial 16S rDNA gene sequence abundance in the feces of female mice. (D) Alpha diversity of fecal 16S rDNA sequencing data from female mice. (E) and (F) Taxonomic distributions of bacteria from fecal 16S rDNA sequencing data in male (E) and female (F) mice. The bubble plot of linear discriminant analysis (LDA) scores of bacterial 16S rDNA sequences in control and PFOS-exposed male (G) and female (H) mouse feces. ; results were presented as the . Summary data can be found in Table S3 (panels B, D, E, and F) and Excel Tables S1 and S2 (Panels G and H, respectively). Wilcoxon Rank-Sum test was used for the differential analysis between two groups. Kruskal-Wallis test was used for the differential analysis between multigroups. Note: F, female; F1–F4, control, , and , respectively; M, male mice; M1–M4, control, , and , respectively; PFOS, perfluorooctane sulfonate; SD, standard deviation. *. **. *** in comparison with the control group.
Specifically, we analyzed the bacterial abundance differences in various groups at the genus level. For example, male mice treated with PFOS had significantly lower abundances of bacteria genera Lactobacillus and Enterococcus by and and had significantly higher abundances of bacteria genera Allobaculum by (Figure 2E; Figure S1D). In female mice, PFOS-exposed mice had significantly lower abundances of Akkermansia by about 100-fold (Figure 2F; Figure S1D), whereas PFOS-exposed mice had significantly higher abundances of bacteria such as Bifidobacterium, Allobaculum and Turicibacter by , , and , respectively (Figure S1D), in comparison with the control mice. More important, PFOS had a sex-selective effect even for the same bacterial genus, as our results suggested that the fecal microbiota profiles were different between male and female mice, especially at the dosage of (Figure S1C). For instance, Akkermansia showed lower abundances only in female mice exposed to PFOS, Bifidobacterium showed higher abundances only in PFOS-treated female mice, and Lactobacillus and Enterococcus showed a significantly lower abundances only in PFOS-treated male mice in comparison with the control mice (Figure S1D).
Untargeted Metabolomic Profiling of Liver and Feces
Untargeted metabolomic analyses were performed on livers and feces to assess metabolic alternations arising from PFOS exposure. The PLS-DA indicated distinct separation of the metabolic profiles between control and PFOS-treated groups for livers and feces (Figure 3A,B; Figure S2A,C). Volcano plots also demonstrated the relative abundances of metabolites in the livers of male (Figure 3C) and female mice (Figure 3D) treated with vehicle or PFOS (). Specifically, our analysis uncovered 116 and 23 differential metabolites associated with PFOS exposure in livers and feces of male mice, respectively. On the other hand, 126 and 16 significantly differentially expressed metabolites were identified in liver and feces of the PFOS-exposed female mice, respectively. An interesting finding was that pathway analysis in livers indicated that the majority of metabolites that contribute to group discrimination were relevant to amino acid metabolism (Figure 3E,F), among which, arginine-related metabolism ranked as the most significant perturbed pathway in livers of both PFOS-exposed male and female mice (Figure 3G,H). In feces, the topology map analysis in male and female mice also revealed that arginine and proline metabolism was one of the most significant altered pathways induced by PFOS exposure (Figure S2B,D–F).
Figure 3.
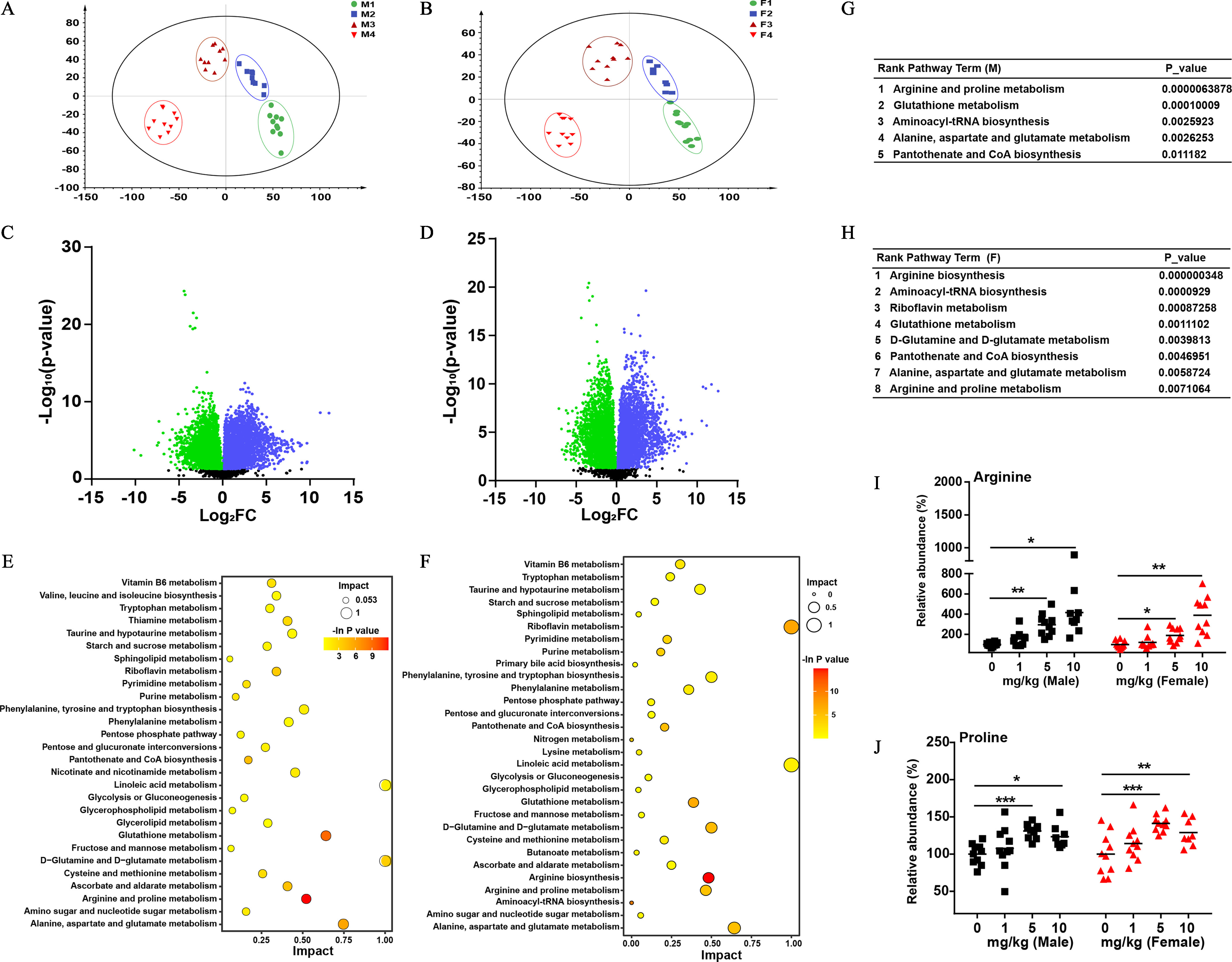
The effects of PFOS on the liver metabolic profiles of male and female mice. PLS-DA score plots for discriminating the metabolome in the livers of male (A) and female (B) mice from control and PFOS treatment groups. Volcano plots showing the relative abundances of metabolites in male (C) and female (D) mice ( dosage). Enrichment analysis of differential metabolites in male (E) and female (F) mice. Rank pathway term analysis of differentially expressed metabolites in male (G) and female (H) mice. Comparison of host concentrations of arginine (I) and proline (J) in livers by LC-MS/MS in the indicated groups. The relative abundance was calculated by the ratio between the level of metabolite in PFOS-exposed group and the average level of control group. . Horizontal lines in I and J represent the mean. Summary data can be found in Table S4 and Excel Tables S3 and S4. Statistical significance was analyzed by one-way ANOVA among multiple groups. Note: ANOVA, analysis of variance; F, female; F1–F4, control, , , and ; LC-MS/MS, liquid chromatography–tandem mass spectrometry; M, male mice; M1–M4, control, , , and ; PFOS, perfluorooctane sulfonate; PLS-DA, partial least squares discrimination analysis. *. **. *** in comparison with the control group.
Targeted Profiling of Amino Acids in the Liver and Feces
The targeted metabolomic profiling analysis of the relevant amino acid levels in the liver and feces from different groups was further conducted. PFOS-exposed mice had significantly higher arginine and proline levels in the liver in a dose-dependent manner when compared with control groups (Figure 3I,J). In feces, we also found a similar variation tendency of arginine levels, whereas the concentrations of proline were markedly lower in a dose-dependent manner in PFOS-treated mice than in control mice (Figure S2G,H), which might be the result of the synthesis of arginine from enteral proline.35
The Effects of ABX Treatment and FMT on PFOS-Induced Liver Injury in Mice
To clarify the contribution of fecal microbiota to PFOS-induced liver injury, we a) treated a cohort of mice with a cocktail of antibiotics to reduce the intestinal bacterial load and b) performed FMT experiments on a second cohort of mice whereby mice were treated orally with feces from a control mouse (Figure 4A). To determine the optimal dose of PFOS with which to treat mice to achieve moderate liver injury, we performed a set of preliminary experiments, treating mice with a range of PFOS doses and examining the resulting hepatosomatic indices. From these preliminary experiments, a dosage of PFOS was selected for further study (Table S1). We then evaluated the fecal bacterial DNA abundance using qPCR analysis. FMT restored fecal DNA to control levels after PFOS administration. ABX treatment with or without PFOS resulted in significantly depleted DNA levels in both male and female mice (Figure S3A and B). Both male and female mice treated with ABX and FMT with PFOS had significantly lower hepatosomatic indices in comparison with the mice treated with PFOS alone (Figure 4B,C). In FMT experiments, the inflammatory cell infiltration and steatosis necrosis induced by PFOS were relieved in male and female mice (Figure 4D,E). Consistently, targeted metabolomic profiling of liver arginine revealed higher arginine concentration in PFOS-exposed mice was blocked by FMT (Figure 4F). However, PFOS-exposed male and female mice had significantly lower proline concentrations unexpectedly in the liver (Figure S3C), whereas and PFOS-exposed male and female mice had significantly higher liver proline levels when compared with control groups in the hepatotoxicity experiment (Figure 3J). The concentrations of arginine in feces were further determined. We found that the arginine levels in feces and liver showed the same trend, and arginine concentrations in mice treated with PFOS and transferred feces were significantly lower than in mice treated with PFOS alone (Figure 4G). However, with ABX treatment, the impacts of PFOS on arginine levels were abolished in the liver and feces (Figure 4F,G), and the treatment of ABX partly reversed the PFOS-induced liver injury (Figure 4D,E).
The Effects of Specific Bacteria on PFOS-Induced Liver Injury in Mice
The associations between the alterations in the fecal microbiota, arginine concentrations, and PFOS-induced liver injury have already been described above. Akkermansia in female mice and Enterococcus and Lactobacillus in male mice were identified as key sex-specific genus responding to PFOS exposure as shown in Figure 2E,F; Figure S1D. Interestingly, we found that PFOS-treated male mice had significantly lower abundances of L. reuteri at the order-family-genus-species levels (Figure 5A1–3; Figure S4A). PFOS-exposed male mice had a significantly lower abundance of Enterococcus at the order-family-genus levels in comparison with control mice (Figure 5A1,4,5). PFOS-exposed female mice had a significantly lower abundance of Akk. muciniphila at the family-genus-species levels, in comparison with the control group (Figure 5A6–8). Furthermore, we confirmed the lower abundances of L. reuteri, E. faecalis, and Akk. muciniphila in respective PFOS-treated mice by qPCR analysis (Figure S4B–D).
To further investigate the relationship between these three bacteria and PFOS-induced liver injury, mice were gavaged once daily with commercial strains of L. reuteri, E. faecalis, and Akk. muciniphila for 5 wk (Figure 5B). Notably, we found that live bacteria-treated mice had significantly lower hepatosomatic indices when compared with the mice treated with PFOS only (Figure 5C). Consistently, histological analysis revealed that the liver injury induced by PFOS in the live bacteria-treated mice was alleviated (Figure 5D,E). In addition, we detected significantly lower abundance of hepatic arginine in mice gavaged with specific live bacteria when compared with the mice treated with PFOS alone (Figure 5F,G). Similar results were also obtained in the fecal samples (Figure S4E,F). Furthermore, heat-killed bacteria also alleviated PFOS-induced liver injury, as demonstrated by the lower hepatic indexes, relieved pathological changes of liver, and the recovery of arginine concentrations (Figure 5C,F,G; Figure S4G–I).
The Impact of PFOS Exposure on the Expressions of mTOR and P70S6K
mTOR plays an important role in liver functions.36,37 We next tested the effects of PFOS on mTOR-P70S6K pathway. With a comparative analysis of immunohistochemistry, we observed that PFOS-exposed male and female mice had a significantly higher expression of the phosphorylation of mTOR, as well as its downstream target P70S6K, in comparison with the control mice (Figure S5A,B). An important finding was that male mice treated with L. reuteri/E. faecalis and female mice treated with Akk. muciniphila all had significantly lower expression of the phosphorylated mTOR and P70S6K in the liver in comparison with the mice treated with PFOS alone (Figure 6A,B), which was consistent with the significantly lower arginine levels in mice. On the other hand, mice treated with PFOS combined with heat-killed bacteria treatment (L. reuteri, E. faecalis and Akk. muciniphila) also showed lower expression of phosphorylated mTOR and P70S6K in comparison with the mice treated with PFOS alone (Figure 6A,B). In contrast, no significant differences in the expression of mTOR and P70S6K were observed in the indicated groups (Figure S6,S7).
Figure 6.
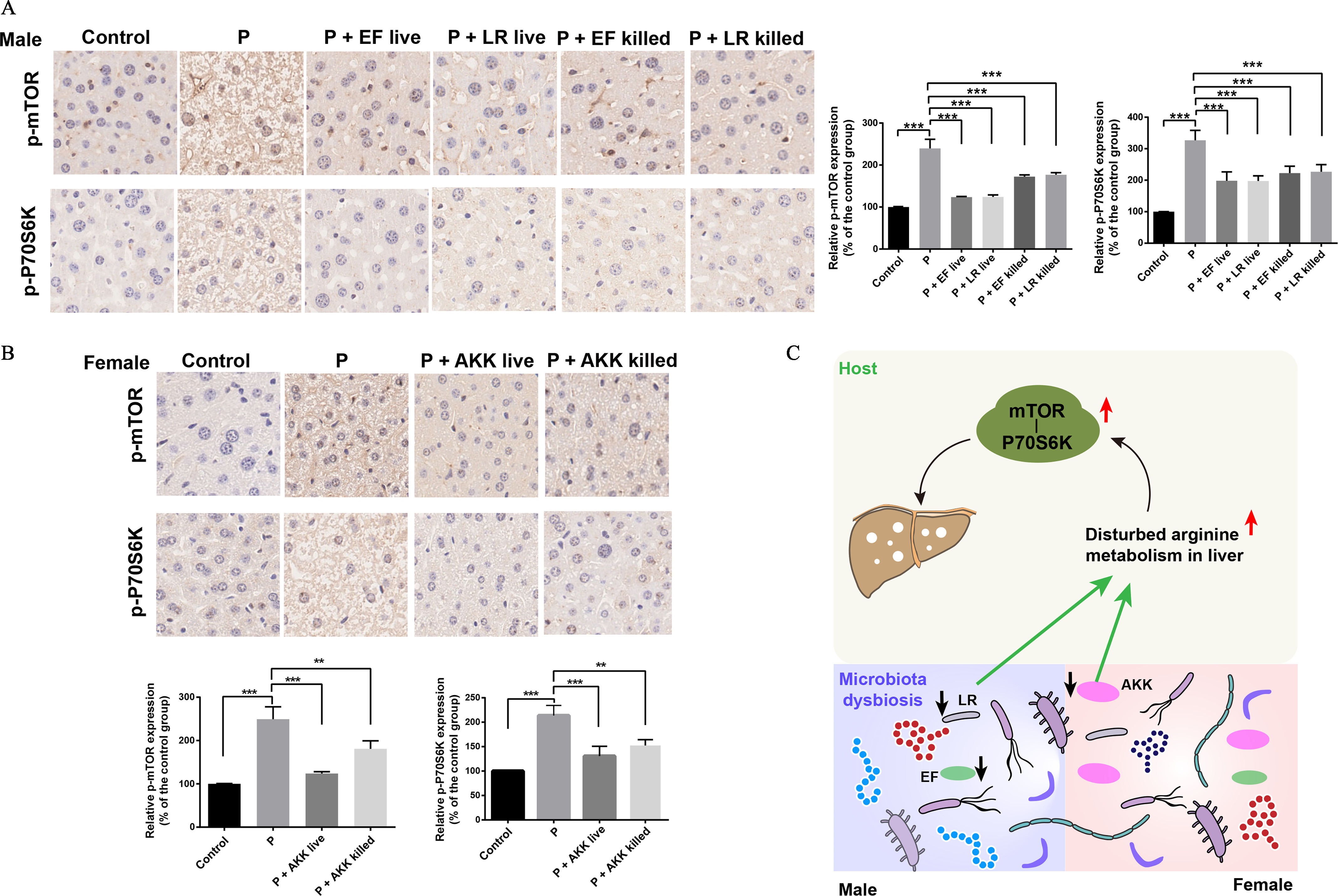
The effects of PFOS on the expressions of mTOR and P70S6K. (A) Expression of phosphorylated mTOR and P70S6K in fixed liver tissues of male mice in the indicated groups. (B) Expression of phosphorylated mTOR and P70S6K in fixed liver tissues of female mice in the indicated groups. (C) Schematic diagram of a potential mechanism by which the fecal microbiota contributes to PFOS-induced liver injury. PFOS regulates the abundances of fecal microbiota, which in turn contribute to the regulation of arginine levels in livers and then result in the activation of mTOR-P70S6K signaling pathway that can cause liver injury. , The relative intensity represents the ratio between the expression level of phosphorylated protein (p-mTOR and p-P70S6K) and the total protein expression level (mTOR and P70S6K). Summary data can be found in Table S7. Statistical significance was analyzed by one-way ANOVA. Results were presented as the . Note: AKK, Akk. muciniphila; ANOVA, analysis of variance; EF, E. faecalis; LR, L. reuteri; mTOR, mammalian target of rapamycin; P and PFOS, perfluorooctane sulfonate; SD, standard deviation. *. **. *** in comparison with the indicated group.
Discussion
The liver is an important organ for detoxification and has been considered a primary site of bioaccumulation of several pollutants.17 Gut microbiota is known to play a pivotal role in host pathophysiological status. For instance, the dysbiosis or imbalance between each bacterium occurred in alcohol-induced liver injury in mice. Impairment of bacterial synthesis of metabolites such as saturated long-chain fatty acids, bile acids and tryptophan may exacerbate liver disease in the mouse model.25,38,39 Here, combining fecal microbiota sequencing and metabolomic analysis, we implicate the critical role of gut–liver axis in mediating PFOS’s liver toxicity.
Given that PFOS was found to alter the fecal microbiota in the present study, we address the question whether PFOS induces liver injury through the remodeling of fecal microbiota. The hypothesis is strongly supported by attenuation of adverse liver effects with the suppression of fecal microbiota using antibiotics and supplementation of bacteria by FMT experiments. Our findings revealed that PFOS caused liver injury partially through fecal microbiota and provided novel mechanistic understanding of liver injury induced by PFOS. However, our data indicated that both FMT and ABX experiments alleviated the liver injury induced by PFOS, which requires further study by using ABX and FMT mouse models alone or in combination to elucidate the underlying mechanisms.
Specifically, we observed that PFOS exposure exhibited different effects on microbiota between sexes. Comparison of fecal microbial compositions between control and PFOS-treated mice revealed a male-specific reduction in the abundances of Lactobacillus and Enterococcus. These taxa are thought to protect against diseases such as obesity40 and nonalcoholic hepatic steatosis in mice.41 PFOS-treated female mice had a dramatically lower abundance of Akkermansia genus in comparison with the control mice. Presence of this species has been shown to be protective in diseases such as type 2 diabetes in the mouse model.42 Similarly, we found that PFOS-exposed mice had significantly lower abundances of L. reuteri, E. faecalis, and Akk. muciniphila at the family-genus-species levels, which were verified by qPCR analysis. Based on this notion, we employed a bacterial administration strategy to demonstrate their protective effects in liver injury and found that intervention with these bacteria could significantly alleviate the liver injury induced by PFOS. Our reports demonstrated that heat-treated bacteria also had protective effects against PFOS-induced liver injury. Previous studies reported that administering autoclaved (i.e., inactivated) bacteria also exhibited effects on diseases like obesity in mice.43 There might be a lack of knowledge of the molecular mechanisms involved, which warrant further study.
A growing number of studies indicated that gut microbiota were involved in metabolite production in cell, rodent, or human models.44 The mouse experiments revealed that the manipulation of the gut microbiota could affect host metabolic energy balance and ameliorate pathological features.26,45 Here, we found the metabolic pathway involving amino acid metabolism strongly associated with PFOS-induced liver injury. Targeted metabolomic analysis revealed a significant difference in the levels of arginine in livers and feces of PFOS-treated mice in comparison with control mice. We further demonstrated that L. reuteri, E. faecalis, and Akk. muciniphila, levels of which were significantly lower in PFOS-exposed animals, had a positive association between liver protective effects and lower arginine levels. These results indicated a potentially causal role of these bacteria in mediating the metabolism of arginine. It seems to be controversial that PFOS-treatment mice had significantly lower proline concentrations in the liver in comparison with control mice, whereas and PFOS-exposed mice had significantly higher liver proline levels. This phenomenon requires further investigation.
Currently, there is little mechanistic insight into how amino acids contribute to PFOS-induced liver injury. A recent study demonstrated that PFOS could perturb several signaling pathways such as mTOR signaling pathway in rodents or human Sertoli cells46; the mTOR pathway has been considered to play a vital role in liver function of mice.47 Our data displayed significantly higher arginine levels in PFOS-exposed mice, whereas arginine was reported to activate mTOR signaling pathway in cultured cells.37,48 We hypothesized that the mTOR signaling pathway might be involved in the mechanisms of PFOS-induced liver injury. Consistently, we observed that PFOS-exposed mice had significantly higher expression of phosphorylated mTOR and P70S6K when compared with the control group. In contrast, bacteria-treated mice had significantly lower expression of the phosphorylated mTOR and P70S6K in comparison with the mice treated with PFOS alone, accompanied by a significantly lower concentration of arginine. Therefore, we speculated that the higher arginine levels associated with PFOS exposure might contribute to liver injury through affecting the mTOR-P70S6K signaling pathway. Future studies are warranted to validate the detailed role of mTOR-P70S6K signaling pathway in PFOS-induced liver injury.
Overall, our data demonstrate that PFOS could induce liver injury by regulating amino acid metabolism via modulation of fecal microbiota (Figure 6C). Several potentially beneficial intestinal bacteria, including L. reuteri, E. faecalis, and Akk. muciniphila, were involved in the protective effects against liver injury. Our findings provide a novel mechanism for PFOS-induced liver injury and should be of value in considering supplementing probiotics as a promising intervention in occupationally or nonoccupationally exposed humans.
Limitations of this Study
The first limitation of our study is that the deep relationship between bacteria alteration and arginine or other amino acid concentrations is insufficiently validated in the current study. Second, it would be interesting to investigate the microbial communities of cecal contents or mucus layer to provide possible additional information that may be key to the understanding of the mechanism of PFOS-induced liver injury. Last, the relationship between PFOS and fecal microbiota needs to be further verified at environmentally relevant concentrations and with long-term exposure.
Supplementary Material
Acknowledgments
The authors thank Z. Zhang from Nanjing Medical University for offering help with Akk. muciniphila culturation. The authors also thank J. Zhao from China Pharmaceutical University for offering help with animal experiments. The authors thank S. Wang from Hong Kong Baptist University for reading our manuscript.
This work was supported by the National Natural Science Foundation of China (21707112 and 21806136).
References
- 1.Olsen GW, Burris JM, Ehresman DJ, Froehlich JW, Seacat AM, Butenhoff JL, et al. 2007. Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ Health Perspect 115(9):1298–1305, PMID: , 10.189/ehp.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giesy JP, Kannan K. 2001. Global distribution of perfluorooctane sulfonate in wildlife. Environ Sci Technol 35(7):1339–1342, PMID: , 10.1021/es001834k. [DOI] [PubMed] [Google Scholar]
- 3.Kannan K, Choi JW, Iseki N, Senthilkumar K, Kim DH, Giesy IP. 2002. Concentrations of perfluorinated acids in livers of birds from Japan and Korea. Chemosphere 49(3):225–231, PMID: , 10.1016/S0045-6535(02)00304-1. [DOI] [PubMed] [Google Scholar]
- 4.Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Tully JS, Needham LL. 2007. Serum concentrations of 11 polyfluoroalkyl compounds in the U.S. population: data from the National Health and Nutrition Examination Survey (NHANES). Environ Sci Technol 41(7):2237–2242, PMID: , 10.1021/es062686m. [DOI] [PubMed] [Google Scholar]
- 5.Inoue K, Okada F, Ito R, Kato S, Sasaki S, Nakajima S, et al. 2004. Perfluorooctane sulfonate (PFOS) and related perfluorinated compounds in human maternal and cord blood samples: assessment of PFOS exposure in a susceptible population during pregnancy. Environ Health Perspect 112(11):1204–1207, PMID: , 10.1289/ehp.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh S, Araki A, Mitsui T, Miyashita C, Goudarzi H, Sasaki S, et al. 2016. Association of perfluoroalkyl substances exposure in utero with reproductive hormone levels in cord blood in the Hokkaido Study on Environment and Children’s Health. Environ Int 94:51–59, PMID: , 10.1016/j.envint.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 7.So MK, Yamashita N, Taniyasu S, Jiang QT, Giesy JP, Chen K, et al. 2006. Health risks in infants associated with exposure to perfluorinated compounds in human breast milk from Zhoushan, China. Environ Sci Technol 40(9):2924–2929, PMID: , 10.1021/es060031f. [DOI] [PubMed] [Google Scholar]
- 8.Kishi R, Nakajima T, Goudarzi H, Kobayashi S, Sasaki S, Okada E, et al. 2015. The association of prenatal exposure to perfluorinated chemicals with maternal essential and long-chain polyunsaturated fatty acids during pregnancy and the birth weight of their offspring: the Hokkaido study. Environ Health Perspect 123(10):1038–1045, PMID: , 10.1289/ehp.1408834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv ZQ, Li GQ, Li YY, Ying CJ, Chen J, Chen T, et al. 2013. Glucose and lipid homeostasis in adult rat is impaired by early-life exposure to perfluorooctane sulfonate. Environ Toxicol 28(9):532–542, PMID: , 10.1002/tox.20747. [DOI] [PubMed] [Google Scholar]
- 10.Wan HT, Zhao YG, Wei X, Hui KY, Giesy JP, Wong CKC. 2012. PFOS-induced hepatic steatosis, the mechanistic actions on β-oxidation and lipid transport. Biochim Biophys Acta 1820(7):1092–1101, PMID: , 10.1016/j.bbagen.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Lai KP, Li JW, Cheung A, Li R, Billah MB, Chan TF, et al. 2017. Transcriptome sequencing reveals prenatal PFOS exposure on liver disorders. Environ Pollut 223:416–425, PMID: , 10.1016/j.envpol.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 12.Keil DE, Mehlmann T, Butterworth L, Peden-Adams MM. 2008. Gestational exposure to perfluorooctane sulfonate suppresses immune function in B6C3F1 mice. Toxicol Sci 103(1):77–85, PMID: , 10.1093/toxsci/kfn015. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Dong GH, Jin YH, He QC. 2009. Immunotoxic changes associated with a 7-day oral exposure to perfluorooctanesulfonate (PFOS) in adult male C57BL/6 mice. Arch Toxicol 83(7):679–689, PMID: , 10.1007/s00204-008-0361-3. [DOI] [PubMed] [Google Scholar]
- 14.Thibodeaux JR, Hanson RG, Rogers JM, Grey BE, Barbee BD, Richards JH, et al. 2003. Exposure to perfluorooctane sulfonate during pregnancy in rat and mouse. I: maternal and prenatal evaluations. Toxicol. Sci 74(2):69–81, PMID: , 10.1093/toxsci/kfg121. [DOI] [PubMed] [Google Scholar]
- 15.Austin ME, Kasturi BS, Barber M, Kannan K, MohanKumar PS, MohanKumar SMJ. 2003. Neuroendocrine effects of perfluorooctane sulfonate in rats. Environ Health Perspect 111(12):1485–1489, PMID: , 10.1289/ehp.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui L, Zhou QF, Liao CY, Fu JJ, Jiang GB. 2009. Studies on the toxicological effects of PFOA and PFOS on rats using histological observation and chemical analysis. Arch Environ Contam Toxicol 56(2):338–349, PMID: , 10.1007/s00244-008-9194-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Wang Y, Liang Y, Li J, Liu YC, Zhang J, et al. 2014. PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci Rep 4:4582, PMID: , 10.1038/srep04582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjork JA, Lau C, Chang SC, Butenhoff JL, Wallace KB. 2008. Perfluorooctane sulfonate-induced changes in fetal rat liver gene expression. Toxicology 251(1–3):8–20, PMID: , 10.1016/j.tox.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Jin R, McConnell R, Catherine C, Xu SJ, Walker DI, Stratakis N, et al. 2020. Perfluoroalkyl substances and severity of nonalcoholic fatty liver in children: an untargeted metabolomics approach. Environ Int 134:105220, PMID: , 10.1016/j.envint.2019.105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratakis N, Conti DV, Jin R, Margetaki K, Valvi D, Siskos AP, et al. 2020. Prenatal exposure to perfluoroalkyl substances associated with increased susceptibility to liver injury in children. Hepatology 72(5):1758–1770, PMID: , 10.1002/hep.31483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Y, Lv S, Liu J, Nie S, Chen J, Dong Q, et al. 2017. Chronic perfluorooctanesulfonic acid exposure disrupts lipid metabolism in zebrafish. Hum Exp Toxicol 36(3):207–217, PMID: , 10.1177/0960327116646615. [DOI] [PubMed] [Google Scholar]
- 22.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. 2014. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 60(5):940–947, PMID: , 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo G. 2015. Gut-liver axis in alcoholic liver disease. Gastroenterology 148(1):30–36, PMID: , 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Y, Pedersen O. 2021. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19(1):55–71, 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 25.Ferrere G, Wrzosek L, Cailleux F, Turpin W, Puchois V, Spatz M, et al. 2017. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol 66(4):806–815, PMID: , 10.1016/j.jhep.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, et al. 2010. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J 24(12):4948–4959, PMID: , 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 27.Juanola O, Hassan M, Kumar P, Yilmaz B, Keller I, Simillion C, et al. 2021. Intestinal microbiota drives cholestasis-induced specific hepatic gene expression patterns. Gut Microbes 13(1):1–20, PMID: , 10.1080/19490976.2021.1911534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang LM, Rimal B, Nichols RG, Tian Y, Smith PB, Hatzakis E, et al. 2020. Perfluorooctane sulfonate alters gut microbiota-host metabolic homeostasis in mice. Toxicology 431:152365, PMID: , 10.1016/j.tox.2020.152365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Sun SS, Wu XB, Yang SR, Wu YM, Zhao JX, et al. 2020. Intestinal environmental disorders associate with the tissue damages induced by perfluorooctane sulfonate exposure. Ecotoxicol Environ Saf 197:110590, PMID: , 10.1016/j.ecoenv.2020.110590. [DOI] [PubMed] [Google Scholar]
- 30.Jiang LL, Hong YJ, Xie GS, Zhang JH, Zhang HN, Cai ZW. 2021. Comprehensive multi-omics approaches reveal the hepatotoxic mechanism of perfluorohexanoic acid (PFHxA) in mice. Sci Total Environ 790:148160, PMID: , 10.1016/j.scitotenv.2021.148160. [DOI] [PubMed] [Google Scholar]
- 31.Li XN, Chung ACK, Li SF, Wu LL, Xu JY, Yu J, et al. 2017. LC-MS-based metabolomics revealed SLC25A22 as an essential regulator of aspartate-derived amino acids and polyamines in KRAS-mutant colorectal cancer. Oncotarget 8(60):101333–101344, PMID: , 10.18632/oncotarget.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. 2018. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 174(2):497, PMID: , 10.1016/j.cell.2018.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. 2018. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 8(4):403–416, PMID: , 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao C, Tang Z, Yan JC, Fang J, Wang HL, Cai ZW. 2017. Bisphenol S exposure modulate macrophage phenotype as defined by cytokines profiling, global metabolomics and lipidomics analysis. Sci Total Environ 592:357–365, PMID: , 10.1016/j.scitotenv.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 35.Tomlinson C, Rafii M, Ball RO, Pencharz PB. 2011. Arginine can be synthesized from enteral proline in healthy adult humans. J Nutr 141(8):1432–1436, PMID: , 10.3945/jn.110.137224. [DOI] [PubMed] [Google Scholar]
- 36.Ericksen RE, Lim SL, McDonnell E, Shuen WH, Vadiveloo M, White PJ, et al. 2019. Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes tumor development and progression. Cell Metab 29(5):1151–1165, PMID: , 10.1016/j.cmet.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyachok J, Earnest S, Iturraran EN, Cobb MH, Ross EM. 2016. Amino acids regulate mTORC1 by an obligate two-step mechanism. J Biol Chem 291(43):22414–22426, PMID: , 10.1074/jbc.M116.732511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, et al. 2015. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology 148(1):203–214. e16, PMID: , 10.1053/j.gastro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrzosek L, Ciocan D, Hugot C, Spatz M, Dupeux M, Houron C, et al. 2021. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut 70(7):1299–1308, PMID: , 10.1136/gutjnl-2020-321565. [DOI] [PubMed] [Google Scholar]
- 40.Quan LH, Zhang CH, Dong M, Jiang J, Xu HD, Yan CL, et al. 2020. Myristoleic acid produced by enterococci reduces obesity through brown adipose tissue activation. Gut 69(7):1239–1247, PMID: , 10.1136/gutjnl-2019-319114. [DOI] [PubMed] [Google Scholar]
- 41.Qiao SS, Bao L, Wang K, Sun SS, Liao MF, Liu C, et al. 2020. Activation of a specific gut Bacteroides-folate-liver axis benefits for the alleviation of nonalcoholic hepatic steatosis. Cell Rep 32(6):108005, PMID: , 10.1016/j.celrep.2020.108005. [DOI] [PubMed] [Google Scholar]
- 42.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. 2014. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63(5):727–735, PMID: , 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 43.Kondoh M, Shimada T, Fukada K, Morita M, Katada K, Higashimura Y, et al. 2014. Beneficial effects of heat-treated Enterococcus faecalis FK-23 on high-fat diet-induced hepatic steatosis in mice. Br J Nutr 112(6):868–875, PMID: , 10.1017/S0007114514001792. [DOI] [PubMed] [Google Scholar]
- 44.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. 2012. Host-gut microbiota metabolic interactions. Science 336(6086):1262–1267, PMID: , 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 45.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, et al. 2015. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun 6:7489, PMID: , 10.1038/ncomms8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao BP, Mruk D, Lian QQ, Ge RS, Li C, Silvestrini B, et al. 2018. Mechanistic insights into PFOS-mediated Sertoli cell injury. Trends Mol Med 24(9):781–793, PMID: , 10.1016/j.molmed.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao PT, Xie ZS, Kuang YJ, Liu SY, Zeng C, Li P, et al. 2021. Discovery of a potent FKBP38 agonist that ameliorates HFD-induced hyperlipidemia via mTOR/P70S6K/SREBPs pathway. Acta Pharm Sin B, PMID: , 10.1016/j.apsb.2021.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris SM Jr. 2016. Arginine metabolism revisited. J Nutr 146(12):2579S–2586S, PMID: , 10.3945/jn.115.226621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.