Abstract
We studied the role of sodium/proton exchanger 8 (NHE8) in retinal pigment epithelium (RPE) and photoreceptor cells of adult mouse retina by using the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease (Cas)9 from Neisseria meningitidis (Nm). Specific single guide RNAs (sgRNAs) were designed to knockdown the Slc9a8 gene, which encodes the NHE8. Nuclease null NmCas9 and sgRNAs were packaged respectively using adeno-associated viral vector (AAV), and delivered into mouse eyes in vivo by subretinal injection on wild-type mice of about four-week-old when mouse retina is fully developed. Eye samples were collected four weeks after injection for phenotype examination. Real-time PCR analysis demonstrated ~38% reduction of NHE8 transcripts in retinas injected with AAV-knockdown sgRNA and AAV-Cas9. Loss of photoreceptor cells was found in eyes injected with AAV-knockdown sgRNA and AAV-Cas9 under either the human rhodopsin promoter or the minimal chicken β-actin promoter, while normal morphology was observed in control eyes injected with AAV-Cas9 and AAV-control sgRNA; immunostaining data showed degenerating photoreceptor cells and RPE cells in eyes injected with knockdown sgRNA and Cas9 AAVs. We further determined that mutant M120K-NHE8 displayed altered intracellular pH regulation in human RPE and primary mouse RPE cells using genetically encoded pH sensor pHluorin and that primary cultured NHE8 mutant RPE cells showed different pH titration curves. These results indicate that NHE8 plays essential function in both RPE and photoreceptor cells. NHE8 dysfunction either in photoreceptor or RPE is sufficient to cause retinal degeneration in adult mice at any age.
Keywords: NHE8, Retinal pigment epithelium, Photoreceptor cells, CRISPR/Cas9, pH regulation
1. Introduction
NHE8, encoded by the Slc9a8 gene, is a member of a large group of monovalent cation/proton antiporters that predominately move Na+ in exchange for H+ and play diverse physiological roles, including the regulation of intracellular pH, absorption of sodium into epithelia, salt tolerance, cell volume, cell adhesion, cell proliferation, organelle bio-genesis and protein trafficking (Orlowski and Grinstein, 2004). Although NHE8 is widely expressed in adult mouse tissues, the most obvious pathological phenotypes of its deletion or mutation are retinal degeneration and male infertility (Jadeja et al., 2015; Oberheide et al., 2017; Xia et al., 2015; Xu et al., 2015). We and others previously reported that mice with either a NHE8 knockout or mutant M120K-NHE8 developed impaired RPE cells and late-onset photoreceptor cell loss, and suggested that NHE8 plays an essential role in RPE cell polarity/function by possibly controlling the homeostasis of pH and sodium in protein trafficking of trans-Golgi network during development (Jadeja et al., 2015; Xia et al., 2015). It remains unclear whether the retinal degeneration is a consequence of earlier eye or retinal developmental defects due to systemic NHE8 knockout or mutation. This work intends to directly evaluate the function of NHE8 in mature mouse retina, and study whether NHE8 plays a role in intracellular pH regulation in RPE cells.
In mouse retina, NHE8 protein expression was observed in the RPE and the photoreceptor cells (Jadeja et al., 2015; Xia et al., 2015). It has been shown that NHE8 proteins co-localized with various intracellular vesicles, and especially with the trans-Golgi network marker in RPE (Xia et al., 2015) and in recycling endosomes (Jadeja et al., 2015). Using genetically encoded fluorescent pH sensor pHluorin (Miesenbock et al., 1998), we studied the intracellular pH homeostasis in human RPE cells and cultured mouse primary RPE cells. By infecting the cells with AAV-NHE8-mCherry-pHluorin, we monitored the fluorescent intensity ratios of pH-sensitive ecliptic pHluorin and pH-insensitive mCherry. Our data suggest that NHE8 plays important roles in the pH homeostasis of RPE and photoreceptor cells.
To disrupt NHE8 function in RPE and photoreceptors of adult mice respectively, we constructed AAV vectors to express nuclease null NmCas9 under the control of a minimal chicken β-actin promoter/CMV enhancer (smCBA) (Pang et al., 2008) or a photoreceptor cell specific human rhodopsin promoter (hRho) (Bennett et al., 1995). Nuclease null NmCas9 fused with transcription repressor KRAB (Agata et al., 1999; Larson et al., 2013; Qi et al., 2013; Urrutia, 2003) was constructed, and specific sgRNAs were selected and designed to knockdown NHE8 expression. Wild-type Cas9 has Ruv C and HNH nuclease like domains with nuclease activities; upon RNA-DNA-Cas9 binding, the Ruv C nuclease will cut the non-complementary DNA strand and the HNH nuclease will cut the complementary DNA strand, resulting in DNA double strand breaks (Jinek et al., 2012); DNA double strand breaks can trigger the cells to repair the DNA mainly by non-homologues end joining (NHEJ), which is error prone and may have the potential to induce photoreceptor damage via DNA insertion and deletion (InDel) as well as result in off-target DNA double strand breaks. Mutagenesis in both Ruv C and HNH domains produced nuclease null Cas9 that binds specific targeted DNA guided by sgRNAs without cutting double strand DNA but works as a “block” to suppress transcription, which is referred as CRISPR interference (CRISPRi) (Qi et al., 2013). In theory, nuclease null Cas9 can recognize the target gene but cannot make DNA double strand breaks, it can therefore reduce the off-target effect of wild-type Cas9 and subsequent nucleotide insertions or deletions (Qi et al., 2013; Larson et al., 2013). The sgRNAs and nuclease null NmCas9-KRAB were packaged into AAV9–2YF (Byrne et al., 2015; Dalkara et al., 2012; Petrs-Silva et al., 2009; Zhong et al., 2008) respectively, and used to knockdown NHE8 expression in RPE or photoreceptor cells by subretinal injection into adult mouse retina. Both the pan-retina knockdown of NHE8 by smCBA-Cas9 and photoreceptor-specific knockdown by hRho-Cas9 displayed severe retinal degeneration. Thus, NHE8 is essential for the survival of photoreceptors in adult animals, presumably due to its function in proton/sodium homeostasis of both RPE and photoreceptors. In addition, by using AAV packaged NHE8-mCherry-pHluorin and cultured human RPE cell line or mouse primary RPE cells in vitro, we have demonstrated that NHE8 is important in intracellular pH regulation.
2. Materials and methods
2.1. Mice
All experimental procedures were approved by the Animal Care and Use Committee (ACUC) at University of California, Berkeley, and were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
2.2. RPE cell culture
For primary mouse RPE culture, a modified method was developed based on a published work (Fernandez-Godino et al., 2016). Briefly, freshly enucleated eyeballs were placed in ice cold HBSS-H- (HBSS- no calcium + 10 mM HEPES, Life Technologies) to dissect away the cornea and lenses, the remaining eyecups were incubated in 1 mg/ml hyaluoronidase at 37 °C with 5% CO2 for 45 min. Eyecups were then transferred to cold HBSS-H+ (HBSS with calcium and Magnesium + 10 mM HEPES, Life Technologies) and incubated on ice for 30 min. In the HBSS-H+, the neural retina was removed, eyecups were transferred to 0.05% Tryspin +0.02% EDTA and incubated at 37 °C with 5% CO2 for 45 min, followed by transferring to 20% FBS in HBSS-H+. The RPE sheets were gently dissociated, and sheets from the same genotype of mice were pooled in 20% FBS/HBSS-H+ solution and centrifuged at 25 °C at 340 g for 2 min. The cell pellets were re-suspended in 1 ml of 0.05% trypsin +0.02% EDTA and incubated at 37 °C for 1 min, RPE medium was added and the cells were centrifuged at 340 g 25 °C for 2 min, the resulting RPE cell pellets were re-suspended in RPE medium, which including 1% Penicillin/Streptomycin, 1% GlutaMax, 1× B-27 (Gibco, cat# 17504–044), 2% fetal bovine serum, 2 μM SB431542 (Stemgent, cat# 04–0010-10) in Gibco Advanced DMEM/F12. Cells were plated into glass bottom dishes pre-coated with 10 μg/ml mouse Laminin (Corning, cat# 354232). Human ARPE-19 cells were cultured in DMEM/F12 medium with 10% fetal calf serum.
2.3. Generation of AAVs for pH titration measurement
We previously generated AAV5-smCBA-(wt, wild-type or r15, mutant M120K) NHE8-SE pHluorin with the pTR-smCBA-NHE8-SE pHluorin plasmids (Xia et al., 2015). To make AAV5-smCBA-NHE8-mCherry-SE pHluorin, we used BamHI to cut the SE pHluorin fragment from the pTR-smCBA-NHE8-SE pHluorin vector and dephosphorylated the remaining vector. We then used the following primers to amplify mCherry and SE pHluorin PCR fragments: mCherry forward (34-mer)-GTGGAGGTGGGGATCCGTGAGCAAGGGCGAGGAG and mCherry reverse (29-mer)- CCTTTACTCTTGTACAGCTCGTCCATGCC; pHluorin forward (36-mer)- GTACAAGAGTAAAGGAGAAGAACTTTTCACTGGAGT and pHluorin reverse (45-mer)- TATCATGTCTGGATCCTTAACCGGTTTTGTATAGTTCATCCATGC. The vector, the mCherry and the pHluorin PCR fragments were mixed together with molecular ratio of 1:1:1 in Cold Fusion reaction enzymes according to manufacturer recommended protocol (Cold Fusion cloning kit, System Biosciences), 2 μl of reaction mixture was used to transform Sure2 competent cells and pTR-smCBA-wt-NHE8 or r15-NHE8-mCherry-SE pHluorin colonies were selected and confirmed. The pTR-smCBA-wt-NHE8 or r15-NHE8-mCherry-SE pHluorin plasmids were packaged into AAV5 and an evolved AAV variant 7m8 (Dalkara et al., 2013), and the following AAVs were generated for transducing cultured cells: AAV5-smCBA-wt-NHE8-mCherry-SE pHluorin (1.72 × 1014 vg/ml), AAV5-smCBA-r15-NHE8-mCherry-SE pHluorin (5.87 × 1013 vg/ml), 7m8-smCBA-wt-NHE8-mCherry-SE pHluorin (3.30 × 1014 vg/ml), and 7m8-smCBA-r15-NHE8-mCherry-SE pHluorin (1.18 × 1014 vg/ml).
2.4. pH titration curves in RPE cells
Human ARPE-19 cells and mouse primary RPE cells expressing wt-NHE8-mCherry-pHluorin or r15-NHE8-mCherry-pHluorin were imaged at 37 °C with a 63x lens for pHluorin (488 nm laser) and mCherry (555 nm laser) fluorescence respectively. For each experiment, six to eight cells on a single plate were imaged for analysis. Cells were first imaged in Live Cell Imaging Solution (ThermoFisher, A14291DJ); for titration, cells were sequentially imaged in buffers at pH 7.5, 6.5, and 5.5 (ThermoFisher, P35379) containing 10 μM nigericin (an ionophore that equilibrates intracellular and extracellular pH), with a 5-min incubation period between each buffer. Titration curves were generated for each cell by plotting the pHluorin/mCherry intensity ratios against the buffer pH values.
2.5. Generation of AAV packaged sgRNA
Specific primers were designed to PCR amplify the two fragments for each sgRNA, using the template NmCas9 specific sgRNA backbone (M-NM-sgRNA, Addgene plasmid #48673, gift from George Church, www.addgene.com) (Esvelt et al., 2013) that includes a U6 promoter and a short form of NmCas9 sgRNA backbone in a TOPO vector. The following primers were used for PCR: 1) for sgRNA Nhe8 KO P1, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- GCC CCG CCC CTG CCC GCA GCG GAA GTT GTA GCT CCC TTT CTC G, fragment 2 forward- TTC CGC TGC GGG CAG GGG CGG GGC GGT GTT TCG TCC TTT CCA C, and fragment 2 reverse- TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG; 2) for sgRNA Nhe8 KO P2, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- GCT AGC CCT GCG GCG CCG AAC TCG TTG TAG CTC CCT TTC TCG, fragment 2 forward- GAG TTC GGC GCC GCA GGG CTA GCG GTG TTT CGT CCT TTC CAC, and fragment 2 reverse- TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG; 3) for sgRNA Nhe8 KO E2, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- GCC TGC TCT CCC GTC TGC ACC GGT TGT AGC TCC CTT TCT CG, fragment 2 forward- CGG TGC AGA CGG GAG AGC AGG CGG TGT TTC GTC CTT TCC AC, and fragment 2 reverse- TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG; 4) for sgRNA Nhe8 KO E5, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- GAA GAA ATG TTT CGC CCC AGT TGT AGC TCC CTT TCT CG, fragment 2 forward- TGG GGC GAA ACA TTT CTT CGG TGT TTC GTC CTT TCC AC, and fragment 2 reverse- TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG; 5) for sgRNA Nhe8 KO E6, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- GCC GAG ATT GCC GTT CCA AAG AGT TGT AGC TCC CTT TCT CG, fragment 2 forward- TCT TTG GAA CGG CAA TCT CGG CGG TGT TTC GTC CTT TCC AC, and fragment 2 reverse- TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG; 6) for control sgRNA, fragment 1 forward- CGG TAC AAT TCA CGC GTG GAG CTC GGA TCC ACT AGT AAC, fragment 1 reverse- TGT GGA AAG GAC GAA ACA CCG TTG TAG CTC CCT TTC TCG, fragment 2 forward- CGA GAA AGG GAG CTA CAA CGG TGT TTC GTC CTT TCC ACA, and fragment 2 reverse-TCC CCA GCA TGC CTG CTA CGA ATT GGG CCC TCT AGA TG. The two PCR fragments of each sgRNA were subcloned into the KpnI and BbsI sites of the scAAV-CB-EGFP vector (Zhong et al., 2008) by Cold Fusion cloning enzyme (SBI, System Biosciences Inc., Mountain View, CA). Correct clones were selected for AAV packaging and purification. Two sgRNAs (sgRNA Nhe8-KO P1 and sgRNA Nhe8-KO P2) were packaged together into scAAV9–2YF (scAAV9–2YF-U6-Nhe8 sgRNA P1/P2), and the other three (sgRNA Nhe8-KO E2, sgRNA Nhe8-KO E5 and sgRNA Nhe8-KO E6) were packaged together (scAAV9–2YF-U6-Nhe8 sgRNA E2/E5/E6) (Dalkara et al., 2012; Jinek et al., 2012). A control sgRNA that only contains NM-sgRNA backbone without any target sequences was also packaged into scAAV9–2YF.
2.6. Construction of the mNmCas9 viral vector
Using a nuclease null mNmCas9 plasmid M-NMn-VP64 (Addgene plasmid #48676, gift of George Church) as template, we PCR amplified the nuclease null mNmCas9 fragment by using a forward primer with the Kozak sequence and the SV40 NLS (5′-tatccatcacactggcggccgcGCCACCatgCCGAAGAAAAAGCGCAAGGTAgccgccttcaagccaac-3′) and a reverse primer with the NLS and HA tag as well as a stop codon (5′-atgtctggatccgcgcggccgcTCAAGCGTAAT CTGGAACATCGTATGGGTAcaccttcctcttcttcttgg-3′); we cloned the pTR-Rho-NLS-mNMm4Cas9-NLS-HA by combining the Cas9 fragment with the pTR-Rhodopsin promoter (by replacing the smCBA promoter and the hGFP in pTR-smCBA-hGFP with the rhodopsin promoter) using the Cold Fusion Cloning enzyme mix (SBI, System Biosciences Inc.) and Sure 2 competent cells (Aligent Technology, Inc.). To reach maximal effects of knockdown, we amplified the KRAB A and B domains of human transcription repressor KOX1 (zinc finger protein 10 or ZNF 10) from human RPE cell cDNA (cDNA provided by Yan Zhang), or the KRAB A domain of mouse transcription repressor KRAZ1 amplified from mouse brain cDNA, and subcloned these domains into pTR-Rho-NLS-mNMm4Cas9-NLS-HA at the C-terminal after mNMm4- NLS-HA to make the pTR-Rho-NLS-mNMm4Cas9-NLS-HA-KRAB(Znf10/KOX1) vector. The pTR-smCBA-NLS-mNMm4Cas9-NLS-HA-KRAB(Znf10/KOX1) vector was generated by replacing the Rho promoter with the smCBA promoter. Primers used for cloning KRAB (Znf1/KOX1) with human RPE cDNA: forward primer- ATGGATGCTAAGTCACTAACTGC and reverse primer- CACCAGCCAGGGCTCTTC; primers for KRAB-A (KRAZ1); with mouse brain cDNA: forward primer- ATGGCTGGTTCT CCATTAATC, and reverse primer- AGACTCCTTTTCAGTCTTCAC.
The pTR-Rho-NLS-mNMm4Cas9-NLS-HA-KRAB(Znf10/KOX1) and the pTR-smCBA-NLS-mNMm4Cas9-NLS-HA-KRAB(Znf10/KOX1) were packaged into AAV9–2YF.
2.7. Subretinal injection of AAVs
C57BL/6J or albino B6-white wild-type mice were anesthetized by intraperitoneally administering a combination of 80 mg/kg ketamine and 12 mg/kg xylazine; mouse pupils were dilated with 2.5% phenylephrine and 0.5% tropicamide, and a topical anesthetic (proparacaine HCl) was administered to the cornea. For subretinal injection, a disposable 30-gauge needle was used to create a hole near the limbus; ~1 μl of virus was delivered into subretinal space with a 10-μl Hamilton syringe with a blunt ended 32-gauge needle through this opening. The AAV injected into each eye is about 108-109 total vector genome, since control AAV at this dosage shows no toxic effect in mouse retina.
2.8. Real time PCR analysis
For quantitative real-time PCR (RT-qPCR) analysis, the following NHE8 primers (amplify a 147 bp fragment from exon 14 to exon 15, spanning 2 exons) were designed and used for RT-qPCR: forward 5′- AAG ACG TCA ACC TCA GCA AG and reverse 5′- TTC AGG TAC TTG GCA TCC AG. The iScript™ Reverse Transcription Supermix (BIO-RAD) was used to make cDNA for RT-qPCR, and quantitative gene expression was determined using SsoAdvanced™ Universal SYBR Green Supermix (BIO-RAD) and CFX96™ Real-Time System (BIO-RAD). For statistical analysis of RT-qPCR results, data was presented as mean ± SD. Experiment and sample collection were performed as follows: wild-type mice at 4 weeks of age were subretinally injected – left eyes with scAAV-control sgRNA and right eyes with AAV-NHE8 knockdown sgRNA as well as AAV-CBA-NMCas9; 4 weeks after injection, retinas were dissected from left and right eyes of each mouse as control or knockdown groups (each sample is one retina). Retinas from un-injected eyes of age matched wild-type mice were also collected as no AAV treatment control. Retinal cDNAs were examined for each group (no AAV, n = 3; control AAV, n = 3; knockdown AAV, n = 4), and triplicate wells for each individual cDNA were repeated on a PCR plate. The NHE8 gene Starting Quantity (SQ) was normalized to the amount of G3PDH and a two-sample Student’s t-Test was used to calculate the P value. The primers for G3PDH were: forward 5′-TGA AGG TCG GTG TGA ACG GAT TTG GC and reverse 5′-CAT GTA GGC CAT GAG GTC CAC CAC.
2.9. Immunofluorescent staining
Immunofluorescent staining of retinal frozen sections and cultured cells was performed as previously described (Xia et al., 2008). Antibodies used in the study: Ezrin rabbit polyclonal antibody (Cell Signaling Technology), NHE8 mouse monoclonal antibody (Chemicon International), TGN46 rabbit polyclonal antibody (Millipore), and HA mouse monoclonal antibody (BioLegend). All images were collected by a Zeiss LSM700 confocal microscope.
3. Results
3.1. Selection of five NHE8 sgRNAs from mouse genome
To select sgRNAs, we used the SnapGene (www.snapgene.com) program to identify PAMs for NmCas9 (NNNNGANN, NNNNGTTN, NNNNGNNT) in the Slc9a8 (NHE8) genomic DNA on mouse chromo-some 2 (Esvelt et al., 2013). Using the software http://www.rgenome.net/cas-offinder (Bae et al., 2014), the 12 nucleotides (Fig. 1A, red color) within the oligos on both DNA strands adjacent to the PAM were used to search for targets in Mus Musculus genome (ftp://ftp.ensembl.org/pub/release-75/fasta/mus_musculus/dna/) (Larson et al., 2013), we identified 68 target hits of oligonucleotides, and further narrowed down to five oligonucleotides with minimal numbers of off-target (Qi et al., 2013). CRISPRi can silence the transcription initiation and elongation; sgRNAs designed to target promoter region need to be close to the transcription start site, and can be either on transcription template and non-template DNA strand; to silence transcription elongation, the sgRNAs need to target non-template strand of exons close to the transcription start site (Qi et al., 2013; Larson et al., 2013). Among our five identified sgRNAs with minimum off-target, sgRNAs P1 and P2 are targeting promoter region, therefore they could be on any strand; sgRNAs E2, E5, and E6 are targeting exons close to transcription start site, they should theoretically target non-template strand (E2 and E6) but E5 is an exception that targets the template strand near the NHE8-M120 coding region since r15 (NHE8-M120K) mutant phenotype was similar to NHE8 knockout phenotype.
Fig. 1. Design sgRNA to knockdown NHE8 in mouse genome.

(A) PAMs (NNNNGANN, NNNNGTTN, NNNNGNNT) for NmCas9 were identified in the Slc9a8 (encoding NHE8) genomic DNA. To design sgRNAs, we chose 20–25 oligonucleotides starting with G, which were adjacent to the PAM, and selected five oligonucleotides with minimal numbers of off-targets. Among them, two oligos (Nhe8-KO P1 and Nhe8-KO P2) were located before transcription start site in the promoter region, three oligos were in exon 2 (Nhe8-KO E2), exon 5 (Nhe8-KO E5) and exon 6 (Nhe8-KO E6), respectively. (B) Locations of five sgRNAs designed to knockdown NHE8 were indicated on the Slc9a8 genomic DNA.
Sequences of these five oligonucleotides (starting with G), adjacent to the PAM on the template DNA strand for oligos Nhe8-KO P1 and Nhe8-KO P2 located before the transcription start site and oligo Nhe8-KO E5 in exon5, and non-template DNA strand for oligo Nhe8-KO E2 in exon 2 and Nhe8-KO E6 in exon 6, were selected as sgRNAs respectively (Fig. 1A). Different combinations of these five sgRNAs with AAV-mediated CRISPR/Cas9 knockdown system was used to inhibit the expression of NHE8 in mature retinas.
3.2. Subretinal injection of AAV9 packaged NmCas9 and sgRNAs
The five sgRNAs were subcloned into the self-complementary recombinant adeno-associated virus (scAAV) vectors that promote efficient transduction independently of DNA synthesis (McCarty et al., 2001) with mammalian U6-driven sgRNA (NMm1) backbone (Esvelt et al., 2013). Among them, two sgRNAs (sgRNA Nhe8-KO P1 and sgRNA Nhe8-KO P2) were packaged together to generate the scAAV9–2YF-U6-Nhe8 sgRNAs P1/P2, while the other three (sgRNA Nhe8-KO E2, sgRNA Nhe8-KO E5 and sgRNA Nhe8-KO E6) were packaged together into AAV9–2YF for the scAAV9–2YF-U6-Nhe8 sgRNA E2/E5/E6 (Dalkara et al., 2012). A control sgRNA (NMm1 backbone) without target sequence was also packaged into AAV9–2YF to generate the scAAV9–2YF-U6-control sgRNA (Esvelt et al., 2013). Two NmCas9 vectors under the control of either the hRho promoter or the smCBA promoter were constructed and packaged into AAV9–2YF, generating the viral vectors AAV9–2YF-hRho-NLS-mNmCas9m4 (mammalian NmCas9 with 4 amino acids mutation)-NLS-HA-KRAB(KOX1) (AAV9-hRho-NmCas9) and AAV9–2YF-smCBA-NLS-mNmCas9m4-NLS-HA-KRAB(KOX1) (AAV9-smCBA-NmCas9) (Agata et al., 1999; Bennett et al., 1995; Pang et al., 2008; Petrs-Silva et al., 2009).
To test the efficacy of NHE8 knockdown in adult mouse retina by NmCas9 with specific sgRNAs, a mixture (1:1:1 ratio) of AAV9-smCBA-NmCas9, scAAV9-Nhe8-sgRNAs-P1/P2 and scAAV9-Nhe8-sgRNAs-E2/E5/E6, was injected into the subretinal space of the right eyes of 4-week-old wild-type mice. As controls, a mixture (1:2 ratio) of AAV9-smCBA-NmCas9 and scAAV9-control sgRNA was injected into the subretinal space of the left eyes of the same mice. Mice were euthanized four weeks after injection and their eyeballs were harvested and evaluated for retinal morphology on the effect of NHE8 knockdown mediated by this CRISPR/Cas9 system.
We first examined the eyes injected with AAV9-smCBA-NmCas9 and scAAV9 packaged with all five NHE8 knockdown sgRNAs. Retinal frozen sections were stained with phalloidin and DAPI to examine the phenotypes. A wild-type eye injected with a mixture of AAV9-smCBANmCas9, scAAV9-Nhe8-sgRNAs-P1/P2 and scAAV9-Nhe8-sgRNAs-E2/E5/E6 displayed severe retinal degeneration in selective areas, presumably resulted from NHE8 knockdown mediated by injected AAVs (Fig. 2A, two right images). Low magnification image covering the whole retina showed several regions (yellow arrows) with loss of photoreceptor cells (Fig. 2B, right image), and a close view revealed regions with only 1–3 layers of photoreceptor cells (Fig. 2A, the middle and right images). The smCBA promoter likely exhibits a pan-retina activity in RPE and other cells in the retina. The control eye from the same wild-type mouse injected with AAV9-smCBA-NmCas9 and scAAV9-control-sgRNAs displayed normal morphology without retinal degeneration or noticeable loss of photoreceptor cells (Fig. 2A and B, the left images). The results were confirmed in more than three litters of mice, with at least three different mice in each litter. The experiments were performed on both female and male mice of the C57BL/6J and B6-albino strains. We did not observe noticeable differences among sexes and strain backgrounds.
Fig. 2. Co-injection of AAV-CBA-NmCas9 and AAV-Nhe8-KO sgRNAs caused retinal degeneration in adult wild-type mice.
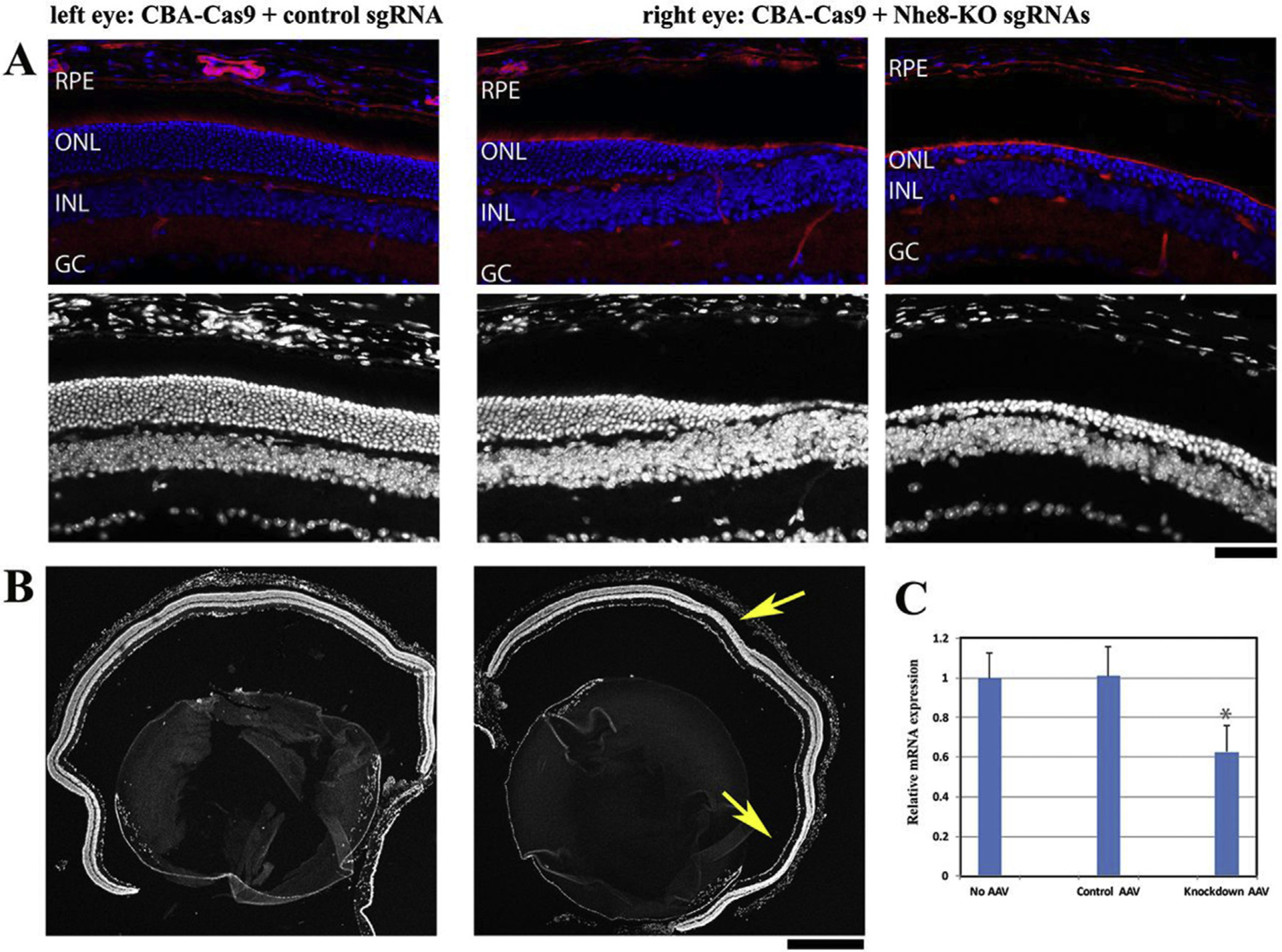
(A) Upper images showed retinal frozen sections stained with DAPI (blue) and phalloidin (red), and lower panels were DAPI stained images corresponding to the upper images. Normal retinal morphology was observed in the control left eye injected with AAV-CBA-Cas9 and AAV-control sgRNA; both middle and right images showed obvious loss of outer nuclear layers from the knockdown eye injected with AAV-CBA-Cas9, AAV-Nhe8-sgRNAs-P1/P2 and AAV-Nhe8-sgRNAs-E2/E5/E6. RPE: retinal pigment epithelium; ONL: photoreceptor outer nuclear layer; INL: inner nuclear layer; GC: ganglion cell layer. Scale bar: 50 μm. (B) Low magnification images stained with DAPI showed whole retinal sections of the control eye (left) and the knockdown eye (right) of a mouse. Two yellow arrows indicate the regions with obvious loss of photoreceptors in the ONL in knockdown retinal section. Scale bar: 500 μm. (C) Quantitative real-time PCR (RT-qPCR) analysis. The retinal cDNAs were examined for each group (no injection, n = 3; control, n = 3; NHE8 knockdown, n = 4), and triplicate wells for each individual cDNA were repeated on a PCR plate. NHE8 Starting Quantity (SQ) was normalized to the amount of G3PDH and a two-sample Student’s t-Test was used to calculate the P value. Statistic calculation revealed about 38% reduction of NHE8 transcripts in the knockdown sgRNAs injected right eyes when compared to the control sgRNA injected left eyes, with a P value of 0.016. Age-matched wild-type retinas were collected as no AAV injection control, and this group of retinas displayed similar NHE8 transcript level to the control AAV-injected retinas.
Since our previous study showed a loss-of-function NHE8 mutant mouse line developed retinal degeneration (Xia et al., 2015), we expect that NmCas9 and knockdown sgRNAs reduce the transcription levels of NHE8 to lead to the retinal phenotype. Thus, NHE8 transcription levels in total retinal RNA samples were determined by real-time PCR analysis. Total retinal RNAs were isolated from the control retinas of left eyes (injected with 1:2 of AAV9-smCBA-NmCas9 and scAAV9-control-sgRNA) and NHE8 knockdown retinas of right eyes (injected with 1:1:1 of AAV9-smCBA-NmCas9, scAAV9-Nhe8-sgRNAs-P1/P2 and scAAV9-Nhe8-sgRNAs-E2/E5/E6) four weeks after subretinal AAV injection; a set of primers that amplify a fragment between NHE8-exons 14 and 15, designed by Integrated DNA Technologies (IDT) PrimerQuest program, were used to examine the transcription level of NHE8 by RT-qPCR. NHE8 Starting Quantity (SQ) was normalized to the amount of G3PDH and a two-sample Student’s t-Test was used to calculate the P value.
Statistic calculation revealed about 38% reduction of NHE8 transcripts in the knockdown sgRNAs injected right eyes when compared to the control sgRNA injected left eyes, with a statistically significant P value of 0.016 (P < 0.05) (Fig. 2C). Age-matched wild-type retinas, collected as no AAV injection control, displayed similar level of NHE8 transcripts as AAV-injected control retinas.
Our previous work reveals that NHE8 plays an essential role in RPE cell polarity during development (Xia et al., 2015). Thus, we further examined the phenotypes of RPE cells in these AAVs injected eyes. The control eyeball, co-injected with AAV-smCBA-Cas9 and scAAV-control-sgRNA, showed that the apical marker ezrin protein (green) was detected as uniform signals at the apical regions of RPE (Fig. 3A and B, left images). In contrast, apical ezrin protein distribution was disrupted in RPE cells of the eyes that were co-injected with AAV-smCBA-Cas9, scAAV-Nhe8-sgRNAs P1/P2 and scAAV-Nhe8-sgRNAs E2/E5/E6 (Fig. 3A and B, the middle two images are from the same mouse as the control, and the right images are from another mouse). In addition, aberrant cells appeared in the subretinal space with a loss of outer nuclear layers of photoreceptors in the retina (Fig. 3B, labeled by white arrows). Regions of NHE8 knockdown retinas displayed separated subretinal spaces (two right images), which might occur during sample fixing or cutting; however, this separation further demonstrated the presence of aberrant cells in the subretinal space.
Fig. 3. Disruption of polarized RPE cells in eyes injected with AAV-CBA-Cas9 and AAV-Nhe8-KO sgRNAs.

Retinal frozen sections of AAV-injected eyeballs were characterized by co-staining with DAPI (blue), phalloidin (red) and anti-Ezrin (green). Left images are controls from AAV-CBA-Cas9 and AAV-control-sgRNA injected eye in low magnification (A) and high magnification (B), which revealed uniform apical distribution of ezrin staining signals in normal polarized RPE. Two middle images of (A) and (B) are from the other eye of the same mouse injected with AAV-CBA-Cas9, AAV-Nhe8-KO sgRNAs-P1/P2 and AAV-Nhe8-KO sgRNAs-E2/E5/E6. The right images of (A) and (B) are from another mouse eye injected with AAV-CBA-Cas9, AAV-Nhe8-KO sgRNAs-P1/P2 and AAV-Nhe8-KO sgRNAs-E2/E5/E6. In addition to a loss of outer nuclear layer, the middle and right images showed disrupted or mislocalized ezrin signals in RPE cells (white arrowheads), as well as aberrant cells (white arrows) in the subretinal space. RPE: retinal pigment epithelium; ONL: outer nuclear layer; INL: inner nuclear layer; GC: ganglion cell layer. Scale bars for all images: 50 μm.
NHE8 proteins are detected in RPE cells as well as in photoreceptor cells, especially in the inner segments (Fig. 4D), we further tested whether a photoreceptor-specific knockdown of NHE8 led to any retinal abnormalities in mature mice. Human rhodopsin promoter driven NmCas9 AAVs were prepared for photoreceptor cell specific knockdown. A mixture (1:1:1 ratio) of AAV9-hRho-NmCas9, scAAV9-Nhe8-sgRNAs-P1/P2 and scAAV9-Nhe8-sgRNAs-E2/E5/E6 was injected subretinally into the right eyes of 4-week old wild-type mice, and scAAV9-control sgRNAs mixed with AAV9-hRho-NmCas9 were injected into the left eyes of the same mice as controls. The results showed that the left control eye displayed normal retinal morphology (Fig. 4A, B, 4C, left images) while the right NHE8-knockdown eye displayed an obvious loss of photoreceptors with some areas only having one layer of photoreceptor cells (Fig. 4A, B, 4C, right images). Since HA-tag was constructed into the C-terminal of the Cas9 vector, the incorporation of AAV-Cas9 in the retina could be evaluated based on HA localization. HA staining signal (red) was localized within the degenerating photoreceptor layer of the right eye injected with a mixture (1:1:1 ratio) of AAV9-hRho-NmCas9, scAAV9-Nhe8-sgRNAs-P1/P2 and scAAV9-Nhe8-sgRNAs-E2/E5/E6 (Fig. 4C, right image). The HA signal was also detected in the control eye (Fig. 4C, left image).
Fig. 4. Photoreceptor specific knockdown of NHE8 by AAV-hRho-NmCas9 and AAV-Nhe8-KO sgRNAs caused retinal degeneration in adult wild-type mice.
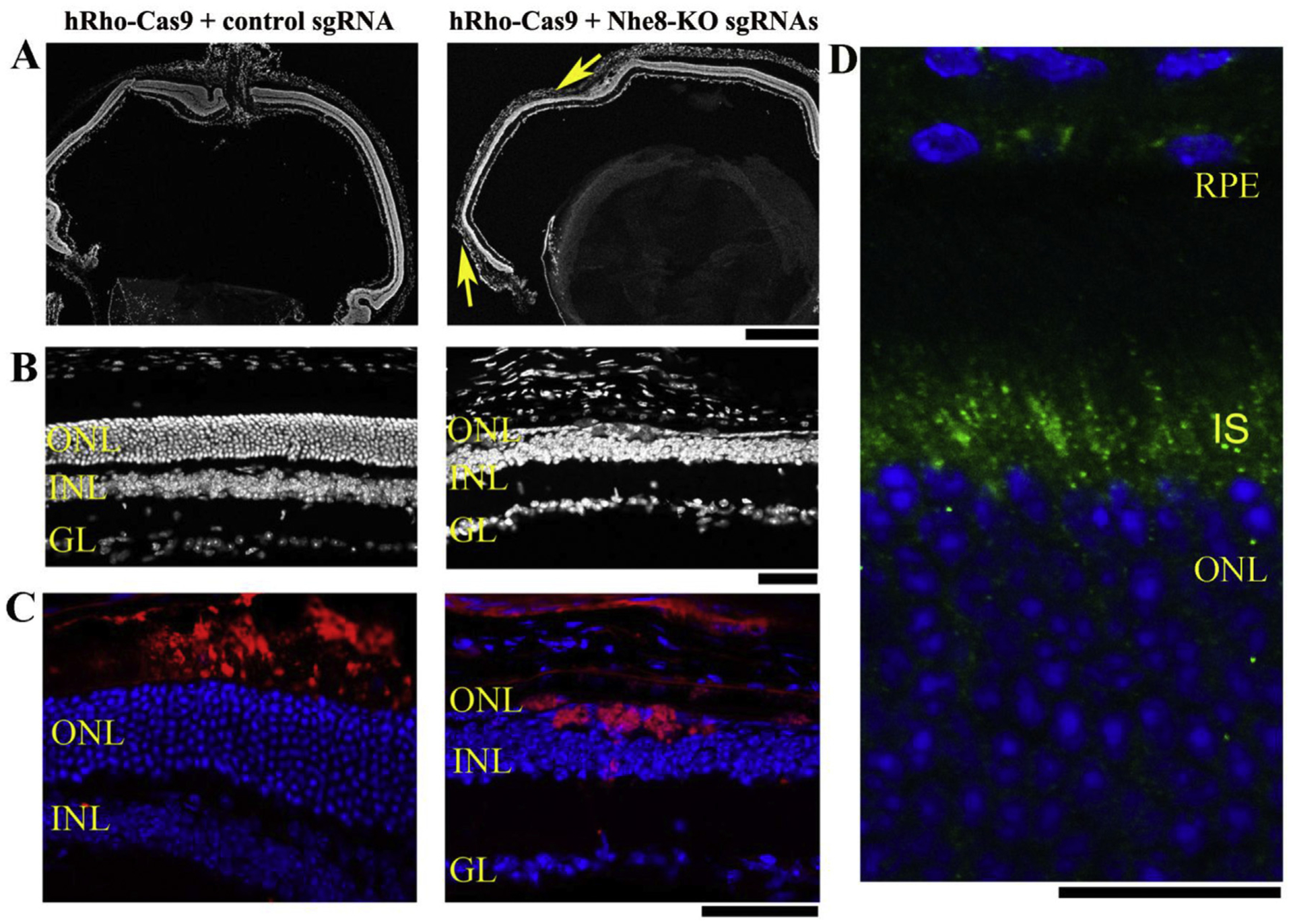
(A–C) The left panels are retinal sections of a control eye injected with a mixture of AAV-hRho-Cas9 and AAV-control sgRNA; the right panels are retinal sections from the right eye of the same mouse injected with AAV-hRho-Cas9, AAV-Nhe8-KO sgRNAs-P1/P2 and AAV-Nhe8-KO sgRNAs-E2/E5/E6. (A) A comparison of low magnification images of DAPI-stained retinal frozen sections between control and knockdown eyes. Two arrows (yellow) in the right image indicate areas with loss of photoreceptor cells. (B) DAPI-stained frozen retinal sections showed control on the left and the knockdown on the right with only one single layer of photoreceptor cells. (C) Images of retinal frozen sections co-stained with anti-HA for Cas9 protein (red) and DAPI (blue). HA tag was constructed in-frame into the C-terminus of Cas9 in the vector, thus the AAV-Cas9 expression can be traced using an antibody recognizing HA tag. ONL: outer nuclear layer; INL: inner nuclear layer; GL: ganglion cell layer. Scale bars: upper panels, 500 μm; middle and lower panels, 50 μm. (D) A retinal section of 3 weeks-old wild-type mouse stained with NHE8 antibody (green) and Dapi (blue). Endogenous NHE8 staining signals were observed in the retinal pigment epithelial cells (RPE) and inner segments (IS) of photoreceptor cells. Scale bar: 20 μm.
3.3. NHE8 protein expressed from recombinant AAV in mouse primary RPE cells
Based on RPE cell defect occurred in the retinas of NHE8 mutant mice reported previously (Jadeja et al., 2015; Xia et al., 2015) and the NHE8 knockdown mice here, we further investigated the role of NHE8 in sodium/pH homeostasis in RPE cells. Both AAV5-CBA-NHE8 and AAV5-CBA-mutant NHE8 were generated to study the potential functions of NHE8 in pH regulation of RPE cells. Distribution of wild-type NHE8 proteins expressed from AAV5 was tested in primary RPE cell cultures. To avoid the potential interference of dark pigments of RPE cells isolated from wild-type C57BL/6J in imaging study, B6-albino (without pigments) mice were used for imaging studies. The distribution of NHE8 proteins delivered by AAV5-CBA-NHE8 was studied in primary RPE cells isolated from wild-type B6-albino mice (Fig. 5A and B). Like the subcellular localization of endogenous NHE8, transiently expressed NHE8 proteins by AAV transduction (red) displayed intracellular vesicular staining pattern, with prominent perinuclear staining, which co-localized with the trans-Golgi complex marker TGN46 (green) (Fig. 5A); while NHE8 proteins showed few not obvious co-localization with the ER marker calreticulin (green) (Fig. 5B). The AAV transduced mutant NHE8 proteins displayed similar intracellular vesicular distribution pattern as wild-type NHE8 proteins. Thus, NHE8 proteins expressed from AAVs resembled the localization of endogenous NHE8 in the trans-Golgi complex, as well as various vesicular structures. AAV-NHE8 expression system provides a useful way to monitor pH changes while the location of NHE8 protein can be visualized in cultured RPE cells. We further constructed AAV5-smCBA-WT-NHE8-mCherry-pHluorin and AAV5-smCBA-mutant-M120K-NHE8-mCherry-pHluorin to study the role of wild-type and mutant NHE8 in intracellular pH regulation of RPE cells.
Fig. 5. Primary RPE cells isolated from wild-type albino B6-white mice. RPE cells were infected with AAV5-CBA-NHE8 and stained with NHE8 and intracellular organelle markers.
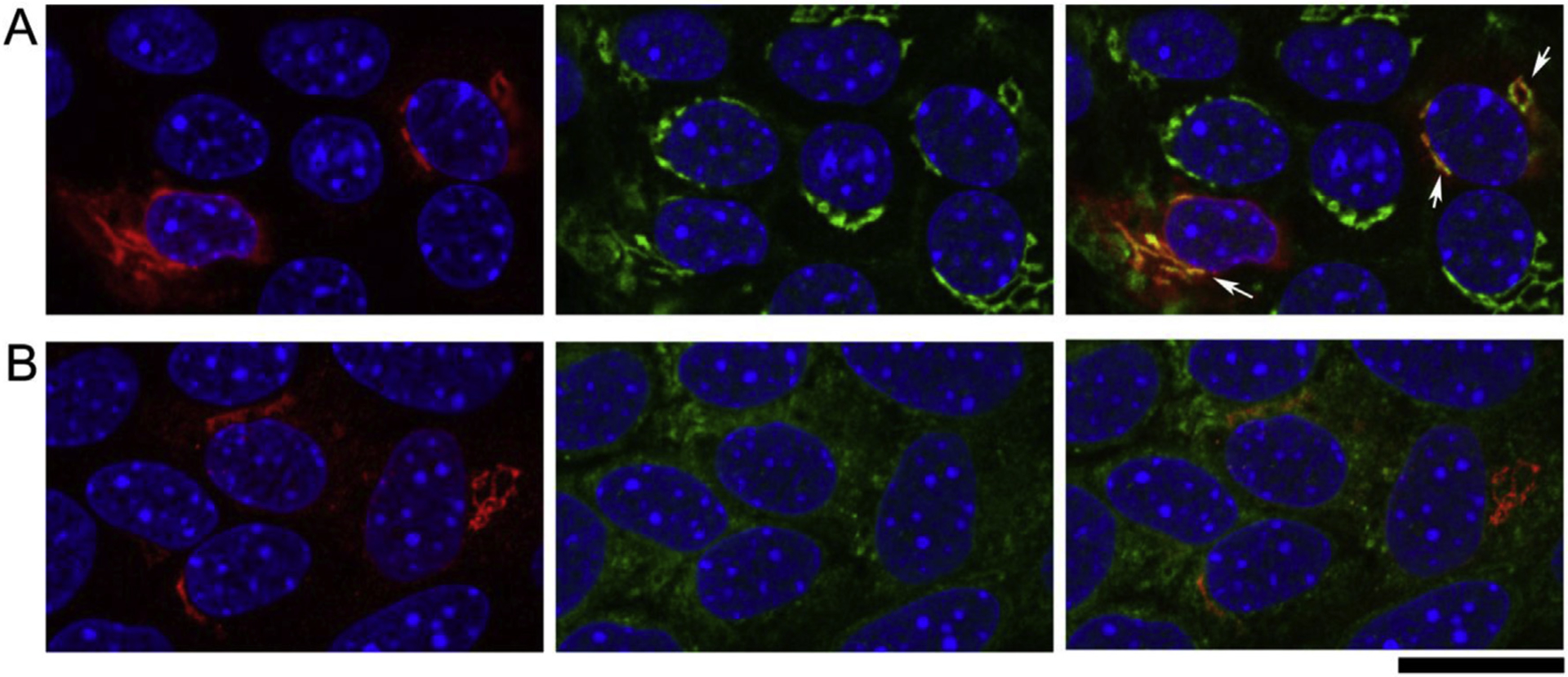
(A) Co-staining of anti-NHE8 (red) and anti-TGN46 (green) revealed co-localization of NHE8 and the Trans-golgi marker. Due to the weak endogenous NHE8 staining, we used the AAV-NHE8 to infect the cells to achieve more robust NHE8 staining. In the two cells (white arrows) with the most intense NHE8 staining, the red NHE8 signals were prominently expressed near nuclear, while the signals were also expressed throughout the cytosol. Co-staining with the Trans-golgi marker TGN46 revealed co-localized peri-nuclear NHE8 expression with the Golgi marker. (B) Co-staining of anti-NHE8 (red) and anti-calreticulin that labels endoplasmic reticulum revealed the prominent peri-nuclear NHE8 signals were not co-localized with ER, although the cytosolic NHE8 might show some co-localization with the ER marker. Scale bar: 20 μm.
3.4. Intracellular pH response curves in hRPE cells
To test a hypothesis that NHE8 plays an essential role in the intracellular pH regulation in RPE cells, and mutant M120K-NHE8 (r15-NHE8) alters its pH regulation to cause retinal degeneration in NHE8 knockout and r15 mutant mice (Xia et al., 2015), we constructed AAV-NHE8 with a pH response marker of the mCherry-pHluorin fusion gene. Fluorescent intensity of pHluorin is sensitive to pH changes while the intensity of mCherry is insensitive. We generated AAV5 recombinant viruses (AAV5-smCBA-wt or r15-NHE8-mCherry-pHluorin) that express NHE8 proteins tagged in-frame with two fluorescent proteins mCherry and pHluorin. These two fluorescent proteins not only can be used to trace the NHE8 protein distribution in live cells, but also can be used to monitor the intracellular pH changes by measuring the fluorescent intensity ratios of pH-sensitive ecliptic pHluorin and pH-insensitive mCherry. We first tested the pH response system in human ARPE-19 (hRPE) cells, almost all cells displayed pHluorin and mCherry fluorescent signals after AAV transduction. The cells were infected with AAVs expressing the wild-type NHE8-mCherry-pHluroin and mutant M120K-NHE8-mCherry-pHluorin, followed by confocal imaging analysis after 3 days of AAV infection. We evaluated whether overexpressed wild-type NHE8 and mutant NHE8-M120K displayed different intracellular pH response with different extracellular pH titrations. Transduced cells were first imaged in live cell imaging solution at pH 7.4; the same cells were sequentially imaged in various pH buffers from pH 7.5, pH 6.5 and pH 5.5. Confocal images of hRPE cells infected with AAV-wt-NHE8-mCherry-pHluorin (Fig. 6A) and AAV-r15-NHE8-mCherry-pHluorin (Fig. 6B) showed that the pHluorin green fluorescent intensity decreased as the pH dropped while the mCherry red fluorescent intensity stayed relatively unchanged in various pH buffers; the cells in imaging solution and pH 7.5 buffer displayed similar intensity of both red and green fluorescent signals (the left two panels in Fig. 6A and B). The pHluorin/mCherry fluorescent intensity ratio, calculated from each cell under each pH buffer, was plotted against the pH value of various buffers to generate the pH-response titration curves (Fig. 6C). Wild-type NHE8 (wt-NHE8) and mutant M120K-NHE8 (r15-NHE8) pH titration curves showed a parallel shift, indicating mutant M120K-NHE8 transfected cells altered intracellular pH responses to the pH titration. The data suggest that NHE8 may play an important role to maintain intracellular pH homeostasis of RPE cells in response to the changes of extracellular pH.
Fig. 6. Wild-type NHE8 and mutant r15(M120K)-NHE8 proteins displayed different pH response curves in cultured human RPE cells.
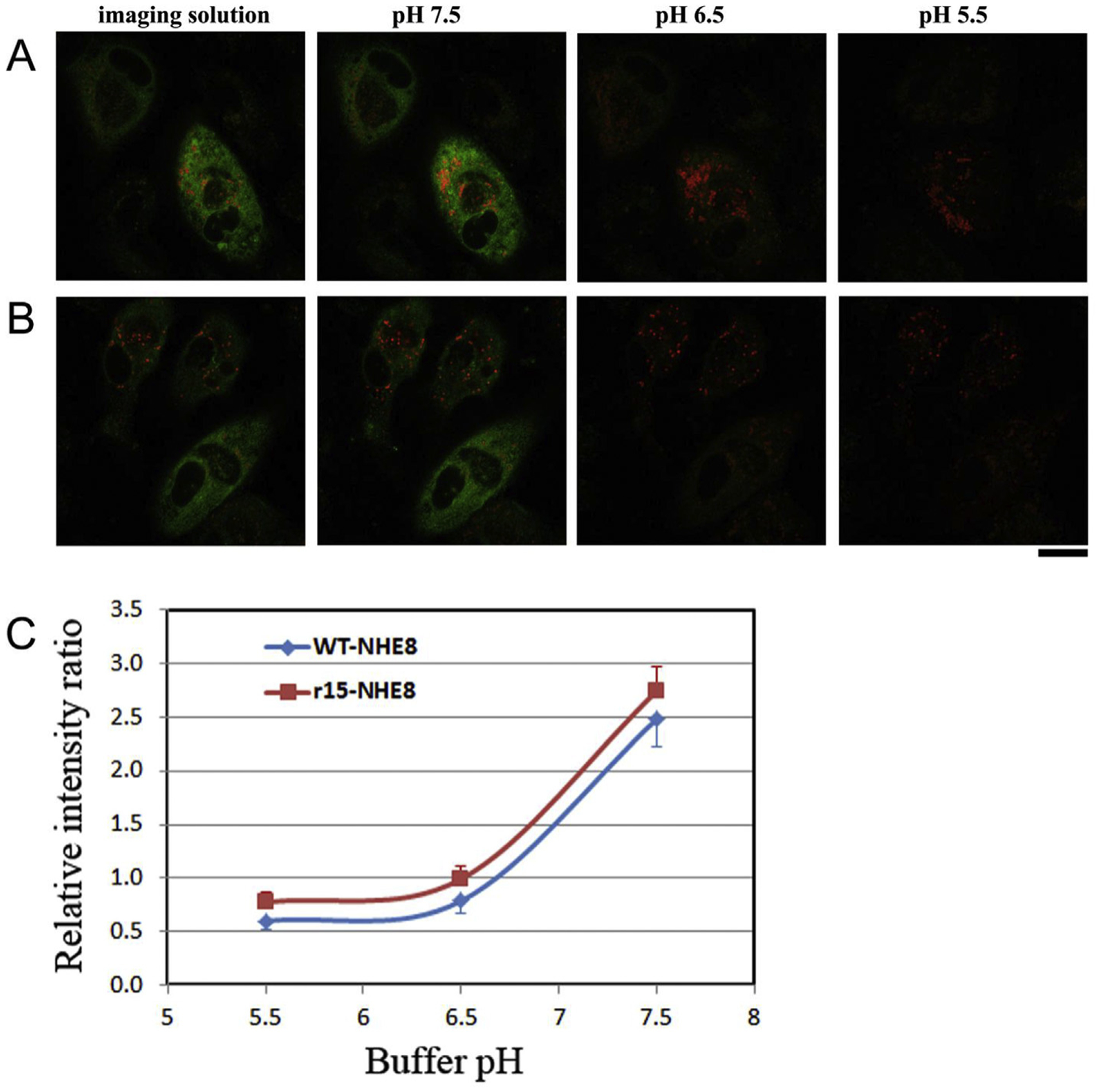
Cells were infected with AAV-wt-NHE8-mCherry-pHluorin and AAV-r15-NHE8-mCherry-pHluorin, respectively. (A) and (B) The cells were imaged in imaging buffer (pH 7.4) first, then subsequently in a series of gradually decreased pH buffers containing nigericin. pHluorin signals are pH sensitive, displaying diminished intensity as the pH decreases (from pH 7.5 to pH 5.5); while the mCherry signals are not sensitive to pH changes, showing relatively stable intensity as the pH drops, although the signals might become weaker due to photo bleaching. (A) The panels show wt-NHE8 AAV infected cells, and (B) the panels are mutant r15-NHE8 AAV infected cells. Scale bar: 20 μm. (C) The r15 mutation affects NHE8 protein’s pH response in hRPE cells. Mutant r15-NHE8 AAV infected cells displayed pH-response curve parallel shifted to the left of wt-NHE8 curve. The fluorescent intensity ratio of pH-sensitive pHluorin and pH-insensitive mCherry was calculated in each cell in various pH buffers containing nigericin, and the ratio was plotted against the buffer pH to generate a pH titration curve. Average ratio values from several cells were calculated and standard errors were shown (n = 3 for wt-NHE8 group, and n = 4 for r15-NHE8 group).
3.5. pH homeostasis in primary RPE cells isolated from wild-type and NHE8 mutant mice
We further performed pH titration experiment in primary cultured mouse RPE cells by using an evolved AAV variant 7m8 (Dalkara et al., 2013), which has better infection efficacy in mouse RPE cells than AAV5. The 7m8-wt-NHE8-mCherry-pHluorin and the 7m8-r15-NHE8-mChery-pHluorin viruses were produced. The pH response curves were generated in primary RPE cells, which were isolated from wild-type B6-albino mice and infected with 7m8-wt-NHE8-mCherry-pHluorin and 7m8-r15-NHE8-mChery-pHluorin viruses respectively (Fig. 7A). Similar to the pH response curves in hRPE cell lines, wt-NHE8 and mutant M120K-NHE8 proteins displayed different responses to pH titration in primary cultured wild-type RPE cells. However, unlike the pH titration curves observed in human RPE cell lines, where the mutant NHE8 caused a parallel shift to the left of the wt-NHE8 curve, the r15 (M120K)-NHE8 caused a shifted in pH 6.5 and pH 7.5, but not in pH 5.5 in the primary RPE cells (Fig. 7C, solid lines). This difference indicated that pH titration curves likely affected by different regulations in different cell types. Therefore, we directly evaluated whether cultured RPE cells of NHE8−/− mice would show different intracellular pH response to pH titration. We performed similar experiment in cultured RPE cells isolated from B6-albino NHE8 knockout mice. Interestingly, the pH titration curves of primary cultured NHE8 knockout RPE cells displayed no obvious shift between 7m8 delivered wt-NHE8 and mutant r15-NHE8 (dashed lines in Fig. 7C). This result further suggested altered intracellular pH regulation in NHE8−/− RPE cells.
Fig. 7. PH-response curves of wt-NHE8 and mutant r15-NHE8 in primary cultured mouse RPE cells isolated from albino wild-type mice and NHE8−/− mice.
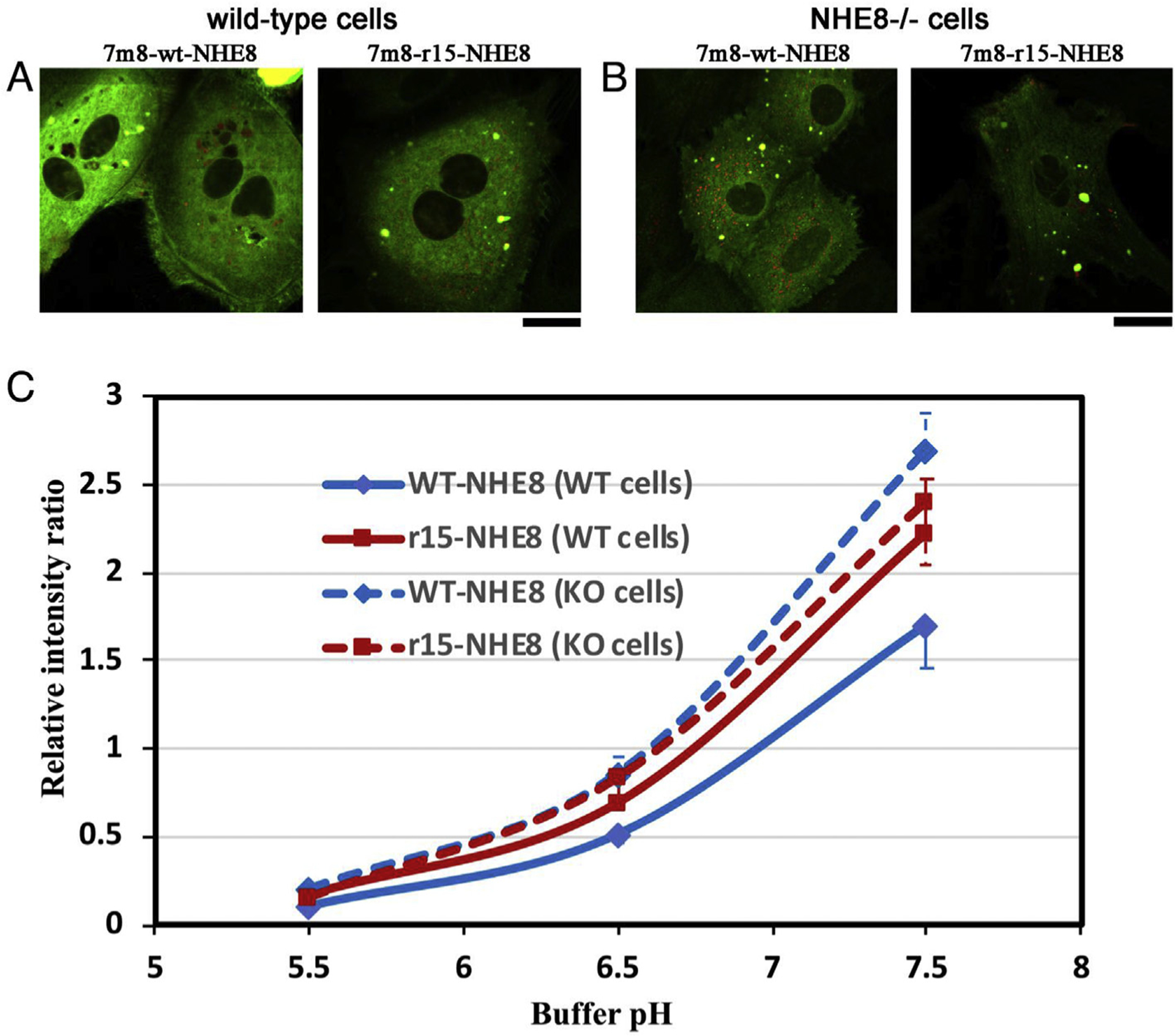
Primary RPE cells were isolated from albino-coated wild-type B6-white mice or NHE8−/− knockout mice, infected with AAVs and imaged in various pH buffers for the pH-titration curve. (A) Cells isolated from wild-type mice were imaged in imaging solution after infecting with the 7m8-wt-NHE8-mCherry-pHluorin or 7m8-r15-NHE8-mCherry-pHluorin. (B) Cells isolated from NHE8 knockout mice were imaged in imaging solution after AAV infection. (C) The pH-response curves of wt-NHE8 and mutant r15-NHE8 in cells isolated from wild-type mice (WT cells, solid lines) and knockout mice (KO cells, dashed lines). Similar to previous hRPE cell line data, in the RPE cells isolated from wild-type mice, mutant r15-NHE8 protein shifted the pH response curve to the left of the wt-NHE8, although it was not parallel since the pH 5.5 points merged possibly due to diminished pHluorin intensity; standard errors were labeled on the curve (n = 4 for both wt- and r15-NHE8 groups). However, in the RPE cells isolated from the NHE8−/− mice, no distinct difference in the pH-titration curves of wt-NHE8 and r15-NHE8 was observed; standard errors were labeled (n = 8 for wt and n = 4 for r15 groups). Scale bar: 20 μm.
4. Discussion
This work indicates that NHE8 is required for the survival of photoreceptor cells as well as for maintaining the intracellular pH homeostasis of RPE cells. Using the CRISPR-Cas9 gene editing system, we have demonstrated that NHE8 knockdown in mature retina could cause a loss of photoreceptor cells. Photoreceptor specific knockdown of NHE8 further suggests that NHE8 is critical for the survival of rod photoreceptor cells in mouse retina. Thus, NHE8 likely plays an essential role for the survival of photoreceptor cells at any given age. Moreover, the pH titration results between wild-type NHE8 and mutant M120K-NHE8 imply that NHE8 plays a role in the regulation of intracellular pH in RPE.
NHE8, encoded by the slc9a8, is a member of the sodium-hydrogen exchanger gene family that has a total of nine members (Slc9a1-Slc9a9), encoding NHE1-NHE9. The primary function of these trans-membrane proteins is to exchange sodium for proton across lipid bi-layers upon their electrochemical gradients. NHE1–5 are localized in the plasma membrane and NHE6–9 are predominantly intracellular isoforms (Fuster and Alexander, 2014). NHE8 is ubiquitously expressed in many other tissues, including intestine, brain and testis (Goyal et al., 2003; Xu et al., 2012). However, NHE8 proteins are detected in the plasma membrane, Golgi apparatus and/or endosomes in different cell types (Jadeja et al., 2015; Nakamura et al., 2005; Xia et al., 2015; Xu et al., 2008; Zhang et al., 2007). NHE8 protein localization has been studied extensively in human gastrointestinal tract (Xu et al., 2008) and in human testis (Xu et al., 2015). NHE8 also seems to play essential roles in endosomes in a previous in vitro study (Lawrence et al., 2010). Recent in vivo studies further demonstrate its functions in different tissues by using NHE8 mutant mice; impaired mucin synthesis and bicarbonate secretion occurs in the colon of NHE8 knockout mice (Xu et al., 2012), and NHE8 knockout male mice are infertile (Jadeja et al., 2015; Xu et al., 2015). Surprisingly, no abnormal kidney function has been found despite of high NHE8 expression levels there (Becker et al., 2007; Goyal et al., 2003).
Current results reveal that the AAV-CRISPR/Cas9 system is an effective way to deliver gene editing in regions of mouse retina and can effectively knock down the function of a given gene at any age. This is the first study to use AAV-nuclease null Cas9 to deliver CRISPRi in the retina of adult mice. AAV packaged nuclease null Cas9 allows the delivery of transcription factors, including enhancer VP64 and repressor KRAB, to any genes in adult animals. Recently, others have used AAV to deliver SpCas9 or SaCas9 for gene therapy in postnatal animals (Chew et al., 2016; Hung et al., 2016; Long et al., 2016; Nelson et al., 2016; Ran et al., 2015; Tabebordbar et al., 2016; Yang et al., 2016). Due to the limitation of AAV package length for targeting vector, we had to use the smaller size NmCas9 from Neisseria meningitides (Esvelt et al., 2013; Hou et al., 2013; Walsh and Hochedlinger, 2013) instead of widely used SpCas9 from Strepotococcus pyogenes (Cong et al., 2013; Esvelt et al., 2013; Jinek et al., 2012, 2013; Mali et al., 2013; Walsh and Hochedlinger, 2013). Using the smCBA or rhodoposin promoters to drive the expression of Cas9, sgRNAs designed to knockdown NHE8, we observed obvious photoreceptor loss. Specific knockdown of NHE8 in mature retina avoids any concerns about retinal phenotypes that might be resulted from early developmental consequences of NHE8 null or point mutations (M120K-NHE8) or even the tissue-specific knockout (Jadeja et al., 2015) occurred in embryonic or neonatal stages.
Photoreceptor cell death is one of the most obvious phenotypes in NHE8 mutant mice. NHE8 is expressed in the retinal pigment epithelial cells, photoreceptor cells and ganglion cells (Jadeja et al., 2015; Xia et al., 2015). Our previous studies have characterized the RPE defect and retinal degeneration in mutant M120K-NHE8 and NHE8 knockout mice (Xia et al., 2015); Another study reveals that RPE-specific knockout of NHE8 seems to be adequate to cause the photoreceptor cell death (Jadeja et al., 2015). However, using human rhodopsin promoter driven Cas9 to disrupt NHE8 function specifically in the photoreceptor cells, we have demonstrated that photoreceptor-specific knockdown of NHE8 is also sufficient to lead to photoreceptor cell degeneration. NHE8 proteins are localized in the inner segments of photoreceptor cells (Fig. 4D) and seem to play an essential role in the survival of photoreceptor cells. Thus, our study indicates that NHE8 probably play essential roles in both photoreceptor and RPE. Intriguingly, the CRISPR-Cas9 mediated NHE8 knockdown, which was induced in the mature retina, displayed more severe phenotypes (photoreceptor loss could be noticed earlier) than that of previous NHE8 mutants, which lack NHE8 function during early retinal development. At current stage, we do not know what causes the fast degeneration in the CRISPR/Cas9 NHE8 knockdown retina. It is possible that slow developing phenotypes of null mutant mice might result from the presence of other compensatory mechanism, such as different NHE isoforms or downstream pathological related molecules during early retinal development; however, such compensatory mechanism is no longer present in mature retina.
Moreover, using an in vitro system of AAV packaged NHE8-mCherry-pHluorin and cultured human cell line or mouse primary RPE cells, we have observed obvious difference of the pH titration curves between wild-type NHE8 and mutant NHE8 proteins expressed in cultured wild-type RPE cells (Fig. 7C). In the wild-type primary RPE cells, overexpressed mutant NHE8 proteins displayed shifted pH response curve, similar to that observed in the human RPE cell line; however, in the NHE8 knockout RPE cells, no obvious difference of pH titration curves was found between wild-type and mutant NHE8 proteins, both wild-type and mutant NHE8 proteins showed different pH response curves, indicating that these knockout cells had altered responses to pH titrations. It is possible that NHE8−/− RPE cells may rely on other unknown compensatory mechanism to control intracellular pH homeostasis. The results further support that NHE8 may play a role in pH homeostasis and mutant-NHE8 affects pH response in RPE cells. It remains important to further investigate how NHE8 regulates pH/sodium homeostasis in the cytosol as well as in the intracellular vesicles such as Golgi apparatus and endosomes in RPE. The NHE8 function in various intracellular vesicles in inner segments of photoreceptor remains unknown at current stage; perhaps, NHE8 plays an important role in pH/sodium homeostasis in vesicular structures in inner segment of photoreceptors. These important questions need to be further addressed in future studies.
Acknowledgements
The authors want to thank Dr. John Flannery for sharing AAV-smCBA, AAV-Rho, scAAV, AAV9-2YF and pHelper plasmids, Dr. Eddie Wang for modifying the software, and Dr. Yan Zhang for providing human RPE cDNA. This study was supported by a grant from the East Bay Community Foundation (XG) and a Core Grant for Vision Research from NIH P30EY003176 (RHK).
Supported by a grant from the East Bay Community Foundation (XG) and a Core Grant for Vision Research (NIH P30EY003176).
Footnotes
The authors declare no competing financial interests.
References
- Agata Y, Matsuda E, Shimizu A, 1999. Two novel Kruppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1beta/KRIP-1). J. Biol. Chem 274, 16412–16422. [DOI] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS, 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AM, Zhang J, Goyal S, Dwarakanath V, Aronson PS, Moe OW, Baum M, 2007. Ontogeny of NHE8 in the rat proximal tubule. Am. J. Physiol. Ren. Physiol 293, F255–F261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J, Sun D, Kariko K, 1995. Sequence analysis of the 5.34-kb 5’ flanking region of the human rhodopsin-encoding gene. Gene 167, 317–320. [DOI] [PubMed] [Google Scholar]
- Byrne LC, Lin YJ, Lee T, Schaffer DV, Flannery JG, 2015. The expression pattern of systemically injected AAV9 in the developing mouse retina is determined by age. Mol. Ther.: J. Am. Soc. Gene Ther 23, 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM, 2016. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, Flannery JG, Schaffer DV, 2013. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med 5 189ra176. [DOI] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Lee T, Hoffmann NV, Schaffer DV, Flannery JG, 2012. Enhanced gene delivery to the neonatal retina through systemic administration of tyrosine-mutated AAV9. Gene Ther. 19, 176–181. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM, 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Godino R, Garland DL, Pierce EA, 2016. Isolation, culture and characterization of primary mouse RPE cells. Nat. Protoc 11, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster DG, Alexander RT, 2014. Traditional and emerging roles for the SLC9 Na+/H + exchangers. Pflugers Archiv.: Eur. J. Physiol 466, 61–76. [DOI] [PubMed] [Google Scholar]
- Goyal S, Vanden Heuvel G, Aronson PS, 2003. Renal expression of novel Na+/H+ exchanger isoform NHE8. American journal of physiology. Ren. Physiol 284, F467–F473. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA, 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A 110, 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SS, Chrysostomou V, Li F, Lim JK, Wang JH, Powell JE, Tu L, Daniszewski M, Lo C, Wong RC, Crowston JG, Pebay A, King AE, Bui BV, Liu GS, Hewitt AW, 2016. AAV-mediated CRISPR/cas gene editing of retinal cells in vivo. Invest. Ophthalmol. Vis. Sci 57, 3470–3476. [DOI] [PubMed] [Google Scholar]
- Jadeja S, Barnard AR, McKie L, Cross SH, White JK, Sanger Mouse Genetics P, Robertson M, Budd PS, MacLaren RE, Jackson IJ, 2015. Mouse slc9a8 mutants exhibit retinal defects due to retinal pigmented epithelium dysfunction. Invest. Ophthalmol. Vis. Sci 56, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J, 2013. RNA-programmed genome editing in human cells. eLife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS, 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc 8, 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SP, Bright NA, Luzio JP, Bowers K, 2010. The sodium/proton exchanger NHE8 regulates late endosomal morphology and function. Mol. Biol. Cell 21, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN, 2016. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, 2013. RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Monahan PE, Samulski RJ, 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8, 1248–1254. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE, 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H, 2005. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem 280, 1561–1572. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA, 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheide K, Puchkov D, Jentsch TJ, 2017. Loss of the Na+/H+ exchanger NHE8 causes male infertility in mice by disrupting acrosome formation. J. Biol. Chem 292, 10845–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J, Grinstein S, 2004. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Archiv.: Eur. J. Physiol 447, 549–565. [DOI] [PubMed] [Google Scholar]
- Pang JJ, Boye SL, Kumar A, Dinculescu A, Deng W, Li J, Li Q, Rani A, Foster TC, Chang B, Hawes NL, Boatright JH, Hauswirth WW, 2008. AAV-mediated gene therapy for retinal degeneration in the rd10 mouse containing a recessive PDEbeta mutation. Invest. Ophthalmol. Vis. Sci 49, 4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H, Dinculescu A, Li Q, Min SH, Chiodo V, Pang JJ, Zhong L, Zolotukhin S, Srivastava A, Lewin AS, Hauswirth WW, 2009. High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol. Ther.: J. Am. Soc. Gene Ther 17, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ, 2016. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R, 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh RM, Hochedlinger K, 2013. A variant CRISPR-Cas9 system adds versatility to genome engineering. Proc. Natl. Acad. Sci. U.S.A 110, 15514–15515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Liu H, Cheung D, Tang F, Chang B, Li M, Gong X, 2015. NHE8 is essential for RPE cell polarity and photoreceptor survival. Sci. Rep 5, 9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Liu H, Cheung D, Wang M, Cheng C, Du X, Chang B, Beutler B, Gong X, 2008. A model for familial exudative vitreoretinopathy caused by LPR5 mutations. Hum. Mol. Genet 17, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chen H, Dong J, Lynch R, Ghishan FK, 2008. Gastrointestinal distribution and kinetic characterization of the sodium-hydrogen exchanger isoform 8 (NHE8). Cell. Physiol. Biochem.: Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 21, 109–116. [DOI] [PubMed] [Google Scholar]
- Xu H, Chen H, Li J, Zhao Y, Ghishan FK, 2015. Disruption of NHE8 expression impairs Leydig cell function in the testes. American journal of physiology. Cell Physiol 308, C330–C338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang B, Li J, Wang C, Chen H, Ghishan FK, 2012. Impaired mucin synthesis and bicarbonate secretion in the colon of NHE8 knockout mice. American journal of physiology. Gastrointest. Liver Physiol 303, G335–G343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM, 2016. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol 34, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bobulescu IA, Goyal S, Aronson PS, Baum MG, Moe OW, 2007. Characterization of Na+/H+ exchanger NHE8 in cultured renal epithelial cells. American journal of physiology. Ren. Physiol 293, F761–F766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Li B, Mah CS, Govindasamy L, Agbandje-McKenna M, Cooper M, Herzog RW, Zolotukhin I, Warrington KH Jr., Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A, 2008. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. U. S. A 105, 7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]


