Abstract

We present an updated analysis of the linker and core histone proteins and their proteoforms in the green microalga Chlamydomonas reinhardtii by top-down mass spectrometry (TDMS). The combination of high-resolution liquid chromatographic separation, robust fragmentation, high mass spectral resolution, the application of a custom search algorithm, and extensive manual analysis enabled the characterization of 86 proteoforms across all four core histones H2A, H2B, H3, and H4 and the linker histone H1. All canonical H2A paralogs, which vary in their C-termini, were identified, along with the previously unreported noncanonical variant H2A.Z that had high levels of acetylation and C-terminal truncations. Similarly, a majority of the canonical H2B paralogs were identified, along with a smaller noncanonical variant, H2B.v1, that was highly acetylated. Histone H4 exhibited a novel acetylation profile that differs significantly from that found in other organisms. A majority of H3 was monomethylated at K4 with low levels of co-occuring acetylation, while a small fraction of H3 was trimethylated at K4 with high levels of co-occuring acetylation.
Introduction
The unicellular green microalga Chlamydomonas reinhardtii (Chlamydomonas) is a model photosynthetic microorganism studied by scientists across many fields and has been used to reveal processes related to photosynthesis, cilia and basal body biogenesis and function, lipid biosynthesis, and cell cycle control.1 As a consequence of decades of basic research, molecular genetic tools developed for Chlamydomonas have made it an attractive reference organism for research on sustainable biofuels and for production of high-value bioproducts.2 The continued development of Chlamydomonas as a green algal model system is partly hindered by lack of knowledge regarding its epigenetic mechanisms that can interfere with consistent high-level transgene expression.3 Epigenetic mechanisms are partly mediated through covalent modifications of DNA (e.g., through methylation) and of histone proteins. The core histones are a family of small, basic proteins that form an octameric cylinder around which ∼147 bp of DNA are wrapped to create a nucleosome. Each histone octamer has two copies each of the four major core histones: H3, H4, H2A, and H2B. Nucleosomes can be further organized by association with linker histones (H1) and other scaffolding proteins into higher order chromatin. Besides their highly structured core regions that interact to form histone octamers, histones also have amino- and/or carboxy-terminal extensions or tails that are unstructured which, along with the globular core region,4 can be subject to post-translational modifications (PTMs). These modifications, in turn, can recruit additional chromatin-modifying enzymes and proteins (aka histone readers) that reshape chromatin structure and accessibility to influence transcriptional activation and repression.5,6
The Chlamydomonas genome has between 32 and 34 copies of each core histone gene arranged as clusters, most with a tandem tail-to-tail pair of H2A–H2B and of H3–H4. Additionally, there are three different genes for the linker histone H1 (Table S1). Most of the core histone genes have a tightly controlled expression profile, with the highest expression during S/M phase of the multiple fission cell cycle.7,8 Core histone proteins arising from these replication-dependent gene clusters are generally referred to as “canonical” core histones. In some cases, the protein sequences arising from these paralogous genes are nearly identical (e.g., histone H4), while in other cases they differ by several amino acids (e.g., histone H2B). In this study, we use the term “canonical variants” to describe these types of histone proteins. In addition to these canonical core histones, there are noncanonical core histone genes that are often found as a single gene outside of the histone gene cluster and may exhibit constitutive, replication-independent gene expression. Additionally, their sequences and expression patterns may deviate significantly from those of canonical core histones and have specialized functions, such as the centromeric histone H3 (e.g., Cre16.g661450 or cenH3.1 in Chlamydomonas). We will refer to these types of histone proteins as “noncanonical variants” throughout this manuscript.
Investigation of Chlamydomonas histones and their PTMs has revealed similarities with those in other organisms. For example, methylation at H3K9 by the SU(VAR)3–9 family of enzymes is known to create and/or maintain silenced heterochromatin in insects, mammals, plants and fungi.9,10 In Chlamydomonas, H3K9 monomethylation, catalyzed by the SU(VAR)3–9 ortholog Set3p, is also involved in silencing.11 Another common histone PTM, acetylation on H3 and H4, is correlated with actively transcribed genes in diverse eukaryotes, including Chlamydomonas.12,13 While many histone PTMs and their impacts on chromatin are conserved across eukaryotes, it is increasingly clear that the histone code is not completely universal.14,15 Even when a PTM is conserved, its epigenetic consequences may be different. For instance, monomethylation on histone H3 at lysine 4 (H3K4me1) is used to prime enhancers in human cells16 but serves as a repressive mark in Chlamydomonas.11,15,17
We previously reported on the most abundant Chlamydomonas core histone modifications from a cell-wall-less (cw) strain using top-down mass spectrometry (TDMS).18 However, cw strains of Chlamydomonas are not suitable for all investigations, especially when considering stress responses that may be modified in wall-less strains could impact chromatin architecture. Subsequently we developed an improved histone extraction method to increase histone yields from cell-walled Chlamydomonas.19 Here, using a further improved histone extraction method for walled strains, advanced instrumentation for TDMS, and custom MS/MS analysis software, we were able to identify Chlamydomonas wild-type strain histone PTMs in greater depth than previously possible. Unlike bottom-up approaches which involve proteolytic digestion before analysis, TDMS allows all modifications on a single histone protein to be detected simultaneously, enabling relationships between different histone marks to be determined without inference. We identified a total of 86 histone proteoforms20 including detection of larger histones H1 and ubiquitylated H2B, complex H4 acetylation isomers, highly acetylated noncanonical variants of H2B and H2A, and a bimodal distribution of H3 where H3K4me1 correlated with H3 hypoacetylation and H3K4me3 correlated with H3 hyperacetylation. Our findings significantly expand the atlas of characterized histone proteoforms in Chlamydomonas and provide context for understanding the constellations of different PTMs that co-occur on individual histone proteins. These data pave the way for further biological studies, such as exploration of PTM dynamics in different growth conditions, cell cycle stages, and in mutants with altered chromatin structure and gene expression.
Methods
Cell Culture and Harvesting
Chlamydomonas cultures were grown and harvested using a method modified from that previously published by our lab.19Chlamydomonas reinhardtii CC-1690 21gr mating type plus (mt+) wild type (WT) were grown in 300 mL of tris-acetate phosphate (TAP) media in 500 mL Erlenmeyer flasks, in temperature-controlled 25 °C water baths, under constant illumination by LEDs with 150 μE each of red (625 nm) and blue (465 nm) light, bubbling with air, to a density of (1–2) × 106 cells/mL. Cells were harvested by centrifugation at 4000g for 10 min at room temperature. Cell pellets were then resuspended in 50 mL of 1× phosphate-buffered saline (PBS), transferred to a 50 mL conical tube, and centrifuged again at 3500g for 5 min, after which the supernatant was discarded. Pelleted cells were immediately flash-frozen in liquid nitrogen and stored at −80 °C.
Nuclei Enrichment
A nuclei enrichment protocol based on previously reported methods was used on frozen cell pellets.18,19 A 2× stock of nuclei isolation buffer (2× NIB) (1.2 M sucrose, 20% v/v glycerol, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 40 mM potassium chloride (KCl), and 40 mM magnesium chloride (MgCl2)) was prepared ahead of time, filter-sterilized using a 0.22 μm pore size poly(ether sulfone) (PES) bottle-top vacuum filter (CellPro, V50022), and stored at 4 °C. At time of use, the 2× NIB stock was diluted to a 1× concentration with water, protease inhibitors, and histone deacetylase inhibitor for a working concentration of 1× NIBA: 1× NIB with working concentrations of 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM dithiothreitol (DTT), 10 mM sodium butyrate, and 0.5× protease inhibitor cocktail (PIC; Roche cOmplete, ETDA-free, 45148300). NIBA and NIBA-containing samples were kept on ice unless otherwise specified.
Frozen cell pellets in 50 mL conical tubes were thawed on ice and resuspended in 5 mL of NIBA + 5% v/v Triton X-100 (CAS No. 9002-93-1), then refrozen dropwise into a pool of liquid nitrogen in a 25 mL capacity stainless steel screw-top grinding jar (Retsch, 014620213). Liquid nitrogen was allowed to boil off, the grinding jar’s matching 15 mm stainless steel ball (Retsch, 053680109) was added to the jar, and then the jar was sealed and submerged in liquid nitrogen to keep the sample frozen. Samples were macerated using a Retsch mixer mill (MM400) at 30 Hz for two rounds of 90 s each, in between which the grinding jar was resubmerged in liquid nitrogen to prevent thawing. These macerated cells were transferred while still frozen, using a small metal spatula, to a 50 mL conical centrifuge tube and resuspended in 20 mL of additional ice-cold NIBA + 5% Triton X-100, thawed on ice, and then incubated on ice for 10 min. Samples were centrifuged at 1500g for 30 min at 4 °C. The resulting nuclei-enriched pellet was gently washed once by adding 20 mL of NIBA without detergent, swirled (but not pipetted) vigorously to resuspend the pellet, and centrifuged again at 1500g for 10 min at 4 °C. Finally, the washed nuclei-enriched pellet was gently resuspended by pipetting in another 1 mL of NIBA without detergent and transferred to a 1.5 mL tube, and pelleted again at 2000g for 10 min at 4 °C. At this step and in all subsequent steps, all 1.5 mL tubes used were low-protein-binding (Life Technologies, 90410). The wash supernatant was aspirated, and the nuclei-enriched pellet was flash-frozen in liquid nitrogen and stored at −80 °C.
Histone Extraction
The nuclei-enriched pellet (typically 100–200 μL) was thawed on ice and then mixed with 1 mL of ice-cold histone extraction buffer (HEB) (2 M CaCl2, 10 mM HEPES, 10 mM sodium butyrate, 5 mM DTT, and 1 mM PMSF, and 0.5×–1× PIC). Nuclei were incubated in HEB, rotating for 1 h at 4 °C, for salt-extraction of the histones from the nuclei-enriched pellet. The sample was acidified by adding 25 μL of concentrated hydrochloric acid (HCl) for a final concentration of approximately 0.3 M HCl and rotated at 4 °C for 20 min. The acidified sample was then centrifuged at 5000g for 10 min at 4 °C to pellet the acid-insoluble material. The ∼1 mL of the salt-extracted and acid-soluble supernatant was transferred to a new tube, and 250 μL of 100% trichloroacetic acid (TCA) (CAS No. 76-03-9, BeanTown Chemical, 144045) was added for a final concentration of 20% TCA and incubated stationary on ice for 1 h to precipitate the protein. The precipitated proteins were pelleted by centrifugation at 14000g for 5 min at 4 °C. Using glass Pasteur pipettes to handle all organic solvents, the pellet was washed once each with 1 mL of ice-cold 20% TCA prepared fresh from a 100% TCA stock, 1 mL of 99.9% HPLC-grade acetone (CAS No. 67-64-1, J.T. Baker 9254-02) with 0.1% HCl (stored at −20 °C), and 1 mL of 100% HPLC-grade acetone (stored at −20 °C) and centrifuged at 14000g for 5 min at 4 °C in between each wash. These TCA and washing steps desalt the proteinaceous pellet, and no further purification (e.g., desalting tip) was done prior to liquid chromatography with tandem mass spectrometry (LC–MS/MS) analysis. The washed histone pellet was dried at room temperature for 10 min to evaporate the acetone. Water-soluble histones were extracted from this washed TCA-precipitated pellet by adding 50 μL of ultrapure water at room temperature, crushing and breaking up the pellet with a glass pestle fashioned by melting the tip of a 2 mL glass Pasteur pipet, centrifuging at 14000g for 5 min at 4 °C to pellet the insoluble material, and transferring the water-soluble proteins in the supernatant to a new tube. This water extraction was repeated a second time on the remaining TCA-precipitated pellet, and the second supernatant was pooled with the first. This 100 μL of pooled water-soluble protein extract were centrifuged again at 14000g or 5 min at 4 °C to pellet any remaining precipitate, and the doubly cleared supernatant was flash-frozen in liquid nitrogen and then either stored at −80 °C or lyophilized for shipment at ambient temperature.
Liquid Chromatography–Mass Spectrometry
Online reversed-phase liquid chromatography (Waters NanoAcquity or Thermo Dionex) coupled with high-resolution mass spectrometry (Thermo Orbitrap Eclipse or Lumos) was used to analyze intact histone samples. Mobile phase A (MPA) was 0.1% formic acid in water, and mobile phase B (MPB) was 0.1% formic acid in acetonitrile. The analytical column (i.d. 75 μm, o.d. 360 μm, length ∼70 cm) was custom packed with C18 particles (Phenomenex Jupiter 3 μm 300 Å). For some samples, a custom-made trap column (Separation Methods Technologies bulk C2 resin BMEB2-3-300, 3 μm 300 Å, i.d. 150 μM, o.d. 360 μm, length ∼5 cm) was used to further desalt the sample on the Waters LC by washing with 1% MPB for 5 min. It should be noted that salt adduction observed was minimal and equivalent in samples run both with or without the trapping column. The gradient of MPB was typically set to 1%, 10%, 20%, 44%, 60%, and 1% at 0, 5, 10, 165, 185, and 190 min, respectively.
To maximize spectral quality, a large number of microscans was used for MS (∼2.6 s per spectrum), and a long maximum injection time was used for MS/MS (one microscan) as follows. MS data were collected between 600 and 1600 m/z with 8 microscans, automatic gain control (AGC) 200% (8 × 105) and a maximum injection time of 50 ms. Three precursors within 600–1150 mass-to-charge ratio (m/z) and charge >5 were selected for both electron transfer dissociation (ETD) and higher-energy collisional dissociation (HCD). ETD had a reaction time of 17 ms, AGC target of 2000% (1 × 106) and maximum injection time of 1.5 s. HCD had stepped energy 30 ± 5%, AGC target of 2000% (1 × 106) and maximum injection time of 0.5 s. Isolation window was set to either 0.4 or 0.6 m/z. Dynamic exclusion was enabled for a duration of 80 s. Single charge state per precursor and exclusion was enabled for the first 90 min of the analysis. After 90 min, undetermined charge states were included to better capture low abundance proteoforms (e.g., for highly modified H3). For data-independent LC–MS/MS experiments, the instrument was configured as above but we incorporated an inclusion list with histone masses and m/z values of interest identified in previous data-dependent runs.
Analysis of TDMS Data (See the Supporting Information for an Expanded Data Analysis Section)
LC–MS/MS Visualization and Database Searching
Each Orbitrap LC–MS .raw data file was converted to the.mzML file format by MSConvert using the default options and no filters.21,22 The .mzML spectrum file was loaded into MASH Explorer23 for MS visualization or TopPIC24 for database searching with a FASTA file of the current, predicted Chlamydomonas nuclear encoded proteome (Phytozome v12). All of the basic and advanced parameters were left as default, with the exception of the maximum mass shift which was set to ±1000 Da. The results were manually reviewed through TopPIC’s web-based viewer. A Feature File map was generated by inputting the .mzML file into LcMsSpectator (results viewer of the Informed-Proteomics package)25 using the default parameters.
Targeted Precursor Mass Search
The .mzML file generated from each LC–MS run was processed into an MSALIGN file by using FLASHDeconv26 or TopFD. The MSALIGN is a text file containing the scan number, precursor monoisotopic m/z and mass, and fragment ion list. Our custom Python script SMC (Search MS and Combine MS/MS) was written in Python 3.8.4 and is available for download at www.github.com/pesavent/SMC. The MSALIGN file was input into SMC and queried for a specific precursor monoisotopic mass with a mass tolerance typically set to ±2–3 Da. The resulting text file included the scan (i.e., spectrum) number, precursor masses, and a combined list of fragment ions per MS/MS activation type (e.g., ETD or HCD). The scan number and precursor mass were cross-referenced with the parent LC–MS data file using Thermo’s QualBrowser software to ensure the precursor masses were all from the same or related proteoform. This parental LC–MS data file and the SMC-generated scan numbers were inputted into TDValidator27 (Proteinaceous, Inc.) for proteoform investigation and PTM localization.
Histone PTM Quantitation
Global and isomeric quantitation of histone PTMs were done as previously described28 and will be described here briefly. An example calculation can be found in the expanded data analysis methods in Supporting Information. Global abundance values of each intact histone mass were generated using MS1 ion intensities from summed LC–MS scans throughout the elution window of the proteoforms quantified. The summed LC–MS scans were deconvoluted in FreeStyle 1.7 (Thermo Scientific) or MASH Explorer to generate each proteoform’s monoisotopic mass and its respective relative ion intensity. Then, intensities of all deconvoluted masses above a signal-to-noise ratio (S/N) of 2 were summed to represent the total proteoform abundance within that LC elution window and the abundance of each mass feature was calculated as a fraction of the total to generate the protein intensity relative ratios (PIRR).28 For mass features comprised of multiple proteoforms (e.g., positional isomers), the fragment ion intensity relative ratios (FIRRs) were used to approximate the abundance of each isomer.28 These values were then multiplied by the PIRR of the corresponding precursor mass to determine global abundance of a specific proteoform. For proteoform abundance values we report with an associated variance, the variance was calculated from three independent biological replicates and represents the standard deviation of that n = 3 population. For proteoform abundance values without standard deviations listed, we could not obtain sufficiently high-quality MS/MS data (when evaluated by either sequence coverage or high S/N fragment ion intensity values) from a sufficient number of biological replicates.
Results and Discussion
Improved Liquid-Chromatography Top-Down Mass Spectrometry of Salt-and-Acid Extracts from C. reinhardtii Nuclei Reveals Many Histone Proteoforms
We recently published a straightforward protocol that generates histone extracts suitable for liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis from walled algae,19 and here, we further improved the method for walled Chlamydomonas by incorporating cryogenic ball milling, which led to higher histone yields (see the Methods for more details and Figures S1 and S2). The increased yields and purity we achieved, when combined with enhanced chromatography, allowed for lower abundance histone proteoforms to be detected without resorting to off-line reversed-phase liquid chromatography (RPLC) purification or online two-dimensional LC. Others have found improved separation of histone proteoforms with different stationary phases such as C329 for TDMS and middle-down (MD) analysis and porous graphitic carbon (PGC)30 for MD and bottom-up (BU) analysis. Herein, we employed a versatile C18 reversed-phase (RP) LC method for global analysis of histone proteoforms (see the Methods). Data-dependent analysis (DDA) via LC–MS was performed with a shallow acetonitrile gradient for histones extracted from asynchronous Chlamydomonas cultures grown in continuously illuminated log-phase conditions (Methods).
Canonical histones, noncanonical histones, and the linker histone H1 were identified after eluting at distinct times during LC–MS (Figure 1A; see Table S1 for a complete list of Chlamydomonas histone genes). Partial oxidation of histones during their purification complicates TDMS analysis by creating multiple mass variants for each proteoform, some with (nearly) isobaric masses. However, the RPLC baseline separation of unoxidized, partially oxidized, and completely oxidized methionine-containing histones allowed for unambiguous assignment of PTMs without the need for additional sample modification (such as intentional oxidation31) for most Chlamydomonas histones. The degree of LC separation for histone oxidation states is exemplified in Figure 1B, where all oxidized H4 proteoforms containing M84ox eluted prior to the unoxidized forms. Even canonical H2B proteins, which contain two methionine residues, show three distinct regions of separation based on three possible oxidation states: 2 oxidized Met, 1 oxidized and 1 unoxidized Met, and 2 unoxidized Met (Figure 1A, box 2: far left, middle and far right mass clusters, respectively). The shallow elution gradient and enhanced separation allowed DDA of approximately 6000–9000 precursor ions per LC–MS analysis. We note that database searching identified many truncated histones (data not shown), generated as proteolytic fragments during protein extraction or from in vivo “histone clipping”32 (Supporting Information). These truncated proteins are apparent as small peptides (<10 000 Da) that follow the same elution profiles as the corresponding intact histones (Figure 1A). We plan to investigate these truncated forms in the future.
Figure 1.
Histone feature map from top-down liquid chromatography mass spectrometry (LC–MS) for all Chlamydomonas core and linker histones shows well-resolved proteoforms. (A) The vertical axis shows the intact mass of each detected polypeptide: the numbered boxes highlight elution regions of linker histone H1 (#1) and core histones H2B (#2), H4 (#3), H2A (#4), and H3 (#5). The color scale to the right of the vertical axis represents log10 ion intensity values. (B) Zooming in on histone H4 (gray box #3 in panel A) illustrates baseline-separation of oxidized versus unoxidized proteoforms and further separation based on methylation/acetylation within each oxidation state.
A New Custom Software Accelerates Hypothesis-Driven Top-Down Analysis of Histone Proteoforms
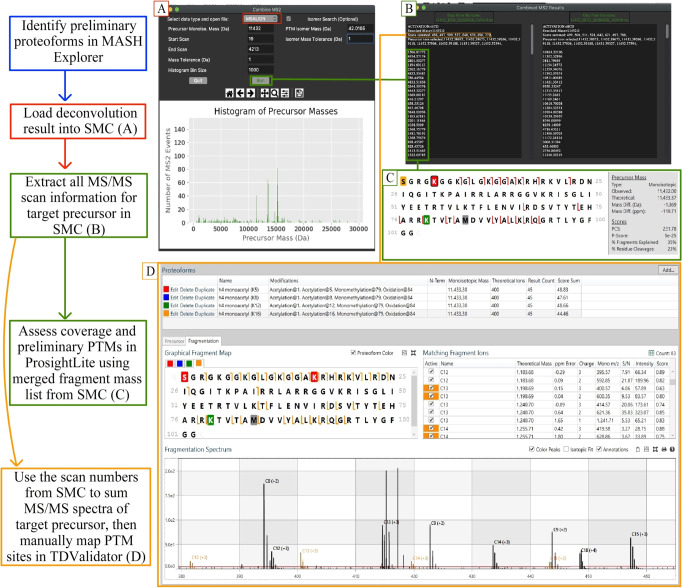
Computational assistance in interpreting mass spectral data is essential for efficient analysis of large data sets, and is exemplified by software tools such as MASH Explorer,23 ProsightPTM,34 and Skyline.35 A discovery-based approach using TopPIC24 and MSPathFinder25 within MASH Explorer to conduct database searches confirmed and expanded the list of detected Chlamydomonas histone proteins,18 as well as nonhistone proteins (mainly ribosomal proteins, other nuclear proteins, and truncated histones: see the Supporting Information). The redundancy in precursor masses selected for ETD and HCD enabled multiple low-intensity MS/MS spectra to be summed for better S/N and visualized, despite using DDA data. Here, we developed a freely available Python script named SMC (Search MS and Combine MS/MS) that accepts deconvoluted LC–MS data as an MSALIGN file (generated through TopFD24 or FLASHDeconv26), searches for MS/MS data from a specified precursor mass (MS) of interest (workflow shown in Figure 2A), and then combines the MS/MS information from all corresponding scans. SMC generates an output text file that includes MS/MS scan numbers, precursor masses and a combined list of fragment ions from the queried precursor mass and all corresponding MS/MS. These fragment ions and the hypothetical protein sequence can be entered into ProsightLite33 to enable quick, hypothesis-driven targeted investigation of the queried precursor mass. SMC allows for some analyses to be done with LC-MS data that can usually only be done using offline-separation and direct infusion of proteins, which includes summing multiple MS/MS scans per precursor ion. Furthermore, it allows for a hybrid approach, where MS/MS data from a range of precursor masses of related proteoforms (or the same form at different charge states) can be summed together to dramatically increase the S/N above that of a single-scan MS/MS event. The data shown in Figure 2 are from a targeted search for a mass of 11 432 Da (hypothesized to be a histone H4 proteoform) using SMC (Figure 2B). Eight MS/MS spectra matched our search query for this precursor mass, and the corresponding ETD fragment ions were inputted into ProsightLite along with a single Chlamydomonas histone H4 amino acid sequence. After PTM assignment, we preliminarily identified this precursor mass as the H4Nα-acK5acK79me1Met84ox proteoform (Figure 2C), but other PTMs were possible suggesting the presence of positional isomers (data not shown). To verify and quantify positional isomers comprising the 11 432 Da precursor mass, the same eight MS/MS spectral scans were summed using the proteoform validation software TDValidator27 (Proteinaceous, Inc.; Figure 2D). Indeed, the enhanced S/N afforded by summing scans revealed additional, low-abundance c ions confirming the presence of multiple monoacetylation isomers, including K12ac and K16ac (represented by orange c ions in Figure 2D). This approach was repeated for multiple precursor ions with each histone and was used to identify 86 histone proteoforms (Table 1). A total of 5 biological replicates were analyzed by LC–MS and in cases where a histone proteoform could be completely characterized in ≥3 of those replicates (such as the monoacetylated histone H4 above), we report its relative abundance with a standard deviation.
Figure 2.
Integration of custom SMC software (Search MS and Combine MS/MS) into a targeted data analysis workflow: investigation of a 11 432 Da mass hypothesized to be monoacetylated histone H4. A directed acyclic graph (left) shows the workflow starting with preliminary proteoform identification by database searching in MASH Explorer (left). (A) The deconvolution file (MSALIGN format) is then loaded into SMC, and the desired precursor mass is queried for tandem MS. (B) If tandem MS was performed, a list including all fragment ions and their respective scan numbers is generated. (C) Copy-pasting the fragment ions generated from (B) into ProsightLite33 allows quick hypothesis-driven proteoform investigation. (D) A deeper analysis can be performed by summing the scans from (C) and visualizing the data with TDValidator,27 which in this case resulted in the detection of isomeric proteoforms (e.g., c123+, c133+, c143+, c153+, orange and black ions).
Table 1. Table of Chlamydomonas Histone Proteoforms Identified by TDMS in This Studya.
| H1 | H1.1Nα-ac | acH1.2Nα-ac | |||
| H2A | H2A.0Nα-ac | H2A.1 | H2A.1Nα-ac | H2A.1Nα-acK5ac | H2A.1Nα-acK188ac |
| H2A. 2 | H2A.2Nα-ac | H2A.2Nα-acK5ac | H2A.2Nα-acK188ac | H2A.3 | |
| H2A.3Nα-ac | H2A.3Nα-acK5ac | H2A.4Nα-ac | H2A.ZNα-ac | H2A.ZNα-acK6acK14ac | |
| H2A.ZNα-acΔ143 | H2A.ZNα-acΔ142-143 | ||||
| H2B | H2BNα-me3.1c | H2BNα-me3.2c | H2BNα-me3.3c | H2BNα-me3.5c | H2BNα-me3.8c |
| H2BNα-me3.9c | H2BNα-me3.11c | H2BNα-me3.12c | H2BNα-me3.13c | H2BNα-me3.14c | |
| H2BNα-me3.15c | H2BNα-me3.v1 | H2BNα-me3.K7ac | H2BNα-me3.v1K11ac | H2BNα-me3.v1K7acK11ac | |
| H2BNα-me3.v1K7ac K11acK12ac | H2BNα-me3.v1K7acK11acK12acK16ac | ||||
| H3 | H3 | H3K4b/9me1 | H3K4me1 + 14 Da (K35-Q86) | H3K4/9me1+ 28 Da (K27-M119) | H3K4me1 + 42 Da (T28-Y40) |
| H3K4me1 + 56 Da (P37–F57) | H3K4me1 + 71 Da (L20-I118) | H3K4me1 + 84 Da (T28-T117) | H3K4me1 + 98 Da (K23-L99) | H3K4me1 + 112 Da (A23-R115) | |
| H3K4me1 + 125 Da (Q54-F77) | H3K4me1 + 140 Da (K23-T44) | H3K4me1 + 155 Da (K23-D105) | H3K4me1 + 167 Da (A25-F83) | H3K4me1 + K18ac +140 Da (Q19-L91) | |
| H3K9me1 + 197 Da (K23–I118) | H3K4/9me1 + 211 Da (G32-F103) | H3K4me3K9acK14acK18ac +70 Da (Q19-I118) | H3K4me3K9acK14ac +128 Da (Q23 - T79) | H3K4/9me1 + 255 Da unlocalized | |
| H3K4me3K9acK14acK18ac +115 Da (Q19-F77) | H3K4me3 + 174 Da (Q19-I118) or H3K4me1 + 283 Da (Q19-I118) | ||||
| H4 | H4Nα-ac | H4Nα-acK79me1 | H4Nα-acR3me1K79me1 | H4Nα-acK5acK79me1 | H4Nα-acK8acK79me1 |
| ac4K12acK79me1 | H4Nα-acK16acK79me1 | H4Nα-acK5acK8acK79me1 | H4Nα-acK5b/8acK12ac K79me1 | H4Nα-acK12acK16ac K79me1 | |
| H4Nα-acK5b/8acK16ac K79me1 | H4Nα-acK5acK8ac K12acK79me1 | H4Nα-acK5acK8ac K16acK79me1 | H4Nα-acK5acK12ac K16acK79me1 | H4Nα-acK8acK12acK16acK79me1 | |
| H4Nα-acK5acK8ac K12acK16acK79me1 | H4Nα-acK5acK12acK16acK20acK79me1 | H4Nα-acK5acK8acK12acK16acK20acK79me1 |
Proteoforms differing in methionine or cysteine oxidation were not considered. α-amino modification is denoted by “Nα-”, while the other histone PTMs follow standardized nomenclature.36 The names of each protein can be cross-referenced to their gene(s) using Table S1. H3 and H4 refer to the H3.1 and H4.1 sequences, respectively.
Indicates ambiguous isomer assignment.
Evidence of monoubiquitylation (see Figure S6).
Our extensive manual analysis, assisted by several software tools (e.g., TDValidator, TopPIC, MASH Explorer, Informed-Proteomics, FLASHDeconv, ProsightLite, and SMC) demonstrate the potential to extract detailed information on histone proteoforms from standard TDMS data sets that are often missed by automated data processing pipelines, such as explicitly defined isomeric proteoforms. Currently, such in-depth analysis still requires significant manual analysis and integration of outputs from multiple software packages. We believe future software development to further automate such analyses will significantly improve the speed and depth of complex proteoform characterization when using TDMS.
Histone H4 Post-translational Modifications Include H4R3me1, Multiple Isomeric Acetylation Profiles, and Ubiquitous H4K79me1
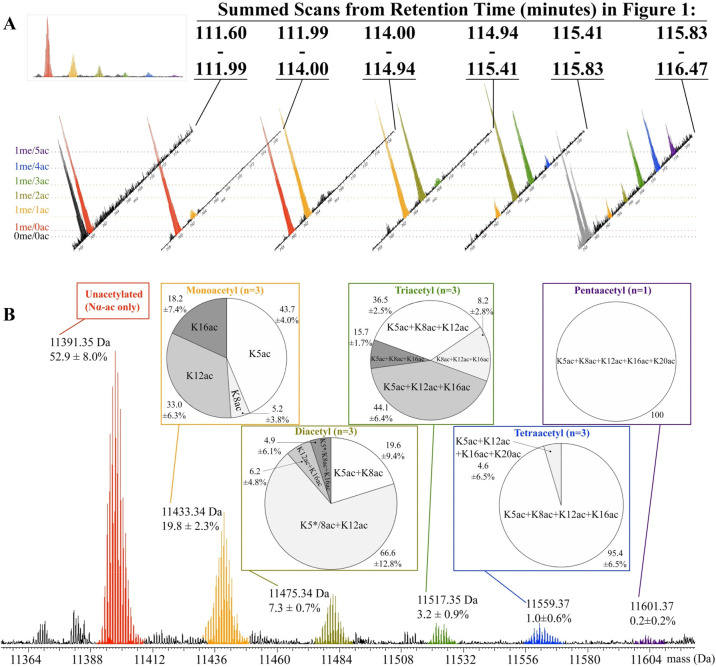
Of the 32 genes encoding H4, 30 encode the same protein sequence (H4.1). The other two H4 genes, H4.2 and H4.3, differ from H4.1 by 1 and 9 substitutions, respectively, and were not detected in our analysis (Table S1). Thus, the term “H4” is used here to specifically mean the H4.1 protein and its proteoforms. Quantification of histone proteoform abundances was established in previous studies,28,37 and involved summing total ion intensity over the LC–MS elution window, using fragment ion information if multiple proteoforms were present in the precursor mass. Label-free quantitation of proteins from TDMS data has been validated for both histone and nonhistone proteins.38−41 The high-quality ETD data for histone H4 from 3 biological replicates provided enough information to report quantitative differences in abundances along with standard deviation of those abundances. Unfortunately, this was not the case for other the histone proteins H3, H2A, H2B and H1 and we report their abundance values from a single LC–MS experiment. The average standard deviation in relative abundance of each H4 positional isomer (5.7%, Figure 3B) is in line with other TDMS studies28,29,42 but was slightly higher than modified histone peptides using bottom-up approaches (e.g., ∼1% for H3 peptides in ref (43)). In some cases, such as the H4NαacK5acK12acK16acK20acK79me1Met84ox proteoform shown in Figure 3B, the relative abundance value (4.6%) is close to its standard deviation (6.5%). Indeed, in some of the replicate LC–MS data, this tetraacetylated H4 form was either undetected or very low abundance. We suspect that the variability in isomeric composition (pie charts in Figure 3B) and precursor mass abundances (Figure 3B) reported here may arise from instrumental and biological sources. For instance, our histone proteoform characterization pipeline generates nanoLC–MS/MS DDA data sets and where low-abundance proteoforms produce highly variable MS/MS data. This may lead to situations where missed or poor S/N fragment ions contribute to variability in the isomeric quantitation because resolving low abundance ions from noise requires averaging more data points (scans). Another factor contributing to variability for intact masses with positional isomers separated by a few amino acid residues (e.g., diacetylated H4) is the ability to quantify these isomers when only one or two fragment ions differentiate them. We expect these instrumental sources of variability to be minimized in future studies, since the “atlas” of intact Chlamydomonas histone masses we report here will inform future data-independent analysis (DIA) experiments with more scans dedicated to targeted masses.
Figure 3.
Identification and quantitation of 15 internally acetylated H4 proteoforms in Chlamydomonas. (A) Top-down mass spectrometric profiles of unoxidized histone H4 (charge state z = 15) summed from the corresponding elution window depicted in (Figure 1B) with the isotopes colored to match the corresponding proteoform with increasing internal acetylation listed on the far left. Dotted horizontal lines connect the individual H4 masses with their corresponding proteoforms shown on the far left. The inset in (A) shows the summed spectra for all unoxidized histone H4 from retention time (tR) 111.5–116.5 min. Isotopes shaded gray (far right MS spectrum) are from a larger, non-H4 protein. (B) Representative summed mass spectrum corresponding to the oxidized H4 eluting from tR 105–110.5 min, also shown in Figure 1B. The six major acetylated forms were found to be internally unacetylated (11 391 Da; red), monoacetylated (11 433 Da; orange), diacetylated (11 475 Da; olive), triacetylated (11 517 Da; green), tetraacetylated (11 559 Da; blue), and pentaacetylated (11 601 Da; purple). The global abundance percentages listed below each mass are calculated from three biological replicates. All H4 proteoforms were found to have Nα-ac, K79me1, and M84ox along with acetylation site composition illustrated by the corresponding pie chart boxed above each peak. Three biological replicates (n = 3) were used in calculating the composition of each acetylation isomer except for pentaacetylated H4. An asterisk (*) indicates K5ac as the predominant second acetylation site (see the main text for more details). The abundance and isomeric composition of each form was estimated from mass spectra as described previously.28
The most abundant histone H4 proteoform had a modification profile that includes Nα-ac (i.e., α-amino acetylation), K79me1, and Met84ox (11 391 Da, Figure 3B). The methionine at position 84 is highly susceptible to oxidation and a significant portion was found to be in its sulfoxide state, as were other methionine residues present in all methionine-containing histones. The pattern of H4 PTMs is nearly identical between unoxidized and oxidized proteins (compare Figure 3A inset MS (unoxidized) with Figure 3B MS (oxidized)), and MS/MS analysis revealed similar proteoform composition between these oxidation states (data not shown). The lack of proteoform bias between oxidation states suggests that a majority of the oxidation is occurring in vitro, which has been documented in other studies.18,44 Because all histone H4 proteoforms were found to have Nα-ac, we will use the term “acetylation” here to specifically refer to internal acetylation (e.g., K5ac, K8ac, etc.).
We previously reported the detection of a unique, highly abundant histone H4 monomethylation at lysine 79, and our current analysis extends this finding to specify that >98% of H4 masses targeted for MS/MS contain K79me1. We did detect H4 lacking K79me1 at low levels (typically <2%), and remarkably, the only H4 proteoform clearly lacking this methylation is the unacetylated H4 form (Figure 3A, MS from tR 111.60–111.99 min). This suggests that K79 methylation may occur prior to the events leading to H4 acetylation, possibly occurring immediately after synthesis during S phase. Experiments involving cell cycle synchronization might assess the addition of methylation to newly synthesized histone H4 to provide clues about the timing of the ubiquitous H4K79me1. Such experiments must employ the TDMS approach, as the connection between all amino-terminally modified H4 proteoforms characterized and their carboxy-terminal K79 methylation would be lost by bottom-up or middle-down approaches (e.g., by GluC digestion). The only modification detected at K79 was monomethylation. H4K79me1 has yet to be detected from many other species including fruit flies, humans, yeast, and maize. However, the diatom P. tricornutum, the brown alga Ectocarpus, and the intracellular parasite T. gondii, each members of the SARs (Stramenopiles, Alveolates, and Rhizaria) supergroup do possess H4K79 mono-, di-, and trimethylation (reported by bottom-up MS).45−47 The extremely high abundance and lateral position of H4K79me1 make it likely to play a role in DNA-histone or nucleosome-nucleosome interactions. The enzyme(s) responsible for H4K79 methylation are currently unknown, so further understanding of this modification awaits their identification and characterization.
Besides H4K79me1, the only other methylation detected on H4 was a minor form (<1%) with monomethylation at R3 (H4R3me1). The H4 proteoform with R3me1 also had Nα-ac, K79me1, and M84ox and coeluted with the corresponding unmethylated R3 form (Mmi 11 405 Da, Figure 3; Figure S3). The low abundance of H4R3me1 and its coelution with more abundant H4 proteoforms often resulted in poor or completely absent MS/MS data for this species. Despite these limitations, we unequivocally identified H4R3me1, which is the first report of its presence in Chlamydomonas despite being identified and heavily studied in other organisms. Enzymes that methylate arginine residues are collectively referred to as protein arginine methyltransferases (PRMT). In humans, protein arginine N-methyltransferase 1 (PRMT1) can methylate H4R3 and leads to enhanced H4 acetylation by activating p300.48 While studies of Chlamydomonas PRMT1 (Cre03.g172550) have not tested for H4R3 methylation activity, they have shown its involvement in asymmetric dimethylation of arginine in proteins involved in flagella resorption in Chlamydomonas.49
Because the TDMS methodology used to quantify Chlamydomonas histone H4 was similar to that done for human HeLa H4,28,37 we compared acetylation abundances and isomeric composition between the two organisms. While the acetylation profiles of unacetylated and multiply acetylated H4 proteoforms in Chlamydomonas (Figure 3B) resemble those in human37 and Drosophila cells,50 there is a striking difference in acetylation site occupancies and global abundances of several acetylated proteoforms. First looking at the monoacetylated isomers of mass 11 433 Da, the most abundant monoacetylated H4 proteoform in Chlamydomonas was found to be acetylated at K5 (9.0%), which is 5-fold greater than K5 acetylation in HeLa (1.7% across all K20 methylation states42). Alternatively, Chlamydomonas K16ac represents only 3.6% of all detected histone H4 (Table 2 and Figure 3B), while in asynchronously grown human HeLa cells it represents the most abundant monoacetylated proteoform at 15% (across all K20 methylation states). Similarly, K12 acetylation was also found to be 5-fold greater in Chlamydomonas than HeLa cells, 6.8% vs 1.3%, respectively. The most abundant diacetylated H4 proteoform was H4Nα-acK5*/K8acK12ac (5.5%), followed by H4Nα-acK5acK8ac (1%) and minor amounts H4Nα-acK12acK16ac and H4Nα-acK5*/K8acK16ac (0.4% each). Analysis of acetylation site occupancies for diacetylated H4 is somewhat confounded by the inability to unambiguously assign K5ac versus K8ac with either K12ac or K16ac without MS/MS/MS.42 Thus, diacetylated H4 denoted by “K5*/K8ac” are predominantly acetylated at K5 over K8 (as denoted by the asterisk). Again, both the isomeric composition and the global abundance of the diacetylated H4 proteoforms are strikingly different than those found in HeLa cells: global abundance of all diacetylated H4 forms in Chlamydomonas account for 7.3% of all H4 while in HeLa it accounts for 2.6%. In Chlamydomonas, four combinations of triacetylated H4 proteoforms were identified, with two the most abundant being H4Nα-acK5acK12acK16ac (1.5%) and H4Nα-acK5acK8acK12ac (1.2%) (Table 2; Figure 3B, green box). The most highly acetylated H4 proteoforms, tetraacetylated and pentaacetylated H4, were found to be less complex, as expected. Of all tetraacetylated proteoforms, the majority were found to be H4Nα-acK5acK8acK12acK16ac (0.9%) with a minor portion being H4Nα-acK5acK12acK16acK20ac (0.1%), being present at a global abundance of 0.9% and 0.1%, respectively. Finally, we also identified a pure pentaacetylated species with acetylations at K5, K8, K12, K16 and K20 (Table 2, Figure 3B, indigo box) at extremely low levels (∼0.2% global abundance). While the intact mass was detected in most LC–MS analysis, either poor ETD fragmentation or lack of selection for MS/MS limited our ability to quantify this proteoform. It should be noted that while H4 acetylation isomers present in Chlamydomonas and HeLa cells are numerous and distinct, each acetylated H4 form (mono, di-, etc.) in S. cerevisiae is a pure species (i.e., no evidence of positional isomers) with the sole species being K16ac, K16ac + K12ac, K16ac + K12ac + K8ac and K16ac + K12ac + K8ac + K5ac for the monoacetylated, diacetylated, triacetylated, and tetraacetylated forms, respectively.51 In sum, we detected all Chlamydomonas H4 acetylation states as found in a previous radio-labeling study that required cycloheximide treatment to detect proteoforms above monoacetylation.52 It is likely these highly acetylated H4 forms are found in the promoter regions of actively transcribing genes and are absent from promoters of silent genes.12
Table 2. Global Abundances of Histone H4 Internal Acetylation Isomersa from Asynchronous Chlamydomonas Cultures.
| unacetylated | unacetylated 52.9 ± 8% | |||
|---|---|---|---|---|
| monoacetylated | K5ac | K8ac | K12ac | K16ac |
| 9.0 ± 1.4% | 1.1 ± 0.8% | 6.8 ± 1.5% | 3.6 ± 1.2% | |
| diacetylated | K5ac + K8ac | K5*/K8ac + K12ac | K12ac + K16ac | K5*/K8ac+K16ac |
| 1.0 ± 0.6% | 5.5 ± 1.4% | 0.4 ± 0.3% | 0.4 ± 0.5% | |
| triacetylated | K5ac + K8ac + K12ac | K5ac + K8ac + K16ac | K5ac + K12ac + K16ac | K8ac + K12ac + K16ac |
| 1.2 ± 0.4% | 0.5 ± 0.2% | 1.5 ± 0.5% | 0.3 ± 0.1% | |
| tetraacetylated | K5ac + K8ac + K12ac + K16ac | K5ac + K12ac + K16ac + K20ac | ||
| 0.9 ± 0.6% | 0.1 ± 0.1% | |||
| pentaacetylated | K5ac + K8ac + K12ac + K16ac + K20ac | |||
| 0.5%b | ||||
All acetylated isomers in this table were also modified at Nα-ac, K79me1, and M84ox. Variances represent the standard deviation from three biological experiments.
Only selected for MS/MS once among four LC–MS DDA experiments.
In animal cells and the yeast S. pombe, H4K20 is found to be heavily methylated (typically dimethylated) and this methylation is implicated in the establishment of heterochromatin, DNA damage repair and chromatin stability (reviewed in53,54). However, H4K20 methylation is not detected in Chlamydomonas, diatoms,46S. cerevisiae,51 or land plants.19,55,56 Instead, Chlamydomonas, Arabidopsis, and diatoms have low levels of H4K20ac.46 This study is the first to report acetylation at H4K20 in green algae, and most importantly, that this modification is extremely rare and is only found on tetra- and pentaacetylated proteoforms. Because bottom-up MS was used in the other studies examining H4K20ac, the relative abundance and other co-occurring PTMs with H4K20 acetylation have not been previously described in detail.
In summary, there is positional bias toward acetylation at K5 and K12 across all acetylated Chlamydomonas H4 proteoforms, a radical departure from the low K5ac and high K16ac levels found in metazoans29.50 A recent paper uncovered that K5ac serves to “bookmark” genes that are silenced during cell division in cultured human cells to enable re-expression in postmitotic daughter cells.57 Compared with the genomes of metazoans, Chlamydomonas has a much higher gene density and a larger portion of it would be transcriptionally active compared to genomes with more noncoding sequences and repeats. Perhaps the global levels of K5 acetylation—low in humans/Drosophila, high in Chlamydomonas—reflects this difference. Alternatively, the higher degrees of K5 and K12 acetylation, which occur on newly synthesized H4 and mark it for nucleosomal deposition,58 might indicate that nucleosomal turnover is higher in Chlamydomonas. This is the first comprehensive study on acetylation states on a green algal histone H4 and may help shed light on the role of this modification in the green lineage.
Chlamydomonas Canonical H2A Are Minimally Modified, while Noncanonical H2A.Z is Multiply Acetylated and C-Terminally Truncated.
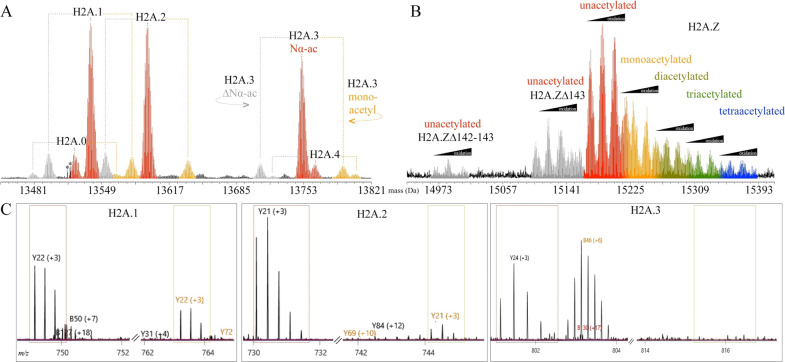
Chlamydomonas canonical H2A proteins are encoded by 26 genes and have four ∼13 kDa sequence variants: H2A.0, H2A.1, H2A.2, H2A.3, and H2A.4 (Table S1). These four proteins eluted slightly before the ∼15 kDa noncanonical H2A variant H2A.Z (Figure 1A, box #4). Other common H2A variants, such as H2A.X and plant-specific H2A.W, were not identified in searches of Chlamydomonas’ predicted proteins using tools in Phytozome.59 The canonical H2A proteoforms lack methionine or cysteine residues, and thus are the only histones that lack any detectable oxidation. The abundance of each variant was proportional to the number of paralogous genes encoding the respective sequence. For example, H2A.0 and H2A.4 are each expressed from a single gene and were the lowest in abundance (Figure 4A, Figure S4). Each canonical variant exhibited a similar pattern of proteoforms: ∼80% were unmodified except for Nα-ac, ∼10% lacked an amino-terminal acetylation (ΔNα-ac) and about 10% were found to have Nα-ac with internal monoacetylation (mainly at K5). Remarkably, ETD fragmentation of monoacetylated H2A.1 and H2A.2 localized a C-terminal acetylation to K118; however, the fragment ion intensity was low (data not shown). HCD fragmentation of the same precursor masses yielded more abundant fragment ions and a diagnostic cleavage product at Pro108 generated an abundant y ion that confirmed the presence of K118ac in H2A.1 and H2A.2, and its absence in H2A.3 (Figure 4C). As the last 13 C-terminal residues contribute to nearly all the sequence variation for Chlamydomonas’ canonical H2A proteins, we suspect that the histone acetyltransferase responsible for H2A K118ac recognizes a specific C-terminal motif present in H2A.1/H2A.2 but lacking in the other H2A.3 variants (Figure S4).
Figure 4.
Canonical H2A proteins exhibit modification profiles similar to each other but deviate significantly from the H2A variant H2A.Z. (A) The canonical H2A proteins H2A.0–4 (z = 17+) have Nα-ac (red) as the most abundant proteoform and minor levels of either an additional internal monoacetylation (orange) or an absent amino-terminal acetylation (ΔNα-ac, gray), as specifically illustrated by H2A.3. The isotopes marked with asterisks (*) are from a non-H2A protein. (B) Three intact H2A.Z molecules (z = 21+) were found to correspond to the entire coding sequence (colored red through blue) or missing one (H2A.ZΔ143) or two (H2A.ZΔ142-143) carboxy-terminal residues (gray). All H2A.Z showed significant oxidation for each acetylation state, as denoted by the black ramp above each mass. The acetylated proteoforms are assigned as in Figure 3. (C) TDValidator outputs showing summed ETD fragmentation data from the monoacetylated precursor masses for H2A.1 (left), H2A.2 (middle), and H2A.3 (right). The K188 diagnostic ion, y22+3, either aligns with an unmodified C-terminus (black label) or, if present, a monoacetylated C-terminus (yellow label).
The high levels of histone H2A lacking Nα-ac are noteworthy as this modification is added co-translationally in other organisms and is thought to be irreversible (mainly because an N-terminal deacetylase remains to be discovered).60 Upon further inspection of other asynchronous Chlamydomonas histone LC–MS replicates, we found the levels of H2AΔNα-ac to be closer to ∼1% (Figure S5). Along with H2A.Z, the other N-terminally acetylated histone, H4, was found to be completely N-terminal acetylated in all of our LC–MS data sets (Figure 3 and data not shown). H4 and H2A.Z have SG as their first two amino acids, while the canonical H2A proteins have AG. Either the S → A substitution is lowering the activity of the amino-terminal acetyltransferase during translation or, perhaps, there exists an amino-terminal deacetylase that is recognizing some moiety specific to the canonical H2As (such as Ala1).
In plants, H2A.Z is found in nucleosomes at the transcriptional start site (TSS) as well as in gene bodies of silenced genes.61,62 However, its roles in these locations seem antagonistic: its presence in the TSS correlates with H3K36me3 and high gene expression, while its presence in the gene body correlates with H3K27me3 and gene silencing (reviewed in ref (63)). In Chlamydomonas, we found the noncanonical H2A variant H2A.Z to be multiply acetylated, heavily oxidized due to having three internal methionine residues, and containing C-terminal truncations (Figure 4B). Unlike the other methionine-containing histones, the various oxidation states of H2A.Z were not well resolved chromatographically. As a consequence, the analysis of H2A.Z proteoforms was impaired and the acetylated proteoforms were not quantified. However, ETD analysis did reveal that acetylation was N-terminal, mainly at residues K6 and K14 (data not shown). Two C-terminal truncated H2A.Z proteoforms coeluted with the intact protein, either lacking the C-terminal amino acid (H2A.ZΔ143), or the last two amino acids (H2A.ZΔ142–143) (Figure 4B, Figure S5). If Chlamydomonas H2A.Z functions similarly to plant H2A.Z, we suspect the highly acetylated forms to be present in the TSS of expressed genes, while the unacetylated forms may be present in the gene bodies of silenced genes. Additionally, the C-terminal truncations we have detected might be involved in H2A.Z turnover in either TSS or gene bodies to regulate gene expression. Follow-up experiments involving chromatin immunoprecipitation (ChIP) targeting unacetylated H2A.Z and multiply acetylated H2A.Z followed by DNA sequencing (ChIP-Seq) and/or mass spectrometry (ChIP-MS or Nucleosome-MS (Nuc-MS)64) would show the genomic locations of these forms and whether they associate with PTMs on other histones (e.g., H3K27me3 or H3K36me3).
Chlamydomonas Expresses Multiple 16 kDa Histone H2B N-Terminal Variants and a Highly Acetylated 13 kDa Variant, H2B.v1
An interesting feature of the Chlamydomonas H2B family is that the amino acids after the amino terminal tail (about residue 70) are 100% conserved across all variants with the exception of the noncanonical variant H2B.v1: all canonical H2B have E100KVATEASKLSR111 (numbered positions based on H2B.9) while H2B.v1 sequence is D68KMANEAVRLAQ79, showing many substitutions. This sequence is toward the end of the histone fold’s second highly conserved α helix, which is embedded deep in the core of the nucleosome65 and may influence intranucleosomal interactions. H2B.v1 was found to be mostly unacetylated, but mono-, di-, tri- and tetraacetylated forms were readily detected (Figure 5A, left). ETD of the acetylated H2B.v1 proteoforms revealed their composition: monoacetylated = ∼50% K7ac and ∼50% K11ac; diacetylated = ∼100% K7ac + K11ac; triacetylated = ∼50% K7ac + K11ac + K12ac and ∼50% K7ac + K11ac + K16ac; tetraacetylated = ∼100% K7ac + K11ac + K12ac + K16ac (Figure 5B). The fragmentation data of monoacetylated and diacetylated H2B.v1 included a small fraction of low abundance ions (<10% relative intensity) that were unassigned, so there may be other minor acetylation isomers present (data not shown). Despite this, the acetylated isomers detected suggest a preference for K7ac and K11ac being acetylated at either position first and then both together, followed by either K12ac and/or K16ac. All of the H2B.v1 proteoforms that underwent ETD fragmentation were also trimethylated on the terminal alpha-amino group (Nα-me3). Previously reported Chlamydomonas H2B amino-terminal acetylation was erroneously assigned due to the small mass difference between trimethylation and acetylation (35 mDa)18 and the lack of detectable levels of amino-terminal mono- and dimethylation, which is often a predictor of trimethylation. The H2B.v1 variant lacks the extended amino-terminal tail found in other Chlamydomonas (Figure 5C) and land plant H2Bs.55,66 Both the size and high degree of acetylation of H2B.v1 is similar to that found in budding yeast, which has only two H2B variants, H2B.1 and H2B.2, that differ by four amino acids (A3S, K4A, T28V, and A36V). Both forms are amino-terminally acetylated and have an average of approximately two internal acetylations per molecule with at least five N-terminal lysine residues being identified as acetylated.51
Figure 5.
Identification and characterization of several Chlamydomonas histone H2B variants. (A) TDMS profiles of the multiply acetylated H2B.v1 variant around 13.3 Da (left) and multiple H2B gene products around 16.5 kDa (right). (B) Graphical fragmentation map showing the localization of H2B.v1 acetylation (left, boxed blue) and essential fragment ions distinguishing between two closely related H2B paralogs H2B.12 and HTB.13 (right, boxed gray). (C) Multiple sequence alignment for the amino-terminal tails of all putative Chlamydomonas H2B variants is shown (consensus sequence on top). Species highlighted in yellow were identified by MS/MS in this study. Amino acids in red text indicate substitutions or changes that deviate from the consensus sequence at that position. The amino termini of the last two H2B variants is shaded blue to indicate it is highly variant and does not align well with the amino termini of other H2B proteins. Monoisotopic masses (without Met1) of the unmodified form of each protein are listed to the far right and the degree of amino acid conservation is shown in the histogram below.
Shortly after the elution of H2B.v1, the entire family of ∼16 kDa H2B proteoforms coeluted, generating the complex MS spectrum shown in Figure 5A (right). Nearly all masses above a S/N of 3 were selected for ETD and HCD fragmentation. Additionally, the enhanced chromatography that was used prior to MS effectively separated the oxidized H2B from unoxidized H2B with very little overlap, making the interpretation of the MS/MS data easier than when the two forms coelute (Figure 1, box 2). Nonetheless, the MS and MS/MS data presented challenges that hindered confident quantification of the H2B proteoforms and gene products. For example, some H2B variants differ in mass by only 2 Da leading to significant overlap of precursor isotopic distributions. Despite these challenges, we conclusively identified 11 out of the possible 16 larger 16 kDa H2B canonical gene products (Figure 5C) and each of these proteins differed only in their amino-terminal sequence (first ∼68 residues). Of these 11 canonical variants, nine differed by only one or two amino acid substitutions, while two had a significantly higher number of substitutions (20 for H2B.15 and 21 for H2B.14). Using tritiated acetate, Waterborg found that the 16 kDa H2B family members have low levels of acetylation,52 so we expected H2B acetylation to be low in our MS investigation. After initial characterization of each cluster of masses corresponding to H2B molecular mass, the unassigned fragment ions from each MS/MS were specifically searched for common PTMs (e.g., methylation, acetylation). In this way, we distinguished overlapping isotopic clusters of nearly isobaric species resulting from an unmodified H2B variant of a higher mass with those of a modified H2B of lower mass. We did not find any evidence of acetylation or other PTMs, suggesting that these forms, if present, have abundances below our detection limit.
As with H2B.v1, all larger H2B histones are trimethylated on the α-amino group of A1 (Nα-me3). The termini of all Chlamydomonas H2B follow the consensus sequence recognized by the human enzyme α-N-methyltransferase NTMT1, which mono-, di-, or trimethylates sequences following a “Xaa-P-K/R” motif.67 While the orthologous enzyme in Chlamydomonas has yet to be identified, a likely candidate based on BLAST-searching the methyltransferase catalytic domain is the protein encoded by the SMM19 gene (Cre06.g274400; Uniprot # A0A2K3DNL8). SMM19 is expressed along with histone genes during S/M phase of the cell cycle, consistent with a role in modifying the N-termini of newly synthesized histones.7 Many of the H2B family members in Arabidopsis contain the “Xaa-P-K/R” motif and are N-terminally mono-, di- and trimethylated, but quantitative information regarding the degree of methylation was not reported.66 However, TDMS analysis of another land plant, sorghum, revealed H2B canonical variants that are nearly completely N-terminally trimethylated.55 Remarkably, Drosophila encode only one H2B protein sequence and its amino-terminal sequence (PPK) follows the consensus recognized by the Drosophila ortholog of NTMT1, dNTM1.68,69 However, there is little evidence of Nα-me3 and the predominant methylation state after treatment with dNTM1 is Nα-me2. Similarly, TDMS analysis of Tetrahymena H2B revealed its amino-terminus to exist in multiple states of methylation.70 Interestingly, yeast and human H2B, which lack the Xaa-P-K/R motif have their N-terminus acetylated.51,71 The blocking of H2B’s N-terminus either by methylation in Chlamydomonas or acetylation as it is in other organisms is a conserved feature, most likely leading to increase stability of H2B.60
H2B is known to be ubiquitylated, and a targeted study confirmed monoubiquitylation of Chlamydomonas H2B most likely at K149.72 While the function of H2B’s ubiquitylation has not been studied in Chlamydomonas, it plays a role in the activation of floral repressor genes in Arabidopsis(73) and may serve as a transcriptional activation mark in algae. Because ubiquitylation was not included in the database search, we used the SMC Python script described above to query masses close to the expected monoubiquitylated H2B eluting close to the family of 16 kDa H2B proteoforms (Figure 1, Box 2 around 25 kDa). The low abundance and large size of monoubiquitylated H2Bs made the fragmentation data sparse. Manual investigation of the ETD and HCD data revealed only a few amino-terminal ions (and no carboxy-terminal ions) that matched to both ubiquitin and H2B.5, in agreement with this modification being on the carboxy terminus (data not shown). The most notable feature is that the abundances of monoubiquitylated H2B mass profiles mirror those of the 16 kDa nonmonoubiquitylated H2B profiles suggesting that the H2B ubiquitylating enzyme does not discriminate among N-terminal variants (compare Figure 5A, right spectra with Figure 6).
Figure 6.
Chlamydomonas reinhardtii histone H3 modifications observed by TDMS analysis. (A) Feature map shows two major populations: oxidized H3 (gray dashed parallelogram) and unoxidized H3 (black dashed parallelogram). (B) Abundance of proteoforms among the unoxidized population of H3. Unmodified canonical H3.1 is represented by proteoform “a” with an observed Mmi of 15 168.49 Da (black text in A). Precursor masses “b”–“q” and “t” (purple text in A) have singly methylated lysine 4 (K4me1) combined with unlocalized additional mass shifts in 14–16 Da increments, from +14 Da to +255 Da. Precursor masses “r”, “s”, “u”, and “v” (green text in A, bold and underlined in B) have K4me3 and multiple acetylations (K9ac, K14ac, and others) combined with unlocalized mass shifts that also increase in 14–16 Da increments. Percent abundance for each proteoform was calculated as the percent feature intensity among all unoxidized H3 proteoforms. (C) MS/MS spectra for several groups of features (and their mass range) were combined using TDValidator, and the intensity of the c41+ ion reporting on H3K4 was used to calculate its methylation site occupancies. H3K4me1 is the most abundant H3 methylation state for masses up to 15 350 Da. H3K4me3 is the most abundant K4 methylation state on precursor masses 15 391 and larger.
Histone H3 Have a Bimodal Distribution Conditioned on K4 Methylation State
The Chlamydomonas genome contains 35 genes annotated as histone H3 or histone H3 variant (Table S1). Thirty of these genes encode the canonical H3 histone we designate here as H3.1, and two genes encode two additional canonical histones, H3.2 and H3.3. Two H3 variants in Chlamydomonas have been identified as homologs of centromeric H3 genes in land plants;74,75 thus, we refer to these proteins as cenH3.1 and cenH3.2, and the remaining noncanonical variant is referred to as H3.v1.
We observed two major populations of histone H3 between the LC retention times (tR) between 149 and 164 min (Figure 6A). The first population to elute, between tR of 149.83 and 158.06 min, contained H3 proteoforms with masses indicating between 1 and 3 oxidations localized to H3C109 and/or H3M119 by HCD (data not shown). The second population to elute, between LC tR of 156.16 and 163.51 min, showed a similar proteoform profile yet appeared to be lacking most oxidation. To avoid redundancy and misidentification of PTMs due to incomplete and variable oxidation levels, we only analyzed the MS/MS data acquired for the unoxidized species, indicated by the groups of features lettered “a”–“v” in Figure 6A.
The only H3 protein identified in our analysis was the canonical H3.1 protein, which is consistent with its gene copy number and transcript abundance being much higher than the other canonical histones, H3.2 and H3.3 (Table S17,8). The noncanonical H3 variants, including the two centromeric H3 proteins, were also not identified here, despite some of these gene variants having transcript abundances similar to that of the identified canonical H3.1 histone.7,8 Like all other core histones, all H3 proteoforms we identified were missing their N-terminal methionine. But unlike the other core histones, H3 lacked any detectable modification on its alpha-amino group (e.g., acetylation or methylation). The combination of low precursor ion abundance and the limited number of MS/MS scans resulted in sparse fragment coverage for many of the H3 proteoforms we analyzed. Due to this low sequence coverage, PTM assignments were restricted to the first few residues on both termini of each proteoform, leaving the remaining mass differences assigned to a broad range of residues on which they may appear (Figure S7 and Table 1).
The majority of H3 proteoforms, represented by features “b”–“q” and “t”, displayed monomethylation on lysine 4 (H3K4me1), or occasionally monomethylation on lysine 9 (H3K9me1) (Figure S7). Most of these H3 molecules with either K4me1 or K9me1 had additional mass shifts ranging from +14 to +255 Da. The fragment data rule out localization of these mass shifts to the N-terminus (e.g., K9, K14, and K18) or to the C-terminus (after M119) but instead suggest localization usually between K23 and I118 (data not shown). For H3 proteoforms up to a mass shift of +84 Da (“b”–“g”), we suspect the unlocalized PTMs to be either multiple methylations and/or a few acetylations, given the consistent pattern of H3 proteoforms increasing in mass in 14–16 Da increments. Additionally, the low-abundance proteoforms with mass shifts from +98 to +225 Da (“h”–“q”) could potentially represent small subpopulations of proteoforms “a”–“j” with the addition of artifactual phosphate or sulfate adducts. These adducts usually appear on residues in the globular region of the molecule as opposed to its termini,55 and typically generate fragment ions representative of low-mass modifications despite the intact precursor mass suggesting larger mass shifts.
The high mass shift proteoforms (>+225 Da) “r”, “s”, “u”, and “v” are all modified with K4me3, frequently in combination with K9ac, K14ac, and additional unlocalized mass shifts >+70 Da between Q19 and I118 (Figure S7). Using a targeted mass list including low-abundance, high-mass histone H3 forms, we investigated the site occupancy of H3K4 methylation as a function of histone H3 mass and degree of modification (Figure 6C). For histone H3 masses 15 168 Da (lowest mass, 0 acetylation equivalents) to 15 285 Da (∼2 acetylation equivalents), H3K4me1 predominated and was the only detectable methylated species. For all species of masses greater than 15 285 Da, which together represent <1.5% of the total H3 population, the relative abundance of H3K4me3 increases dramatically, reaching ∼70% of all H3 in the highest mass range (∼7 acetylation equivalents). Interestingly, H3K4me2 was detected only in the mass ranges that also have H3K4me3 and its abundance remained low (2.7% to 14.2%) and relatively stable across these mass ranges. This suggests that H3K4me2 is a transient intermediate in the H3K4 trimethylation pathway. The small population of highly acetylated H3K4me3 contrasts with the more abundant and minimally modified H3 proteoforms with K4me1 that lack additional amino-terminal (prior to K23) modifications. This bimodality of H3 modification state suggests that monomethylation on N-terminal lysine(s) precludes the accumulation of multiple acetylations on the same residues, as suggested previously.76,77 We suspect the unlocalized PTMs on these trimethylated proteoforms may include H3K27ac, H3K27me1/2/3, and/or H3K36ac, since others have shown those marks colocalized to the same promoter regions as H3K4me3 in Chlamydomonas.15,78 In Chlamydomonas, H3 proteins occupying repressed promoter regions displayed high levels of H3K4me1 or H3K9me1, low levels of H3 acetylation, and low levels of H3K4me3.12,77,79,80 On the other hand, prior work studying transcriptional activation marks in Chlamydomonas found H3K4me3 and multiacetylated H3 occupying transcriptionally active promoter regions.12,81 Overall, our data show low abundance of H3 with K4me3 and multiple acetylations (0.9% of total H3) and high abundance of H3 with monomethylation on K4 or K9 (99.1% of total H3). This trend is similar to prior reports that ∼80% of Chlamydomonas H3 histones are monomethylated at K4 and ∼16% are monomethylated at K9,76,79 while ∼20% are dynamically multiacetylated.52 Additionally, those same studies report that dimethyl- and trimethyllysines on histone H3 were not detected, most likely due to their low global abundances (<1.5%) These prior observations noting high global abundance of repressive marks and low global abundance of activation marks is in contrast with transcriptional studies showing that a majority of the Chlamydomonas genome is active at most cell cycle stages7,8 and with the Chlamydomonas genome’s high gene density suggesting a low proportion of inactive intergenic regions.82 This contrast suggests the possibility that H3 monomethylation on its own is unlikely to be a silencing mark, but it may be required in combination with other modifications for active silencing of transcription at specific loci. Further studies will be needed to parse the global roles versus promoter-specific roles of individual and combinatorial H3 modifications in Chlamydomonas under a variety of growth conditions.
Here, we report 22 partially characterized H3 proteoforms in Chlamydomonas building upon the three H3 proteoforms identified previously.18 Additional optimization of the LC and mass spectrometry methods (such as more comprehensive target mass lists and fine-tuned fragmentation parameters) may increase proteoform purity and sequence coverage in the future. This will allow us to localize combinations of PTMs further into the globular region of Chlamydomonas H3, as well as explore the hypothesized competitive relationship between lysine monomethylation and acetylation, possibly governed by H3K4 methylation status.
Two Linker Histone H1 Gene Products Were Found to be Minimally Modified
Of the three predicted histone H1 genes in Chlamydomonas, we identified protein products from two of them: Cre06.g275900 (HON2) and Cre13.g567450 (HON1), with monoisotopic mass of 24588.56 and 27249.91 Da, respectively, each being N-terminally acetylated but lacking any other detectable modifications. The third H1 gene, Cre13.g562300, was not detected in our analysis, most likely due to low expression8.7 As per suggested histone paralog naming conventions83HON2 refers to the protein product H1.2, while HON1 refers to the protein product H1.1 (Table S1). Besides full length H1s, we also detected many truncated forms (Supporting Information).
Conclusions
In this study, we report 86 Chlamydomonas histone proteoforms whose detection was made possible by the development of new software, improved chromatographic separation, and high resolution/rich fragmentation provided by state-of-the-art top-down mass spectrometry. We also demonstrated that manual analysis, assisted by multiple TDMS data visualization and processing tools, is necessary for characterizing low abundance and isomeric proteoforms. Future software development will make this process more robust for high throughput TDMS analysis of proteoforms. These technical improvements of top-down analysis enabled us to create a new and valuable resource for algal epigenetics that describes a significantly expanded number of algal PTMs and their co-occurrence on different histone variants. This reference resource, along with our improved methods, can facilitate characterization of the histone dynamics in various growth conditions, cell cycle states, and mutant strains. Examination of the histone PTM landscape in strains from the Chlamydomonas Library Project (CLiP) insertion mutant library84 could be combined with these methods to improve annotations of currently putative gene models, especially putative histone modifying enzymes, to further the elucidation of the Chlamydomonas and algal histone codes.
Acknowledgments
This material is based partially upon work supported by the National Science Foundation under Grant No. MCB-1943493 (J.J.P. and H.S.) and under Grant No. DBI-1827534 for acquisition of the Orbitrap Fusion Lumos LC-MS/MS (B. J.J.P. and H.S. were further supported by funding from the Department of Education HSI STEM Award (PR Award P031C160168). A portion of the research was performed using EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Biological and Environmental Research program. Work in the laboratory of J.G.U. was supported by National Institutes of Health Grant No. R01 GM126557 and National Science Foundation Grant Nos. MCB 1515220 and MRI 1427621.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.1c00029.
The authors declare no competing financial interest.
Notes
This paper was intended for the Top-Down Proteomics: Technology Advances and Biomedical Applications Focus, published as the June 2021 issue of J. Am. Soc. Mass Spectrom. (Vol. 32, No. 6).
Supplementary Material
References
- Blaby I. K.; Blaby-Haas C. E.; Tourasse N.; Hom E. F. Y.; Lopez D.; Aksoy M.; Grossman A.; Umen J.; Dutcher S.; Porter M.; King S.; Witman G. B.; Stanke M.; Harris E. H.; Goodstein D.; Grimwood J.; Schmutz J.; Vallon O.; Merchant S. S.; Prochnik S. The Chlamydomonas genome project: a decade on. Trends Plant Sci. 2014, 19, 672–680. 10.1016/j.tplants.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scranton M. A.; Ostrand J. T.; Fields F. J.; Mayfield S. P. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015, 82, 523–531. 10.1111/tpj.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H.; Ma X.; Msanne J.; Repas T. RNA-Mediated Silencing in Algae: Biological Roles and Tools for Analysis of Gene Function. Eukaryotic Cell 2011, 10, 1164–1172. 10.1128/EC.05106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersfelder E. L.; Parthun M. R. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006, 34, 2653–2662. 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G. D.; Poirier M. G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015, 115, 2274–2295. 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart S. B.; Strahl B. D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta, Gene Regul. Mech. 2014, 1839, 627–643. 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones M. J.; Blaby I. K.; Merchant S. S.; Umen J. G. High-Resolution Profiling of a Synchronized Diurnal Transcriptome from Chlamydomonas reinhardtii Reveals Continuous Cell and Metabolic Differentiation. Plant Cell. 2015, 27, 2743–2769. 10.1105/tpc.15.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D.; Schmollinger S.; Gallaher S. D.; Salomé P. A.; Purvine S. O.; Nicora C. D.; Mettler-Altmann T.; Soubeyrand E.; Weber A. P. M.; Lipton M. S.; Basset G. J.; Merchant S. S. Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 2374–2383. 10.1073/pnas.1815238116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G.; Ebert A.; Reuter G. SU(VAR)3–9 is a conserved key function in heterochromatic gene silencing. Genetica 2003, 117, 149–158. 10.1023/A:1022923508198. [DOI] [PubMed] [Google Scholar]
- Zhou H.; Liu Y.; Liang Y.; Zhou D.; Li S.; Lin S.; Dong H.; Huang L. The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci. 2020, 29, 1120–1137. 10.1002/pro.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano J. A.; van Dijk K.; Eisenhart J.; Cerutti H. SET3p monomethylates histone H3 on lysine 9 and is required for the silencing of tandemly repeated transgenes in Chlamydomonas. Nucleic Acids Res. 2007, 35, 939–950. 10.1093/nar/gkl1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D.; Schmollinger S.; Schroda M. Heat shock factor 1 counteracts epigenetic silencing of nuclear transgenes in Chlamydomonas reinhardtii. Nucleic Acids Res. 2013, 41, 5273–5289. 10.1093/nar/gkt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J.; Gallaher S. D.; Lu Y.; Strenkert D.; Segal N.; Barahimipour R.; Fitz-Gibbon S. T.; Schroda M.; Merchant S. S.; Bock R. An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat. Commun. 2020, 11, 6269. 10.1038/s41467-020-19983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B. A.; Hake S. B.; Diaz R. L.; Kauer M.; Morris S. A.; Recht J.; Shabanowitz J.; Mishra N.; Strahl B. D.; Allis C. D.; Hunt D. F. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007, 282, 7641–7655. 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Ngan C. Y.; Wong C.-H.; Choi C.; Yoshinaga Y.; Louie K.; Jia J.; Chen C.; Bowen B.; Cheng H.; Leonelli L.; Kuo R.; Baran R.; García-Cerdán J. G.; Pratap A.; Wang M.; Lim J.; Tice H.; Daum C.; Xu J.; Northen T.; Visel A.; Bristow J.; Niyogi K. K.; Wei C.-L. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat. plants. 2015, 1, 15107. 10.1038/nplants.2015.107. [DOI] [PubMed] [Google Scholar]
- Kang H.; Shokhirev M. N.; Xu Z.; Chandran S.; Dixon J. R.; Hetzer M. W. Dynamic regulation of histone modifications and long-range chromosomal interactions during postmitotic transcriptional reactivation. Genes Dev. 2020, 34, 913–930. 10.1101/gad.335794.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. Good News for Nuclear Transgene Expression in Chlamydomonas. Cells 2019, 8, 1534. 10.3390/cells8121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.; Eikani C. K.; Khan H.; Iavarone A. T.; Pesavento J. J. Characterization of Chlamydomonas reinhardtii Core Histones by Top-Down Mass Spectrometry Reveals Unique Algae-Specific Variants and Post-Translational Modifications. J. Proteome Res. 2018, 17, 23–32. 10.1021/acs.jproteome.7b00780. [DOI] [PubMed] [Google Scholar]
- Wong A. L.; Totah N. N.; Iavarone A. T.; Pesavento J. J. Quick and facile preparation of histone proteins from the green microalga Chlamydomonas reinhardtii and other photosynthetic organisms. Methods 2020, 184, 102–111. 10.1016/j.ymeth.2020.01.019. [DOI] [PubMed] [Google Scholar]
- Smith L. M.; Kelleher N. L. Consortium for Top Down Proteomics: Proteoform: a single term describing protein complexity. Nat. Methods 2013, 10, 186–7. 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D.; Chambers M.; Burke R.; Agus D.; Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M.-Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.; Guner H.; Gregorich Z. R.; Chen A. J.; Ayaz-Guner S.; Peng Y.; Valeja S. G.; Liu X.; Ge Y. MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics. Mol. Cell. Proteomics. 2016, 15, 703–714. 10.1074/mcp.O115.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Q.; Xun L.; Liu X. TopPIC: A software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 2016, 32, 3495–3497. 10.1093/bioinformatics/btw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Piehowski P. D.; Wilkins C.; Zhou M.; Mendoza J.; Fujimoto G. M.; Gibbons B. C.; Shaw J. B.; Shen Y.; Shukla A. K.; Moore R. J.; Liu T.; Petyuk V. A.; Tolić N.; Paša-Tolić L.; Smith R. D.; Payne S. H.; Kim S. Informed-Proteomics: open-source software package for top-down proteomics. Nat. Methods 2017, 14, 909–914. 10.1038/nmeth.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K.; Kim J.; Gaikwad M.; Hidayah S. N.; Heikaus L.; Schlüter H.; Kohlbacher O. FLASHDeconv: Ultrafast, High-Quality Feature Deconvolution for Top-Down Proteomics. Cell Syst. 2020, 10, 213–218. 10.1016/j.cels.2020.01.003. [DOI] [PubMed] [Google Scholar]
- Fornelli L.; Srzentić K.; Huguet R.; Mullen C.; Sharma S.; Zabrouskov V.; Fellers R. T.; Durbin K. R.; Compton P. D.; Kelleher N. L. Accurate Sequence Analysis of a Monoclonal Antibody by Top-Down and Middle-Down Orbitrap Mass Spectrometry Applying Multiple Ion Activation Techniques. Anal. Chem. 2018, 90, 8421–8429. 10.1021/acs.analchem.8b00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento J. J.; Mizzen C. A.; Kelleher N. L. Quantitative Analysis of Modified Proteins and Their Positional Isomers by Tandem Mass Spectrometry: Human Histone H4. Anal. Chem. 2006, 78, 4271–4280. 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- Holt M. V.; Wang T.; Young N. L. One-Pot Quantitative Top- and Middle-Down Analysis of GluC-Digested Histone H4. J. Am. Soc. Mass Spectrom. 2019, 30, 2514. 10.1007/s13361-019-02219-1. [DOI] [PubMed] [Google Scholar]
- Janssen K. A.; Coradin M.; Lu C.; Sidoli S.; Garcia B. A. Quantitation of Single and Combinatorial Histone Modifications by Integrated Chromatography of Bottom-up Peptides and Middle-down Polypeptide Tails. J. Am. Soc. Mass Spectrom. 2019, 30, 2449–2459. 10.1007/s13361-019-02303-6. [DOI] [PubMed] [Google Scholar]
- Pesavento J. J.; Garcia B. A.; Streeky J. A.; Kelleher N. L.; Mizzen C. A. Mild Performic Acid Oxidation Enhances Chromatographic and Top Down Mass Spectrometric Analyses of Histones. Mol. Cell. Proteomics. 2007, 6, 1510–1526. 10.1074/mcp.M600404-MCP200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H.; Kirmizis A.; Nelson C.; Bartke T.; Saksouk N.; Cote J.; Kouzarides T. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 2009, 16, 17–22. 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellers R. T.; Greer J. B.; Early B. P.; Yu X.; Leduc R. D.; Kelleher N. L.; Thomas P. M. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15, 1235–1238. 10.1002/pmic.201400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamdborg L.; LeDuc R. D.; Glowacz K. J.; Kim Y. Bin; Viswanathan V.; Spaulding I. T.; Early B. P.; Bluhm E. J.; Babai S.; Kelleher N. L. ProSight PTM 2.0: Improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 2007, 35, 701–706. 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino L. K.; Searle B. C.; Bollinger J. G.; Nunn B.; MacLean B.; MacCoss M. J. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020, 39, 229–244. 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. M. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 2005, 12, 110–112. 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- Pesavento J. J.; Yang H.; Kelleher N. L.; Mizzen C. A. Certain and Progressive Methylation of Histone H4 at Lysine 20 during the Cell Cycle. Mol. Cell. Biol. 2008, 28, 468–486. 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Holt M. V.; Young N. L. Early butyrate induced acetylation of histone H4 is proteoform specific and linked to methylation state. Epigenetics 2018, 13, 519–535. 10.1080/15592294.2018.1475979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntai I.; Kim K.; Fellers R. T.; Skinner O. S.; Smith A. D.; Early B. P.; Savaryn J. P.; Leduc R. D.; Thomas P. M.; Kelleher N. L. Applying label-free quantitation to top down proteomics. Anal. Chem. 2014, 86, 4961–4968. 10.1021/ac500395k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur M. T.; Cardasis H. L.; Spellman D. S.; Liaw A.; Yates N. A.; Hendrickson R. C. Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 7728–7733. 10.1073/pnas.0910776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X.; Singh A.; Spetman B. D.; Nolan K. D.; Isaacs J. S.; Dennis J. H.; Dalton S.; Marshall A. G.; Young N. L. Label-Free Relative Quantitation of Isobaric and Isomeric Human Histone H2A and H2B Variants by Fourier Transform Ion Cyclotron Resonance Top-Down MS/MS. J. Proteome Res. 2016, 15, 3196–3203. 10.1021/acs.jproteome.6b00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento J. J.; Bullock C. R.; LeDuc R. D.; Mizzen C. A.; Kelleher N. L. Combinatorial Modification of Human Histone H4 Quantitated by Two-dimensional Liquid Chromatography Coupled with Top Down Mass Spectrometry. J. Biol. Chem. 2008, 283, 14927–14937. 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G.; DiMaggio P. A; Chan E. Y; Zee B. M; Blanco M; Bryant B.; Flaniken I. Z; Liu S.; Kang Y.; Trojer P.; Garcia B. A A quantitative atlas of histone modification signatures from human cancer cells. Epigenet. Chromatin 2013, 6, 20. 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M. V.; Wang T.; Young N. L. High-Throughput Quantitative Top-Down Proteomics: Histone H4. J. Am. Soc. Mass Spectrom. 2019, 30, 2548–2560. 10.1007/s13361-019-02350-z. [DOI] [PubMed] [Google Scholar]
- Nardelli S. C.; Che F.-Y.; Silmon de Monerri N. C.; Xiao H.; Nieves E.; Madrid-Aliste C.; Angel S. O.; Sullivan W. J.; Angeletti R. H.; Kim K.; Weiss L. M. The Histone Code of Toxoplasma gondii Comprises Conserved and. mBio 2013, 4, 1–12. 10.1128/mBio.00922-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy A.; Rastogi A.; Lin X.; Lombard B.; Murik O.; Thomas Y.; Dingli F.; Rivarola M.; Ott S.; Liu X.; Sun Y.; Rabinowicz P. D.; McCarthy J.; Allen A. E.; Loew D.; Bowler C.; Tirichine L. An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 2015, 16, 102. 10.1186/s13059-015-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdareau S.; Tirichine L.; Lombard B.; Loew D.; Scornet D.; Wu Y.; Coelho S. M.; Cock J. M. Histone modifications during the life cycle of the brown alga Ectocarpus. Genome Biol. 2021, 22, 1–27. 10.1186/s13059-020-02216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Huang Z. Q.; Xia L.; Feng Q.; Erdjument-Bromage H.; Strahl B. D.; Briggs S. D.; Allis C. D.; Wong J.; Tempst P.; Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science (Washington, DC, U. S.) 2001, 293, 853–857. 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Werner-Peterson R.; Sloboda R. D. Methylation of structural components of the axoneme occurs during flagellar disassembly. Biochemistry 2013, 52, 8501–9. 10.1021/bi4011623. [DOI] [PubMed] [Google Scholar]
- Yang H.; Pesavento J. J.; Starnes T. W.; Cryderman D. E.; Wallrath L. L.; Kelleher N. L.; Mizzen C. A. Preferential Dimethylation of Histone H4 Lysine 20 by Suv4–20. J. Biol. Chem. 2008, 283, 12085–12092. 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.; Smith J. N.; Anderson S. L.; Ma P.; Mizzen C. A.; Kelleher N. L. Global Assessment of Combinatorial Post-translational Modification of Core Histones in Yeast Using Contemporary Mass Spectrometry. J. Biol. Chem. 2007, 282, 27923–27934. 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- Waterborg J. H. Dynamics of histone acetylation in Chlamydomonas reinhardtii. J. Biol. Chem. 1998, 273, 27602–27609. 10.1074/jbc.273.42.27602. [DOI] [PubMed] [Google Scholar]
- Jørgensen S.; Schotta G.; Sørensen C. S. Histone H4 Lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuland R.; Gozani O. Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol. Cell. Proteomics. 2016, 15, 755–764. 10.1074/mcp.R115.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.; Malhan N.; Ahkami A. H.; Engbrecht K.; Myers G.; Dahlberg J.; Hollingsworth J.; Sievert J. A.; Hutmacher R.; Madera M.; Lemaux P. G.; Hixson K. K.; Jansson C.; Paša-Tolić L. Top-down mass spectrometry of histone modifications in sorghum reveals potential epigenetic markers for drought acclimation. Methods 2020, 184, 29. 10.1016/j.ymeth.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Sridhar V. V.; Zhu J.; Kapoor A.; Zhu J. K. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One 2007, e1210. 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R.; Nakamura T.; Fu Y.; Lazar Z.; Spector D. L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 2011, 13, 1295–304. 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo A.; Wangh L.; Allfrey V. Processing of newly synthesized histone molecules. Science (Washington, DC, U. S.) 1975, 190, 117–128. 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- Goodstein D. M.; Shu S.; Howson R.; Neupane R.; Hayes R. D.; Fazo J.; Mitros T.; Dirks W.; Hellsten U.; Putnam N.; Rokhsar D. S. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriadou C.; Koufaris C.; Kirmizis A. Histone N-alpha terminal modifications: Genome regulation at the tip of the tail. Epigenet. Chromatin 2020, 13, 1–13. 10.1186/s13072-020-00352-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D.; Zilberman D. Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. PLoS Genet. 2012, 8, e1002988. 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R.; Stroud H.; Holec S.; Zhou K.; Feng S.; Zhong X.; Muthurajan U. M.; Nie X.; Kawashima T.; Groth M.; Luger K.; Jacobsen S. E.; Berger F. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in arabidopsis. Cell 2014, 158, 98–109. 10.1016/j.cell.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B.; Berger F. H2A Variants in Arabidopsis: Versatile Regulators of Genome Activity. Plant Commun. 2020, 1, 100015. 10.1016/j.xplc.2019.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachner L. F.; Jooß K.; Morgan M. A.; Piunti A.; Meiners M. J.; Kafader J. O.; Lee A. S.; Iwanaszko M.; Cheek M. A.; Burg J. M.; Howard S. A.; Keogh M.; Shilatifard A.; Kelleher N. L. Decoding the protein composition of whole nucleosomes with Nuc-MS. Nat. Methods 2021, 18, 303–308. 10.1038/s41592-020-01052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K.; Mäder A. W.; Richmond R. K.; Sargent D. F.; Richmond T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Bergmüller E.; Gehrig P. M.; Gruissem W. Characterization of Post-Translational Modifications of Histone H2B-Variants Isolated from Arabidopsis thaliana. J. Proteome Res. 2007, 6, 3655–3668. 10.1021/pr0702159. [DOI] [PubMed] [Google Scholar]
- Wu R.; Yue Y.; Zheng X.; Li H. Molecular basis for histone N-terminal methylation by NRMT1. Genes Dev. 2015, 29, 2337–2342. 10.1101/gad.270926.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Garea A.; Forne I.; Vetter I.; Kremmer E.; Thomae A.; Imhof A. Developmental regulation of N-terminal H2B methylation in Drosophila melanogaster. Nucleic Acids Res. 2012, 40, 1536–49. 10.1093/nar/gkr935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R.; Tanguay R. M. Methylation of Drosophila histones at proline, lysine, and arginine residues during heat shock. J. Biol. Chem. 1988, 263, 4686–4692. 10.1016/S0021-9258(18)68837-4. [DOI] [PubMed] [Google Scholar]
- Medzihradszky K. F.; Zhang X.; Chalkley R. J.; Guan S.; McFarland M. A.; Chalmers M. J.; Marshall A. G.; Diaz R. L.; Allis C. D.; Burlingame A. L. Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS). Mol. Cell. Proteomics. 2004, 3, 872–86. 10.1074/mcp.M400041-MCP200. [DOI] [PubMed] [Google Scholar]
- Siuti N.; Roth M. J.; Mizzen C. A.; Kelleher N. L.; Pesavento J. J. Gene-Specific Characterization of Human Histone H2B by Electron Capture Dissociation. J. Proteome Res. 2006, 5, 233–239. 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- Shimogawara K.; Muto S. Purification of Chlamydomonas 28-kDa ubiquitinated protein and its identification as ubiquitinated histone H2B. Arch. Biochem. Biophys. 1992, 294, 193–199. 10.1016/0003-9861(92)90157-R. [DOI] [PubMed] [Google Scholar]
- Xu L.; Ménard R.; Berr A.; Fuchs J.; Cognat V.; Meyer D.; Shen W. H. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- Cui J.; Zhang Z.; Shao Y.; Zhang K.; Leng P.; Liang Z. Genome-wide identification, evolutionary, and expression analyses of histone H3 variants in plants. BioMed Res. Int. 2015, 1. 10.1155/2015/341598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draizen E. J.; Shaytan A. K.; Mariño-Ramírez L.; Talbert P. B.; Landsman D.; Panchenko A. R. HistoneDB 2.0: A histone database with variants - An integrated resource to explore histones and their variants. Database 2016, 2016, baw014. 10.1093/database/baw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg J. H.; Robertson A. J.; Tatar D. L.; Borza C. M.; Davie J. R. Histones of Chlamydomonas reinhardtii. Synthesis, acetylation, and methylation. Plant Physiol. 1995, 109, 393–407. 10.1104/pp.109.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano J. A.; Jeong B.-R.; Xu J.; Moriyama H.; Cerutti H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6486–6491. 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver S.; Casas-Mollano J. A.; Cerny R. L.; Cerutti H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics 2010, 5, 301–312. 10.4161/epi.5.4.11608. [DOI] [PubMed] [Google Scholar]
- van Dijk K.; Marley K. E.; Jeong B.; Xu J.; Hesson J.; Cerny R. L.; Waterborg J. H.; Cerutti H. Monomethyl histone H3 lysine 4 as an epigenetic mark for silenced euchromatin in Chlamydomonas. Plant Cell. 2005, 17, 2439–2453. 10.1105/tpc.105.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T.; Miyasaka H.; Ohama T. Unstable RNAi effects through epigenetic silencing of an inverted repeat transgene in Chlamydomonas reinhardtii. Genetics. 2008, 180, 1927–1944. 10.1534/genetics.108.092395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D.; Schmollinger S.; Schroda M. Protocol: methodology for chromatin immunoprecipitation (ChIP) in Chlamydomonas reinhardtii. Plant Methods 2011, 7, 35. 10.1186/1746-4811-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S. S.; Prochnik S. E.; Vallon O.; Harris E. H.; Karpowicz S. J.; Witman G. B.; Terry A.; Salamov A.; Fritz-Laylin L. K.; Marechal-Drouard L.; Marshall W. F.; Qu L.-H.; Nelson D. R.; Sanderfoot A. A.; Spalding M. H.; Kapitonov V. V.; Ren Q.; Ferris P.; Lindquist E.; Shapiro H.; Lucas S. M.; Grimwood J.; Schmutz J.; Cardol P.; Cerutti H.; Chanfreau G.; Chen C.-L.; Cognat V.; Croft M. T.; Dent R.; Dutcher S.; Fernandez E.; Fukuzawa H.; Gonzalez-Ballester D.; Gonzalez-Halphen D.; Hallmann A.; Hanikenne M.; Hippler M.; Inwood W.; Jabbari K.; Kalanon M.; Kuras R.; Lefebvre P. A.; Lemaire S. D.; Lobanov A. V.; Lohr M.; Manuell A.; Meier I.; Mets L.; Mittag M.; Mittelmeier T.; Moroney J. V.; Moseley J.; Napoli C.; Nedelcu A. M.; Niyogi K.; Novoselov S. V.; Paulsen I. T.; Pazour G.; Purton S.; Ral J.-P.; Riano-Pachon D. M.; Riekhof W.; Rymarquis L.; Schroda M.; Stern D.; Umen J.; Willows R.; Wilson N.; Zimmer S. L.; Allmer J.; Balk J.; Bisova K.; Chen C.-J.; Elias M.; Gendler K.; Hauser C.; Lamb M. R.; Ledford H.; Long J. C.; Minagawa J.; Page M. D.; Pan J.; Pootakham W.; Roje S.; Rose A.; Stahlberg E.; Terauchi A. M.; Yang P.; Ball S.; Bowler C.; Dieckmann C. L.; Gladyshev V. N.; Green P.; Jorgensen R.; Mayfield S.; Mueller-Roeber B.; Rajamani S.; Sayre R. T.; Brokstein P.; Dubchak I.; Goodstein D.; Hornick L.; Huang Y. W.; Jhaveri J.; Luo Y.; Martinez D.; Ngau W. C. A.; Otillar B.; Poliakov A.; Porter A.; Szajkowski L.; Werner G.; Zhou K.; Grigoriev I. V.; Rokhsar D. S.; Grossman A. R. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science (Washington, DC, U. S.) 2007, 318, 245–250. 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B.; Ahmad K.; Almouzni G.; Ausiá J.; Berger F.; Bhalla P. L.; Bonner W. M.; Cande W. Z.; Chadwick B. P.; Chan S. W. L.; Cross G. A. M.; Cui L.; Dimitrov S. I.; Doenecke D.; Eirin-Lápez J. M.; Gorovsky M. A.; Hake S. B.; Hamkalo B. A.; Holec S.; Jacobsen S. E.; Kamieniarz K.; Khochbin S.; Ladurner A. G.; Landsman D.; Latham J. A.; Loppin B.; Malik H. S.; Marzluff W. F.; Pehrson J. R.; Postberg J.; Schneider R.; Singh M. B.; Smith M. M.; Thompson E.; Torres-Padilla M. E.; Tremethick D. J.; Turner B. M.; Waterborg J. H.; Wollmann H.; Yelagandula R.; Zhu B.; Henikoff S. A unified phylogeny-based nomenclature for histone variants. Epigenet. Chromatin 2012, 5, 1–19. 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Zhang R.; Patena W.; Gang S. S.; Blum S. R.; Ivanova N.; Yue R.; Robertson J. M.; Lefebvre P. A.; Fitz-Gibbon S. T.; Grossman A. R.; Jonikas M. C. An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 2016, 28, 367–387. 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.