Abstract
Photoreceptor degeneration leads to irreversible vision loss in humans with retinal dystrophies such as retinitis pigmentosa. Whereas photoreceptor loss is permanent in mammals, zebrafish possesses the ability to regenerate retinal neurons and restore visual function. Following acute damage, Müller glia (MG) re-enter the cell cycle and produce multipotent progenitors whose progeny differentiate into mature neurons. Both MG reprogramming and proliferation of retinal progenitor cells require reactive microglia and associated inflammatory signaling. Paradoxically, in zebrafish models of retinal degeneration, photoreceptor death does not induce the MG to reprogram and regenerate lost cells. Here, we used male and female zebrafish cep290 mutants to demonstrate that progressive cone degeneration generates an immune response but does not stimulate MG proliferation. Acute light damage triggered photoreceptor regeneration in cep290 mutants but cones were only restored to prelesion densities. Using irf8 mutant zebrafish, we found that the chronic absence of microglia reduced inflammation and rescued cone degeneration in cep290 mutants. Finally, single-cell RNA-sequencing revealed sustained expression of notch3 in MG of cep290 mutants and inhibition of Notch signaling induced MG to re-enter the cell cycle. Our findings provide new insights on the requirements for MG to proliferate and the potential for immunosuppression to prolong photoreceptor survival.
SIGNIFICANCE STATEMENT Inherited retinal degenerations (IRDs) are genetic diseases that lead to the progressive loss of photoreceptors and the permanent loss of vision. Zebrafish can regenerate photoreceptors after acute injury by reprogramming Müller glia (MG) into stem-like cells that produce retinal progenitors, but this regenerative process fails to occur in zebrafish models of IRDs. Here, we show that Notch pathway inhibition can promote photoreceptor regeneration in models of progressive degeneration and that immunosuppression can prevent photoreceptor loss. These results offer insight into the pathways that promote MG-dependent regeneration and the role of inflammation in photoreceptor degeneration.
Keywords: cep290, microglia, Müller cell, regeneration, zebrafish
Introduction
In humans and mammals, the loss of retinal neurons from injury or inherited retinal degeneration (IRD) is irreversible because mammals do not mount a regenerative response to lost or dying neurons. In contrast, fish possess the capability to regenerate lost neurons and restore visual function from populations of endogenous stem cells (Johns, 1977; Johns and Easter, 1977; Raymond and Rivlin, 1987; Raymond et al., 1988; Hitchcock and Raymond, 1992; Vihtelic and Hyde, 2000; Otteson and Hitchcock, 2003; Goldman, 2014; Hyde and Reh, 2014; Lenkowski and Raymond, 2014). Numerous groups have revealed the identity of retina stem cell populations (Yurco and Cameron, 2005; Fausett and Goldman, 2006), identified the sequence of events that underpin regeneration (Nelson et al., 2012, 2013), and dissected molecular pathways that promote and inhibit regeneration (Ramachandran et al., 2010, 2012; Morris et al., 2011; Hoang et al., 2020; Nagashima et al., 2020).
In zebrafish, Müller glia (MG) constitute an inducible stem cell population capable of regenerating lost neurons. In response to widespread acute retinal damage MG undergo cellular reprogramming and produce multipotent retinal progenitors that proliferate and differentiate into all retinal cell types (Goldman, 2014; Gorsuch and Hyde, 2014). If damage is limited to a small number of rod photoreceptors, proliferation of rod precursors located in the outer nuclear layer (ONL) increases without a noticeable increase in MG proliferation (Morris et al., 2005; Vihtelic et al., 2006; Montgomery et al., 2010). Conversely, cone photoreceptors can only be regenerated from MG-derived retinal progenitors (Bernardos et al., 2007; Thummel et al., 2008).
In response to photoreceptor degeneration in mammals, microglia become activated, release proinflammatory cytokines, and phagocytize photoreceptors (Zeng et al., 2005; Yoshida et al., 2013; L. Zhao et al., 2015). Zebrafish also exhibit a robust inflammatory response following acute retinal injury, with activation of microglia/macrophages and the release of proinflammatory cytokines (Nelson et al., 2013; White et al., 2017; Mitchell et al., 2018, 2019). While microglia-mediated inflammation is generally considered neurotoxic in mammals, inflammation in zebrafish appears critical for the reprogramming of MG and proliferation of MG-derived progenitors (Nelson et al., 2013; Silva et al., 2020). Immunosuppression with dexamethasone or depletion of microglia with PLX3397 impaired proliferation of MG-derived progenitors, thereby restricting regeneration (White et al., 2017; Conedera et al., 2019; Silva et al., 2020). As immunomodulation has been posited as a potential therapy for retinal degeneration (Silverman and Wong, 2018; Silverman et al., 2019), it is important to consider the impact of immunosuppression on regeneration in zebrafish models of progressive retinal degeneration.
Although it is typically assumed that zebrafish regenerate retinal neurons following damage and disease, knowledge about regeneration comes primarily from studies where the retina was acutely damaged by light, mechanical injury, or injection of toxins such as ouabain (Raymond et al., 1988; Vihtelic and Hyde, 2000; Ramachandran et al., 2010; Gorsuch and Hyde, 2014). Multiple reports exist, however, of adult zebrafish models with progressive photoreceptor degeneration with little evidence for cone regeneration from MG-derived progenitors (Li and Dowling, 1997; Stenkamp et al., 2008; Liu et al., 2015; M. Yu et al., 2016; Lu et al., 2017; S. Yu et al., 2017; Iribarne et al., 2019; Song et al., 2020). Here, we use a zebrafish cep290 mutant (centrosomal protein 290) to determine the immune response in a model of progressive retinal degeneration. We demonstrate that progressive retinal degeneration triggers a transcriptional and cellular immune response but inflammation alone is insufficient to induce MG reprogramming. Whereas immunosuppression by dexamethasone inhibited proliferation of rod precursor cells, blocking glucocorticoid signaling with RU486 did not stimulate regeneration. Loss of irf8 in addition to cep290 resulted in chronic immunosuppression and prevented cone degeneration. Finally, we show that upregulation of notch3 in MG of cep290 mutants suppresses their proliferation, and that Notch inhibition releases that constraint. Collectively, our results indicate that therapies to suppress microglia function may prolong photoreceptor survival and that inducing regeneration requires more than inflammation alone.
Materials and Methods
Animal maintenance
Adult zebrafish were maintained and housed in 1.5-, 3.0-, and 10-l tanks in an Aquatic Habitats recirculating system (Pentair) with a 14/10 h light/dark cycle with ambient room lighting. Zebrafish lines used in this study included the mutant lines cep290fh297 (Lessieur et al., 2019) and irf8st95 (Shiau et al., 2015), the transgenic reporter lines Tg(Xla.Rho.eGFP)fl1 (Fadool, 2003), Tg(gfap:eGFP)mi2002 (Bernardos and Raymond, 2006), and Tg(gfap:stat3-eGFP)mi35 (X.F. Zhao et al., 2014), as well as the transgenic line Tg(rho:YFP-Eco.NfsB)gmc500 (herein referred to as Tg(rho:ntr-YFP)) to ablate rod photoreceptors (White and Mumm, 2013). Animals from heterozygous crosses of cep290 or irf8 lines were genotyped using high-resolution melt analysis (HRMA) or PCR using specified primer sets (Extended Data Table 1-1). Homozygote, heterozygote, and wild-type siblings were identified for each cross. Transgenic lines were confirmed by PCR using primers specific for GFP (Extended Data Table 1-1). Fin clips were added to 50 µl of a 50 mm NaOH solution heated to 95°C for 10 min and then neutralized with 5 µl of 1 m Tris pH 8.0 before cooling to room temperature. Experiments included animals of both sexes and were used at the specified ages. All animal procedures were done with approval by the Institutional Animal Care and Use Committee (IACUC) at the Cleveland Clinic and in accordance with relevant guidelines and regulations, including the ARVO statement for the use of animals in research.
EdU labeling
To label proliferating cells, animals were anesthetized with tricaine methanesulfonate (0.4 mg/ml) and placed on a wet towel and injected intraperitoneally with 20 µL of a 20 mm EdU solution (PBS). Animals were injected on two consecutive days, 24 h apart, and then allowed to recover for an additional 24 h before enucleation. Eyes were subsequently processed for immunohistochemistry.
Light damage
Light damage experiments were performed using a protocol adapted from Thomas and Thummel (Thomas and Thummel, 2013). Adult zebrafish were first dark adapted for 36–42 h. Up to five animals were placed in a 250-ml glass beaker with system water that was seated inside a 1-l glass beaker with Milli-Q water and exposed to high-intensity light from a 120-W X-CITE series 120Q metal halide lamp (Excelitas) for 30 min and then exposed to 14,000 lux light from an illumination cage for 50 min. Animals were allowed to recover in system water for either 3 or 30 d. To label proliferating cells during early stages of regeneration, animals were injected intraperitoneally with 20 µl of a 20 mm EdU solution (PBS) at 2 d postinjury (dpi) and eyes were enucleated 24 h later (3 dpi). To assess regeneration, animals were allowed to recover for 30 dpi before enucleation.
Eye lesions
Eye lesions were performed in a manner similar to that previously described (Senut et al., 2004). Briefly, animals were anesthetized with tricaine methanesulfonate (0.4 mg/ml) and placed on a wet towel on the stage of a stereomicroscope and illuminated with a fiber optic light source. The eye was gently raised and poked through the sclera in the back of the eye with a 30-gauge needle to the depth of the bevel (∼5 mm) in both the dorsal and ventral hemispheres, near the dorsoventral meridian and as close to the optic nerve as possible. Animals were returned to fresh system water. After 4 d of recovery, animals were intraperitoneally injected with EdU (20 mm) and euthanized 4 h later. Eyes were removed and processed for immunohistochemistry.
Immunohistochemistry
Adult zebrafish were deeply anesthetized with tricaine methanesulfonate (0.4 mg/ml) and decapitated with a razor blade. Eyes were rapidly enucleated, immersed for 2 h in 4% paraformaldehyde and washed in PBS. Samples were equilibrated in 5% sucrose/PBS for 3 h at room temperature and transferred to 30% sucrose/PBS overnight at 4°C. Eyes were washed in a 1:1 solution of 30% sucrose:tissue freezing medium overnight at 4°C and embedded for cryosectioning.
Transverse cryosections sections (10 µm) were cut and mounted on Superfrost Plus slides and dried at room temperature overnight. Slides were washed 3 × 10 min in PBS and then incubated in blocking solution (PBS + 2% BSA, 5% goat serum, 0.1% Tween 20, 0.1% DMSO) for 1 h. The following primary antibodies were used: mouse monoclonal zpr1 [1:100, Zebrafish International Resource Center (ZIRC)], mouse monoclonal zpr3 (1:100, ZIRC), mouse monoclonal 4C4 (1:1000, a gift from Peter Hitchcock, University of Michigan), rabbit polyclonal L-plastin (1:1000, GeneTex, GTX124420), mouse monoclonal PCNA (1:100, Sigma, clone PC-10), peanut agglutinin (PNA)-lectin conjugated to Alexa Fluor-568 (1:100, ThermoFisher). EdU labeling was detected with the Click-iT Edu Alexa Fluor-555 Imaging kit (ThermoFisher). Alexa-conjugated secondary antibodies were used at 1:500 in blocking buffer and incubated for at least 1 h. Slides were counterstained with 4,5-diamidino-2-phenylendole (DAPI) to stain nuclei.
Image acquisition and quantification
Z-stacked images of 5- to 15-µm immunostained cryosections were imaged on a Zeiss Imager Z.2 equipped with an ApoTome using 10× dry, 20× dry, or 63× oil immersion lenses (Zeiss). Images were acquired with Zen2 software and postprocessed in ImageJ. All imaging and quantitative analysis was performed on dorsal retina sections, which contained or were immediately adjacent to the optic nerve. Cells or labels of interest (e.g., PNA, EdU, PCNA) were manually quantified in maximum-projection z-stacks of the dorsal retina and densities or ratios were calculated by measuring the curvilinear distance of retina in each section using ImageJ. Each data point represents the ratio or density from the dorsal region of a central retinal section from one eye.
Lineage tracing
Retinal sections from Tg(XOPS:eGFP); cep290 fish that were injected with EdU four weeks before killing were immunostained with zpr1 and imaged with a Zeiss Imager.Z2 with Apotome.2 attachment. Labeled cells were identified in single optical sections. Using DAPI staining as a guide, regions of interest (ROIs) were drawn around the nucleus of cells labeled with either zpr1 or EdU. The GFP fluorescence in each ROI was normalized to the average GFP fluorescence in the nuclei of five random GFP+ cells in the ONL from the same image. Thus, rods will have a fluorescence intensity near 1, whereas cones will have an intensity near 0. As a negative control, we also measured background GFP fluorescence of random nuclei in the INL, which does not contain photoreceptors.
Metronidazole (MTZ) treatment
To ablate photoreceptors using the Tg(rho:ntr-YFP) line, adult zebrafish were incubated in system water containing 10 mm MTZ (Sigma) for 24 h in a 28°C incubator. Animals were transferred to fresh system water and dark-adapted for 18 h before euthanasia. The eyes were removed and retinas were dissected as described above.
Dexamethasone and RU486 treatments
Immunosuppression by dexamethasone treatment was performed similar to previous protocols (Kyritsis et al., 2012; Silva et al., 2020). Briefly, a stock solution (12.5 mg/ml) of dexamethasone was prepared by adding 500-mg dexamethasone (Sigma-Aldrich) to 40 ml methanol (vehicle). Animals were housed in 500 ml of system water containing a final concentration of 15 mg/l dexamethasone or 0.12% methanol for 14 d. Inhibition of glucocorticoid signaling by the glucocorticoid receptor (GR) antagonist RU486 was performed using a protocol previously shown to partially phenocopy a GR mutant (Ziv et al., 2013). A 20 mm solution of RU486 (mifepristone, Sigma) was prepared by dissolving 100 mg RU486 into 11.5 ml methanol (vehicle). Animals were housed in 500 ml of system water containing a 1.25-µm solution of RU486 or 0.12% methanol for 7 d. Solutions containing dexamethasone, RU486, or vehicle were changed daily and animals were fed daily with flake food ∼2 h before solution changes. At the end of the treatment, animals were euthanized and eyes were removed by enucleation and processed for immunohistochemistry.
Notch inhibition
Adult wild-type or cep290 mutants were injected intraperitoneally with 20–25 µl of 1 mm RO4929097 (2,2-dimethyl-N-((S)−6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)-N'-(2,2,3,3,3-pentafluoro-propyl)-malonamide) or 1% DMSO in PBS (vehicle) using a 30-gauge needle. Animals were injected every 12 h for 4 d and allowed to recover for 24 h before euthanasia.
RNA-seq analysis
Total RNA was isolated from adult zebrafish between the age of 6–7 mpf. Animals were dark-adapted for 16–18 h and euthanized in the dark before being moved to room light for enucleation and retina dissection Both retinas from each fish were pooled and four animals per genotype were used as biological replicates (n = 4). RNA was extracted using TRIzol with chloroform and isopropanol used to separate aqueous phases. Glycoblue (ThermoFisher) was added before precipitation to enhance visibility of the pellets. Nucleic acid pellets were resuspended in nuclease-free water and treated with TURBO DNase (ThermoFisher) for 15 min. RNA was subsequently precipitated with LiCl, washed with 70% ethanol and resuspended in nuclease-free water. RNA concentration and purity was analyzed by Qubit Fluorometer (Invitrogen) and Agilent Bioanalyzer 2100. All samples had RNA integrity number (RIN) values >8.5. A total of 500 ng of each sample were submitted for Illumina RNA-seq. All library preparations, quality control steps, and next-generation sequencing was performed by the Lerner Research Institute (LRI) Genomics Core at the Cleveland Clinic. Sequencing was done on an Illumina NovaSeq 6000 with 150-bp pair-end read sequencing runs.
scRNA-seq
Six-month-old Cep290 and heterozygote sibling fish were used for all scRNA-seq experiments. Whole retinas from five fish were dissected and dissociated as previously described (Hoang et al., 2020). Briefly, retinas were placed in Leibowitz media and incubated with hyaluronidase. Retinas were then incubated in Papain solution at 28°C with vigorous shaking for 30 min. Dissociated cells were then centrifuged at 1500 × g for 10 min, and resuspended in PBS containing Leupeptin and DNase. Dissociated cells were then fixed using the SplitBio single nuclei fixation kit, and samples processed using the SplitBio single-cell whole transcriptome kit per the manufacturer's instructions. Libraries were then sequenced by following the manufacturer instruction (Read1: 76, i7 Index: 6, Read2: 86) to obtain ∼25,000 reads/cell. Sequencing files were processed using the SplitBio pipeline with reads mapped to the zebrafish genome GRCz11. Further processing and analysis was performed in Seurat 4.0. Cells with fewer than 200 UMIs, >2500 UMIs, or >5% mitochondrial RNA were excluded from analysis. Known markers were used to identify retinal cell types. For differential expression analysis genes must be expressed in at least 10% of the cells within a group, have a log fold change of 0.25 or greater, and an adjusted p-value <0.05 to be considered significant.
qRT-PCR
A total of 500 ng of purified RNA remaining from RNA-seq experiments was used for reverse transcription and qPCR. Reverse transcription and qPCR were done per manufacturer's instructions using the Bio-Rad iScript cDNA synthesis kit and the Bio-Rad SsoFast EvaGreen Supermix kit, respectively. Reactions were run on a Bio-Rad CFX96 Touch Real-Time PCR detection system and analyzed using CFX Manager. Primer sequences are listed in Extended Data Table 1-1. Four biological replicates were prepared for each sample and three technical replicates were performed on each biological sample. Fold changes were calculated by the ΔΔC(t) method, with 18S rRNA used for normalization.
Statistics and data analysis
All data were analyzed and graphed using GraphPad Prism (v8). Data sets were first tested for normal distribution using Prism. Normally distributed datasets were subsequently analyzed by Student's t tests or one-way ANOVA with Dunnet T3 correction for multiple comparisons. Where necessary, nonparametric Kruskal–Wallace tests were used. The statistical tests and p values for each experiment are provided in Extended Data Table 1-2.
Results
Proliferating progenitor cells differentiate into rod photoreceptors in cep290 mutants
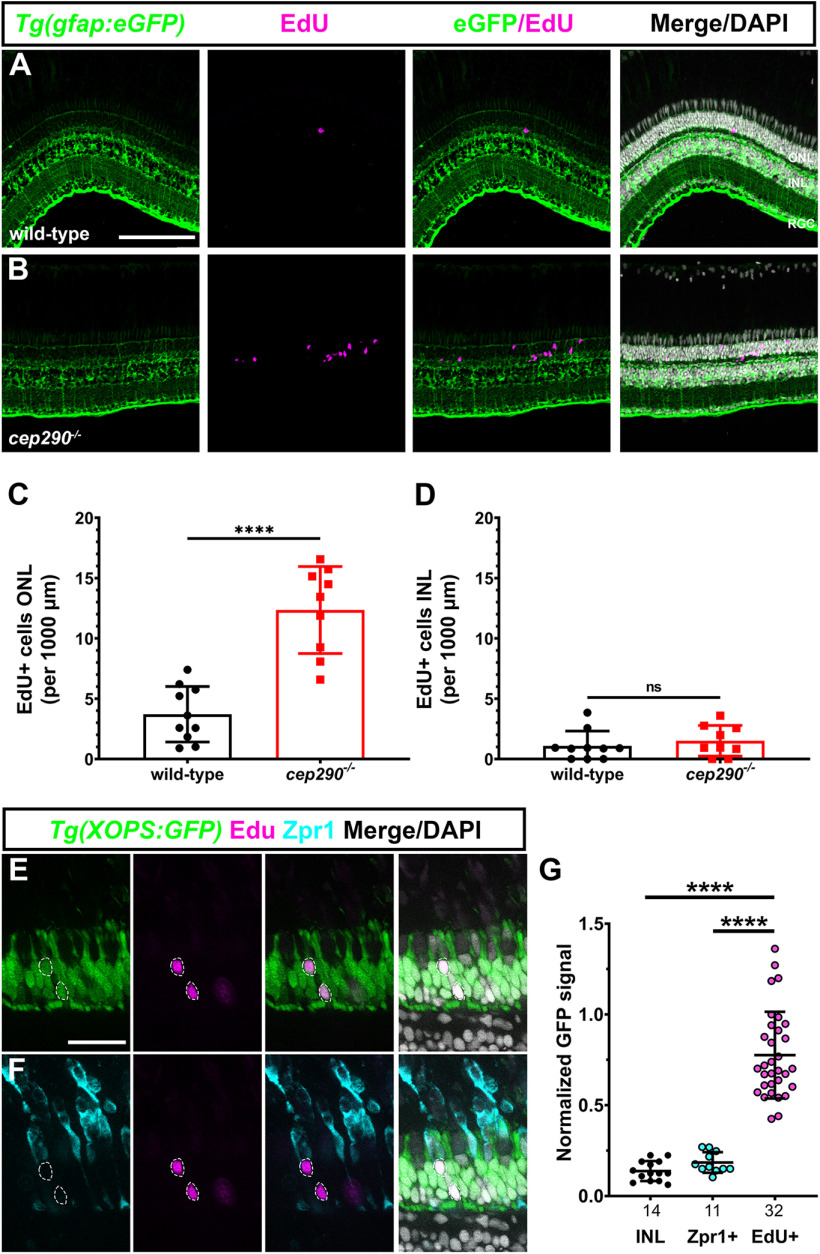
We previously reported that the cep290 mutant undergoes progressive cone degeneration and noted an increased number of cells in the ONL that stained positive for the proliferation marker PCNA (Lessieur et al., 2019). This suggested that rod photoreceptors also degenerated but were replaced by unipotent rod precursors (Raymond and Rivlin, 1987; Stenkamp, 2011) and that degeneration of photoreceptors remained insufficient to induce MG proliferation in cep290 mutants. To investigate the identity of the proliferating cells and to confirm that MG were not undergoing mitosis, the cep290 mutant was crossed into the transgenic reporter line Tg(gfap:eGFP)mi2002, which labels MG with GFP (Bernardos and Raymond, 2006). At six months postfertilization (mpf), cep290 mutants and wild-type siblings were injected with EdU on two consecutive days and eyes were collected for immunohistochemistry 24 h later. In wild-type retinas, a few EdU+ cells were present in the ONL or inner nuclear layer (INL; Fig. 1A). In cep290 mutants, a 3.3-fold increase in EdU+ cells was observed in the ONL (Fig. 1B,C). Few of the EdU+ cells colocalized with GFP+ MG nuclei in the INL (Fig. 1D). These results strongly suggest that proliferation is limited to ONL-localized rod precursors in cep290 mutants. We next sought to confirm that the proliferating cells gave rise exclusively to rod photoreceptors. To unambiguously label rod nuclei, the cep290 mutation was crossed with the transgenic reporter line Tg(Xla.Rho:eGFP)fl1, also known as Tg(XOPS:eGFP) (Fadool, 2003), and animals were injected with EdU. Retinal cryosections were processed and imaged for EdU, GFP fluorescence (rods), and with the antibody Zpr-1 (red/green double cone photoreceptors). EdU colocalized with GFP from rod photoreceptors (Fig. 1C) and failed to localize to Zpr-1+ cones (Fig. 1D). The GFP fluorescence intensity was measured in individual EdU+ cells, in cells located in the INL, and in Zpr-1+ cells and normalized to a sample of GFP+ cells in the ONL from the same image (Fig. 1E). Cells in the INL and all Zpr-1+ cells were EdU–, whereas all EdU+ cells exhibited GFP signal above background. These data indicate that proliferation of rod precursors in cep290 mutants contributes to regeneration of rods and likely explains why rod degeneration is not observed in cep290 mutants.
Figure 1.
Cell proliferation occurs by rod precursors in cep290 mutants. A, B, EdU labeling (magenta) in retinas from 6 mpf wild-type and cep290 mutants on the transgenic reporter line Tg(gfap:eGFP)mi2002 to label MG (green). Almost all EdU+ cells in cep290 mutants were located in the ONL. C, D, Quantification of EdU+ cells in the ONL and INL, respectively, of wild-type and cep290 mutants. Data are plotted as mean ± SD and p-values were generated by Welch's t test (ONL) or Mann–Whitney test (INL). ****p < 0.0001, ns = not significant. E, F, Single optical slices showing EdU labeling (magenta) and Zpr1 immunoreactivity (cyan) in retinas from 6 mpf cep290 mutants on the transgenic reporter line Tg(XOPS:eGFP)fl1, which labels rod photoreceptors (green). EdU+ cells (dashed circles) colocalized with rod nuclei but not in Zpr1+ cells. G, The normalized GFP fluorescence signal from the Tg(XOPS:eGFP)fl1 transgene was quantified in cells located the INL, in Zpr1+ cells, and EdU+ cells. N values are indicated on the graph. GFP signal was significantly higher in EdU+ cells than in Zpr1+ cells or in cells in the INL. Data are plotted as mean ± SD and p-values were generated by Welch's ANOVA test with Dunnett's T3 multiple comparisons test. ****p < 0.0001. ONL = outer nuclear layer; INL = inner nuclear layer; RGC = retinal ganglion cell layer. Scale bars: 100 µm (A, B) and 25 µm (C, D).
Progressive retinal degeneration in cep290 mutants leads to an immune cell response
Retinal injury and cell death in zebrafish results in rapid activation of resident microglia and increased expression of inflammatory cytokines, such as TNFα (Nelson et al., 2013; White et al., 2017; Mitchell et al., 2018). Activation of microglia is associated with proliferation of MG and techniques that ablate or deplete microglia attenuate retinal regeneration following damage (White et al., 2017; Conedera et al., 2019). To determine whether cep290 mutants exhibited signs of inflammation, retinas from 6 mpf animals were stained with the antibody 4C4 to label microglia/macrophages (Mazzolini et al., 2020), and anti-L-plastin antibodies to label leukocytes (Herbomel et al., 1999; Mitchell et al., 2018). In wild-type retinas, ramified microglia/macrophage were found in the nerve fiber layer, the inner plexiform layer (IPL) and outer plexiform layer (OPL), and in the subretinal space between the outer segments and RPE (Fig. 2A). In contrast, significant numbers of amoeboid-shaped, activated microglia/macrophage accumulated in the subretinal space of cep290 mutants (Fig. 2B). These cells were both 4C4+ and L-plastin+, which is consistent with resident microglia/macrophages responding to photoreceptor degeneration. Similar to prior results from rod ablation studies in larvae (White et al., 2017), no evidence was found that peripheral macrophages (e.g., 4C4–,L-plastin+ cells) entered the retinas of cep290 mutants. In both wild-type and cep290 mutants, microglia/macrophages in the OPL remained ramified (Fig. 2C,D), although an 80% reduction in microglia/macrophages was observed in the IPL of cep290 mutants (Fig. 2C,D, cyan arrows; quantified in Fig. 2E). This resembles what occurs in humans with retinitis pigmentosa and in rodent models of IRD, where death of photoreceptors promotes the translocation of activated microglia/macrophages from the inner retina to the subretinal space and the release of inflammatory cytokines, including TNFα (Silverman and Wong, 2018; Karlen et al., 2020). Combined with the results above, these data indicate that although cep290 mutants exhibited immune system activation and proliferation of rod precursors, the level of immune reactivity was insufficient to induce the mutant MG to enter the cell cycle.
Figure 2.
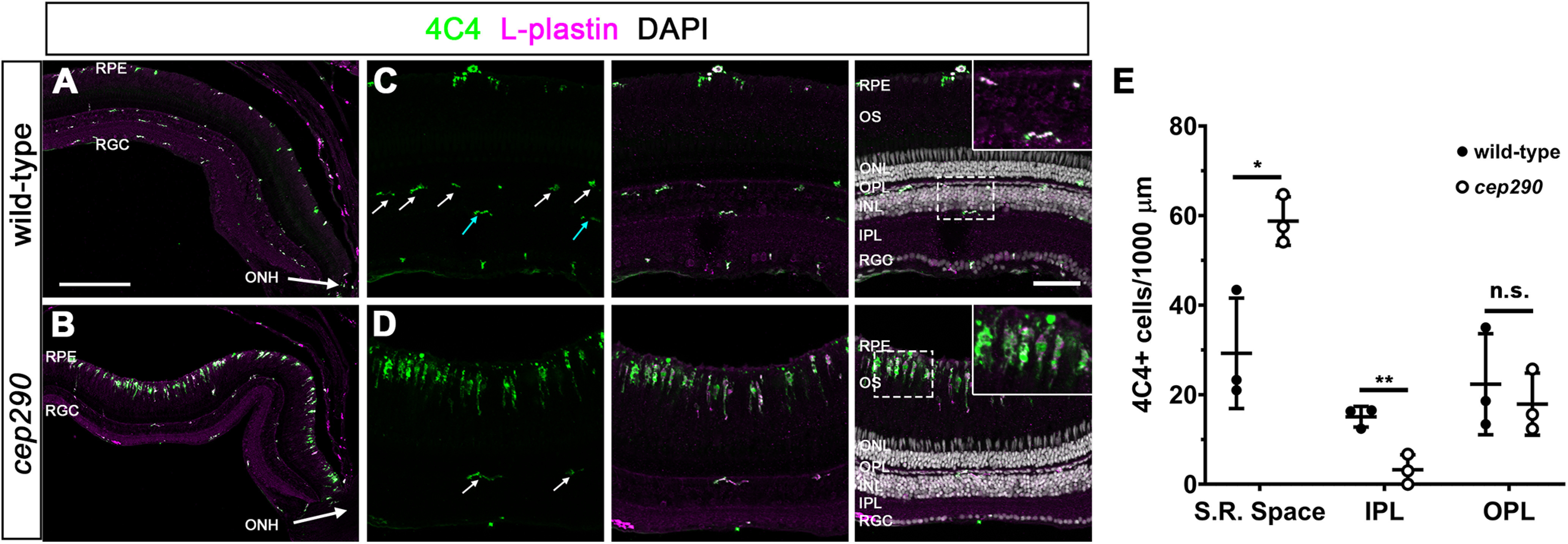
Immune cells accumulate in the subretinal space of cep290 mutants. A, B, Immunohistochemistry with the monoclonal antibody 4C4 (green) and anti-L-plastin (magenta) label microglia/macrophage in the dorsal retina of 6 mpf wild-type and cep290 mutants. The ONH is located at the bottom right corner of each image. C, D, Higher magnification images show the accumulation of activated microglia/macrophage in the subretinal space of cep290 mutants. Ramified microglia/macrophage (4C4+/L-plastin+) were seen in the OPL (white arrows) of both wild-type and cep290 mutants but only in the IPL of wild-type retinas (cyan arrows). Insets, Higher magnification of boxed regions illustrates the ramified morphology of quiescent microglia/macrophage in the plexiform layers of wild-type retinas and the elongated and amoeboid shape of microglia/macrophage in the subretinal space of cep290 mutants. E, Quantification of 4C4+ cells in different regions of the retina in 6 mpf fish (n = 3 per genotype). Data are plotted as mean ± SD and p-values were generated by Welch's t tests. *p < 0.05, ****p<0.01; n.s. = not significant. RPE = retinal pigment epithelium, RGC = retinal ganglion cell layer, ONH = optic nerve head, OS = outer segments, ONL = outer nuclear layer, INL = inner nuclear layer, S.R. space = subretinal space, IPL = inner plexiform layer, OPL = outer plexiform layer. Scale bars: 200 µm (A, B) and 50 µm (C, D).
Transcriptome analysis of cep290 mutant retinas
To gain a better understanding of transcriptional changes that occur during photoreceptor degeneration and that may promote regeneration, we performed RNA-seq analysis on retinas from 6 mpf cep290 mutants (n = 4) and wild-type siblings (n = 4). Pairwise comparisons of all cep290 mutant and wild-type transcriptomes revealed 1379 differentially expressed genes (DEGs; padj < 0.05; Extended Data Table 3-1). Of these, 235 DEGs were upregulated at least 2-fold (padj < 0.05). Pairwise comparisons of all samples were tested for enriched gene sets by Gene Set Enrichment Analysis (GSEA) and 11 pathways were upregulated in cep290 mutants (Table 1). The unfolded protein response, TP53, and apoptosis pathways were upregulated in cep290 mutants, which likely reflects cellular stress and degeneration of photoreceptors. Upregulation of the interferon-α, interferon-γ, IL-6/JAK/STAT3 signaling and TNFα signaling pathways were evidence of immune system activation in cep290 mutants. These signaling pathways promote MG reprogramming and cell division following acute injury in healthy adult zebrafish (Nelson et al., 2013; Wan et al., 2014; X.F. Zhao et al., 2014), yet MG did not proliferate in cep290 mutants. In injury models, Müller cell reprogramming involves TNFα-dependent expression and activation of the transcription factor Stat3 (Kassen et al., 2007; Nelson et al., 2012, 2013). Expression of stat3 increases following retinal injury and suppression of stat3 expression significantly inhibits regeneration (Nelson et al., 2012). Activated phospho-Stat3 (p-Stat3) enters the nucleus and drives expression of reprogramming genes, including ascl1a and lin28 (Wan et al., 2014; X.F. Zhao et al., 2014). In our RNA-seq dataset, expression of stat3 was increased 1.86-fold (p < 0.0001) in cep290 mutant retinas. To validate this result, we performed qRT-PCR on several factors essential for Müller cell reprogramming, including tnfa, stat3, hmga1a, and yap1 (Nelson et al., 2013; Hoang et al., 2020). As a positive control, we used the Tg(rho:YFP-Eco.NfsB)gmc500 transgenic line that expresses a YFP-nitroreductase fusion protein in rod photoreceptors. Exposing this line to MTZ results in a selective ablation of rods and robust MG activation (White and Mumm, 2013; White et al., 2017). MTZ-induced rod ablation resulted in a significant increase in the expression of all regeneration-associated genes (Fig. 3A–D). In cep290 mutants, we confirmed a 2.1-fold increase (p < 0.05) of stat3 and a 1.6-fold increase (p < 0.01) in yap1 but detected no difference in expression of tnfa or hmga1a when compared with wild-type adults (Fig. 3A–D). The transcriptomic analysis suggests that photoreceptor degeneration stimulates inflammatory and upstream Stat3 signaling pathways, but this is insufficient to trigger MG reprogramming and proliferation.
Table 1.
GSEA upregulated pathways
| Gene set | NES | FDR |
|---|---|---|
| Interferon-α response | 2.38 | 0.000 |
| Interferon-γ response | 2.23 | 0.000 |
| IL-6/JAK/STAT3 signaling | 2.12 | 0.000 |
| Unfolded protein response | 1.76 | 0.007 |
| TP53 pathway | 1.70 | 0.012 |
| Myc targets | 1.59 | 0.030 |
| TNFα signaling via NFκB | 1.59 | 0.027 |
| Complement | 1.59 | 0.025 |
| Allograft rejection | 1.58 | 0.023 |
| mTORC1 signaling | 1.57 | 0.026 |
| Apoptosis | 1.54 | 0.029 |
NES = normalized enrichment score.
FDR = false discovery rate.
See Extended Data Table 3-1 for complete results.
Figure 3.
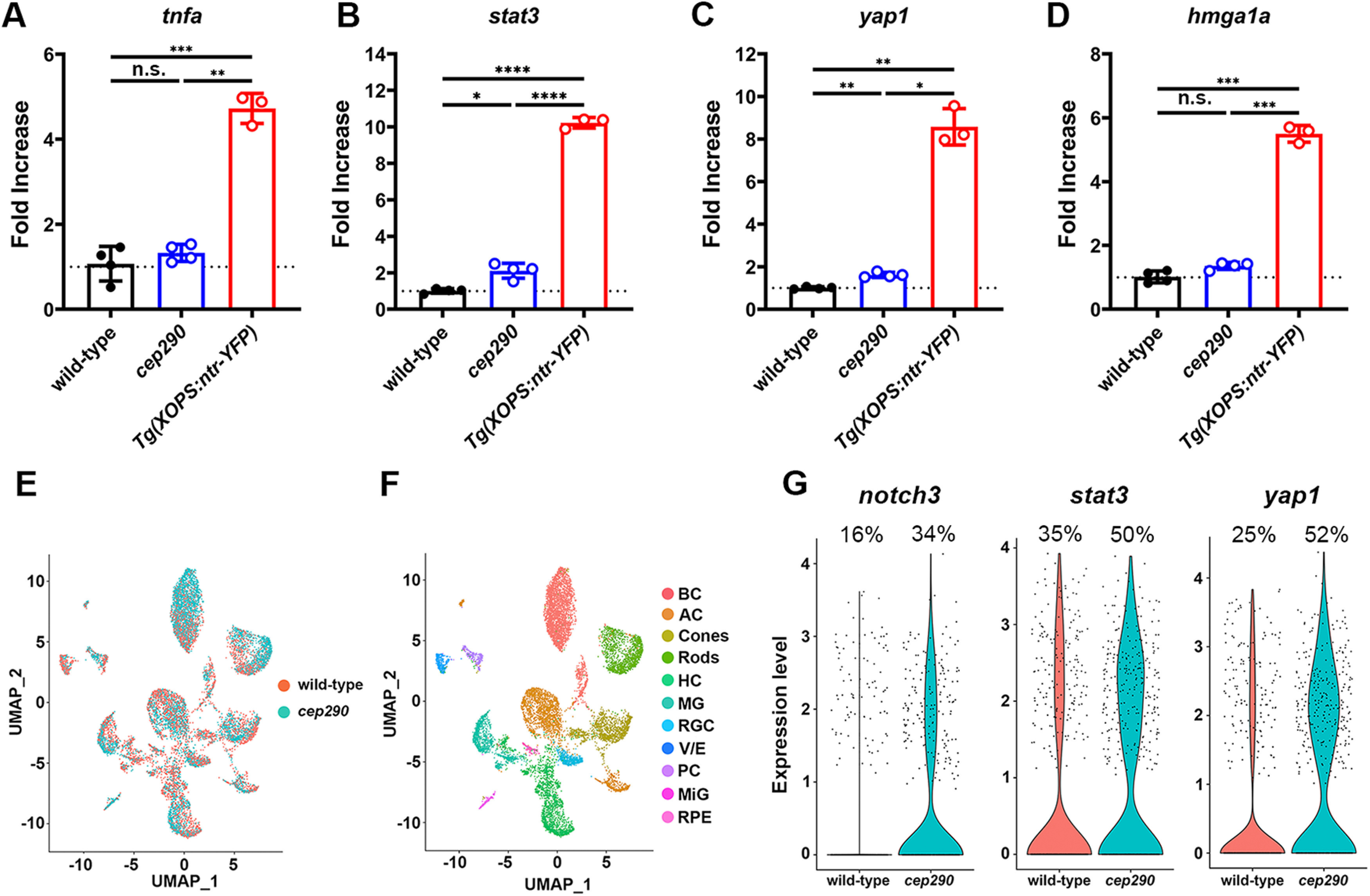
Expression of proregeneration genes was only modestly upregulated in 6 mpf cep290 mutants. A–D, qPCR for tnfa, stat3, yap1, and hmga1a in 6 mpf wild-type and cep290 mutant zebrafish. Gene expression in MTZ-treated Tg(rho:ntr-YFP)gmc500 zebrafish was used as a positive control for MG reactivity. Fold changes were calculated by the ΔΔC(t) method, with 18S rRNA used for normalization. No significant difference (n.s.) was observed between wild-type and cep290 mutants for tnfa (p > 0.63) or hmga1a (p > 0.06). cep290 mutants upregulated expression of stat3 2-fold (*p < 0.02) and yap1 1.6-fold (**p < 0.002) compared with wild-type animals. Expression of all genes was significantly upregulated in MTZ-treated Tg(rho:ntr-YFP)gmc500 fish compared with both wild-type and cep290 fish. Welch's ANOVA with Dunnett T3 multiple comparisons test; *p < 0.02, **p < 0.002, ***p < 0.0002, ****p < 0.0001. E, UMAP plot showing all cells obtained from sequencing with cells colored by sample. F, UMAP plot showing identified cell types from sequencing data, including bipolar cells (BP), amacrine cells (AC), horizontal cells (HC), retinal ganglion cells (RGC), vascular/endothelial cells (V/E), pericytes (PC), microglia (MiG), and retinal pigment epithelium (RPE). G, Violin plots showing expression of notch3, stat3, and yap1 in MG between sample groups. See Extended Data Tables 3-1, 3-2, 3-3, 3-4 for complete sequencing data.
Resources and reagents used. Download Table 1-1, DOCX file (21.8KB, docx) .
Extended data Table 1-2. Download Table 1-2, DOCX file (16KB, docx) .
DEGs. Download Table 3-1, XLSX file (161.8KB, xlsx) .
Muller glia DEGs. Download Table 3-2, XLSX file (48.7KB, xlsx) .
State DEGs. Download Table 3-3, XLSX file (13.8KB, xlsx) .
Regulatory DEGs. Download Table 3-4, XLSX file (10.5KB, xlsx) .
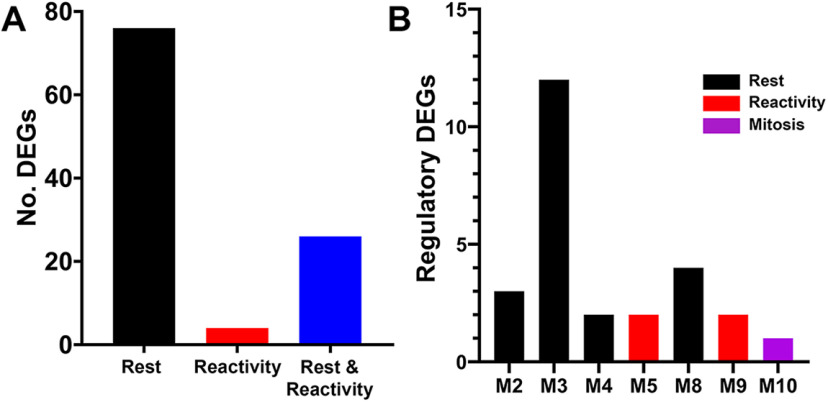
To determine whether the observed transcriptomic changes are specific to MG, we performed scRNA-seq with cep290 mutants and their heterozygote siblings (Fig. 3E,F). Differential expression analysis comparing MG from the two sample groups identified 561 significantly upregulated genes in cep290 MG (Extended Data Table 3-2). Similar to whole retina RNA-seq, significantly more MG were observed to increase expression of stat3 and yap1 (Fig. 3G). Increased expression of notch3 was also observed specifically in MG, which was not detected by whole retina RNA-seq. To build on the scRNA-seq analysis, we compared the 561 DEGs to previously published transcriptomic data (Hoang et al., 2020), which used pseudotime analysis of the regenerating retina to identify 3 MG states: resting, reactive, or proliferating. By comparing the 561 DEGs to known markers of each of these states, we identified 106 DEGs in cep290 MG that were associated with any of these states (Extended Data Table 3-3). Of those 106 DEGs, 76 were associated with resting MG (Fig. 4A). This suggested an enrichment in rest-associated genes in cep290 MG. We also compared the 561 DEGs to a predicted regulatory network within MG of the regenerating zebrafish retina (Hoang et al., 2020). Of the 26 genes that belong to the predicted regulatory network (Extended Data Table 3-4), 21 belong to rest-associated modules (Fig. 4B, M2, M3, M4, M8) that were previously described (Hoang et al., 2020).
Figure 4.
Distribution of genes by single-cell RNA-sequencing of cep290 mutants and heterozygote siblings. A, Plot showing the number of DEGs associated with each pseudotime state of rest (black), reactivity (red), or both rest and reactivity (blue). B, Plot showing the number of DEGs belonging to each regulatory module based on previously published work (Hoang et al., 2020). Colors correspond to each regulatory module.
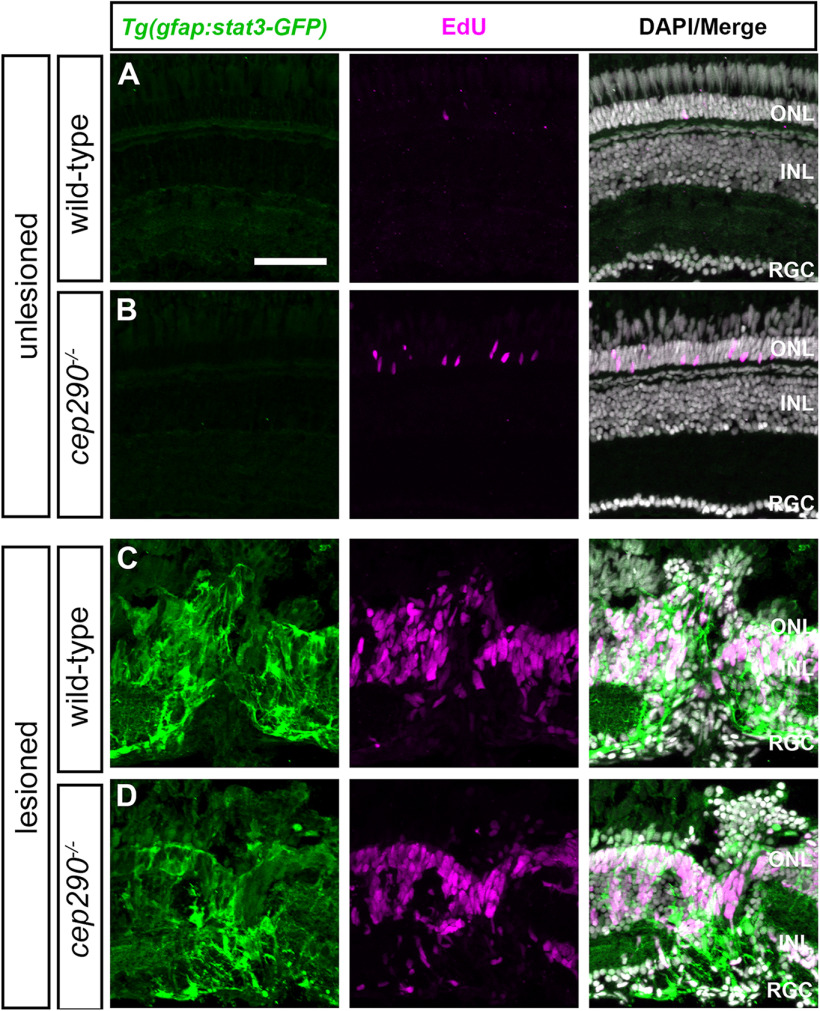
To further assess Stat3 activation, the transgenic reporter line Tg(gfap:stat3-GFP)mi35Tg (Zhao et al., 2014) was crossed into the cep290 background. The Tg(gfap:stat3-GFP) line constitutively expresses stat3-gfp mRNA in MG, but the unphosphorylated Stat3-GFP protein is degraded and remains undetectable in undamaged retinas (Zhao et al., 2014). Following retinal injury, however, activated p-Stat3-GFP accumulates specifically in proliferating MG-derived progenitors. Animals were administered a single intraperitoneal injection of EdU and allowed to recover for 24 h. In 6 mpf wild-type and cep290 mutant retinas, Stat3-GFP was not detected and EdU+ cells were only observed in the ONL of cep290 mutants (Fig. 5A,B). As a positive control, animals were first injected with EdU and mechanically injured by a needle poke. Stat3-GFP accumulated at the site of injury and localized to EdU+ cells in both wild-type and cep290 mutants (Fig. 5C,D). We conclude that cep290 mutants could activate Stat3 in response to acute injury. Surprisingly, the ongoing photoreceptor degeneration and inflammation observed in cep290 mutants remains insufficient to activate a p-Stat3 that can be detected in vivo.
Figure 5.
Injury is required for Stat3 activation in cep290 mutants. Retinal cryosections of uninjured (A) wild-type and (B) cep290 mutants on the Tg(gfap:stat3-GFP) background at 6 mpf were immunolabeled with anti-GFP antibodies (green) to label Stat3-GFP and processed for EdU labeling (magenta) to identify proliferating cells. In the absence of injury, Stat3-GFP expression was undetectable and the only proliferating cells were observed in the ONL. C, D, At 4 d following mechanical injury (e.g., unilateral needle poke), Stat3-GFP expression and cell proliferation increased in both wild-type and cep290 mutants at the site of injury. ONL = outer nuclear layer; INL = inner nuclear layer, RGC = retinal ganglion cell layer. Scale bar: 50 µm.
Light-induced photoreceptor death stimulates regeneration in cep290 mutants
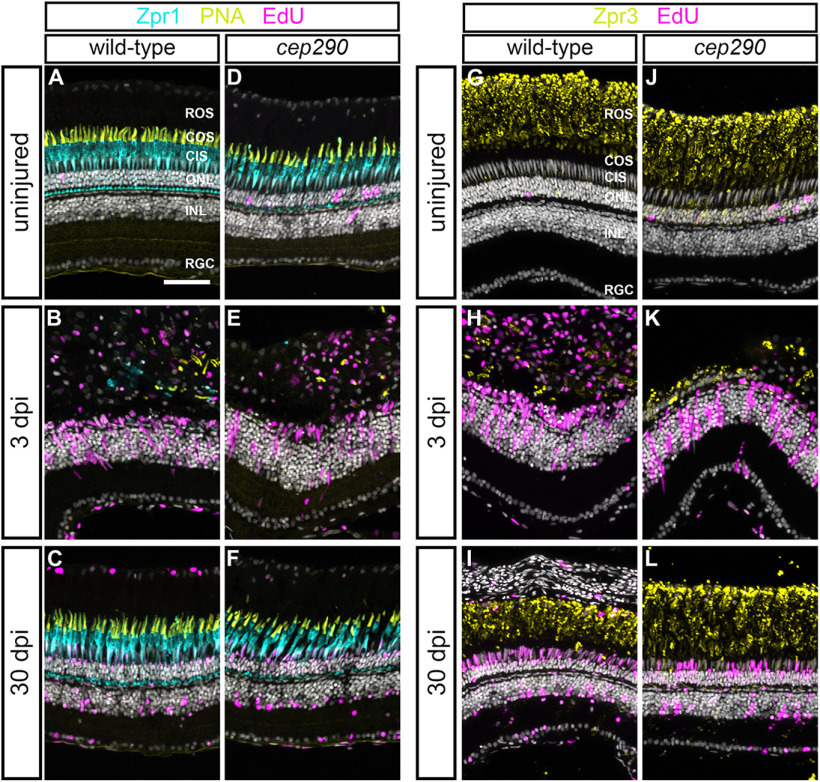
To determine whether regeneration by MG was possible in cep290 mutants, we used high-intensity light to ablate photoreceptors (Vihtelic and Hyde, 2000; Thomas and Thummel, 2013). Following light-induced photoreceptor cell death, MG dedifferentiate, re-enter the cell cycle, and produce neuronal progenitors that migrate to the ONL and differentiate to replace lost photoreceptors (Vihtelic and Hyde, 2000; Bernardos et al., 2007; Thomas and Thummel, 2013). To track proliferation of MG, the transgenic reporter line Tg(gfap:eGFP)mi2002 (Bernardos and Raymond, 2006) was crossed into the cep290 background. At 48 h postinjury (hpi), animals were injected with EdU. In both wild-type and cep290 mutant retinas at 72 hpi, EdU+ cells were observed in the INL and ONL (Fig. 6A,B). Within the INL, the EdU signal colocalized with GFP fluorescence from the Tg(gfap:eGFP) transgene, establishing the proliferating cells as MG. Importantly, there was no difference in the number of EdU+ cells in either the INL or ONL in light-damaged retinas from wild-type or cep290 mutants (Fig. 6C). We next asked whether MG-derived progenitors in cep290 mutants could regenerate photoreceptors following light damage. Photoreceptor degeneration and regeneration were assessed using Zpr1 and PNA, or Zpr3. At 3 dpi, cones were ablated in light damaged wild-type and cep290 mutants, and proliferation of neural precursors was observed in the INL (Fig. 7A,B,D,E). By 30 dpi, cones had regenerated in both wild-type and cep290 mutants (Fig. 7C,F). Similarly, rod photoreceptors degenerated by 3 dpi but had regenerated by 30 dpi in both wild-type and cep290 mutants (Fig. 7G–L).
Figure 6.
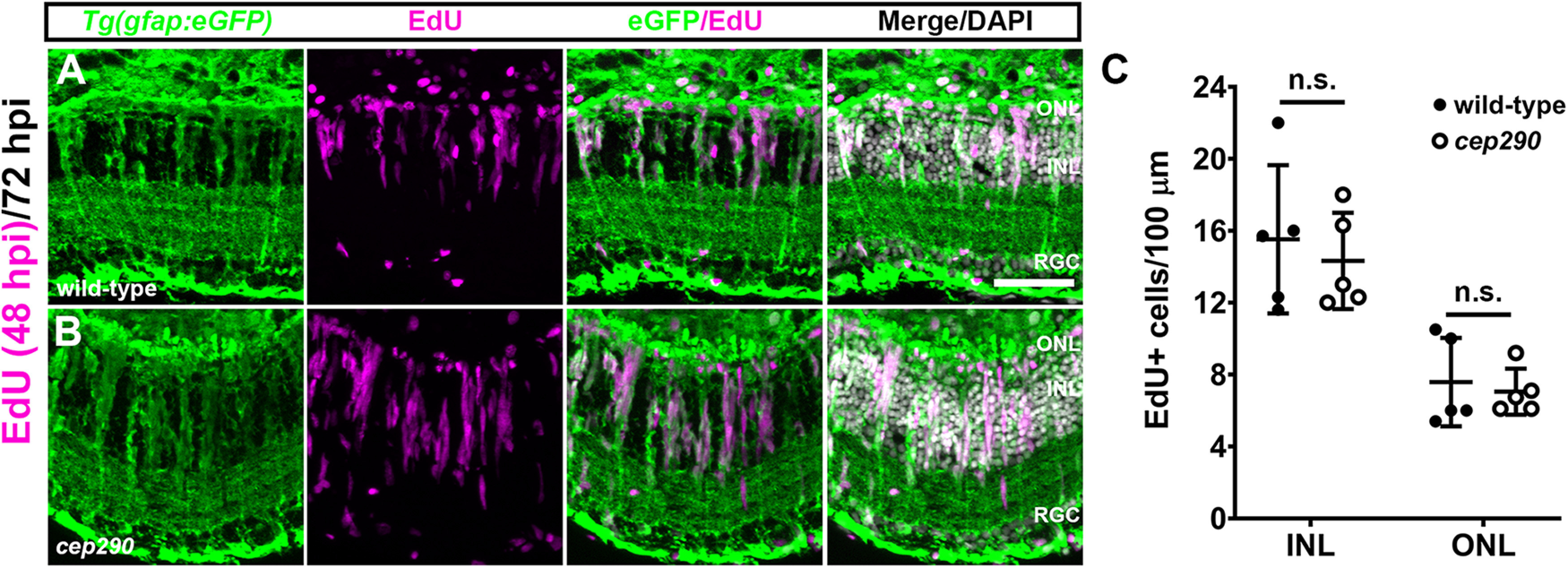
MG in cep290 mutants respond to acute light damage. Retinal cryosections of 6 mpf wild-type (A) or cep290 mutants (B) carrying the Tg(gfap:eGFP)mi2002 transgenic reporter line were immunolabeled with anti-GFP (green) antibodies to visualize MG and processed for EdU labeling (magenta) to identify MG-derived progenitor cells at 72 hpi. C, The number of EdU+ cells was not statistically different (n.s.) between wild-type and cep290 mutants (INL: p > 0.99; ONL: p > 0.66; Mann–Whitney tests). Data are plotted as mean ± SD ONL = outer nuclear layer; INL = inner nuclear layer, RGC = retinal ganglion cell layer. Scale bar: 50 µm.
Figure 7.
Photoreceptors regenerate in 6 mpf cep290 mutants following acute light damage. A–F, Views of cryosections of the dorsal retina of 6 mpf wild-type (A–C) or cep290 mutants (D–F) immunolabeled with the red/green cone photoreceptor marker, Zpr1 (cyan), PNA (yellow) to visualize cone outer segments, and EdU (magenta) to detect cells that had undergone proliferation in undamaged retinas or at 3 or 30 dpi. G–L, Immunolabeling of retinas from 6 mpf wild-type (G–I) or cep290 mutants (J–L) with the rod marker Zpr3 (yellow) and EdU (magenta) in undamaged retinas or at 3 or 30 dpi. ROS = rod outer segments; COS = cone outer segments; CIS = cone inner segments; ONL = outer nuclear layer; INL = inner nuclear layer, RGC = retinal ganglion cell layer. Scale bar: 50 µm.
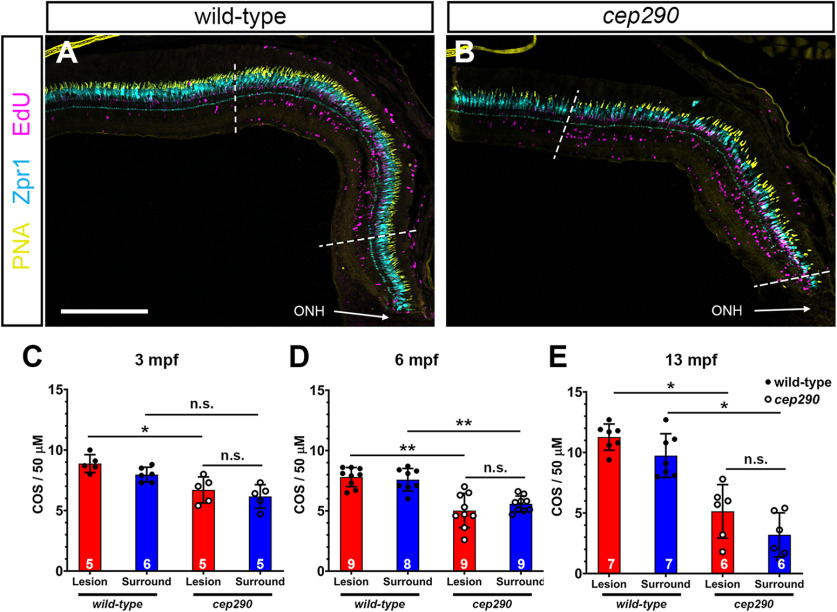
Following light damage, regeneration can restore the cone density of wild-type retinas to predamage levels (Vihtelic et al., 2006). Acute light damage triggers photoreceptor death in the central region of the dorsal retina, with the dorsal periphery and ventral retina areas largely spared from cell death and serving as an internal measure of prelesion cone densities (Vihtelic et al., 2006). EdU incorporation during proliferation of neural progenitors marked the location of light damage in wild-type and cep290 mutants, while few EdU+ cells were observed in the undamaged periphery (Fig. 8A,B). To determine whether the density of regenerated cones in cep290 mutant retinas was similar to that found in wild-type densities, we quantified cones in the damaged (“lesion”) and undamaged areas (“surround”) after one month of recovery in 3, 6, and 13 mpf fish. Cone density in the undamaged cep290 retinas was consistently less than wild-type at 6 and 13 mpf (Fig. 8C–E, blue bars). In both wild-type and cep290 mutants regeneration restored cone density only to predamage levels (Fig. 8C–E, compare blue and red bars). That is, the total number of regenerated cones in cep290 mutants remained lower than age-matched wild-type fish. These results indicate that zebrafish restore cones only to prelesion densities.
Figure 8.
Cone regeneration is limited by prelesion cone density in cep290 mutants. A, B, Views of the dorsal retina in cryosections of 6 mpf wild-type and cep290 mutant retinas at 30 dpi following light damage. Cryosections were immunolabeled with the red/green cone photoreceptor marker, Zpr1 (cyan), PNA (yellow) to visualize cone outer segments, and EdU (magenta) to detect cells that had undergone proliferation. Dotted lines indicate the regenerated area (i.e., lesion) based on highest density of EdU+ cells. C–E, Quantification of cone outer segment density in the lesioned (red bars) and surrounding areas (blue bars) of wild-type (filled circles) and cep290 retinas (open circles) at 30 dpi in animals aged 3, 6, and 13 mpf. Each data point represents quantification from an individual animal. Data are provided as mean ± SD (3 mpf: *p < 0.05, Kruskal–Wallis test with Dunn's multiple comparisons test; 6 mpf: **p < 0.005, Welch ANOVA test with Dunnett's T3 multiple comparisons test; 13 mpf: *p < 0.05, Kruskal–Wallis test with Dunn's multiple comparisons test; n.s. = not significant). ONH = optic nerve head. Scale bar: 200 µm.
Pharmacological modulation of glucocorticoid signaling impairs regeneration
Inflammation and microglia/macrophages are considered essential during the early proliferation phase of retinal regeneration in zebrafish (White et al., 2017; Silva et al., 2020; Nagashima and Hitchcock, 2021). Recent reports found that pharmacological ablation of microglia/macrophages before injury inhibited proliferation of MG-derived progenitors (White et al., 2017; Conedera et al., 2019). Dexamethasone, a synthetic corticosteroid and agonist of the GR, is a potent anti-inflammatory agent. Pretreatment with dexamethasone also inhibits MG proliferation following injury (White et al., 2017; Silva et al., 2020). Conversely, blocking endogenous GR activity with the antagonist RU486 after NMDA-induced retinal degeneration increased proliferation MG-derived progenitor cells in the chick retina (Gallina et al., 2014). Given these observations, we tested what effect treating cep290 mutants with RU486 or dexamethasone had on microglia/macrophage reactivity, and the proliferation of Müller-glia derived progenitors or ONL rod precursors, respectively. As noted previously, the cep290 mutants exhibit significantly more 4C4+ microglia/macrophages in the subretinal space compared with wild-type siblings (Fig. 9A,D; quantified in Fig. 9G). In both wild-type and cep290 mutants, exposure to dexamethasone significantly reduced the number of 4C4+ microglia/macrophages in the subretinal space and in the inner retina as compared with control animals exposed to vehicle (Fig. 9A–H). Exposure to RU486 had no effect on the number of microglia/macrophage in either the subretinal space or the inner retina (Fig. 9G,H), consistent with the findings that RU486 had no effect on microglia/macrophage reactivity in chick (Gallina et al., 2014). Next, retinas were stained with PCNA to determine the effects of immunosuppression on cell proliferation. PCNA was chosen because intraperitoneal injections of EdU increased mortality in animals treated by dexamethasone, presumably because of immunosuppression. Dexamethasone treatment significantly reduced the number of proliferating cells in the ONL of cep290 mutants (Fig. 9L–O). Although dexamethasone appeared to reduce the number of PCNA+ cells in the ONL of wild-type retinas, the results were not statistically significant, as few PCNA+ cells are found in undamaged wild-type retinas. As expected, treatment with RU486 had no effect on the number of PCNA+ cells in wild-type retinas. However, RU486 also had no effect on the number of PCNA+ cells found in the ONL or INL of cep290 mutants (Figs. 9P,O). This is in contrast to previous findings in chick where RU486 treatment increased the number of proliferating MG-derived progenitors nearly 6-fold (Gallina et al., 2014). These results suggest that immunosuppression inhibits proliferation of rod precursors in cep290 mutants but that inhibition of the GR, i.e., blocking endogenous anti-inflammatory activity, is insufficient to stimulate proliferation of either MG-derived retinal progenitors or rod precursors in cep290 mutants.
Figure 9.
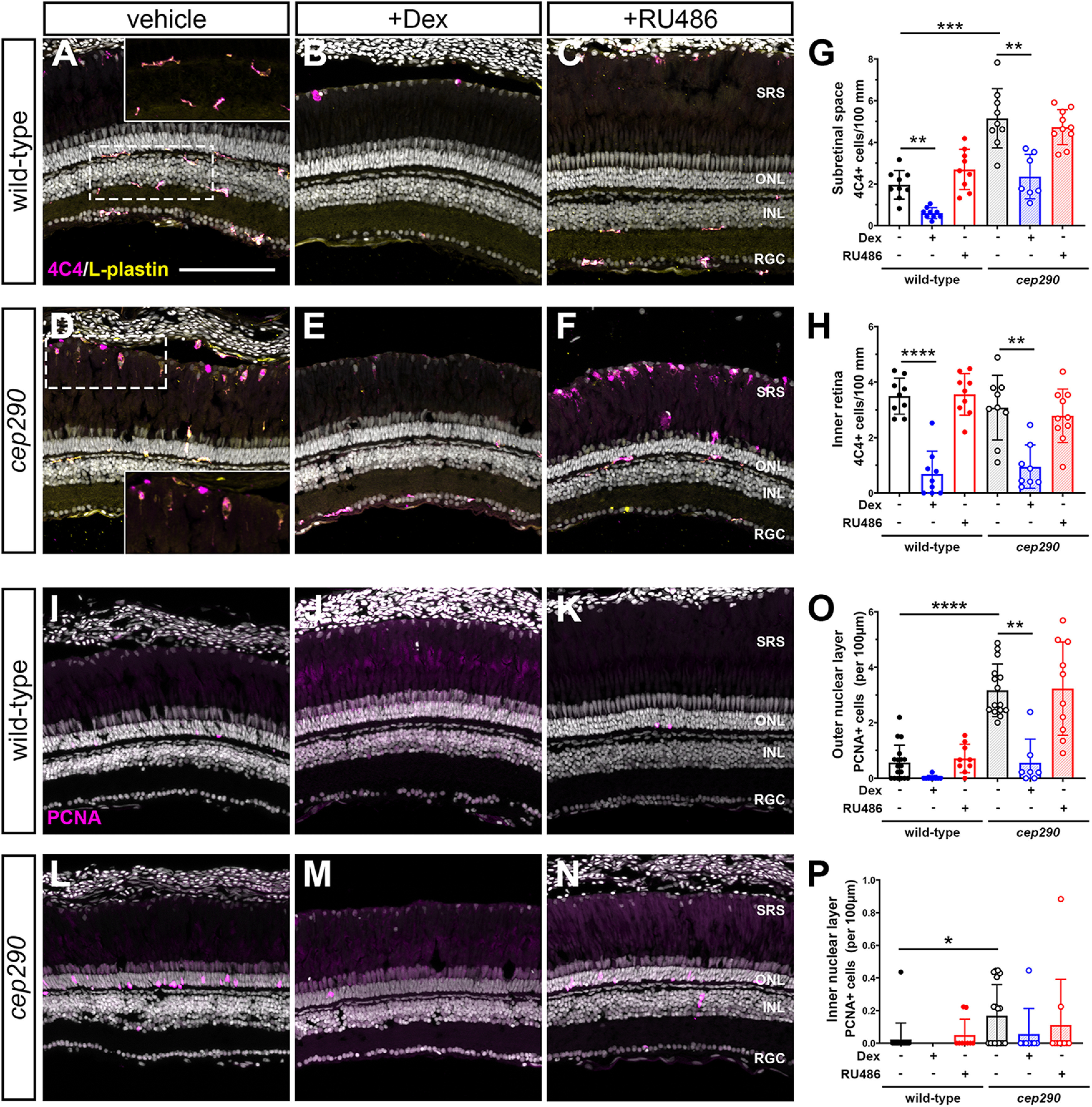
Immunosuppression with dexamethasone inhibits proliferation and microglia/macrophage activation in cep290 mutants. A–F, Immunohistochemistry of the dorsal retina with markers for microglia/macrophage (4C4, magenta) and leukocytes (L-plastin, yellow) in 6 mpf wild-type and cep290 mutant animals treated with methanol (vehicle/left column), dexamethasone (middle column) or RU486 (right column). Insets in A, D, Higher magnification of boxed regions illustrates the ramified morphology of quiescent microglia/macrophage in the plexiform layers of wild-type retinas and the elongated and amoeboid shape of microglia/macrophage in the subretinal space of cep290 mutants. G, H, Quantification of 4C4+ cells in the subretinal space and inner retina, respectively, of wild-type (filled circles/open bars) and cep290 mutants (open circles/hashed bars). I–N, Immunohistochemistry of the dorsal retina with PCNA (magenta) in 6 mpf wild-type and cep290 mutant animals. O, P, Quantification of PCNA+ cells in the ONL and INL, respectively, of wild-type (filled circles/open bars) and cep290 mutants (open circles/hashed bars). Counts following dexamethasone treatment are plotted in blue, whereas counts following RU486 treatment are plotted in red. Each data point represents counts from the dorsal retina of one eye. For graphs in G, H, the significance in differences was determined using Welch ANOVA test with Dunnett's T3 multiple comparisons test (**p < 0.01, ***p < 0.001, ****p < 0.0001). For graphs in O, P, the significance in differences was determined by Kruskal–Wallis test with Dunn's multiple comparisons test (*p < 0.05, **p < 0.01, ****p < 0.0001). SRS = subretinal space; ONL = outer nuclear layer; INL = inner nuclear layer, RGC = retinal ganglion cell layer. Scale bar: 100 µm.
Chronic immunosuppression rescues cone photoreceptors
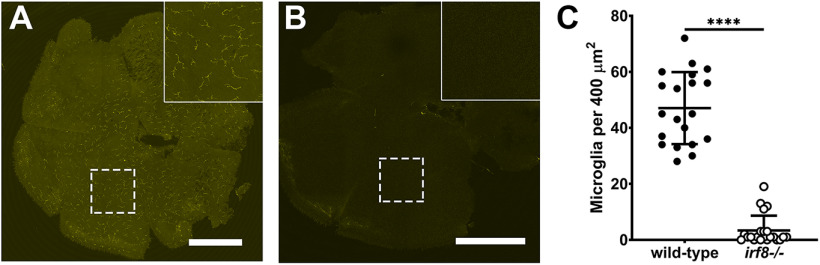
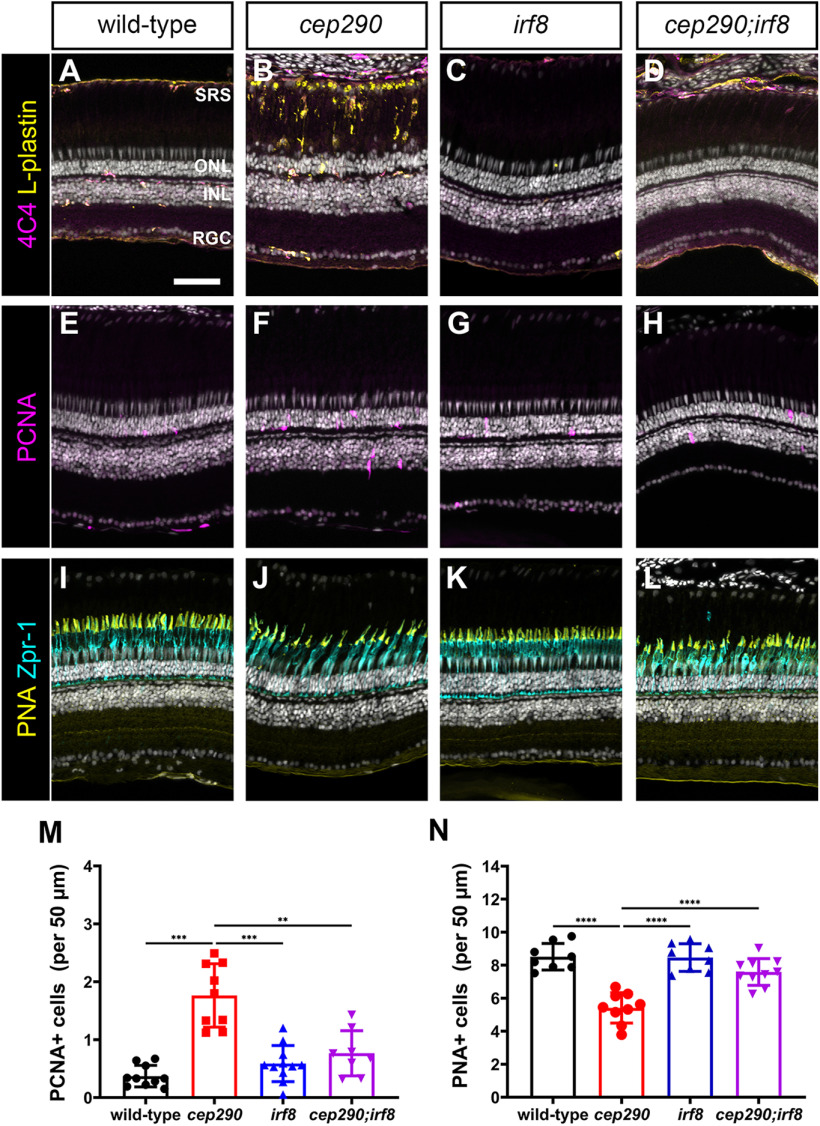
We next asked whether sustained immunosuppression could improve cone survival. While the immune response appears critical for regeneration, chronic inflammation also contributes to ongoing neuronal degeneration. When dexamethasone was provided after injury, cone survival was enhanced in mmp9−/− zebrafish mutants (Silva et al., 2020). Furthermore, genetic ablation of microglia in the rd10 mouse improved long-term survival of photoreceptors (L. Zhao et al., 2015). We hypothesized that although sustained immunosuppression may inhibit rod cell regeneration, it may prolong cone photoreceptor survival in cep290 mutants. To test this, we used the zebrafish irf8 mutant line (Shiau et al., 2015). The transcription factor interferon regulatory factor 8 (IRF8) is essential for the development of all embryonic macrophages and microglia in zebrafish (Shiau et al., 2015). Adult irf8 mutants are viable and lack most resident microglia in the CNS (Earley et al., 2018). Quantification of microglia/macrophage in flat-mounted retinas found a 92.8% reduction in microglia/macrophage density in irf8 mutants compared with wild-type siblings (Fig. 10A–C). We next generated cep290;irf8 double mutants and compared the phenotypes to cep290 and irf8 single mutants at 6 mpf. Whereas ramified microglia/macrophages were observed in the plexiform layers of wild-type animals (Fig. 11A) and reactive microglia/macrophages adopted a more amoeboid shape and accumulated in the subretinal space of cep290 mutants (Fig. 11B), there was a striking absence of microglia/macrophages the retinas of either irf8 mutants or the cep290;irf8– mutants (Fig. 11C,D). To assess proliferation, the retinas of 6 mpf animals were examined by immunohistochemistry with PCNA. While proliferation of rod precursors in the ONL was minimal in wild-type animals (Fig. 11E) and significantly greater in cep290 mutants (Fig. 11F), few proliferating cells were observed in the irf8 mutants (Fig. 11G). Consistent with data showing that immunosuppression impaired regeneration, the number of proliferating ONL cells in cep290;irf8 mutants significantly lower than in cep290 mutants but not statistically different from wild-type or irf8 mutants (Figs. 11M, 11E–H). Compared with wild-type siblings, the number of cones was reduced in cep290 mutants but not irf8 mutants, which indicated that loss of irf8 does not impair cone survival (Fig. 11I–K,N). When cone density was quantified in cep290;irf8 mutants, the number of cones was significantly greater than cep290 mutants but not statistically different from wild-type siblings (Figs. 11L,N). These results indicate that immunosuppression can prolong cone survival in cep290 mutants. The lack of proliferation in the INL or ONL of the cep290;irf8 mutants also indicates that the increased number of cones is most likely because of cone preservation and not enhanced regeneration. Taken together, these data indicate that inflammation resulting from cell death stimulates ONL cell proliferation in cep290 mutants to maintain rod cell numbers, but that the absence of microglia/macrophage promotes cone survival in cep290 mutants.
Figure 10.
Mutation of irf8 reduces the number of microglia/macrophages. A, B, Confocal microscopy of flat-mounted retinas from wild-type and irf8 mutant adults (6 mpf) immunostained with 4C4 (yellow) to label microglia/macrophage. Inset, Higher magnification of boxed region illustrates the ramified morphology of quiescent microglia/macrophage in wild-type retinas and the significant reduction of microglia/macrophage in irf8 mutants. C, Quantification of 4C4+ cell density in wild-type (n = 20, filled circles) and cep290 mutants (n = 22, open circles) found a significant decrease in microglia/macrophage. Each data point represents quantification from one complete 400 × 400 μm region of retina with five retinas per genotype analyzed. irf8−/− = 3.36 ± 1.13 (n = 22); wild-type = 47.05 ± 2.88 (n = 20); mean ± SEM. Statistical significance was determined by Mann–Whitney test (****p < 0.0001).
Figure 11.
Mutation of irf8 reduces the number of activated microglia/macrophage and proliferating cells and promotes cone survival in cep290 mutants. A–D, Immunohistochemistry of the dorsal retina with markers for microglia/macrophage (4C4, magenta) and leukocytes (L-plastin, yellow) in 6 mpf wild-type, cep290 mutants, irf8 mutants, and cep290;irf8 mutants. E–H, Immunohistochemistry of the dorsal retina at 3 d postinjection with PCNA (magenta) to mark proliferating cells in 6 mpf wild-type, cep290 mutants, irf8 mutants, and cep290;irf8 mutants. I–L, Immunohistochemistry of the dorsal retina with markers for cone inner segments (Zpr-1, cyan) and cone outer segments (PNA, yellow) in 6 mpf wild-type, cep290 mutants, irf8 mutants, and cep290;irf8 mutants. M, N, Quantification of PCNA+ cells in the ONL and PNA+ outer segments in wild-type (filled black circles) and cep290 mutants (filled red circle), irf8 mutants (filled blue triangles), and cep290;irf8 mutants (filled purple triangles). Each data point represents counts from the dorsal retina of one eye. For the graph in M, the significance in differences was determined using Welch ANOVA test with Dunnett's T3 multiple comparisons test (**p < 0.01, ***p < 0.001). For the graph in N, the significance in differences was determined by one-way ANOVA with Tukey's multiple comparisons test (***p < 0.001, ****p < 0.0001). SRS = subretinal space; ONL = outer nuclear layer; INL = inner nuclear layer, RGC = retinal ganglion cell layer. Scale bars: 400 µm (A, B) and 50 µm (D–O).
Notch inhibition induces MG to re-enter the cell cycle in cep290 mutants
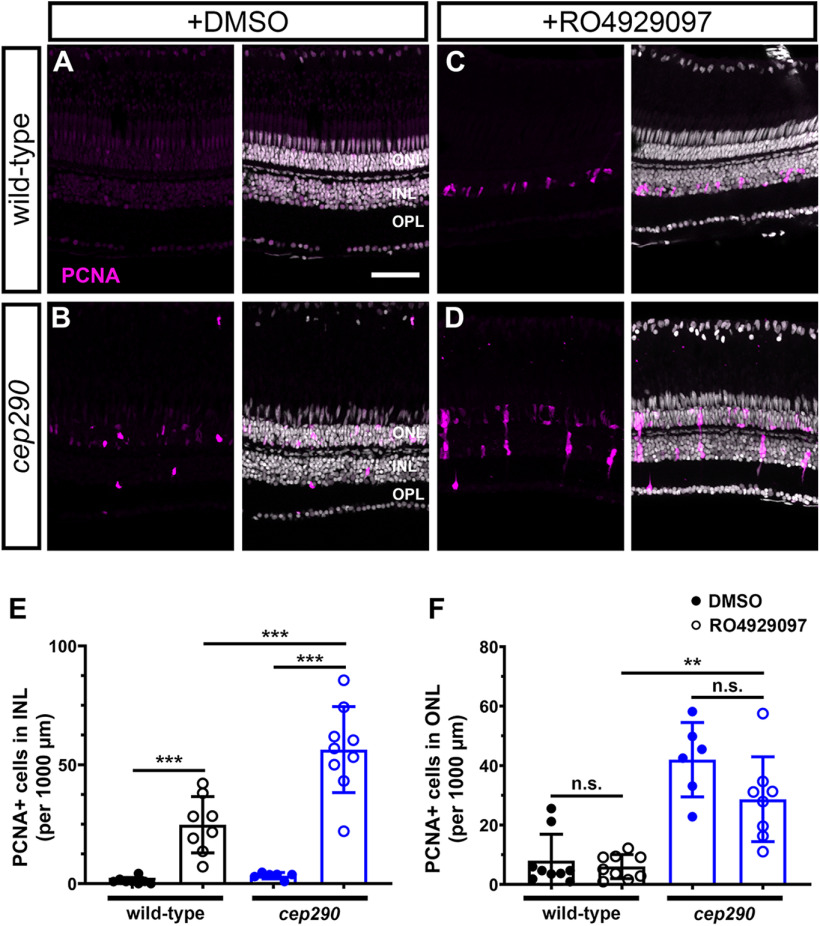
In zebrafish, Notch signaling suppresses regeneration by maintaining MG in a quiescent state (Conner et al., 2014; Wan and Goldman, 2017; Lee et al., 2020) and proliferation requires the downregulation of notch3 expression in MG (Campbell and Hyde, 2017; Campbell et al., 2021). As more MG in cep290 mutants maintain expression notch3 (Fig. 3G), we hypothesized that persistent Notch signaling maintained MG quiescence in cep290 mutants despite chronic degeneration. In DMSO-control retinas, very few PCNA+ cells were observed in wild-type animals (Fig. 12A), while PCNA+ cells were limited primarily to the ONL in cep290 mutants (Fig. 12B). Following RO4929097 treatment, a significant increase in PCNA+ cells were observed in the INL of both wild-type and cep290 mutant retinas (Fig. 12C,D). Importantly, Notch inhibition resulted in a significantly greater number of PCNA+ cells in the INL of cep290 mutants than in wild-type retinas (Fig. 12E), roughly matching the 2-fold increase in the number of notch3-expressing MG observed in cep290 mutants by scRNA-seq (Fig. 3G). The PCNA+ cells were often observed in clusters within the INL of cep290 mutants rather than the single PCNA+ cells observed in wild-type retinas, consistent with proliferation of MG and closely associated MG-derived progenitor cells. This is similar to the synergistic stimulation of proliferation seen following co-injection of RO4929097 and TNFα in the uninjured retina (Conner et al., 2014). Interestingly, Notch suppression had no effect on the proliferation of rod precursors in the ONL (Fig. 12F). When combined with our transcriptomic data, these results suggest that the regenerative response of MG in cep290 mutants is suppressed by Notch signaling but that repressing Notch signaling in the chronic degenerative/inflammatory environment of the cep290 retina is sufficient to induce MG proliferation.
Figure 12.
Inhibition of Notch signaling by injection of RO4929097 induces more robust MG proliferation in 6 mpf cep290 mutant retinas than in wild-type retinas. Wild-type (top row) and cep290 mutant fish (bottom row) were injected intraperitoneally with 1% DMSO or 1 mm RO4929097 for four consecutive days. A, B, Immunofluorescent images of dorsal retinas from DMSO injected fish or (C, D) RO4929097-injected fish stained with PCNA (magenta). E, Quantification of PCNA+ cells in the INL or (F) ONL of DMSO-treated wild-type (filled black circles) and cep290 mutants (filled blue circles) or RO4929097-treated wild-type (open black circles) and cep290 mutants (open blue circles). Each data point represents counts from the dorsal retina of one eye and graphs represent mean ± SD. Significance in differences were determined using Mann–Whitney tests except for comparisons between RO4929097-treated wild-type to cep290 mutants, which used t test with Welch's correction (**p < 0.005, ***p < 0.001; n.s. = not significant). ONL = outer nuclear layer; INL = inner nuclear layer, OPL = outer plexiform layer. Scale bar: 50 µm.
Discussion
Strategies that aim to stimulate endogenous repair in the retina are an attractive therapeutic approach for progressive retinal degenerative diseases. Identifying both the extrinsic cues and the intrinsic molecular mechanisms necessary for MG reprogramming and proliferation, and which regulate the differentiation of MG-derived retinal progenitors, remain central to achieving these goals. Much of the foundational knowledge on retinal regeneration has come from studying how zebrafish respond to acute damage (Iribarne, 2021; Nagashima and Hitchcock, 2021). Interestingly, several zebrafish mutants exhibiting progressive photoreceptor degeneration do not initiate a robust regenerative response to chronic cell loss (Lessieur et al., 2019; Song et al., 2020; Noel et al., 2021). Here, characterization of the zebrafish cep290 mutant, which undergoes progressive photoreceptor degeneration, has revealed that the molecular pathways required for Müller cell reprogramming remain intact in the diseased retina and helped refine the views of inflammation in driving regeneration. Specifically, we report that slow, progressive photoreceptor degeneration results in a sustained immune response exemplified by reactive microglia/macrophages and upregulation of inflammation-related genes in cep290 mutants. Further, we show that immune suppression protects cone cells from degeneration, and that sustained expression of Notch signaling and quiescence-associated genes in MG prevents the reentry into the cell cycle even in the presence of chronic inflammatory signals. This study is the first to show that in response to chronic degeneration, zebrafish MG adopt a novel state characterized by the simultaneous expression of genes associated with quiescence and reactivity.
Like the response to photoreceptor degeneration in mammals, cep290 mutant retinas showed evidence of microglia/macrophage activation and increased inflammation, as well as an absence of a MG-based regenerative response. The immune cells accumulated in the subretinal space and the number of immune cells in the INL was reduced, similar to what has been observed in mouse models of RP (L. Zhao et al., 2015; O'Koren et al., 2019). By using RNA-seq to explore whole-retina gene expression, we noted the upregulation of several pathways associated with inflammation, including interferon-α, interferon-γ, and Jak/Stat3 signaling. We also noted the upregulation of the recruitment factors for neutrophils (cxcl8, cxcl18a.1, cxcl18b) and macrophages (il34), all of which were also upregulated in a zebrafish model of RPE damage (Leach et al., 2021). These data suggest that a similar inflammatory response exists to both chronic degeneration and acute damage within the zebrafish retina. Given the observed immune response, it is surprising that MG did not proliferate in cep290 mutants. Several studies have documented a strong correlation between inflammation and MG proliferation following retinal cell death (White et al., 2017; Mitchell et al., 2018, 2019; Conedera et al., 2019; Silva et al., 2020; Z. Zhang et al., 2020). In fact, injection of the inflammatory cytokine TNFα into undamaged retinas is sufficient to induce proliferation (Conner et al., 2014). In zebrafish lacking matrix metalloproteinase-9 (mmp-9), TNFα expression was elevated over wild-type in both undamaged and damaged retinas and acute light injury triggered an overproduction of MG-derived progenitors in mmp-9 mutants (Silva et al., 2020). Dexamethasone, a potent anti-inflammatory agent, suppresses MG proliferation following injury (White et al., 2017; Silva et al., 2020). Ablation of microglia by the drug PLX3397 also reduces proliferation of MG following laser injury (Conedera et al., 2019). Our results indicate, however, that inflammation alone was insufficient to induce MG proliferation in a model of progressive photoreceptor degeneration. The loss of cep290 function was likely not a factor as photoreceptor death also fails to induce MG proliferation in the bbs2 mutant (Song et al., 2020) or the arylhydrocarbon interaction protein like 1b (aipl1b) mutant (Iribarne et al., 2019). Furthermore, acute light damage triggered MG proliferation in both cep290 and bbs2 mutants, indicating that both mutants could respond to acute damage. The results from scRNA-seq analysis suggest that sustained expression of notch3 in MG of cep290 mutants inhibits proliferation even in a chronic inflammatory environment. MG maintain an “injury-response threshold” to limit regeneration and this threshold is controlled by Notch signaling (Sahu et al., 2021). Several studies have confirmed that Notch signaling maintains MG quiescence (Conner et al., 2014; Wan and Goldman, 2017; Lee et al., 2020; Campbell et al., 2021). Following acute injury, expression of notch3 is rapidly downregulated to permit expression of regeneration-associated genes (Campbell et al., 2021). Notch3 reportedly signals through the transcriptional repressor Hey1 to limit chromatin accessibility and establish the injury threshold. Morpholino knock-down of either notch3 or hey1 significantly increases MG proliferation following injury (Campbell et al., 2021; Sahu et al., 2021). The link between inflammation and Notch inhibition remains unclear. TNFα expression and RO4929097-mediated Notch suppression act synergistically to induce MG proliferation but it is not known whether these factors are mechanistically inked or functionally independently. Why MG in cep290 mutants maintain Notch3 expression despite inflammatory signals remains unknown. Additional studies are also needed to identify the factors that inhibit Notch following acute injury.
Our results also show that proliferation of rod precursors depends on inflammation but not Notch signaling. The rod precursors are derived from MG and are maintained as a population of slowly dividing unipotent stem cells in the ONL that proliferate rapidly in response to rod death (Morris et al., 2005; Stenkamp, 2011). Our data show that the proliferating cells in the ONL of cep290 mutants differentiate exclusively into rods, confirming them as rod precursors. We demonstrated that immunosuppression with dexamethasone or with an irf8 mutant impaired rod precursor proliferation, indicating that rod precursors respond to inflammatory signals. As cone survival was increased in cep290;irf8 mutants, it is possible that rod degeneration was also mitigated. Thus, the reduced proliferation of rod precursors likely reflected the decrease in rod cell death, which would also reduce inflammation. Interestingly, injection of RO4929097 did not impact rod precursor proliferation. A previous report found that morpholino-induced knock-down of notch3 did not increase the number of PCNA+ cells within the ONL at 96 h following light damage (Campbell et al., 2021). Taken together, these results suggest that Notch signaling does not influence the behavior of rod precursors.
Inflammatory signals enhance the regenerative response to retinal damage in zebrafish (Nelson et al., 2013; Iribarne et al., 2019; Silva et al., 2020) but studies also show that inflammation potentiates photoreceptor degeneration in mouse models of retinitis pigmentosa (L. Zhao et al., 2015; Silverman and Wong, 2018) and inflammatory signals from microglia limit regeneration in mammals (Todd et al., 2020). As immunomodulation has been posited as a potential therapy for retinitis pigmentosa (Silverman and Wong, 2018; Silverman et al., 2019), we investigated the effects of chronic immunosuppression on photoreceptor regeneration and survival. When retinal microglia/macrophages were depleted in cep290;irf8 double mutants, cone photoreceptors were protected from degeneration. Although rod precursor proliferation was reduced in cep290;irf8 double mutants, we found no evidence of accelerated rod degeneration, suggesting that rods were also spared. This was unexpected as loss of Cep290 leads to cell-autonomous photoreceptor death by destabilizing the architecture and function of the connecting cilium (Rachel et al., 2012). Lowering inflammation may mitigate the cellular stress associated with trafficking defects and slow photoreceptor death. Genetic and pharmacological inhibition of microglia function was reported to improve photoreceptor survival in mouse IRD models (C. Zhang et al., 2004; Peng et al., 2014; L. Zhao et al., 2015; Zabel et al., 2016; Wang et al., 2017). Cone survival following regeneration also requires resolution of the inflammatory response, suggesting that prolonged inflammation exacerbates photoreceptor degeneration (Silva et al., 2020). It is reasonable to speculate that prolonged inflammation may partially explain why cones only regenerate to prelesion densities following light damage in cep290 and bbs2 models. Perhaps the inability to resolve higher levels of inflammation limits cone survival following light damage. Alternatively, chronic inflammation could decrease survival of MG-derived progenitors.
The goal of studying zebrafish models of retinal degeneration is to determine how the mechanisms that promote regeneration function in different contexts. In zebrafish, both acute injury and progressive degeneration activate microglia/macrophages and increase inflammation (White et al., 2017; Mitchell et al., 2018, 2019; Song et al., 2020). In contrast, microglia activation appears to inhibit MG-associated regeneration following acute injury in mice (Todd et al., 2020). The role of Notch is also complex and context-dependent. The Notch pathway remains active in MG following damage in the mouse (Hoang et al., 2020) and in zebrafish with progressive degeneration (this study), but Notch is downregulated in zebrafish MG following widespread acute injuries (Hoang et al., 2020; Campbell et al., 2021). Furthermore, Notch inhibition combined with overexpression of the key reprogramming factors Ascl1a and Lin28 did not exert the same proregenerative response in mouse MG as was observed following similar expression studies in zebrafish (Elsaeidi et al., 2018). Developing strategies for successful regeneration of photoreceptors in diseases like retinitis pigmentosa or age-related macular degeneration will require a better understanding of how inflammation and Notch signaling function in both injury and disease states.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants R01-EY017037 and R01-EY030574 (to B.D.P.) and U01-EY027267 (to D.R.H. and S.B.), a Knights Templar Eye Foundation Career Initiation Grant (J.F.), and a Doris and Jules Stein Professorship Award from Research to Prevent Blindness (B.D.P.). Additional support was provided by the NIH P30 Core Grant P30-EY025585, a Foundation Fighting Blindness (FFB) Center Grant, and an Unrestricted Award from Research to Prevent Blindness to the Cole Eye Institute. The LRI Genomics Core is supported by the NIH P30 Core Grant P30-CA043703. We thank Dr. Peter Hitchcock, Dr. Daniel Goldman, Dr. Jim Fadool, Dr. Ryota Matsuoka, and Dr. William Talbot for providing fish lines and reagents. We also thank the animal care staff of the Biological Resources Unit at the LRI.
The authors declare no competing financial interests.
References
- Bernardos RL, Raymond PA (2006) GFAP transgenic zebrafish. Gene Expr Patterns 6:1007–1013. 10.1016/j.modgep.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA (2007) Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci 27:7028–7040. 10.1523/JNEUROSCI.1624-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LJ, Hyde DR (2017) Opportunities for CRISPR/Cas9 gene editing in retinal regeneration research. Front Cell Dev Biol 5:99. 10.3389/fcell.2017.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LJ, Hobgood JS, Jia M, Boyd P, Hipp RI, Hyde DR (2021) Notch3 and DeltaB maintain Müller glia quiescence and act as negative regulators of regeneration in the light-damaged zebrafish retina. Glia 69:546–566. 10.1002/glia.23912 [DOI] [PubMed] [Google Scholar]
- Conedera FM, Pousa AMQ, Mercader N, Tschopp M, Enzmann V (2019) Retinal microglia signaling affects Müller cell behavior in the zebrafish following laser injury induction. Glia 67:1150–1166. 10.1002/glia.23601 [DOI] [PubMed] [Google Scholar]
- Conner C, Ackerman KM, Lahne M, Hobgood JS, Hyde DR (2014) Repressing notch signaling and expressing TNFα are sufficient to mimic retinal regeneration by inducing Müller glial proliferation to generate committed progenitor cells. J Neurosci 34:14403–14419. 10.1523/JNEUROSCI.0498-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley AM, Graves CL, Shiau CE (2018) Critical role for a subset of intestinal macrophages in shaping gut microbiota in adult zebrafish. Cell Rep 25:424–436. 10.1016/j.celrep.2018.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaeidi F, Macpherson P, Mills EA, Jui J, Flannery JG, Goldman D (2018) Notch suppression collaborates with Ascl1 and Lin28 to unleash a regenerative response in fish retina, but not in mice. J Neurosci 38:2246–2261. 10.1523/JNEUROSCI.2126-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadool JM (2003) Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev Biol 258:277–290. 10.1016/s0012-1606(03)00125-8 [DOI] [PubMed] [Google Scholar]
- Fausett BV, Goldman D (2006) A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J Neurosci 26:6303–6313. 10.1523/JNEUROSCI.0332-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina D, Zelinka C, Fischer AJ (2014) Glucocorticoid receptors in the retina, Müller glia and the formation of Müller glia-derived progenitors. Development 141:3340–3351. 10.1242/dev.109835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D (2014) Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci 15:431–442. 10.1038/nrn3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsuch RA, Hyde DR (2014) Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exp Eye Res 123:131–140. 10.1016/j.exer.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C (1999) Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735–3745. 10.1242/dev.126.17.3735 [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA (1992) Retinal regeneration. Trends Neurosci 15:103–108. 10.1016/0166-2236(92)90020-9 [DOI] [PubMed] [Google Scholar]
- Hoang T, et al. (2020) Gene regulatory networks controlling vertebrate retinal regeneration. Science 370:eabb8598. 10.1126/science.abb8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde DR, Reh TA (2014) The past, present, and future of retinal regeneration. Exp Eye Res 123:105–106. 10.1016/j.exer.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Iribarne M (2021) Inflammation induces zebrafish regeneration. Neural Regen Res 16:1693–1701. 10.4103/1673-5374.306059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iribarne M, Hyde DR, Masai I (2019) TNFα induces Müller glia to transition from non-proliferative gliosis to a regenerative response in mutant zebrafish presenting chronic photoreceptor degeneration. Front Cell Dev Biol 7:296. 10.3389/fcell.2019.00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns PR (1977) Growth of the adult goldfish eye. III. Source of the new retinal cells. J Comp Neurol 176:343–357. 10.1002/cne.901760304 [DOI] [PubMed] [Google Scholar]
- Johns PR, Easter SS Jr (1977) Growth of the adult goldfish eye. II. Increase in retinal cell number. J Comp Neurol 176:331–341. 10.1002/cne.901760303 [DOI] [PubMed] [Google Scholar]
- Karlen SJ, Miller EB, Burns ME (2020) Microglia activation and inflammation during the death of mammalian photoreceptors. Annu Rev Vis Sci 6:149–169. 10.1146/annurev-vision-121219-081730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, T Burket C, Liu C-G, Vihtelic TS, Hyde DR (2007) Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol 67:1009–1031. 10.1002/dneu.20362 [DOI] [PubMed] [Google Scholar]
- Kyritsis N, Kizil C, Zocher S, Kroehne V, Kaslin J, Freudenreich D, Iltzsche A, Brand M (2012) Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 338:1353–1356. 10.1126/science.1228773 [DOI] [PubMed] [Google Scholar]
- Leach LL, Hanovice NJ, George SM, Gabriel AE, Gross JM (2021) The immune response is a critical regulator of zebrafish retinal pigment epithelium regeneration. Proc Natl Acad Sci U S A 118:e2017198118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Wan J, Goldman D (2020) Tgfb3 collaborates with PP2A and notch signaling pathways to inhibit retina regeneration. Elife 9:e55137. 10.7554/eLife.55137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkowski JR, Raymond PA (2014) Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res 40:94–123. 10.1016/j.preteyeres.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessieur EM, Song P, Nivar GC, Piccillo EM, Fogerty J, Rozic R, Perkins BD (2019) Ciliary genes arl13b, ahi1 and cc2d2a differentially modify expression of visual acuity phenotypes but do not enhance retinal degeneration due to mutation of cep290 in zebrafish. PLoS One 14:e0213960. 10.1371/journal.pone.0213960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dowling JE (1997) A dominant form of inherited retinal degeneration caused by a non-photoreceptor cell-specific mutation. Proc Natl Acad Sci U S A 94:11645–11650. 10.1073/pnas.94.21.11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chen J, Yu S, Raghupathy RK, Liu X, Qin Y, Li C, Huang M, Liao S, Wang J, Zou J, Shu X, Tang Z, Liu M (2015) Knockout of RP2 decreases GRK1 and rod transducin subunits and leads to photoreceptor degeneration in zebrafish. Hum Mol Genet 24:4648–4659. 10.1093/hmg/ddv197 [DOI] [PubMed] [Google Scholar]
- Lu Z, Hu X, Liu F, Soares DC, Liu X, Yu S, Gao M, Han S, Qin Y, Li C, Jiang T, Luo D, Guo AY, Tang Z, Liu M (2017) Ablation of EYS in zebrafish causes mislocalisation of outer segment proteins, F-actin disruption and cone-rod dystrophy. Sci Rep 7:46098. 10.1038/srep46098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini J, Le Clerc S, Morisse G, Coulonges C, Kuil LE, van Ham TJ, Zagury JF, Sieger D (2020) Gene expression profiling reveals a conserved microglia signature in larval zebrafish. Glia 68:298–315. 10.1002/glia.23717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DM, Lovel AG, Stenkamp DL (2018) Dynamic changes in microglial and macrophage characteristics during degeneration and regeneration of the zebrafish retina. J Neuroinflammation 15:163. 10.1186/s12974-018-1185-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DM, Sun C, Hunter SS, New DD, Stenkamp DL (2019) Regeneration associated transcriptional signature of retinal microglia and macrophages. Sci Rep 9:4768. 10.1038/s41598-019-41298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, Hyde DR (2010) A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J Comp Neurol 518:800–814. 10.1002/cne.22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Schroeter EH, Bilotta J, Wong RO, Fadool JM (2005) Cone survival despite rod degeneration in XOPS-mCFP transgenic zebrafish. Invest Ophthalmol Vis Sci 46:4762–4771. 10.1167/iovs.05-0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Forbes-Osborne MA, Pillai LS, Fadool JM (2011) Microarray analysis of XOPS-mCFP zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration. Invest Ophthalmol Vis Sci 52:2255–2266. 10.1167/iovs.10-6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, Hitchcock PF (2021) Inflammation regulates the multi-step process of retinal regeneration in zebrafish. Cells 10:783. 10.3390/cells10040783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, D'Cruz TS, Danku AE, Hesse D, Sifuentes C, Raymond PA, Hitchcock PF (2020) Midkine-a is required for cell cycle progression of Müller glia during neuronal regeneration in the vertebrate retina. J Neurosci 40:1232–1247. 10.1523/JNEUROSCI.1675-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR (2012) Stat3 defines three populations of Müller glia and is required for initiating maximal Müller glia proliferation in the regenerating zebrafish retina. J Comp Neurol 520:4294–4311. 10.1002/cne.23213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O'Hayer P, Bailey TJ, Gorsuch RA, Hyde DR (2013) Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Müller glia proliferation during zebrafish retinal regeneration. J Neurosci 33:6524–6539. 10.1523/JNEUROSCI.3838-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel NCL, MacDonald IM, Allison WT (2021) Zebrafish models of photoreceptor dysfunction and degeneration. Biomolecules 11:78. 10.3390/biom11010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Koren EG, Yu C, Klingeborn M, Wong AYW, Prigge CL, Mathew R, Kalnitsky J, Msallam RA, Silvin A, Kay JN, Bowes Rickman C, Arshavsky VY, Ginhoux F, Merad M, Saban DR (2019) Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity 50:723–737.e7. 10.1016/j.immuni.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF (2003) Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res 43:927–936. 10.1016/s0042-6989(02)00400-5 [DOI] [PubMed] [Google Scholar]
- Peng B, Xiao J, Wang K, So KF, Tipoe GL, Lin B (2014) Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci 34:8139–8150. 10.1523/JNEUROSCI.5200-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel RA, Li T, Swaroop A (2012) Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia 1:22. 10.1186/2046-2530-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D (2010) Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol 12:1101–1107. 10.1038/ncb2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D (2012) Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat Cell Biol 14:1013–1023. 10.1038/ncb2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Rivlin PK (1987) Germinal cells in the goldfish retina that produce rod photoreceptors. Dev Biol 122:120–138. 10.1016/0012-1606(87)90338-1 [DOI] [PubMed] [Google Scholar]
- Raymond PA, Reifler MJ, Rivlin PK (1988) Regeneration of goldfish retina: rod precursors are a likely source of regenerated cells. J Neurobiol 19:431–463. 10.1002/neu.480190504 [DOI] [PubMed] [Google Scholar]
- Sahu A, Devi S, Jui J, Goldman D (2021) Notch signaling via Hey1 and Id2b regulates Müller glia's regenerative response to retinal injury. Glia 69:2882–2898. 10.1002/glia.24075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut MC, Gulati-Leekha A, Goldman D (2004) An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J Neurosci 24:7663–7673. 10.1523/JNEUROSCI.2281-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau CE, Kaufman Z, Meireles AM, Talbot WS (2015) Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLoS One 10:e0117513. 10.1371/journal.pone.0117513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NJ, Nagashima M, Li J, Kakuk-Atkins L, Ashrafzadeh M, Hyde DR, Hitchcock PF (2020) Inflammation and matrix metalloproteinase 9 (Mmp-9) regulate photoreceptor regeneration in adult zebrafish. Glia 68:1445–1465. 10.1002/glia.23792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SM, Wong WT (2018) Microglia in the retina: roles in development, maturity, and disease. Annu Rev Vis Sci 4:45–77. 10.1146/annurev-vision-091517-034425 [DOI] [PubMed] [Google Scholar]
- Silverman SM, Ma W, Wang X, Zhao L, Wong WT (2019) C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa. J Exp Med 216:1925–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Fogerty J, Cianciolo LT, Stupay R, Perkins BD (2020) Cone photoreceptor degeneration and neuroinflammation in the zebrafish Bardet-Biedl syndrome 2 (BBS2) mutant does not lead to retinal regeneration. Front Cell Dev Biol 8:578528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL (2011) The rod photoreceptor lineage of teleost fish. Prog Retin Eye Res 30:395–404. 10.1016/j.preteyeres.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL, Satterfield R, Muhunthan K, Sherpa T, Vihtelic TS, Cameron DA (2008) Age-related cone abnormalities in zebrafish with genetic lesions in sonic hedgehog. Invest Ophthalmol Vis Sci 49:4631–4640. 10.1167/iovs.07-1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Thummel R (2013) A novel light damage paradigm for use in retinal regeneration studies in adult zebrafish. J Vis Exp (80):e51017. 10.3791/51017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR (2008) Characterization of Müller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res 87:433–444. 10.1016/j.exer.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd L, Finkbeiner C, Wong CK, Hooper MJ, Reh TA (2020) Microglia suppress Ascl1-induced retinal regeneration in mice. Cell Rep 33:108507. 10.1016/j.celrep.2020.108507 [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR (2000) Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol 44:289–307. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Soverly JE, Kassen SC, Hyde DR (2006) Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res 82:558–575. 10.1016/j.exer.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Wan J, Goldman D (2017) Opposing actions of Fgf8a on notch signaling distinguish two Müller glial cell populations that contribute to retina growth and regeneration. Cell Rep 19:849–862. 10.1016/j.celrep.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhao XF, Vojtek A, Goldman D (2014) Retinal injury, growth factors, and cytokines converge on β-catenin and pStat3 signaling to stimulate retina regeneration. Cell Rep 9:285–297. 10.1016/j.celrep.2014.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao L, Zhang Y, Ma W, Gonzalez SR, Fan J, Kretschmer F, Badea TC, Qian HH, Wong WT (2017) Tamoxifen provides structural and functional rescue in murine models of photoreceptor degeneration. J Neurosci 37:3294–3310. 10.1523/JNEUROSCI.2717-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DT, Mumm JS (2013) The nitroreductase system of inducible targeted ablation facilitates cell-specific regenerative studies in zebrafish. Methods 62:232–240. 10.1016/j.ymeth.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DT, Sengupta S, Saxena MT, Xu Q, Hanes J, Ding D, Ji H, Mumm JS (2017) Immunomodulation-accelerated neuronal regeneration following selective rod photoreceptor cell ablation in the zebrafish retina. Proc Natl Acad Sci U S A 114:E3719–E3728. 10.1073/pnas.1617721114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Ikeda Y, Notomi S, Ishikawa K, Murakami Y, Hisatomi T, Enaida H, Ishibashi T (2013) Clinical evidence of sustained chronic inflammatory reaction in retinitis pigmentosa. Ophthalmology 120:100–105. 10.1016/j.ophtha.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Yu M, Liu Y, Li J, Natale BN, Cao S, Wang D, Amack JD, Hu H (2016) Eyes shut homolog is required for maintaining the ciliary pocket and survival of photoreceptors in zebrafish. Biol Open 5:1662–1673. 10.1242/bio.021584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Li C, Biswas L, Hu X, Liu F, Reilly J, Liu X, Liu Y, Huang Y, Lu Z, Han S, Wang L, Yu Liu J, Jiang T, Shu X, Wong F, Tang Z, Liu M (2017) CERKL gene knockout disturbs photoreceptor outer segment phagocytosis and causes rod-cone dystrophy in zebrafish. Hum Mol Genet 26:2335–2345. 10.1093/hmg/ddx137 [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA (2005) Responses of Müller glia to retinal injury in adult zebrafish. Vision Res 45:991–1002. 10.1016/j.visres.2004.10.022 [DOI] [PubMed] [Google Scholar]
- Zabel MK, Zhao L, Zhang Y, Gonzalez SR, Ma W, Wang X, Fariss RN, Wong WT (2016) Microglial phagocytosis and activation underlying photoreceptor degeneration is regulated by CX3CL1-CX3CR1 signaling in a mouse model of retinitis pigmentosa. Glia 64:1479–1491. 10.1002/glia.23016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng HY, Zhu XA, Zhang C, Yang LP, Wu LM, Tso MO (2005) Identification of sequential events and factors associated with microglial activation, migration, and cytotoxicity in retinal degeneration in rd mice. Invest Ophthalmol Vis Sci 46:2992–2999. 10.1167/iovs.05-0118 [DOI] [PubMed] [Google Scholar]
- Zhang C, Lei B, Lam TT, Yang F, Sinha D, Tso MO (2004) Neuroprotection of photoreceptors by minocycline in light-induced retinal degeneration. Invest Ophthalmol Vis Sci 45:2753–2759. 10.1167/iovs.03-1344 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hou H, Yu S, Zhou C, Zhang X, Li N, Zhang S, Song K, Lu Y, Liu D, Lu H, Xu H (2020) Inflammation-induced mammalian target of rapamycin signaling is essential for retina regeneration. Glia 68:111–127. 10.1002/glia.23707 [DOI] [PubMed] [Google Scholar]
- Zhao XF, Wan J, Powell C, Ramachandran R, Myers MG Jr, Goldman D (2014) Leptin and IL-6 family cytokines synergize to stimulate Müller glia reprogramming and retina regeneration. Cell Rep 9:272–284. 10.1016/j.celrep.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zabel MK, Wang X, Ma W, Shah P, Fariss RN, Qian H, Parkhurst CN, Gan WB, Wong WT (2015) Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med 7:1179–1197. 10.15252/emmm.201505298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv L, Muto A, Schoonheim PJ, Meijsing SH, Strasser D, Ingraham HA, Schaaf MJ, Yamamoto KR, Baier H (2013) An affective disorder in zebrafish with mutation of the glucocorticoid receptor. Mol Psychiatry 18:681–691. 10.1038/mp.2012.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Resources and reagents used. Download Table 1-1, DOCX file (21.8KB, docx) .
Extended data Table 1-2. Download Table 1-2, DOCX file (16KB, docx) .
DEGs. Download Table 3-1, XLSX file (161.8KB, xlsx) .
Muller glia DEGs. Download Table 3-2, XLSX file (48.7KB, xlsx) .
State DEGs. Download Table 3-3, XLSX file (13.8KB, xlsx) .
Regulatory DEGs. Download Table 3-4, XLSX file (10.5KB, xlsx) .