Abstract
Aromatic compound degradation in six bacteria representing an ecologically important marine taxon of the α-proteobacteria was investigated. Initial screens suggested that isolates in the Roseobacter lineage can degrade aromatic compounds via the β-ketoadipate pathway, a catabolic route that has been well characterized in soil microbes. Six Roseobacter isolates were screened for the presence of protocatechuate 3,4-dioxygenase, a key enzyme in the β-ketoadipate pathway. All six isolates were capable of growth on at least three of the eight aromatic monomers presented (anthranilate, benzoate, p-hydroxybenzoate, salicylate, vanillate, ferulate, protocatechuate, and coumarate). Four of the Roseobacter group isolates had inducible protocatechuate 3,4-dioxygenase activity in cell extracts when grown on p-hydroxybenzoate. The pcaGH genes encoding this ring cleavage enzyme were cloned and sequenced from two isolates, Sagittula stellata E-37 and isolate Y3F, and in both cases the genes could be expressed in Escherichia coli to yield dioxygenase activity. Additional genes involved in the protocatechuate branch of the β-ketoadipate pathway (pcaC, pcaQ, and pobA) were found to cluster with pcaGH in these two isolates. Pairwise sequence analysis of the pca genes revealed greater similarity between the two Roseobacter group isolates than between genes from either Roseobacter strain and soil bacteria. A degenerate PCR primer set targeting a conserved region within PcaH successfully amplified a fragment of pcaH from two additional Roseobacter group isolates, and Southern hybridization indicated the presence of pcaH in the remaining two isolates. This evidence of protocatechuate 3,4-dioxygenase and the β-ketoadipate pathway was found in all six Roseobacter isolates, suggesting widespread abilities to degrade aromatic compounds in this marine lineage.
The Roseobacter lineage in the α-proteobacteria is abundant in southeastern U.S. estuaries (3, 15) and other coastal environments (27, 46). In the expansive salt marshes of the southeastern United States, where many Roseobacter strains have been isolated, organic matter is strongly influenced by naturally occurring aromatic compounds in the form of lignin and humic substances (25, 26). Studies of isolate Sagittula stellata E-37 (16) and preliminary screens of other cultured Roseobacter isolates revealed capabilities for the transformation of synthetic lignin and degradation of lignin-related aromatic monomers. Because the Roseobacter lineage is one of the few dominant marine clades that is amenable to culturing (13), the group presents a unique opportunity to investigate the catabolism of aromatic compound catabolism by a cluster of bacteria that is ecologically important and exclusively marine.
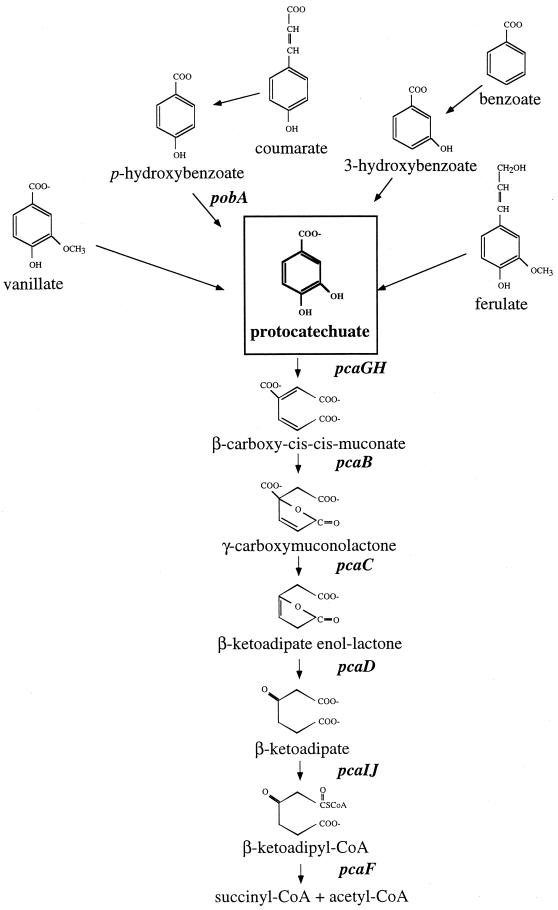
Despite the vast array of aromatic compounds in aquatic and terrestrial environments, the degradation of different compounds usually proceeds through a limited number of metabolic pathways. Most aromatic compounds are first converted to one of several di- or trihydroxylated substrates, such as catechol or protocatechuate (Fig. 1), whose aromatic ring can be enzymatically cleaved (2). In the β-ketoadipate pathway, a primarily chromosomally encoded catabolic route that is widely distributed in soil bacteria and fungi, catechol and protocatechuate are cleaved between their two hydroxyl groups by catechol 1,2-dioxygenase (1,2-CTD) or protocatechuate 3,4-dioxygenase (3,4-PCD), respectively (20). This branched pathway converges, and the ring cleavage products of either catechol or protocatechuate are converted to β-ketoadipate, the metabolite for which the pathway was named. Two additional steps complete the conversion of β-ketoadipate to tricarboxylic acid cycle intermediates.
FIG. 1.
Schematic of the protocatechuate branch of the β-ketoadipate pathway in some prokaryotes (20). Gene designations are shown in italics. CoA, coenzyme A.
Studies of soil bacteria have revealed that the catechol and protocatechuate branches of the β-ketoadipate pathway contain analogous enzymes that are similar in sequence (20). Both 3,4-PCD and 1,2-CTD belong to a large class of nonheme iron-containing dioxygenases (17). 3,4-PCD is composed of equimolar amounts of two nonidentical α and β subunits that are encoded by the usually cotranscribed pcaG and pcaH genes. The PcaG and PcaH proteins are similar to each other at both the structural and amino acid sequence levels, with approximately 30% of their aligned residues being identical (20, 29, 48). Significant sequence similarity is also observed between these proteins and the subunits of 1,2-CTD, which are usually composed of homodimers encoded by the catA gene. The similar sequences of these proteins from soil bacteria such as Pseudomonas putida, Agrobacterium tumefaciens, and Acinetobacter species suggest that the PcaG, PcaH, and CatA proteins all arose from a common ancestor that diverged fairly recently (39).
Although aromatic compounds are abundant in coastal marshes and estuaries, investigations of the aerobic degradation of these compounds by marine bacteria have been rare. A spate of recent articles indicates increasing interest in this topic (10, 11, 22, 34), although most of the current knowledge is still based primarily on studies of soil bacteria. Here we describe investigations of the protocatechuate branch of the β-ketoadipate pathway, a route that in coastal marine environments may be used to degrade aromatic monomers that arise during the decay of lignin and other vascular plant components (e.g., vanillate, coumarate, cinnamate, ferulate, benzoate, and p-hydroxybenzoate [POB]) (4, 21, 35) (Fig. 1). Some environmental aromatic pollutants may be degraded by this pathway as well (2). In this report, evidence is provided that a key enzyme of this pathway, 3,4-PCD, is present in six marine bacteria affiliated with the Roseobacter group. We suggest that aromatic ring cleavage may be characteristic of this ecologically important lineage.
MATERIALS AND METHODS
Bacterial isolation and phylogenetic analysis.
Isolates used in this study were cultured from seawater or sediments collected in the estuaries or coastal waters of the southeastern United States, either isolated from lignin or aromatic monomer enrichments (Sagittula stellata E-37, Sulfitobacter sp. strain EE-36, isolate Y3F, isolate IC4, and isolate S25com04) or cultured directly from coastal seawater using nonselective, low-nutrient seawater plates (isolate GAI-16) (15, 16). All isolates were shown to be members of the Roseobacter group by sequencing of rRNA regions as previously described (15). Full 16S rDNA sequences were previously reported for S. stellata E-37 (U58356), GAI-37 (AF007260), and Sulfitobacter sp. strain EE-36 (AF007254). The complete 16S rDNA sequence for Y3F and partial sequences for IC4 and S25com04 were determined using universal primers as outlined by González et al. (14).
Screening of Roseobacter group isolates for growth on aromatic compounds.
Isolates were initially grown on marine basal medium (MBM) (100 mM NaCl, 25 mM MgSO4, 5 mM KCl, 5 mM CaCl2, 25 mM FeEDTA [pH 7.5]) agar plates amended with 0.4% yeast extract. Fresh cultures were transferred to MBM plates supplemented with 4 mM aromatic monomer as the sole carbon source (anthranilate, benzoate, POB, salicylate, vanillate, ferulate, protocatechuate, or coumerate) and 0.1% vitamins (16). The plates were incubated at 28 to 30°C in the dark for up to 14 days.
Preparation of cell extracts and enzyme assays.
Isolates were grown in 100 ml of MBM with 4 mM POB or sodium acetate as the sole carbon source at 28 to 30°C for 48 to 96 h, with shaking and in the dark. The cells were harvested by centrifugation, washed once with sterile deionized water, and stored at −20°C. Cell pellets were suspended in 100 to 200 μl of breaking buffer [50 mM Tris-HCl, 10% glycerol, 5 mM (NH4)2SO4, 2.5 mM EDTA, 1 mM dithiothreitol (pH 7.5)]. Cell extracts were prepared as previously described (43), and 3,4-PCD activity was determined spectrophotometrically by measuring the decrease in absorbance at 290 nm (44). Protein concentrations were determined by the method of Bradford (1) or Lowry et al. (24).
Detection and isolation of catabolic genes from S. stellata E-37 and isolate Y3F.
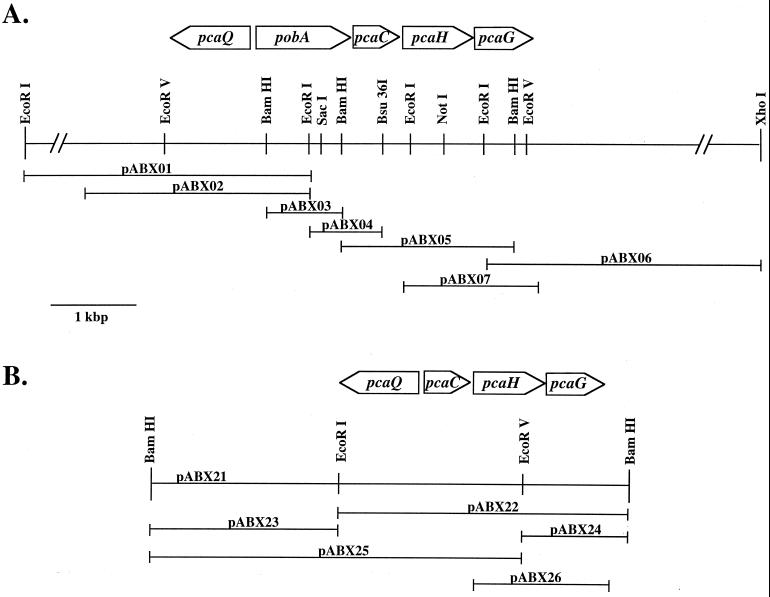
pcaHF1 (5′GARRTRTGGCARGCSAAYGC3′) was designed based on conserved regions found within the amino acid alignments of PcaH from Acinetobacter sp. strain ADP1 (M33798), Pseudomonas putida ATCC 23975 (L26294), and Burkholderia cepacia DB01 (M30791), where R = A + G, S = G + C, and Y = C + T. The degenerate oligonucleotide corresponds to residues 74 to 79 of PcaH from Acinetobacter sp. strain ADP1. Chromosomal digestions of S. stellata E-37 were used in Southern hybridization analysis (42) with the degenerate oligonucleotide. Nonradioactive probes made by 3′ tailing of the degenerate oligonucleotide with digoxigenin (DIG) were used in hybridizations (Genius system; Roche Molecular Biochemicals, Indianapolis, Ind.). Adjacent fragments in S. stellata E-37 were identified by Southern hybridization analysis using DIG-labeled fragments generated from distal ends of the primary fragment (Fig. 2A).
FIG. 2.
Restriction map of the chromosomal pca regions from S. stellata E-37 (A) and Y3F (B). The locations of genes and their transcriptional directions are shown relative to selected restriction endonuclease recognition sites. Horizontal lines indicate the DNA regions contained on recombinant plasmids, whose designations are shown above the corresponding line.
The catabolic gene cluster was initially identified in isolate Y3F by PCR amplification of a portion of pcaG using S. stellata E-37-specific primers P34O1321R (5′GGATGTCGAAGCGGT3′), designed from a conserved region spanning residues 183 to 187 in PcaG (S. stellata E-37 numbering), and pcaHF1ND (5′GAGGTCTGGCAGGCCAAT3′), a nondegenerate version of pcaHF1. The PCR mixture contained 1× buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.3]), 2 mM deoxynucleoside triphosphates, 30 ng of template DNA, 36 pmol of P34O1321R primer, 50 pmol of pcaHF1ND primer, and 1 U of Taq DNA polymerase. PCR was performed in a DNA thermal cycler model 480 (Perkin-Elmer Corp., Foster City, Calif.) with an initial cycle of 10 min at 95°C followed by 30 cycles of 1 min at 95°C, 1 min at 47°C, and 1 min at 72°C. The PCR product from isolate Y3F was labeled with DIG by a random priming reaction and used in Southern hybridizations with isolate Y3F chromosomal digestions.
Hybridizations with random-primed DIG-labeled probes were carried out at 42°C. The hybridization temperatures for the DIG-labeled oligonucleotide probes were determined empirically for each probe and ranged between 55 and 65°C. In lower-stringency hybridizations, all steps were done at a temperature 5 to 10°C below the empirically determined optimal temperature.
DNA preparation and plasmid construction.
DNA fragments of the sizes corresponding to positive hybridization signals in Southern blot analysis were gel excised and ligated into either the pZERO (Invitrogen Corp., Carlsbad, Calif.) or pT7Blue-2 (Novagen Inc., Madison, Wis.) vector. Colony hybridization of genomic libraries with DIG-labeled DNA probes identified positive clones.
Expression of Roseobacter group isolate DNA in E. coli.
To express the pcaHG genes from S. stellata E-37 and isolate Y3F under control of the lac promoter in Escherichia coli, pABX07 and pABX26 were constructed using PCR primers that would introduce restriction sites for optimal positioning in the expression vector pCYB1 (New England Biolabs). A NdeI cleavage site was introduced just before the ATG start codon of the pcaH gene, and a HindIII cleavage site was introduced downstream of the pcaG stop codon by PCR amplification using the high-fidelity pwo DNA polymerase (Roche Molecular Biochemicals). The 1.3-kbp NdeI-HindIII pcaHG DNA fragments were then ligated into the corresponding sites on the pCYB1 vector. The correct sequences of the resultant recombinant plasmids were confirmed. Luria broth cultures (100 ml) of plasmid-carrying E. coli Top10F′ cells (Invitrogen) were grown at 30°C for 13 h. At the time of inoculation, 100 or 800 μM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to cultures of S. stellata E-37 and isolate Y3F, respectively.
Detection of putative pcaH in Roseobacter isolates.
A degenerate PCR primer pair was designed based on conserved PcaH regions: P34OIDf (5′YTIGTIGARRTITGGCARGCIAAYGC3′) and P34OIDr (5′ICYIAIRTGIAYRTGIGCIGGICKCCA3′). Genomic DNA was prepared from each isolate by scraping fresh colonies (ca. 50 mg) from an agar plate. The colonies were washed twice with deionized H2O and then boiled in 500 μl of deionized H2O for 7 to 10 min. Cellular debris was collected by centrifugation at 15,000 × g for 2 min. The supernatant fluid was drawn off and used directly in PCR amplifications using the MasterAmp PCR optimization kit (Epicentre Technologies, Madison, Wis.). To each 50 μl of reaction mixture, 50 pmol of each primer, 1 U of Taq polymerase (Roche Molecular Biochemicals), and 3 μl of cell lysate were added. PCR was performed with an initial cycle of 4 min at 98°C followed by 30 cycles of 95°C for 1 min, 49°C for 45 s, and 72°C for 45 s. Amplification products of the appropriate size were ligated into the pCR 2.1 vector (Invitrogen) and sequenced.
Sequence determination and analysis.
DNA sequences were determined with double-stranded templates and primers that recognized the cloning vector. When necessary, new oligonucleotide primers were made based on previously sequenced regions. Either an ABI373A or an ABI310 automated DNA sequencer (PE Applied Biosystems) was used. Both strands of reported genes were sequenced. Sequences were analyzed using the Genetics Computer Group program package 8.0 (5). Homology searches (BLAST) were carried out at the network server of the National Center for Biotechnology Information.
Phylogenetic trees were constructed for PcaGH sequences with the PHYLIP package (8) by using evolutionary distances (Kimura distances) and the neighbor-joining method.
Nucleotide sequence accession numbers.
Sequences were deposited into the GenBank database (AF253465, AF253466, AF253538, AF253539, AF253467, AF254098, and AF254099).
RESULTS
Choice of marine strains.
The six isolates chosen for this study are part of a larger collection of Roseobacter group strains cultured from the vascular-plant-dominated coastal marshes of the southeastern United States (14, 15). Four of the six isolates (S. stellata E-37, Sulfitobacter sp. strain EE-36, isolate Y3F, and isolate IC4) were obtained by enrichment with lignin-rich pulp mill effluent (Indulin) as the sole carbon and energy source. Three of these Indulin-grown isolates (S. stellata E-37, Sulfitobacter sp. strain EE-36, and isolate Y3F) originated in seawater from Georgia intertidal marshes (Skidaway River and Duplin River), while the fourth (isolate IC4) was from marine sediments collected near the outfall of a pulp wood plant in St. Mary's Estuary, Ga. Isolate S25com04, from the Satilla River, was obtained by selection for growth on POB. Finally, isolate GAI-16 was obtained directly from coastal seawater near Skidaway, Ga., by nonselective culturing (15). These isolates thus originate from four different coastal regions of the southeastern United States and were obtained by three different culture approaches.
Growth on aromatic compounds.
The ability of the marine bacteria to grow on aromatic substrates was tested on solid media containing as the sole carbon source one of eight single-ring-containing compounds known to be degraded by soil microbes via the β-ketoadipate pathway. These compounds, arising in nature from the decay of vascular plant material, may be converted either to catechol or to protocatechuate. Compounds such as protocatechuate, POB, vanillate, ferulate, and coumerate are typically degraded through the protocatechuate branch of the β-ketoadipate pathway, whereas benzoate is converted to catechol by some soil microbes and to protocatechuate by others. All six Roseobacter group isolates were able to grow well on at least two of the aromatic monomers provided as a sole carbon source, and one strain, Sulfitobacter sp. strain EE-36, was able to grow at the expense of all compounds tested (Table 1). All isolates grew well on POB, a characteristic typical of organisms containing the β-ketoadipate pathway. Most also grew on protocatechuate and coumerate. Of the two compounds typically converted to catechol by soil microbes, anthranilate was a good growth substrate for most of the Roseobacter group isolates and salicylate was a good growth substrate for half of these strains (Table 1).
TABLE 1.
Growth on aromatic compounds by Roseobacter group isolates
| Isolate | Growtha of isolate on:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Anthranilate | Benzoate | POB | Salicylate | Vanillate | Ferulate | Protocatechuate | Coumarate | |
| S. stellata E-37 | ± | − | + | − | − | + | + | + |
| Y3F | + | − | + | − | − | − | + | − |
| GAI-16 | + | ± | + | − | − | − | − | ± |
| Sulfitobacter sp. strain EE-36 | + | + | + | + | + | + | + | + |
| S25com04 | ± | + | + | + | + | + | + | + |
| IC4 | ± | + | + | + | + | + | + | + |
Growth was determined on MBM plates containing 4 mM growth substrate and 0.1% vitamins. +, growth within 4 days; ±, growth within 14 days; −, no growth within 14 days.
3,4-PCD enzyme assays.
All six Roseobacter isolates grew on POB, and since this compound might generate protocatechuate during catabolism, 3,4-PCD enzyme activity was assayed. 3,4-PCD activity was detected in cell extracts of four of the isolates (S. stellata E-37, GAI-16, S25com04, and IC4) (Table 2). This activity appeared to be inducible by growth on POB since it was not detected when the isolates were grown on acetate, a nonaromatic growth substrate. No 3,4-PCD activity was detected when Y3F or Sulfitobacter sp. strain EE-36 was grown on POB or when Y3F was grown on vanillate.
TABLE 2.
Specific activity of 3,4-PCD in cell extracts of Roseobacter group isolatesa
| Isolate | Sp act (nmol/min/mg protein)b in extract from:
|
|
|---|---|---|
| POB | Acetate | |
| S. stellata E-37 | 460 ± 110 | <5 |
| GAI-16 | 310 ± 60 | <5 |
| S25com04 | 730 ± 90 | <5 |
| IC4 | 880 ± 120 | <5 |
All isolates were grown in MBM with 4 mM substrate.
Identification of catabolic genes in S. stellata E-37 and isolate Y3F.
To identify the pcaGH genes from the Roseobacter group isolates, we relied on available DNA sequences of soil bacteria. Efforts focused on two isolates that grew well on POB and protocatechuate, one having detectable 3,4-PCD enzyme activity in cell extracts (S. stellata E-37) and the other not having this activity (isolate Y3F). Three degenerate DNA probes for 3,4-PCD were designed based on sequence alignments of the genes encoding the α and β subunits of 3,4-PCD from Acinetobacter sp. strain ADP1, P. putida ATCC 23975, and Burkholderia cepacia DB01. In dot blot hybridizations, one of the three probes, pcaHF1, successfully hybridized with DNA from Acinetobacter sp. strain ADP1, P. putida, and S. stellata E-37 but not with DNA from Y3F or the other Roseobacter strains (Table 3). DNA from E. coli, which does not contain the 3,4-PCD enzyme, did not hybridize with this probe. Southern blot analysis revealed that a 1.7-kbp BamHI fragment from S. stellata E-37 hybridized with the pcaHF1 probe, as did the expected 2.4-kbp HindIII fragment from Acinetobacter sp. strain ADP1 (18). Under low-stringency conditions, a 1.5-kbp BamHI fragment from isolate GAI-16 also gave a positive hybridization signal with the pcaHF1 probe.
TABLE 3.
Indicators of 3,4-PCD in the six Roseobacter isolates
| Isolate | Isolation substrate | Growth on POB | 3,4-PCD activitya | Hybridization with pcaH probe | pcaGH retrieved | pcaGH expression in E. coli | pcaH fragment amplifiedb |
|---|---|---|---|---|---|---|---|
| S. stellata E-37 | Indulin | + | + | +c | + | + | + |
| Y3F | Indulin | + | − | +d | + | + | − |
| GAI-16 | Yeast extract | + | + | +c | NAe | NA | − |
| Sulfitobacter sp. strain EE-36 | Indulin | + | − | − | NA | NA | + |
| S25com04 | POB | + | + | +d | NA | NA | − |
| IC4 | Indulin | + | + | − | NA | NA | + |
Enzyme activity was detected spectrophotometrically in cell extracts of POB-grown cultures.
A 159-bp PCR fragment was amplified using the P34OID primer set.
Positive Southern hybridization signal with the pcaHF1 probe was obtained.
Positive Southern hybridization signal with the pcaHF1ND probe was obtained.
NA, not attempted.
Isolation, cloning, and sequencing of the fragment from S. stellata E-37 genomic DNA showed it contained an open reading frame (ORF) of 723 bp immediately followed by an incomplete ORF of 429 bp. Homology searches with sequences from the database revealed that these two ORFs had highest similarity to the two subunits of 3,4-PCD (Table 4), and we designated these ORFs pcaH and pcaG. Southern hybridizations were carried out to identify and isolate the adjacent DNA fragment containing the terminal portion of the putative pcaG. ORFs showing significant similarity to the catabolic genes γ-carboxymuconolactone decarboxylase (pcaC) and p-hydroxybenzoate hydroxylase (pobA) and to a LysR transcriptional regulator (pcaQ) were also isolated and identified on adjacent DNA fragments from S. stellata E-37 (Fig. 2A and Table 4).
TABLE 4.
β-Ketoadipate pathway genes identified in S. stellata E-37 and Y3F
| Gene designation | Putative function of gene product | Size of gene (bp) | % G+C | Size of deduced gene product
|
Most similar gene, bacterium (% amino acid similarity/identity) GenBank/SwissProt accession no. | |
|---|---|---|---|---|---|---|
| No. of residues | kDa | |||||
| S. stellata E-37 | ||||||
| pobA | p-Hydroxybenzoate hydroxylase | 1,179 | 63.6 | 393 | 44.1 | pobA, P. fluorescens (70/61) (640342) |
| pcaC | γ-Carboxymuconolactone decarboxylase | 393 | 64.9 | 131 | 14.4 | pcaL, S. coelicolor (55/44) (AL079355) |
| pcaG | α subunit of 3,4-PCD | 603 | 65.1 | 201 | 21.9 | pcaG, Acinetobacter sp. strain ADP1 (61/52) (M33798) |
| pcaH | β subunit of 3,4-PCD | 723 | 66.7 | 241 | 26.7 | pcaH, Pseudomonas sp. strain HR199 (70/61) (Y18527) |
| pcaQ | LysR-type transcriptional regulator | 957 | 65.7 | 319 | 34.4 | pcaQ, A. tumefaciens A348 (49/39) (P526681) |
| Y3F | ||||||
| pcaC | γ-Carboxymuconolactone decarboxylase | 396 | 67.6 | 132 | 14.3 | pcaL, S. coelicolor (50/43) (AL079355) |
| pcaG | α subunit of 3,4-PCD | 621 | 64.2 | 207 | 22.9 | pcaG, Acinetobacter sp. strain ADP1 (67/59) (M33798) |
| pcaH | β subunit of 3,4-PCD | 729 | 63.9 | 243 | 26.5 | pcaH, P. putida NCIMB 9869 (69/63) (U96339) |
| pcaQ | LysR-type transcriptional regulator | 927 | 69.1 | 309 | 32.9 | pcaQ, A. tumefaciens A348 (50/43) (P526681) |
Although Southern hybridization of genomic DNA from isolate Y3F did not yield a detectable signal with the pcaHF1 probe, the possibility remained that this isolate could encode 3,4-PCD. To test this possibility, a new oligonucleotide probe (pcaHF1ND) was designed to anneal to a position approximately 350 nucleotides into pcaH, based on the sequence information obtained from S. stellata E-37. This probe did hybridize with DNA from isolate Y3F. A 2.0-kbp BamHI fragment from isolate S25com04 also gave a positive signal with the pcaHF1ND probe (Table 3). This oligonucleotide was then used in a PCR amplification as the forward primer, with a reverse primer based on the sequence of a terminal region in the S. stellata E-37 pcaG gene. Using either S. stellata E-37 or Y3F template DNA and the same amplification conditions, S. stellata E-37 yielded the expected product of approximately 950 bp while isolate Y3F yielded a significantly smaller product of 390 bp. Sequence determination of this smaller fragment and comparisons with known pcaGH sequences indicated that the pcaHF1ND primer had not annealed in Y3F to pcaH as predicted but, rather, to the homologous portion of pcaG. A DIG-labeled probe made from the PCR product was used in Southern blot analysis and detected a genomic BamHI fragment of approximately 3.0 kbp. This fragment was isolated on plasmid pABX20 after appropriately sized fragments were ligated into a standard cloning vector. A series of subclones facilitated the sequencing of the fragment (Fig. 2B). Sequence determination revealed ORFs with homology to pcaG and pcaH, as well as two additional ORFs with homology to pcaC and pcaQ (Table 4).
Sequence analysis of pcaG and H from S. stellata E-37 and isolate Y3F.
The deduced sequences of PcaG and PcaH, the putative α and β subunits of 3,4-PCD, indicated molecular masses of 21.9 and 26.7 kDa in S. stellata E-37 and 22.9 and 26.5 kDa in isolate Y3F. These values correspond well to those reported for 3,4-PCDs in Acinetobacter sp. strain ADP1, P. putida ATCC 23975, P. marginata ATCC 10248, B. cepacia DB01, Azotobacter vinelandii, Brevibacterium fuscum, and a Moraxella sp. (i.e., 23.2 ± 1.1 kDa for PcaG and 28.8 ± 5.1 kDa for PcaH) (40).
Pairwise comparisons of the amino acid sequences deduced from pcaG and pcaH from the two Roseobacter isolates were carried out with the corresponding sequences from the soil microbes Acinetobacter sp. strain ADP1, B. cepacia DB01, P. putida ATCC 23975, and Rhodococcus opacus 1CP. The percentages of identical residues in PcaG were 39 to 59% for isolate Y3F and 42 to 52% for S. stellata E-37. Comparisons of PcaH sequences revealed 47 to 62% identity for isolate Y3F and 47 to 61% identity for S. stellata E-37. In these alignments, residues known to be important for catalytic function (31) were well conserved. The close arrangement of the pcaG and pcaH genes, which are separated by 1 bp in isolate Y3F and overlap by 1 bp in S. stellata E-37, are consistent with the likelihood that these genes are cotranscribed and could be translationally coupled to form nonidentical subunits of the same enzyme (28). Pairwise comparisons with all PcaGH sequences in the database revealed that the deduced amino acid similarity was highest between S. stellata E-37 and isolate Y3F (i.e., 67% similarity and 60% identity). High similarity was also evident at the nucleotide level, with pcaGH from the two Roseobacter group isolates showing 66% identity.
Expression of the pcaGH genes in E. coli.
The pcaGH genes from the two Roseobacter group isolates were expressed from an IPTG-inducible promoter in E. coli, a bacterium that does not encode 3,4-PCD. The presence of pcaGH from either marine bacterium resulted in IPTG-inducible 3,4-PCD activity in cell extracts of the plasmid-bearing E. coli strains. The Y3F-encoded activity was lower than that of S. stellata E-37 (19 ± 8 and 113 ± 10 nmol/min/mg, respectively) and required different IPTG induction conditions. Nevertheless, in each case this activity was inducible and linear with respect to the amount of cell extract added in the assay, indicating that the pcaGH genes of both marine isolates encode 3,4-PCD. No activity was detected in E. coli with the cloning vector or with the recombinant plasmids in the absence of IPTG.
Detection of pcaH in other members of the Roseobacter group.
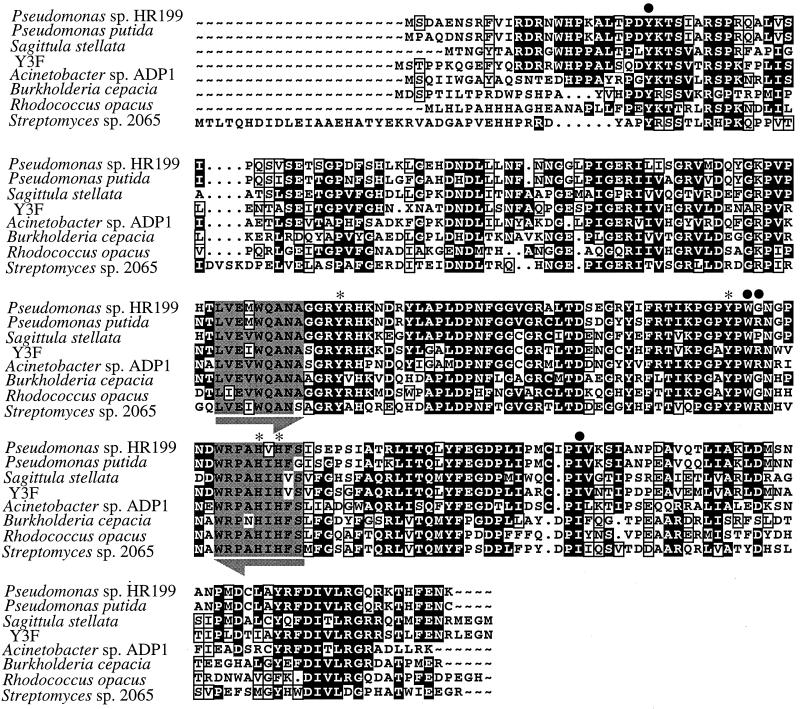
Southern hybridization analysis using two different pcaH probes failed to yield a positive hybridization signal with DNA from two of the six Roseobacter isolates, IC4 and Sulfitobacter sp. strain EE-36. To investigate further the possible presence of pcaGH in these strains, a PCR-based approach was used. A degenerate primer pair was designed from alignments of the deduced amino acid sequences of PcaH (Fig. 3). This primer pair successfully amplified a 159-bp product from both Sulfitobacter sp. strain EE-36 and isolate IC4. In addition, a PCR fragment of the expected size was amplified from DNA of S. stellata E-37. The sequence of these PCR fragments indicated homology to pcaH. Comparisons of the deduced amino acid sequence over this 53-residue stretch of PcaH showed significant conservation within this region among the Roseobacter group isolates, as well as among other bacteria in the database (Table 5). The entire pcaH regions of the marine bacteria were studied only for isolate Y3F and S. stellata E-37, but the full sequences of pcaH and pcaG from these strains, as well as analysis of the adjacent genetic regions, support the presence of the β-ketoadipate pathway in bacteria of the Roseobacter group.
FIG. 3.
Protein sequence alignments of PcaH. Aligned residues that are identical or similar are shown with black backgrounds or boxed, respectively. Residues presumed to be involved in substrate specificity are indicated by a dot, and those demonstrated to be involved in catalysis and Fe2+ binding in P. putida are indicated by an asterisk (31). Gray regions underscored with arrows indicate residues used for the design of the P34OID degenerate PCR primers, with identical and similar residues shaded gray and boxed, respectively. Alignments are shown for P. putida ATCC 23975 (L14836), Acinetobacter sp. strain ADP1 (M33798), B. cepacia DB01 (M30799), R. opacus 1CP (AF003947), and Streptomyces sp. strain 2056 (AF019386). Alignments were conducted using the PILEUP program of the Genetics Computer Group package.
TABLE 5.
Similarity among corresponding 53-amino-acid regions of PcaHa
| Organism | % Similarity (% identity) tob:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC4 | Y3F | EE36 | HR199 | PP | E-37 | ADP1 | BC | RO | 2065 | |
| IC4 | 100 (100) | |||||||||
| Y3F | 100 (98) | 100 (100) | ||||||||
| EE36 | 96 (94) | 96 (94) | 100 (100) | |||||||
| HR199 | 96 (94) | 96 (94) | 96 (96) | 100 (100) | ||||||
| PP | 94 (93) | 94 (93) | 93 (91) | 93 (91) | 100 (100) | |||||
| E-37 | 81 (76) | 81 (76) | 79 (74) | 81 (76) | 85 (77) | 100 (100) | ||||
| ADP1 | 79 (70) | 79 (72) | 79 (70) | 77 (68) | 79 (72) | 79 (62) | 100 (100) | |||
| BC | 70 (66) | 70 (66) | 72 (68) | 72 (68) | 72 (66) | 68 (60) | 93 (91) | 100 (100) | ||
| RO | 70 (66) | 70 (66) | 70 (66) | 72 (70) | 72 (68) | 72 (60) | 76 (72) | 76 (68) | 100 (100) | |
| 2065 | 68 (62) | 68 (62) | 68 (62) | 66 (60) | 68 (60) | 64 (57) | 66 (57) | 72 (66) | 66 (59) | 100 (100) |
Similarities and identities are shown for residues encoded by the 159 bp located between the two P34OID primers.
IC4, isolate IC4 (AF253539); Y3F, isolate Y3F (AF253466); EE36, Sulfitobacter sp. strain EE36 (AF253538); HR199 Pseudomonas sp. strain HR199 (418527); PP, P. putida ATCC 23975 (L14836); E-37, S. stellata E-37 (AF253465); ADP1 Acinetobacter sp. strain ADP1 (M33798); BC, B. cepacia DB01 (M30791); RO, R. opacus 1CP (AF003947); 2065, Streptomyces sp. strain 2065 (AF019386).
Genes adjacent to pcaGH that may encode proteins of the β-ketoadipate pathway.
The two 390-bp ORFs immediately upstream of pcaH in both S. stellata E-37 and isolate Y3F were homologous to a portion of pcaL from Streptomyces coelicolor A3(2) (23) (Table 4). In the gram-positive bacteria S. coelicolor A3(2) and Rhodococcus opacus 1CP, β-ketoadipate enol-lactone hydrolase and 4-carboxymuconolactone decarboxylase, termed PcaC and PcaD, respectively, in gram-negative soil microorganisms (Fig. 1), appear to have fused into one protein demonstrating both activities, designated PcaL (7, 23). The ORFs of the two Roseobacter group strains were homologous to the decarboxylase (PcaC-like) segment of PcaL, which comprises the C-terminal third of the protein, and no similarity was found to the hydrolase portion of the protein. In light of the size and sequence of the ORFs, we designated the Roseobacter genes pcaC. Their deduced amino acid sequences were 35 to 54% identical to the PcaC sequences from soil bacteria Acinetobacter sp. strain ADP1 (P20370), Bradyrhizobium japonicum USDA110 (Y10223), and P. putida ATCC 23975 (P0081). The pcaC genes from the two Roseobacter group isolates showed high similarity to each other at the nucleic acid (77% identity) and amino acid (80% similarity; 74% identity) levels. The stop codon of pcaC in S. stellata E-37 is located 14 bp upstream of the initiation codon of pcaH, while 1 bp separates pcaC and pcaH in isolate Y3F.
In S. stellata E-37, an 1,179-bp ORF, designated pobA, was 53 bp upstream of and transcribed in the same direction as pcaC. It may encode a hydroxylase for the conversion of POB to protocatechuate (Fig. 1). Its deduced sequence was 56 to 61% identical to PobA sequences from soil bacteria Azotobacter chroococcum (AF019891), Rhizobium leguminosarum bv. viciae (L23969), P. fluorescens (640432), P. aeruginosa (P20586), and Acinetobacter sp. strain ADP1 (Q03298). The regions associated with flavin adenine dinucleotide and substrate binding were highly conserved among all the sequences. The 21 amino acids involved with flavin adenine dinucleotide binding in P. fluorescens PobA showed 100% identity to the deduced protein from S. stellata E-37, while seven of the eight amino acids associated with substrate binding were also identical (47). As with PobA from Acinetobacter sp. strain ADP1, the single discrepancy is at residue 124, where the alanine residue that typically occupies this position is replaced by valine in S. stellata E-37 and by serine in Acinetobacter sp. strain ADP1. The putative protein in S. stellata E-37 appears to contain an additional 3 residues between the completely conserved amino acids at position 127 (Val) and position 137 (Pro) of the corresponding proteins from soil bacteria.
Upstream of and divergently transcribed from the pca catabolic genes in both S. stellata E-37 and isolate Y3F were ORFs that we designated pcaQ on the basis of the similarity of their deduced amino acid sequences to the LysR-type transcriptional activator protein PcaQ from A. tumefaciens A348 (37). These putative Roseobacter regulatory proteins showed 39 to 43% identity to PcaQ from A. tumefaciens A348 and 18 to 24% identity to several LysR-type regulators of the catechol branch of the β-ketoadipate pathway, including CatR from P. putida (P20667) and BenM and CatM from Acinetobacter sp. strain ADP1 (AF009224). The region of highest similarity among the PcaQ proteins was in the N terminus, an area presumed to comprise a helix-turn-helix motif for DNA binding (49). In S. stellata E-37, pcaQ was 78 bp upstream of pobA, while in isolate Y3F, it was 86 bp upstream of pcaH. By analogy to PcaQ of A. tumefaciens A348 and other LysR-type regulators, the Roseobacter PcaQ proteins might be expected to regulate their own synthesis and also control the expression of genes downstream of and divergently transcribed from pcaQ (36).
DISCUSSION
Despite the ecological importance of the β-ketodipate pathway for the degradation of aromatic compounds in a variety of environments, studies of this pathway have focused primarily on soil microorganisms, with an emphasis on bacterial groups associated with plants (e.g., Pseudomonas, Agrobacterium, and Rhizobium). Here we report the identification of a key enzyme of this pathway, 3,4-PCD, in members of a marine lineage in the α-proteobacteria. Although no single method was successful with all isolates, multiple approaches using DNA probes and PCR primer sets indicated that all six Roseobacter group members encode 3,4-PCD (Table 3). The identification of a cluster of ORFs in S. stellata E-37 and isolate Y3F with homology to genes encoding proteins of the β-ketodipate pathway in addition to 3,4-PCD strengthens the argument for the presence of the pathway. These initial characterizations of the marine bacteria provide a framework for comparisons of catabolism with their well-studied soil counterparts. Moreover, the presence of the β-ketoadipate pathway in marine Roseobacter isolates, including an isolate cultured using nonselective methods, suggests that these bacteria are contributing to the degradation of aromatic components of vascular plant material in coastal rivers and marshes.
3,4-PCD in Roseobacter group isolates.
The level of 3,4-PCD activity in cell extracts from four of the Roseobacter isolates (Table 2) was comparable to that observed in similar studies of POB-grown soil microbes (32, 51). It was surprising, therefore, that this activity was not detectable in POB- or vanillate-induced cell extracts of the Y3F isolate, from which the entire pcaGH genes were isolated, or in POB-induced extracts of Sulfitobacter sp. strain EE-36, from which a portion of the pcaH gene was characterized. It may be that some of these environmental isolates are less sensitive to our method of cell lysis and/or that the functional enzymes were damaged by heat during sonication. Since the pcaGH genes of Y3F resulted in a sixfold-lower 3,4-PCD specific activity when expressed in E. coli than was found in comparable experiments with S. stellata E-37, the Y3F enzyme could be more heat labile. It seemed unlikely that a protocatechuate 4,5-dioxygenase, rather than 3,4-PCD, cleaved protocatechuate during growth on POB, since no activity for this enzyme was detected in cell extracts from any of the six isolates (A. Buchan, E. L. Neidle, and M. A. Moran, unpublished data). The activity of another enzyme known to cleave protocatechuate, protocatechuate 2,3-dioxygenase, was not determined, although this enzyme has been identified in only one isolate (50). The possibility that 3,4-PCD failed to be induced by the presence of POB (and vanillate in the case of Y3F) because metabolism proceeds via a ring cleavage substrate other than protocatechuate for these particular compounds in Y3F or Sulfitobacter sp. strain EE-36 was not investigated.
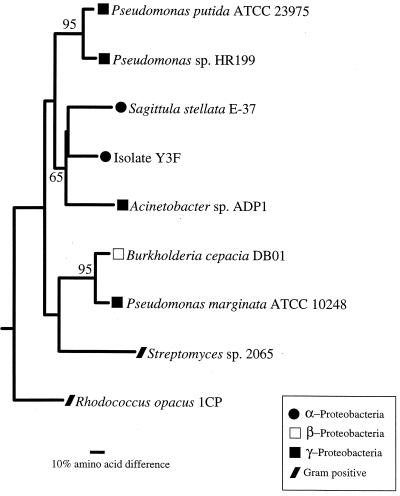
Phylogenetic analysis of the α and β subunits of 3,4-PCD obtained from S. stellata E-37 and isolate Y3F indicated that the genes from the Roseobacter isolates cluster with low bootstrap values with the genes from Acinetobacter sp. strain ADP1 (Fig. 4). The six residues shown to be involved in ligand binding and the four residues demonstrated to be involved in the active site of P. putida (29) are conserved in both Roseobacter isolates. The only exception is in S. stellata E-37, for which the ligand binding residue in PcaH located at position 148 (S. stellata E-37 numbering) is replaced by proline, where typically either an arginine or a glycine is found (Fig. 3).
FIG. 4.
Phylogenetic tree of PcaGH protein sequences. The tree is based on the deduced amino acids encoded by the pcaGH genes and is unrooted, with CatA from Acinetobacter sp. strain ADP1 (Z36909) as the outgroup. Bootstrap values greater than 50% are indicated at branch nodes. The scale bar indicates the amount of genetic change measured as the number of amino acid substitutions per site.
The α and β subunits of S. stellata E-37 show a high degree of similarity to one another (45% similarity, 37% identity), as do the α and β subunits of isolate Y3F (36% similarity, 42% identity). This indicates a common ancestry of the two subunits, as previously suggested for the α and β subunits of other 3,4-PCD enzymes (29). Among soil bacteria, and now marine Roseobacter isolates, the amino acids in the β subunit (PcaH) are more highly conserved than are those in the α subunit (PcaG), a fact that has been attributed to the presence of the four Fe ligand residues within the β subunit (31, 48).
Genetic organization.
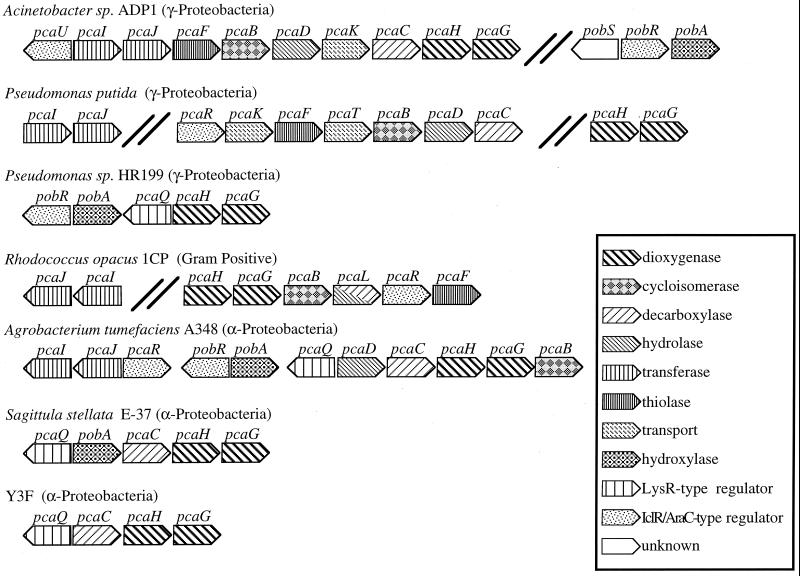
The pcaGH genes in S. stellata E-37 and isolate Y3F were linked to other genes likely to be involved in protocatechuate degradation in a manner similar to that of previously characterized bacteria (Fig. 5). Genes of the β-ketoadiapate pathway are generally clustered on the chromosome in supraoperonic units, i.e., arrangements of linked genes and operons having related physiological functions (7, 20, 30, 32). In Acinetobacter sp. strain ADP1, the operon units needed for POB, protocatechuate, shikimate, and quinate degradation are within a 20-kbp region of the chromosome (12), although in other organisms these operons can be separated by more than 10 kbp, as is the case for P. putida and R. opacus 1CP (20). The pcaG and pcaH genes are always found adjacent to each other, most probably because they encode a two-subunit enzyme and hence would be expected to evolve as a unit (20). The genetic arrangement in S. stellata E-37 differs from that of isolate Y3F in having pobA between pcaQ and pcaC.
FIG. 5.
Organization of gene clusters for protocatechuate metabolism in selected bacteria. Arrows indicate the direction of transcription. Bold double lines indicate genes separated by <10 kbp. The information was compiled from the following sources: Acinetobacter strain ADP1 (12), P. putida (7, 9, 20, 39), Pseudomonas sp. strain HR199 (32), R. opacus 1CP (7), A. tumefaciens A348 (39), and S. stellata E-37 and isolate Y3F (this study).
At the level of nucleic and deduced amino acids, the catabolic genes identified from S. stellata E-37 and isolate Y3F are more similar to each other than to analogous genes from other organisms. Whether these genes were obtained before the Roseobacter isolates and other α-proteobacteria diverged or whether they were obtained by horizontal gene transfer at a later time is unclear. The discontinuities in evolutionary distances of pca genes relative to presumably stable markers such as 16S rRNA (Fig. 4) sequences make these questions difficult to address but clearly portray the dynamic nature of this group of catabolic genes.
Regulation.
This dynamic nature is also reflected in a variety of different classes of transcriptional regulators that have been found to modulate gene expression in the β-ketoadipate pathway of Acinetobacter sp. strain ADP1, P. putida PRS2000, R. opacus 1CP, and A. tumefaciens A348 (6, 12, 19, 33, 39, 41) (Fig. 5). Investigations of the LysR-type regulator PcaQ in A. tumefaciens A348 (36), as well as studies demonstrating the widespread distribution of PcaQ homologs among members of the family Rhizobiaceae, prompted Parke (38) to suggest that the absence of PcaQ may be the exception rather than the rule for the protocatechuate branch of the pathway within the α-proteobacteria. This suggestion is further supported by the identification of PcaQ homologs in our two Roseobacter group isolates (Table 4). Nevertheless, a putative pcaQ was recently found directly upstream of pcaHG in a Pseudomonas strain (a member of the γ-proteobacteria) (32), perhaps indicating a less recent dispersal of this regulator.
In S. stellata E-3, pobA, encoding a putative enzyme for the conversion of POB to protocatechuate, is located immediately upstream of the pca catabolic genes (Fig. 5). Thus far, the pcaQ-pobA orientation in S. stellata E-37 appears to be unique and raises the possibility that in this organism, pobA is under the regulatory control of pcaQ and is regulated in conjunction with the other pca genes identified. In Acinetobacter sp. strain ADP1 and A. tumefaciens A348, pobA is activated by a transcriptional regulator, PobR, that responds to POB and is not a member of the LysR-type transcriptional regulatory family (6, 39). A PobR homolog has also been found directly upstream of pobA in Pseudomonas sp. strain HR199 (32) (Fig. 5).
Ecological significance.
Marine bacteria affiliated with the Roseobacter clade have recently been found to be abundant in the estuaries of the southeastern United States, where they can contribute up to 30% of the bacterioplankton 16S rRNA genes (15). To date, the Roseobacter clade is one of only a few dominant lineages of marine bacteria known to be readily amenable to culturing (13), making laboratory studies of its physiology and genetics particularly important.
Five of the six Roseobacter isolates examined in this study were cultured following enrichment or selection for growth on lignin. Although not common in many marine ecosystems, lignin can be an important source of organic matter in vascular-plant-dominated coastal marshes, including the Spartina alterniflora marshes from which these isolates were obtained (26). As vascular plant material decays, the lignin is converted to smaller aromatic compounds such as benzoate, catechol, POB, cinnamate, p-coumarate, vanillate, ferulate, quinate, and shikimate (45). Many soil bacteria convert these lignin-related monomers to protocatechuate and then degrade them via the β-ketoadipate pathway (4, 21, 35). Marine bacteria in the Roseobacter clade apparently degrade lignin-related compounds in the same manner. The presence of ring cleavage dioxygenases and associated genes of the β-ketoadipate pathway in marine Roseobacter isolates provides a valuable resource for comparative studies on the regulation and ecology of aromatic compound degradation in diverse environments.
ACKNOWLEDGMENTS
José González kindly provided some of the Roseobacter isolates and advised on culturing methods. Nathaniel Cosper helped with attempts to complement Acinetobacter mutants and participated in helpful discussions concerning the 3,4-PCD enzyme.
This work was supported by the NSF through grants from the Biological Oceanography Program (OCE-9730745 to M.A.M.) and the Molecular and Cellular Biosciences Program (MCB-9808784 to E.L.N.) and a traineeship (to A.B.) provided through a research training grant in prokaryotic diversity (BIR-9413235).
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Dagley S. Biochemistry of aromatic hydrocarbon degradation in pseudomonads. In: Sokatch J R, editor. The bacteria. Vol. 10. New York, N.Y: Academic Press, Inc.; 1986. pp. 527–555. [Google Scholar]
- 3.Dang H, Lovell C R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis 16S rRNA genes. Appl Environ Microbiol. 2000;66:467–475. doi: 10.1128/aem.66.2.467-475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delneri D, Degrassi G, Rizzo R, Bruschi C V. Degradation of trans-ferulic acid and p-courmaric acid by Acinetobacter calcoaceticus DSM 586. Biochim Biophys Acta. 1995;1244:363–367. doi: 10.1016/0304-4165(95)00021-3. [DOI] [PubMed] [Google Scholar]
- 5.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR, and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eulberg D, Lakner S, Golovleva L A, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J Bacteriol. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Frazee K W, Livingston D M, LaPorte D C, Lipscomb J D. Cloning, sequencing, and expression of Pseudomonas putida protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1993;175:6194–6202. doi: 10.1128/jb.175.19.6194-6202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia E N, Siegert I G, Suarez P. Toxicity assays and naphthalene utilization by natural bacteria selected in marine environments. Bull Environ Contam Toxicol. 1998;61:370–377. doi: 10.1007/s001289900772. [DOI] [PubMed] [Google Scholar]
- 11.Geiselbrecht A D, Hedlund B P, Tichi M A, Staley J T. Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl Environ Microbiol. 1998;64:4703–4710. doi: 10.1128/aem.64.12.4703-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerischer U, Segura A, Ornston L N. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannoni S, Rappé M. Evolution, diversity, and molecular ecology of marine prokaryotes. In: Kirchman D, editor. Microbial ecology of the oceans. New York, N.Y: Wiley Interscience; 2000. pp. 47–84. [Google Scholar]
- 14.González J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 17.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 18.Hartnett C, Neidle E L, Ngai K L, Ornston L N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990;172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood C S, Nichols N N, Kim M K, Ditty J L, Parales R E. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol. 1994;176:6479–6488. doi: 10.1128/jb.176.21.6479-6488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins A R, Lamb H K, Moore J D, Charles I G, Roberts C F. The pre-chorimsate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects. J Gen Microbiol. 1993;139:2891–2899. doi: 10.1099/00221287-139-12-2891. [DOI] [PubMed] [Google Scholar]
- 22.Hedlund B P, Geiselbrecht A D, Blair T J, Staley J T. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphtovorans gen. nov., sp. nov. Appl Environ Microbiol. 1999;65:251–259. doi: 10.1128/aem.65.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwagami S G, Keqian Y, Davies J. Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl Environ Microbiol. 2000;66:1499–1508. doi: 10.1128/aem.66.4.1499-1508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Moran M A, Hodson R E. Formation and bacterial utilization of dissolved organic carbon derived from detrital lignocellulose. Limnol Oceanogr. 1989;35:1744–1756. [Google Scholar]
- 26.Moran M A, Hodson R E. Dissolved humic substances of vascular plant origin in a coastal marine environment. Limnol Oceanogr. 1994;39:762–771. [Google Scholar]
- 27.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 28.Normark S, Bergstrom S, Edlund T, Grundstrom T, Jaurin B, Lindberg F P, Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 29.Ohlendorf D H, Orville A M, Lipscomb J D. Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15Å resolution. J Mol Biol. 1994;244:586–608. doi: 10.1006/jmbi.1994.1754. [DOI] [PubMed] [Google Scholar]
- 30.Ornston L N, Houghton J, Neidle E L, Gregg L A. Subtle selection and novel mutation during evolutionary divergence of the β-ketoadipate. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis and evolving biotechnology. Washington, D.C.: American Society for Microbiology; 1990. pp. 207–225. [Google Scholar]
- 31.Orville A M, Lipscomb J D, Ohlendorf D H. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe super (3+) ligand displacement in response to substrate binding. Biochemistry. 1997;36:10052–10066. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
- 32.Overhage J A, Kresse U, Priefert H, Sommer H, Krammer G, Rabenhorst J, Steinbüchel A. Molecular characterization of the genes pcaG and pcaH, encoding protocatechate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR 199. Appl Environ Microbiol. 1999;65:951–960. doi: 10.1128/aem.65.3.951-960.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parales R E, Harwood C S. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J Bacteriol. 1993;175:5829–5838. doi: 10.1128/jb.175.18.5829-5838.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardos J, Pelletier E, Siron R, Delille D. Fate of a new silicone-based oil treating agent and its effect on marine microbial communities. Environ Toxicol Chem. 1999;18:819–827. [Google Scholar]
- 35.Parke D, Rynne F, Glenn A. Regulation of phenolic catabolism in Rhizobium leguminosarum biovar trifolii. J Bacteriol. 1991;173:5546–5550. doi: 10.1128/jb.173.17.5546-5550.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parke D. Positive regulation of phenolic catabolism in Agrobacterium tumefaciens by the pcaQ gene in response to β-carboxy-cis-cis-muconate. J Bacteriol. 1993;175:3529–3535. doi: 10.1128/jb.175.11.3529-3535.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parke D. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J Bacteriol. 1996;178:266–272. doi: 10.1128/jb.178.1.266-272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parke D. Conservation of PcaQ, a transcriptional activator of pca genes for catabolism of phenolic compounds, in Agrobacterium tumefaciens and Rhizobium species. J Bacteriol. 1996;178:3671–3675. doi: 10.1128/jb.178.12.3671-3675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parke D. Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1997;146:3–12. [Google Scholar]
- 40.Petersen E I, Zuegg J, Ribbons D W, Schwab H. Molecular cloning and homology modeling of protocatechuate 3,4-dioxygenase from Pseudomonas marginata. Microbiol Res. 1996;151:359–370. doi: 10.1016/s0944-5013(96)80004-8. [DOI] [PubMed] [Google Scholar]
- 41.Romero-Steiner S, Parales R E, Harwood C S, Houghton J E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Shanley M S, Neidle E L, Parales R E, Ornston L N. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J Bacteriol. 1986;165:557–563. doi: 10.1128/jb.165.2.557-563.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanier R Y, Ingraham J L. Protocatechuic acid oxidase. J Biol Chem. 1954;210:799–808. [PubMed] [Google Scholar]
- 45.Subba Rao P V, Nambudiri A M D, Ghat J V. Microbial degradation of phenylpropanoid compounds. J Sci Ind Res. 1971;30:663–679. [Google Scholar]
- 46.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Berkel W, Westphal A, Eschrich K, Eppink M, Dekok A. Substitution of Arg214 at the substrate-binding site of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Eur J Biochim. 1992;210:411–419. doi: 10.1111/j.1432-1033.1992.tb17436.x. [DOI] [PubMed] [Google Scholar]
- 48.Vetting M W, D'Argenio D A, Ornston L N, Ohlendorf D H. Structure of Acinetobacter sp. ADP1 protocatechuate 3,4-dioxygenase at 2.2 Å resolution: implications for the mechanisms of an intradiol dioxygenase. Biochemistry. 2000;39:7943–7955. doi: 10.1021/bi000151e. [DOI] [PubMed] [Google Scholar]
- 49.Viale A M, Kobayashi H, Akazawa T, Henikoff S. rcbR, a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J Bacteriol. 1991;173:5224–5229. doi: 10.1128/jb.173.16.5224-5229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolgel S A, Dege J E, Perkinsolson P E, Juarezgarcia C H, Crawford R L, Munck E, Lipscomb J D. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans—a new extradiol catecholic dioxygenase. J Bacteriol. 1993;175:4414–4426. doi: 10.1128/jb.175.14.4414-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zylstra G J, Olsen R H, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989;171:5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]