Abstract
In silico predictions combined with in vitro, in vivo, and in situ observations collectively suggest that mouse adaptation of the severe acute respiratory syndrome 2 virus requires an aromatic substitution in position 501 or position 498 (but not both) of the spike protein’s receptor binding domain. This effect could be enhanced by mutations in positions 417, 484, and 493 (especially K417N, E484K, Q493K, and Q493R), and to a lesser extent by mutations in positions 486 and 499 (such as F486L and P499T). Such enhancements, due to more favorable binding interactions with residues on the complementary angiotensin-converting enzyme 2 interface, are, however, unlikely to sustain mouse infectivity on their own based on theoretical and experimental evidence to date. Our current understanding thus points to the Alpha, Beta, Gamma, and Omicron variants of concern infecting mice, whereas Delta and “Delta Plus” lack a similar biomolecular basis to do so. This paper identifies 11 countries (Brazil, Chile, Djibouti, Haiti, Malawi, Mozambique, Reunion, Suriname, Trinidad and Tobago, Uruguay, and Venezuela) where targeted local field surveillance of mice is encouraged because they may have come in contact with humans who had the virus with adaptive mutation(s). It also provides a systematic methodology to analyze the potential for other animal reservoirs and their likely locations.
Keywords: AlphaFold, animal reservoir, COVID-19, in silico, in vitro, in vivo, mouse adaptation, SARS-CoV-2, variants
INTRODUCTION
The novel coronavirus disease (COVID-19) has resulted in significant global morbidity and mortality on a scale similar to the influenza pandemic of 1918.1 The ongoing pandemic has been sustained through the activities of human beings, who are the largest reservoir of the causative severe acute respiratory syndrome virus 2 (SARS-CoV-2). This RNA virus in a new host (human beings) is evolving rapidly, accumulating mutations, and existing as a cloud of variants with quasispecies diversity.2 Last year, the world witnessed the risk of this virus acquiring additional reservoirs (such as minks) and new mutations of consequence (such as “Cluster 5”), which could increase transmissibility and lead to a potentially weaker antibody response.3
Both the original form of the virus (known as D614) and the subsequent more transmissible G614 variant, which has replaced it almost entirely in circulation,4,5 did not infect mice because their angiotensin-converting enzyme 2 (ACE2) receptor did not bind the viral spike protein effectively to allow entry into cells. Because mouse (Mus musculus) is a popular animal model of infection, the virus had to be adapted through techniques such as sequential passaging in mouse lung tissues and modifying the receptor binding domain (RBD).6–8 Other strategies for infecting mice with the original form of the virus or the G614 included transgenic mice expressing human ACE2 (hACE2) and sensitizing the mouse respiratory tract through transduction with adenovirus or adeno-associated virus expressing hACE2.9–12 Recently, virus variants of concern (VOC) originating from Brazil, South Africa, and the United Kingdom (which contain the common mutation N501Y in the RBD) were shown to infect mice.13,14 This ability of SARS-CoV-2 VOC to infect mice is unsettling because of the potential to establish additional reservoirs in a species that is in close contact with people and companion animals, especially because its population is hard to vaccinate or control.
METHODOLOGY
In this paper, we have combined our structural predictions from biomolecular modeling with available experimental evidence to understand how specific mutations in VOC and mouse-adapted strains, especially in the RBD, have enabled this virus to infect mice. To do this, we compared the spatial interactions of these key mutations with the corresponding regions in both mouse ACE2 (mACE2) and hACE2 using homology molecular models based on cryogenic electron microscopy data of the ACE2/RBD interface (protein database entry number 6 M17).15 We have assumed the coordinates of ACE2 and RBD in the 6 M17 structure to be reasonable starting positions for our respective models, with root-mean-square deviation (RMSD) scripts alignment based on C-Alpha carbon protein backbones. Because the protein prediction software AlphaFold was recently released,16 we also recalculated RBD structures, finding AlphaFold predictions to be in excellent agreement with our initial homology models, with less than 0.9 Å RMSD average alignment. Amino acid side chain conformations of the RBD models were adapted from AlphaFold predictions because they were deemed more reliable.
By introducing the ACE2 models and variants into molecular dynamics simulations to optimize the interactions, similar to our recent work,17,18 we qualitatively identified the key interactions of side chain residues at the ACE2/RBD interface. Further details are provided under “Supplementary Methods for Molecular Modeling.” We then compared our in silico findings with experimental results reported by different research groups with mouse-adapted strains/isolates6–8,13,21–29 and VOC14,30–34 (Tables 1 and 2). Taken together, we were able to gain valuable insights into the likely effects of different mutations of consequence.
Table 1.
Key mutations in 14 “mouse adapted strains” and 3 “variants of concern” of SARS-CoV-2 known to infect mice, with further characterization as outlined in Table 2 (Omicron VOC not included as experimental evidence is not yet available)
|
Table 2.
Description of the characteristics of the “mouse adapted strains” generated from the in vivo studies discussed in Table 1. *Strain = Mouse adapted strain name, Dose = Dose of SARS-CoV-2 mouse adapted strain provided to the mice stated in plaque forming units (PFU) or the Tissue culture infective dose (TCID50), Shedding & Transmission = Any evidence of viral shedding and viral transmission observed in the mice
| Species and age of mice (Study reference), Strain, dose, shedding and transmission* | Evidence of productive SARS-CoV-2 replication in the respiratory tract | Clinical outcomes, histopathological evidence of disease |
|---|---|---|
|
Species and age of mice: Aged (9 months old) and young (6 weeks old) BALB/c mice studied.6 Strain name: MASCp66 Dose: Intranasal inoculation with 1.6 × 104 PFU Shedding: Viral shedding observed in the feces at day 3,5 and 7 after inoculation. Transmission: No information |
High amounts of viral RNAs in the lungs and tracheas were detected at 3, 5 and 7 days after inoculation in all aged mice, with peak viral RNA loads of ~ 1010 copies/g at 3 days after inoculation. Viral RNAs were also detected in heart, liver, spleen, and brain. Similar tissue distribution of SARS-CoV-2 RNA was also seen in the MASCp6 infected young mice. |
Clinical: No visible clinical symptoms or body weight loss were observed in both the young and aged mice. No fatalities recorded. Histopathological: Led to interstitial pneumonia and inflammatory responses in both young and aged mice, however the lung damage in the aged mice was more severe. |
|
Species and age of mice: Young (8-week-old) and aged (9 months old) male and female BALB/c mice and C57BL/6 mice studied.21 Strain name: MASCp3621 Dose: Intranasal inoculation with varying doses including: 1.2 PFU, 12 PFU, 120 PFU, 1200 PFU, 12000 PFU Shedding: No information Transmission: No information |
qRT-PCR results of aged BALB/c and C57BL/6 mice challenged with 12,000 PFU of MASCp36 showed that high levels of SARS-CoV-2 subgenomic RNAs were persistent in the lung and tracheas till 4 dpi. A similar tissue distribution of SARS-CoV-2 was observed in the young BALB/c and C57BL/6 mice. |
Clinical: All the aged BALB/c and C57BL/6 mice challenged with high doses (1200 or 12,000 PFU) of MASCp36 developed typical respiratory symptoms and exhibited features like ruffled fur, hunched back, and reduced activity. Young BALB/c and C57BL/6 mice were resistant to MASCp36 challenge, and only one animal that received 12,000 PFU challenge died during the observation period. Histopathological: Necrotizing pneumonia and extensive diffuse alveolar damages observed on 4 dpi in aged mice. Milder pathology observed in the young mice. |
|
Species and age of mice: Young adult (12 weeks old) and aged (One year old) BALB/c mice studied.7 Strain name: IC-MA17 Dose: Intranasal inoculation with 105 PFU Shedding: No information Transmission: No information |
High-titre virus replication was also observed in lung tissue at 2 dpi. This cleared by 4 dpi in the young adult mice but continued to persist at 4 dpi in the aged mice, suggesting that there was increased viral replication in the aged mice. MA1 was also observed in the upper airway and viral antigen was present in nasal turbinate epithelium at 2 dpi in young adult mice; viral antigen was found in conducting airway epithelium, interstitium and nasal epithelium in aged mice at 4 dpi. |
Clinical: No overt clinical signs such as weight loss observed in young adult mice but a significant decrease in body weight was observed in aged mice. Whole-body plethysmography on mice indicated impaired lung function at 2 dpi, with the extent of impairment significantly worse in the aged mice. Histopathological: Interstitial congestion, epithelial damage, inflammatory infiltrate and peribronchial lymphocytic inflammation surrounding airways at 2 dpi. Histopathological effects seen in the aged mice are more severe than those observed in young adult mice. |
|
Species and age of mice: Young (10-week-old) and aged (1 year old) BALB/c mice studied.8 Strain name: IC-MA108 Dose: Doses ranging from 102, 103, 104 or 105 PFU were tested. However, 104 PFU and 103 PFU were determined as the optimal doses for analysis in young and aged mice respectively. Shedding: Both old and young mice exhibited viral titers in the nasal cavity over the first 3 days of infection. Transmission: No information |
Virus replication in the lungs of young mice peaked 1–2 dpi and was absent in most surviving mice by 7 dpi. Viral replication in the upper respiratory tract of young mice remained high on 1–3 dpi but was undetectable in most mice by 5 dpi. A similar trend was observed in the aged mice, but the viral replication was detectable for a much longer period. |
Clinical: Young mice rapidly lost weight and reached maximum weight loss at day 4 post inoculation. On day 5, the collective mortality rate was approximately 15%. Significant weight loss was also observed in aged mice. Mortality rates of 20% and 60% were recorded for infection with 104 and 105 PFU, respectively in the young mice. Histopathological: At the time of necropsy, acute stage lung damage was noted in the young mice. Analysis at 2, 4, and 7 dpi revealed early multifocal damage to conducting airway epithelia and hallmarks of diffuse alveolar damage (DAD) was observed. The aged mice had severe DAD and higher acute lung injury scores. Overall, a dose- and age-related increase in pathogenesis was observed. |
|
Species and age of mice: C57Bl6, BALB/c and 129S1/SVMJ mice were studied Young (6–8-weeks) or aged (52 weeks old) C57Bl6 mice were used to study the effect of obesity, obesity-associated diabetes, and advanced age on clinical outcomes.13 Strain name: WA1-MA-P1113 Dose: Intranasal inoculation with 2.5⨯104 PFU Shedding: No information Transmission: No information |
Detectable virus titres in lungs and nasal turbinates of 129S1/SVMJ mice and lungs of male and female C57Bl6 and BALB/c mice, obtained at different days post infection |
Clinical: Weight loss detected in the 129S1/SVMJ mice but not in the C57Bl6 and BALB/c mice. Effect of increased age, comorbidities: Both obesity/diabetes and advanced age in mice result in higher morbidity during SARS-CoV-2 infection. |
|
Species and age of mice: Groups of young (4–6-week-old) female BALB/c mice, young (4–6-week-old female) C57BL/6 J mice and aging (8–9-month-old) male BALB/c mice studied.23 Strain name: HRB26M23 Dose: Intranasal inoculation with 104.4 PFU in a volume of 50 μL. Shedding: No information Transmission: No direct evidence of transmission provided. However, the presence of efficient viral replication in the upper and lower respiratory tracts indicates that transmission could be possible. |
In young BALB/c mice, viral RNA was detected in the nasal turbinates on day 3, 5, and 7 p.i. (post infection) and the infectious virus was detected on day 3 and 5 p.i. The viral RNA was also detected in the heart, liver, kidney, and spleen on day 3 p.i. In C57BL/6 J mice, high viral loads in the nasal turbinates and lungs on days 3, 5 and 7 p.i. Similar observations made in the aging mice. However, when compared to the young mice, the average PFU titres in the aging mice were 3 times higher and 10 times higher in the nasal turbinates and lungs respectively. |
Clinical: Similar to the young mice, the aging adult mice showed transient weight loss on days 2 and 3 p.i. and recovered thereafter. Histopathological: Mild pathological changes were observed in the respiratory tract of young BALB/c mice infected. Moderate-to-severe pathological changes in the lungs after infection in the aging mice. |
|
Species and age of mice: Young (4–6-week-old) BALB/c mice studied.[Female 12-month-old BALB/c mice only used for the initial generation of the mouse adapted strain, not for any analysis of the mouse adapted strain].26 Strain name: Hu-1-WBP-126 Dose: Intranasally infected with 10-fold serial dilutions of the WBP-1 virus (i.e., 102 to 105 PFU). Subsequently, detailed analysis on mice was done by infecting mice with 50 μL of 2 LD50 [The LD50 of WBP-1 is 103.8 PFU]. Shedding: No information Transmission: No information |
In mice given the 50 μL of 2 LD50: High viral loads were observed in the trachea and lung of infected mice, with lung tissues having the highest number of viral RNA copies. Trace amounts of viral nucleic acid were detectable in turbinate, heart, and spleen. |
Clinical: In the groups of mice infected with the 10-fold serial dilutions: All groups of infected mice began to lose weight at 2 dpi and the weight loss was dose dependent. Bristled fur and depressed spirits in mice was also observed. The group of mice receiving a dose of 105 PFU WBP-1 virus all died (5/5) by 5 dpi. Two out of five mice survived in the group administered 104 PFU virus. In the mice given the 50 μL of 2 LD50: Clinical signs of infection were observed in infected mice at 2 dpi. Histopathological: In mice given the 50 μL of 2 LD50, Acute stage lung damage was noted. Moderate interstitial pneumonia with thickened alveolar septa occurred at 3 dpi, which progressed to severe interstitial pneumonia on 5 dpi. Inflammatory cell infiltration was observed in the lung tissue. |
|
Species and age of mice: 10-week-old female BALB/c mice).27 Strain name: IC-MA4 Dose: Mice infected with 105 PFU Shedding: No information Transmission: No information |
Evidence of robust viral replication in mouse lungs. |
Clinical: No information Histopathological: No information |
|
Species and age of mice: 10-week-old female BALB/c mice.27 Strain names: IC-CMA127, IC-CMA227, IC-CMA327 Dose: Mice intranasally inoculated with 105 PFU Strain names: IC-CMA3p20 [IC-CMA3 after passage 20], IC-CMA3p20 [IC-CMA3 after passage 20] Dose: 105 PFU was the main dose administered, but doses 104 and 106 PFU also administered to some mice. |
All three mutants (CMA1, CMA2, CMA3) produced approximately, 105 PFU per lobe at day 2 post infection. However, no virus was detected at day 4, suggesting rapid clearance by the host. In mice given IC-CMA3p20, robust viral replication was observed. There was a significantly greater viral load in the lung compared to IC-CMA3p20 at 2 dpi. |
Clinical: None of the three mutants (CMA1, CMA2, CMA3) induced major disease, although both CMA2 and CMA3 caused more weight loss than CMA1. Mice infected with IC-CMA3p20 had a 10% weight loss by day 3 and showed signs of disease including ruffled fur and hunched posture. Histopathological: Mice infected with CMA3p20 had significant lung infiltration and inflammation characterized by peribronchiolar, perivascular cuffing, and perivasculitis by day 2 post infection. At 4 dpi, collapsed airways and interstitial pneumonia was observed in the mice. |
|
Species and age of mice: Young (2 months old) and aged (12-month-old) BALB/c mice.28 Strain name: LG Dose: 1 × 105 TCID50 in a volume of 50 μL. Shedding: No transmission Transmission: No information |
Effective viral replication in the lungs and trachea of the young and aged mice, although there was more viral replication in the aged mice. |
Clinical: Weight of the aged mice was slightly decreased, but there were no evident changes in the weight of young mice. Histopathological: Lung lesions of the aged BALB/c mice were obvious, but those of young mice were not, with only slight lymphocyte exudation. |
Subsequently, we queried the world’s largest public database called Global Initiative on Sharing All Influenza Data (GISAID) and looked for these mutations and equivalent mutations of consequence. These “big data,” comprising circa 2.4 million and 4 million SARS-CoV-2 genome sequences as of July 21 and October 4, 2021, respectively, formed the basis of our in situ analysis.29 Over 99.9% of these sequences are from viruses sequenced from human hosts. In brief, sequences were aligned to the back to the SARS-CoV-2 reference (EPI_ISL_402124, denoted as the wild-type [WT]) to generate a file containing all mutations in the variant call format, which is a concise way for storing gene sequence variations. We then calculated the ratio of the frequency at which the WT allele vs the mutant allele were observed across a rolling 14-day window. Given the data are discrete, highly variable in size across countries, and contain background noise, such a window of time is essential from our experience to reduce distortions and glean meaningful insights. For example, if 14 WT and 28 mutant sequences were observed within a specified 14-day window, then the frequencies were 1/day and 2/day, respectively. The ratio of mutant to WT frequency is therefore 2, indicating the mutant allele is appearing twice as frequently as the WT allele within that period. This was used to create a heatmap for the key mutations, both individually and in combination (as discussed below). For clarity, we only considered countries where the mutant to WT ratio exceeded 1.2 across the entire time the mutant was sampled or within any given 14-day window. To reduce the noise further from low-sampled countries, we also instituted a minimum threshold of 10 WT and 10 mutant samples over at least 14 days to suggest possible spread locally. For this reason, a country where a mutant had been recorded on only a single day or where only 9 mutant samples were recorded overall was not included in our analysis. Our heatmap scale was also truncated at 2.00 for ease of visual comparison.
RESULTS AND DISCUSSION
In Silico Results
In silico comparison of the interface residues of the RBD/ACE2 complex in human and mouse models reveals 30 ACE2 residues in close contact, of which 19 are conserved between mACE2 and hACE2 (approximately 63% identity for the contacting ACE2 residues). The RBD mutations associated with the mouse adaptations listed in Table 1 can be grouped loosely into 3 regions by their positions on the ACE2/RBD interface as follows: region 1 (RBD positions 498, 499, and 501) centered around the highly conserved ACE2 residue tyrosine 41 (Y41); region 2 (RBD positions 417, 493) centered around ACE2 residue 34; and region 3 (RBD positions 484 and 486) close to a cluster of ACE2 residues 78 to 82. ACE2 residues within 5 Å (chosen to account for molecular fluctuations) of each RBD adaptation are listed in Table 3, with dissimilar hACE2/mACE2 residues highlighted in yellow. Figure 1 further illustrates these residues visually, and how they are relatively positioned in 3 regions. Figure 2 aligns the hACE2 and mACE2 to highlight the key differences at the contact points with the RDB (shown in yellow).
Table 3.
SARS-CoV-2 spike RBD mutations associated with mouse adaptation and their close contact residues in respective ACE2 proteins. Positions with dissimilar human/mouse residues are highlighted in yellow
|
Figure 1.
The RBD/ACE2 interface. The receptor binding domain (RBD) is shown in cyan, and the ACE2 is shown in yellow. Pink spheres indicate relative positions of mouse adapting mutations on the RBD, while the blue spheres represent interface residues that differ between human and mouse ACE2 sequences, shown in green and orange, respectively.
Figure 2.
Sequence alignment of human and mouse ACE2 highlighting contact points with the SARS-CoV-2 spike receptor binding domain in yellow. Differences between contact points are highlighted in green (human) and orange (mouse), and common contact residues are highlighted in cyan.
Modeling the N501Y mutation at the ACE2/RBD interface reveals a close interaction with the highly conserved Y41 residue in ACE2, through attractive, non-covalent bonding between aromatic amino acids known as π-stacking interactions. In mACE2, π-stacking can be enhanced by the proximal substitution of histidine (H) in place of lysine (K), which is present at position 353 of hACE2. Our modeling also shows similar π-stacking enhancement with the conserved Y41, through the substitution of RBD glutamine at position 498 (Q498) to either histidine (H) or tyrosine (Y). These aromatic π-stacking interactions appear to be reasonably strong because N501Y and Q498H can each sustain mouse adaptation on its own in experiments.6,13,23,28 Counterintuitively, our modeling predicts that simultaneous aromatic mutations at RBD positions 498 and 501 are detrimental to mouse adaptation due to local π-stacking distortion to the binding interface. This could explain why very few simultaneous aromatic mutations at these positions have been observed. In over 2.4 million entries on GISAID as of July 21, 2021, we detected only a single co-occurrence, that of an adapted isolate from mouse lung homogenate where N501Y occurred with glutamate 498 to arginine.29 However, when we recently re-ran our in situ model with over 4 million human-origin GISAID entries, we detected 0, 2, and 14 instances of Y501 coexisting with Y498, H498, and R498, respectively. The 2 instances of Y501-H498 are low-quality and/or low-coverage sequences; the 14 instances of Y501-R498 have been reported from France, Netherlands, South Africa, Spain, United Kingdom, and United States between March and September 2021. Although arginine is not aromatic, it is frequently associated with π-stacking interactions with inherent conformational flexibility compared with tyrosine or histidine. Therefore, it is likely that the Y501-R498 combination is more tolerated (eg, with the recent Omicron variant of concern).37
In region 1 (Table 3), we also note the proline 499 to threonine (P499T) substitution in 2 infectious clones, presumably engineered to enhance Y498. Modeling provides the following insight on this: the change from P to T will relax the backbone constraints of proline and allow conformational rearrangement of threonine to contact the conserved ACE2 residues Y41 and L45. However, we did not find any strong molecular modeling basis for this mutation to sustain mouse adaptation on its own or evolve naturally alongside adaptive mutations at positions 498 or 501; this is borne out by experimental evidence to date. In other words, in silico predictions combined with in vitro and in vivo evidence collectively suggest that mouse adaptation requires an aromatic substitution in either position 501 or position 498 (but not both); additional mutations, especially in region 2 and region 3 of the RBD as summarized below, enhance ACE2-binding interactions and specificity in mice. These predictions are further supported by AlphaFold, which assigns very high confidence scores (>93 in a scale of 0–100) for the structural predictions involving these key mutations of the individual proteins; however, predictions of the RBD/ACE2 complex are currently suboptimal. With time, the quality of AlphaFold predictions of protein complexes will improve thanks to additional experimental observations, and thus it is expected to play a more central role in structural interpretation during pandemics such as COVID-19 and future “disease-X.”
From Table 1, we see that mouse-adapted strains sometimes carry the Q493K/R mutation (polar glutamine to basic lysine or arginine). Modeling predicts this as being enabled through favorable salt-bridge interactions with both glutamic acid 35 or aspartic acid 38, both of which are conserved in hACE2 as well as mACE2 (see Table 3, region 2). The K417N substitution (lysine to asparagine), which is another experimental observation from Table 1, is also predicted by modeling to be advantageous for mouse adaptation due to favorable amide hydrogen bond interactions with interfacial mACE2 residues asparagine 30 and glutamine 34; such amide hydrogen bonding is not possible in hACE2 because it has non-amide lysine (K) and histidine (H) residues, as also noted by other researchers.21 With the Gamma variant of concern, it is unclear whether the K417T enhances the role of N501Y in mouse adaptation in a similar manner. The “Delta plus” variant of concern has the K417N mutation, but there is no molecular modeling basis to believe that it can infect mice without an aromatic change in position 498 or 501 as described above. It would be worthwhile to further investigate any interfering role of glycosylation at this interface region, because hACE2 contains N-linked glycosylation at asparagine 90, whereas mACE2 does not (its analogous residue is threonine T90, according to Uniprot references Q8R0I0 and Q9BYF1).
In region 3, the K484 residue is not positioned directly at the interface and not observed to interact strongly with any ACE2 residues; however, our model shows occasional salt bridges can be formed with relatively close glutamic acid residues in positions 35 and 75 that are conserved in both hACE2 and mACE2. Our simulations show that the distance between K484 and these glutamic acid residues fluctuates dynamically from 3 to 20 Å, with salt bridges more likely when distances are approximately 3 Å. Thus, the E484K, which is present in the Beta and Gamma VOC, and more recently in some Alpha isolates as well, is likely to have an enhancing role through transient salt bridges. The same cannot be said about E484Q seen in the Delta variant of concern because salt bridge formation is unlikely with the polar glutamine (Q) residue. With no accompanying aromatic change in positions 498 or 501, we believe that the E484Q in Delta, and additionally the K417N in “Delta plus,” cannot sustain mouse infectivity on their own based on current biomolecular understanding. It also follows that the Omicron variant of concern is expected to infect WT mice because it has the essential and enhancing mutations. The residues 75 to 82 in mACE2 are significantly different from hACE2 (Table 3); therefore, any mutation in the corresponding RBD interface is worth investigation. We could find one from experimental observations, the engineered substitution F486L,27 and consider it to have at best an enhancing role. Because it was observed simultaneously with Q498Y (which is likely to sustain mouse infectivity on its own), the contribution of F486L to the overall mouse adaptation remains to be ascertained.
Comparison With In Vitro, In Vivo, and In Situ Observations
Early in silico predictions based on comparative structural analysis of ACE2 suggested that mouse has a very low probability of being infected.35,36 Although correct about mouse, those analyses also made inconsistent and erroneous predictions that ferrets would not be susceptible, pigs would be susceptible, etc, thus exposing the need for experimental inputs into the model. Therefore, this paper takes into account a range of experimental observations to cross-check our in silico predictions through biomolecular modeling. Wan et al reasoned that “mouse or rat ACE2 contains a histidine at the 353 position which does not fit into the virus-receptor interaction as well as a lysine does.”35 Although this is true of the original Wuhan strain containing N501 in the RBD, our modeling indicates why the tyrosine 501 mutation enables mouse infectivity, even on its own. In hACE2, lysine 353 creates a salt bridge with conserved aspartic acid 38. In mACE2, lysine 353 is replaced by the aromatic histidine to complete the salt bridge as well as contribute to π-stacking with the Y501 variant. The N501Y mutation will also lead to favorable π-stacking with the highly conserved Y41 residue in mammalian ACE2, as suggested by Starr et al38 with deep scanning of RBD mutations and hACE2 affinity assays. These authors highlighted enhanced affinity of F501 (as it had the highest score), followed by Y501, V501, W501, and T501, in that order. But Y501 and T501 require only a single nucleotide change and have been observed more frequently in situ (Supplementary Figure S1a)—compared with F501, V501, and W501, which require 2, 2, and 3 nucleotide changes, respectively. It is unsurprising that the latter variants requiring 2 or more changes were rarely observed in situ regardless of their high in vitro affinity scores from Starr et al.38
From the above and Table 4, we see that in silico analysis can provide valuable insights to interpret and bridge in vitro, in vivo, and in situ observations on the RBD position 501. A similar analysis is possible with the alternative essential mutation for mouse adaptation at RBD position 498, where the in vitro affinity enhancement order is H498, Y498, F498, and W498 according to Starr et al.38 Of these, H498 (on its own) and Y498 (with enhancing RBD mutations) have been shown to result in mouse adaptation in vivo (Table 1)7,8,23–28; glutamate 498 to arginine was also reported once, unusually in combination with N501Y, isolated from mouse lung after 30 passages. In humans, in situ observations of these variants have been limited to R498 (57 occurrences) and H498 (8 occurrences) so far. Thus, it is clear from in vitro, in vivo, and in situ analyses (Table 4) that H498, R498, and Y498 are possible but not yet common. This is consistent with our in silico predictions because H498 and Y498 are aromatic (enabling π-stacking with ACE2 Y41; similar to Y501), and R498 has conformational flexibility and can still be associated with π-stacking interactions.37 H498 and R498 observed in situ require a single nucleotide change from Q498, whereas Y498 requires 2 nucleotide changes (or 1 change from H498).
Table 4.
Comparison of in silico, in vitro, in vivo and in situ observations of key mutations in Table 1
| Essential position in RBD | Enhancing position(s) in RBD for mouse adaptation | ||||||
|---|---|---|---|---|---|---|---|
| Analysis | Q498 | N501 | K417 | E484 | F486 | Q493 | P499 |
| In silico a | H Y F We | Y H F We | N Qh | K Q | L | K L R | T |
| in vitro b | H Y F W | F Y V W T | – | R K T Q | – | M A Y F K L V | – |
| in vivo c | H R Y | Y | M N T | K Q | L | K R | T |
| In situ d | R Hf | Y T H Fg | N T Mi | K Q R Tj | Lk | K L Rl | Tm |
aPredicted by our modeling. For E484, F486, Q493, and P499, our work only reconfirmed in vivo observations.
bArranged in descending order of affinity binding from in vitro experiments reported by Starr et al.38
cObserved in vivo with studies summarized in Table 1 (alphabetical order). Note L486 and P499 were engineered.
dObserved on GISAID as of July 21, 2021, after removing noise from low-sample countries. Only key trends are presented.
eThese aromatic residues are predicted to have similar interactions with Y41 of ACE2.
fF W Y not observed yet. Since early 2021, R observed in several countries, and H in Slovenia and USA (albeit the latter are of low quality and/or low coverage).
gV W not observed yet. H observed in several countries (mid 2020), T in Spain’s Canary Islands (early 2021), and F in Colombia, Germany, Mexico, and Sweden (mid-2021). Y observed in several countries (Supplementary Figure 1).
hModeled mutations predict favorable amide hydrogen bonding. N and Q mutations each require a single nucleotide change, but there are 2 ways to get from N to K.
iQ observed in the USA (August 2021). N (mutation G22813T) and T observed in several countries (Supplementary Figure 2a and b). Interestingly, N (alternative mutation G22813C) has a sporadic presence in UK and USA (since late 2020).
jK observed in several countries (Supplementary Figure 2c). Sporadic presence of Q (several countries from March 2020 to August 2021), R (Angola, Brazil, South Africa, South Korea, UK, USA, since late 2020), and T (USA in mid-2021) observed.
kSporadic presence of different synonymous mutations (T23018C in several countries; T23020G in USA and 1 instance in Turkey) observed since late 2020.
lA, F, M, V, Y not observed yet. Sporadic presence observed for K (Italy since early 2020), L (Trinidad and Tobago in mid-2021), and R (several countries mid-2020).
mSporadic presence since late 2020 in Bulgaria, Canada, Netherlands, and USA.
Similar insights are also possible for the enhancing RBD mutations (see Tables 1 and 4). From in vivo and in situ observations, we see that K417M is less common than K417N or K417T (Supplementary Figure S2a and b), although in vitro studies did not predict any enhancement.38In silico predictions show that all these require a single nucleotide change but that N417 (and Q417) would benefit from amide hydrogen bonding. E484K is present in Beta and Gamma (Supplementary Figure S2c) and increasingly in Alpha VOC, whereas E484Q is present in the Kappa variant of interest that is related to the Delta VOC. Compared with these 2 substitutions and notwithstanding higher in vitro affinity scores, R484 and T484 are infrequently observed in situ, which is consistent with our in silico predictions because they each require 2 nucleotide changes from E484 or 1 change from K484. The F486L and P499T were originally engineered in vivo but have had sporadic in situ presence in human populations. In silico predictions suggest that the F486L mutation (accessible by 3 possible ways of a single nucleotide change) can aid mACE2 adaptation due to the human-mouse differences in ACE2 at the 78–82 region; the P499T is also a single nucleotide change (but only 1 way from P to T) and predicted to be rare in comparison. Finally, in silico predictions for Q493 substituted by K, L, or R (each a single nucleotide change) are borne out in vivo and in situ, although their affinity scores from in vitro experiments are low. The affinity scores from Starr et al38 were developed for hACE2 (not mACE2), so we expected a greater correlation than what has been observed in situ in human populations, but perhaps it is still early in the pandemic to assess this definitively. It also looks likely that there are factors other than enhancement of ACE2 binding that determine susceptibility—as seen from Starr et al38 and Table 4, from our own inconclusive attempts at correlating free energy binding affinities using in silico methods (not shown), and from a more detailed in silico model by Piplani et al39 that counter-intuitively predicts lower binding free energy for dogs compared with more susceptible animals such as monkeys, hamsters, ferrets, cats, and tigers.10
Some mutations in the essential as well as enhancing positions can lead to other mutations. For example, N501Y, the key mutation common to the Alpha, Beta, and Gamma VOC, can lead to F501 with a further nucleotide change. Similarly, the enhancing E484K mutation can also lead to R484 or T484 with a further nucleotide change. We examined whether in situ observations are consistent with or contrary to our in silico predictions. Indeed, F501 was observed in Sweden (April 28, 2021), Germany (May 7, 2021), Mexico (June 22, 2021), and Colombia (June 30, 2021), once in each of these 4 countries, while the Y501 has been observed in these countries since March 12, 2020; October 21, 2020; January 31, 2021; and September 19, 2020, respectively. The United Kingdom reported E484R in August 2020, followed by Angola in April 2021, Brazil and the United States in May 2021, South Korea in June 2021, and South Africa in July 2021. In each case, the E484K was detected prior to E484R—the former mutation circulating in the United Kingdom, Angola, Brazil, United States, South Korea, and South Africa from April, August, April, March, December, and August 2020, respectively. Similarly, E484T was only recently detected in the United States in June 2021, 15 months after the first report of E484K in that country. All 11 instances are thus consistent with our prediction—whether this link is causal or a coincidence is worthy of investigation with local epidemiological data. Although bioinformatics tools can provide useful insights, only 1 sample out of 53 COVID-positive cases is on average sent for virus genome sequencing (as of October 4, 2021), with huge variations across time and locations and a lot of missing meta-data.40,41 This means we are more confident about ruling in (eg, when a variant has been detected in a location) than ruling out the possibility of a mutation circulating purely based on in silico data, even if the latter is statistically large (4 million as of October 4, 2021).
Our analysis is not just of theoretical interest; it has huge practical applications because mice can be kept as pets or come into contact with other pets like cats, which are known to be susceptible. Also, mouse plague can occur in areas of COVID-19 outbreaks or endemicity, as is currently the case in New South Wales and adjacent states of Australia.42 To help public health and animal health professionals, Figure 3a and b show the 11 countries where the key essential and enhancing mutations listed in Tables 1 and 4 (viz. N501Y, E484K, and K417N/T) have co-occurred. The underlying raw data, down to regional counts for these combinations, are available.43 We believe that this information will help locate areas at risk (especially in Brazil, Chile, Djibouti, Haiti, Malawi, Mozambique, Reunion, Suriname, Trinidad and Tobago, Uruguay, and Venezuela) for appropriate mitigation measures.
Figure 3.
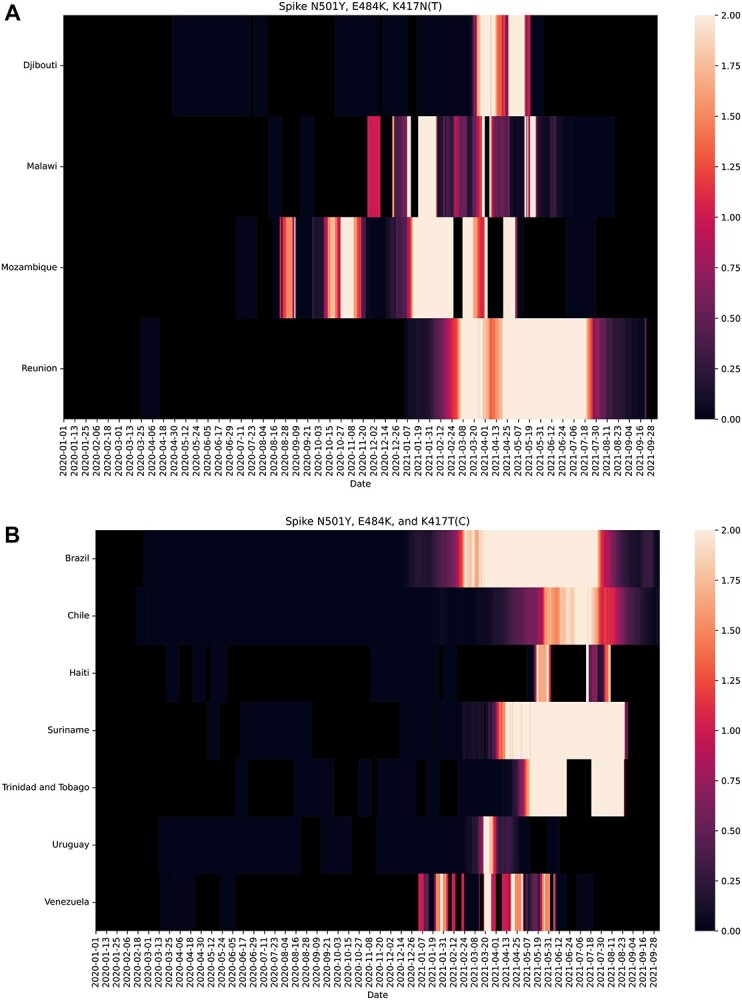
Significant occurrences on GISAID of (a) N501Y, E484K, and K417N (nucleotide change G22813T) triple mutations and (b) N501Y, E484K, and K417T triple mutations at the virus receptor binding domain (RBD) since the start of the COVID-19 pandemic (December 31, 2019). These countries are encouraged to perform targeted field surveillance.
CONCLUSION AND FURTHER ANALYSES
Assessing the risk of viruses adapting to new hosts requires careful interpretation of all available data from in silico, in vivo, in vitro, and in situ sources. Understanding host adaptation at a molecular level via modeling helps reconcile seemingly conflicting, experimental, and clinical observations while a pandemic is still in progress. We have demonstrated this with the SARS-CoV-2 virus adapting to mice. Our conclusions come with humility, because they are based on best available evidence up to this point but allow us and others to refine when more evidence becomes available. Armed with our collective understanding from different approaches and bolstered by bioinformatics and emerging artificial intelligence technologies such as AlphaFold, we have shown how to position ourselves better to predict and mitigate virus host adaptations, not just for this pandemic but also for future disease-X. Further analyses pertaining to COVID-19 should focus on the role of mutations beyond the spike and RBD; experimentally prove that the Omicron variant of concern can infect WT mice; improve computational modeling of binding affinities to explore correlations with susceptibility, if any; assess the actual risk of transmission for this virus through aerosol vs other routes; and study other hosts such as rats and other potential reservoir species (even those that previously exhibited low receptor activities) that will be hard to vaccinate or control.35,36,44
SUPPLEMENTARY METHODS FOR MOLECULAR MODELING
Molecular simulations were performed using NAMD2.1445 with CHARM36m46 forcefield employing a “TIP3” water model. The SARS-CoV-2 spike/ACE2 model was a homology model based on one of the best protein database structures available at the time of our analysis, viz. 6 M17, which is deemed to be of sufficient quality for our purpose.15 Variant models of the SARS-CoV-2 spike RBD domain (residues 330 to 530) containing the mouse adapted mutations were constructed by mutating residues in the NAMD build scripts, but later found to be in very close agreement with the same model constructed using AlphaFold16 (less than 0.9 Å RMSD difference to C-Alpha backbone atoms). A truncated mACE2 consisting of residues 19 to 600 was built using Swiss modeller,47 and similarly found to be in close agreement with the equivalent AlphaFold model. Amino acid side chain conformations predicted by AlphaFold were used in all initial RBD model starting conformations. Glycosylation of the spike and mACE2 protein was manually constructed using Visual Molecular Dynamics (VMD). Simulations were run with Periodic Boundary Conditions “PBCs” using the “NPT” isothermal-isobaric ensemble at 310 K and 1 bar pressure employing Langevin dynamics. The PBCs were constant in the XY dimensions. Long-range Coulomb forces were computed with the Particle Mesh Ewald method with a grid spacing of 1 Å. 2 fs timesteps were used with non-bonded interactions calculated every 2 fs and full electrostatics every 4 fs while hydrogens were constrained with the “SHAKE” algorithm. The cut-off distance was 12 Å with a switching distance of 10 Å and a pair-list distance of 14 Å. Pressure was controlled to 1 atmosphere using the Nosé-Hoover Langevin piston method employing a piston period of 100 fs and a piston decay of 50 fs. Trajectory frames were captured every 100 ps. Eleven variant models were constructed representing the mouse-adapted variants observed in Table 1 as well as the original Wuhan RBD model with both mouse and hACE2. Models were simulated for 300 nanoseconds. Trajectories were visualized with VMD and Nanome. Modelling data shall be made available on the CSIRO Data access portal (https://data.csiro.au/).
Supplementary Material
Acknowledgments
This work was supported by funding (Principal Investigator: S.S.V.) from Australia’s Department of Finance, CSIRO Future Science Platforms, National Health and Medical Research Council (MRF2009092), and United States Food and Drug Administration (FDA) Medical Countermeasures Initiative contract (75F40121C00144). The article reflects the views of the authors and does not represent the views or policies of the funding agencies including the FDA. We are grateful for support from our colleagues at the Australian Centre for Disease Preparedness (https://www.grid.ac/institutes/grid.413322.5) (especially Simran Chahal, Trevor Drew, Alexander McAuley, and Nagendrakumar Singanallur) and the Transformational Bioinformatics Group (especially Denis Bauer, Yatish Jain, Brendan Hosking, and Aidan Tay). L.O.W.W. acknowledges grant funding from the Australian Academy of Science and Australia’s Department of Industry, Science, Energy and Resources. The title is from the poem “To a Mouse: On Turning Her up in Her Nest, with the Plough, November 1785” by Scotland’s national poet, Robert Burns, in which he says that the mouse is not alone in proving foresight may be vain as the best-laid schemes of mice and men go oft awry (But, Mousie, thou art no thy-lane, In proving foresight may be vain: The best-laid schemes o’ Mice an’ Men Gang aft agley).
Contributor Information
Michael J Kuiper, CSIRO Data61, Dockands, Victoria, Australia.
Laurence O W Wilson, CSIRO Transformational Bioinformatics Group, North Ryde, New South Wales, Australia.
Shruthi Mangalaganesh, Monash University, Clayton, Victoria, Australia; CSIRO Australian Centre for Disease Preparedness, Geelong, Victoria, Australia.
Carol Lee, CSIRO Transformational Bioinformatics Group, North Ryde, New South Wales, Australia.
Daniel Reti, CSIRO Transformational Bioinformatics Group, North Ryde, New South Wales, Australia; Centre for Population Genomics, Garvan Institute of Medical Research, Sydney, New South Wales, Australia; Murdoch Children’s Research Institute, Melbourne, Victoria, Australia.
Seshadri S Vasan, CSIRO Australian Centre for Disease Preparedness, Geelong, Victoria, Australia; Department of Health Sciences, University of York, York, UK.
Author contributions
Conceptualization, methodology, and funding acquisition, S.S.V.; in silico analysis, M.J.K.; in vitro analysis, M.J.K. and S.S.V.; in vivo analysis, S.M. and S.S.V.; in situ analysis, L.O.W.W., D.R., and C.L.; writing: original draft preparation, S.S.V, M.J.K., and S.M.; writing: review and editing, all authors.
Potential conflicts of interest. All authors: No reported conflicts.
References
- 1. Dong E, Du H, Gardern L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020; 20(5):533–4. 10.1016/S1473-3099(20)30120-1. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauer DC, Tay AP, Wilson LOW, et al. . Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS-CoV-2 outbreak. Transbound Emerg Dis. 2020; 67(4):1453–62. 10.1111/tbed.13588. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lassaunière R, Fonager J, Rasmussen, M, et al. . Working paper on SARS-CoV-2 spike mutations arising in Danish mink, their spread to humans and neutralization data. 2020. https://files.ssi.dk/Mink-cluster-5-short-report_AFO2. Accessed December 1, 2020.
- 4. Korber B, Fischer WM, Gnanakaran S, et al. . Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020; 182(4):812–27. 10.1016/j.cell.2020.06.043. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J, Cai Y, Xiao T, et al. . Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021; 372(6541):525–30. 10.1126/science.abf2303. Accessed May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu H, Chen Q, Yang G, et al. . Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020; 369(6511):1603–1607. 10.1101/2020.05.02.073411v1. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dinnon KH, Leist SR, Schäfer A, et al. . A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020; 586:560–6. 10.1038/s41586-020-2708-8. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leist SR, Dinnon KH, Schäfer A, et al. . A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020; 183:1070–85. 10.1016/j.cell.2020.09.050. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan L, Tang Q, Cheng T, et al. . Animal models for emerging coronavirus: progress and new insights. Emer Microb Infect. 2020; 9(1):949–61. 10.1080/22221751.2020.1764871. Accessed May 15, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muñoz-Fontela C, Dowling WE, Funnell SGP, et al. . Animal models for COVID-19. Nature. 2020; 586:509–15. 10.1038/s41586-020-2787-6. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeiss CJ, Compton S, Veenhuis RT. Animal models of COVID-19. I. Comparative virology and disease pathogenesis. ILAR J. 2021; ilab007. 10.1093/ilar/ilab007. Accessed May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veenhuis RT, Zeiss CJ. Animal models of COVID-19 II. Comparative immunology. ILAR J. 2021; ilab010. 10.1093/ilar/ilab010. Accessed May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rathnasinghe R, Jangra S, Cupic A, et al. . The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. 20212021.01.19.21249592; 10.1101/2021.01.19.21249592. Accessed February 1, 2021. [DOI] [Google Scholar]
- 14. Yao W, Wang Y, Ma D, et al. . Circulating SARS-CoV-2 variants B.1.1.7, 501Y.V2, and P.1 have gained ability to utilize rat and mouse Ace2 and altered in vitro sensitivity to neutralizing antibodies and ACE2-Ig. bioRxiv. 20212021.01.27.428353. 10.1101/2021.01.27.428353. Accessed May 1, 2021. [DOI] [Google Scholar]
- 15. Yan R, Zhang Y, Li Y, et al. . Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020; 367:1444–8. 10.1126/science.abb2762. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021. 10.1038/s41586-021-03819-2. Accessed July 20, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McAuley AJ, Kuiper MJ, Durr PA, et al. . Experimental and in silico evidence suggests vaccines are unlikely to be affected by D614G mutation in SARS-CoV-2 spike protein. Npj Vaccines. 2020; 5:96. 10.1038/s41541-020-00246-8. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riddell S, Goldie S, McAuley A, et al. . Live virus neutralisation of the 501Y.V1 and 501Y.V2 SARS-CoV-2 variants following INO-4800 vaccination of ferrets. bioRxiv. 20212021.04.17.440246. 10.1101/2021.04.17.440246. Accessed May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tai W, He L, Zhang X. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020; 17:613–20. 10.1038/s41423-020-0400-4. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974; 185(4154):862–4. 10.1126/science.185.4154.862. Accessed December 1, 2020. [DOI] [PubMed] [Google Scholar]
- 21. Sun S, Gu H, Cao L, et al. . Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Commun 2021; 12: 5654. Available at: https://www.nature.com/articles/s41467-021-25903-x. Accessed September 30, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fagre A, Lewis J, Eckley M, et al. . SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for spillback to new world rodents. PLoS Pathog. 2021; 17(5):e1009585. 10.1371/journal.ppat.1009585. Accessed May 24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Shuai L, Wang C, et al. . Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell. 2020; 11:776–82. 10.1007/s13238-020-00767-x. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Z, Zheng H, Lin H, et al. . Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. 2020; 94(17):e00790–20. 10.1128/JVI.00790-20. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou H, Chen X, Hu T, et al. . A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020; 30:2196–203. 10.1016/j.cub.2020.05.023. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang K, Zhang Y, Hui X, et al. . Q493K and Q498H substitutions in spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021; 67:103381. 10.1016/j.ebiom.2021.103381. Accessed May 24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muruato A, Vu MN, Johnson BA, et al. . Mouse adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. bioRxiv. 20212021.05.03.442357. 10.1101/2021.05.03.442357. Accessed May 15, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Huang K, Wang T, et al. . SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J Virol. 2021; 95(11):e02477–20. 10.1128/JVI.02477-20. Accessed May 24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Noorden R. Scientists call for fully open sharing of coronavirus genome data. Nature. 2021; 590:195–6. 10.1038/d41586-021-00305-7. Accessed March 1, 2021. [DOI] [PubMed] [Google Scholar]
- 30. Rambaut A, Loman N, Pybus O, et al. . Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological 2020. Available at: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563. Accessed March 1, 2021. [Google Scholar]
- 31. Li Q, Nie J, Wu J, et al. . SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021; 184(9):2362–71. 10.1016/j.cell.2021.02.042. Accessed May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montagutelli X, Prot M, Levillayer L, et al. . The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 20212021.03.18.436013. 10.1101/2021.03.18.436013. Accessed May 1, 2021. [DOI] [Google Scholar]
- 33. Faria NR, Claro IM, Candido D, et al. . Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological 2021. Available at: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586. Accessed March 1, 2021. [Google Scholar]
- 34. Lucaci AG, Zehr JD, Shank SD, et al. . RASCL: rapid assessment of SARS-COV-2 clades enabled through molecular sequence analysis and its application to B.1.617.1 and B.1.617.2. Virological 2021. Available at: https://virological.org/t/rascl-rapid-assessment-of-sars-cov-2-clades-enabled-through-molecular-sequence-analysis-and-its-application-to-b-1-617-1-and-b-1-617-2/709. Accessed May 29, 2021. [Google Scholar]
- 35. Wan Y, Shang J, Graham R, et al. . Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020; 94(7):e00127–0. 10.1128/JVI.00127-20. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Damas J, Hughes GM, Keough C, et al. . Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. PNAS. 2020; 117(36):22311–22. 10.1073/pnas.2010146117. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flocco MM, Mowbray SL. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol. 1994; 235(2):709–17. 10.1006/jmbi.1994.1022. Accessed December 1, 2020. [DOI] [PubMed] [Google Scholar]
- 38. Starr TN, Greaney AJ, Hilton SK, et al. . Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020; 182(5):1295–1310.e20. 10.1016/j.cell.2020.08.012. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piplani S, Singh PK, Winkler DA, et al. . In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Sci Rep. 2021; 11:13063. 10.1038/s41598-021-92388-5. Accessed October 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bauer DC, Metke-Jimenez A, Maurer-Stroh S, et al. . Interoperable medical data: The missing link for understanding COVID-19. Transbound Emerg Dis 2021; 68(4):1753–1760. 10.1111/tbed.13892. Accessed July 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Priyadarshini S. Massive coronavirus sequencing efforts urgently need patient data. Nature India (special issue #13 on COVID-19 crisis). 2020; 11–3. 10.1038/nindia.2020.75. Available at: https://www.natureasia.com/en/nindia/pdf/special-issues/13/Nature-India-COVID-19-Crisis.pdf, https://go.nature.com/2y7kUIw. Accessed December 1, 2020. [DOI] [Google Scholar]
- 42. New South Wales Government . Help for regional communities impacted by the mouse plague. 2021. Available at: https://www.nsw.gov.au/initiative/mouse-control-support-program. Accessed May 24, 2021.
- 43. Kuiper MJ, Wilson LOW, Mangalaganesh S, et al. But mouse you are not alone: On some severe acute respiratory syndrome coronavirus 2 variants infecting mice. bioRxiv. 2021; 8(4):455042. Available at: https://www.biorxiv.org/content/10.1101/2021.08.04.455042v2.supplementary-material. Accessed November 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao X, Chen D, Szabla R, et al. . Broad and differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J Virol 2020; 94(18):e00940–e00920. 10.1128/JVI.00940-20. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Phillips JC, Braun R, Wang W, et al. . Scalable molecular dynamics with NAMD. J Comput Chem. 2005; 26:1781–802. 10.1002/jcc.20289. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang J, Rauscher S, Nawrocki G, et al. . CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017; 14:71–3. 10.1038/nmeth.4067. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waterhouse A, Bertoni M, Bienert S, et al. . SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–303. 10.1093/nar/gky427. Accessed December 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



