Abstract
We identified an operon in Listeria monocytogenes EGD with high levels of sequence similarity to the operons encoding the OpuC and OpuB compatible solute transporters from Bacillus subtilis, which are members of the ATP binding cassette (ABC) substrate binding protein-dependent transporter superfamily. The operon, designated opuC, consists of four genes which are predicted to encode an ATP binding protein (OpuCA), an extracellular substrate binding protein (OpuCC), and two membrane-associated proteins presumed to form the permease (OpuCB and OpuCD). The operon is preceded by a potential SigB-dependent promoter. An opuC-defective mutant was generated by the insertional inactivation of the opuCA gene. The mutant was impaired for growth at high osmolarity in brain heart infusion broth and failed to grow in a defined medium. Supplementation of the defined medium with peptone restored the growth of the mutant in this medium. The mutant was found to accumulate the compatible solutes glycine betaine and choline to same extent as the parent strain but was defective in the uptake of l-carnitine. We conclude that the opuC operon in L. monocytogenes encodes an ABC compatible solute transporter which is capable of transporting l-carnitine and which plays an important role in osmoregulation in this pathogen.
The food-borne pathogen Listeria monocytogenes represents a problem for the food industry because it is found ubiquitously in nature, it can grow at refrigeration temperatures, and it is capable of growth over a wide range of osmotic pressures (8, 9). Like many organisms, L. monocytogenes responds to hyperosmotic stress by accumulating osmotically active compounds, which are not inhibitory to enzymatic processes within the cell; these compounds are the so-called compatible solutes. The accumulation of these solutes, termed osmoadaptation, serves to counteract the outward flow of water, thereby maintaining cell turgor (6, 10, 19). In L. monocytogenes, the compatible solutes betaine and carnitine are also thought to play a role in cold adaptation. The rate of betaine accumulation is found to be maximal at about 10°C, and growth is stimulated almost twofold at 4°C when betaine is included in the growth medium (20). Carnitine accumulates to higher levels (three- to fourfold) at 4°C than at 30°C (20) and is transported at significant rates at 7°C (31).
The compounds used by bacteria as compatible solutes include amino acids, such as proline and glutamate; quaternary amines, such as betaine and carnitine; sugars (e.g., trehalose); and small peptides (e.g., N-acetyl-glutaminyl-glutamine amide and prolyl-glycyl-glycine). From among these, L. monocytogenes is known to derive osmoprotective effects from betaine (N,N,N-trimethylglycine [20]), carnitine (β-hydroxy-γ-[trimethyloammonio]butyrate [4]), and small proline-containing peptides (2). The transport of betaine and carnitine by L. monocytogenes has been studied in some detail. The extent of betaine accumulation is directly dependent on the osmolarity of the growth medium (20, 27, 28). The intracellular concentration of betaine increases approximately 20-fold when the external osmolarity is raised by 1.5 M with NaCl (20). The transport rate is osmotically activated and, at least in part, sodium driven (11). The transport of carnitine in L. monocytogenes is ATP dependent and is mediated by a high-affinity uptake system, with a Km of 10 μM and a Vmax of 48 nmol min−1 mg of protein−1 (31). This system appears to be specific for l-carnitine and the related compounds acetylcarnitine and butyrobetaine; the presence of either proline or betaine in excess has little effect on carnitine uptake (31). The accumulation of carnitine is influenced by the osmolarity of the medium, and uptake is subject to negative regulation by preaccumulated solute (32).
For other gram-positive organisms, such as Bacillus subtilis, Corynebacterium glutamicum, and Lactococcus lactis, a number of compatible solute transporters have now been well characterized. B. subtilis has at least five solute transporters (each given the designation Opu, for osmoprotectant uptake). OpuE is a single-component proline transporter and a member of the sodium/solute symporter family (34). OpuD is a single-component glycine betaine transporter (15). OpuA, OpuB, and OpuC are all closely related transporters which belong to the ATP binding cassette (ABC) superfamily of transporters and which can transport proline betaine and glycine betaine (OpuA); choline (OpuB); and ectoine, crotonobetaine, γ-butryobetaine, carnitine, choline-O-sulfate, choline, proline betaine, and glycine betaine (OpuC) (19). In C. glutamicum, BetP has been identified as the main glycine betaine transporter. It belongs to same family of transporters as OpuD from B. subtilis and CaiT (carnitine transport) and BetT (choline transport) from Escherichia coli (29). In a recent study, a glycine betaine transporter was identified for L. lactis and designated BusA (betaine uptake system). It is an ABC transporter which is closely related to the OpuA system from B. subtilis (26).
Despite several studies on the physiology of compatible solute uptake in L. monocytogenes, not until recently have genetic approaches led to the identification of two betaine transporters in this organism. BetL is a high-affinity secondary betaine transporter that is required for optimal growth in high-osmolarity medium (30). It is a member of a small family of transport proteins including BetT and CaiT from E. coli, BetP from C. glutamicum (29), and OpuD from B. subtilis (19). A second transporter, designated Gbu, was identified recently by screening of a mutant library for mutants defective in betaine-mediated osmoprotection (21). This transporter is encoded by three genes (gbuABC) which are closely related to the opuAA, opuAB, and opuAC genes of B. subtilis (18, 21). Recently, a membrane vesicle system was used to demonstrate that the Gbu transporter can be activated either by an osmotic gradient or by low temperatures (12). Gbu belongs to the ABC superfamily of transporters, which includes the ProU betaine transporter from E. coli. It is not yet clear whether the BetL and Gbu transporters represent the only betaine transporters in L. monocytogenes or whether one or more others remain unidentified.
No carnitine transporter has yet been identified at the genetic level for L. monocytogenes. Here we report the identification and characterization of an operon from L. monocytogenes encoding a new member of the ABC superfamily. We show that this operon is required for efficient growth under conditions of elevated osmolarity and demonstrate that a mutation in this operon leads to a defect in the ability to transport the solute l-carnitine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type L. monocytogenes serotype 1/2a strain EGD was used throughout. It was grown in brain heart infusion (BHI) broth (Difco), defined medium (DM) (2), or DM supplemented with 0.5% (wt/vol) type I peptone (Sigma) (DMP). Cultures were grown in 25 ml of medium (in 125-ml flasks) at 30°C with aeration. Cell growth was monitored spectrophotometrically (Ultrospec 4050; LKB, Biochrom) by measuring the optical density at 600 nm (OD600) of 1-ml samples taken at suitable intervals during growth. The opuC::pAULA mutant was maintained on BHI agar plates with 2 μg of erythromycin ml−1. In liquid medium, erythromycin selection was not used, as it was found to reduce the growth rate. PCR was used to confirm that the opuC::pAULA integration was stable even in the absence of selection. The E. coli strain DH5α was used routinely for cloning experiments. It was cultured in Luria broth at 37°C. Strains were maintained long term at −80°C in 1-ml aliquots after the addition of 7% (vol/vol) dimethyl sulfoxide (Sigma) to overnight cultures.
Mutant construction.
The opuCA gene was insertionally inactivated using the temperature-sensitive suicide vector pAULA. This 9.2-kb plasmid carries an erythromycin resistance marker, the pUC19 multicloning site, and a temperature-sensitive replication origin which enables chromosomal integration events to be selected at a nonpermissive temperature (7). Oligonucleotide primers directed against the opuCA gene were used to amplify a 400-bp region internal to the gene by PCR. The PCR primers (P121 [5′-TGACGGATCCACATCATGGCGGAAGATC-3′] and P117 [5′-TGACGGATCCGCTTCATCCATATCATGG-3′]) used included at their 5′ ends BamHI recognition sites (underlined) which facilitated the cloning of the PCR product into BamHI-digested pAULA. The resulting clone, designated pCOB26, was isolated from E. coli DH5α and used to electrotransform L. monocytogenes EGD. Chromosomal integration of pCOB26 was selected by repeated plating at 42°C with selection for erythromycin (2 μg ml−1) resistance as described previously (7). The integration of pCOB26 into the opuC locus in EGD was confirmed by PCR, and the resulting strain was designated EGD opuC::pAULA. The vector-specific For (position −20) and Rev primers (5′-TGTAAAACGACGGCCAGT-3′ and 5′-CAGGAAACAGCTATGAC-3′, respectively) were used to confirm the absence of independently replicating pCOB26 in the integrant strain. The PCR primers P125 and P114 (5′-CACACGTGCCACAAGTACG-3′ and 5′-CACGAGTAACAATTCCGACAAG-3′, respectively), which amplify the entire wild-type opuCA gene, were used to confirm the insertion of pAULA in the integrant strain (no PCR product was detected from the integrant strain). The primers P125 and For were used for further confirmation of this integration (a PCR product was detected only from the integrant strain).
The presence of a transcriptional terminator downstream from the erythromycin resistance gene on pAULA (as well as the fact that pAULA is 9.2 kb), which is oriented with the direction of opuCA transcription, suggests that the insertion mutation is polar, thereby inactivating the entire opuC operon. The stability of the pAULA insertion was confirmed by PCR analysis of cultures grown without erythromycin selection at 30°C. Even after repeated subculturing, no plasmid excision was detected.
Cloning and sequencing.
A 4.3-kb HindIII fragment from L. monocytogenes ScottA (which was being investigated because a Tn917 insertion conferring acid sensitivity mapped to this region) was sequenced and found to carry two full open reading frames (ORFs) and a partial ORF spanning 2.3 kb at the 3′ end of the fragment. These ORFs showed high levels of sequence similarity to the opuC and opuB operons of B. subtilis. The sequence obtained from the 4.3-kb clone was used to design PCR primers for the amplification of this region from L. monocytogenes EGD. The remainder of the operon was amplified by inverse PCR. Southern analysis of the opuC region in L. monocytogenes EGD indicated that the 3′ end of the operon was carried on a 4.2-kb ClaI fragment (data not shown). Chromosomal DNA isolated from EGD was digested with ClaI, purified on a Wizzard column (Promega), and ligated with T4 DNA ligase (Roche). The ligated chromosomal DNA was then used as a template for PCR with oligonucleotide primers directed against the known opuC sequences. The 2.5-kb inverse PCR product obtained was sequenced by primer walking. The sequence was obtained from both strands using a BigDye Terminator cycle sequencing kit (PE Applied Biosystems). The DNA sequence was determined using a Perkin-Elmer Applied Biosystems 377 automated sequencer. The DNA sequence obtained was analyzed using DNASTAR Inc. software on a Power Macintosh computer.
Betaine and carnitine transport assays.
The uptake of betaine and carnitine was measured using the method of Verheul et al. (31), modified as follows. Cells were grown overnight in DMP supplemented with 0.04% (wt/vol) glucose. Glucose (0.4% [wt/vol]) was added to the overnight culture, and growth was continued to an OD600 of 0.4. This culture was then used to inoculate 100 ml of DMP (0.5% [vol/vol] inoculum). Cells were grown at 30°C with shaking to mid-exponential phase (OD600 = 0.4). Cells were harvested by centrifugation for 10 min at 12,000 rpm and 4°C. The supernatant was removed, and cells were washed twice with ice-cold assay buffer (50 mM potassium phosphate [pH 6.9], 5 mM magnesium sulfate) containing chloramphenicol (50 μg ml−1). For studying uptake under high-osmolarity conditions, 0.5 M NaCl was added to the assay buffer. After being washed, cells were resuspended in the assay buffer to an OD600 of 1. The transport assay was performed with 5 ml of cell suspension in the presence of 0.4% (wt/vol) glucose at 30°C with stirring. Radiolabeled l-[14C]betaine (ICN) or l-[3H]carnitine hydrochloride (Amersham) was added to a final concentration of 20 μM. Samples of 100 μl were removed, immediately filtered through glass microfiber filters (Whatman International Ltd.), and then washed twice with 2 ml of ice-cold buffer. Filters were dried, and the radioactivity was determined by scintillation counting. Samples (100 μl) were removed and transferred to filters, without filtering, and dried as standards for determination of betaine or carnitine specific activity. The total cell protein values used to calculate solute pools were 170 μg ml−1 per OD600 unit at low osmolarity and 130 μg ml−1 per OD600 unit at high osmolarity, as previously described (2).
Measurement of steady-state solute pools.
Steady-state carnitine or betaine pools were measured using the method of Koo and Booth (22). Cells were grown in DMP as described above for the uptake assays. Cultures (25 ml) in DMP (0.4% [wt/vol] glucose) containing 200 μM betaine, carnitine, or choline were grown to an OD600 of 0.2. A 2.5-ml aliquot was transferred to a sterile 25-ml test tube containing 10 nCi of l-[14C]betaine, 30 nCi of l-[3H]carnitine, or 600 nCi of [14C]choline (Amersham, Life Sciences). Incubation was continued at 30°C with shaking. When growth in the control flask reached an OD600 of 0.4, the test tube was removed from the incubator. Three 0.5-ml samples were taken from the test tube, filtered through glass microfiber filters, and washed with 3 ml of DMP (lacking betaine, carnitine, or choline). The filters were dried, and the radioactivity was determined by scintillation counting. The remaining 1 ml of culture in the test tube was transferred to a cuvette, and the OD600 was measured. Then, 50-μl samples were removed from the cuvette and transferred to filters, without filtering, and dried as standards for determination of betaine, carnitine, or choline specific activity. Final solute pool values were corrected for background radioactivity (i.e., radioactivity which was associated with the cells and which was not removed during the washing step) as follows. Cells were grown to an OD600 of 0.4 as described above, and then 2.5 ml was permeabilized by the addition of 5% (vol/vol) butanol and incubation at 30°C for 30 min. Radiolabeled solute was added to the cells, and incubated was continued for a further 30 min. Samples were removed, filtered, and washed as described above for nonpermeabilized cells. Background radioactivity was measured by scintillation counting, and the values were used to correct the final solute pool calculations. The total cell protein values used to calculate uptake rates were 170 μg ml−1 per OD600 unit at low osmolarity and 130 μg ml−1 per OD600 unit at high osmolarity, as previously described (2).
Nucleotide sequence accession number.
The GenBank accession number for the sequence described in this paper is AF249729.
RESULTS
Identification of the L. monocytogenes opuC operon.
A newly identified ORF was being investigated in L. monocytogenes ScottA. This ORF was cloned on a 4.3-kb HindIII fragment, and the entire region was sequenced (C. P. O'Byrne, unpublished data). Downstream of the locus being investigated, two full ORFs and a partial ORF with high levels of sequence homology to the opuC and opuB operons of B. subtilis were identified. These ORFs were further investigated, as they represented ORFs for a newly identified putative solute transporter in L. monocytogenes. The opuC and opuB operons of B. subtilis encode ABC transporters capable of transporting a variety of compatible solutes into the cell in response to increased osmolarity (15, 16, 17, 19, 24). The full sequence of the corresponding operon in L. monocytogenes EGD was obtained by a combination of PCR, inverse PCR, and primer walking as described in Materials and Methods. EGD was the strain of choice for this analysis, as the genome sequence for this strain is scheduled for public release by September 2000 (see http://www.pasteur.fr/recherche/unites/gmp/Gmp_projects.html#lm). The ScottA and EGD sequences were 94.5% identical over the first two ORFs, although this figure may be an underestimate of their relatedness, as the ScottA sequence was obtained from one strand only in this region and therefore may have contained sequencing errors.
Sequence analysis.
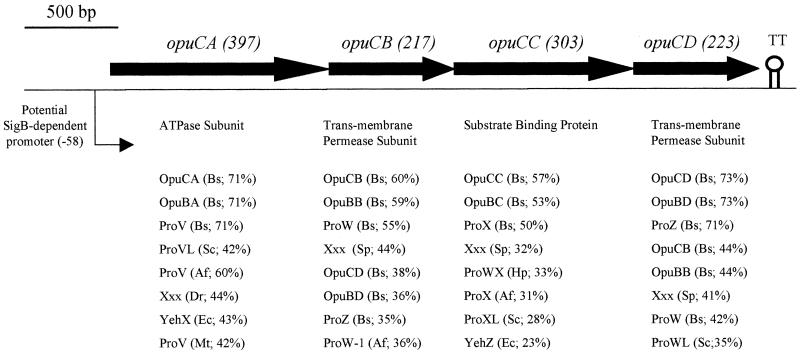
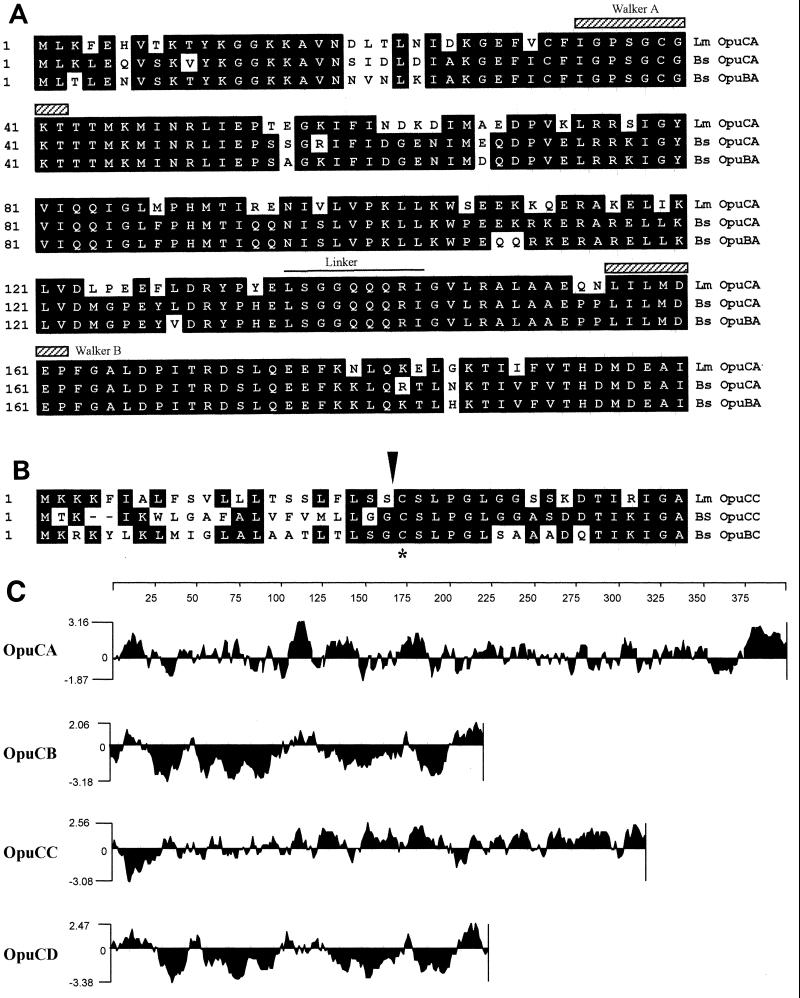
A 3,761-bp region of the EGD chromosome was sequenced (GenBank accession number AF249729), and four ORFs were identified (Fig. 1). ORF1, hereafter designated opuCA, is predicted to encode a hydrophilic protein of 397 amino acid residues (Fig. 2C) and with close sequence similarity (71% identical, 85% similar) to OpuCA from B. subtilis. In B. subtilis, OpuCA encodes an ATPase subunit associated with the OpuC ABC solute transporter. The Walker A and B motifs (35) and the linker peptide (5) are well conserved (Fig. 2A). The initiation codon of opuCA was identified as GTG, based on homology to the opuCA gene from B. subtilis and because this codon is preceded by a strong candidate Shine-Dalgarno sequence (−16 to −10; 5′-ATGGAGG-3′). A potential SigB promoter was identified 58 bp upstream from the initiation codon of ORF1 (5′-GTTTAA-N14-GGGAAA-3′), based on a comparison with the promoter sequences of genes known to be SigB regulated in B. subtilis and L. monocytogenes. The 3′ end of opuCA was found to be separated from the initiation codon of ORF2 by 6 bp, including the TAA stop codon.
FIG. 1.
Organization of the opuC operon in L. monocytogenes EGD depicted schematically. ORFs are shown as solid arrows. The numbers in parentheses above the ORFs indicate predicted sizes (in amino acid residues) of the protein products. A potential transcriptional terminator is indicated (TT). The potential SigB-dependent promoter is indicated by an angled arrow. The predicted amino acid sequence for each ORF was used to perform BLASTP searches (1) against the nonredundant databases (performed at http://www.ncbi.nlm.nih.gov/BLAST/). The eight proteins with the highest levels of sequence similarity (lowest E value [Expect value]) are listed beneath the corresponding ORF, followed in parentheses by the organism abbreviation and the percent identity. The organism abbreviations are as follows: Bs, B. subtilis; Mt, Mycobacterium tuberculosis; Ec, E. coli; Sp, Streptococcus pneumoniae; Dr, Deinococcus radiodurans; Af, Archaeoglobus fulgidus; Hp, Helicobacter pylori; Sc, Streptomyces coelicolor. Xxx, no assigned name.
FIG. 2.
Features of the OpuC protein subunits. (A) The first 200 amino acid residues of OpuCA from L. monocytogenes (Lm) were aligned by consensus with the corresponding N-terminal regions of OpuCa and OpuBA from B. subtilis (Bs), and identical residues are indicated by shading. Alignments were performed with a Clustal algorithm (14) using MegAlign software (DNASTAR Inc). The conserved “linker peptide” is overlined, and both Walker motifs (35) are overlined with hatched boxes. (B) The first 40 residues of the substrate binding protein (OpuCC) from L. monocytogenes (Lm) were aligned with the corresponding regions of OpuCC and OpuBC from B. subtilis (Bs). The predicted processing site for the pro-OpuCC lipoprotein is indicated with an arrowhead, and the conserved N-terminal cysteine residue is marked with an asterisk. (C) Kyte-Doolittle (23) hydrophobicity profiles for each of the four protein subunits of OpuC. The amino acid residue number is indicated above the plots. The profiles were obtained with a window of nine residues.
ORF2, designated opuCB, is predicted to encode a hydrophobic protein (Fig. 2C) of 218 residues and with strong sequence homology to the product of the opuCB gene of B. subtilis (60% identical, 80% similar). In B. subtilis, OpuCB is a membrane-spanning protein (217 amino acids long) which acts as a permease subunit in the OpuC solute transporter. The 3′ end of opuCB was separated from the initiation codon of ORF3 by 4 bp, including the TAA stop codon.
ORF3 is predicted to encode a hydrophilic protein (Fig. 2C) of 308 residues and closely related to the OpuCC protein from B. subtilis (57% identical, 75% similar). OpuCC is a 303-residue protein which is believed to act as an extracellular substrate binding subunit of the OpuC transporter. It has an N-terminal signal sequence with the characteristic features of lipoproteins. After processing, the N-terminal cysteine residue is tethered to the cell membrane via a lipid modification (17). The residues required for this modification are conserved in OpuCC from L. monocytogenes (Fig. 2B), indicating that this substrate binding protein is also likely to be tethered to the cytoplasmic membrane. The 3′ end of opuCC is separated from the initiation codon of the final ORF by 17 bp, including the TAA stop codon.
ORF4 encodes a hydrophobic polypeptide (Fig. 2C) predicted to be 223 residues long and to show a high level of sequence homology to the product of the corresponding gene of the opuC operon from B. subtilis, opuCD (73% identical; 89% similar). In B. subtilis, the OpuD protein is membrane associated and is believed to act as the second permease subunit in the OpuC solute transporter, in conjunction with OpuCB.
An inverted repeat sequence which could function as a rho-independent transcriptional terminator was identified 6 bp downstream from the stop codon of opuCD. The next ORF detected was on the same strand but was 112 bp downstream from the end of opuCD (data not shown). Together, these factors suggest that in L. monocytogenes, the opuCA, opuCB, opuCC, and opuCD genes are likely to form an operon which shares a high degree of sequence and organizational similarity with the opuC operon from B. subtilis.
Little is known about the regions immediately upstream and downstream of the opuC operon. The ORF immediately 5′ of opuCA in L. monocytogenes EGD, including the first 366 bp of opuCA, has recently been identified in a study aimed at identifying eukaryotic cell adhesion determinants (25). The gene, which is on the same DNA strand as opuCA, appears to play a role in adherence to the melanoma-derived SK-Mel 28 cell line (25). Downstream of the opuC operon (115 bp downstream from the opuCD stop codon), we have sequenced an incomplete ORF whose product has 50% sequence identity over 194 amino acid residues with a manganese transport protein from Xylella fastidiosa. This protein belongs to the natural resistance-associated macrophage protein (NRAMP) family of transporters, which includes several bacterial manganese transporters. The partial ORF is also found on the same DNA strand as the opuC operon.
Growth characteristics of an opuC mutant.
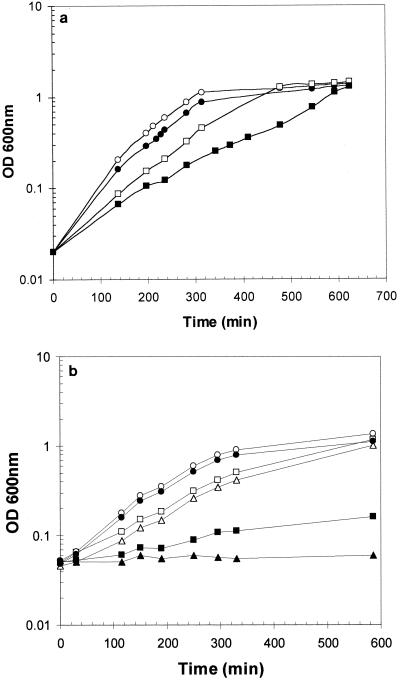
To test whether the opuC operon in L. monocytogenes played any role in osmoregulation, it was mutated by insertional inactivation of the opuCA gene as described in Materials and Methods. The mutant was found to grow normally in BHI broth (Fig. 3a). Supplementation of the medium with 4% NaCl led to a decrease in the growth rates of the wild type and the mutant. However, in the presence of added NaCl, the mutant was found to grow with a specific growth rate approximately half that of the wild type (0.41 and 0.90 h−1, respectively; Fig. 3a). In addition, the mutant formed very small colonies on BHI agar plates supplemented with 4% NaCl, whereas growth appeared normal on BHI agar plates without added NaCl (data not shown). Thus, inactivation of the opuC operon in L. monocytogenes led to reduced osmotolerance when cells were grown on BHI medium.
FIG. 3.
Growth of EGD (open symbols) and EGD opuCA::pAULA (solid symbols). (a) Cultures were grown in BHI in either the presence (squares) or the absence (circles) of 4% (wt/vol; 0.684 M) added NaCl. (b) Cultures were grown in DM without supplementation (triangles), with 0.5% (wt/vol) peptone (circles), or with 0.1% (wt/vol) Casamino Acids (squares).
Attempts to grow the opuC mutant strain in DM were unsuccessful, despite the fact that the wild-type parent grows well in this medium (Fig. 3b). The reason for this phenotype was unclear, but supplementation of the medium with 0.5% (wt/vol) peptone was found to enable the mutant to grow, whereas supplementation of DM with Casamino Acids (0.1% [wt/vol]) only partially rescued the growth defect (Fig. 3b). The osmoprotective effect of the peptides present in peptone (2) may contribute to this rescuing effect. Several peptides were examined in growth experiments to determine if any could individually restore the growth defect of the mutant in DM. None was found to relieve the growth defect to the same extent as peptone (data not shown). The growth defect in DM was not due to the presence of plasmid genes in the mutant strain (which were present as a result of the insertional inactivation using the suicide plasmid pAULA), as the wild-type strain transformed with the pAULA vector grew normally in this medium (data not shown). The growth defect was also unlikely to result from a secondary mutation, as several independently isolated integrants were found to display the same phenotype.
Carnitine transport is abolished in an opuC mutant.
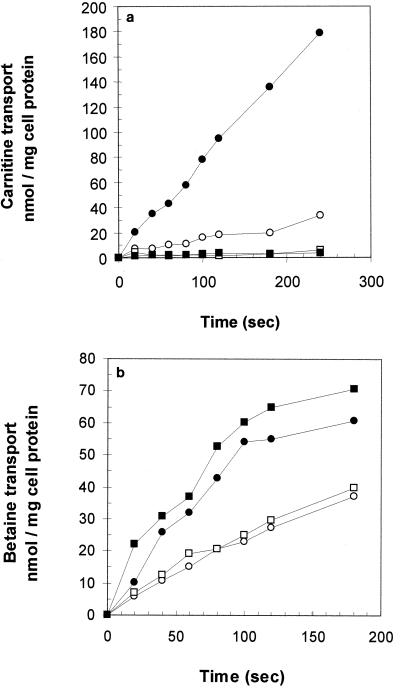
The close sequence similarity between OpuC from B. subtilis and OpuC from L. monocytogenes prompted us to investigate whether the L. monocytogenes transporter was capable of transporting carnitine (in B. subtilis, OpuC is the sole uptake route for this compatible solute). Previous studies had indicated that carnitine transport was ATP dependent, suggesting the involvement of an ABC transporter. The kinetics of carnitine transport were studied using cells grown in DMP and assayed for solute uptake in potassium phosphate buffer, with or without added NaCl (0.5 M), in the presence of the protein synthesis inhibitor chloramphenicol (50 μg ml−1). Under these conditions, the parent strain was found to transport l-carnitine at a rate of approximately 10 nmol min−1 mg of cell protein−1, and this rate increased to approximately 50 nmol min−1 mg of cell protein−1 when uptake was assayed in the presence of added NaCl (Fig. 4a). In contrast, the opuC mutant strain showed no detectable carnitine uptake over the same time course, either with or without added NaCl. These data indicate that carnitine transport is stimulated by hyperosmotic shock and that both basal transport and osmotically stimulated transport are abolished in a strain lacking the OpuC transporter.
FIG. 4.
Compatible solute uptake in EGD (circles) or in the opuCA::pAULA mutant derivative (squares). Cultures were grown in DMP prior to the assay. The assay was performed with potassium phosphate buffer with (solid symbols) or without (open symbols) 0.5 M added NaCl. The assay was performed in the presence of the protein synthesis inhibitor chloramphenicol (50 μg ml−1) and with 0.4% (wt/vol) added glucose. l-[3H]carnitine hydrochloride (a) or l-[14C]betaine (b) was added to a final concentration of 20 μM.
The transport of betaine, a related compatible solute, was also studied using this assay. In the absence of added NaCl, both the wild-type and the opuC mutant strains were found to accumulate betaine at approximately 15 nmol min−1 mg of cell protein−1 (Fig. 4b). When 0.5 M NaCl was added to the assay medium, the rate increased to approximately 35 nmol min−1 mg of cell protein−1. The presence of the opuC::pAULA insertion mutation therefore had no effect on the rate of betaine accumulation under these assay conditions, suggesting that OpuC is not a major route of betaine uptake in L. monocytogenes.
Steady-state solute pools in an opuC mutant.
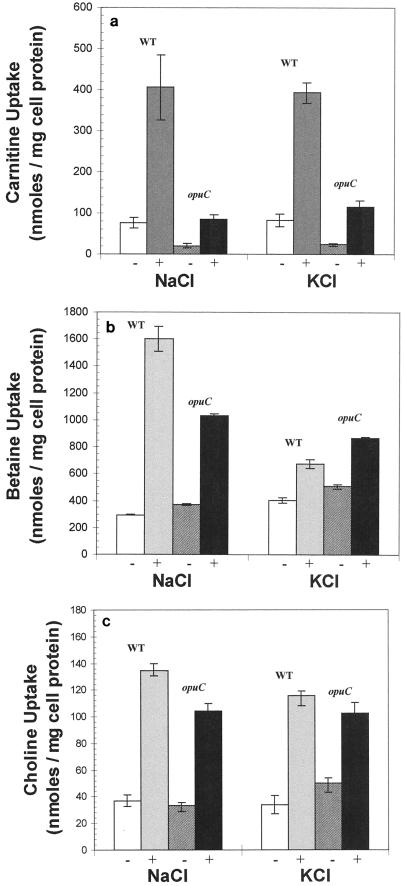
Steady-state measurements of carnitine, betaine, and choline cytoplasmic pools were also obtained with cultures grown in DMP. Cultures were grown to mid-exponential phase in the presence or absence of 0.5 M NaCl, and the steady-state accumulation of radiolabeled solutes was measured by removing samples for filtration and scintillation counting as described in Materials and Methods. When carnitine accumulation was examined, the mutant was found to accumulate only a fraction of the levels accumulated by the parent strain (Fig. 5a). Wild-type cells grown in DMP without the addition of 0.5 M NaCl accumulated approximately four times as much carnitine as opuC mutant cells. The addition of NaCl (0.5 M) stimulated carnitine accumulation to approximately 400 nmol mg of cell protein−1 in the wild type, whereas the level increased only to 80 nmol mg of cell protein−1 in the mutant (Fig. 5a). Almost identical data were obtained when KCl was used as the osmolyte. Together, these data indicate that OpuC is required for the accumulation of high levels of carnitine under conditions of osmotic stress. The data also suggest the presence of an alternative route for carnitine uptake.
FIG. 5.
Compatible solute pools measured at steady state. Intracellular pools of carnitine (a), betaine (b), and choline (c) were measured as described in Materials and Methods. Cells of EGD (WT) or the opuCA::pAULA mutant (opuC) were grown in DMP either with (+) or without (−) 0.5 M NaCl or 0.5 M KCl. The error bars represent the standard deviation from the mean (n = 3).
The parent strain (EGD) was found to accumulate betaine to approximately 1,600 nmol mg of cell protein−1 when grown in medium containing 0.5 M NaCl. This level was approximately fivefold higher than the level accumulated in the absence of added salt. Similarly, the opuC mutant strain accumulated betaine to a higher level when grown in the presence of 0.5 M NaCl, although there was only a threefold increase in the level compared to that in the untreated control (Fig. 5b). This difference in the betaine pools between the parent and the opuC mutant when cultures were grown in DMP containing 0.5 M NaCl was small but reproducible. It is possible that this difference is related to the apparent growth defect of the mutant in DM (Fig. 3b). The mutant may derive some nutrients (i.e., peptides or amino acids) from the peptone in DMP which allow it to overcome the apparent auxtrophy in DM. This accumulation of nutrients may alter the intracellular solute pools of the mutant enough to affect the betaine requirement and therefore the final betaine pool in this medium. When 0.5 M KCl was used as the osmolyte, only a slight increase in betaine accumulation was observed, and the parent and the mutant accumulated similar levels (Fig. 5b). This finding is consistent with the known stimulatory effect of Na+ on betaine transport in L. monocytogenes (11). Taken together with the kinetic data (Fig. 4b), these data suggest that in L. monocytogenes, OpuC does not play a major role in betaine accumulation.
Given the high levels of sequence homology between OpuC and OpuB in B. subtilis, it was also important to test whether OpuC in L. monocytogenes was capable of transporting choline, the only compatible solute known to be accumulated via OpuB in B. subtilis. We examined the steady-state accumulation of choline in the mutant and wild-type strains grown in DMP with or without the addition of 0.5 M NaCl (Fig. 5c). In the absence of added NaCl, both the mutant and the wild type accumulated choline at approximately 40 nmol mg of cell protein−1. In the presence of added NaCl, choline accumulation increased approximately threefold in both the wild type and the mutant. Similarly, when the osmolarity of the growth medium was raised by 0.5 M with KCl instead of NaCl, an increase in choline accumulation was detected for both strains (Fig. 5c). These data indicate that choline is accumulated to a low level (compared to carnitine and betaine; see Fig. 5a and b) by L. monocytogenes and that this uptake is subject to osmotic stimulation but is not dependent on the OpuC transporter. The accumulation of choline by L. monocytogenes has not previously been demonstrated, but the low level of transport is consistent with the fact that choline acts poorly as an osmoprotectant. Patchett et al. (27) reported that the presence of choline (1 mM) in plates prepared with DM and supplemented with 4% (wt/vol) NaCl provides only a slight osmoprotective effect.
DISCUSSION
Physiological studies of L. monocytogenes have previously shown that the uptake of carnitine is ATP dependent, and transport was therefore predicted to occur via an ABC transporter (31). Here we have confirmed that an ABC transporter (designated OpuC, based on its close homology to the corresponding transporter in B. subtilis) belonging to the binding protein-dependent subgroup is the major carnitine uptake system in L. monocytogenes EGD. The ATP binding domains (which include Walker motifs [35]), which are the most characteristic feature of the ABC transporters (5, 13), are well conserved in the OpuCA protein (Fig. 2A). OpuC appears not to play a role in the accumulation of betaine or choline, compatible solutes that are chemically related to carnitine. This notion is consistent with the finding that betaine and choline have little inhibitory effect on this transporter, even when present at a 100-fold excess (31). It is interesting that in B. subtilis, OpuC does have a role, albeit a minor one, in betaine accumulation (16). The small degree of sequence divergence between the three related transporters (L. monocytogenes OpuC, B. subtilis OpuC, and B. subtilis OpuB) is therefore enough to account for considerable diversity in the substrate specificity. The determinant of substrate specificity in binding protein-dependent ABC transporters remains unclear, but the extracellular substrate binding subunit as well as the permease subunits are likely to play a role (13). In this respect, it is perhaps significant that among the four transporter subunits, the greatest divergence is seen in the substrate binding protein (Fig. 1).
The rate of carnitine transport in L. monocytogenes EGD is shown here to be osmotically stimulated (Fig. 4a). This stimulation is not dependent on increased levels of OpuC expression, as the uptake assays were performed in the presence of chloramphenicol. The presence of an osmotic gradient is therefore sufficient to stimulate the activity of the carnitine uptake system in the cell. This finding is in contrast to that of an earlier study showing that the initial rate of uptake of carnitine or betaine was independent of the assay medium osmolarity for cells cultured in DM (32). In that study, the authors also showed that osmotic stimulation was observed only when the cells were allowed to preaccumulate either betaine or carnitine, suggesting that the accumulation of either compatible solute could inhibit the activity of the transporter(s) and that this inhibition could be overcome by hyperosmotic shock. In the present study, cells were grown in DMP prior to the assay of carnitine uptake. It is possible that the preaccumulation of peptides (supplied in peptone), known to have an osmoprotective effect on L. monocytogenes (2), leads to inhibition of the carnitine transporter(s) and that this inhibition is reversed by hyperosmotic shock. In order to test this idea, we examined rates of carnitine transport by wild-type cells cultured in DM. The initial uptake rate was approximately 110 nmol mg−1 min−1 (10-fold faster than in DMP), and this rate was found to be independent of the assay medium osmolarity (data not shown). It seems likely, therefore, that the activity of the carnitine transporter(s) is inhibited by preaccumulated peptides or amino acids from the DMP growth medium and that this inhibition can be overcome, at least partially, by osmotic stimulation. The mechanisms underlying this regulation remain to be elucidated.
The identification of a putative SigB-dependent promoter upstream from the opuCA gene may indicate a role for this stress-inducible (3) sigma factor in the regulation of opuC expression. The sequence identified (at position −58) both closely resembles the consensus −10 and −35 boxes identified for other SigB-dependent promoters (3, 34) and shows a good match to the conserved spacing (14 ± 1 bp) between these elements (Fig. 6). Sleator et al. (30) have also identified a potential SigB-dependent promoter upstream of the betL gene in L. monocytogenes. It is interesting that two genes in B. subtilis that encode compatible solute transporters are also preceded by potential SigB-dependent promoters: opuD, which encodes a betaine transporter, and opuE, which encodes a proline transporter (Fig. 6) (34). It is noteworthy that in L. monocytogenes, a sigB mutant is impaired in its ability to use both betaine and carnitine as osmoprotectants. In addition, betaine transport is defective in a sigB mutant (3), although the transport of carnitine has not been studied directly with a strain lacking this sigma factor. Becker et al. (3) have also shown that the transcription of sigB is strongly induced by elevated osmolarity. Taken together, these observations suggest that opuC may be regulated at the transcriptional level by SigB. We are currently investigating this hypothesis.
FIG. 6.
Alignment of the putative opuC SigB promoter with known SigB promoters from L. monocytogenes and B. subtilis. The sequence identified upstream from opuCA in L. monocytogenes is shown aligned with other known or predicted SigB promoter sequences. Genes indicated with an asterisk have a potential SigB promoter which has not yet been confirmed experimentally. Position is given as the distance of the final base shown in the −10 box from the initiation codon. The alignment of promoters was adapted and updated from the literature (3, 34).
Here we have identified the opuC operon initially on the basis of sequence homology with opuC in B. subtilis. A recent study has also identified the opuC operon in L. monocytogenes by screening a bank of insertion mutants to isolate a carnitine uptake mutant (C. Hill, personal communication). In that study, a mutant was selected that was unable to grow on high-osmolarity medium supplemented with carnitine. The chromosomal region surrounding the plasmid insertion in this strains was sequenced and found to be almost identical to the sequences described here for the opuCA and opuCB genes. It is interesting to note that in that study, the mutant strain was also found to have defective growth in the DM used (Hill, personal communication). The reason for this growth defect remains unclear. It is possible that the OpuC transporter is also capable of transporting some component of DM (e.g., an amino acid) and that the inactivation of OpuC therefore leads to some auxotrophy. Provision of this component in an alternative form in the added peptone (e.g., a short peptide) may be sufficient to overcome this auxotrophy. Further studies are under way to address this possibility.
Given the complexity of solute transport in the related gram-positive organism B. subtilis, where at least five solute transporters are known to be involved in osmoregulation, it seems possible that other compatible solute transporters remain to be identified in L. monocytogenes. Recently, another solute-transporting ABC transporter was identified in L. monocytogenes: the Gbu betaine transporter, which is encoded by only three genes and which is related to the OpuA transporter in B. subtilis (21). Evidence also exists to indicate that peptide transport in L. monocytogenes, which is known to relieve hyperosmotic stress (2), is also dependent on an ATP-dependent transporter (33). There is substantial sequence similarity between OpuC and Gbu as well as between OpuC and OpuA, and it is possible that these transporters have evolved from a common ancestor, as seems likely for OpuC and OpuB in B. subtilis (17). It is possible that other ABC solute transporters in L. monocytogenes are responsible for transporting other solutes or accumulating solutes under other growth conditions. In this respect, it is interesting to note that even in the opuC mutant, there is still a detectable carnitine pool (approximately 20 nmol mg of cell protein−1 compared to approximately 70 nmol mg of cell protein−1 in the parent). Furthermore, this residual level of accumulation can be stimulated by the addition of NaCl (approximately three- to fourfold) (Fig. 5a). These data suggest that in L. monocytogenes, an alternative route for carnitine accumulation must exist. The availability of the L. monocytogenes genome sequence, which is due to be released shortly (see http://www.pasteur.fr/recherche/unites/gmp/Gmp_projects.html#lm), should allow other potential solute transporters to be identified and classified for this important food-borne pathogen.
ACKNOWLEDGMENTS
We thank Colin Hill for sharing data prior to publication, Phil Carter for help with the DNA sequencing, and Ian Booth and Txaro Amezaga for helpful discussions. We also thank Trinad Chakraborty for generously providing the plasmid pAULA.
C.P. O'Byrne is supported by an ACT(R) University of Aberdeen fellowship. This work was funded in part by a Unilever Research CASE award.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amezaga M-R, Davidson I, McLaggan D, Verheul A, Abee T, Booth I R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology. 1995;141:41–49. doi: 10.1099/00221287-141-1-41. [DOI] [PubMed] [Google Scholar]
- 3.Becker L A, Sevket Cetin M, Hutkins R W, Benson A K. Identification of the gene encoding the alternative sigma factor ςB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180:4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beumer R R, Te Giffel M C, Cox L J, Rombouts F M, Abee T. Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl Environ Microbiol. 1994;60:1359–1363. doi: 10.1128/aem.60.4.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boos W, Lucht J M. Periplasmic binding protein dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1175–1209. [Google Scholar]
- 6.Booth I R, Pourkomailian B, McLaggan D, Koo S-P. Mechanisms controlling compatible solute accumulation: a consideration of the genetics and physiology of bacterial osmoregulation. J Food Eng. 1994;22:381–397. [Google Scholar]
- 7.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole M, Jones M, Holyoak C. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J Appl Microbiol. 1990;69:63–72. doi: 10.1111/j.1365-2672.1990.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 9.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galinski E A, Truper H G. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol Rev. 1994;15:95–108. [Google Scholar]
- 11.Gerhardt P N M, Smith L T, Smith G M. Sodium-driven, osmotically activated glycine betaine transport in Listeria monocytogenes membrane vesicles. J Bacteriol. 1996;178:6105–6109. doi: 10.1128/jb.178.21.6105-6109.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhardt P N M, Smith L T, Smith G M. Osmotic and chill activation of glycine betaine porter II in Listeria monocytogenes membrane vesicles. J Bacteriol. 2000;182:2544–2550. doi: 10.1128/jb.182.9.2544-2550.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 14.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 15.Kappes R M, Kempf B, Bremer E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol. 1996;178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappes R M, Bremer E. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and γ-butyrobetaine via the ABC transport system OpuC. Microbiology. 1998;144:83–90. doi: 10.1099/00221287-144-1-83. [DOI] [PubMed] [Google Scholar]
- 17.Kappes R M, Kempf B, Kneip S, Boch J, Gade J, Meier-Wagner J, Bremer E. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol Microbiol. 1999;32:203–216. doi: 10.1046/j.1365-2958.1999.01354.x. [DOI] [PubMed] [Google Scholar]
- 18.Kempf B, Bremer E. OpuA, an osmotically regulated binding-protein dependent transport system for the osmoprotectant in Bacillus subtilis. J Biol Chem. 1995;270:16701–16713. doi: 10.1074/jbc.270.28.16701. [DOI] [PubMed] [Google Scholar]
- 19.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolarity environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 20.Ko R, Tombras Smith L, Smith G M. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol. 1994;176:426–431. doi: 10.1128/jb.176.2.426-431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko R, Tombras Smith L. Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl Environ Microbiol. 1999;65:4040–4048. doi: 10.1128/aem.65.9.4040-4048.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo S P, Booth I R. Quantitative analysis of growth stimulation by glycine betaine in Salmonella typhimurium. Microbiology. 1994;140:617–621. doi: 10.1099/00221287-140-3-617. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Hansen J N. Characterization of a chimeric proU operon in a subtilin-producing mutant of Bacillus subtilis 168. J Bacteriol. 1995;177:6874–6880. doi: 10.1128/jb.177.23.6874-6880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milohanic E, Pron B, Berche P, Gaillard J-L the European Listeria Genome Consortium. Identification of new loci involved in adhesion of Listeria monocytogenes to eukaryotic cells. Microbiology. 2000;146:731–739. doi: 10.1099/00221287-146-3-731. [DOI] [PubMed] [Google Scholar]
- 26.Obis D, Guillot A, Gripon J-C, Renault P, Bolotin A, Mistou M-Y. Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactis reveals a new functional organization within bacterial ATP-binding cassette transporters. J Bacteriol. 1999;181:6238–6246. doi: 10.1128/jb.181.20.6238-6246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patchett R A, Kelly A F, Kroll R G. Effect of sodium chloride on the intracellular solute pools of Listeria monocytogenes. Appl Environ Microbiol. 1992;58:3959–3963. doi: 10.1128/aem.58.12.3959-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patchett R A, Kelly A F, Kroll R G. Transport of glycine-betaine by Listeria monocytogenes. Arch Microbiol. 1994;162:205–210. doi: 10.1007/BF00314476. [DOI] [PubMed] [Google Scholar]
- 29.Peter H, Burkovski A, Kramer R. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J Bacteriol. 1996;178:5229–5234. doi: 10.1128/jb.178.17.5229-5234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleator R D, Gahan C G M, Abee T, Hill C. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl Environ Microbiol. 1999;65:2078–2083. doi: 10.1128/aem.65.5.2078-2083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verheul A, Rombouts F M, Beumer R R, Abee T. An ATP-dependent l-carnitine transporter in Listeria monocytogenes ScottA is involved in osmoprotection. J Bacteriol. 1995;177:3205–3212. doi: 10.1128/jb.177.11.3205-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verheul A, Glaasker E, Poolman B, Abee T. Betaine and l-carnitine transport by Listeria monocytogenes ScottA in response to osmotic signals. J Bacteriol. 1997;179:6979–6985. doi: 10.1128/jb.179.22.6979-6985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verheul A, Rombouts F M, Abee T. Utilization of oligopeptides by Listeria monocytogenes ScottA. Appl Environ Microbiol. 1998;64:1059–1065. doi: 10.1128/aem.64.3.1059-1065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Blohn C, Kempf B, Kappes R M, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 35.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]