Abstract
Rat genomic tools have been slower to emerge than for those of humans and mice and have remained less thorough and comprehensive. The arrival of a new and improved rat reference genome, mRatBN7.2, in late 2020 is a welcome event. This assembly, like predecessor rat reference assemblies, is derived from an inbred Brown Norway rat. In this “user” survey we hope to provide other users of this assembly some insight into its characteristics and some assessment of its improvements as well as a few caveats that arise from the unique aspects of this assembly. mRatBN7.2 was generated by the Wellcome Sanger Institute as part of the large Vertebrate Genomes Project. This rat assembly has now joined human, mouse, chicken, and zebrafish in the National Center for Biotechnology Information (NCBI)’s Genome Reference Consortium, which provides ongoing curation of the assembly. Here we examine the technical procedures by which the assembly was created and assess how this assembly constitutes an improvement over its predecessor. We also indicate the technical limitations affecting the assembly, providing illustrations of how these limitations arise and the impact that results for this reference assembly.
Keywords: evaluation, genome assembly, Rattus norvegicus, reference
INTRODUCTION
A long-sought goal of the rat genomics community has been improved genomic resources equivalent to those available for the mouse and human. The creation of a new rat reference assembly using the same inbred Brown Norway strain that was used for prior rat reference assemblies offers an advanced resource for the investigation of rat genetics and genomics. We are pleased by and grateful for the accomplishment of the mRatBN7.2 assembly and are indebted to the scientists who committed material and intellectual resources and technical skills to its creation. The authors of this survey constitute a “users group” that includes rat geneticists, bioinformaticians, and sequencing experts who were not involved in its creation. Together we have been assessing this new reference assembly and note both important improvements as well as some unexpected features. Here we present a summary of the production process and the features of this new genome assembly and indicate its important new strengths as well as some outcomes that result from the technical methods used in the assembly. We anticipate that this increased insight into the properties of the assembly will increase its utility to the rat genomics community.
THE UTILITY OF RAT GENETIC MODELS UNDERPINS THE NEED FOR AN IMPROVED REFERENCE GENOME ASSEMBLY
The rat has played an important role in cardiovascular research because of the greater accessibility of its cardiovascular system to investigation than is possible in the much smaller mouse. Several rat models of cardiovascular disease, including high blood pressure, stroke, and hypertensive renal injury have been created. These models have resulted from selective breeding such that naturally occurring genetic variation has been fixed, resulting in disease traits that are usually polygenic, as they are in the human population. Five models of hypertension and hypertension-related end organ disease have been developed from Wistar stocks (spontaneously hypertensive rat, SHR; spontaneously hypertensive stroke-prone rat, SHRSP, Dahl, Milan, New Zealand, and Sabra), and have added to the discovery of genetic variation that contributes to hypertension-associated pathology including renal disease and stroke (1–10). Furthermore, the pathogenic mechanism arising from this disease-causing genetic variation can much more readily be functionally linked in animal models than in humans. For example, the mechanistic understanding of the role of ApoL1 variation in humans in creating risk of hypertension-associated renal disease has been advanced by manipulation of ApoL1 function in rodent models (11).
Rats also provide essential and widely used behavioral and drug addiction research models (12). In part, this is because the rat expresses a rich repertoire and complexity of behaviors, including social behaviors. This has permitted studies that have been insightful for modeling and understanding behavioral, psychiatric, and substance misuse in humans. Reflecting this, the National Institutes of Health (NIH) has released an RFA entitled “The Rat Opioid Genome Project” (RFA-DA-20-010). In addition, the NIH/National Institute on Drug Abuse (NIDA) Animal Genomics Program (PAR-21-244) has provided strong support for rat studies. Rat models in which natural genetic variation promotes disease risk phenotypes continue to be useful in cancer research (13, 14), and several models of autoimmune disease have been developed in rats (15, 16). Rat strains also provide an important model organism for metabolic syndrome, manifesting features such as glucose intolerance/insulin resistance/hyperglycemia, elevated triglycerides, hypertension, increased adiposity, and altered cholesterol to high-density lipoprotein ratios (17).
The creation of heterogenous stocks (HSs) derived by crossing eight inbred rat lines has created an important genetic resource now being exploited to link natural genetic variation to medically relevant traits (18, 19). By crossing these inbred founder strains meiotic recombination events result in individual HS lines that capture a unique combination of random mosaic chromosomal haplotypes from multiple founders. This allows a genome-wide association approach to be used in which traits are analyzed across multiple individuals. The presence of chromosomal marker associations with a trait can be made in these admixed populations. Genetic variants contributing to trait variance can be isolated for future identification in as few as 700 individuals using the HS approach (20). The rat is now being used to create a hybrid rat diversity panel (HRDP), created from 35 inbred rat strains and over 60 recombinant inbred lines created from two pairs (SHR/OlaIpcv x BN-Lx/Cub and F344/Stm x LE/Stm) of founders (21).
RECENT ADVANCES IN GENOME SEQUENCING AND ASSEMBLY AND THEIR APPLICATION TO THE BROWN NORWAY GENOME
The past 3 years have seen the introduction of advanced sequencing and assembly pipelines that can yield genome assemblies of extremely high contiguity and completeness at much lower cost than was previously accessible. Ultimately this has resulted in the completion of a “perfect” telomere to telomere assembly of a haploid human cell line derived from a hydatidiform mole, which includes accurate assembly of highly repetitive sequences at the telomeres and centromeres (22). A foundational advance that has driven improved genome assemblies, has been the introduction of long-read sequencing. This is possible as the result of single-molecule sequencing methods introduced by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). Although they utilize different sequencing modalities, both are remarkable in being able to sequence across far greater lengths than previously attainable. Long reads offer the attraction of simpler assembly tasks (bigger jigsaw puzzle pieces capable of spanning complex and repetitive regions that confound short read assembly) and so provide much better assembly contiguity and much lower rate of misassemblies. An important limitation is the high error rate associated with both methods compared with short-read next-generation sequencing technologies. However, innovations such as PacBio circular consensus sequencing (CCS; HiFi) of genomes and ongoing improvements in ONT technology are reducing error rates and improving base-level accuracy of genome assemblies derived from long reads.
THE NEW RAT REFERENCE GENOME, mRatBN7.2
As a result of initiatives proposed by investigators associated with the Rat Genome Database at the Medical College of Wisconsin, the genome of the rat has been sequenced and assembled as part of the Vertebrate Genomes Project (VGP). This project aims to generate high-quality sequence assemblies of all extant vertebrate species (23). The material submitted was derived from a close male relative of the original female Brown Norway rat (BN/NHsdMcwi) used for earlier assemblies. This strain has been brother-sister mated since 1958 to produce a fully inbred, haploid genome. The inbreeding state of the genome was initially confirmed in 2000 when the heterozygosity rate of 200 genome-wide microsatellite markers was observed to be nil (24). In 2008, a panel of 17,006 rat single nucleotide polymorphisms (SNPs) was genotyped in this strain, and the heterozygosity rate was so low that it approximated the error rate of the genotyping platform (25).
The recent mRatBN7.2 assembly was produced from PacBio continuous long reads (CLRs; 26) with 92x coverage depth of the genome. The assembly pipeline and major features have been reported by its creators (27). Assembly naming follows the VGP nomenclature: m indicating the class Mammalia; Rat indicating species (strictly following VGP standards should require RatNor); BN indicates the strain, Brown–Norway; and 7 indicates succession to the Rnor_6 reference. The UCSC Genomics Institute has also adopted Rn7 to identify this reference. Assembly of the CLRs used an assembly pipeline incorporating the FALCON/FALCON unzip assembly software (28) whose procedures anticipate an outbred, diploid genome (27). Such an assembler is suitable for outbred genomes typical of the VGP, which are often collected from wild-caught species. However, the fully inbred BN/NHsdMcwi has a haploid genome and other assemblers are more suitable for inbred strains (29). Coupled with error-prone CLRs, the assembler can sometimes mistake CLR sequence errors for the presence of two haplotypes (maternal and paternal) in the genome and this can contribute to misassemblies and the inclusion of base-level errors in the sequence. The result is that the assembler creates two outputs, a primary assembly (principal pseudohaplotype of diploid) and an alternate assembly (alternate pseudohaplotype of diploid). The presence of a primary assembly and an alternate assembly is a new occurrence for the rat reference. Many users may not fully understand the origin and significance of the alternate assembly.
The alternate assembly of mRatBN7.2 comprises a little over 10% of the size of the primary assembly (Table 1). Most of the alternate assembly can be aligned to the primary assembly and comprises ∼8,000 blocks of sequence termed “haplotigs” of 33 kb average length. Some regions of the genome have multiple haplotig alignments ranging up to sixfold depth of haplotig alignment for genome regions longer than 25 kb (Supplemental Table S1). These alternate haplotypes may contain sequences that represent inaccuracies in CLRs in which the diploid assembler has identified assembled sequences as related to the primary assembly, but arising from a different parental haplotype. However, as a fully inbred strain, Brown Norway has only a single parental haplotype.
Table 1.
Comparison of primary and alternative haplotypes of the mRatBN7.2 assemblies
| Value | |
|---|---|
| Primary haplotype assembly size | 2,647,915,728 |
| Alternate haplotype assembly size | 286,931,382 |
| Total alternate haplotype mapped to primary | 263,480,807 |
| Unique alternate haplotigs | 7,915 |
| Mean alternate haplotig length | 33,639 |
| Alignment depth range to assembled chromosomes | 0–6 |
Except for alignment depth, all values are in bases. Mapping of alternative to primary haplotype used Minimap2.
Assembling from CLRs can create difficulty in correctly assembling segmental duplications (SDs). Such duplications are an important means of genomic evolution. They may be especially important in immune gene evolution in which host-pathogen competition may be favored when evolution of the host responses begins with duplication of copies of existing genes. Segmental duplications most commonly occur in tandem and sometimes may have more than a single repetition. The alternative haplotype of mRatBN7.2 suggests, based on alignment depth of the alternative to primary assembly, that there exist regions of assembled chromosomes that may have been duplicated as many as five times over (i.e., 6 copies in total, Table 1). The FALCON/FALCON unzip assembler may not distinguish that the CLRs from such duplications represent separate authentic components of the genome. These are removed from the primary by a software tool that seeks to place in the primary assembly the best single representation of the duplicates (30). The output of this first step in the sequence assembly of CLRs are overlapping sequences termed contigs. In the VGP sequence assembly pipeline leading to mRatBN7.2, the assembler generated 757 contigs with an N50 (the size at which 50% of the entire genome is assembled in elements of this size or larger) of 29.2 Mbases (Mb).
The assembled contigs were extended and connected with three distinct scaffolding steps that seek to connect and correctly orient contigs: scaffolding by 10X Chromium linked reads, Bionano optical mapping (31), and Hi-C super scaffolding (32). Optical mapping provides a means of fluorescently tagging high molecular weight genomic DNA using a proprietary enzyme to couple a fluorescent tag to a six base sequence motif (33). This motif occurs ∼15 times per 100 kb. The physical distance between tags is estimated by passing the linearized DNA molecules through nanofluidic channels in a flow cell. The fluorescence data captured from these tagged molecules can then be used to make a genome assembly based on fluorescent marker overlaps which, in the case of the mRatBN7.2 assembly, had a N50 of 78.0 Mb. The assembled optical map proposes a complete genome size of 2,857 Mb. The mRatBN7.2 optical genome map represented the whole genome in 249 scaffolds with a read depth of tagged molecules of ∼140X. This large-scale scaffolding can then be used to place assembled contig sequences that are of a smaller scale onto the genomic optical map. This is achieved by in silico marking of the sequence-based contig to identify overlaps with the marks of the optical map. Hi-C is a chromosomal conformation analysis method that uses ligation to estimate the frequency of contact between different regions of a chromosome. These frequencies then can be used to infer positional relationships between sequences by taking advantage of the intrinsic ordering of DNA folding within the nucleus. Nearby sequences will be frequently found together, whereas distant sequences will not, thus providing an additional orthogonal parameter reflecting chromosomal position through sequence (34) that allows further correction and refinement of the assembly.
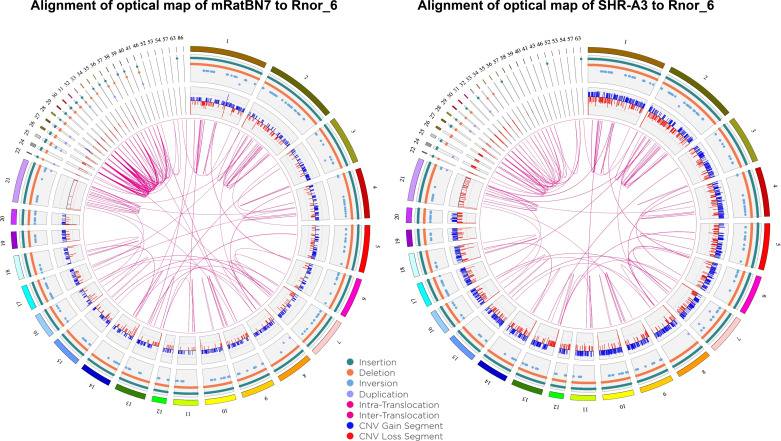
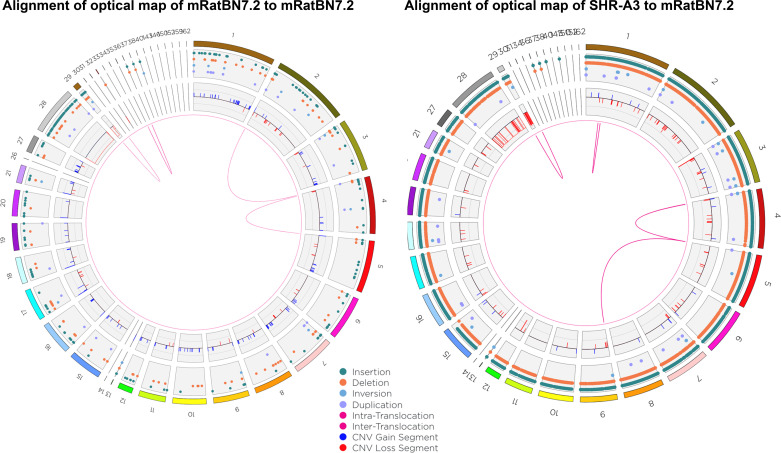
The use of these large-scale assembly methods has greatly enhanced the contiguity of the rat genome assembly and resulted in an mRatBN7.2 genome constructed from 176 scaffolds with a scaffold N50 of 135 Mb. In comparison, Rnor_6 comprised 1,395 scaffolds with an N50 of 15.0 Mb. Thus, the key characteristic of assembly contiguity is much improved. More importantly, the use of long reads contributed significantly to the correction of misassemblies, which have been previously identified to be present in Rnor_6 (35). These can be seen in Fig. 1 in which the optical map of mRatBN7.2 is aligned to the in silico optical map of Rnor_6 using the Bionano Solve software suite. Although the methods used to generate the optical map of mRatBN7.2 provides prima facie evidence of improved assembly over Rnor_6, the presence of similar misassemblies when the Bionano optical map of the SHR-A3 rat strain is aligned to Rnor_6 confirms that the assembly errors are in Rnor_6. Further, when optical maps of BN/NHsdMcwi and SHR-A3 (Rat Genome Database strain ID No. 8142383) are aligned to mRatBN7.2, the larger scale misassemblies are mostly lost (Fig. 2) and remaining differences may be predominantly due to authentic differences between the genomes. Similar observations were made for alignment to mRatBN7.2 of optical maps for SHR-B2 (ID No. 8142385), and Wistar Kyoto (WKY, ID No. 1581635) (data not shown).
Figure 1.
Circos plot in which the optical map of mRatBN7.2 is aligned to the in silico optical map of Rnor_6 using Bionano Solve software (left). The outer numbered bars are the rat chromosomes and unplaced scaffolds (22–63) of the Rnor_6 assembly. The key indicates the type and distribution of detected variation between the assemblies. The right-side circos plot shows the alignment to Rnor_6 of the optical map of SHR-A3. A similar pattern of variation is observed across the genome.
Figure 2.
Circos plot in which the optical map of mRatBN7.2 produced in the VGP is aligned to the in silico map of mRatBN7.2 assembly (left), and alignment of SHR-A3 optical map to the in silico map of mRatBN7.2 (right). Both alignments show much greater concordance. As expected, small-scale variations (insertions and deletions) are more notable in the SHR-A3/mRatBN7.2 comparison, whereas larger-scale structural variations are markedly reduced in both alignments. VGP, Vertebrate Genomes Project.
ASSESSMENT OF GENOME SEQUENCE ACCURACY
Potential errors in the base sequence accuracy of mRatBN7.2 have emerged as the new assembly has been scrutinized by rat geneticists. The first indication arose from observations that the new assembly contained a large number of SNP and short indel variants that were observed when mapping short read whole genome sequence (WGS) data from BN/NHsdMcwi, the reference strain. Our extended mapping to mRatBN7.2 of WGS from 38 inbred rats [sequencing data available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), SRA repository accession numbers are provided in Supplemental Table S2A, complete variant listing in Supplemental Table S2B] and including several BN/NHsdMcwi rats, revealed that there are 135,495 SNPs and indels that are shared by at least 35 of these strains. The most parsimonious explanation of such differences is that the SNP/indel variations are errors in mRatBN7.2 attributable to the error rate associated with CLRs. These errors may be one characteristic that contributes to the formation of alternative assemblies. The assembler may propose two possible haplotypes at a given locus, one of which may correctly (partially or fully) reflect the actual haploid sequence of the genome at this locus. The haplotype selected for the primary assembly may include sequencing errors, whereas the alternative haplotype at the locus may be correct, or vice versa. Alternatively, errors may be mixed so that within these homologous haplotigs there may be regions that are correct and other regions within the same haplotig that contain errors.
When we examined the relationship between the 135,495 variants between the 38 inbred strains and mRatBN7.2 and the variation that occurs between the primary and alternative “haplotypes,” we found that 44.3% of these variants occur at the same locations as a variation between the primary and alternative haplotypes. Thus, variants were identified in the genome at a rate of 1 variant per 4,553 bases in the ∼10% of the genome in which there is an overlapping alternative haplotype, but at a rate of 1 in 32,742 bases in the ∼90% of the genome that is not overlapped by the alternative haplotype.
LONG READ SEQUENCING AND BASE LEVEL ERRORS AFFECTING CODING GENES
The mRatBN7.2 reference assembly has been annotated using information from rat gene transcript sequences and gene models. The inclusion of the mRatBN7.2 assembly in the National Center for Biotechnology Information (NCBI) Genome Reference Consortium (GRC) now provides curation services to improve the reference. RefSeq gene annotation resulted in ∼210 coding sequence genes whose exons were disrupted in the new assembly and have been identified as requiring additional curation.
The relatively low accuracy of PacBio CLRs sequencing is not, in theory, an absolute barrier to accurate sequence assembly so long as errors are random, in which case sufficient depth of coverage should wash out many such errors. Error randomness is largely the case for CLRs, but there is an exception for errors within homopolymers (sequences comprised of repetitions of a single base) and two base microsatellite repeats (36, 37). We have investigated the unresolved issues identified by GRC (https://www.ncbi.nlm.nih.gov/grc/rat/issues) that disrupt genes to determine whether a coherent explanation could be obtained for disrupted genes. Our investigation began by using Minimap2 (38) to align the reads of the alternative haplotype to the primary haplotype. This allowed us to identify how and where in the genome the alternative assembly aligns. The most important finding is that almost all of the alternative assembly aligns well to the primary assembly, as would be expected if it comprised authentic alternative parental haplotypes. These alignments exist between both coding and noncoding regions of the genome. They also exist on the Y and X chromosomes from the male mRatBN7.2 genome. The alignment reveals the nature and distribution of sequence differences between the primary and alternative haplotypes. A list of positions in the primary assembly, at which variation is present between the primary and alternative assembly, is provided as supplemental data (Supplemental Table S3, alt_var.txt).
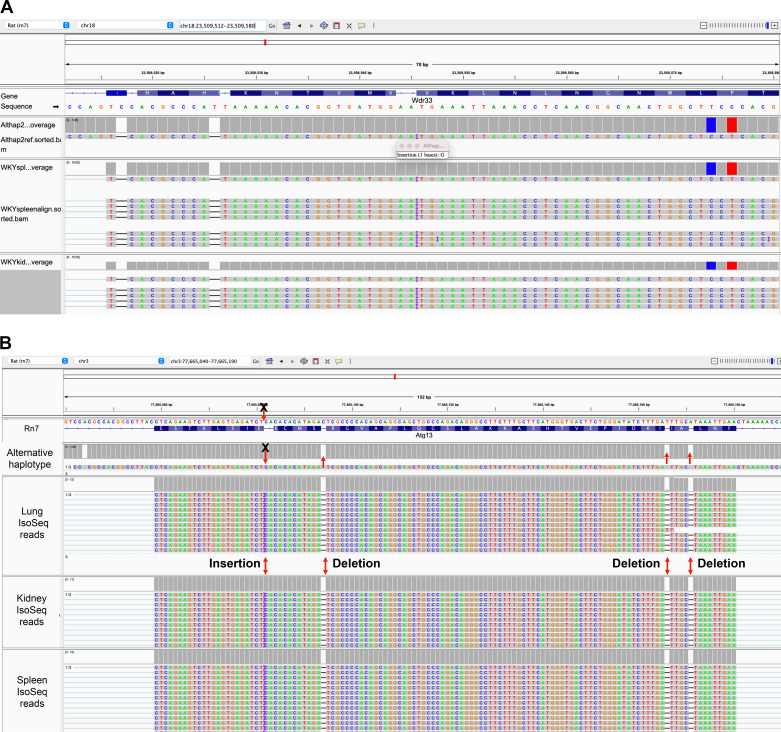
To clarify the role of sequencing errors in disrupting GRC gene annotations, we used single-molecule transcript expression data (PacBio IsoSeq) from several tissues representing the genomes of the Wistar-Kyoto and the SHR-A3 strains and aligned them to the new reference. Although these reads do not directly arise from BN and so would not identify genes that are actually disrupted in the BN genome, they can provide a means to reconcile differences between the BN assembly, the RefSeq sequence that has provided gene annotation of the assembly (which is often based on genomic sequence from prior BN reference assemblies and BN cDNA sequences) and the transcript sequence of other rat strains. Thus, a disrupted gene, which has an undisrupted RefSeq sequence and for which two other rat strains also lack disrupted coding sequences, is likely disrupted as a result of an error in the current assembly. The IsoSeq sequencing mode utilizes circular consensus sequencing (CCS), which provides higher sequencing accuracy than CLRs (37). A complete list of disrupted genes in mRatBN7.2 is provided by the NCBI Genome Reference Consortium and can be obtained at their website for the rat reference genome (https://www.ncbi.nlm.nih.gov/grc/rat/issues). Figure 3 shows examples of mRatBN7.2 assembly problems disrupting 2 genes. Disruptions of the annotated genes are indicated as gaps in the RefSeq gene annotation track. Alignment of the alternative assembly indicates the presence of variations that coincide with the error such that the alternative assembly would result in no gene disruption. The IsoSeq reads further resolve the reference sequence as containing likely errors. To systematize this analysis, we have examined GRC noted issues associated with genes on chromosome 1, and we find explanations for most that are similar to the examples in Fig. 3. Of 133 gene-specific issues identified from the GRC on chromosome 1, the affected genome locus was represented in the alternative haplotype in 119 of the issues. Available IsoSeq data was able to resolve 107 of these, with no IsoSeq data for 22 issues affecting 6 genes (Supplemental Table S4, Chr1_issues.xlsx). The resolved issues were largely attributable to homopolymer sequences, resulting in insertion/deletion errors. Some unresolved issues (i.e., affecting Dmbt1 and Ifit1) may arise from inaccuracies in the RefSeq gene sequence or the translation used for annotation rather than the assembly. The analyzed issues affect only coding exons, the same rate of base errors can be expected in intergenic and intragenic noncoding sequences, thereby affecting investigation of traits arising from noncoding variation.
Figure 3.
A: this exon of Wdr33 has three breaks, the leftmost is an insertion in the mRatBN7.2 sequence that is absent from the aligned alternative haplotig and the IsoSeq reads. The next is another mRatBN7.2 insertion absent from the alt haplotig. The next is a single base deletion in the reference that appears as an insertion in the alternative haplotig, and finally there are two synonymous SNPs shared by the alternative haplotype and the IsoSeq reads. B: the exon of Atg13 reveals an assembly wherein errors are present in both the primary and alternative assemblies. All IsoSeq reads indicate an insertion at the location of the leftmost disruption of the exon; however, this is present neither in the primary nor alternative haplotypes (this could constitute an actual mutation in BN since the IsoSeq reads do not come from BN). The remaining three disruptions of this exon are attributable to single-base insertions in the primary reference sequence. BN, Brown Norway; SNPs, single nucleotide polymorphisms.
SEGMENTAL DUPLICATIONS AND REPETITIVE PERI-CENTROMERIC SEQUENCES
Assembly from error-prone CLRs limits the ability to detect and assemble SDs. Segmental duplications are frequent in the human genome with over 200 Mb of the genome comprised of duplications > 20 kb that share 98% base level identity (36). The presence of collapsed duplications is apparent from increased read depth, and heterozygous variation across collapsed segments of the genome when sequencing reads are aligned to the reference. However, high rates of base-level errors relative to actual variation between the duplicates result in the assembler being unable to distinguish the constituent units of the SDs from one another. In the VGP assembly pipeline a software tool, purge_dups, was developed to identify such regions and to remove the excess read depth to the alternative haplotype leaving a span representing the best assembly of a single unit of the duplicated region (30).
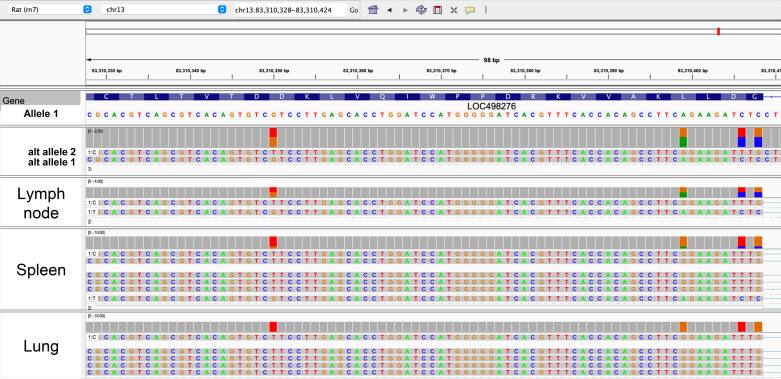
In Fig. 4, we provide an illustration of collapsed SD highlighting a region of mRatBN7.2 Chr13:87 Mb. This locus contains members of a family of receptors, which bind the Fc region of antibodies, that are expressed largely on macrophages and initiate macrophage immune responses to antibody/antigen complexes. Copy number polymorphisms affecting the Fc receptor encoded by Fcgr3a have been shown to predispose to autoimmune disease in both rats and humans (39). LOC498276 is annotated in the Rat Genome Database as Fcgr2b. The alternative haplotype contains two haplotigs that overlap this gene. The two haplotigs contain SNPs that in one case differ from and in the other are isogenic with the reference sequence. Examination of IsoSeq expression data from this region shows that some tissues contain transcripts that are identical in sequence to one of the two alternative haplotigs, but differ from the sequence of the primary haplotigs (lung). In contrast, spleen expresses transcripts that are identical to one of the alternative haplotigs and a lesser number that are identical to the primary assembly. Finally, lymph node shows equivalent expression of both haplotypes. This reveals the presence of a segmental duplication in the BN rat that creates two duplicate copies of Fcgr2b for a total of three copies. Furthermore, IsoSeq data indicate that at least two and potentially all three copies are transcribed.
Figure 4.
Two alternative copies of LOC498276, annotated by the Rat Genome Database as Fcgr2b, are present in the alternative haplotype. IsoSeq indicates expression of at least two of the duplicated copies in WKY and further suggests that usage of the duplicated copies may be tissue specific. Note that IsoSeq data have been processed to represent unique isoforms; each isoform track is representative of at least two identical copies of the transcript in the data set, and the track representing the most abundant unique isoform is typically present in multiple copies in the data set.
Duplicated regions that are not represented fully in the assembly can also be analyzed by alignment of the PacBio CLR sequencing reads back to the assembly created from them. This allows the regions that have been purged of duplication to be assessed. For example, Fig. 5 shows a region on Chr1 in which the CLRs align with approximately, about fourfold greater depth than the genome-wide average, indicating that the reference assembly process has stripped duplicated genomic material and does not include a complete representation of the genome in this region. It may be that the smaller size of the mRatBN7.2 assembly compared with its predecessors (e.g., mRatBN7.2 2.648 Gb vs. Rnor_6 2.870 Gb) reflects a greater absence of SD’s and other repetitive sequences from the assembly.
Figure 5.
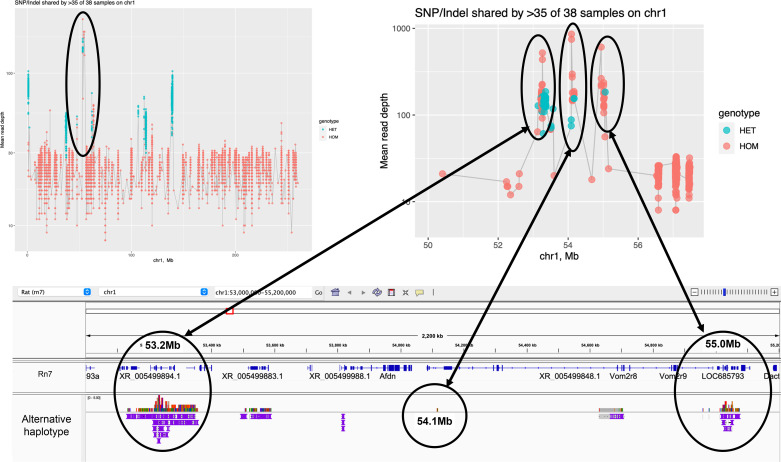
Multiple segmental duplication in the outer peaks at Chr1:53.2-55.0 Mb corresponds to haplotig alignment depths up to five and three. The middle peak was subsequently shown to correspond to a small (1,129 bp) sequence that was repeated 35 times in the rat genome, based on our own draft HiFi assembly (unpublished observations), but only once in the reference. This repeat was present across a location corresponding to the Chr1 centromere. This copy number corresponds well with the amplification of read depth from a baseline of ∼30 to ∼1,000 in this region. However, all copies are collapsed in the mRatBN7.2 to a single copy.
Centromeric and telomeric regions of the genome are comprised largely of repetitious sequence information that makes assembly a significant challenge. Rat has chromosomes that are a mixture of telocentric (Chr2, 4-10 and X) and metacentric (40). Successful assembly across such repetitive sequences using CLRs appears unlikely. A more likely outcome is that the primary assembly will have aligned regions in the alternative assembly where duplicate reads have been removed. In addition, alignment of the raw CLRs to the assembly should reveal these pileups as increased read depth. The centromere of Chr1 located at approximately Chr1:53.2 in mRatBN7.2 provides an illustration. Examination of short-read sequence alignments here indicates the presence of increased read depth up to ∼1,000-fold coverage, or ∼35-fold greater than the genome-wide average (Fig. 5). These read pileups show peaks that correspond to the alignment of multiple alternative haplotigs. Furthermore, alignment of the VGP CLRs used to make the assembly, show similar regions of increased read depth with regions up to 12 times the genome-wide average depth extending from Chr1:53.09-53.41 Mb. These pileups cannot be fully resolved but are likely to reflect centromeric repeat sequences that have been collapsed to the regions adjacent to the centromere. Although centromeric and telomeric regions are unlikely to contain genetic variation driving traits observed in inbred rat models of disease, it is clear that methods are now emerging that do permit the resolution of these extremely complex and repetitive regions of the genome (41, 42).
COMPLETENESS OF THE mRatBN7.2 ASSEMBLY
Another essential characteristic of a genome assembly is completeness. This can be broadly assessed using the Benchmarking Universal Single-copy Orthologs (BUSCO) tool (43). The principal of this tool is to assemble a collection of gene orthologs conserved in single-copy number in members of a phylogenetic group, for example members of the class Mammalia or the clade Glires. This tool can then determine what fraction of these orthologs are represented in the assembly, as well as what fraction are present as duplications and fragments. Table 2 provides BUSCO scores for mRatBN7.2 and compares scores for the prior reference Rnor_6. These scores indicate an improved level of completeness in mRatBN7.2 relative to the prior reference (43, 44).
Table 2.
BUSCO analysis of Rnor_6 and mRatBN7.2 using ortholog databases for the class Mammalia and for the clade Glires
| Assembly | Ortholog Database | Complete BUSCOs (C) | Complete and Single-Copy BUSCOs (S) | Complete and Duplicated BUSCOs (D) | Fragmented BUSCOs (F) | Missing BUSCOs (M) | Total BUSCO Groups Searched |
|---|---|---|---|---|---|---|---|
| Rnor_6 | glires_odb10 | 13,110 | 12,711 | 399 | 148 | 540 | 13,798 |
| mRatBN7.2 | glires_odb10 | 13,244 | 12,932 | 312 | 102 | 452 | 13,798 |
| Rnor_6 | mammalia_odb10 | 8,650 | 8,382 | 268 | 144 | 432 | 9,226 |
| mRatBN7.2 | mammalia_odb10 | 8,880 | 8,678 | 202 | 84 | 262 | 9,226 |
BUSCO version is: 5.2.2, database versions 2021-02-19. BUSCO, Benchmarking Universal Single-copy Orthologs.
IMPROVING THE RAT REFERENCE ASSEMBLY
As we have illustrated earlier, despite the large improvement of mRatBN7.2 over Rnor_6, sequence level errors reflecting the overall high error rate of CLRs, and the propensity of CLRs to misread homopolymers have led to an assembly in which over 200 genes have disruption of coding sequence, largely due to such errors. Although many of these can be corrected by alignment of accurate single-molecule transcript sequences (IsoSeq) to the assembly, this approach is limited to the 1% of the genome containing expressed genes and by the availability of sufficiently diverse collections of IsoSeq data so that tissue-specific gene expression does not significantly impact successful correction. For the remaining 99% of the genome, these errors cannot be readily corrected.
The production of a more accurate rat reference assembly is attainable with current sequencing and assembly methods. Increasing read lengths and throughput of single-molecule sequencing chemistry and instrumentation have allowed circular consensus sequencing (CCS) of genomic DNA, resulting in “HiFi” reads (CCS reads with >99% accuracy). Effectively this allows inserts ranging in size from 15 to 20 kbases, similar to those previously restricted to CLR sequencing, to be recursively read multiple times, thus increasing accuracy within a single read as well as allowing multiple coverage of the same genomic region from distinct sequencing constructs. When combined with the publicly available optical map and Hi-C chromosome conformation capture map of Brown-Norway rat, the resulting assemblies should provide greater accuracy at the base level and more fully represent both segmentally duplicated genes and repetitive genomic regions such as centromeres and telomeres. Until such an assembly is completed, we hope that our analysis of mRatBN7.2 will assist in exploiting this new reference resource as effectively as possible.
CONCLUSIONS
The recently released rat reference assembly represents a substantial improvement over prior assemblies, most notably in its increased contiguity and in its completeness of representation of conserved orthologs. However, it presents an alternative pseudohaplotype that is a novel representation of this inbred strain’s genome and is potentially a source of confusion and misunderstanding to the rat genomics community. Here we provide an explanation and show how this pseudohaplotype may occasionally be able to resolve base-level sequencing errors that have arisen from error-prone PacBio CLR sequencing. The unexpectedly small size of the genome compared with most prior rat reference assemblies may reflect removal of authentic primary assembly sequences to the alternative haplotype and may also result from purging of authentically duplicated sequences from the primary to the alternative assembly. Furthermore, the inability of CLRs to resolve highly repetitive segments of the genome may have resulted in an under-representation of highly repetitive centromeric and telomeric sequences in the assembly.
SUPPLEMENTAL DATA
Supplemental Table S1 (Alternative_genome_coverage_bedtools.xlsx): https://doi.org/10.5281/zenodo.6040085.
Supplemental Table S2A (38_Rat_WGS_Samples_SRA.xlsx): https://doi.org/10.5281/zenodo.6040087.
Supplemental Table S2B (variants_shared_by_35_of_38_samples.tab): https://doi.org/10.5281/zenodo.6473845.
Supplemental Table S3 (alt_var.txt): https://doi.org/10.5281/zenodo.5770288.
Supplemental Table S4 (Chr1_issues.xlsx): https://doi.org/10.5281/zenodo.6040093.
GRANTS
This work was supported by National Institutes of Health Award Numbers NIH R01HG011252 (to P.A.D./M.L.S./T.S.K.), R01DK081866 (to P.A.D.), and U01DA047638 (to H.C.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.V.d.J., H.C., and P.A.D. prepared figures; P.A.D. drafted manuscript; T.V.d.J., H.C., M.L.S., T.S.K., and P.A.D. edited and revised manuscript; T.V.d.J., H.C., W.A.B., K.J.K., A.E.H., Y.Z., I.S.D., E.A.H., M.H.S., M.L.S., T.S.K., and P.A.D. approved final version of manuscript.
REFERENCES
- 1.Fan F, Geurts AM, Pabbidi MR, Ge Y, Zhang C, Wang S, Liu Y, Gao W, Guo Y, Li L, He X, Lv W, Muroya Y, Hirata T, Prokop J, Booz GW, Jacob HJ, Roman RJ. A mutation in γ-adducin impairs autoregulation of renal blood flow and promotes the development of kidney disease. J Am Soc Nephrol 31: 687–700, 2020. doi: 10.1681/ASN.2019080784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rangel-Filho A, Lazar J, Moreno C, Geurts A, Jacob HJ. Rab38 modulates proteinuria in model of hypertension-associated renal disease. J Am Soc Nephrol 24: 283–292, 2013. doi: 10.1681/ASN.2012090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangel-Filho A, Sharma M, Datta YH, Moreno C, Roman RJ, Iwamoto Y, Provoost AP, Lazar J, Jacob HJ. RF-2 gene modulates proteinuria and albuminuria independently of changes in glomerular permeability in the fawn-hooded hypertensive rat. J Am Soc Nephrol 16: 852–856, 2005. doi: 10.1681/ASN.2005010029. [DOI] [PubMed] [Google Scholar]
- 4.Flister MJ, Hoffman MJ, Lemke A, Prisco SZ, Rudemiller N, O’Meara CC, Tsaih S-W, Moreno C, Geurts AM, Lazar J, Adhikari N, Hall JL, Jacob HJ. SH2B3 is a genetic determinant of cardiac inflammation and fibrosis. Circ Cardiovasc Genet 8: 294–304, 2015. doi: 10.1161/CIRCGENETICS.114.000527. [DOI] [PubMed] [Google Scholar]
- 5.Dhande IS, Cranford SM, Zhu Y, Kneedler SC, Hicks MJ, Wenderfer SE, Braun MC, Doris PA. Susceptibility to hypertensive renal disease in the spontaneously hypertensive rat is influenced by two loci affecting blood pressure and immunoglobulin repertoire. Hypertension 71: 700–708, 2018. doi: 10.1161/HYPERTENSIONAHA.117.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhande IS, Kneedler SC, Zhu Y, Joshi AS, Hicks MJ, Wenderfer SE, Braun MC, Doris PA. Natural genetic variation in Stim1 creates stroke in the spontaneously hypertensive rat. Genes Immun 21: 182–192, 2020. doi: 10.1038/s41435-020-0097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhande IS, Kneedler SC, Joshi AS, Zhu Y, Hicks MJ, Wenderfer SE, Braun MC, Doris PA. Germ-line genetic variation in the immunoglobulin heavy chain creates stroke susceptibility in the spontaneously hypertensive rat. Physiol Genomics 51: 578–585, 2019. doi: 10.1152/physiolgenomics.00054.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhande IS, Zhu Y, Kneedler SC, Joshi AS, Hicks MJ, Wenderfer SE, Braun MC, Doris PA. Stim1 polymorphism disrupts immune signaling and creates renal injury in hypertension. J Am Heart Assoc 9: e014142, 2020.doi: 10.1161/JAHA.119.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett MR, Korstanje R. Using genetic and species diversity to tackle kidney disease. Trends Genet 36: 499–509, 2020. doi: 10.1016/j.tig.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson AC, Wu W, Attipoe EM, Sasser JM, Taylor EB, Showmaker KC, Kyle PB, Lindsey ML, Garrett MR. Loss of Arhgef11 in the dahl salt-sensitive rat protects against hypertension-induced renal injury. Hypertension 75: 1012–1024, 2020. doi: 10.1161/HYPERTENSIONAHA.119.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman DJ, Pollak MR. APOL1 and kidney disease: from genetics to biology. Annu Rev Physiol 82: 323–342, 2020. doi: 10.1146/annurev-physiol-021119-034345. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, Palmer AA. Behavioral genetic studies in rats. Methods Mol Biol 2018: 319–326, 2019. doi: 10.1007/978-1-4939-9581-3_16. [DOI] [PubMed] [Google Scholar]
- 13.Devapatla B, Sanders J, Samuelson DJ. Genetically determined inflammatory-response related cytokine and chemokine transcript profiles between mammary carcinoma resistant and susceptible rat strains. Cytokine 59: 223–227, 2012. doi: 10.1016/j.cyto.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan H, Kendziorski CM, Haag JD, Shepel LA, Newton MA, Gould MN. Genetic loci controlling breast cancer susceptibility in the Wistar-Kyoto rat. Genetics 157: 331–339, 2001. doi: 10.1093/genetics/157.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behmoaras J, Bhangal G, Smith J, McDonald K, Mutch B, Lai PC, Domin J, Game L, Salama A, Foxwell BM, Pusey CD, Cook HT, Aitman TJ. Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat Genet 40: 553–559, 2008. doi: 10.1038/ng.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitarokoili K, Ambrosius B, Gold R. Lewis rat model of experimental autoimmune encephalomyelitis. Curr Protoc Neurosci 81: 9.61.1–9.61.20, 2017. doi: 10.1002/cpns.36. [DOI] [PubMed] [Google Scholar]
- 17.Kwitek AE. Rat models of metabolic syndrome. Methods Mol Biol 2018: 269–285, 2019. doi: 10.1007/978-1-4939-9581-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solberg Woods LC, Palmer AA. Using heterogeneous stocks for fine-mapping genetically complex traits. Methods Mol Biol 2018: 233–247, 2019. doi: 10.1007/978-1-4939-9581-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods LC, Mott R. Heterogeneous stock populations for analysis of complex traits. Methods Mol Biol 1488: 31–44, 2017. doi: 10.1007/978-1-4939-6427-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keele GR, Prokop JW, He H, Holl K, Littrell J, Deal A, Francic S, Cui L, Gatti DM, Broman KW, Tschannen M, Tsaih S-W, Zagloul M, Kim Y, Baur B, Fox J, Robinson M, Levy S, Flister MJ, Mott R, Valdar W, Solberg Woods LC. Genetic fine-mapping and identification of candidate genes and variants for adiposity traits in outbred rats. Obesity (Silver Spring) 26: 213–222, 2018. doi: 10.1002/oby.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabakoff B, Smith H, Vanderlinden LA, Hoffman PL, Saba LM. Networking in biology: the hybrid rat diversity panel. Methods Mol Biol 2018: 213–231, 2019. doi: 10.1007/978-1-4939-9581-3_10. [DOI] [PubMed] [Google Scholar]
- 22.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, et al. The complete sequence of a human genome. Science 376: 44–53, 2022. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746, 2021. doi: 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley AW, Stoll M, Greene AS, Kaldunski ML, Roman RJ, Tonellato PJ, Schork NJ, Dumas P, Jacob HJ. Genetically defined risk of salt sensitivity in an intercross of Brown Norway and Dahl S rats. Physiol Genomics 2: 107–115, 2000. doi: 10.1152/physiolgenomics.2000.2.3.107. [DOI] [PubMed] [Google Scholar]
- 25.STAR Consortium, Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet 40: 560–566, 2008. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlin K, Koren S, Chin C-S, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630, 2015. [Erratum in Nat Biotechnol 33: 1109, 2015]. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 27.Howe K, Dwinell M, Shimoyama M, Corton C, Betteridge E, Dove A, Quail MA, Smith M, Saba L, Williams RW, Chen H, Kwitek AE, McCarthy SA, Uliano-Silva M, Chow W, Tracey A, Torrance J, Sims Y, Challis R, Threlfall J, Blaxter M. The genome sequence of the Norway rat, Rattus norvegicus Berkenhout 1769. Wellcome Open Res 6: 118, 2021. doi: 10.12688/wellcomeopenres.16854.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, Cramer GR, Delledonne M, Luo C, Ecker JR, Cantu D, Rank DR, Schatz MC. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13: 1050–1054, 2016. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736, 2017. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36: 2896–2898, 2020. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bocklandt S, Hastie A, Cao H. Bionano genome mapping: high-throughput, ultra-long molecule genome analysis system for precision genome assembly and haploid-resolved structural variation discovery. Adv Exp Med Biol 1129: 97–118, 2019. doi: 10.1007/978-981-13-6037-4_7. [DOI] [PubMed] [Google Scholar]
- 32.Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Phillippy AM, Koren S. Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Comput Biol 15: e1007273, 2019. doi: 10.1371/journal.pcbi.1007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichler EE. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med 381: 64–74, 2019. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Dixon JR. Genome reconstruction and haplotype phasing using chromosome conformation capture methodologies. Brief Funct Genomics 19: 139–150, 2020. doi: 10.1093/bfgp/elz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramdas S, Ozel AB, Treutelaar MK, Holl K, Mandel M, Woods LCS, Li JZ. Extended regions of suspected mis-assembly in the rat reference genome. Sci Data 6: 39, 2019. doi: 10.1038/s41597-019-0041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurk S, Walenz BP, Rhie A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res 30: 1291–1305, 2020. doi: 10.1101/gr.263566.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wenger AM, Peluso P, Rowell WJ, Chang P-C, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, Töpfer A, Alonge M, Mahmoud M, Qian Y, Chin C-S, Phillippy AM, Schatz MC, Myers G, DePristo MA, Ruan J, Marschall T, Sedlazeck FJ, Zook JM, Li H, Koren S, Carroll A, Rank DR, Hunkapiller MW. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 37: 1155–1162, 2019. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100, 2018. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851–855, 2006. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 40.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521, 2004. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 41.Altemose N, Logsdon GA, Bzikadze AV, Sidhwani P, Langley SA, Caldas GV, et al. Complete genomic and epigenetic maps of human centromeres. Science 376: eabl4178, 2022. doi: 10.1126/science.abl4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585: 79–84, 2020. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35: 543–548, 2018. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38: 4647–4654, 2021. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1 (Alternative_genome_coverage_bedtools.xlsx): https://doi.org/10.5281/zenodo.6040085.
Supplemental Table S2A (38_Rat_WGS_Samples_SRA.xlsx): https://doi.org/10.5281/zenodo.6040087.
Supplemental Table S2B (variants_shared_by_35_of_38_samples.tab): https://doi.org/10.5281/zenodo.6473845.
Supplemental Table S3 (alt_var.txt): https://doi.org/10.5281/zenodo.5770288.
Supplemental Table S4 (Chr1_issues.xlsx): https://doi.org/10.5281/zenodo.6040093.