Abstract
Background/purpose
During orthodontic tooth movement, mechanical forces induce the osteogenic differentiation of periodontal ligament stem cells (PDLSCs), which contributes to alveolar bone remodeling. MicroRNAs (miRNAs) are involved in regulating PDLSC osteogenic differentiation. Therefore, we intended to explore the role of miR-34a and miR-146a in osteogenic differentiation of PDLSCs under cyclic stretch.
Materials and methods
Phenotypic identification of PDLSCs was determined by flow cytometry analysis. PDLSCs were incubated with osteogenic differentiation medium for 3 weeks and the osteogenic differentiation capability was detected by Alizarin Red staining. To mimic the orthodontic forces, cyclic mechanical stretch was applied to PDLSCs. Alkaline phosphatase (ALP) activity assay and ALP staining were used for evaluating the ALP activity. The expression of osteogenesis markers in PDLSCs was assessed by western blotting and qRT-PCR. The binding between miR-34a (or miR-146a) and CUGBP Elav-like family member 3 (CELF3) was validated by luciferase reporter assay.
Results
Cyclic stretch elevated ALP activity and the expression of osteogenesis markers, osteopontin (OPN), runt-related transcription factor 2 (RUNX2), type I collagen (COL1), ALP, osteocalcin (OCN) and osterix (OSX), in PDLSCs. MiR-146a and miR-34a were downregulated in PDLSCs under cyclic stretch. Either overexpressing miR-146a and miR-34a reduced ALP activity and the expression of osteogenesis markers. CELF3 was a target of both miR-146a and miR-34a. CELF3 silencing attenuated while CELF3 overexpression enhanced ALP activity and the expression of osteogenesis markers.
Conclusion
MiR-34a and miR-146a repress cyclic stretch-induced osteogenic differentiation of PDLSCs via regulating the expression of CELF3.
Keywords: Periodontal ligament stem cells, Osteogenic differentiation, microRNA-34a, microRNA-146a, CELF3
Introduction
Orthodontic tooth movement (OTM) is induced by mechanical stimuli and promoted by the remodeling of the alveolar bone and periodontal ligament (PDL).1 The force exerted on the teeth is transmitted to the alveolar bone through the PDL, resulting in bone deposition at tension side and bone resorption at pressure side.2,3 Periodontal ligament stem cells (PDLSCs) extracted from PDL tissues have mesenchymal stem cell (MSC) properties.4 PDLSCs possess the potential for osteogenic differentiation and the capacity for self-renewal.5 Previous studies have demonstrated that osteogenic differentiation induced by orthodontic force plays a crucial role in bone formation at tension side during the process of OTM.6, 7, 8 PDLSCs can differentiate into osteoblasts to participate in alveolar bone remodeling or cementoblasts to synthesize cementum/PDL-like structures, providing skeletal support for teeth.9 Even though numerous studies have explored the influence of mechanical strain on PDLSC osteogenic differentiation, the molecular regulatory mechanism during the process of PDLSC osteogenic differentiation is poorly understood.
MicroRNAs (miRNAs) are small non-coding RNAs, which modulate gene expression at the post-transcriptional level, thereby influencing cellular physiological or pathological processes.10 MiRNAs exert regulatory roles on gene expression by pairing with the 3′-untranslated regions (3′-UTR) of mRNAs.11 Many miRNAs were reported to engage in PDLSC osteogenic differentiation via regulating their target genes. For example, the gradually upregulated miR-383-5p during PDLSC osteogenic differentiation enhances the expression of osteogenic markers, Alkaline phosphatase (ALP) activity and mineral node formation by inhibiting the expression of histone deacetylase 9, thereby facilitating PDLSC osteogenic differentiation.12 MiR-132 overexpression attenuates osteoblast activity, reduces ALP and ARS intensity, and decreases levels of osteogenic markers by targeting growth differentiation factor 5, which shows the inhibition of miR-132 on PDLSC osteogenesis.13 MiR-24-3p level is discovered notably decreased in osteogenic-differentiated PDLSCs and miR-24-3p knockdown promotes the formation of the mineralized nodules, ALP activity and the expression of osteogenic differentiation markers.14 Previously, miR-146a and miR-34a were predicted to be involved in modulating PDLSC osteogenic differentiation.15 However, the detailed function of both miRNAs in PDLSC osteogenic differentiation is not clear. Besides, by using TargetScan V7.1 (http://www.targetscan.org/vert_71/), CUGBP Elav-like family member 3 (CELF3) was discovered as the only common target gene of miR-146a and miR-34a. Thus, we proposed a hypothesis that miR-146a and miR-34a play a regulatory role in PDLSC osteogenic differentiation via targeting CELF3.
We intend to investigate whether miR-34a and miR-146a regulate cyclic mechanical stretch-induced PDLSC osteogenic differentiation via targeting CELF3. This study might clarify the molecular mechanism involved in the differentiation of osteoblasts induced by cyclic mechanical stretch and provide novel insights for improving orthodontic treatment.
Materials and methods
Identification and culture of PDLSCs
Ten disease-free premolars, extracted for orthodontic treatment, were obtained from 6 healthy volunteers (female: male = 1:1, aged 12–24 years) at Aerospace Center Hospital (Beijing, China). Prior to tooth extraction, all participants provided written informed consent. All experimental procedures were performed according to the National Institutes of Health guidelines concerning the use of human tissues and approved by the Medical Ethical Committee of Aerospace Center Hospital (Beijing, China).
PDLSCs were isolated and cultured following the previously described procedure.16 The extracted premolars were kept in Eagle's minimum essential medium (Sigma–Aldrich, St. Louis, MO, USA) and washed with phosphate-buffered saline (PBS). Periodontal ligament (PDL) tissues were gently scraped off from the middle third of the premolar root and minced into smaller pieces using an aseptic scalpel, followed by 1 h digestion in a mixed solution supplemented with 8 mg/ml dispase (Sigma–Aldrich) and 6 mg/ml collagenase type I (Sigma–Aldrich). Then, single cell suspension was harvested and centrifuged to remove the supernatant. Cells were cultured in alpha-modified Eagle's Medium (α-MEM, Hyclone, Logan, UT, USA) containing 10% (V/V) fetal bovine serum (FBS; Hyclone) supplemented with antibiotics at 37 °C with 5% CO2. Cells were passaged after reaching 90% confluence. PDLSCs at passages 3–6 were selected for the following experiments.
Cell transfection
Short hairpin RNA (shRNA) for CELF3 (sh-CELF3), miR-34a mimics, miR-146a mimics and their relative negative controls (sh-NC, NC mimics) were bought from GenePharma (Shanghai, China). To overexpress CELF3, the full length of CELF3 was synthesized by Sangon (Shanghai, China) and cloned into pcDNA3.1 vector (GenePharma) to generate the overexpressing plasmid, which was named pcDNA3.1/CELF3. The empty vector pcDNA3.1 was regarded as the negative control. PDLSCs were transfected with the above plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. After 48 h, cells were harvested, followed by the extraction of total RNA to detect the transfection efficiency of plasmids using qRT-PCR.
Colony formation assay
For evaluating the colony-forming efficiency of PDLSCs, colony formation assay was performed. Single cell suspensions (500 cells/dish) were seeded into 100-mm culture dishes with α-MEM growth medium. After 14 days, the dishes were washed twice with PBS, fixed with ice-cold methanol and stained with Giemsa stain (Sigma–Aldrich). Photographs of cell aggregates were taken with a microscope and colonies were then counted.
Flow cytometry analysis
Cell phenotype of PDLSCs was determined by flow cytometry analysis. Cultured PDLSCs were detached as single cell suspensions and resuspended in blocking buffer for 30 min. Then, 5 × 105 cells were incubated with PE-conjugated monoclonal antibodies specific for CD14, CD31, CD29, CD90, CD146 or STRO-1 (Thermo Fisher Scientific, Waltham, MA, USA) in the dark at 4 °C for 1 h. Cells were analyzed using a flow cytometer (BD Bioscience, San Jose, CA, USA) after washing thrice with PBS.
Osteogenic induction and alizarin red staining
To determine the osteogenic differentiation potential, PDLSCs were plated in growth medium on 24-well plates 2 × 104 cells/cm.2 After reaching 100% confluence, the growth medium was changed to osteogenic differentiation medium (Sigma–Aldrich) containing 10% FBS, 10 nM dexamethasone (MedChemExpress, Monmouth Junction, NJ, USA), 10 mM β-glycerophosphate (MedChemExpress) and 50 mg/l ascorbic acid (Sigma–Aldrich). The medium was replaced every 3 days. After 21 days of osteogenic culture, cells were stained with Alizarin red dyes (Sigma–Aldrich) after fixed with 4% formalin to detect the calcium deposition.
Application of cyclic stretch
Equal amounts of PDLSCs were seeded into 6-well, flexible-bottomed plates coated with type I collagen (COL1; Sigma–Aldrich). After reaching 80% confluence, the osteo-inductive medium was added. Then, cyclic mechanical stretch (10% deformation) was applied to PDLSCs at a frequency of 0.1 Hz (5s stretch and 5s relaxation) using Flexercell FX-4000 Strain Unit (Flexcell International Corporation, Hillsborough, NC, USA). PDLSCs cultivated under identical conditions without mechanical stretch were used as the control group. Cells were harvested for mRNA or protein extraction after 24 h of stretch application and for ALP activity detection after 72 h of stretch application.
Quantitative real-time PCR (qRT-PCR)
Total RNA from PDLSCs was extracted using TRIzol reagent (Invitrogen) and a NanoDrop 2000 spectrometer (Thermo Fisher Scientific) was applied to quantify its concentration. Two μg of the total RNA was reversely transcripted to cDNA using miRCURY LNA RT Kit (Qiagen, Dusseldorf, Germany) or PrimeScript RT master mix (Takara, Tokyo, Japan) for miRNA or mRNA reverse transcription. Subsequently, qRT-PCR was conducted in Touch real-time PCR Detection system (Bio-Rad, Hercules, CA, USA) using SYBR Green qPCR Master Mix (Takara). Primer sequences are displayed in Table 1. GAPDH served as the internal control for CELF3, osteopontin (OPN), COL1, alkaline phosphatase (ALP), osterix (OSX), runt-related transcription factor 2 (RUNX2) and osteocalcin (OCN), and U6 served as the internal control for miR-34a and miR-146a. Relative gene expression was quantified following the 2−ΔΔCT method.
Table 1.
Primer sequences for qRT-PCR. CELF3, CUGBP Elav-like family member 3; OPN, osteopontin; RUNX2, runt-related transcription factor 2; COL1, type I collagen; ALP, alkaline phosphatase; OSX, osterix; OCN, osteocalcin; GADPH, glyceraldehyde-3-phosphate dehydrogenase; miR-34a, microRNA-34a; miR-146a, microRNA-146a; U6, U6 small nuclear RNA.
| Gene | Primer sequences |
|---|---|
| CELF3 | Forward: 5′-ATCAACACCCTTCACAGCA-3′ |
| Reverse: 5′-CAGCAAACTTCACCACCAG-3′ | |
| OPN | Forward: 5′-GAAGTTTCGCAGACCTGACAT-3′ |
| Reverse: 5′-GTATGCACCATTCAACTCCTCG-3′ | |
| RUNX2 | Forward: 5′-TTATTCTGCTGAGCTCCGG-3′ |
| Reverse: 5′-GTGAAACTCTTGCCTCGTC-3′ | |
| COL1 | Forward: 5′-CCCCCTCCCCAGCCACAAAG-3′ |
| Reverse: 5′-TCTTGGTCGGTGGGTGACTCT-3′ | |
| ALP | Forward: 5′-ACTGGTACTCAGACAACGAGAT-3′ |
| Reverse: 5′-ACGTCAATGTCCCTGATGTTATG-3′ | |
| OSX | Forward: 5′-GAGGCAACTGGCTAGGTGG-3′ |
| Reverse: 5′-CTGGATTAAGGGGAGCAAAGTC-3′ | |
| OCN | Forward: 5′-CACTCCTCGCCCTATTGGC-3′ |
| Reverse: 5′-CCCTCCTGCTTGGACACAAAG -3′ | |
| GADPH | Forward: 5′-CCCACATGGCCTCCAAGGAGTA-3′ |
| Reverse: 5′-GTGTACATGGCAACTGTGAGGAGG-3′ | |
| miR-34a | Forward: 5′-CGCGTGGCAGTGTCTTAGCT-3′ |
| Reverse: 5′-AGTGCAGGGTCCAGG GTATT-3′ | |
| miR-146a | Forward: 5′-GTGCAGGGTCCGAGGT-3′ |
| Reverse: 5′-CAACACCAGTCGATGGGCTGT-3′ | |
| U6 | Forward: 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse: 5′-AACGCTTCACGAATTTGCGT-3′ |
ALP staining and ALP activity assay
Both assays were conducted for the assessment of early osteogenesis ability of PDLSCs. PDLSCs were plated in 6-well plates and treated with osteogenic inducing medium. After an incubation of 21 days, cells were harvested, fixed with 4% paraformaldehyde and washed thrice with PBS. First, a NBT/BCIP staining kit (Beyotime, Shanghai, China) was applied to visualize ALP staining in PDLSCs. Staining images were captured with a phase-contrast microscope and the depth of dark blue reflects ALP activity. Additionally, the quantification of ALP activity was performed using an ALP assay kit (Sigma–Aldrich). Briefly, cells were lysed in 0.2% Triton X-100 lysis buffer (Sigma–Aldrich). ALP activity level (U/ml) and the total protein concentration (mg/ml) were measured using an ALP activity assay kit and an BCA assay kit (Thermo Fisher Scientific), respectively. ALP activity level was normalized to the total protein content.
Western blotting
Total protein was extracted from PDLSCs with radio immunoprecipitation assay lysis buffer (Sigma–Aldrich). Nuclear protein was extracted using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime). The protein content was determined using a Bicinchoninic Acid Protein Assay kit (Beyotime). Then, 20 μg of total protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). The membranes were incubated overnight at 4 °C with primary antibodies: anti-CELF3 (bs-13830R; 1:1000; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China), anti-OPN (ab8448; 1:1000; Abcam, Cambridge, UK), anti-RUNX2 (#12556; 1:1000; Cell Signaling Technology, Danvers, MA, USA), anti-COL1 (ab96723; 1:1000; Abcam), anti-ALP (ab229126; 1:1000; Abcam), anti-OSX (ab209484; 1:1000; Abcam), anti-OCN (ab133612; 1:1000; Abcam) and anti-GAPDH (ab181603; 1:10,000; Abcam) after blocked with 5% BSA for 2 h. The next day, the membranes were supplemented with secondary antibodies at room temperature for 2 h. Finally, the membranes were washed thrice with TBS-Tween 20 (Sigma–Aldrich), and protein band was visualized with an enhanced chemiluminescence kit (Beyotime).
Luciferase reporter assay
The potential binding site of miR-34a (or miR-146a) on CELF3 was predicted using TargetScan V7.1 and further validated using luciferase reporter assay. The 3′UTR of CELF3 containing miR-34a (or miR-146a) target sites was synthesized by GenePharma and was inserted into pmirGLO luciferase vectors (Promega, Madison, WI, USA). Then, pmirGLO vectors loaded with CELF3 3′UTR-WT (MUT) were introduced into PDLSCs with cotransfection of miR-34a (or miR-146a) mimics or NC mimics using Lipofectamine 3000. After 48 h, the Dual-Luciferase Reporter Assay System (Promega) was used for the detection of Renilla and firefly luciferase activities.
Statistical analysis
Statistical analyses were performed using the SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). All data are expressed as the mean ± standard deviation. Differences between two groups were analyzed by Student's t-test and variances among multiple groups were analyzed by one-way analysis of variance followed by Tukey's post-hoc test. All experiments were repeated in triplicate independently. Statistical significance was set at p < 0.05.
Results
Identification of PDLSCs
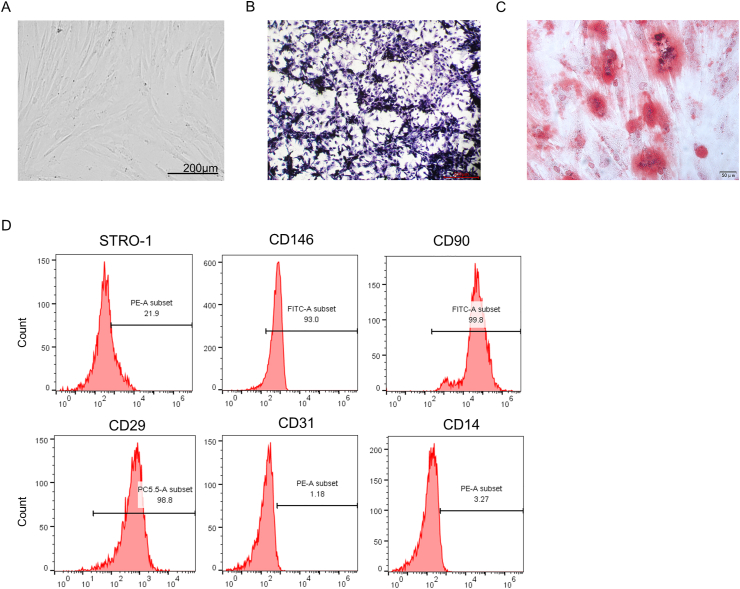
PDLSCs extracted from human periodontal ligament tissues were observed under a microscope, which displayed a spindle-shaped morphology (Fig. 1A). Colony formation assay also demonstrated that PDLSCs could grow in an adherent manner, followed by the formation of clonogenic cell clusters (Fig. 1B). The formation of Alizarin Red-stained calcium deposits was observed after 3 weeks of culture of PDLSCs with osteogenic differentiation medium (Fig. 1C). Additionally, we examined the MSC phenotype of PDLSCs through flow cytometric analysis, which revealed that PDLSCs were positive to MSC-specific surface markers CD29, CD90, CD146 and Stro-1, and negative to hematopoietic markers CD14 and CD31 (Fig. 1D). These results suggested that PDLSCs can differentiate into osteoblasts.
Figure 1.
Identification of PDLSCs. (A) The morphology of PDLSCs was observed under a microscope. (B) Colonies formed by PDLSCs after 14 days of culture were stained with Giemsa stain. (C) Alizarin Red staining of calcium deposits formed by PDLSCs after incubation in osteogenic induction medium for 3 weeks. (D) Flow cytometric analysis was applied for examining the mesenchymal stem cell (MSC) phenotype of PDLSCs.
Cyclic stretch induces PDLSC osteogenic differentiation
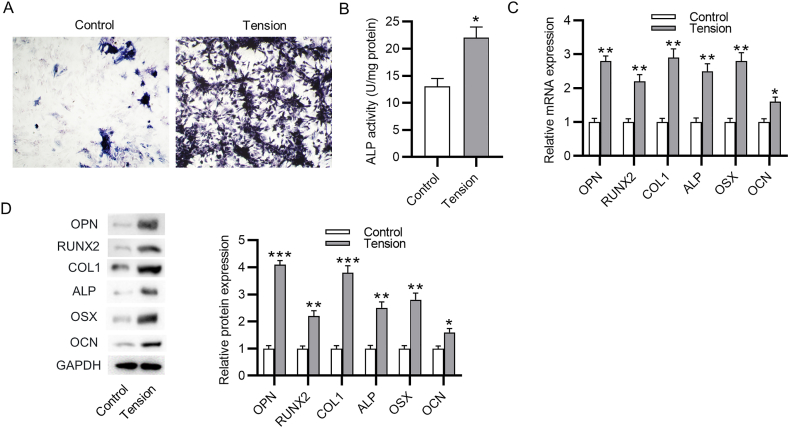
Next, we investigated the influence of cyclic tension on PDLSC osteogenic differentiation. PDLSCs were incubated in the osteogenic differentiation medium without or with cyclic tensile force for 3 days. First, ALP staining as well as ALP activity assay revealed that the ALP activity was significantly enhanced in PDLSCs after cyclic stretch compared to the control cells, which indicated that cyclic stretch triggered the osteogenic differentiation of PDLSCs (Fig. 2A–B). Furthermore, the mRNA expression of osteogenesis markers, including OPN, RUNX2, COL1, ALP, OCN and OSX in PDLSCs, showed a great upregulation after cyclic tension (Fig. 2C). The findings of western blotting analysis also revealed an obvious elevation in the protein levels of these markers (Fig. 2D). Overall, cyclic stretch promotes PDLSC osteogenic differentiation.
Figure 2.
Cyclic stretch induces osteogenic differentiation of PDLSCs. (A–B) ALP staining and ALP activity assay of the ALP activity in PDLSCs after cyclic stretch. (C–D) PCR analysis and western blotting analysis of the influence of cyclic stretch on the mRNA expression and protein level of osteogenesis markers. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
MiR-146a and miR-34a suppress osteoblastic differentiation of PDLSCs under cyclic stretch
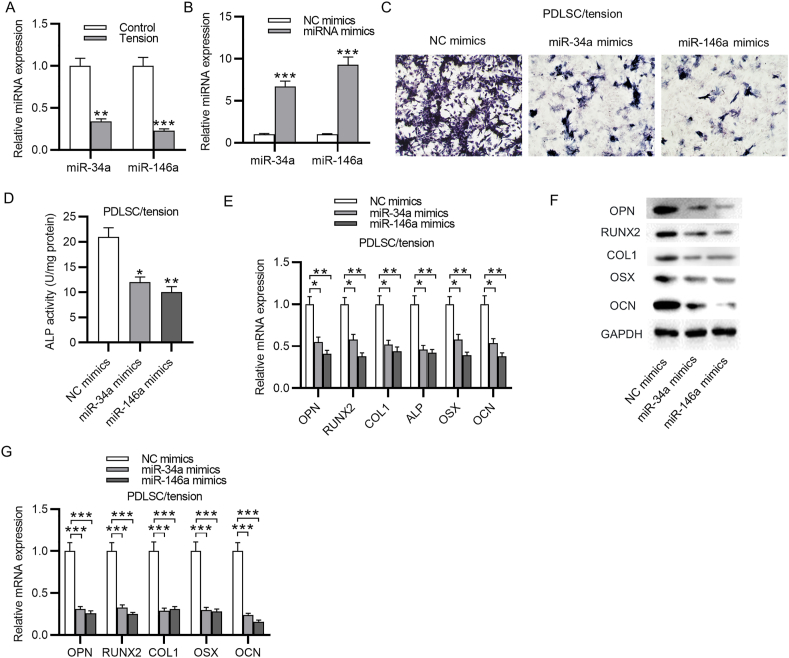
MiR-146a and miR-34a were reported to participate in the modulation of PDLSC osteoblastic differentiation, herein, we detected whether these two miRNAs affect cyclic stretch-induced PDLSC osteoblastic differentiation. PCR analysis demonstrated that both miR-146a and miR-34a expression was markedly downregulated in PDLSCs subjected to cyclic stretch (Fig. 3A). The overexpression efficiency of miR-146a and miR-34a was also evaluated by qRT-PCR (Fig. 3B). As revealed by ALP staining and ALP activity assay, either miR-34a overexpression or miR-146a overexpression remarkably attenuated the ALP activity in PDLSCs, which was previously enhanced after cyclic stretch (Fig. 3C–D). Additionally, a significant downregulation in the mRNA expression and protein level of osteogenesis markers was observed in PDLSCs after overexpressing miR-146a or miR-34a (Fig. 3E–G). In summary, miR-146a and miR-34a inhibit PDLSC osteoblastic differentiation induced by cyclic stretch.
Figure 3.
MiR-34a and miR-146a suppress cyclic stretch-induced osteoblastic differentiation of PDLSCs. (A) PCR analysis of miR-34a and miR-146a level in PDLSCs with or without cyclic stretch application. (B) PCR analysis of miR-34a and miR-146a level in PDLSCs transfected with miR-34a mimics or miR-146a mimics compared to NC mimics. (C–D) ALP staining and ALP activity assay of ALP activity in PDLSCs under cyclic stretch after overexpressing miR-34a or miR-146a. (E–G) PCR analysis and western blotting analysis of the mRNA expression and protein level of osteogenesis markers in PDLSCs under cyclic stretch after overexpressing miR-34a or miR-146a. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
CELF3 is a target of miR-146a and miR-34a
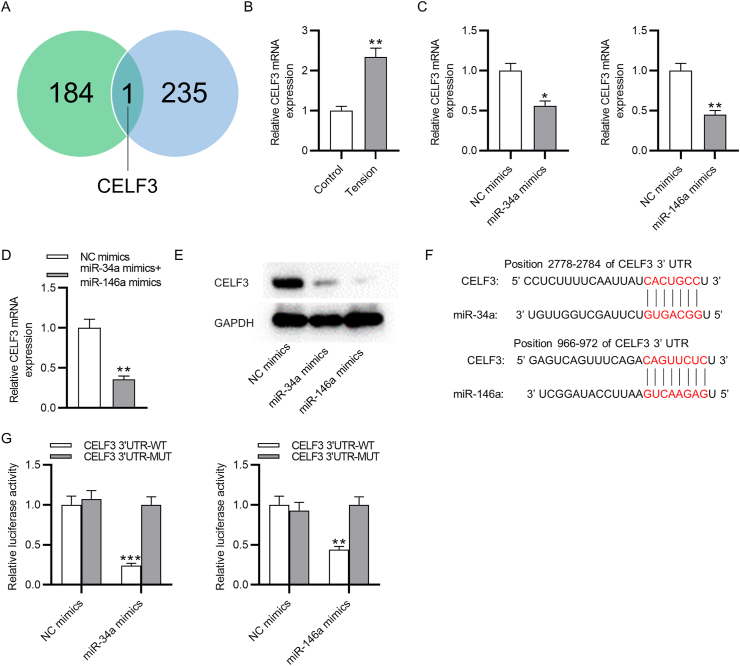
To further determine the regulatory mechanism of miR-146a and miR-34a on cyclic stretch-induced osteoblastic differentiation of PDLSCs, we predicted the target gene of both miRNAs using TargetScan database. CELF3 was discovered as the only common target gene of both miRNAs (Fig. 4A). As shown by PCR analysis, cyclic tension markedly elevated CELF3 mRNA expression in PDLSCs (Fig. 4B). In PDLSCs transfected with miR-34a mimics, miR-146a mimics or miR-34a mimics + miR-146a mimics, the mRNA expression of CELF3 was significantly downregulated (Fig. 4C–D). Furthermore, either miR-34a overexpression or miR-146a overexpression reduced the protein level of CELF3 in PDLSCs (Fig. 4E). The binding position of miR-146a and miR-34a on CELF3 3′UTR was obtained by TargetScan database (Fig. 4F) and was further verified by luciferase reporter assay. MiR-146a mimics or miR-34a mimics could specifically lessen only the luciferase activity of wild-type CELF3, but exerted no apparent influence on the luciferase activity of mutant CELF3 (Fig. 4G). Therefore, there are direct binding effects between miR-34a (or miR-146a) and CELF3 in PDLSCs.
Figure 4.
CELF3 is a target of miR-34a and miR-146a. (A) Target genes of miR-34a and miR-146a predicted at TargetScan database. (B) PCR analysis of CELF3 expression in PDLSCs with or without cyclic stretch application. (C–D) PCR analysis of CELF3 mRNA expression in PDLSCs after transfection with miR-34a mimics, miR-146a mimics or miR-34a mimics + miR-146a mimics. (E) Western blotting analysis of CELF3 protein level in PDLSCs after miR-34a or miR-146a overexpression. (F) The binding position of miR-34a (or miR-146a) on CELF3 3′UTR predicted at TargetScan database. (G) Luciferase reporter assay of the luciferase activity of vectors containing wild-type or mutant CELF3 in PDLSCs transfected with miR-34a (or miR-146a) mimics compared to NC mimics. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Silencing of CELF3 suppresses cyclic stretch-induced osteoblastic differentiation of PDLSCs
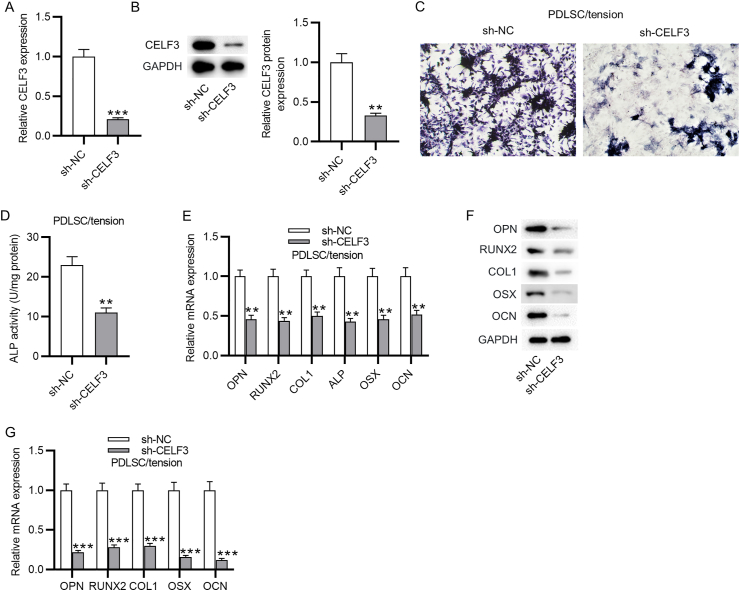
Since CELF3 was the common target of miR-34a and miR-146a, we then assessed whether CELF3 affects cyclic stretch-induced PDLSC osteoblastic differentiation. The interfering efficiency of CELF3 was examined by PCR analysis and western blotting analysis, which showed a marked decrease in the expression and protein level of CELF3 in PDLSCs transfected with sh-CELF3 (Fig. 5A–B). The detection of PDLSC osteoblastic differentiation was performed as above. CELF3 silencing significantly restrained the ALP activity in cyclic stretch-stimulated PDLSCs (Fig. 5C–D). Besides, cyclic stretch-stimulated PDLSCs displayed upregulating mRNA expression and protein level of osteogenesis markers after CELF3 depletion (Fig. 5E–G). In general, these findings suggested an inhibitory effect of CELF3 silencing on cyclic stretch-induced PDLSC osteoblastic differentiation.
Figure 5.
Silencing of CELF3 suppresses cyclic stretch-induced PDLSC osteoblastic differentiation. (A) PCR analysis of CELF3 mRNA expression in PDLSCs transfected with sh-CELF3 or sh-NC. (B) Western blotting analysis of CELF3 protein level in PDLSCs after downregulating CELF3. (C–D) ALP staining and ALP activity assay of ALP activity in cyclic stretch-stimulated PDLSCs after knocking down CELF3. (E–G) PCR analysis and western blotting analysis of the mRNA expression and protein level of osteogenesis markers in cyclic stretch-stimulated PDLSCs after CELF3 silencing. ∗∗p < 0.01, ∗∗∗p < 0.001.
Overexpression of CELF3 facilitates cyclic stretch-induced osteoblastic differentiation of PDLSCs
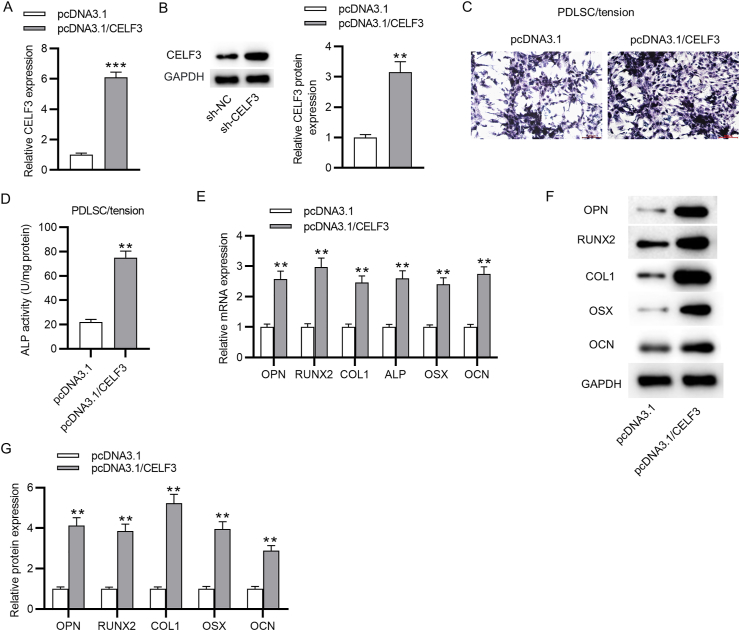
On the other hand, whether overexpressing CELF3 facilitates the osteoblastic differentiation of PDLSCs was also investigated. The overexpressing vector for CELF3, pcDNA3.1/CELF3, was transfected into PDLSCs, and the overexpression efficiency was assessed by qRT-PCR and western blotting. The results indicated that both the mRNA and protein expression level of CELF3 in PDLSCs transfected with pcDNA3.1/CELF3 were upregulated (Fig. 6A–B). The CELF3-overexpressed PDLSCs were incubated in the osteogenic differentiation media with cyclic stretch for 3 days and then were harvested for detecting their potential of osteoblastic differentiation. We discovered that ALP activity in CELF3-overexpressed PDLSCs was significantly elevated (Fig. 6C–D). Additionally, the mRNA and protein expression levels of osteogenesis-related genes were markedly increased in the cyclic stretch-stimulated PDLSCs after overexpressing CELF3 (Fig. 6E–G). Taken together, overexpression of CELF3 enhances the cyclic stretch-induced osteoblastic differentiation of PDLSCs.
Figure 6.
Overexpression of CELF3 facilitates cyclic stretch-induced osteoblastic differentiation of PDLSCs. (A) PCR analysis of CELF3 mRNA expression in PDLSCs transfected with pcDNA3.1 or pcDNA3.1/CELF3. (B) Western blotting analysis of CELF3 protein level in PDLSCs after overexpressing CELF3. (C–D) ALP staining and ALP activity assay of ALP activity in cyclic stretch-stimulated PDLSCs after CELF3 overexpression. (E–G) PCR analysis and western blotting analysis of the mRNA expression and protein level of osteogenesis markers in cyclic stretch-stimulated PDLSCs after overexpressing CELF3. ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
The osteogenic differentiation of PDLSCs induced by mechanical stretch contributes to alveolar bone remodeling plays a pivotal role during orthodontic treatment.17 MicroRNAs have been demonstrated to regulate PDLSC osteogenic differentiation.18,19 In this study, the role of miR-146a and miR-34a in cyclic stretch-induced PDLSC osteogenic differentiation was investigated. We discovered that miR-146a and miR-34a were downregulated in PDLSCs under cyclic mechanical stretch, suggesting the downregulation of miR-146a and miR-34a in osteogenic-differentiated PDLSCs. Importantly, miR-146a and miR-34a overexpression suppressed PDLSC osteogenic differentiation by downregulating CELF3.
During the process of OTM, mechanical forces induce the osteogenic differentiation of PDLSCs, thereby promoting PDL tissue repair and regeneration as well as alveolar bone remodeling.20 ALP, as an indicator of bone formation, is identified as an early marker of MSC osteogenic differentiation in tension force environment.21 RUNX2 is a crucial transcription factor, playing a key role in modulating MSC osteogenic differentiation and bone formation.22 RUNX2 has also been demonstrated to modulate the expression of other osteoblast-specific genes, such as OPN, ALP, COL1, OSX and OCN.23 It was reported that deficiency of RUNX2 leads to the lack of bone formation in mice.24 In addition, as a target of mechanical signals in osteoblastic cells, RUNX2 can also be upregulated by mechanical stimuli.25 Previously, numerous studies have demonstrated that cyclic tension force could facilitate PDLSC osteogenic differentiation. For example, cyclic tension force results in an upregulation of the RUNX2, OCN and ALP in PDLSCs, indicating the promotive effects of mechanical stimulation on PDLSC osteogenic differentiation.7 Static mechanical strain enhances ALP activity, facilitates mineralized nodule formation, and elevates RUNX2 and ALP expression in PDLSCs.26 In our study, cyclic stretch was also applied to PDLSCs to mimic the force on both sides of teeth during OTM. The results also revealed that the ALP activity as well as the expression of RUNX2, OCN, ALP, OSX, OCN and COL1 in PDLSCs were enhanced after cyclic stretch, indicating that cyclic stretch promoted PDLSC osteogenic differentiation. This is in consistent with the findings in previous studies.
Increasing studies have indicated that miRNAs with aberrant expression in PDLSCs affect osteogenic differentiation through regulating their target genes.27, 28, 29 Previously, miR-146a and miR-34a were predicted to be involved in PDLSC osteogenic differentiation.15 Both miRNAs were reported to modulate MSC osteogenic differentiation. For example, in osteoporosis which is mainly caused by decreased osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs), miR-34a is upregulated during BMSC osteogenic differentiation and miR-34a overexpression facilitates BMSC osteogenic differentiation via inhibiting bone morphogenetic protein 3.30 MiR-34a overexpression represses osteoblast differentiation of BMSCs by targeting the cellular glycolysis mediated by lactate dehydrogenase A.31 In traumatic femoral head necrosis, miR-146a level is lower in BMSCs of necrosis group than in those of normal group, and miR-146a overexpression leads to increased calcium deposition as well as elevated mRNA expression of OCN and ALP, suggesting that overexpressing miR-146a promotes BMSC osteogenic differentiation.32 MiR-146a downregulation stimulates of human adipose-derived stem cell osteogenic differentiation though increasing the expression of SMAD family member 4.33 In our study, we first discovered that miR-146a and miR-34a were both downregulated in osteogenic-differentiated PDLSCs under cyclic mechanical stretch. Furthermore, overexpressing either miR-34a or miR-146a attenuated ALP activity and downregulated the expression of osteogenesis markers. Therefore, miR-34a or miR-146a overexpression suppressed PDLSC osteogenic differentiation.
CELF3 is a member of the CUGBP and ELAV-like factor (CELF) family RNA-binding proteins, which are highly conserved in animals and can modulate the life cycle of mRNAs from transcription to translation.34,35 The role of CELF3 in human diseases is poorly explored and elucidated. Some studies demonstrated that CELF3 is involved in neural tube development.36,37 In the rat model with sciatic nerve injury, CELF3 expression is significantly increased and CELF3 is closely correlated to the sciatic nerve regeneration.38 In the present study, by using the informatic tool, CELF3 was discovered as the only common target gene of miR-34a or miR-146a, harboring binding sites with both miRNAs. Their binding capacity was further verified by a luciferase reporter assay. Then, we found that CELF3 was remarkably upregulated in PDLSCs under cyclic mechanical stretch, indicating the potential role of CELF3 in PDLSC osteogenic differentiation. Either miR-34a or miR-146a overexpression downregulated the expression of CELF3 in PDLSCs. Importantly, CELF3 silencing significantly attenuated the ALP activity and the expression of osteogenesis markers in cyclic stretch-stimulated PDLSCs, suggesting the inhibition of CELF3 depletion on PDLSC osteogenic differentiation. Overall, miR-34a and miR-146a repress PDLSC osteogenic differentiation induced by cyclic stretch via downregulating CELF3.
Despite novel findings in our study, some limitations are still required to be improved in future studies. First, our study performed in vitro experiments to figure out the role of miR-34a and miR-146a in PDLSC osteogenic differentiation. In vivo experiment for further validation of the findings in our studies are needed. Second, the precise molecular mechanism regarding how miR-146a and miR-34a modulate CELF3 expression remains unclear.
In summary, we explored the role of the miR-34a/miR-146a-CELF3 axis in the cyclic stretch-induced PDLSC osteogenic differentiation. We discovered that miR-34a and miR-146a suppress cyclic mechanical stretch-induced PDLSC osteogenic differentiation by downregulating CELF3. Although there exist some limitations, this finding might provide novel therapeutic targets for improving orthodontic treatment.
Declaration of competing interest
The authors have no conflicts of interest relevant to this article.
Acknowledgements
Not applicable.
References
- 1.Asiry M.A. Biological aspects of orthodontic tooth movement: a review of literature. Saudi J Biol Sci. 2018;25:1027–1032. doi: 10.1016/j.sjbs.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masella R.S., Meister M. Current concepts in the biology of orthodontic tooth movement. Am J Orthod Dentofacial Orthop. 2006;129:458–468. doi: 10.1016/j.ajodo.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Wise G.E., King G.J. Mechanisms of tooth eruption and orthodontic tooth movement. J Dent Res. 2008;87:414–434. doi: 10.1177/154405910808700509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang G.T., Gronthos S., Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadipiralla K., Yochim J.M., Bahuleyan B., et al. Osteogenic differentiation of stem cells derived from human periodontal ligaments and pulp of human exfoliated deciduous teeth. Cell Tissue Res. 2010;340:323–333. doi: 10.1007/s00441-010-0953-0. [DOI] [PubMed] [Google Scholar]
- 6.Pinkerton M.N., Wescott D.C., Gaffey B.J., Beggs K.T., Milne T.J., Meikle M.C. Cultured human periodontal ligament cells constitutively express multiple osteotropic cytokines and growth factors, several of which are responsive to mechanical deformation. J Periodontal Res. 2008;43:343–351. doi: 10.1111/j.1600-0765.2007.01040.x. [DOI] [PubMed] [Google Scholar]
- 7.Shen T., Qiu L., Chang H., et al. Cyclic tension promotes osteogenic differentiation in human periodontal ligament stem cells. Int J Clin Exp Pathol. 2014;7:7872–7880. [PMC free article] [PubMed] [Google Scholar]
- 8.Wescott D.C., Pinkerton M.N., Gaffey B.J., Beggs K.T., Milne T.J., Meikle M.C. Osteogenic gene expression by human periodontal ligament cells under cyclic tension. J Dent Res. 2007;86:1212–1216. doi: 10.1177/154405910708601214. [DOI] [PubMed] [Google Scholar]
- 9.Kawarizadeh A., Bourauel C., Götz W., Jäger A. Early responses of periodontal ligament cells to mechanical stimulus in vivo. J Dent Res. 2005;84:902–906. doi: 10.1177/154405910508401006. [DOI] [PubMed] [Google Scholar]
- 10.Asa'ad F., Garaicoa-Pazmiño C., Dahlin C., Larsson L. Expression of microRNAs in periodontal and peri-Implant diseases: a systematic review and meta-analysis. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21114147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzeszinski J.Y., Wei W., Huynh H., et al. Retraction note: miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2020;582:134. doi: 10.1038/s41586-020-2273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L., Wu D. MicroRNA-383-5p regulates osteogenic differentiation of human periodontal ligament stem cells by targeting histone deacetylase 9. Arch Oral Biol. 2021;129 doi: 10.1016/j.archoralbio.2021.105166. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y., Ren C., Zhao X., Wang W., Zhang N. MicroRNA-132 inhibits osteogenic differentiation of periodontal ligament stem cells via GDF5 and the NF-κB signaling pathway. Pathol Res Pract. 2019;215 doi: 10.1016/j.prp.2019.152722. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., Sun Y., Cao S., Zhang J., Wei J. Downregulation of miR-24-3p promotes osteogenic differentiation of human periodontal ligament stem cells by targeting SMAD family member 5. J Cell Physiol. 2019;234:7411–7419. doi: 10.1002/jcp.27499. [DOI] [PubMed] [Google Scholar]
- 15.Gu X., Li M., Jin Y., Liu D., Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18 doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang M., Lin H., Luo M., Wang J., Han G. Integrated miRNA and mRNA expression profiling of tension force-induced bone formation in periodontal ligament cells. In Vitro Cell Dev Biol Anim. 2015;51:797–807. doi: 10.1007/s11626-015-9892-0. [DOI] [PubMed] [Google Scholar]
- 17.Yoo J.H., Lee S.M., Bae M.K., et al. Effect of orthodontic forces on the osteogenic differentiation of human periodontal ligament stem cells. J Oral Sci. 2018;60:438–445. doi: 10.2334/josnusd.17-0310. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Feng C., Li M., Zhang Z., Liu J., Wei F. Analysis of lncRNAs-miRNAs-mRNAs networks in periodontal ligament stem cells under mechanical force. Oral Dis. 2021;27:325–337. doi: 10.1111/odi.13530. [DOI] [PubMed] [Google Scholar]
- 19.Wei F., Yang S., Guo Q., et al. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci Rep. 2017;7 doi: 10.1038/s41598-017-16720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C., Gu W., Sun B., et al. CTHRC1 promotes osteogenic differentiation of periodontal ligament stem cells by regulating TAZ. J Mol Histol. 2017;48:311–319. doi: 10.1007/s10735-017-9729-0. [DOI] [PubMed] [Google Scholar]
- 21.Ku S.J., Chang Y.I., Chae C.H., et al. Static tensional forces increase osteogenic gene expression in three-dimensional periodontal ligament cell culture. BMB Rep. 2009;42:427–432. doi: 10.5483/bmbrep.2009.42.7.427. [DOI] [PubMed] [Google Scholar]
- 22.Wen Q., Jing J., Han X., et al. Runx2 regulates mouse tooth root development via activation of WNT inhibitor NOTUM. J Bone Miner Res. 2020;35:2252–2264. doi: 10.1002/jbmr.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L., Zeng Z., Kang N., Yang J.C., Wei X., Hai Y. Circ-VANGL1 promotes the progression of osteoporosis by absorbing miRNA-217 to regulate RUNX2 expression. Eur Rev Med Pharmacol Sci. 2019;23:949–957. doi: 10.26355/eurrev_201902_16981. [DOI] [PubMed] [Google Scholar]
- 24.Schroeder T.M., Jensen E.D., Westendorf J.J. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- 25.Ziros P.G., Basdra E.K., Papavassiliou A.G. Runx2: of bone and stretch. Int J Biochem Cell Biol. 2008;40:1659–1663. doi: 10.1016/j.biocel.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Li Q., Liu S., et al. Periodontal ligament stem cells in the periodontitis microenvironment are sensitive to static mechanical strain. Stem Cell Int. 2017;2017 doi: 10.1155/2017/1380851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H., Jia P. MiR-153-3p inhibits osteogenic differentiation of periodontal ligament stem cells through KDM6A-induced demethylation of H3K27me3. J Periodontal Res. 2021;56:379–387. doi: 10.1111/jre.12830. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y.H., Li S.Y., Yuan S.J., Pan Y.X., Hua Y., Liu J.Y. MiR-375 promotes human periodontal ligament stem cells proliferation and osteogenic differentiation by targeting transducer of ERBB2, 2. Arch Oral Biol. 2020;117 doi: 10.1016/j.archoralbio.2020.104818. [DOI] [PubMed] [Google Scholar]
- 29.Yan G.Q., Wang X., Yang F., et al. MicroRNA-22 promoted osteogenic differentiation of human periodontal ligament stem cells by targeting HDAC6. J Cell Biochem. 2017;118:1653–1658. doi: 10.1002/jcb.25931. [DOI] [PubMed] [Google Scholar]
- 30.Zeng H.B., Dong L.Q., Huang Y.L., Xu C., Zhao X.H., Wu L.G. USF2 reduces BMP3 expression via transcriptional activation of miR-34a, thus promoting osteogenic differentiation of BMSCs. J Bone Miner Metabol. 2021;39:997–1008. doi: 10.1007/s00774-021-01254-x. [DOI] [PubMed] [Google Scholar]
- 31.Hong M., Zhang X.B., Xiang F., Fei X., Ouyang X.L., Peng X.C. MiR-34a suppresses osteoblast differentiation through glycolysis inhibition by targeting lactate dehydrogenase-A (LDHA) In Vitro Cell Dev Biol Anim. 2020;56:480–487. doi: 10.1007/s11626-020-00467-0. [DOI] [PubMed] [Google Scholar]
- 32.Kong Y., Chen Z.T. MiR-146a regulates osteogenic differentiation and proliferation of bone marrow stromal cells in traumatic femoral head necrosis. Eur Rev Med Pharmacol Sci. 2019;23:441–448. doi: 10.26355/eurrev_201901_16853. [DOI] [PubMed] [Google Scholar]
- 33.Wan S., Wu Q., Ji Y., Fu X., Wang Y. Promotion of the immunomodulatory properties and osteogenic differentiation of adipose-derived mesenchymal stem cells in vitro by lentivirus-mediated mir-146a sponge expression. J Tissue Eng Regen Med. 2020;14:1581–1591. doi: 10.1002/term.3113. [DOI] [PubMed] [Google Scholar]
- 34.Dasgupta T., Ladd A.N. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscipl Rev RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladd A.N. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol Cell Neurosci. 2013;56:456–464. doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veeman M.T., Newman-Smith E., El-Nachef D., Smith W.C. The ascidian mouth opening is derived from the anterior neuropore: reassessing the mouth/neural tube relationship in chordate evolution. Dev Biol. 2010;344:138–149. doi: 10.1016/j.ydbio.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Yu J., Mu J., Guo Q., et al. Transcriptomic profile analysis of mouse neural tube development by RNA-Seq. IUBMB Life. 2017;69:706–719. doi: 10.1002/iub.1653. [DOI] [PubMed] [Google Scholar]
- 38.Mao S., Zhang S., Zhou Z., et al. Alternative RNA splicing associated with axon regeneration after rat peripheral nerve injury. Exp Neurol. 2018;308:80–89. doi: 10.1016/j.expneurol.2018.07.003. [DOI] [PubMed] [Google Scholar]