Abstract
Conventional drug discovery and development is tedious and time-taking process; because of which it has failed to keep the required pace to mitigate threats and cater demands of viral and re-occurring diseases, such as Covid-19. The main reasons of this delay in traditional drug development are: high attrition rates, extensive time requirements, and huge financial investment with significant risk. The effective solution to de novo drug discovery is drug repurposing. Previous studies have shown that the network-based approaches and analysis are versatile platform for repurposing as the network biology is used to model the interactions between variety of biological concepts. Herein, we provide a comprehensive background of machine learning and deep learning in drug repurposing while specifically focusing on the applications of network-based approach to drug repurposing in Covid-19, data sources, and tools used. Furthermore, use of network proximity, network diffusion, and AI on network-based drug repurposing for Covid-19 is well-explained. Finally, limitations of network-based approaches in general and specific to network are stated along with future recommendations for better network-based models.
Keywords: Drug repurposing, Network analysis, Machine learning, Deep learning, Network proximity, Network diffusion, AI on networks
Graphical Abstract
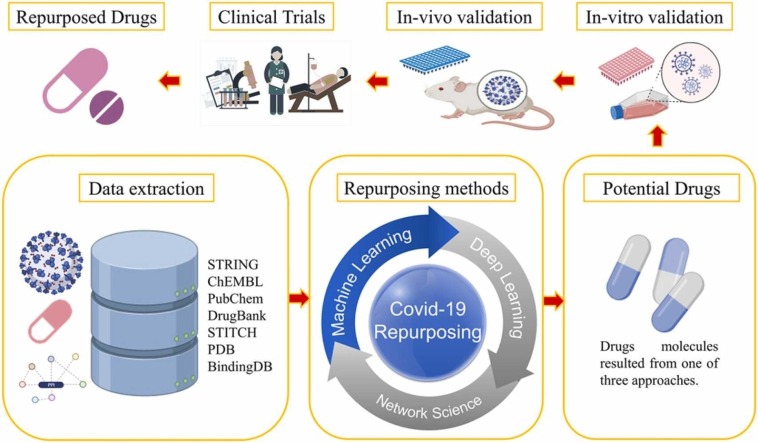
Process of drug repurposing in Covid-19. The data related to drugs, diseases, proteins, and genes etc. are extracted from different data sources (databases) followed use of different repurposing methods. Repurposing methods are used to find out the potential drugs which are then validated in in-vitro (2D or 3D) cell culture and in-vivo animal models. Finally, the shortlisted drugs are forwarded to clinical trials and successful drugs are repurposed
1. Introduction
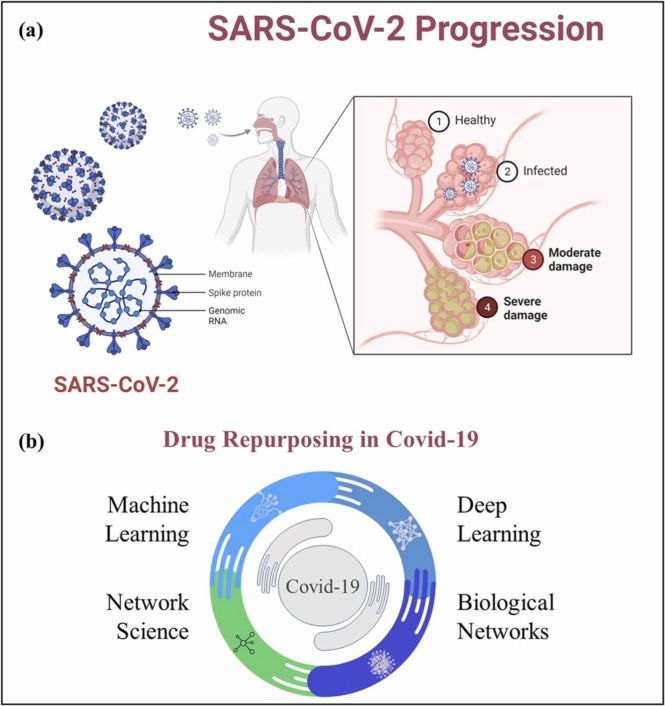
A novel coronavirus (CoV) first appeared by the end of 2019 in Wuhan, China and spread to over 70,000 individuals with 1800 death cases within the matter of first 50 days of the disease [1]. Public health emergency of international health concern was declared by the World Health Organization (WHO) on January 30, and it scaled up to pandemic as of March 11, 2020 [2]. The analytical methods like amino acid and nucleotide sequencing confirmed the relation of novel CoV to same class as severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) [3]. This new version of coronavirus was named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the international committee on the taxonomy of viruses and similarly, the disease was named as Covid-19/SARS-CoV-2 [4]. SARS-CoV-2 is a variant of coronavirus with enveloped single-stranded RNA which bears a corona like structure of spikes as shown in Fig. 1(a). The main host protein for the attachment of spikes has identified as angiotensin converting enzyme 2 (ACE2) receptor. This variant of coronavirus evolved to become the most contagious and infectious among coronavirus family. It enters the lungs by inhaling the contaminated air with viral bodies and infects distal airway cells leading to further spread towards alveolar epithelial cells and causes the damage starting from infection to severe damage as shown in Fig. 1(a). As of May 25, 2021, total number of Covid-19 cases reported have surpassed the 30 million cases globally with 1.6 % in US only [5]. Owing to its high mortality, morbidity, and contagiousness there is dire need for the effective treatments.
Fig. 1.
Workflow the current study. a) the schematics of Sar-CoV-2 virus structure and its pathogenesis in alveoli with different stages, and b) different drug repurposing methods for Sar-CoV-2 discussed in the present study.
Whereas, drug development is complex, time taking, and expensive process. On average it takes around 12–15 years with $2–$3 billion of investment to take a de novo drug from concept to a market ready product [6]. The total number of drugs which are getting the FDA approval are less than 10 % and this rate is declining with time [7]. As a result, pharmaceuticals research suffers from decrease in productivity and faces a persistent gape between the need of treatments and available response [8], [9]. Repurposing the already approved drugs is a quick and efficient to provide the new treatments by finding out novel uses of the drugs that have well known safety and efficacy profiles. Many drugs have already been repurposed based on variety of the methods and have been approved for new indication [10]. While repurposing can be done at the any stage during drug discovery and development, but its actual potential can be seen in case of already approved drugs [11]. Thus, recently studies have been shown to increasingly use the computational methods to predict the novel drug-target interactions for repurposing the drug or target. One of the methods widely used for drug repurposing is network-based approach. Network based approaches are in-silico approaches which are faster with lower-cost as compared to experimental approaches and serve as a robust strategy to identify the novel drug target interactions or repurposed compound among a pool of many.
Moreover, in the wake of current situation novel Covid-19 (severe acute respiratory syndrome coronavirus 2) has infected around 526.7 million people and have caused more than 6 million deaths globally [5]. In an emergency situations like Covid-19 de novo drug development is not the feasible option since there is urgent need to find the treatments which can be immediately prescribed to prevent the mortality and morbidity. So, to face the crisis because of pandemic drug repurposing seems a tangible strategy for fighting against Covid-19 and other such viral diseases. Additionally, a lot of methods, applications, and databases have been proposed in literature motivating for information rich studies for furthering in unprecedented yet potential direction. Recently, multiple review articles have been published focusing on artificial intelligence (AI) algorithms [12] such as deep learning models [13], and network-based methods [14]. Moreover, survey targeting the drug repurposing have also been performed [15], [16] and these include comments and opinion focusing the drug design [17] and development process [18]. Additionally, articles summarizing the covid-19 drug repurposing using machine learning [19] and deep learning [20], [21] methods are published as well. To the best of our knowledge, until now there is no review study until now has published which summarizes the drug repurposing studies for Covid-19 with special emphasis on network-based drug repurposing approaches to Covid-19.
As new studies are reported widely almost every day, so it is not applicable to provide a wide and comprehensive overview of all the available repurposing studies. Thus, this review is mainly focused to bridge the gap by providing overview of machine learning and deep learning approaches to drug repurposing with focus on recent developments in network-based drug repurposing methods, tools, and databases used in Covid-19 as shown in Fig. 1(b). In this connection, first we present an overview to computational drug repurposing studies for Covid-19 along with drug and disease related databases followed by recent machine learning and deep learning-based drug repurposing studies. Next, the overview of different network methods and recent state of art studies categorized for each network type is explained. Moreover, recent network-based drug repurposing studies for Covid-19 are discussed and their limitations, benefits, and tools used are summarized. Finally, limitations and future recommendation for improved outcome of network-based drug repurposing in Covid-19 and in general are discussed followed by conclusion.
2. Computational approaches to drug repurposing
Generally, drug repurposing principals can be classified in to two major categories, drug-based or disease-based repurposing. As per the common and widely used hypothesis a drug can cure two diseases if there exist some similarity or the interdependence between two diseases. Similarly, a drug can be associated with various targets and targets owing to confounding nature of the drugs [22], [23]. Additionally, when it comes to computational drug repurposing, a major challenge is to distinguish between the drug targets and additional gene products which are indirectly participating in the activity of the targets. Unfortunately, traditional methods are limited in their ability to recognize the molecular targets of drugs in a vast population of gene products followed using small datasets originated from different platforms and environmental settings. This might cause inaccuracies in the findings reported in some studies [22]. Small datasets and networks may give the partial knowledge of the living systems.
The amount of biomedical and pharmaceutical data has seen the tremendous rise in the recent years so the improvement in the computational drug repurposing approaches is also visible. Computational approaches like data mining, machine and deep learning, and network analysis have been taken to depth to systematically repurpose the drugs to overcome the limitations faced by the traditional statistical approaches.
Newly adopted computational methods and approaches for demystifying the relationships among different types of biological networks have benefitted the drug repurposing to a greater length. Different biological entities such as genes, proteins, drugs, and diseases and interaction can uncover the potential therapeutic opportunities. The overview of the computational drug repurposing studies with emphasis on network-based approaches is given in Table 5, Table 6, Table 7.
Table 5.
Network Proximity based drug repurposing studies in Covid-19 along with datasets used, number of repurposed drugs, evaluation criteria, and tools used.
| Title | Datasets used | Repurposed Drugs | Evaluation Criteria | Tools used | Ref. | |
|---|---|---|---|---|---|---|
| 1 | Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2 | GEO, CMAP, DrugBank, Uniprot | 16 Drugs And 4 Drug combination |
Network Proximity measures, Gene enrichment |
Gephi, Enricher, KEGG, GO | [164] |
| 2 | Designing a Network Proximity-Based Drug Repurposing Strategy for COVID-19 | DrugBank, GEO, GeneCards |
– | Network proximity, Functional analysis |
VarElect | [169] |
| 3 | SAveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19 | DrugBank, Uniprot, TTD, Phenopedia, CMAP |
24 Drugs, 5 Drug combinations |
Network Proximity measures, Gene enrichment, ROC performance Curve |
KEGG Pathway analysis, GSEA | [170] |
| 4 | Network-Based Approach to Repurpose Approved Drugs for COVID-19 by Integrating GWAS and Text Mining Data | GWAS, LINCS, DrugBank |
13 Drugs | – | GWAS | [175] |
| 5 | Network medicine framework for identifying drug-repurposing opportunities for COVID-19 | 13 Datasets, DrugBank, STRING | 989 Drugs, 77 Validated in VeroE6 Cells, 76/77 validated in Human Cells | Network proximity, network diffusion, Network AI | Experimental, Ensembl algorithmic prediction | [176] |
| 6 | Integrative resource for network-based investigation of COVID-19 combinatorial drug repositioning and mechanism of action | STRING, DrugBank, ChEMBL, PubChem, GO, KEGG, ClinicalTrials.gov, SIDER, Uniprot, SNAP | 867 FDA approved Drugs and Combinations | Proximity, Gene ontology | – | [177] |
| 7 | A Network Medicine Approach to Investigation and Population-based Validation of Disease Manifestations and Drug Repurposing for COVID-19 | CMAP, DrugBank, STRING, GEO |
34 Drugs | Network Proximity measures | GSEA, RNA-Sequencing | [168] |
Table 6.
AI Based drug repurposing application on networks.
| Title | Datasets used | Repurposed Drugs | Evaluation Criteria | Tools used | Ref. | |
|---|---|---|---|---|---|---|
| 1 | Network medicine framework for identifying drug-repurposing opportunities for COVID-19 | 13 Datasets, DrugBank, STRING | 989 Drugs, 77 Validated in VeroE6 Cells, 76/77 validated in Human Cells | Network proximity, network diffusion, Network AI | Experimental, Ensembl algorithmic prediction | [176] |
| 2 | Drug repurposing for COVID-19 using graph neural network and harmonizing multiple evidence | CTDbase, STRING, Hetionet, DrugBank | 22 drugs and drug combinations | Validation with multiple sources, In-vitro screening | GSEA, Plotly, Gephi, GNN | [178] |
| 3 | COVID-19 Multi-Targeted Drug Repurposing Using Few-Shot Learning | Chemical Molecules, ZINC, ChEMBL, | – | Loss function and metrics | Few-shot learning | [179] |
| 4 | Machine learning and network medicine approaches for drug repositioning for COVID-19 | STRING, ChEMBL, DrugBank | – | In-vitro, In-vivo, clinical trials, and CMAP | – | [180] |
| 5 | Using informative features in machine learning based method for COVID-19 drug repurposing | STRING, DrugBank, Uniprot | 80 % of the predictions were verified | Statistical and clinical evidence | Enrichment analysis, | [182] |
| 6 | An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19 | Literature, DrugBank, CTD, Uniprot, CMap | 41 Drugs | wet-lab, Expression profiles of patients and drug perturbation cells, clinical trials data | Molecular docking | [183] |
| 7 | Network-based repurposing identifies anti-alarmins as drug candidates to control severe lung inflammation in COVID-19 | Uniprot, STRING, CMap, LINCS, GEO | – | Gene expression profiling, CMap ranking | LINCS tools | [184] |
| 8 | Drug repurposing for COVID-19 via knowledge graph completion | PubMed and Covid-19 research literature, ClinicalTrials.gov | 5 Drugs | ClinicalTrials.gov | – | [185] |
| 9 | Multiscale interactome analysis coupled with off-target drug predictions reveals drug repurposing candidates for human coronavirus disease | PolypharmDB, DrugBank, STRING | 26 Drugs | Infectivity assays, combination of human coronavirus entry and infection assays | MatchMaker | [186] |
Table 7.
Network diffusion based drug repurposing studies for Covid-19.
| Title | Datasets used | Repurposed Drugs | Evaluation Criteria | Tools used | Ref. | |
|---|---|---|---|---|---|---|
| 1 | Designing a Network Proximity-Based Drug Repurposing Strategy for COVID-19 | BioGRID | – | Network proximity/ Network Diffusion | Cytoscape, VarElect tool |
[169] |
| 2 | Network medicine framework for identifying drug-repurposing opportunities for COVID-19 | 13 Datasets, DrugBank, STRING | 989 Drugs, 77 Validated in VeroE6 Cells, 76/77 validated in Human Cells | Network proximity, network diffusion, Network AI | Experimental, Ensembl algorithmic prediction | [176] |
| 3 | Drug repurposing for coronavirus (SARS-CoV-2) based on gene co-expression network analysis | DGIDb, gene co-expression, DrugBank | 5 Drugs | Gene enrichment analysis for Genes and miRNA | Node degree and centralities | [195] |
| 4 | Network-based repurposing identifies anti-alarmins as drug candidates to control severe lung inflammation in COVID-19 | Uniprot, STRING, CMap, LINCS, GEO | – | Gene expression profiling | CMap ranking | [184] |
| 5 | Integrative In Silico Investigation Reveals the Host-Virus Interactions in Repurposed Drugs Against SARS-CoV-2 | STITCH, KEGG, BioGRID, PubChem, IID, | – | Enrichment analysis, Molecular docking | DAVID, GOplot, AutoDock Vina, Cytoscape, Ligplot+ | [196] |
| 6 | Discovery of Potential Therapeutic Drugs for COVID-19 Through Logistic Matrix Factorization with Kernel Diffusion | – | 4 Drugs | 5-fold cross validations, AUC, AUPRs, recall, similarity diffusion | Molecular docking | [197] |
| 7 | HeTDR: Drug repositioning based on heterogeneous networks and text mining | Mesh, DrugBank, PubMed | 10 Drugs | AUPR, AUROC, F1-measure | – | [198] |
| 8 | MNBDR: A Module Network Based Method for Drug Repositioning | STRING, Cmap, LINCS, GEO, | – | AUC, FPR, Specificity, FDR | GSEA, KEGG enrichment | [199] |
2.1. Machine learning
First attempt to drug repurposing started around two decades ago using simple similarity measure with single source of biological and biomedical data utilization and finally reached to machine learning [24], [25] break through. Artificial and machine learning technology play a crucial role which is modernized with the advent of deep learning and neural network algorithms. Machine learning algorithm can learn from the data and mimic the cognitive human behavior [26]. These days, biological scientists have integrated the machine learning in drug design and discovery processes for improved outcomes [8], [27], [28], [29].
The applications of the machine learning in drug discovery and development are numerous which include but are not limited to virtual screening (both primary and secondary), toxicity monitoring of the drug, drug efficacy prediction, dosage predictions, drug repurposing, and drug target interactions (DTIs) predictions [25]. Some of the machine learning methods used for drug repurposing in Covid-19 include k-nearest neighbors [30], random forest [31], support vector machine [32], [33], and more. Thus, over the past decade AI based applications have helped the drug discovery efforts [34], [35]. Moreover, with the invention of deep learning and its ability to extract the features from raw data automatically, has increased the impact of AI approaches as compared to other Insilico methods [36], [37]. Various deep learning algorithms utilized in finding treatment against Covid-19 includes artificial neural networks (ANN), convolutional neural networks (CNN), and long short-term memory (LSTM).
Apart from this, to accelerate the process of drug development for Covid-19, machine learning based molecular docking frameworks were utilized on approved as well as new chemical entities. ML enabled molecular docking has been utilized a lot in virtual screening and repurposing [38], [39], [40], [41]. ML enabled molecular docking methods help to find out the bioactive molecules against disease protein of interest.
2.1.1. Drug repurposing based on classification
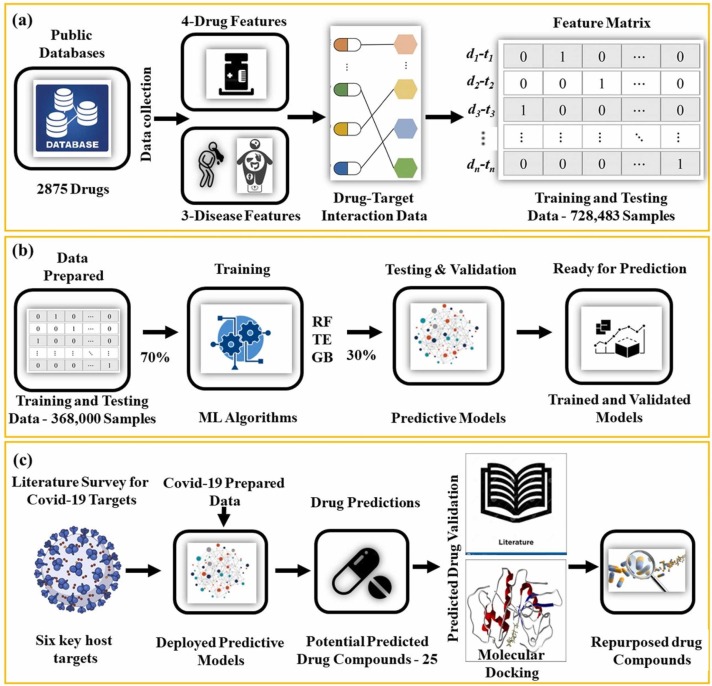
There are a lot of studies which have taken the drug repurposing task as binary classification or classification task. Drug-target interaction (DTI) prediction or drug-disease association prediction can help to reduce the volume of redundant compounds and thus serve as an important step in drug development. In our recent work, we developed a feature enriched drug repurposing framework named SperoPredictor [42] which consisted multiple state of art machine learning algorithms as shown in Fig. 2(b). The algorithms were individually trained and later stacked together (Fig. 2(b)) followed by molecular docking-based validation (Fig. 2(c)). SperoPredictor training was performed on the feature enriched drug-disease association data of 1430 FDA approved drugs collected from various databases and then unified into binary strings as shown in Fig. 2(a). The training data contained four drug features and three disease features all unified in binary strings which enabled the reduction in training time and space required. The machine learning models were tested on the performance parameters such as accuracy, precision, f1 score, Mathew’s correlation coefficient (MCC), sensitivity, and specificity. The testing accuracies of the models were reported to be 99.3 % for Random Forest (RF) and 99.03 % for Tree Ensemble (TE). SperoPredictor was deployed to predict the potential drugs to be repurposed for Covid-19, NASH, and renal fibrosis. In case of Covid-19, 25 potential candidate drugs were predicted. Literature survey of peer reviewed journals, databases, and patents suggested 12/25 drugs (48 %) were already used for Covid-19. Additionally, molecular docking validation results suggested that four drugs (Balaglitazone, Cortivazol, Velusetrag, and 16 alpha Bromoepiandrosterone) can be used for preclinical validation and clinical trials prior to use in patients. The SperoPredictor results testify the usefulness of having such platforms that can be readily deployed in case of emergent situations like Covid-19. Moreover, the importance of the DTI prediction is highlighted in many studies such as in [43]. These studies to predict the drug-disease associations or drug target interactions require various resources such as done in [44]. Drug-drug interaction based on structural similarity and side effects and similarly disease-disease based on gene expression or disease phenotype have been used. Many online databases such as DrugBank [45], CTD [46], and ChEMBL [47] are providing the open-source drug-target data. The other databases for the various type of drug and disease related data are mentioned in the Tables 1 and 2 along with other information. Whereas Table 1 provides information related Drug specific databases followed by Table 2 providing information on disease information contained databases. Additionally, drug-drug, disease-disease similarity measures were used here as classification features followed using logistic regression. These interactions are normally used in ML and DL studies as shown in Fig. 3(a-b). Moreover, in the mentioned study classifier was trained to distinguish between the true and false drug-disease association leading to prediction of undiscovered drug-disease associations. Another study designed to predict the DTIs in Covid-19 is given in [48]. The study avails the proteins and drugs provided in their structured format. The physical and chemical features were extracted from the protein amino-acid sequences of drug targets and from SMILES forms of the drug structures [49], [50], [51], [52]. Then the developed model was compared with existing studies using k-fold cross validation (Fig. 3(b)). To deploy the model covid-19 was selected as a case study. Prediction model was deployed as shown in Fig. 3(c) on the proteins known to have been involved in the Covid-19 to predict the possible interactions or drug-target interactions. The model was further deployed to drugs active in Covid-19 and the drugs were predicted with 100 % accuracy. The interaction of the drugs was shown with ACE2 [53], [54], [55], [56] self-membrane protein, a key player in Covid-19 enabling disease infection.
Fig. 2.
SperoPredictor framework for drug repurposing and a) shows how drug and disease related information is collected from various databases and transformed into a feature matrix of binary strings. Whereas b) depicts the development, training, testing and validation of machine learning algorithms and finally c) demonstrates the procedure for the deployment of SperoPredictor followed by the literature and molecular docking-based validation [42].
Table 1.
Drug related databases and their information of the data type, data statistics, availability, and programmatic access are summarized.
| Database name | Type | Statistic | Availability | Programmatic access | |
|---|---|---|---|---|---|
| 1 | PubChem | Structural and common data | PubChem contains 237 million bioactivities for three million compounds, determined in over 1.2 million biological assay experiments | Open source | Rest API |
| 2 | ChEMBL | Structural and common data | more than 2.2 million distinct compound structures, with 5.4 million activity values from more than 1.5 million assays. | Open source | Rest API |
| 3 | STITCH | DTIs | 0.5 million Chemicals, 9.6 million proteins, and 1.6 billion interactions | Open source | Rest API |
| 4 | TTD | DTIs | 33598 drug-like agents, 1102 targets and their interactions. | Open source | Django Rest API |
| 5 | DrugBank | Common resource | Total small molecules 11937, Biotech drugs 2687, approved drugs 4243, approved small molecules 2725 | Open source and commercial | – |
| 6 | BindingDB | DTIs | containing ∼20 000 experimentally determined binding affinities of protein–ligand complexes, for 110 protein targets including isoforms and mutational variants, and ∼11 000 small molecule ligands | Open source and commercial | Rest API |
| 7 | DT-Web | DTIs | – | Open source | R-CMD |
| 8 | TCGA | Perturbation Data | TCGA generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data | Open source | – |
| 9 | SIDER | Side effect data | Side effects of 1400 FDA approved Drugs | Open source | – |
| 10 | PharmaGKB | Drug-Disease associations | – | – | API |
| 11 | GEO | Perturbation Data | Contains around 4348 datasets, 23,172 platforms, and 5,029,933 samples. | Open source | – |
| 12 | LINCS | Perturbation Data | Gene expressions of thousands of perturbations | Open source | – |
Table 2.
Various databases related to diseases, genes, proteins, and drugs and other information about their data type, statistics, availability, and programmatic access.
| Database name | Type | Statistic | Availability | Programmatic access | |
|---|---|---|---|---|---|
| 1 | OMIM-21 | Gene–disease association | Autosomal – 24,944, X linked – 1331, Y linked – 63, mitochondrial – 71 | Upon request | – |
| 2 | miR2disease – 22 | miRNA–disease association | Number of miRNAs: 349 Number of diseases: 163 Number of entries: 3273 |
Open source | – |
| 3 | STITCH-23 | Chemical–Protein interaction | 0.5 million Chemicals, 9.6 million proteins, and 1.6 billion interactions | Open source | Rest API |
| 4 | HPRD-24 | Protein–protein interaction (PPI) | PPIs – 41,327, Protein expressions – 112,158, PubMed Links 453,521 | Open source | – |
| 5 | BioGRID-25 | PPI/genetic interaction | 86,000 + genes, 745 different cell lines, 127 different cell types, and 23 different phenotypes across 4 different organisms | Open source | Rest API, Blast, and more |
| 6 | DIP-26 | PPI | – | Upon request | – |
| 7 | TRED-27 | TF–gene interaction | It contains 11,660 target genes and around 14,908 target promoters | – | Sequence tool analysis |
| 8 | TRANSFAC-28 | TF–gene interaction | – | Paid | MATCH Suite |
| 9 | miRbase-29 | MicroRNA–gene interaction | 38,589 miRNA entries | Open source | Blast |
| 10 | TargetScan-30 | MicroRNA–gene interaction | – | Open source | – |
| 11 | Signaling Network-31 | Human signaling network | – | Open source | Multiple tools |
| 12 | GO-32 | Functional annotation | More than 700,000 experimentally supported | Open Source | Multiple tools |
| 13 | CMAP-33 | Microarray | library containing over 1.5 M gene expression profiles from ~5000 small-molecule compounds, and ~3000 genetic reagents, tested in multiple cell types | Open source | Rest API, multiple tools |
| 14 | Arrayexpress-34 | Microarray | It is consisted of 75,570 experiments and 25,96,304 assays | Open Source | Rest API, Blast, Multiple tools |
| 15 | GEO-35 | Microarray | Contains around 4348 datasets, 23,172 platforms, and 5,029,933 samples. | Open source | – |
| 16 | DCDB-39 | Drug combination–disease association | It contains the drug combinations of 330 approved and 1033 investigational drugs. | Open source | – |
| 17 | KEGG-40 | Pathway,disease, drug | Information can extract and stats are different for different KEGG databases | Open source | Fasta, Blast |
| 18 | ClinicalTrial.gov | Drug–disease association | It contains around 414,049 studies conducted in 220 countries | Open source | – |
| 19 | Drugs@FDA | Drug–disease association | – | Open source | – |
Fig. 3.
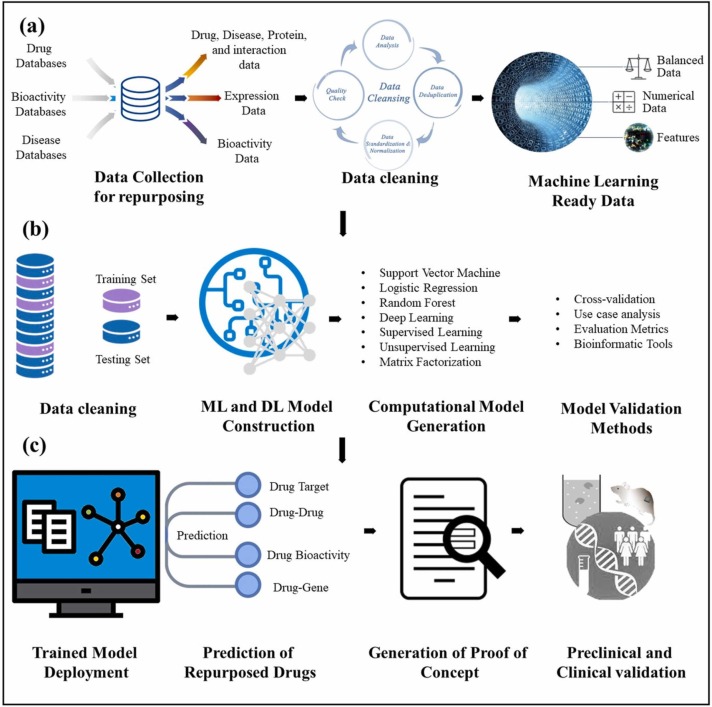
Machine learning and deep learning-based drug repurposing methods. a) preprocessing of the data accessing different databases following by data cleaning and then making the data machine learning ready. b) splitting of the training and then training and testing of the machine and deep learning models is carried out. Different machine learning and deep learning models are described followed by model validation methods and finally c) shows the deployment of the trained models and prediction validation.
2.1.2. Drug repurposing based on collaborative or similarity filtering
There are many studies which have applied the machine learning based collaborative filtering for predicting novel drug-disease associations or repurposed indications backed by trends found in the gene expressions of different samples [22], [57], [58], [59]. One such study is present in [60], multiple drug similarity datasets were constructed for finding the therapeutic classes of FDA approved drugs. In this method any mismatches between known and unknow associations were considered as potential repurposed indication. Drug-drug similarity matrices were constructed based on gene expression, structural, and molecular target-based similarity. The prepared data was used to train the support vector machine (SVM) classifier followed by the collaborative filtering to predict novel drug-disease associations. Moreover, a unified computational framework was proposed [61] based upon integration of various heterogenous data sources to foresee the drug-drug, disease-disease, and drug-disease similarity (known) based data set for building drug-disease network. The problem set forth in the form of drug-disease network was optimally solved by using block coordinate descent (BCD) strategy. A causal inference-probabilistic matrix factorization (CI-PMF) approach was followed in [61] for classification of novel drug-disease associations. Many biomedical and biological sources were leveraged to build a causal network which contained multiple nodes such as drugs, genes, pathways, and diseases together. Probabilistic matrix factorization (PMF) model was constructed following the network creation to classify drug-disease associations in different groups followed by drug ranking leading to identification of novel drug-disease associations. Additionally, somehow similar studies on larger scale were carried out in [62], [63], [64] to infer off-target drug interactions for undiscovered drug candidates’ identification. Machine learning plays a key role in drug repurposing against Covid-19 and pace of research in the field has skyrocketed since the emergence of Covid-19. However, there are still some issues to be addressed. For example, lack of standardized frameworks, because without standardization the results received from machine learning models cannot be translated to clinical research. Additionally, there is lack of replicability and reproducibility, despite these, benefits offered by ML methods are numerous within the field of drug repurposing.
2.2. Deep learning
Robust and efficient computational methods making use deep learning are widely required for drug discovery against Covid-19 [65]. The overall combined workflow of ML and DL is shown in Fig. 3 and few recent well-known studies along with further information are summarized in Table 3. Compared to machine learning (ML), deep learning (DL) methods enable the automatic feature extraction and feature engineering. Convolutional neural networks and recursive neural networks have been widely used in bioinformatics studies with successful results such as in Covid-19 [21], [66], [67], [68]. Since deep learning models are good at finding the intricate underlying patterns in data, thus they are suitable to large scale unstructured data such as electronics health records (EHR), whole proteome, and human genome leading to high chances and suitability to be applied in life sciences applications such as drug repurposing [65]. In comparison to state of art machine learning methods, deep learning is distinguished by the flexibility of neural networks [36]. Deep learning methods have also been widely used in drug repurposing against Covid-19. A few well-known studies are given in Table 3.
Table 3.
Machine learning and deep learning based drug repurposing studies for Covid-19.
| Study | ML Models | Dataset | Targets | Drugs | Ref. | |
|---|---|---|---|---|---|---|
| 1 | AI approach fighting COVID-19 with repurposing drugs | Deep neural network | Drugs with known profiles | FIP and Covid-19 | 13/80 Drugs showed in-vitro activity | [80] |
| 2 | Deep Learning based prediction of Commercially available drugs | Deep learning-based, Molecule Transformer-Drug Target Interaction (MT-DTI) |
SMILES and DTIs | Covid-19 associated 3CLpro, RdRp, helicase, 3′-to-5′ exonuclease, endoRNAse, and 2′-O-ribose methyltransferase | 8 Drugs | [70] |
| 3 | machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection | mechanistic model | Drug targets, pathway genes, Gene expression | Covid-19 related signaling pathways and transduction circuits | – | [81] |
| 4 | Open Data to Discover Therapeutics for COVID-19 Using Deep Learning | Deep learning-based knowledge graph | Knowledge graph of 15 million edges across 39 types of relationships | Covid-19 | 41 Drugs | [72] |
| 5 | Repositioning of 8565 Existing Drugs for COVID-19 | 2-D fingerprint, GBDT model, Recurrent Neural Network (RNN) | Largest available experimental data set for SARS-CoV-2 or SARS-CoV 3CL (main) protease inhibitors | Covid-19 associated 3CLpro | 40 Drugs | [82] |
| 6 | Large scale virtual screening of ligand using DNN | Deep neural network enabled ChemAI | 220 M data points across 3.6 M molecules from three public drug-discovery databases | Covid-19 related 3CLpro, PLP | 20 Drugs | [83] |
| 7 | Deep learning enabled docking to rapidly identify the potential Covid-19 inhibitors | Deep learning-based Docking Platform | Purchasable compounds obtained from databases. (1.3 billion compounds from Zinc) | Covid-19 associated Mpro | 1000 Drugs | [78] |
| 8 | Prediction of potential commercially inhibitors against SARSCoV-2 by multi-task deep model | Multi-task deep learning model for classification and regression | Covid-19 related viral protein dataset | Covid-19 related 3CLpro | 10 Drugs | [84] |
| 9 | DeepPurpose: a deep learning library for drug–target interaction prediction | Convolutional neural network (CNN), Deep purpose | DTIs | Covid-19 related 3CLpro | 13 Drugs | [85] |
| 10 | Screening of Therapeutic Agents for COVID-19 Using Machine Learning and Ensemble Docking Studies | Random forest (RF) regression algorithm, ensemble docking | DTIs | S, S-ACE2 complex | 187 Drugs | [86] |
| 11 | SARS-CoV-2 using ML from a > 10 million chemical space | Support vector machine (SVM), Random Forest (RF) | assay data of 65 Covid-19 host targets and purchasable 14 million chemicals | Covid-19 host targets | 58 Drugs | [87] |
A recent work involving drug repurposing in Covid-19 using deep learning is given in [69] and are given in Table 3. Here, deep learning model named as Molecule Transformer-Drug Target Interaction (MT-DTI) was used for predicting novel drug target interactions (DTIs). It utilized chemical structure SMILES and amino acid sequences (as a chemical-protein pairs) as an input obtained from BindingDB and Drug Target Commons (DTC) databases. MT-DTI predicted several commercially available or in practice anti-viral drugs having potency against Covid-19 protein targets [70] confirmed by the pre-clinical validation assays of the Covid-19. MT-DTI model was deployed for the selection of compounds from 1400 FDA approved drugs having the strong affinity against the host cellular targets for viral diseases [71]. Another deep learning-based drug repurposing study done in [72] used deep learning on a massive knowledge graph to identify and select promising anti-Covid-19 drug candidates. The knowledge graph consisted around 15 million edges with 39 types of relationship connecting the whole bunch of networks together. Deep learning was primarily used in this study to learn representation of graph entities leading to 41 repurposed drug candidates. These drugs include dexamethasone (an anti-inflammatory) agent having bioactivity against Covid-19 and Niclosamide [73] validated by proteomics of Covid-19 infected cell and gene expression.
Some studies have adopted hybrid strategy where deep learning is coupled with molecular simulation to further prioritize or performing the screening procedure for the predicted drug target interactions as shown in Fig. 3(b-c). One such study is done in [74] which focused on RNA-dependent RNA polymerase (RdRp) targeting candidate drug molecules from 1906 FDA approved drugs. Drug candidate such as pralatrexate [75] and azithromycin [76] predicted from this study were validated in in-vitro testing and found to inhibit Covid-19. First drug was active once the virus entered host cells while the later mentioned drug has efficacy in before and after stages.
Additionally, software tools (MolAICal) are also developed which combine the deep learning models along with classical algorithms to find the drugs which interact effectively with 3D pockets of proteins [77] ( Fig. 4(c)). Deep learning enabled framework, MolAICal, were trained using the approved drug fragment data available in PDBbind and drug like, bioactive molecules available in ZINC. To demonstrate the drug design features of the MolAICal, membrane protein glucagon receptor and Covid-19 main protease were modelled [72]. Other tools include Deep Docking (DD) [78] platform developed using deep learning which avails the QSAR model good at performing virtual screening to find the potential drug candidate or lead like molecule from ZINC database. Using the proposed method in study [78] led to identification of 1000 top ranked potential drug compound supposed to have efficacy against Covid-19 Mpro.
Fig. 4.
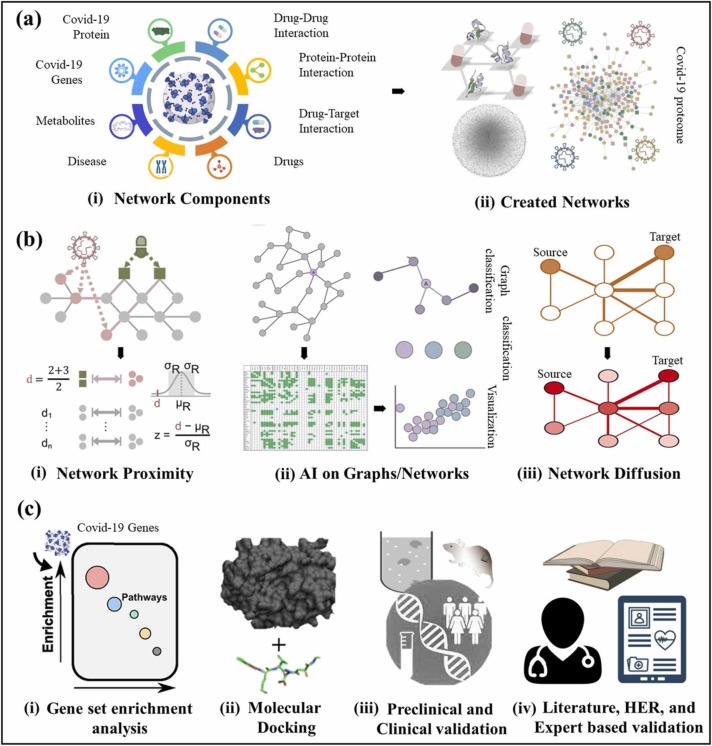
Network based approaches to drug repurposing for Covid-19. a) Different biological entities related to Covid-19 disease pathogenesis and drugs in general which are later used to create a network. b) Network created can be analyzed based on network proximity, AI on graphs, and network diffusion for repurposing the potential drugs. Finally, c) shows the different Insilico, in-vitro, and in-vivo validation methods for results obtained.
Deep learning methods (such as Fig. 3(b)) has number of advantages in comparison to traditional methods such as ability of DL models to model and learn complex features automatically. DL has helped in finding the repurposed drugs against Covid-19 and in other pathogens however certain challenges still exist. First to mention is the black box nature of DL models, lack of interpretability, output is dependent on the input data, and large standardized and dataset of biochemical data is necessary to gain the better learning and performance outcome of the system. Although there is huge amount of data already available but mining and making the standard data ready for ML and DL applications is still challenging. Hopefully, the large dataset which will be developed in future will make it possible to ensure the standard characterization of the deep learning-based drug discovery and drug repurposing models [65], [79].
2.3. Network analysis
Networks are empirical and versatile data structures and now a days are widely used in drug repurposing studies as shown in Fig. 4. Myriad number of concepts in the field of biology are represented using networks and graphs [88]. In these approaches data is collected and the networks are created (Fig. 4(a)) to infer the associations by using myriad number of statistical and computational approaches (Fig. 4(b)). Considerable research interest in network analysis on structural and functional level to has been developed to extract the relationships between entities in networks and their underlying biological properties [89]. Thus, various promising insights comes from this emerging field of network medicine [90], [91] where tools and concepts from network theory are applied. Network interaction concept is heavily used in biology since there exist the interactions between different biological phenomenon [92]. Networks are composed of two components: nodes and edges. Nodes can be drugs, proteins, genes, and complexes etc. while the edges can be the interaction between them. Network edges can be directed [93], [94], [95], undirected [96] or weighted [97], [98]. Mostly the quantitative information derived from high-throughput screening is used to construct the weighted networks where edge is represented by some numerical values. In directed networks, traversing is only done in the direction of the edge where in case of undirected networks traversing is possible in any direction unless it is connected.
Molecular networks are good at providing insights into contextual understanding of drug target and its corresponding mechanism of action [99]. Network algorithms can easily achieve such targets by visualizing the various already added interactions in the network along with addition of newly found interactions with superimposition of additional properties [100]. The networks can handle the heterogenous data collected from various data sources. Thus, analyzing the network has now become a diverse area to integrate multiple sources like publicly available datasets and high throughput data [101].
The perfection of these approaches is still partial since these methods suffer from lack of knowledge on molecular interactome often leading to false results [6], [99], [102]. Moreover, the available interactomes provide the static results of the biological process in contrast to dynamic systems in real [99]. Despite these limitations previous efforts have demonstrated the potential and efficacy of network-based approaches and has been proved many times in studies like drug-target interaction predictions. Thus, they can also be used as drug repurposing studies. Moreover, networks can be categorized based on the data source used to construct them such as molecular interaction networks, gene regulatory networks, metabolic networks, and protein-protein interaction networks etc. [103].
2.3.1. Molecular interaction networks
In network biology, interactions between various biological concepts are presented using networks. Networks are made of nodes and edges, nodes can be various components such as genes, proteins, complex phenotypes such as diseases, and atoms etc. whereas edges can be interactions between proteins, genes, or complex phenotypes to show relationship or functional interaction and similarity between them. Moreover, multiple relationships (Fig. 4(a)) between two nodes can also be represented using edges at the same time [104], [105]. Further, the nodes and edges can be directed [106], [107], undirected [108], and weighted [109] with qualitative and quantitative information to emphasize a proper concept [100]. Systems biology contains various molecular interaction which are used to represent distinct molecular interactions depending upon the nature of interactions. Molecular networks are good at providing scenario based valuable insights into drug-targets interact and thus can be used to understand the mechanism of action of a drug [99].
Moreover, numerous studies have demonstrated the practical use of networks in drug repurposing [110]. Network algorithms are developed to achieve tasks such as visualization of the graphs and interaction between them while adding and imposing the newly discovered relationships, additional properties, and connectivity over the primary component and their known interactions [100]. Various type of data collected from heterogeneous data sources can be represented in one network using multiple node and edge arrangement. Due to the diversity and abundance of information contained in networks the analysis of the networks has become a versatile platform for integrating and analyzing multiple sources of high-throughput data for useful insights [101]. One survey providing the detailed use of graph theoretical techniques is provided in [111].
2.3.2. Gene regulatory networks
Gene regulatory networks use the transcriptomic data which provides the information and insights into drug function by capturing the dynamic properties of the cell [99]. Few of the most recent gene regulatory network-based studies are summarized in Table 4. Gene expression-based drug repurposing studies hypothesize that drugs with similar gene expression signatures would target the same proteins [14]. Moreover, Gene expressions change substantially when disease occur specifically the amount of RNA transcript for dysregulated genes changes considerably. Differential gene expression analysis can be performed to analyze the difference between genes expressions in disease and control samples [112]. With the help of technologies now available such as microarrays, gene expression profiles rich source of disease data is easily available which can be used for multiple purposes. Differential gene expression profiles are used as input data in many drugs repurposing studies to prioritize the drug targets [113] because many drug targets work as transcription factors that are likely to regulate the expression of the genes.
Table 4.
Network based repurposing studies using gene regulatory networks, DTI, Drug–disease, and side-effect association.
| Study name | Method | Datasets | Evaluation criteria | |
|---|---|---|---|---|
| Gene regulatory networks and Gene expression Data | ||||
| 1 | DTI prediction for repositioning [113] | Network propagation, scoring based on neighborhood, random walks | GEO repository for Gene expression data | AUC - ROC |
| 2 | A network flow approach for repurposing with case study on prostate cancer [115] | Maximum Flow | OMIM, KEGG, PGDB, DrugBank | Precision, Mean, Position |
| 3 | NFFinder [119] | Statistical method for analysis | Cmap, DrugMatrix, and GEO | – |
| 4 | System biology approach developed by a novel knowledge-driven method [121] | Bayesian network-based approach | Gene-gene interaction | – |
| 5 | Functional Module Method with case study of Prostate Cancer [145] | Functional linkage network | TCGA, LINCS, GEO, OMIM | AUC - ROC |
| 6 | computational DR using Kolmogorov– Smirnov enrichment testing for [123] | Enrichment (Kolmogorov–Smirnov) | GEO and CTD | – |
| Protein–protein interaction networks | ||||
| 7 | Network Analysis for potential DTI identification [146] | SVM, Logistic regression, L1-regularization, KNN | STRING, DrugBank, Gene cards | Z score and Standard deviation |
| 8 | Comprehension of Complex disease using PPIN [112] | – | Gene expression omnibus (GEO) | Harmonic mean and Precision |
| 9 | Drug repurposing shared network of PPIs and genes [147] | Similarity | STRING and DrugBank | – |
| 10 | PPINs and MMP cellular model [148] | Cross talk by analysis of betweenness centrality | KEGG, OMIM and iRef Index database | – |
| Drug–target interactions | ||||
| 11 | DTI prediction using Probabilistic soft logic [135] | Soft probabilistic logic | DrugBank, KEGG, DCDB, and Matador | AUC, Precision, AUPR |
| 12 | Network-based inference for prediction of DTI [136] | – | DrugBank | AUC and Precision |
| 13 | Bayesian matrix factorization-based DTI prediction[111] | Prediction based on Bayesian algorithm | DrugBank and DTIs | AUC |
| 14 | DTI prediction from integration of chemical and genomic spaces [138] | Bi-partite graph ML | DrugBank and DTIs | AUC |
| 15 | Bipartite model for DTI prediction [139] | Supervised method of network inference | DrugBank and DTIs | AUC-ROC, AUPR |
| Drug–disease and side-effect association | ||||
| 16 | Drug repositioning methods based on network inference [149] | – | CTD | AUC-ROC |
| 17 | PREDICT, a method to infer the new indications [44] | Logistic regression | DrugBank, SIDER, KEGG (Drug), DCDB, Expression Atlas, OMIM | AUC, precision |
| 18 | Clustering of Heterogeneous networks to repurpose the drugs [150] | Clustering | NCBI Gene, KEGG Medicus | – |
| 19 | Use of Side effects in Network based approaches to drug repurposing [151] | Statistical analysis | SIDER, FDA approved drugs | – |
There are many studies who used gene expression data for drug repurposing based on network-based approaches as shown in Table 4. Additionally, one such study which used neighborhood scoring, interconnectivity, and propagation random walks methods is presented in [113]. Gene expression data used in this study was downloaded from Gene Expression Omnibus (GEO) [114] repository. The conducted study was confined to case studies such as scleroderma, different types of cancer and type 1 diabetes with results evaluated using area under the curve (AUC). Another network-based study using gene expression data used maximum flow method to repurpose the drugs for prostate cancer [115]. The data developing network was downloaded from DrugBank, OMIM [116], KEGG [117], and PGDB [118]. The evaluation criteria were set to mean, precision, and average position (AP). NFFinder [119] presented statistical analysis method to repurpose the drugs for neurofibromin and the data for this study was downloaded from GEO, CMAP [120], and DrugBank. Other studies include Bayesian networks [121], virtual gene technique Bayesian networks [122], Kolmogorov Smirnov enrichment [123], and functional linkage network [124] etc. developed for repurposing in different disease domains.
Despite being effective, gene regulatory network-based drug repurposing suffers from some limitations. First, it is difficult to define and set a robust gene signature because of the random nature of some gene expression data which ultimately results in biased extracted responsive networks [125]. Second, the drug targeted genes and regulatory genes are not always found differentially expressed. So, the assumption that rests on the notion that mathematical optimum from the responsive network coincides with maximal biological relevance does not stand true. Third, proved from the previous studies, network topological characteristic points out that there is no significant relevance between potential target proteins and significant points in network [115]. Fourth and final, the biological network and living organism’s response are complex to map thus estimated gene networks are often used to rely on [126]. These limitations can be overcome by integration of information from multiple sources and finally a robust drug repurposing framework can be developed based on gene regulatory networks.
2.3.3. Metabolic networks
Metabolic networks are interconnected pathways depicting biochemical reactions taking place within living cells through involvement of metabolites with necessary compounds [127]. In metabolic networks, metabolite and chemical compounds are represented as nodes. Whereas the edges represent the relation between the metabolites and compounds. Direct edges show the reactions done by one or more enzymes. In some other networks, nodes denote the compounds and metabolites whereas edges capture the reaction between nodes. Thus, a bipartite graph with directed edges is provided connected two distinct entities (nodes). Implying, if a metabolite is direct result of a reaction, then a directed edge from compound to metabolite using reaction notation denotes the relationship. In other words, if there is an edge from metabolite to reaction it implies that reaction is activated by metabolite. Once represented, metabolite networks are subject to topological feature analysis to capture different kind of relations and uncover repurposing opportunities. Additionally, positive change in compound concentration caused by specific enzymes may be the cause of disease and drugs are used to regulate and adjust the concentration of compounds cause for disease [128], [129].
Moreover, in metabolic networks flux-based analysis (FBA) methods are used to identify the drug targets. FBA and other similar methods aim at prediction of essential enzymes critical to survival and growth of pathogens. In practical, on metabolic networks FBA model was developed in [128] to predict the DTIs. The data used in the two-step study was retrieved from KEGG database followed by drug target prediction for human hyperuricemia. Another study done in [130] employed the greedy heuristic search approach to predict the DTIs for lunch cancer using metabolic networks. Whereas the methods describing anti-microbial DTIs identification were reviewed in [131] that used FBA modelling on metabolic networks. Advantages and disadvantages of methods are discussed clearly in.
2.3.4. Protein-protein interaction networks
Protein-protein interactions (PPIs) are required in almost every process in cell which emphasizes that the need for understanding is crucial for cell physiology in normal and disease states. It is essential in drug development because drugs have effect on PPIs. Protein interaction networks have proved useful in drug repurposing studies over the time (Table 4), since their interactions determine molecular and cellular mechanisms, which control healthy and disease states in organism [132]. Therefore, with the help of such networks pathogenic mechanism of disease can also be understood. Some PPINs based repurposing studies are listed in Table 4. Moreover, confining this study to drug repurposing it can be stated that PPIs depicts the interactions between known druggable targets and other proteins. This interaction between the proteins can be direct or indirect [101]. The central assumption used in drug repurposing when focusing on PPI networks is that proteins which are targeted by similar drugs are functionally related in networks or if the drugs are similar, they are supposed to treat same disease and vice versa. It is also believed that the topological analysis of the PPIN can provide useful insights in drug-target predictions as proteins function in form of interaction mostly [133].
Owing to the importance of PPIs, many repurposing studies have been successfully done. One such study using PPIN for drug repurposing is done in [133] which utilizes human PPIN obtained from UniHI and maps it with DTIs obtained from DrugBank and GeneCards databases. SVM, along with L1-regularized logistic regression and k-nearest neighbors’ methods are used for drug repurposing. Z-score and standard deviation are used as an evaluation criterion to judge the performance of developed models. Other study is presented in [112] predicting the drugs for metastasis in colon cancer. Here data was downloaded from GEO and set cover-based formulation of coordinate dysregulation in complex phenotypes was used followed by harmonic mean method as an evaluation of the predictions. Additionally, Parkinson’s disease was predicted by analyzing crosstalk in the network by betweenness centrality. The data here was downloaded from KEGG, OMIM, and iRef databases.
Despite progress and proving useful, PPIN based drug repurposing studies experiences some disadvantages such as, PPIs obtained as the part of experimental sources contains links which are not fully characterized in detail. Moreover, the data obtained are noisy and incomplete thus there are higher chances for network being ended up as biased extracted network.
2.3.5. Drug-target interactions
DTI prediction is the basis of drug discovery and drug repurposing as shown in (Fig. 4(a)). Many of the studies show that the drugs have off-target effects and the process of drug repurposing become simplified if the targets are predicted accurately. Computational prediction of DTI makes discovery process fast, economical, and maximizes the chances of success whereas experimental validation of DTI is both time consuming and expensive. This emphasizes the necessity of development of computational potential methods to predict DTI accurately [134] and some of the well-known DTI based repurposing studies are listed in Table 4. Many network-based methods for predicting DTI are present. Bipartite or tripartite interaction models are created where nodes denote the drugs and targets, and edges show interaction between them in bipartite networks and in case of tripartite networks nodes represent drugs, targets, and disease and edges denote interaction or relationship between them.
Some of the notable network-based DTI prediction studies are described here in detail (Table 4). One such study is done in [135] which presents a probable soft logic method for DTI network analysis. The data was collected from the DrugBank, KEGG Drug, and DCDB databases (Table 1 and Table 2). The evaluation of the network performance was judged using the AUC, AUPR, precision (Fig. 3(b)), and other bioinformatics tools (Fig. 4(c)). Additionally, recommendation algorithm was used in [136], [137] where drug-based, target-based, and network-based similarity inferences were drawn to predict novel DTI for drug repurposing. The predicted drugs were pre-clinically validated in in-vitro binding assays. Another study used supervised bipartite graph learning approach [138] to predict enzyme interaction and GPCRs interaction network. For DTI prediction kernel regression-based method was used and method performance parameters used in this study were ROC and AUC etc. Other network-based DTI prediction studies for drug repurposing include bipartite local model (BLM) [139], neighbor-based BLM integrated inference [140], statistical analysis of topological measures [141], network consistency-based prediction method (NetCBP) [142] and more.
Network-based DTI prediction methods have widely been used because of their good performance and various methods as discussed in above paragraph. Instead, there are some limitations of these approaches such as in-ability or poor performance of the models in case of novel DTI prediction. Other limitations include, low amount of known drug-target interaction data available, poor dataset balance, selection of negative samples is difficult or not possible since there is either no or less negative DTI data available from experiments [142]. Possible solutions for the said problems are suggested by many studies such as in [143] where they initiated the process by targeting, drug targets of highly similar drugs to new targets and vice versa for the drugs. Solving a problem leads another problem in this study, which is how to define the highly similar drugs or highly similar targets. Additionally, the problem of scarce data samples was solved by introducing the randomization technique for balancing data samples as in [144].
2.3.6. Drug-drug interactions
Similarity based drug-drug interaction shows the association between two drugs and talks about the aftermaths of the interaction. Drug-drug similarity is used in network-based drug repurposing to augment the network. Drug-drug interaction similarity can be constructed either as biological effect similarity, chemical similarity, side effect-based similarity and more. Specifically, structural similarity can be developed by different method, either by using 2D fingerprint similarity or 3D conformation of the structures. Having said that, let’s say, two drugs having similar expression patterns in cultured cells or lead to similar side effects then the drugs can be said to have interaction between them. Moreover, drug repurposing opportunities can be found using the chemical structures of the drugs. The hypothesis rests on the notion that, two drugs which are structurally similar are supposed to treat the same indication by targeting the similar protein targets and disease pathways. Whereas this hypothesis does not always hold true, however structural similarity have been used in successful drug repurposing studies [152] which motivates to deeply investigate these approaches in future.
Moreover, one drug-drug similarity-based study is given in [153] where two chemical compounds were compared based upon the 2D graphs. Atoms in this study were designated as nodes while bonds between atoms served as edges. This study was based on the basic hypothesis that two structurally similar drugs share the same bioactivity [154]. Chemical similarity-based studies often extract multiple structural features and then compare the drugs one-on-one based on these features. Thus, if the hypothesis returns true, drug repurposing opportunities can thus be availed using chemical association, biological means, side effects, or by searching some known biological features such as known targets [152]. Another similarity-based drug repurposing study using network approach is given in [155] which presents a method called SITAR and utilizes five similarities between drugs and targets followed by MANTRA [156] which studies the mechanism of action of the drug using network-based approach.
Uncertainty in drug-drug similarity and chemical properties of the known drug compounds leads to limitation of the drug-drug interaction-based drug repurposing studies. Additionally, there are some physiological effects which are hard to capture while using the structural features alone thus it leads to the use of multiple similarity measures to give a broader perspective like done in [157]. The similarity-based strategy is typically employed by drug-drug interaction prediction with most similarity-based algorithms relying only on nearest similarities overlooking some of the important similarity measures.
3. Network based studies for Covid-19
Human coronaviruses (HCoV) include severe acute respiratory syndrome coronavirus (SARS-CoV) and novel 2019 coronavirus (Covid-19 also referred to as SARS-CoV-2). SARS-CoV is pathogenic coronavirus that is known to emerge from zoonotic reservoir leading to global dissemination of the virus [158]. Recently, novel coronavirus (2019 nCoV) has caused global pandemic with high morbidity and mortality rate. Instead of highly accelerated efforts in the field of drug research, until now there is only drug approved for the treatment of Covid-19.
Moreover, de novo drug development is time taking, costly and risky process. It takes 12–15 years with $2~$3 billion of the investment to develop and new drug and bring it to market [158], [159], [160]. An effective and timely possible approach to quickly response the drug needs in case of emergencies is known as drug repurposing. Drug repurposing is the process of finding the alternative uses of already approved drugs [161], [162], [163]. Over the period, multiple techniques based on machine learning, deep learning, mining, and network or graph-based approaches have been widely used to repurpose the drugs for a Covid-19.
Herein, we provide the comprehensive review along with benefits, challenges, and future directions of network science-based drug repurposing studies specifically for the Covid-19. The studies are divided in to three categories based on network proximity, network diffusion, and artificial intelligence ( Table 5, Table 6, Table 7).
3.1. Network proximity-based drug repurposing in Covid-19
Network proximity-based drug repurposing studies use the proximity measures such as network centralities, node degree, and more for identifying and providing the key protein-protein, drug-drug, or drug-target associations to repurpose the drugs (Fig. 4a (i)). One of the recent studies done in [164] presented a powerful network-based drug methodologies for identification of potential repurposable drugs and their combination targeting Covid-19. Presented anti-viral and integrated repurposing methodology mainly quantified the interplay between HCoV-host proteins and drug targets in human protein-protein interactions (PPI). Use of network proximity analysis as shown in Fig. 4(b) (i) between HCoV-host targets and drug targets in human PPI led to prioritization of 16 anti-HCoV drugs potentially repurposable. Further validation [165] was performed by enrichment analysis (Fig. 4c (i)) of drug gene signatures (Table 5). Moreover, potential drug combinations were captured by complementary exposure pattern. Hypothesis of the study was that, for successful replication of viruses and infection host cellular factors are required, thus systematic study of human PPI (Fig. 4a (ii)) and virus hosts can present an effective method toward understanding the viral mechanism [166], [167]. Despite successful results, this study suffered from limitations such as, lack of known host proteins on Covid-19, a low binding affinity value of 10 μM was used to specify the DTI. Whereas lower binding energy up to 1 μM or less could be a more effective cut off. Another network proximity-based drug repurposing study is given in [168] which used network medicine-based approach to repurpose drugs in Covid-19. This study enriched the PPI with transcriptomics and proteomics and then performed network proximity measurement where broach Covid-19 associated underlying pathogenesis of disease manifestation was explored. A further step was added, and network-based predictions were prioritized for treatment by combining predictions with propensity score (PS) matching 26,779 individuals from observational study. Here, the associations between 6 different diseases (consisting of autoimmune, malignant, cancer, cardiovascular, metabolic, neurological, pulmonary) and Covid-19 were created. Created network was analyzed followed by the single-cell RNA sequencing to uncover the underlying pathobiological and pathophysiological relationships between Covid-19 and its associated treatments. Moreover, like [164] in this study z-score was used to measure the closest proximity distance between pair of interaction proteins and all those with z < −1.5 were considered closest ones. Although not significant, network proximity results in this study showed that rheumatoid arthritis has small network proximities (negative Z scores) across all 5 SARS-CoV-2 datasets. For checking the potential of repurposed drugs to reverse the expression at proteomics and transcriptomic level mutated by viruses Gene set enrichment analysis was performed to evaluate each drug. Next, the drugs were prioritized based on combination of factors such as strength of network based and bio-informatic based predictions, literature evidence of the prioritized drugs, meaningful evaluation by enough patient data and more. This type of muti-layered validation of the network predicted drugs find out the key repurposable drugs supposed to have potential to reverse the expression of Covid-19 expressed genes. In total 34 drugs (Z< −1.5 and P< 0.05, permutation test) (Fig. 4b (i)) were computationally repurposed with the SARS-CoV-2 datasets (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI) using the above-mentioned criteria. These drugs were significantly proximal to 2 or more SARS-CoV-2 host protein sets. We manually curated their reported antiviral profiles. Additionally, Knowledge of the complex interplays between SARS-CoV-2 targets and human diseases indicate possibilities of drug repurposing, as the drugs that target other diseases could potentially target SARS-CoV-2 through the shared functional PPI networks. Moreover, further information about the databases, toots, and techniques used in these studies are listed in Table 5. Some studies are combining different network approaches for improved results such as done in [169] which aims to captures the bigger picture of human molecular landscape of Covid-19 infection by exploiting the network medicine approach as strategy to repurpose the drugs for Covid-19. As a starting point, experimentally validated host proteins of the Covid-19 known to have viral interaction, tissue specific gene expression data, were explored by use of techniques such as connectivity significance, network diffusion, and network propagation. This process was followed to rank genes followed repurposing drugs targeting the ranked genes as a treatment for Covid-19. The prioritized collection of genes which served as key drug targets for repurposed anti-Covid-19 drugs responsible for causing pharmacological effect resulted from normalization and aggregation of different scoring methods. Ranking of genes focused on the contrasting the different phases of the virus infection and viral replication of the cycle. Further, in this study, drug repurposing was carried out by drug-gene interaction data obtained from DrugBank database. DrugBank was manually investigated to find out the drugs having direct and indirect interaction to virus host proteins. A set of criteria was made to find the restricted list of genes (18 genes) and related drugs. Criteria include top ranked genes in VarElect ranks of groups G124, G12345 etc. followed by main cellular location of target proteins, RNA replication, and translation, and finally the existence of drugs already approved as potential candidates for repositioning. Moreover, another study called SAveRUNNER [170] presents a novel network similarity-based drug repurposing framework. It first creates a bipartite drug-disease network between drug-targets and disease-causing genes, and it uses network based new similarity measure that prioritizes the associations sharing the same neighborhood. PPI data gathered from 15 commonly used databases was retrieved from [171]. The data contained 217,160 PPIs between 15,970 unique proteins. Additionally, disease gene associations were obtained from Phenopedia [172] disease-centered view of genetic association studies by online Human Genome Epidemiology (HuGE) encyclopedia [173]. Moreover, a total of 1875 FDA approved drugs were selected for analysis from DrugBank and Therapeutic Target Database (TTD) [174] databases. Whereas presented algorithm is six step procedure which takes the list of drug-targets and disease genes as input followed by the proximity and similarity computation which leads to proximal drug-disease associations. Clustering was performed on the drugs and quality score was computed to adjust the network similarity and finally normalization of similarity score was carried out using sigmoid function. This study utilized many useful steps from creating the network to outcome of repurposed drugs which ensure the robustness and usefulness of the approach. Additionally, another network-based repurposing approach for Covid-19 done in [175] was conducted by integrating GWAS and text mining the data. Here lung-specified gene subnetwork enriched with Covid-19 gene associated data obtained from GWAS was created to screen 220 FDA approved drugs obtained from DrugBank database. Their drug-perturbed gene expression profiles were obtained from LINCs and lung-targeted, drug-focused gene modules were created. This led to prioritization of the 13 FDA approved drugs as a potential treatment for Covid-19. Findings provide the timely and valuable insights to repurposed use options of the drugs to be further explored in Covid-19. Another very distinguished and interesting work in the field of network science and its applications to drug repurposing (Covid-19) was done in [176]. Here in this study multiple layers or approaches normally applied individually on the networks were fused into a single algorithm. Deployed algorithms consist of artificial intelligence, network diffusion, and network proximity. Each of them was utilized to prioritize the 6340 drugs for expected efficacy in Covid-19. This led to a successful approach as a network medicine framework for identifying repurposing opportunities in Covid-19. In practice one cannot find a single algorithm that consistently performs across all the datasets which served as a motivation for this study. A fundamental hypothesis was created that multimodal approach that combines the predictions from all approaches, based on the consensus among all the approaches, combined approaches always exceed the individually best performing models. In result of this approach 918 drugs were prioritized based upon the predictions from all algorithms and were subsequently tested in wet lab experiment. As 77/918 showed the anti-viral effects in cultured non-human primate cells. As a step further all those 77 drugs were tested in human-derived cells and 76/77 drugs showed the efficacy against viral disease. 96 % of the drugs tested in the study were FDA approved which is beneficial as these drugs could be directly moved to clinical trials. A real predictive power of the approach was demonstrated by the positive outcomes of the second human screen with success rate of 62 % followed by identification of six drugs potentially easily repurposed for Covid-19. All in all, the approach presented here did not only provided an effective strategy and results in the repurposing of Covid-19 drugs but also provided an algorithmic toolset as well. The toolset could be readily deployed in future for diseases left untouched due to huge cost and time constraints associated with it. As only 918 of the 6340 drugs prioritized by CRank were screened, a selection driven by compound availability, many potentially efficacious FDA-approved drugs remain to be tested. Finally, it is also possible that some drugs that lacked activity in VeroE6 cells may nevertheless show efficacy in human cells, like loratadine. Additionally, despite the tremendous progress in the repurposing of the drugs, repurposing a combination of drugs with varying mechanism of actions remains an undefeated feat. Since drug combinations are widely used to fight many deadly diseases effectively. Severe and rapidly emerging replications of the Covid-19 made the world realize for the combination therapies that were expected to response more effectively. The ability to repurpose the drug combination was hampered by the availability of large range of possible pairs. To address this gap a study was done in [177] where an integrative network pharmacology inspired web-based tool named COVID-CDR was developed for in-silico repurposing of the drug combinations. Deployed web-based tool can visualize the interaction in the interactome on the cellular level involved in the mechanism of action of the drugs involved. COVID-CDR could prioritize Covid-19 repurposable drug combination for synergistic effect. In this approach, drugs with targets sharing the topological proximity with Covid-19 targets are used to prioritize the drug combinations based on drug-target separation. Previously it was hypothetical presented that different drug-targets lead to different footprints. Combination sharing the commentary exposure can be considered as putatively effective in this case. Whereas, successfully evaluated drug combinations because of COVID-CDR were evaluated using the network computation methods. Network proximity measures are calculated using the subnetworks of protein interactome that were active context of interest. Since this platform is not automated thus it can be further improved by automating the screening of drug-pairs created by multi-object optimizers. Finally, in the light of studies presented, it can be said that network proximity-based approaches have proved very useful for drug repurposing applications, possessing high flexibility and diversity. However, mere use of proximity measures to analyze the network may or may not present significant results but when combined with additional validation approaches such as GSEA, molecular docking, and pathway enrichment could present more putative yet diverse framework.
3.2. AI on network-based drug repurposing in Covid-19
Network-based approaches play very important role in drug repurposing studies and few of the recent studies are listed in Table 6. Artificial Intelligence algorithms are widely applied on network data for variety of prediction tasks as shown in Fig. 4(b) (ii). These tasks include node prediction, link prediction, or graph prediction (Fig. 4b (ii)). These predictions can be applied to variety of applications and drug repurposing is one among them. In this section some of the recent studies, where AI on networks has been applied to repurpose the drugs, are presented. One among these studies is done in [178] in which a Covid-19 based knowledge graph was built. The knowledge graph consists of virus baits, host genes, pathways, and other information. To infer the representation of the candidate drugs, deep graph neural network approach based on biological interaction was used. Candidate drugs were later ranked based on the clinical trial history (Fig. 4c (iv)) to prioritize them followed by the validation using the genetic profiles and in vitro experimental efficacy (Fig. 4c (iii)). Use of deep neural networks and integration of the diverse interactions helped in quick identification of drugs repurposable for Covid-19. This study suffered from the limitations of missing the effective drugs possibly during the initial drug ranking. Since the initial group of 3635 drugs were taken as candidate. The validation of the drugs was carried out using the population based retrospective EHRs analysis and EHRs are incomplete and prone to errors in comparison to randomized experimental data. Other limitations include the inconsistency between the gene sets. Another Covid-19 based drug repurposing study using the Few-Shot learning is done in [179]. This study introduces the multi-targeted drug repurposing strategy which is taken as most efficient approach to diseases such as Covid-19. Multi-targeted compound screening is hampered by the lack of good quality experimental evidence and information extraction from molecules. Recognizing this gap, MOLGNN, a deep learning model was proposed and presented which enabled the prediction of molecular property. Chemical molecule embedding was performed by using graph neural networks (GNNs). GNNs were used to model the ability of the drugs to target proteins and their interaction. Let’s say G = (V, E) is a graph representing the nodes connected with edges. V represents the vertices or the nodes whereas E represents edges between the nodes. In connection with the present study, molecules are represented as graphs (G), nodes are items and bonds between them are the edges. Following this, an input data comprising molecules as graphs in the form of adjacency matrix A, was constructed for this study. This dataset contained more than 2 million unlabeled molecules obtained from ZINC database. The deployment of the trained models and downstream classification (Fig. 3(c)), Covid-19 related chemical molecules were used. The models trained with MolGNN performed well when applied to a small labeled fine-tuned dataset. This led to a potentially powerful method called few shots learning. This study provided a powerful tool for future research in drug repurposing specifically in case of poorly known diseases such as Covid-19. Instead of these benefits, there are certain limitations such as uncertainties in the datasets used and lack of validation of the predicted drugs in corresponding in-vitro and in-vivo models for efficacy testing in Covid-19. Moreover, multiple machine learning models are also used disease agnostic fashion for network datasets as done in [180] for repurposing targeted drugs in Covid-19. Used models are complementary covering Covid-19 and host factors. It starts with ranking of drugs matrix factorization on anti-viral first followed using graph kernels to rank drugs based on perturbation induced in subnetworks. Manually curated datasets containing drug-virus associations with known status obtained from [181] were used. Broad-spectrum analysis that started from in-vitro validation spanned over the animal models and clinical study. Furthermore, results suggest that the drugs ranked using anti-viral of the top predicted broad spectrum were found in clinical practice for Covid-19 and those from graph kernels were found experimentally validated. Additionally, online repurposing tools are also widely deployed recently and have proven useful in many cases. Similarly, here Covid-19 drug repurposing explorer abbreviated as CoREx was deployed as an interactive tool. This can be used to observe and understand the interaction between drugs and host Covid-19 targets in context of biological networks, clinical use of drugs, protein functioning, and cMAP. Despite the contribution and benefits of CoREx tool there some limitations such as the matrix decomposition approach can only be applied to the drugs for which association with viral disease is known. Main limitation of the study is the annotation of the drug-virus pairs based on the developmental stages and the related data is hard to find. Similarly, the other part of the study (Network medicine) has limitation that only those drugs for which targets are known can be used. Moreover, feature information is very important machine learning is involved. In this connection study emphasizing the informative features of network in machine learning based drug repurposing are conducted in [182]. Human PPI, Covid-19 specific network, and DTIs were first created like most of the studies and later essential proteins were passed through a set of algorithms (Machine learning). ML step was followed by clustering which served as an important step in extraction of informative features. Enrichment analysis was then performed for these informative features to validate them in case of Covid-19. This approach helps identify key information followed by validation which minimizes the false positive or false negative rate of the final prediction. In addition, integrative approaches are used in drug repositioning and are found to perform more effective as compared to single or standalone approach. This follows the notion that, weaker when joined come together stronger. One such graph machine learning based drug repurposing study in Covid-19 is presented in [183] which avails the ML and statistical approaches to map and mine the knowledge graph, transcriptomic and literature data on the large scale to uncover the potential drugs for Covid-19. The computational approach in combination with pre-clinical validation indicated that a poly-ADP-ribose polymerase 1 (PARP1) inhibitor, CVL218, currently in Phase I clinical trial, may be repurposed to treat COVID-19. To observe all these predictions virus-centered knowledge graph was created and mined using the mining algorithm. In realization of the network seven different networks were considered consisting of DTI network, an interaction network of drug-target (virus), a human protein-virus protein, and more. The databases such as DrugBank, Uniprot, TTD, ChEMBL, PubChem, and IUPHAR_BPS [187], Binding DB [188], and GHDDI [189] were used. One other study that used DrugBank and other databases along with graph convolutional networks (GCN) is published as [186] but it consists of analysis of multiscale virus host interactome resulting in off-target prediction of the drugs. Coupled with cell-based experimental validation this study came up with several clinically efficient drugs as potential repurposing candidates found as a result GCN application to Covid-19 related data. Specifically, as a part of the mentioned study, capmatinib (MET inhibitor) was found to have potent and wide range antiviral activity in multiple coronaviruses. The antiviral activity was observed in MET-independent manner and additional roles of capmanitib for host cell proteins promoting the infection caused by human coronavirus thus it can inform further drug discovery studies. Thus, AI assisted network-based drug repurposing strategies present effective strategy along with many benefits such as time and cost efficiency and high reliability. Additionally, the use of GNN and GCN have opened the new doors of opportunities in network-based drug repurposing strategies and at the same time these approaches also suffer from limitation. Since like the other AI methods, GNN and GCN also need data and in most cases balanced data to be trained properly and biases can be avoided without compromising on the accuracy of the models. Additionally, there is no doubt huge amount of the data is being generated and is publicly available but there are chances for the data to contain uncertain information ultimately leading to improper and vague model training. Realizing these problems and tactfully handling them can unveil the unfathomable power of the GNN and GCN approaches being applied to network-based approaches to drug repurposing in Covid-19 and in general as well.
3.3. Network diffusion-based drug repurposing in Covid-19
Diffusion is a process through which information related to genes, proteins, viruses, and other entities diffuse across the drug-target, drug-drug, disease-disease, or social networks [190], [191], [192], [193], [194] as shown in Fig. 4(b) (iii). Some of the related studies are given in Table 7. A simple notion behind this concept is that behaviors such as information spread in environment, modeled by construction of simple graphs called Regular Graphs (GRID-based). There are multiple significant studies utilizing this approach to repurpose the drugs in Covid-19 and other diseases. Once recent Covid-19 based drug repurposing study is done in [195] where gene co-expression-based network for 1441 genes was constructed and analysis was performed. The analysis resulted in two significant gene co-expression network modules and then data related to miRNAs and transcription factors targeting co-expressed modules’ genes were gathered from reputable databases led to the establishment of two subnetworks consisted of transcription factors and miRNAs. Subsequently, the list of drugs targeting the obtained subnetworks was gained from DGIDb database and two networks were constructed again. Finally, because of proposed network analysis, five drugs were repurposed as candidate repurposable drugs for Covid-19 treatment. Additionally, around 10 miRNAs were found to be significant to treat Covid-19. This study using diffusion has the capacity to integrate and analyze the heterogeneous data to support the predictions. Where similar integrative approach is followed in [196] to perform validation of repurposed drugs using a comparatively different framework which contains KEGG enrichment, molecular docking, STITCH search tool, and virus-host-drug interactome. These techniques were used to investigate the interaction network linked with Covid-19 and chloroquine and hydroxychloroquine were utilized as reference drugs to further get into the network. As a result, 47 drug targets were found enriched in TLR pathway and were found overlapping with Covid-19. Another step of molecular docking and dynamics in investigation led to confirmation of binding affinity of TLR9 to the mentioned drugs. Additional insights from human-drug protein map identified and further confirmed the interaction of Covid-19 with TLR pathway will provide key information for Covid-19 itself and its treatment. Thus, ensemble or integrative approaches aimed at investigation are useful and results in very fruitful insights. Moreover, all the tools, databases, and evaluation matrices used in the studies for proximity, AI, and diffusion-based networks are given in Tables 5 , 6, and 7 respectively. Furthermore, combination of diffusion and matrix factorization methods is also used [197] to prioritize the Anti-Covid-19 drugs. The drug prioritization is based on integration of multiple informative features such as virus sequences, established viral-drug interactions, drug chemical structures, and the application of logistic matrix factorization coupled with diffusion kernel on the graph data. Molecular docking was used for prioritization or ranking of the compounds (Fig. 4c (ii)). Here, the gaussian models were first built followed by virus sequence and chemical structure similarity kernel are designed based on the biological features of identity matrix. Then the gaussian and similarity kernel are diffused and finally the logistic matrix factorization model with kernel diffusion is proposed to identify potential Covid-19 drugs. Finally, to validate the drugs molecular docking was performed to find the binding affinity of the drugs potentially repurposable with Covid-19 spike and ACE2 proteins. The algorithm is named as VDA-KLMF is compared with recent relevant studies (prediction and classical models) based on the 5-fold cross-validation. This cross-validation was performed on viruses, drugs, and VDAs and as a result best results (recalls, AUCs, AUPRs), significantly outperforming the other studies were obtained. Thus, it leads to conclusion that integration of various biological data features and suitable models, or kernels lead to development of tools that may assist in drug development for Covid-19.
4. Benefits of drug repurposing methods in clinical practice
Drug repurposing has proved as an effective alternative to costly, time consuming, and risky process of de-novo drug development. Due to the sudden emergence of Covid-19 pandemic drug repurposing efforts helped greatly to make the treatments available to the patients faster. A large community of the researchers and scientists have been working on the different type of approaches to drug repurposing such as earlier defined: machine learning based, deep learning based, and network-based repurposing. These methods have made it possible for the drugs to enter the clinical trials to fight against Covid-19.
For example, a network-based drug repurposing study done in [164] predicted the 16 repurposable drugs with anti-viral evidence obtained from the literature and clinical trial data. Among these predicted drugs Sirolimus (originally approved as an immunosuppressant) is registered in phase-II clinical trials for Covid-19 [200]. Another work done in [182] applied machine learning on networks and repurposed 14 potential repurposable drugs for Covid-19, of which Chloroquine is registered in Phase-II & III [201] and Dexamethasone is registered in Phase-IV clinical trials [202] as mentioned in Table 8. Similarly, a machine learning and network medicine drug repurposing work [180] retrieved a list of 23 FDA approved drugs as potential therapeutics for Covid-19. Many of these drugs are already in clinical trials for Covid-19.
Table 8.
List of repurposed drugs in clinical trials for Covid-19.
| DrugBank ID | Drug Name | Original indication | Trial Phase – Covid-19 | Reference | |
|---|---|---|---|---|---|
| 1 | DB00877 | Sirolimus | Immunosuppressant | II | [164] |
| 2 | DB00608 | Chloroquine | Antimalarial drugs | II and III | [182] |
| 3 | DB01234 | Dexamethasone | Inflammatory conditions | IV | [182] |
| 4 | DB12610 | Ebselen | Type 1 and 2 diabetes condition | II | [42] |
| 5 | DB13132 | Artemisinin | antimalarial drug | II | [42] |
| 6 | DB01054 | Nitrendipine | hypertension | IV | [42] |
Additionally, in our recent work of machine learning and molecular docking-based drug repurposing framework [42], 12 potential drugs were suggested as potential therapeutic for Covid-19. Out of 12 potential drugs, Ebselen was found under trial in Phase-II [203] whereas Artemesian and Nitrendipine are in Phase-IV clinical trials [204] as shown in Table 8. Similarly, multiple deep learning techniques have also been used to identify the repurposable drug candidates for Covid-19. One of the deep learning-based studies, is done in [205] where deep learning-based drug screening was performed followed by a study done in [206] where a resource based on deep learning is created to enable the fast repurposing of the drug candidates for Covid-19. Finally, in the light of the examples tabulated in Table 8, it can be concluded that the drug repurposing methods are quite helpful and quick to response in case Covid-19 and related emergent situations. Moreover, as the time is passing, current methods are being improved and advanced while newer methods are introduced. Thus, drug repurposing methods will continue to help in fighting against pandemics.
5. Limitations and future recommendations
There is no doubt that network medicine approaches are gaining more attention and have successfully been deployed in several studies to repurpose the drugs for Covid-19 (Table 5, Table 6, Table 7). Still there are some limitations which needs to be highlighted along with future recommendations to overcome them for improved results in the future.
The limitations in general include the inconsistency in subnetwork extraction in biological networks due to the parameter and thresholding adjustment. Modified genes and gene expression changes are not captured which limits the graphs to estimate the off-target effects of drugs on the genes. Additionally, genes used as pharmacological targets do not always show the observable signature changes which contrast with the commonly adopted notion of mathematical optimum of response networks. Similarly, inability of networks to comprehend pharmacokinetic modifications and noisy side effects data of drugs make it difficult for structural similarities alone to be utilized for drug repurposing.
Moreover, there are different limitations and challenges associated with different types of networks. Such as problem of functional links that are possibly not defined as they determined through various experimental background, is associated with PPINs. So, the constructed network will be a biased network because of limited and noisy data. Furthermore, there is no easy method to link a response network to an entire organism’s reactions. Following other networks, DTI networks suffer from the limitations such as various structural and chemical properties of known compounds are inaccurate limiting the drug repurposing studies based on drug-drug similarity to unlock the true potential. Furthermore, structural traits alone cannot predict many physiological outcomes. To address these constraints, a variety of similarity measures, such as the one employed, can be used.
To overcome these limitations, vast biological data including molecular, pathological, structural, and online health community data should be utilized to construction of networks with multiple interactions to capture the bigger picture. To construct them, heterogenous data from various sources must be utilized to improve the reliability and reach of repurposing methods. Since the biological systems of complex and cannot be reflected by one aspect only, thus, one type of feature or data present the singular aspect of biological system. Single data type-based systems may not be reliable and use of integration of heterogenous data to capture the multiple aspects of biological system may omit the major limitations. Finally, for the standardized evaluation of the network-based approaches to drug repurposing a reference body should be created.
6. Conclusion
The Coronavirus disease (Covid-19) is an infectious disease caused by SARS-CoV-2 virus. Started in November 2019 in the Wuhan-China, Covid-19 was soon declared as public health emergency of international health concern by the World Health Organization (WHO) on January 30, and it scaled up to pandemic as of March 11, 2020. Due to the severity of disease and its volatile speed drug research was accelerated to help the patients and hamper the growing health concerns. Since de-novo drug development is costly and time consuming and on average it takes around 12–15 years with $2~$3 billion of investment to take a de novo drug from concept to market ready product. Thus, drug repurposing emerged as an efficient and scalable approach to find the treatment for Covid-19. In this review, a comprehensive overview of drug repurposing studies for Covid-19 is provided. The review spans to machine learning, deep learning, and network-based approaches while specifically network-based approaches to drug repurposing in Covid-19 are focused. Network based approaches for Covid-19 drug repurposing are further divided in three categories: Network diffusion, network proximity, and AI on networks. The databases, tools, and other resources followed by in-silico, in-vitro, and in-vivo validation studies used are also summarized. Moreover, in simplest form machine learning studies take the different type of drug and disease related data, perform feature extraction, create predictive models, and then models are deployed for the prediction in Covid-19 and other diseases. Despite successful application in drug repurposing for Covid-19, machine learning approaches struggle to handle the diverse and sparse biological data but are easy to interpret which is essential for studies like drug repurposing. In addition, deep learning approaches have also been widely adopted for repurposing studies have gained attention. Since deep learning models are effective in terms of heterogenous and sparse data handling and have been deployed to predict repurposed candidate drugs for Covid-19. Despite being successful, deep learning approaches suffer from lack of interpretability of predictive model and lack of data quality. Additionally, network-based methods have also been widely and effectively deployed for Covid-19 repurposing. Networks as per the evidence and analysis from literature have proved best tools for discovering the treatments by linking and investigating protein-protein, molecular, phenotype, or gene-expression or other data types. Studies have demonstrated that the network models have the potential to generate amazing and timely results and their true potential can be uncovered by overcoming the limitations of data inconsistency, problem of functional links, inability to capture gene expression variation, gene regulation, and lack of structure data etc. Despite these limitations, network models offer many advantages such as the interpretation of the prediction results that can be validated and visualized by mathematical means. The biological network-based studies in Covid-19 covered multiple pharmacological aspects such as gene regulations, protein interactions, drug-target or drug-drug interactions, drug-side effect, and metabolic interactions etc. The integration of such diverse biological entities extracted from many different databases and experiments in network helps to mimic the complex biological systems good for drug repurposing specific in Covid-19 and in general. Finally, in the light of recent literature network-based drug repurposing methods offer more advantages than traditional method by simplifying and accelerating the process of drug repurposing instead of the challenges and limitations.
CRediT authorship contribution statement
Faheem Ahmed: Conceptualization, Formal Analysis, original draft, Writing – review & editing, and images. Afaque Manzoor Soomro: Conceptualization, Writing – review & editing, Chethikkattuveli Salih Abdul Rahim: review & editing, Anupama Samantasinghar: managing the revision and editing, Arun Asif: managing the revision and editing, In Suk Kang: review & editing, Kyung Hyun Choi: Conceptualization, review & editing, Resources, Supervision, Funding acquisition.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the 2022 scientific promotion program funded by Jeju National University.
References
- 1.Hui D.S., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264. doi: 10.1016/J.IJID.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.“WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020.” 〈https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020〉 (accessed May 23, 2022).
- 3.P. Zhou et al., “Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin”, bioRxiv, p. 2020.01.22.914952, Jan. 2020, 10.1101/2020.01.22.914952. [DOI]
- 4.Gorbalenya A.E., et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.“COVID-19 Map - Johns Hopkins Coronavirus Resource Center.” 〈https://coronavirus.jhu.edu/map.html〉 (accessed May 23, 2022).
- 6.Emmert-Streib F., Tripathi S., de R., Simoes M., Hawwa A.F., Dehmer M. The human disease network. Syst. Biomed. 2013;1(1):20–28. doi: 10.4161/sysb.22816. [DOI] [Google Scholar]
- 7.Weng L., Zhang L., Peng Y., Huang R.S. Pharmacogenetics and pharmacogenomics: a bridge to individualized cancer therapy. Pharmacogenomics. 2013;14(3):315. doi: 10.2217/PGS.12.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodos R.A., Kidd B.A., Shameer K., Readhead B.P., Dudley J.T. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8(3):186–210. doi: 10.1002/WSBM.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asif A., et al. Zika virus: Immune evasion mechanisms, currently available therapeutic regimens, and vaccines. Viral Immunol. 2017;30(10):682–690. doi: 10.1089/vim.2017.0046. [DOI] [PubMed] [Google Scholar]
- 10.Paranjpe M.D., Taubes A., Sirota M. Insights into computational drug repurposing for neurodegenerative disease. Trends Pharmacol. Sci. 2019;40(8):565–576. doi: 10.1016/J.TIPS.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanseau P., Koehler J. Editorial: computational methods for drug repurposing. Brief. Bioinform. 2011;12(4):301–302. doi: 10.1093/BIB/BBR047. [DOI] [PubMed] [Google Scholar]
- 12.D’Souza S., Prema K.V., Balaji S. Machine learning models for drug-target interactions: current knowledge and future directions. Drug Discov. Today. 2020;25(4):748–756. doi: 10.1016/J.DRUDIS.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T., Leng J., Liu Y. Deep learning for drug-drug interaction extraction from the literature: a review. Brief. Bioinform. 2020;21(5):1609–1627. doi: 10.1093/BIB/BBZ087. [DOI] [PubMed] [Google Scholar]
- 14.Lotfi Shahreza M., Ghadiri N., Mousavi S.R., Varshosaz J., Green J.R. A review of network-based approaches to drug repositioning. Brief. Bioinform. 2018;vol. 19(5):878–892. doi: 10.1093/BIB/BBX017. [DOI] [PubMed] [Google Scholar]
- 15.Tanoli Z., Vähä-Koskela M., Aittokallio T. Artificial intelligence, machine learning, and drug repurposing in cancer. Expert Opin. Drug Discov. 2021;16(9):977–989. doi: 10.1080/17460441.2021.1883585. [DOI] [PubMed] [Google Scholar]
- 16.Tanoli Z., Seemab U., Scherer A., Wennerberg K., Tang J., Vähä-Koskela M. Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief. Bioinform. 2021;22(2):1656–1678. doi: 10.1093/BIB/BBAA003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider P., et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020;19(5):353–364. doi: 10.1038/S41573-019-0050-3. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira L.L.G., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today. 2019;24(5):1157–1165. doi: 10.1016/J.DRUDIS.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Lv H., et al. Application of artificial intelligence and machine learning forCOVID-19 drug discovery and vaccine design. Brief. Bioinform. 2021;22(6):1–10. doi: 10.1093/BIB/BBAB320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X., et al. Deep learning for drug repurposing: methods, databases, and applications. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2022 doi: 10.48550/arxiv.2202.05145. [DOI] [Google Scholar]
- 21.Lee C.Y., Chen Y.P.P. New insights into drug repurposing for COVID-19 using deep learning. IEEE Trans. Neural Netw. Learn. Syst. 2021;32(11):4770–4780. doi: 10.1109/TNNLS.2021.3111745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarada T.N., Rokne J.G., Alhajj R. A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions. J. Chemin. 2020;12(1):1–23. doi: 10.1186/S13321-020-00450-7/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004 38. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez S., et al. Machine learning identifies candidates for drug repurposing in Alzheimer’s disease. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-21330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta R., Srivastava D., Sahu M., Tiwari S., Ambasta R.K., Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol. Divers. 2021;25(3):1315–1360. doi: 10.1007/S11030-021-10217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peyvandipour A., Saberian N., Shafi A., Donato M., Draghici S. A novel computational approach for drug repurposing using systems biology. Bioinformatics. 2018;34(16):2817–2825. doi: 10.1093/BIOINFORMATICS/BTY133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong F., et al. Artificial intelligence in drug design. Sci. China Life Sci. 2018;61(10):1191–1204. doi: 10.1007/S11427-018-9342-2. [DOI] [PubMed] [Google Scholar]
- 28.Brown N., Ertl P., Lewis R., Luksch T., Reker D., Schneider N. Artificial intelligence in chemistry and drug design. J. Comput. Mol. Des. 2020 347. 2020;34(7):709–715. doi: 10.1007/S10822-020-00317-X. [DOI] [PubMed] [Google Scholar]
- 29.Harrer S., Shah P., Antony B., Hu J. Artificial intelligence for clinical trial design. Trends Pharmacol. Sci. 2019;40(8):577–591. doi: 10.1016/J.TIPS.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Shen M., Xiao Y., Golbraikh A., Gombar V.K., Tropsha A. Development and validation of k-nearest-neighbor QSPR models of metabolic stability of drug candidates. J. Med. Chem. 2003;46(14):3013–3020. doi: 10.1021/JM020491T. [DOI] [PubMed] [Google Scholar]
- 31.Susnow R.G., Dixon S.L. Use of robust classification techniques for the prediction of human cytochrome P450 2D6 inhibition. J. Chem. Inf. Comput. Sci. 2003;43(4):1308–1315. doi: 10.1021/CI030283P. [DOI] [PubMed] [Google Scholar]
- 32.Suykens J.A.K. Support vector machines and kernel-based learning for dynamical systems modelling. IFAC Proc. 2009;42(10):1029–1037. doi: 10.3182/20090706-3-FR-2004.00171. [DOI] [Google Scholar]
- 33.Asif A., Khalid M., Manzoor S., Ahmad H., Rehman A.U. Role of purinergic receptors in hepatobiliary carcinoma in Pakistani population: an approach towards proinflammatory role of P2X4 and P2X7 receptors. Purinergic Signal. 2019;15(3):367–374. doi: 10.1007/s11302-019-09675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleming N. How artificial intelligence is changing drug discovery spotlight /631/45 /639/705/117 /631/154 /706/703/559 n/a. Nature. 2018;557(7707):S55–S57. doi: 10.1038/D41586-018-05267-X. [DOI] [PubMed] [Google Scholar]
- 35.Lavecchia A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discov. Today. 2019;24(10):2017–2032. doi: 10.1016/J.DRUDIS.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Chen H., Engkvist O., Wang Y., Olivecrona M., Blaschke T. The rise of deep learning in drug discovery. Drug Discov. Today. 2018;23(6):1241–1250. doi: 10.1016/J.DRUDIS.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 37.Zhavoronkov A., et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019;37(9):1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 38.Keshavarzi Arshadi A., et al. Artificial intelligence for COVID-19 drug discovery and vaccine development. Front. Artif. Intell. 2020;3 doi: 10.3389/FRAI.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Heal. 2020;2(12):e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mottaqi M.S., Mohammadipanah F., Sajedi H. Contribution of machine learning approaches in response to SARS-CoV-2 infection. Inform. Med. Unlocked. 2021;23 doi: 10.1016/J.IMU.2021.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccialli F., di Cola V.S., Giampaolo F., Cuomo S. The role of artificial intelligence in fighting the COVID-19 pandemic. Inf. Syst. Front. 2021;23(6):1467–1497. doi: 10.1007/S10796-021-10131-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.F. Ahmed et al., “SperoPredictor: An integrated machine learning and molecular docking-based drug repurposing framework with use case of Covid-19”, Front. Public Heal., vol. 0, p. 1484, 1AD, 10.3389/FPUBH.2022.902123. [DOI] [PMC free article] [PubMed]
- 43.Xu L., Ru X., Song R. Application of machine learning for drug–target interaction prediction. Front. Genet. 2021;12:1077. doi: 10.3389/FGENE.2021.680117/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011;vol. 7 doi: 10.1038/MSB.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.“DrugBank Release Version 5.1.9 | DrugBank Online.” 〈https://go.drugbank.com/releases/latest#biotech-sequences〉 (accessed May 23, 2022).
- 46.Grondin C.J., et al. Predicting molecular mechanisms, pathways, and health outcomes induced by Juul e-cigarette aerosol chemicals using the Comparative Toxicogenomics Database. Curr. Res. Toxicol. 2021;2:272–281. doi: 10.1016/j.crtox.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendez D., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47(D1):D930–D940. doi: 10.1093/NAR/GKY1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Behery H., Attia A.F., El-Feshawy N., Torkey H. Efficient machine learning model for predicting drug-target interactions with case study for Covid-19. Comput. Biol. Chem. 2021;93 doi: 10.1016/J.COMPBIOLCHEM.2021.107536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirohara M., Saito Y., Koda Y., Sato K., Sakakibara Y. Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinforma. 2018;19(19):83–94. doi: 10.1186/S12859-018-2523-5/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David L., Thakkar A., Mercado R., Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J. Chemin. 2020;12(1):1–22. doi: 10.1186/S13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quirós M., Gražulis S., Girdzijauskaitė S., Merkys A., Vaitkus A. Using SMILES strings for the description of chemical connectivity in the Crystallography Open Database. J. Chemin. 2018;10(1):1–17. doi: 10.1186/S13321-018-0279-6/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weininger D. SMILES, a chemical language and information system: 1: introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28(1):31–36. doi: 10.1021/CI00057A005. [DOI] [Google Scholar]
- 53.Ni W., et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24(1):1–10. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhalla V., Blish C.A., South A.M. A historical perspective on ACE2 in the COVID-19 era. J. Hum. Hypertens. 2020;35(10):935–939. doi: 10.1038/s41371-020-00459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin J. Severe acute respiratory syndrome-coronavirus 2 infection: role of angiotensin-converting enzyme 2. Korean J. Med. 2020;95(4):232–235. doi: 10.3904/KJM.2020.95.4.232. [DOI] [Google Scholar]
- 56.Kragstrup T.W., et al. Plasma ACE2 predicts outcome of COVID-19 in hospitalized patients. PLoS One. 2021;16(6) doi: 10.1371/JOURNAL.PONE.0252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le B.L., et al. Transcriptomics-based drug repositioning pipeline identifies therapeutic candidates for COVID-19. Sci. Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-91625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dönertaş H.M., Fuentealba Valenzuela M., Partridge L., Thornton J.M. Gene expression‐based drug repurposing to target aging. Aging Cell. 2018;17(5) doi: 10.1111/ACEL.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H., Huang J., Zhong Y., Huang Q. DrugSig: a resource for computational drug repositioning utilizing gene expression signatures. PLoS One. 2017;12(5) doi: 10.1371/JOURNAL.PONE.0177743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Napolitano F., et al. Drug repositioning: a machine-learning approach through data integration. J. Chemin. 2013;5(6):1–9. doi: 10.1186/1758-2946-5-30/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J., Li Z., Fan X., Cheng Y. Drug-disease association and drug-repositioning predictions in complex diseases using causal inference-probabilistic matrix factorization. J. Chem. Inf. Model. 2014;54(9):2562–2569. doi: 10.1021/CI500340N. [DOI] [PubMed] [Google Scholar]
- 62.Lim H., et al. Large-scale off-target identification using fast and accurate dual regularized one-class collaborative filtering and its application to drug repurposing. PLOS Comput. Biol. 2016;12(10) doi: 10.1371/JOURNAL.PCBI.1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozsoy M.G., Özyer T., Polat F., Alhajj R. Realizing drug repositioning by adapting a recommendation system to handle the process. BMC Bioinforma. 2018;19(1):1–14. doi: 10.1186/S12859-018-2142-1/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asif A., Kim K.H., Jabbar F., Kim S., Choi K.H. Real-time sensors for live monitoring of disease and drug analysis in microfluidic model of proximal tubule. Microfluid. Nanofluidics. 2020;24(6) doi: 10.1007/s10404-020-02347-1. [DOI] [Google Scholar]
- 65.Zhang Y., Ye T., Xi H., Juhas M., Li J. Deep learning driven drug discovery: tackling severe acute respiratory syndrome coronavirus 2. Front. Microbiol. 2021;12:3195. doi: 10.3389/FMICB.2021.739684/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lipinski C.F., Maltarollo V.G., Oliveira P.R., da Silva A.B.F., Honorio K.M. Advances and perspectives in applying deep learning for drug design and discovery. Front. Robot. AI. 2019;6:108. doi: 10.3389/FROBT.2019.00108/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hooshmand S.A., Zarei Ghobadi M., Hooshmand S.E., Azimzadeh Jamalkandi S., Alavi S.M., Masoudi-Nejad A. A multimodal deep learning-based drug repurposing approach for treatment of COVID-19. Mol. Divers. 2021;25(3):1717–1730. doi: 10.1007/S11030-020-10144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pham T.H., Qiu Y., Zeng J., Xie L., Zhang P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat. Mach. Intell. 2021;3(3):247–257. doi: 10.1038/s42256-020-00285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., Xi H., Juhas M. Biosensing detection of the SARS-CoV-2 D614G mutation. Trends Genet. 2021;37(4):299. doi: 10.1016/J.TIG.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/J.CSBJ.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zahradník J., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6(9):1188–1198. doi: 10.1038/S41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 72.Zeng X., et al. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 2020;19(11):4624–4636. doi: 10.1021/ACS.JPROTEOME.0C00316/ASSET/IMAGES/LARGE/PR0C00316_0004.JPEG. [DOI] [PubMed] [Google Scholar]
- 73.Xu J., Shi P.Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6(5):909–915. doi: 10.1021/ACSINFECDIS.0C00052/ASSET/IMAGES/LARGE/ID0C00052_0002.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi Y., Shin B., Kang K., Park S., Beck B.R. Target-centered drug repurposing predictions of human angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine subtype 2 (TMPRSS2) interacting approved drugs for coronavirus disease 2019 (COVID-19) treatment through a drug-target interaction deep learning model. Viruses. 2020;12(11) doi: 10.3390/V12111325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwok K.K., Vincent E.C., Gibson J.N. Antineoplastic drugs. Pharmacol. Ther. Dent. Seven. Ed. 2017:530–562. doi: 10.1016/B978-0-323-39307-2.00036-9. [DOI] [Google Scholar]
- 76.Banjanac M., et al. Anti-inflammatory mechanism of action of azithromycin in LPS-stimulated J774A.1 cells. Pharmacol. Res. 2012;66(4):357–362. doi: 10.1016/J.PHRS.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 77.Bai Q., Tan S., Xu T., Liu H., Huang J., Yao X. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinform. 2021;22(3):1–12. doi: 10.1093/BIB/BBAA161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inf. 2020;39(8) doi: 10.1002/MINF.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang F., et al. Machine learning applications in drug repurposing. Interdiscip. Sci. Comput. Life Sci. 2022;14(1):15–21. doi: 10.1007/S12539-021-00487-8/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ke Y.Y., et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020;43(4):355–362. doi: 10.1016/J.BJ.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loucera C., Esteban-Medina M., Rian K., Falco M.M., Dopazo J., Peña-Chilet M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal Transduct. Target. Ther. 2020;5(1):1–3. doi: 10.1038/s41392-020-00417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao K., Duy Nguyen D., Chen J., Wang R., Wei G.-W. Repositioning of 8565 existing drugs for COVID-19. J. Phys. Chem. Lett. 2020;11:5373–5382. doi: 10.1021/acs.jpclett.0c01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hofmarcher M., et al. Large-scale ligand-based virtual screening for SARS-CoV-2 inhibitors using deep neural networks. SSRN Electron. J. 2020 doi: 10.48550/arxiv.2004.00979. [DOI] [Google Scholar]
- 84.Hu F., Jiang J., Yin P. Prediction of potential commercially inhibitors against SARS-CoV-2 by multi-task deep model. 2020 doi: 10.48550/arxiv.2003.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang K., Fu T., Glass L.M., Zitnik M., Xiao C., Sun J. DeepPurpose: a deep learning library for drug–target interaction prediction. Bioinformatics. 2021;36(22–23):5545–5547. doi: 10.1093/BIOINFORMATICS/BTAA1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Batra R., Chan H., Kamath G., Ramprasad R., Cherukara M.J., Sankaranarayanan S.K.R.S. Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking studies. J. Phys. Chem. Lett. 2020;11 doi: 10.1021/acs.jpclett.0c02278. [DOI] [PubMed] [Google Scholar]
- 87.Kowalewski J., Ray A. Predicting novel drugs for SARS-CoV-2 using machine learning from a >10 million chemical space. 2017 doi: 10.1016/j.heliyon.2020.e04639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emmert-Streib F., Tripathi S., de R., Simoes M., Hawwa A.F., Dehmer M. The human disease network. Ceased. 2013;1(1):20–28. doi: 10.4161/SYSB.22816. [DOI] [Google Scholar]
- 89.O. Mason and M. Verwoerd, “Graph Theory and Networks in Biology”, 2006. [DOI] [PubMed]
- 90.Silverman E.K., et al. Molecular networks in network medicine: development and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020;12(6) doi: 10.1002/WSBM.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2010;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alaimo S., Pulvirenti A. Network-based drug repositioning: approaches, resources, and research directions. Methods Mol. Biol. 2019;1903:97–113. doi: 10.1007/978-1-4939-8955-3_6. [DOI] [PubMed] [Google Scholar]
- 93.Foster J.G., Foster D.V., Grassberger P., Paczuski M. Edge direction and the structure of networks. Proc. Natl. Acad. Sci. U. S. A. 2010;107(24):10815–10820. doi: 10.1073/PNAS.0912671107/SUPPL_FILE/PNAS.0912671107_SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Hernandez J.E., et al. Network-based approaches reveal potential therapeutic targets for host-directed antileishmanial therapy driving drug repurposing. Microbiol. Spectr. 2021;9(2) doi: 10.1128/SPECTRUM.01018-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.“A Directed Graph Neural Network-based Drug-Repurposing Approach to Identify a Lead Combination of Drugs for Alzheimer’s Disease - healthylongevitychallenge.org.” 〈https://healthylongevitychallenge.org/winners/a-directed-graph-neural-network-based-drug-repurposing-approach-to-identify-a-lead-combination-of-drugs-for-alzheimers-disease/〉 (accessed May 23, 2022).
- 96.M. Langberg and M. Effros, “Edge removal in undirected networks”, May 2020, doi: 10.48550/arxiv.2005.10315. [DOI]
- 97.Barrat A., Barthélemy M., Pastor-Satorras R., Vespignani A. The architecture of complex weighted networks. Proc. Natl. Acad. Sci. U. S. A. 2004;101(11):3747–3752. doi: 10.1073/PNAS.0400087101/ASSET/7970A6FD-0C68-49D2-AB37-938124DCEAFA/ASSETS/GRAPHIC/ZPQ0080439150007.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumpula J.M., Onnela J.P., Saramäki J., Kaski K., Kertész J. Emergence of communities in weighted networks. Phys. Rev. Lett. 2007;99(22) doi: 10.1103/PHYSREVLETT.99.228701/FIGURES/5/MEDIUM. [DOI] [PubMed] [Google Scholar]
- 99.Dai Y.F., Zhao X.M., Chang H.T. A survey on the computational approaches to identify drug targets in the postgenomic era. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/239654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arrell D.K., Terzic A. Network systems biology for drug discovery. Clin. Pharmacol. Ther. 2010;88(1):120–125. doi: 10.1038/CLPT.2010.91. [DOI] [PubMed] [Google Scholar]
- 101.Azuaje F. Drug interaction networks: an introduction to translational and clinical applications. Cardiovasc. Res. 2013;97(4):631–641. doi: 10.1093/CVR/CVS289. [DOI] [PubMed] [Google Scholar]
- 102.Modiano P. Chapter 15. Dora Bruder. 2019;1878:62–66. doi: 10.1525/9780520962026-015. [DOI] [Google Scholar]
- 103.Somolinos F.J., León C., Guerrero-Aspizua S. Drug repurposing using biological networks. Process. 2021;9(6):1057. doi: 10.3390/PR9061057. [DOI] [Google Scholar]
- 104.Zou J., Zheng M.W., Li G., Su Z.G. Advanced systems biology methods in drug discovery and translational biomedicine. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/742835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.“Systems Biology: Integrative Biology and Simulation Tools - Google Books.” 〈https://books.google.co.kr/books?hl=en&lr=&id=uVuRAAAAQBAJ&oi=fnd&pg=PR6&ots=gXldCs1pHd&sig=WhzO2ot7q9UmniBx87ILM86z9g4&redir_esc=y#v=onepage&q&f=false〉 (accessed May 23, 2022).
- 106.Castelletti F., La Rocca L., Peluso S., Stingo F.C., Consonni G. Bayesian learning of multiple directed networks from observational data. Stat. Med. 2020;39(30):4745–4766. doi: 10.1002/SIM.8751. [DOI] [PubMed] [Google Scholar]
- 107.Vandersickel N., et al. Directed networks as a novel way to describe and analyze cardiac excitation: directed graph mapping. Front. Physiol. 2019;10:1138. doi: 10.3389/FPHYS.2019.01138/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Askar M., Cañadas R.N., Svendsen K. An introduction to network analysis for studies of medication use. Res. Soc. Adm. Pharm. 2021;17(12):2054–2061. doi: 10.1016/J.SAPHARM.2021.06.021. [DOI] [PubMed] [Google Scholar]
- 109.H. Wu, Z. Fu, and Y. Wang, “A medical network clustering method with weighted graph structure:”, https://doi.org/10.1177/0020294020952469, vol. 53, no. 9–10, pp. 1751–1759, Sep. 2020, 10.1177/0020294020952469. [DOI]
- 110.Ye H., Liu Q., Wei J. Construction of drug network based on side effects and its application for drug repositioning. PLoS One. 2014;9(2) doi: 10.1371/JOURNAL.PONE.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gönen M. Predicting drug–target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics. 2012;28(18):2304–2310. doi: 10.1093/BIOINFORMATICS/BTS360. [DOI] [PubMed] [Google Scholar]
- 112.Koyutürk M. vol. 45. 2012. Using Protein Interaction Networks to Understand Complex Diseases; pp. 31–38. (Computer (Long. Beach. Calif)). [DOI] [Google Scholar]
- 113.Emig D., et al. Drug target prediction and repositioning using an integrated network-based approach. PLoS One. 2013;8(4) doi: 10.1371/JOURNAL.PONE.0060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.“Home - GEO - NCBI.” 〈https://www.ncbi.nlm.nih.gov/geo/〉 (accessed May 23, 2022).
- 115.Yeh S.H., Yeh H.Y., Soo V.W. A network flow approach to predict drug targets from microarray data, disease genes and interactome network - case study on prostate cancer. J. Clin. Bioinforma. 2012;2(1):1–11. doi: 10.1186/2043-9113-2-1/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.“OMIM - Online Mendelian Inheritance in Man.” 〈https://www.omim.org/〉 (accessed May 23, 2022).
- 117.“KEGG: Kyoto Encyclopedia of Genes and Genomes.” 〈https://www.genome.jp/kegg/〉 (accessed May 23, 2022).
- 118.Li L.C., Zhao H., Shiina H., Kane C.J., Dahiya R. PGDB: a curated and integrated database of genes related to the prostate. Nucleic Acids Res. 2003;31(1):291–293. doi: 10.1093/NAR/GKG008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Setoain J., et al. NFFinder: an online bioinformatics tool for searching similar transcriptomics experiments in the context of drug repositioning. Nucleic Acids Res. 2015;43(W1):W193–W199. doi: 10.1093/NAR/GKV445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.“Connectivity Map (CMAP) | Broad Institute.” 〈https://www.broadinstitute.org/connectivity-map-cmap〉 (accessed May 23, 2022).
- 121.Chang R., Shoemaker R., Wang W. A novel knowledge-driven systems biology approach for phenotype prediction upon genetic intervention. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2011;8(5):1170–1182. doi: 10.1109/TCBB.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Imoto S., Tamada Y., Savoie C.J., Miyano S. Analysis of gene networks for drug target discovery and validation. Methods Mol. Biol. 2007;360:33–56. doi: 10.1385/1-59745-165-7:33. [DOI] [PubMed] [Google Scholar]
- 123.Brown A.S., Kong S.W., Kohane I.S., Patel C.J. ksRepo: a generalized platform for computational drug repositioning. BMC Bioinforma. 2016;17(1):1–5. doi: 10.1186/S12859-016-0931-Y/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.“A network based approach to drug repositioning identifies candidates for breast cancer and prostate cancer.” 〈https://open.bu.edu/handle/2144/19066〉 (accessed May 23, 2022). [DOI] [PMC free article] [PubMed]
- 125.Wu Z., Wang Y., Chen L. Network-based drug repositioning. Mol. Biosyst. 2013;9(6):1268–1281. doi: 10.1039/C3MB25382A. [DOI] [PubMed] [Google Scholar]
- 126.Jiang Z., Zhou Y. Using gene networks to drug target identification. J. Integr. Bioinform. 2005;2(1):48–57. doi: 10.1515/JIB-2005-14. [DOI] [Google Scholar]
- 127.Dräger A., Planatscher H. Metabolic networks. Encycl. Syst. Biol. 2013:1249–1251. doi: 10.1007/978-1-4419-9863-7_1277. [DOI] [Google Scholar]
- 128.Z. Li, R.S. Wang, and X.S. Zhang, “Two-stage flux balance analysis of metabolic networks for drug target identification”, BMC Syst. Biol., vol. 5, no. SUPPL. 1, pp. 1–11, Jun. 2011, doi: 10.1186/1752-0509-5-S1-S11/FIGURES/3. [DOI] [PMC free article] [PubMed]
- 129.Kim Y.S., Asif A., Chethikkattuveli Salih A.R., Lee J.W., Hyun K.N., Choi K.H. Gravity-based flow efficient perfusion culture system for spheroids mimicking liver inflammation. Biomedicines. 2021;9(10) doi: 10.3390/biomedicines9101369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Folger O., Jerby L., Frezza C., Gottlieb E., Ruppin E., Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011;7(1):501. doi: 10.1038/MSB.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chavali A.K., D’Auria K.M., Hewlett E.L., Pearson R.D., Papin J.A. A metabolic network approach for the identification and prioritization of antimicrobial drug targets. Trends Microbiol. 2012;20(3):113–123. doi: 10.1016/J.TIM.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Taylor I.W., Wrana J.L. Protein interaction networks in medicine and disease. Proteomics. 2012;12(10):1706–1716. doi: 10.1002/pmic.201100594. [DOI] [PubMed] [Google Scholar]
- 133.Dezso Z., Ceccarelli M. Machine learning prediction of oncology drug targets based on protein and network properties. BMC Bioinforma. 2020;21(1):1–12. doi: 10.1186/S12859-020-3442-9/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dai Y.F., Zhao X.M., Chang H.T. A survey on the computational approaches to identify drug targets in the postgenomic era. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/239654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fakhraei S., Huang B., Raschid L., Getoor L. Network-based drug-target interaction prediction with probabilistic soft logic. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2014;11(5):775–787. doi: 10.1109/TCBB.2014.2325031. [DOI] [PubMed] [Google Scholar]
- 136.Cheng F., et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLOS Comput. Biol. 2012;8(5) doi: 10.1371/JOURNAL.PCBI.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou T., Ren J., Medo M., Zhang Y.C. Bipartite network projection and personal recommendation. Phys. Rev. E - Stat. Nonlinear Soft Matter Phys. 2007;76(4) doi: 10.1103/PHYSREVE.76.046115/FIGURES/4/MEDIUM. [DOI] [PubMed] [Google Scholar]
- 138.Yamanishi Y., Araki M., Gutteridge A., Honda W., Kanehisa M. Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24(13):i232–i240. doi: 10.1093/BIOINFORMATICS/BTN162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bleakley K., Yamanishi Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics. 2009;25(18):2397–2403. doi: 10.1093/BIOINFORMATICS/BTP433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mei J.P., Kwoh C.K., Yang P., Li X.L., Zheng J. Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics. 2013;29(2):238–245. doi: 10.1093/BIOINFORMATICS/BTS670. [DOI] [PubMed] [Google Scholar]
- 141.Cline M.S., et al. Integration of biological networks and gene expression data using cytoscape. Nat. Protoc. 2007;2(10):2366–2382. doi: 10.1038/NPROT.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen H., Zhang Z. A semi-supervised method for drug-target interaction prediction with consistency in networks. PLoS One. 2013;8(5) doi: 10.1371/JOURNAL.PONE.0062975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alaimo S., Giugno R., Pulvirenti A. Recommendation techniques for drug–target interaction prediction and drug repositioning. Methods Mol. Biol. 2016;1415:441–462. doi: 10.1007/978-1-4939-3572-7_23. [DOI] [PubMed] [Google Scholar]
- 144.Ding H., Takigawa I., Mamitsuka H., Zhu S. Similarity-based machine learning methods for predicting drug–target interactions: a brief review. Brief. Bioinform. 2014;15(5):734–747. doi: 10.1093/BIB/BBT056. [DOI] [PubMed] [Google Scholar]
- 145.Chen H.R., Sherr D.H., Hu Z., Delisi C. A network based approach to drug repositioning identifies plausible candidates for breast cancer and prostate cancer. BMC Med. Genom. 2016;9(1) doi: 10.1186/S12920-016-0212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.“Analysis of Network Topological Features for Identifying Potential Drug Targets | Semantic Scholar.” 〈https://www.semanticscholar.org/paper/Analysis-of-Network-Topological-Features-for-Drug-Zhang-Huan/107c8b7d3c77b144f9c5c684787bc93a69e85845〉 (accessed May 23, 2022).
- 147.Fukuoka Y., Takei D., Ogawa H. A two-step drug repositioning method based on a protein-protein interaction network of genes shared by two diseases and the similarity of drugs. Bioinformation. 2013;9(2):89. doi: 10.6026/97320630009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keane H., Ryan B.J., Jackson B., Whitmore A., Wade-Martins R. Protein-protein interaction networks identify targets which rescue the MPP+ cellular model of Parkinson’s disease. Sci. Rep. 2015;5(1):1–12. doi: 10.1038/srep17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen H., Zhang H., Zhang Z., Cao Y., Tang W. Network-based inference methods for drug repositioning. Comput. Math. Methods Med. 2015;2015 doi: 10.1155/2015/130620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu C., Gudivada R.C., Aronow B.J., Jegga A.G. Computational drug repositioning through heterogeneous network clustering. BMC Syst. Biol. 2013;7(Suppl. 5):1–9. doi: 10.1186/1752-0509-7-S5-S6/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye H., Liu Q., Wei J. Construction of drug network based on side Effects and its application for drug repositioning. PLoS One. 2014;9(2) doi: 10.1371/JOURNAL.PONE.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dudley J.T., Deshpande T., Butte A.J. Exploiting drug–disease relationships for computational drug repositioning. Brief. Bioinform. . 2011;12(4):303–311. doi: 10.1093/BIB/BBR013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hattori M., Okuno Y., Goto S., Kanehisa M. Heuristics for chemical compound matching. Genome Inform. 2003;14:144–153. doi: 10.11234/GI1990.14.144. [DOI] [PubMed] [Google Scholar]
- 154.van Laarhoven T., Marchiori E. Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS One. 2013;8(6) doi: 10.1371/JOURNAL.PONE.0066952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Perlman L., Gottlieb A., Atias N., Ruppin E., Sharan R. Combining drug and gene similarity measures for drug-target elucidation. J. Comput. Biol. 2011;18(2):133–145. doi: 10.1089/CMB.2010.0213/SUPPL_FILE/SUPP_DATA.PDF. [DOI] [PubMed] [Google Scholar]
- 156.Iorio F., et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. U. S. A. 2010;107(33):14621–14626. doi: 10.1073/PNAS.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu X., et al. In silico target fishing: addressing a ‘Big Data’ problem by ligand-based similarity rankings with data fusion. J. Chemin. 2014;6(1) doi: 10.1186/1758-2946-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mouchlis V.D., et al. Advances in de novo drug design: from conventional to machine learning methods. Int. J. Mol. Sci. 2021;22(4):1676. doi: 10.3390/IJMS22041676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Domenico A., Nicola G., Daniela T., Fulvio C., Nicola A., Orazio N. De novo drug design of targeted chemical libraries based on artificial intelligence and pair-based multiobjective optimization. J. Chem. Inf. Model. 2020;60(10):4582–4593. doi: 10.1021/ACS.JCIM.0C00517/SUPPL_FILE/CI0C00517_SI_002.PDF. [DOI] [PubMed] [Google Scholar]
- 160.Wang M., et al. Deep learning approaches for de novo drug design: an overview. Curr. Opin. Struct. Biol. 2022;72:135–144. doi: 10.1016/J.SBI.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 161.Talevi A. Drug repurposing. Ref. Modul. Biomed. Sci. 2021 doi: 10.1016/B978-0-12-820472-6.00108-0. [DOI] [Google Scholar]
- 162.Rudrapal M., Khairnar S.J., Jadhav A.G. Drug repurposing (DR): an emerging approach in drug discovery. Drug Repurposing - Hypothesis Mol. Asp. Ther. Appl. 2020 doi: 10.5772/INTECHOPEN.93193. [DOI] [Google Scholar]
- 163.Pushpakom S. Chapter 1: introduction and historical overview of drug repurposing opportunities. RSC Drug Discov. Ser. 2022;2022(82):1–13. doi: 10.1039/9781839163401-00001. [DOI] [Google Scholar]
- 164.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Asif A., et al. Microphysiological system with continuous analysis of albumin for hepatotoxicity modeling and drug screening. J. Ind. Eng. Chem. 2021;98:318–326. doi: 10.1016/j.jiec.2021.03.035. [DOI] [Google Scholar]
- 166.Yang S., Fu C., Lian X., Dong X., Zhang Z. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems. 2019;4(2) doi: 10.1128/MSYSTEMS.00303-18/SUPPL_FILE/MSYSTEMS.00303-18-SF006.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu C., et al. Computational network biology: data, models, and applications. Phys. Rep. 2020;846:1–66. doi: 10.1016/J.PHYSREP.2019.12.004. [DOI] [Google Scholar]
- 168.Zhou Y., et al. A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol. 2020;18(11) doi: 10.1371/JOURNAL.PBIO.3000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stolfi P., Manni L., Soligo M., Vergni D., Tieri P. Designing a network proximity-based drug repurposing strategy for COVID-19. Front. Cell Dev. Biol. 2020;8:1021. doi: 10.3389/FCELL.2020.545089/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fiscon G., Conte F., Farina L., Paci P. SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLOS Comput. Biol. 2021;17(2) doi: 10.1371/JOURNAL.PCBI.1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cheng F., et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yu W., Clyne M., Khoury M.J., Gwinn M. Phenopedia and genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26(1):145–146. doi: 10.1093/BIOINFORMATICS/BTP618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yu W., Gwinn M., Clyne M., Yesupriya A., Khoury M.J. A navigator for human genome epidemiology. Nat. Genet. 2008;40(2):124–125. doi: 10.1038/NG0208-124. [DOI] [PubMed] [Google Scholar]
- 174.Wang Y., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031. doi: 10.1093/NAR/GKZ981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liang S., et al. Network-based approach to repurpose approved drugs for COVID-19 by integrating GWAS and text mining data. Processes. 2022;10(2):326. doi: 10.3390/PR10020326/S1. [DOI] [Google Scholar]
- 176.Gysi D.M., et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2021;118(19) doi: 10.1073/PNAS.2025581118/SUPPL_FILE/PNAS.2025581118.SD12.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Azad A.K.M., Fatima S., Capraro A., Waters S.A., Vafaee F. Integrative resource for network-based investigation of COVID-19 combinatorial drug repositioning and mechanism of action. Patterns (N. Y., N. Y.) 2021;2(9) doi: 10.1016/J.PATTER.2021.100325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hsieh K., et al. Drug repurposing for COVID-19 using graph neural network and harmonizing multiple evidence. Sci. Rep. 2021;11(1):1–13. doi: 10.1038/s41598-021-02353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu Y., Wu Y., Shen X., Xie L. COVID-19 multi-targeted drug repurposing using few-shot learning. Front. Bioinforma. 2021:18. doi: 10.3389/FBINF.2021.693177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de S., Santos S., Torres M., Galeano D., del M., Sánchez M., Cernuzzi L., Paccanaro A. Machine learning and network medicine approaches for drug repositioning for COVID-19. Patterns. 2022;3(1) doi: 10.1016/J.PATTER.2021.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Andersen P.I., et al. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis. 2020;93:268–276. doi: 10.1016/J.IJID.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Aghdam R., Habibi M., Taheri G. Using informative features in machine learning based method for COVID-19 drug repurposing. J. Chemin. 2021;13(1):1–14. doi: 10.1186/S13321-021-00553-9/TABLES/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ge Y., et al. An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. Signal Transduct. Target. Ther. 2021;6(1):1–16. doi: 10.1038/s41392-021-00568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Desvaux E., et al. Network-based repurposing identifies anti-alarmins as drug candidates to control severe lung inflammation in COVID-19. PLoS One. 2021;16(7) doi: 10.1371/JOURNAL.PONE.0254374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang R., Hristovski D., Schutte D., Kastrin A., Fiszman M., Kilicoglu H. Drug repurposing for COVID-19 via knowledge graph completion. J. Biomed. Inform. 2021;115 doi: 10.1016/J.JBI.2021.103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sugiyama M.G., et al. Multiscale interactome analysis coupled with off-target drug predictions reveals drug repurposing candidates for human coronavirus disease. Sci. Rep. 2021 111. 2021;11(1):1–18. doi: 10.1038/s41598-021-02432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pawson A.J., et al. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014;42(Database issue) doi: 10.1093/NAR/GKT1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44(D1):D1045–D1053. doi: 10.1093/NAR/GKV1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.“Targeting COVID-19 Portal.” 〈https://ghddi-ailab.github.io/Targeting2019-nCoV/CoV_Experiment_Data/〉 (accessed May 23, 2022).
- 190.S. Godin, “Unleashing the Ideavirus”, Accessed: May 23, 2022. [Online]. Available: 〈www.ideavirus.com〉.
- 191.Acm Reference Format: Leskovec J., Adamic L.A., Huberman B.A. The dynamics of viral marketing. ACM Trans. Web. 2007;5:39. doi: 10.1145/1232722.1232727. [DOI] [Google Scholar]
- 192.Lloyd A.L., May R.M. How viruses spread among computers and people. N. Ser. 2001;292(5520):1316–1317. doi: 10.1126/science.1061076. [DOI] [PubMed] [Google Scholar]
- 193.López-Pintado D. Diffusion in complex social networks. Games Econ. Behav. 2008;62(2):573–590. doi: 10.1016/J.GEB.2007.08.001. [DOI] [Google Scholar]
- 194.Pastor-Satorras R., Vespignani A. Epidemic spreading in scale-free networks. Phys. Rev. Lett. 2001;86(14):3200. doi: 10.1103/PhysRevLett.86.3200. [DOI] [PubMed] [Google Scholar]
- 195.MotieGhader H., Safavi E., Rezapour A., Amoodizaj F.F., asl Iranifam R. Drug repurposing for coronavirus (SARS-CoV-2) based on gene co-expression network analysis. Sci. Rep. 2021 111. 2021;11(1):1–15. doi: 10.1038/s41598-021-01410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yu W., Bai Y., Raha A., Su Z., Geng F. Integrative in silico investigation reveals the host-virus interactions in repurposed drugs against SARS-CoV-2. Front. Bioinforma. 2022:81. doi: 10.3389/FBINF.2021.763540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tian X., et al. Discovery of potential therapeutic drugs for covid-19 through logistic matrix factorization with kernel diffusion. Front. Microbiol. 2022;13:455. doi: 10.3389/FMICB.2022.740382/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jin S., et al. HeTDR: drug repositioning based on heterogeneous networks and text mining. Patterns (N. Y., N. Y.) 2021;2(8) doi: 10.1016/J.PATTER.2021.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen H.G., Zhou X.H. Vol. 12. 2020. MNBDR: a module network based method for drug repositioning; pp. 1–12. (Genes (Basel)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.“Efficacy and Safety of Sirolimus in COVID-19 Infection - Full Text View - ClinicalTrials.gov.” 〈https://clinicaltrials.gov/ct2/show/NCT04461340〉 (accessed Jun. 22, 2022).
- 201.“Efficacay of Chloroquine or Hydroxychloroquine in COVID-19 Treatment - Full Text View - ClinicalTrials.gov.” 〈https://clinicaltrials.gov/ct2/show/NCT04353336〉 (accessed Jun. 22, 2022).
- 202.“Dexamethasone for COVID-19 - Full Text View - ClinicalTrials.gov.” 〈https://clinicaltrials.gov/ct2/show/NCT04707534〉 (accessed Jun. 22, 2022).
- 203.“SPI-1005 Treatment in Moderate COVID-19 Patients - Full Text View - ClinicalTrials.gov.” 〈https://clinicaltrials.gov/ct2/show/NCT04484025〉 (accessed Jun. 22, 2022).
- 204.“A Study to Evaluate the Safety and Efficacy of Artemisinin- a Herbal Supplement on COVID-19 Subjects - Full Text View - ClinicalTrials.gov.” 〈https://clinicaltrials.gov/ct2/show/NCT05004753〉 (accessed Jun. 22, 2022).
- 205.Saravanan K.M., Zhang H., Hossain M.T., Reza M.S., Wei Y. Deep learning-based drug screening for COVID-19 and case studies. Methods Pharmacol. Toxicol. 2021:631–660. doi: 10.1007/7653_2020_58/COVER/. [DOI] [Google Scholar]
- 206.Redka D.S., MacKinnon S.S., Landon M., Windemuth A., Kurji N., Shahani V. PolypharmDB, a Deep Learning-Based Resource, Quickly Identifies Repurposed Drug Candidates for COVID-19. 2020 doi: 10.26434/HEMRXIV.12071271.V1. [DOI] [Google Scholar]