Abstract
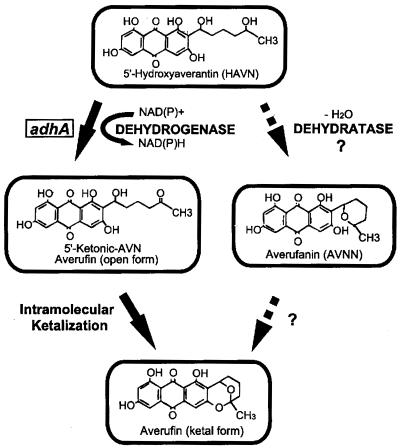
Two routes for the conversion of 5′-hydroxyaverantin (HAVN) to averufin (AVF) in the synthesis of aflatoxin have been proposed. One involves the dehydration of HAVN to the lactone averufanin (AVNN), which is then oxidized to AVF. Another requires dehydrogenation of HAVN to 5′-ketoaverantin, the open-chain form of AVF, which then cyclizes spontaneously to AVF. We isolated a gene, adhA, from the aflatoxin gene cluster of Aspergillus parasiticus SU-1. The deduced ADHA amino acid sequence contained two conserved motifs found in short-chain alcohol dehydrogenases—a glycine-rich loop (GXXXGXG) that is necessary for interaction with NAD+-NADP+, and the motif YXXXK, which is found at the active site. A. parasiticus SU-1, which produces aflatoxins, has two copies of adhA (adhA1), whereas A. parasiticus SRRC 2043, a strain that accumulates O-methylsterigmatocystin (OMST), has only one copy. Disruption of adhA in SRRC 2043 resulted in a strain that accumulates predominantly HAVN. This result suggests that ADHA is involved in the dehydrogenation of HAVN to AVF. Those adhA disruptants that still made small amounts of OMST also accumulated other metabolites, including AVNN, after prolonged culture.
Aflatoxins constitute a family of toxic and carcinogenic secondary metabolites that are produced by some members in Aspergillus section Flavi, particularly Aspergillus flavus, Aspergillus parasiticus, and Aspergillus nomius (26). They have been found as contaminants in corn, peanut, cottonseed, and tree nuts. Aflatoxin B1 (AFB1) is the most carcinogenic member of this family. The biosynthesis of AFB1 from acetyl coenzyme A is a complex process which involves more than 20 enzymatic steps (4, 14, 25). All genes encoding the biosynthetic enzymes are located in a gene cluster (5, 28, 31, 34).
The early steps in the synthesis of AFB1 involve the conversion of norsolorinic acid (NOR) to averantin (AVN), which is oxidized to 5′-hydroxyaverantin (HAVN) and then to averufin (AVF). Yabe et al. (32) showed that HAVN is an intermediate in the enzymatic conversion of AVN to AVF. Yu et al. (35) found that the avnA gene encodes a cytochrome P-450-type monooxygenase necessary for the conversion of AVN to HAVN. However, the enzyme involved in the oxidation of HAVN was not identified. Two possible routes from HAVN to AVF have been proposed (Fig. 1). In one, dehydration of HAVN occurs first to give the lactone, averufanin (AVNN), which is then oxidized to AVF (4). In the second route, the 5′-hydroxyl of HAVN is oxidized to give 5′-ketoaverantin, the open-chain form of AVF, which then spontaneously cyclizes to AVF (14, 25, 32). As evidence for the first route, McCormick et al. (24) found that radiolabeled [14C]AVNN was efficiently converted to [14C]AFB1 by wild-type A. parasiticus as well as to aflatoxin precursors by blocked A. parasiticus strains. Also, Prieto et al. (27) observed AVNN buildup in AVF-accumulating mutants. AVNN has been isolated from A. flavus, Aspergillus versicolor, and Bipolaris sp. (2, 16, 17). However, in none of these cases is there direct evidence showing that accumulation of AVNN is necessary for AVF formation.
FIG. 1.
Postulated mechanisms for the conversion of HAVN to AVF. The adhA gene product, a short-chain alcohol dehydrogenase, proposed to convert HAVN to AVF, is depicted.
In this study, we isolated the adhA gene from the known aflatoxin gene cluster of A. parasiticus. We disrupted this gene in A. parasiticus SRRC 2043, and the resulting clones accumulated predominantly HAVN. We conclude that direct dehydrogenation of HAVN to the open-chain form of AVF is the most likely enzymatic step in the aflatoxin pathway and that AVNN is probably a by-product formed by nonenzymatic dehydration of HAVN.
MATERIALS AND METHODS
Fungal strains and media.
A. parasiticus SU-1 (ATCC 56775, SRRC 143), an aflatoxin-producing strain, and A. parasiticus SRRC 2043, a strain that accumulates O-methylsterigmatocystin (OMST) were maintained on V8 agar plates (5% V-8 vegetable juice and 2% agar, pH 5.2). A. parasiticus RHN1, a NiaD− (nitrate reductase) mutant derived from SRRC 2043, served as the recipient strain for fungal transformation. Czapek solution agar (Difco, Detroit, Mich.) supplemented with 0.6 M KCl was used as the protoplast regeneration medium. Cove's medium (12) containing 10 mM ammonium or 10 mM nitrite was used as aflatoxin conducive and nonconducive medium, respectively, in preparation of mycelia for total RNA isolation for Northern blot analysis. Transformants were grown on potato dextrose agar (PDA) (Difco) to detect precursor accumulation. Adye and Mateles (A&M) (1) medium was used to grow submerged cultures for analysis of aflatoxin precursors, preparation of genomic DNA, and total RNA for reverse transcription-PCR of adhA cDNA.
Isolation of adhA.
The 2.5-kb region downstream of aflR in plasmid pHX (8) was completely sequenced by Sanger's dideoxy chain termination method and analyzed by using the Basic Local Alignment Search Tool. The DNA sequence proximate to the XbaI site encoded a short stretch of 48 amino acids, including a glycine-rich loop that had approximately 40 to 50% identity to various dehydrogenases and ketoreductases that belong to the short-chain alcohol dehydrogenase family. The genomic DNA, a 0.8-kb XbaI fragment, beyond the XbaI site was subcloned from plasmid pHH (8). Nucleotide and deduced amino acid sequences indicated that the majority of an open reading frame, designated adhA, was present in this 0.8-kb region.
Sequencing of adhA cDNA.
First-strand cDNA was synthesized with 1 μg of total RNA prepared from a 48-h culture with a 1ST-STRAND cDNA Synthesis kit (Clontech, Palo Alto, Calif.). One-tenth (10 μl) of the resultant first-strand cDNA reaction mixture was used in PCR with UlTma polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) and primers 5′-AAAAGCTTAAGGCAATAGGCATACCGTA-3′ and 5′-AAAGAATTCTGGATACAACCGTTGAC-3′. The PCR products were directly cloned with a TA Cloning kit (Invitrogen Corp., San Diego, Calif.). Dideoxy sequencing was performed on several cloned inserts from both directions with a sequencing kit (Amersham Pharmacia Biotech, Piscataway, N.J.).
Construction of the adhA disruption vector and fungal transformation.
The adhA disruption vector, pADD, was constructed using a three-step procedure. First, a 369-bp portion of adhA was cloned into pUC18. To accomplish this, PCR primers were constructed matching nucleotides (nt) 147 to 168 and nt 497 to 516 in the adhA gene. In the forward primer (ATATAAGCTTCTAGAGACGGGGCAGAACATC), a HindIII site (boldface type) was placed in front of the gene's XbaI site (underlined), and in the reverse complementary primer (ACAAGTCACTGATCCTCATGGTCGACAATT), a SalI site was added 5′ to the sequence. The PCR product was digested with HindIII and SalI and cloned into the same sites in pUC18. Second, a 282-bp SalI-XbaI fragment of adhA (nt 742 to 1024) was cloned into the SalI and XbaI sites of the above construct. Third, a 6.7-kb XbaI fragment containing the niaD gene from A. parasiticus (10) was cloned into the unique SalI site by blunt-end ligation. The resulting adhA disruption construct lacked a 225-bp region including the sequence that encodes the YPASK motif involved in the active site of the putative enzyme. Plasmid pADD was linearized with XbaI to release the portion derived from pUC18 before fungal transformation, which was carried out as previously described (19).
Sequence determination of adhA from A. parasiticus SRRC 2043 and the duplicated copy in A. parasiticus SU-1.
The PCR primers 5′-TAAGGGCGGAGAGAGAGAGAGAGAG-3′ and 5′-GGATATGAATGGTAGACTATGCTCA-3′, corresponding to the N-terminal and C-terminal ends of the deduced coding region of the adhA gene, were used with either SRRC 2043 genomic DNA or cosmid Ver2 (21) as templates. The PCR products were directly sequenced with an ABI Prism 377 fluorescent dye automatic sequencer.
Northern blot analysis of the adhA transcript.
A. parasiticus SU-1 spores (∼107) were inoculated into 100 ml of Cove's medium (12) containing either 10 mM ammonium or 10 mM nitrite as the sole nitrogen source. We isolated total RNA from fungal cultures grown at 30°C for 3 days with orbital shaking (150 rpm) by using a hot phenol extraction protocol (23), and hybridization was performed using adhA cDNA as a probe.
TLC.
Mycelia from fungal cultures were extracted with acetone and chloroform (4:1, vol/vol), and the pigments were separated by silica gel thin-layer chromatography (TLC) with toluene-ethyl acetate-acetic acid (50:30:4, vol/vol/vol) as previously described (24). Compounds on the scale-up analytical TLC plates were scraped and eluted from the silica gel matrix by immersion for 4 h in acetone. The acetone solutions were filtered through a 0.2-μm-pore-size PTFE filter prior to mass spectrometry.
Mass spectrometric analysis.
Two mass spectrometers were used for structural characterizations. For electron impact (EI) and chemical ionization (CI) analyses a Micromass Autospec EBE magnetic sector instrument (Altrincham, Manchester, United Kingdom) was used. Samples were directly inserted into the source region using a heated probe. EI mass spectrometry was conducted at 70 eV, and methane gas was used for CI mass spectrometry. Electrospray ionization mass spectra were obtained using a Micromass Quattro II triple quadrupole mass spectrometer (Altrincham). Samples were first dissolved in acetone before being mixed with methanol in a 1:1 volume ratio and directly infused at a flow rate of 5 μl/min using a Harvard Apparatus syringe driver (South Natick, Mass.). The analyses were carried out in both positive-ion and negative-ion mode.
Nucleotide sequence accession number.
The GenBank accession number for the adhA gene of A. parasiticus SU-1 is U76621.
RESULTS
Isolation and characterization of adhA.
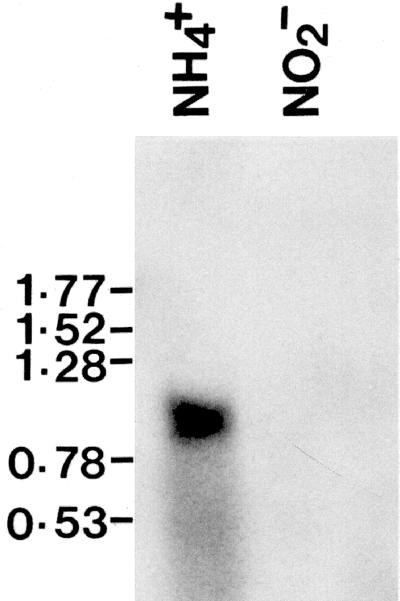
Chromosome walking between aflR and norA in the aflatoxin gene cluster in A. parasiticus SU-1 (6, 26, 34) yielded an open reading frame, named adhA, that was transcribed in the same direction as norA and located approximately 2.5 kb from aflR. This gene contains no introns based on a comparison of the genomic and cDNA sequences. Expression of this gene in A. parasiticus SU-1 grown in aflatoxin-conducive, ammonium-containing medium gave an approximately 1-kb mRNA, but expression could not be detected in a nonconducive medium containing nitrite as the sole nitrogen source (Fig. 2). This result was the same as that for known aflatoxin biosynthetic genes, such as nor1, ver1, and omtA (unpublished results). A search of the GenBank DNA sequence database with the Basic Local Alignment Search Tool protocol indicated that adhA encoded a protein with 50 to 65% amino acid sequence similarity to proteins in the short-chain alcohol dehydrogenase family. Two functional motifs are conserved in the short-chain alcohol dehydrogenase family (13, 18). The first motif was GGASGIG (residues 22 to 28) in ADHA, a glycine-rich loop required for interaction with NAD+-NADP+, and the second motif was YPASK (residues 187 to 191), a conserved portion of the alcohol dehydrogenase catalytic active site. The short-chain alcohol dehydrogenase family consists of a wide variety of dehydrogenases, including those with a specific C-terminal extension, and with a reductase C terminus. ADHA is a typical short-chain alcohol dehydrogenase based on Pfam (Protein families database of alignments and HMMs) analysis (3). In contrast, besides the two conserved motifs, NOR1 encoded by the A. parasiticus nor1 (stcE in Aspergillus nidulans) (5, 30) contains a not-well-defined Pfam-B-7 domain. VER1 encoded by A. parasiticus ver1 (stcU in A. nidulans) (5, 29) contains a reductase C terminus. NOR1 and VER1 both function as a ketoreductase (29, 30, 36). A comparison of the adhA sequence to that of the 60-kb sterigmatocystin gene cluster (GenBank accession number U34740) suggests that A. nidulans stcG is a homolog of adhA.
FIG. 2.
Northern blot analysis of total RNA of A. parasiticus SU-1 prepared from medium containing either ammonium or nitrite as the sole nitrogen source. The membrane was hybridized with a radiolabeled adhA cDNA probe. The positions of the relevant RNA molecular size standards (0.16- to 1.77-kb RNA ladder; GIBCO-BRL, Rockville, Md.) are indicated.
Sequence analysis of the duplicated adhA gene of SU-1.
Gene duplication has been reported for some of the aflatoxin biosynthetic genes in a few A. parasiticus strains (20, 21). An approximately 12-kb region of the known 80-kb aflatoxin gene cluster (28, 31, 34) is duplicated in A. parasiticus SU-1 (21, 31). To determine if the second copy of adhA has a nucleotide change(s) that could affect the function of deduced ADHA in SU-1, we used PCR to amplify the gene from the cosmid Ver2 (21), which contains the partially duplicated region of the aflatoxin gene cluster. The duplicated adhA (adhA1) had eight nucleotide substitutions; three of them are silent mutations and five that would result in amino acid substitutions are at positions 89 (A→G; Q→R), 94 (A→G; K→E), 124 (G→T; G→C), 344 (C→T; A→V), and 417 (C→A; D→E). One of these, the replacement of guanine by thymine at nt 124, would result in conversion of Gly26 to Cys in the glycine-rich loop, which might affect the binding of NAD+-NADP+.
Disruption of adhA in A. parasiticus SRRC 2043.
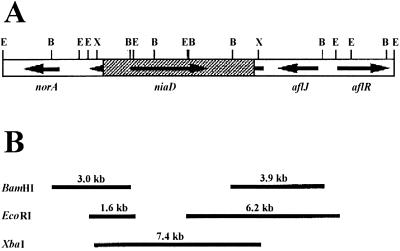
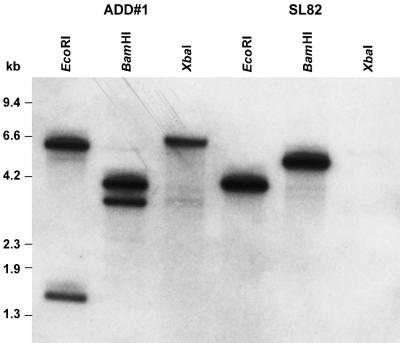
Previous studies of norA and aflR have suggested that the region containing these two genes is not duplicated in SRRC 2043 (7; unpublished results). The adhA gene of SRRC 2043 is identical to the adhA gene of SU-1. We used the vector pADD to disrupt the adhA gene in SRRC 2043. Eight of more than 200 transformants produced bright-yellow pigment(s) on PDA plates, indicative of the occurrence of gene disruption events. These strains were otherwise morphologically identical to the recipient strain and to transformants that did not accumulate the bright-yellow pigment(s). PCR amplification with primers encompassing the adhA coding region, or the niaD vector and the adhA flanking regions beyond the XbaI sites of pADD (Fig. 3), indicated that the adhA gene in the four pigmented transformants we examined was disrupted (data not shown). We also made Southern analyses with DNA from one of the disruptants, ADD#1, and from the niaD transformant, SL82. The restriction patterns of ADD#1 obtained with BamHI and EcoRI were different from those of SL82 (Fig. 4); the adhA probe hybridized to a 5.0-kb BamHI and a 4.0-kb EcoRI fragment, respectively, in SL82. In contrast, the adhA probe hybridized to 3.0- and 3.9-kb BamHI fragments as well as to 1.6- and 6.2-kb EcoRI fragments in ADD#1. These restriction patterns are consistent with a double crossover between the linearized pADD disruption vector and the resident adhA gene of SRRC 2043, thereby introducing restriction sites from the niaD insert (Fig. 3).
FIG. 3.
Schematic diagram depicting disruption of the adhA gene of A. parasiticus SRRC 2043. (A) Resulting disrupted adhA gene in the genome of SRRC 2043. (B) Expected BamHI, EcoRI, and XbaI fragments derived from the genome of an adhA disruptant. Abbreviations: B, BamHI; E, EcoRI; X, XbaI.
FIG. 4.
Southern blot analysis of genomic DNA of the adhA-disrupted strain ADD#1 and the recipient strain transformed with the niaD vector (SL82). The membrane was hybridized with a radiolabeled adhA cDNA probe. The 0.8-kb XbaI fragment derived from the genomic DNA of SL82 is not shown here. The molecular size standards are HindIII-digested lambda and HaeIII-digested φX174 DNA fragments.
Identification of metabolites accumulated by adhA disruptants.
The four adhA disruptants examined accumulated mainly HAVN. OMST also was produced by the adhA disruptants, but the amount of OMST was about one-tenth of that produced by the parental strain RHN1 when equal volumes of extracts were loaded on the TLC plate (Fig. 5). HAVN was not detectable in the colonies of untransformed RHN1. The yellow band at Rf = 0.44 comigrated with an authentic HAVN standard (32). After the TLC plate was sprayed with a 1% solution of Naturstoffreagenz A (NA) (β-aminodiethylester of diphenylboric acid) in methanol, the putative HAVN spot turned purple, a color change characteristic of anthraquinones with an uncyclized side chain at position 2. The identification of this metabolite as HAVN (molecular weight [MW] = 388) was confirmed by mass spectrometry of the band eluted into acetone. Using electrospray negative-ion mass spectrometry, a peak was found at m/z 387, corresponding to the [M−H]− ion of HAVN.
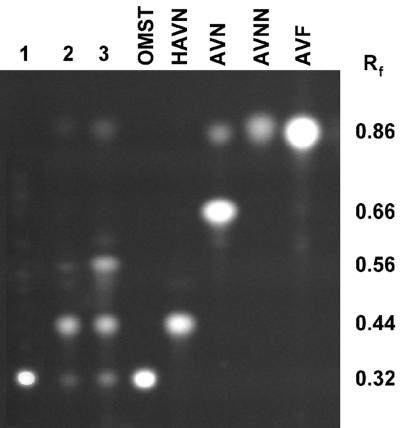
FIG. 5.
TLC of metabolites from RHN1 and an adhA disruptant (ADD#1). Metabolites were isolated from 2- and 5-day-old cultures on PDA plates by extraction for 2 h in acetone-chloroform (4:1, vol/vol). The sample was then diluted fourfold with water, an aliquot of the bottom layer was spotted on a Silica Gel 250 TLC plate, and the plate was developed in toluene-ethyl acetate-acetic acid (50:30:4, vol/vol/vol). Authentic standards were obtained from previous studies (AVNN, AVN, and AVF) or from Sigma (OMST). HAVN was a gift from K. Yabe. Lane 1: RHN1, 5 days; lane 2: ADD#1, 2 days; lane 3: ADD#1, 5 days.
In 5-day-old cultures of the adhA disruptants grown on PDA plates or in A&M medium, two other metabolites appeared at Rf = 0.56 and Rf = 0.86 (Fig. 5). The Rf = 0.86 band migrated approximately the same as the AVNN standard but slightly ahead of the AVF standard. Upon being sprayed with NA reagent, the spot turned pink rather than purple, a color change characteristic of anthraquinones with cyclized side chains. This compound had a mass spectrum consistent with its identification as AVNN. That is, the electrospray negative-ion mass spectrum indicated the presence of the [M−H]− ion at m/z 369 (AVNN; MW = 370), the CI mass spectrum (MS) indicated the presence of the [M+H]+ ion at m/z 371, and the EI MS showed a molecular ion (M+) at m/z 370. In contrast, AVF (MW = 368) had a molecular peak corresponding to the [M−H]− ion at m/z 367 as determined by electrospray MS, a molecular peak corresponding to the [M+H]+ ion at m/z 369 as determined by CI MS, and an M+ ion at m/z 368 as determined by EI MS. Besides AVNN, an unknown compound (Rf = 0.56 [Fig. 5, lane 3]) gave an [M−H]− band on negative-ion electrospray mass spectrometry at m/z 357, indicating a metabolite with a mass of 358. This metabolite accumulated in larger amounts in the 5-day-old cultures than in the 2-day-old culture. On being sprayed with NA reagent, the spot turned purple rather than pink, suggesting that it is an anthraquinone with a noncyclized hydrocarbon side chain.
DISCUSSION
The transcription pattern of adhA is consistent with that of other aflatoxin biosynthetic genes; i.e., transcription occurs when the fungus is grown in aflatoxin conducive medium containing glucose and ammonium but is suppressed in nonconducive medium in which ammonium is replaced by nitrite or nitrate (9).
The deduced ADHA protein contains two distinct motifs, GXXXGXG and YXXXK, found exclusively in short-chain alcohol dehydrogenases. Disruption of the adhA gene results in the accumulation of mainly HAVN in 2-day-old cultures. These results strongly suggest that adhA encodes a dehydrogenase that is involved in dehydrogenation of HAVN. Oxidation of the 5′-hydroxyl moiety of HAVN would lead to formation of 5′-ketoaverantin, the open-chain form of AVF (Fig. 1). Spontaneous intramolecular ketalization between the resultant ketone and the two adjacent hydroxyls then could lead to formation of the closed form of AVF [(1′S,5′R)-averufin] (14, 32). Dutton (14) proposed that the enzyme responsible for oxidation of the 5′-hydroxy moiety of HAVN is the same dehydrogenase that catalyzes the reversible reaction of NOR⇆AVN. The results from this study argue against this hypothesis, since the adhA disruptants do not accumulate NOR.
The accumulation of a small amount of OMST in the adhA disruptants suggests that other enzymes or alternative pathways may exist in the fungi for the conversion of HAVN to AVF. Yabe et al. (32) showed that when A. parasiticus was cultured in aflatoxin nonconducive YEP (yeast extract-peptone) medium, which normally inhibits the transcription of aflatoxin biosynthetic genes (15, 22), no enzymatic activities responsible for the conversion of NOR⇆AVN→HAVN were detected from the cell extract, but a limited conversion of HAVN to AVF was observed in the presence of NAD+ when the cytosol fraction of the extract was used. Their findings might mean that enzyme activities similar to that of ADHA, but not associated with aflatoxin biosynthesis, convert HAVN to AVF. This hypothesis, however, does not exclude the possibility that adhA expression might not be completely repressed in YEP medium. Overlapping enzymatic activities have been implicated in another aflatoxin biosynthetic step, the conversion of NOR to AVN (6, 11, 30). Using NOR-agarose chromatography, Chuturgoon and Dutton (11) isolated a 140-kDa dehydrogenase that carries out the conversion of NOR⇆AVN. Yet another gene, norA, in the aflatoxin gene cluster encodes a 43-kDa protein that demonstrates a reductase activity similar to that of NOR1 (6). Disruption of the single copy of nor1 in A. parasiticus, a gene which encodes a 31-kDa ketoreductase that reduces NOR to AVN, resulted in strains that accumulated NOR and aflatoxins, although in reduced amounts compared to the strains with intact nor1 (30).
In addition to OMST, AVNN and another metabolite (MW = 358) accumulated in the cultures depending on the length of time of incubation. The 5-day-old cultures accumulated small amounts of AVNN, but the 2-day-old adhA disruptants produced very little AVNN in A&M medium and on PDA plates. McCormick et al. (24) proposed that AVNN is an intermediate in the conversion of AVN to AVF. However, Yabe et al. (32) did not detect the conversion of AVNN to aflatoxins in their feeding experiments; they also reported the lack of formation of AVNN in the conversion of NA to AVF with cell extracts. Based on the study of McCormick et al. (24), Dutton (14) suggested that AVNN is derived from HAVN by dehydration and acts as a side shunt at the HAVN level. Yabe et al. (33), on the other hand, noticed that rapid drying of HAVN results in AVNN. In spite of the controversy with respect to placement of AVNN in the aflatoxin biosynthetic pathway, it has been agreed upon by all researchers that dehydration of HAVN is essential for the formation of AVNN (14, 24, 25, 26, 32, 33).
No conclusions can be drawn from this study with regard to AVNN, i.e., if it is produced enzymatically or formed spontaneously. Nonetheless, based on the presence of conserved motifs and adhA gene disruption, we conclude that ADHA is a dehydrogenase involved in the dehydrogenation of the 5′-hydroxy moiety of HAVN in the formation of AVF. Since OMST is still formed by the adhA disruptants, other dehydrogenases, either belonging to the aflatoxin pathway or made for other processes, may be able to oxidize HAVN to AVF, thereby avoiding the adhA genetic block. The fact that no dehydratase activity including aflatoxin-specific activity or activity of nonspecific origin has been shown to convert HAVN to AVNN and that no gene encoding a dehydratase has been found in the aflatoxin pathway gene cluster suggests that AVNN is probably a nonenzymatic by-product.
ACKNOWLEDGMENT
We thank J. E. Linz for providing the cosmids that made this study possible.
REFERENCES
- 1.Adye J, Mateles R I. Incorporation of labeled compounds into aflatoxin. Biochim Biophys Acta. 1964;86:418–420. doi: 10.1016/0304-4165(64)90077-7. [DOI] [PubMed] [Google Scholar]
- 2.Aucamp P J, Holzapfel C W. Polyhydroxyanthraquinones from Aspergillus versicolor, Aspergillus nidulans and Bipolaris sp. Their significance in relation to biogenetic theories of aflatoxin B1. S Afr Chem Inst J. 1970;23:40–56. [Google Scholar]
- 3.Bateman A, Birney E, Durbin R, Eddy S R, Finn R D, Sonnhammer E L L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatnagar D, Ehrlich K C, Cleveland T E. Oxidation-reduction reactions in biosynthesis of secondary metabolites. In: Bhatnagar D, Lillehoj E B, Arora D K, editors. Handbook of applied mycology. 5. Mycotoxins in ecological systems. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 255–286. [Google Scholar]
- 5.Brown D W, Yu J H, Kelkar H S, Fernandes M, Nesbitt T C, Keller N P, Adams T H, Leonard T J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary J W, Wright M, Bhatnagar D, Lee R, Chu F S. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl Environ Microbiol. 1996;62:360–366. doi: 10.1128/aem.62.2.360-366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cary J W, Barnaby N, Ehrlich K C, Bhatnagar D. Isolation and characterization of experimentally induced, aflatoxin biosynthetic pathway deletion mutants of Aspergillus parasiticus. Appl Microbiol Biotechnol. 1999;51:808–812. doi: 10.1007/s002530051466. [DOI] [PubMed] [Google Scholar]
- 8.Chang P-K, Cary J, Bhatnagar D, Cleveland T E, Bennett J W, Linz J E, Woloshuk C P, Payne G A. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang P-K, Ehrlich K C, Yu J, Bhatnagar D, Cleveland T E. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl Environ Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang P-K, Ehrlich K C, Bhatnagar D, Linz J E, Cleveland T E, Bennett J W. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr Genet. 1996;30:68–75. doi: 10.1007/s002940050102. [DOI] [PubMed] [Google Scholar]
- 11.Chuturgoon A A, Dutton M F. The affinity purification and characterization of a dehydrogenase from Aspergillus parasiticus involved in aflatoxin B1 biosynthesis. Prep Biochem. 1991;21:125–140. doi: 10.1080/10826069108018008. [DOI] [PubMed] [Google Scholar]
- 12.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 13.de Pouplana L, Fothergill-Gilmore L A. The active site architecture of a short-chain dehydrogenase defined by site-directed mutagenesis and structure modeling. Biochemistry. 1994;33:7045–7055. doi: 10.1021/bi00189a005. [DOI] [PubMed] [Google Scholar]
- 14.Dutton M F. Enzymes and aflatoxin biosynthesis. Microbiol Rev. 1988;52:274–295. doi: 10.1128/mr.52.2.274-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng G H, Chu F S, Leonard T J. Molecular cloning of genes related to aflatoxin biosynthesis by differential screening. Appl Environ Microbiol. 1992;58:455–460. doi: 10.1128/aem.58.2.455-460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heathcote J G, Dutton M F. New metabolite of Aspergillus flavus. Tetrahedron. 1969;25:1497–1500. [Google Scholar]
- 17.Holker J S E, Kagal S A, Mulheirn L J, White P M. Some new metabolites of Aspergillus versicolor and a revised structure of averufin. J Chem Soc Sect D. 1966;24:911–913. [Google Scholar]
- 18.Holm L, Sander C, Murzin A. Three sisters, different names. Nat Struct Biol. 1994;1:146–147. doi: 10.1038/nsb0394-146. [DOI] [PubMed] [Google Scholar]
- 19.Horng J S, Chang P-K, Pestka J J, Linz J E. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol Gen Genet. 1990;224:294–296. doi: 10.1007/BF00271564. [DOI] [PubMed] [Google Scholar]
- 20.Kusumoto K, Mori K, Nogata Y, Ohta H, Manbe M. Homologs of the aflatoxin biosynthetic gene ver-1 in strains of Aspergillus oryzae and related species. J Ferm Bioeng. 1996;82:161–164. [Google Scholar]
- 21.Liang S-H, Skory C D, Linz J E. Characterization of the function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1996;62:4568–4575. doi: 10.1128/aem.62.12.4568-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B-H, Chu F S. Regulation of aflR and its product, AflR, associated with aflatoxin biosynthesis. Appl Environ Microbiol. 1998;64:3718–3723. doi: 10.1128/aem.64.10.3718-3723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maramatsu M. Preparation of RNA from animal cells. Methods Cell Biol. 1973;7:23–51. doi: 10.1016/s0091-679x(08)61770-7. [DOI] [PubMed] [Google Scholar]
- 24.McCormick S P, Bhatnagar D, Cleveland T E. Averufanin is an aflatoxin B1 precursor between averantin and averufin in the biosynthetic pathway. Appl Environ Microbiol. 1987;53:14–16. doi: 10.1128/aem.53.1.14-16.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minto R E, Townsend C A. Enzymology and molecular biology of aflatoxin biosynthesis. Chem Rev. 1997;97:2537–2555. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]
- 26.Payne G A, Brown M P. Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- 27.Prieto R, Yousibova G L, Woloshuk C P. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl Environ Microbiol. 1996;62:3567–3571. doi: 10.1128/aem.62.10.3567-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva J C, Minto R E, Barry III C E, Holland K A, Townsend C A. Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. Expansion of the aflatoxin B1 biosynthetic gene cluster. J Biol Chem. 1996;271:13600–13608. doi: 10.1074/jbc.271.23.13600. [DOI] [PubMed] [Google Scholar]
- 29.Skory C D, Chang P-K, Cary J, Linz J E. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1992;58:3527–3537. doi: 10.1128/aem.58.11.3527-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trail F, Chang P-K, Cary J, Linz J E. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl Environ Microbiol. 1994;60:4078–4085. doi: 10.1128/aem.60.11.4078-4085.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trail F, Mahanti N, Mehigh R, Liang S-H, Zhou R, Rarich M, Linz J E. A physical and transcriptional map of the aflatoxin gene cluster in Aspergillus parasiticus and the functional disruption of a gene involved early in the aflatoxin pathway. Appl Environ Microbiol. 1995;61:2665–2673. doi: 10.1128/aem.61.7.2665-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabe K, Nakamura Y, Nakajima H, Ando Y, Hamasaki T. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl Environ Microbiol. 1991;57:1340–1345. doi: 10.1128/aem.57.5.1340-1345.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yabe K, Matsuyama Y, Ando Y, Nakajima H, Hamasaki T. Stereochemistry during aflatoxin biosynthesis: conversion of norsolorinic acid to averufin. Appl Environ Microbiol. 1993;59:2486–2492. doi: 10.1128/aem.59.8.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Chang P-K, Cary J W, Wright M, Bhatnagar D, Cleveland T E, Payne G A, Linz J E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol. 1995;61:2365–2371. doi: 10.1128/aem.61.6.2365-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Chang P-K, Bhatnagar D, Cleveland T E. avnA, encoding a putative P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1997;63:1349–1356. doi: 10.1128/aem.63.4.1349-1356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou R, Linz J E. Enzymatic function of the Nor-1 protein in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1999;65:5639–5641. doi: 10.1128/aem.65.12.5639-5641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]