Abstract
Gastric ulcerations in dolphins have been reported for decades. Some of these lesions were associated with parasitic infections. However, cases of nonparasitic gastric ulcers with no clearly defined etiology also have been reported in wild and captive dolphins. Considerable speculation exists as to whether dolphins have Helicobacter-associated gastritis and peptic ulcer disease. The stomachs of seven stranded Atlantic white-sided dolphins, Lagenorhynchus acutus, and 1 common dolphin, Delphinus delphis, were assessed for the presence of Helicobacter species. Novel Helicobacter species were identified by culture in the gastric mucosa of two of the eight dolphins studied and by PCR in seven of the eight dolphins. The gram-negative organisms were urease, catalase, and oxidase positive. Spiral to fusiform bacteria were detected in gastric mucosa by Warthin Starry staining. Histopathology revealed mild to moderate diffuse lymphoplasmacytic gastritis within the superficial mucosa of the main stomach. The pyloric stomach was less inflamed, and bacteria did not extend deep into the glands. The lesions parallel those observed in Helicobacter pylori-infected humans. Bacteria from two dolphins classified by 16S rRNA analysis clustered with gastric helicobacters and represent a novel Helicobacter sp. most closely related to H. pylori. These findings suggest that a novel Helicobacter sp. may play a role in the etiopathogenesis of gastritis and gastric ulcers in dolphins. To our knowledge this represents the first isolation and characterization of a novel Helicobacter sp. from a marine mammal and emphasizes the wide host distribution and pathogenic potential of this increasingly important genus.
The genus Helicobacter has been expanded considerably in recent years, and several species have been isolated from the gastrointestinal tract of humans and a wide range of different animals (8–11, 14–16, 25, 28, 33, 34, 37). In humans, Helicobacter pylori is an important pathogen which causes chronic gastritis and peptic ulcers and has been associated with gastric adenocarcinoma and lymphoma (8–11, 25, 37). H. pylori has been identified worldwide and is considered to be responsible for the most common infection in humans (24, 27, 37). In animals, other Helicobacter species are linked to gastritis with and without ulcers in their respective hosts (11, 15, 34, 37).
Gastric ulcers have been reported for decades in wild and captive dolphins (1, 19, 20, 26, 31, 32, 36). Clinical signs include inappetance, anorexia, abdominal tenderness, depression, and occasional unresponsiveness (20, 36). Complete blood count may reveal a leukocytosis and anemia if bleeding is present. This clinical syndrome is consistent with gastritis and peptic ulcer disease (PUD).
The dolphin stomach has three divisions; forestomach, main stomach, and pylorus, which joins the duodenal ampulla. The forestomach has a keratinized stratified squamous epithelium, and it is the only nonglandular portion of the dolphin's stomach. The main and pyloric portions of the stomach are glandular. The main stomach consists of neck cells, chief cells, and parietal cells, whereas the pylorus has columnar mucous cells and argentaffin cells. The duodenal ampulla is an extension of the duodenum from which the common bile duct exits from the liver (22, 31, 32).
Lesions in the forestomach and cranial portion of the main stomach can be visualized by endoscopy. However, the distal portions of the main stomach and pyloric stomach are not accessible by endoscopic examination. A number of cases of dolphins with gastric ulcers have been associated with parasitic infections and foreign bodies (6, 18, 23, 32, 35, 36). Reports of non-parasitic gastric ulcers with no clearly defined etiology also have been noted in the forestomach, main stomach, and pyloric stomach of dolphins (5, 20, 35, 36). As a consequence, considerable speculation exists as to whether dolphins have Helicobacter-associated gastritis and PUD.
White-sided dolphins, Lagenorhynchus acutus, are found in cool temperate waters in the northern Atlantic Ocean. Their distribution ranges from southern Greenland to northern Virginia, and they are commonly found in New England waters. They range in size from 6 to 9 ft and travel in pods of up to 1,000 individuals. They commonly strand themselves on the eastern seashores of the United States, but fortunately their species is not endangered or threatened (19). The common dolphin, Delphinus delphis, is seen worldwide in tropical, subtropical, and warm temperate waters. They are deep-water dolphins and travel in groups containing 20 to several thousand animals (4, 29).
The goal of this study was to determine if Helicobacter spp. could be isolated from the stomachs of stranded dolphins, as well as to determine whether the gastritis and ulcers that are noted in dolphins could be attributed to the presence of helicobacters.
MATERIALS AND METHODS
Animals.
A total of eight stranded dolphins, all of which died on the beach, were evaluated. The animals were divided into two groups (A and B). Group A was comprised of six Atlantic white-sided dolphins (numbers 1 to 6), which stranded in March 1999 in Wellfleet Cape Cod, Mass. Group B consisted of two animals (one Atlantic white-sided dolphin and one common dolphin), numbered 7 and 8 (Table 1). Both groups consisted of dolphins that were stranded on the shores of Cape Cod in the fall of 1999.
TABLE 1.
Culture and PCR results for the eight dolphins presented by animal number and sample sitea
| Animal no. (group) | Speciesa | MIT accession no.b | Sample sitec | Resultsd
|
|
|---|---|---|---|---|---|
| Culture | PCR | ||||
| 1 (A) | WSD | 99-5664 | Forestomach | − | + |
| 99-5665 | Main stomach | + | + | ||
| 99-5656 | Main stomach | + | + | ||
| 2 (A) | WSD | 99-5658 | Forestomach | − | + |
| 99-5657 | Main stomach | + | + | ||
| 3 (A) | WSD | 99-5666 | Forestomach | Mixed | + |
| 99-5659 | Main stomach | − | + | ||
| 4 (A) | WSD | 99-5661 | Forestomach | − | + |
| 99-5660 | Main stomach | − | + | ||
| 5 (A) | WSD | 99-5662 | Forestomach | − | − |
| 99-5663 | Main stomach | − | + | ||
| 6 (A) | WSD | 99-5668 | Forestomach | − | − |
| 99-5667 | Main stomach | − | − | ||
| 7 (B) | CD | 99-7442-1 | Jct fore- and main stomachs | Mixed | + |
| 99-7442-2 | Main stomach | − | + | ||
| 99-7442-3 | Jct main and pyloric stomachs | − | + | ||
| 99-7442-4 | Pyloric stomach | Mixed | + | ||
| 99-7442-5 | Jct pyloric and duodenal ampulla | Mixed | + | ||
| 8 (B) | WSD | 99-7443-1 | Jct fore- and main stomachs | − | + |
| 99-7443-2 | Main stomach | − | + | ||
| 99-7443-3 | Jct main and pyloric stomachs | Mixed | |||
| 99-7443-4 | Pyloric stomach | − | − | ||
| 99-7443-5 | Jct pyloric and duodenal ampulla | − | + | ||
WSD, Atlantic white-sided dolphin; CD, common dolphin.
MIT, Massachusetts Institute of Technology, Cambridge, Mass.
Jct, junction (of).
Mixed denotes mixed bacterial growth on artificial media that amplified a 1,200-bp Helicobacter species-specific PCR product.
Sample collection.
Two stomach samples (forestomach and main stomach from the six Atlantic white-sided dolphins from Group A) were obtained for culture and PCR. No histological samples were collected from the animals in Group A. In the two animals from Group B, five gastric samples were collected for culture, PCR, and histology (junction of the forestomach and main stomach, main stomach, junction of the main stomach and pyloric stomach, pyloric stomach, and junction of the pyloric stomach and duodenal ampulla). Each sample measured 2 cm by 2 cm and was collected under field conditions.
Microaerobic culture and biochemical characterization.
The specimens were gently rinsed with sterile physiological saline and placed in individual vials with 3 ml of 20% glycerol in brucella broth. The vials were maintained at −70°C prior to culture. Media used for culture were Trypticase soy agar with 5% sheep blood and TVP (trimethoprim, vancomycin, and polymyxin) and CVA (cefoperazone, vancomycin, and amphotericin B) antibiotic-impregnated media (Remel Laboratories, Lenexa, Kans.). In addition, selective antibiotic medium (ABM) contained the following components: blood agar base (Oxoid and Remel), 5% horse blood (Remel), 50 μg of amphotericin B per ml, 100 μg of vancomycin per ml, 3.3 μg of polymyxin B per ml, 200 μg of bacitracin per ml, and 10.7 μg of nalidixic acid per ml (Sigma Chemical Co., St. Louis, Mo.). A small amount of stomach tissue was homogenized in 1 ml of brucella broth (Difco Laboratories, Detroit, Mich.) containing 5% fetal calf serum (Summit Technologies, Fort Collins, Colo.) in a glass tissue grinder. Approximately 100 μl of sample was applied directly to TVP, CVA, and ABM media. Half of the remaining sample was filtered through a 0.45-μm-pore-size filter onto a blood agar plate. The plates were incubated at 37°C under microaerobic conditions for 2 to 4 weeks in vented jars containing N2, H2, and CO2 (80:10:10). Biochemical and morphological analyses following a previously described protocol were performed on isolated bacteria (28).
Electron microscopy.
Strain MIT 99-5665 was examined by electron microscopy. Cells grown on blood agar plates were centrifuged and suspended in 10 μM Tris-HCL buffer (pH 7.4) at a concentration of about 108 cells per ml. One percent (wt/vol) phosphotungstic acid (pH 6.5) was used to negatively stain the samples. The specimens were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
DNA extraction and PCR analysis.
DNA was extracted from bacteria with the High Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, Ind.) and from tissues by using modifications of the same method. Helicobacter-specific primer pairs C97 and C05 were used to generate 16S rRNA amplicons of 1,200 bases (16). Ten microliters of the DNA preparation was used for PCR. The PCR mixture contained 1× Taq polymerase buffer, 0.5 μM each of the two primers, 200 μM each deoxynucleotide, and 200 μg of bovine serum albumin per ml. The samples were heated at 94°C for 4 min, centrifuged briefly, and cooled to 58°C, and 0.5× Taq polymerase (Roche Molecular Biochemicals) was added. Amplification conditions were as follows: denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 3 min. Thirty-five cycles were completed before a final elongation step at 72°C for 8 min. A 15-μl aliquot of the PCR product was electrophoresed through a 1% agarose gel separation matrix prior to ethidium bromide staining and viewing under a UV light.
Purification of PCR products for 16S rRNA sequencing from tissue samples.
A 1.2-kb piece of amplified DNA from biopsy samples was purified by precipitation with polyethylene glycol 8000. After removal of Ampliwax, 0.6 volume of 20% polyethylene glycol 8000 (Sigma) in 2.5 M NaCl was added and the mixture was incubated at 37°C for 10 min. The sample was centrifuged for 15 min at 15,000 × g, and the pellet was washed with 80% ethanol and pelleted as described before. The pellet was air dried, and dissolved in 30 μl of distilled water, and used for cycle sequencing as described below.
Cloning and sequencing of 16S rRNA PCR products.
A pGEM-T vector was used for cloning the PCR products (Promega, Madison, Wis.). The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) from low-melting-point agarose gel. Fifty nanograms of purified PCR product was ligated with 50 ng of pGEM-T vector at 4°C overnight and then transferred into competent JM109 cells. Ampicillin plates with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) were used to select positive clones. Plasmid DNA was isolated from E. coli with a QIAprep Spin Miniprep Kit (Qiagen). The 1,200-base sequences of two PCR products from Helicobacter-specific primers were obtained by cycle sequencing (16).
Amplification of 16S rRNA cistrons by PCR and purification of PCR products from isolates.
The 16S rRNA cistrons were amplified with bacterial universal primers F24 and F25 (7). PCR was performed in thin-walled tubes with a Perkin-Elmer 9700 Thermocycler. One microliter of the DNA template was added to a reaction mixture (50-μl final volume) containing 20 pmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 unit of Taq 2000 polymerase (Stratagene, La Jolla, Calif.) in buffer containing Taqstart antibody (Sigma). In a hot-start protocol, samples were preheated at 95°C for 8 min followed by amplification under the following conditions: denaturation at 95°C for 45 s, annealing at 60°C 45 s, and elongation for 1.5 min with an additional 5 s for each cycle. A total of 30 cycles were performed, followed by a final elongation step at 72°C for 10 min. The results of PCR amplification were examined by electrophoresis in 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light.
16S rRNA sequencing.
Purified DNA from PCR was sequenced with an ABI prism cycle-sequencing kit (BigDye Terminator Cycle Sequencing kit with AmpliTaq DNA polymerase FS; Perkin-Elmer). The primers used for sequencing were the same ones previously described by us (7). Quarter-dye chemistry was used with 80 μM primers and 1.5 μl of PCR product in a final volume of 20 μl. Cycle sequencing was performed with an ABI 9700 DNA sequencer with 25 cycles of denaturation at 96°C for 10 s and annealing and extension at 60°C for 4 min. Sequencing reactions were run on an ABI 377 DNA sequencer.
16S rRNA data analysis.
Sequence data were entered into RNA, a program set for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction for 16S rRNA in Microsoft QuickBasic for use with personal computers and were aligned as previously described (30). Our database contains over 1,000 sequences obtained in our laboratory and over 500 obtained from GenBank. Dendrograms were constructed by the neighbor-joining method.
Southern blot analysis.
Southern blot analysis was performed with a 16S ribosomal DNA Helicobacter probe. Fifteen microliters of amplicons was electrophoresed through a 1% agarose gel and transferred onto a Hybond N nylon membrane as outlined by the manufacturer (Amersham, Arlington Heights, Ill.). DNA was then UV cross-linked. The fixed DNA was then hybridized with a Helicobacter probe generated by PCR amplification of H. pylori DNA using primers C97 and C05. The probe was labeled with horseradish peroxidase, hybridized overnight to the nylon membrane at 42°C, and exposed in the presence of luminol to Hyperfilm-ECL as outlined by the manufacturer (Amersham).
RFLP analysis.
DNA fragments of 1.2 kb from the bacterial isolates were subjected to restriction fragment length polymorphism (RFLP) analysis. DNA digestion was accomplished by addition of 10 U of restriction endonucleases AluI and HhaI (New England Biolabs, Beverly, Mass.) and 2 μl of restriction buffer (New England Biolabs) to 16 μl of DNA and incubation at 37°C for 2 h. The samples were then electrophoresed through a 6% Visigel separation matrix followed by ethidium bromide staining and viewed by UV illumination.
Histopathology.
The tissue was fixed in neutral buffered 10% formalin, processed by standard methods, and embedded in paraffin. Five-micrometer sections were stained with hematoxylin and eosin and Warthin-Starry silver stains. These sections were examined by light microscopy for evidence of lesions and for the presence of a bacterium with morphology consistent with members of the genus Helicobacter.
RESULTS
Biochemical characterization of Helicobacter species.
Helicobacter species were cultured from the glandular mucosa in two of the eight animals (Table 1). Helicobacter spp. were not isolated by culture from the forestomach. A total of three pure cultures of Helicobacter spp. were isolated from the main stomach of two different Atlantic white-sided dolphins from Group A; two cultures were isolated from animal 1 and one culture was isolated from animal 2 (99-5656, 99-5657, and 99-5665, respectively). The biochemical characteristics were determined for the three pure cultures from Group A (Table 2). The bacteria were oxidase, catalase, and urease positive. The bacteria were negative for nitrate reduction, alkaline phosphatase hydrolysis, and indoxyl acetate hydrolysis. All isolates grew at 37°C and 42°C and were susceptible to both cephalothin and nalidixic acid (Table 2). In Group B, no pure cultures of Helicobacter spp. were isolated. However, four mixed cultures containing Helicobacter spp. were isolated as ascertained by PCR analysis (Table 1).
TABLE 2.
Biochemical and phenotypic characteristics of the novel Helicobacter species from dolphins and other gastric Helicobacter speciesa
| Taxon | Catalase production | Nitrate reduction | Alkaline phosphatase hydrolysis | Urease activity | Indoxyl acetate hydrolysis | Gamma-glutamyl transpeptidase activity | Growth at 42°C | Growth with 1% glycine | Susceptibility to:
|
Periplasmic fibers | No. of flagella | Distribution of flagella | G+C content (mol%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nalidixic acid (30-μg disc) | Cephalothin (30-μg disc) | |||||||||||||
| Helicobacter sp. from dolphin | + (3/3) | − (3/3) | − (3/3) | + (3/3) | − (3/3) | + (3/3) | + (3/3) | + (3/3) | S (3/3) | S (3/3) | ||||
| H. pylori | + | − | + | + | − | + | − | − | R | S | − | 4–8 | Bipolar | 35–37 |
| H. nemestrinae | + | − | + | + | − | ND | + | − | R | S | − | 4–8 | Bipolar | 24 |
| H. acinonychis | + | − | + | + | − | + | − | − | R | S | − | 2–5 | Bipolar | 30 |
| H. felis | + | + | + | + | − | + | + | − | R | S | + | 14–20 | Bipolar | 42 |
| H. mustelae | + | + | + | + | + | + | + | − | S | R | − | 4–8 | Peritrichous | 36 |
Symbols and abbreviations: +, positive reaction; −, negative reaction; S, susceptible; R, resistant; I, intermediate; ND, not determined. Numbers in parentheses are numbers of strains with the indicated result or number of strains tested.
Ultrastructure.
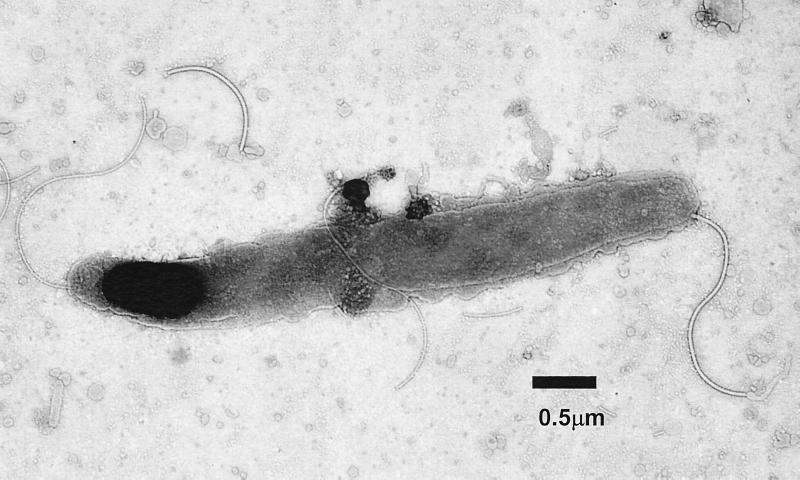
The bacteria isolated from animal 1 (99-5665) measured 0.6 μm by 4 μm and are slightly spiral with bipolar sheathed flagella, which are laterally located at the end of the bacteria (Fig. 1).
FIG. 1.
Transmission electron micrograph of the novel Helicobacter species (MIT 99-5665) isolated from an Atlantic white-sided dolphin, Lagenorhynchus acutus. The bacterium is fusiform to slightly spiral and has several sheathed flagella at each end. Bar, 0.5 μm.
Phylogenetic analysis.
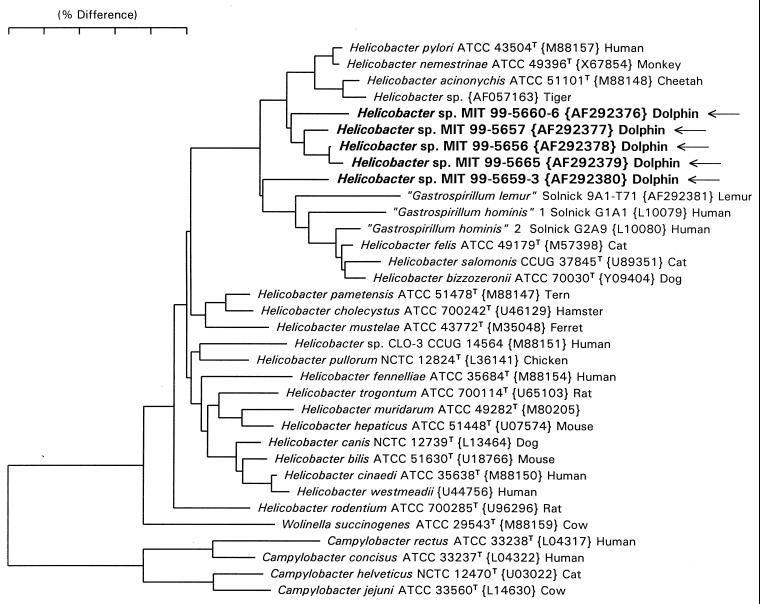
Full 16S rRNA sequences of about 1,490 bases were obtained for the three bacterial isolates from two dolphins in Group A (99-5657, 99-5656, 99-5665), and partial sequences of two 1,200-base PCR products were obtained from dolphin stomach tissue. A neighbor-joining tree for these sequences is shown in Fig. 2. The branch position and branch length for the two partial sequences (99-5660-6 and 99-5659-3) are probably distorted (due to the 290 bases of missing data); however, it is highly probable that full sequences would have fallen in nearly the same position in the tree. The dolphin sequences cluster with the gastric helicobacters (H. pylori, H. nemestrinae, H. acinonychis, H. felis, H. salmonis, H. bizzozeronii, and the “gastrospirillia”). The dolphin isolates, except MIT 99-5659-3, are most closely related to H. pylori, differing by 2 to 3%. Strain MIT 99-5659-3 may be closest to the “gastrospirillum” side of the gastric cluster, but its branching is slightly uncertain due to its being a partial sequence. These isolates and PCR products represent novel Helicobacter species.
FIG. 2.
Phylogenetic tree depicting the phylogenetic location of the novel Helicobacter sp. constructed on the basis of 16S rRNA sequence similarity values. The scale bar is equal to a 5% difference in nucleotide sequences as determined by measuring the lengths of the horizontal lines connecting two species.
PCR identification of Helicobacter spp. in tissue.
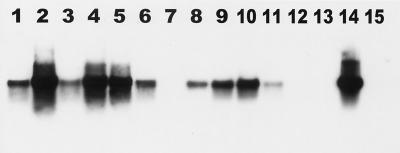
DNA from stomach tissue of eight dolphins from Groups A and B was amplified with a Helicobacter-specific primer set (Roche Molecular Biochemicals). Of the 23 samples analyzed, 18 were positive on the basis of confirmation by Southern blot analysis (Table 1 and Fig. 3). In Group A, animals 1, 2, 3, and 4 were positive for the presence of Helicobacter spp. in both the forestomach and the main stomach. In samples from animal 5, Helicobacter spp. were identified by PCR in the main stomach but not in the forestomach. Animal 6 was negative in both the main stomach and the forestomach. In Group B, animals 7 and 8 were positive in all five parts of the stomach with the exception of animal 8, which was negative in the pyloric stomach (Table 1).
FIG. 3.
Southern blot hybridization of the 1,200-bp PCR products of Helicobacter-specific primers hybridized with helicobacter 16S rRNA gene PCR product. Lanes 1 to 13, DNA extracted from stomach tissue of six dolphins (animals 1 to 6, accession numbers 99-5656 to 99-5668); lane 14, Helicobacter positive control; lane 15, reagent control.
RFLP.
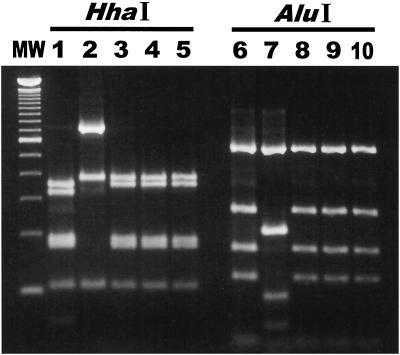
Five 1,200-bp Helicobacter-specific PCR products from four dolphins in Group A (99-5656, 99-5657, 99-5659, 99-5660, and 99-5665) were subjected to RFLP analysis by HhaI and AluI digestion. Two RFLP patterns were observed. Strains MIT 99-5656 and MIT 99-5665 were isolated from the same dolphin (animal 1) and had identical RFLP patterns. The RFLP patterns of the dolphin PCR products corresponded with the 16S rRNA sequence data (Fig. 4).
FIG. 4.
PCR-RFLP patterns of the 1,200-bp species-specific Helicobacter PCR product from the dolphin isolates and from stomach tissue. Lanes 1 and 2, 99-5660 and 99-5659 DNA from dolphin stomach tissue digested by HhaI; lanes 3 to 5, 99-5656, 99-5657, and 99-5665 DNA from dolphin isolates digested by HhaI; lanes 6 and 7, 99-5660 and 99-5659 DNA from dolphin stomach tissue digested by AluI; lanes 8 to 10, 99-5656, 99-5657, and 99-5665 DNA from dolphin isolates digested by AluI. Two patterns were observed in the HhaI digests, and two patterns were observed in the AluI digests.
Histopathology.
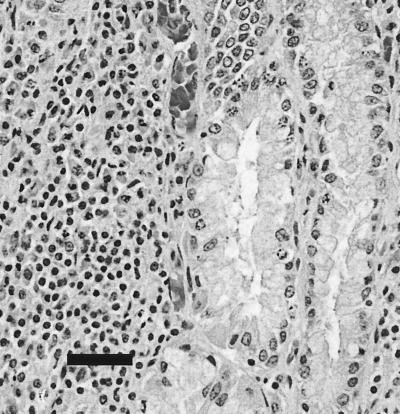
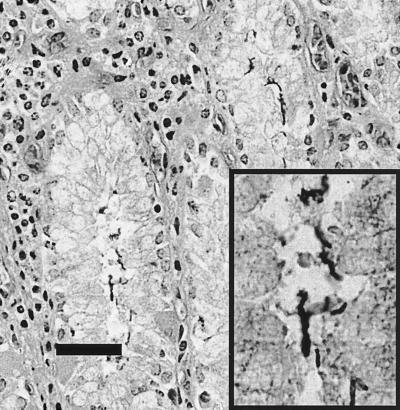
Within the superficial lamina propria of the main stomach of the Group B common dolphin, there was a mild to moderate diffuse lymphoplasmacytic infiltrate with occasional focal aggregates (Fig. 5). Many small (<5 μm), rod- to comma-shaped, silver-positive bacteria were closely associated with the epithelial lining of the gastric pits and, to a lesser extent, superficial areas of the fundic glands (Fig. 6). Similar infiltrates and organisms were observed in the main stomach of the Group B white-sided dolphin, although the tissues were considerably more autolyzed. The pyloric stomachs of both dolphins had minimal lymphoplasmacytic infiltrates in the superficial lamina propria and occasional silver-positive bacteria closely associated with the epithelium lining the stomach lumen and the apical portions of the gastric pits. In addition, the white-sided dolphin had multifocal moderately sized oval lymphocytic aggregates within the deep lamina propria and submucosa, where they were often perivascular. Rare trematodes observed in the main deep stomach mucosa of the common dolphin were not associated with inflammation.
FIG. 5.
Common dolphin (MIT 99-7442), main stomach, hematoxylin and eosin stain. Within the superficial lamina propria of the main stomach there is a focus of lymphoplasmacytic infiltration, and bacteria are associated with the epithelial surfaces of adjacent gland lumens. Bar, 20 μm.
FIG. 6.
Common dolphin (MIT 99-7442), main stomach, Warthin-Starry stain. Plump comma-shaped bacteria that are positive with Warthin-Starry stain are closely associated with the epithelial surfaces lining gastric lumens within the main stomach. The inset provides an additionally magnified image of bacteria. Bar, 20 μm.
DISCUSSION
In this study we have isolated and identified novel a Helicobacter species from the main stomachs of two stranded Atlantic white-sided dolphins. In addition, we have identified by PCR, using Helicobacter-specific primers, gastric helicobacters in seven dolphin stomachs. It is not surprising that we were unable to grow organisms from the nonglandular portion of the stomach (forestomach) given the fact that gastric helicobacters colonize only glandular tissue. However, we identified by PCR the presence of Helicobacter spp. in both the glandular and nonglandular stomachs; the latter probably reflects gastric reflux into the more proximal forestomach. The Helicobacter spp. isolated from the dolphins' glandular stomachs were urease, catalase, and oxidase positive, which is biochemically consistent with the properties of other gastric helicobacters. Ultrastructurally, helicobacters are typically fusiform to spiral and possess various numbers of sheathed flagella, which can be distributed in a polar or bipolar manner. The novel dolphin Helicobacter sp. is slightly spiral with bipolar sheathed flagella. Phylogenetically, the bacteria cluster with H. pylori differing by only 2 to 3%.
The microbial floras of human and animal stomachs have been the focus of considerable research since the discovery that H. pylori in humans caused a variety of gastric diseases (8–13, 15, 25, 27, 34). Helicobacter spp. have been isolated from the stomachs of a wide variety of mammals, including dogs, cats, ferrets, pigs, monkeys, and cheetahs, all of which have been associated with various degrees of gastritis in their hosts (9, 11, 37). Helicobacter species also have been isolated from the intestinal tracts of humans, other animals, and birds. There are currently at least 30 formally named species of the genus Helicobacter, and the majority have been proven to be or are suspected to be gastrointestinal pathogens (37). Furthermore, several of these organisms have zoonotic potential (9). Every year additional Helicobacter species are identified, some of which have been determined to be important pathogens in humans and animals.
Most of the reports on the epizootiology of helicobacter infections in animals have been done with H. mustelae in ferrets. These studies have focused on assessing the age of acquisition, mode of transmission, and susceptibility to natural or experimental reinfection following eradication (15). These data support the concept that H. mustelae (and perhaps other gastric helicobacters) transmission is fecal-oral (3, 12, 13). H. pylori also has been cultured from feces and may survive in water in a nonculturable but viable coccoid form. The other proposed route is oral-oral (27, 37), which is supported by the observation that H. pylori has been cultured from saliva and dental plaque in humans and from the saliva of H. pylori-infected cats (2, 17, 21, 27).
Gastric ulcers have been reported in wild and captive cetaceans. In certain cases the ulcerations were reported in dolphins and whales, such as Delphinus delphis, Stenella coeruleoalba, Tursiops truncatus, Phocoena phocoena, and Globicephala mela (1, 5, 23, 32, 35). Many of these cases were associated with parasitic infections, e.g., Anisakis simplex, Pholeter gastrophilus, and Braunina cordiformis (1, 5, 23, 26). The lesions were noted in the forestomach and the main stomach. However, incidences of nonparasitic gastric ulcers with no clearly defined etiology have also been noted in various species such as Delphinapterus leucas, Tursiops truncatus, and Phocoena phocoena (6, 20, 35, 36). However, there are shortcomings in the published reports regarding cetacean gastric ulcers. First, there was a lack of microbiological analysis associated with the lesions, and in many instances the poor quality of the tissue available precluded any in-depth histological analysis. Furthermore, most of the gastric lesions were defined primarily by gross observation.
Histological samples were unfortunately not taken from animals in Group A due to circumstances associated with the timing and location of the stranding. The histological sections analyzed in the two dolphins from Group B had gastric lesions which parallel those observed in H. pylori-infected humans, including multifocal lymphoplasmacytic gastritis. The Warthin-Starry stain clearly showed fusiform to slightly spiral bacteria closely associated with gastric epithelia and in areas of gastritis.
Our findings suggest that a novel Helicobacter sp. plays a role in the etiopathogenesis of gastritis in the main and pyloric stomachs of dolphins and may play a role in the development of gastric ulcers. To our knowledge this represents the first isolation and characterization of a novel Helicobacter sp. from a marine mammal and emphasizes the wide host distribution and pathogenic potential of this increasingly important genus. Further work is required to confirm whether these microorganisms play a role in etiopathogenesis of PUD in wild and captive dolphins.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants R01-AI37750 and T32-RR07036.
We thank the New England Aquarium Marine Animal Rescue Team for providing us access to the stranded animals and helping with the sample collection. We also thank Belinda Rubinstein and Greg Early from the New England Aquarium, Ruth Ewing from the National Oceanic and Atmospheric Administration and National Marine Fisheries in Florida, and Gregory Bossart and Julia Zaias from the Division of Comparative Pathology, School of Medicine, University of Miami.
REFERENCES
- 1.Abollo E. Long-term recording of gastric ulcers in cetaceans stranded in the Galician (NW Spain) coast. Dis Aquat Org. 1998;32:71–73. doi: 10.3354/dao032071. [DOI] [PubMed] [Google Scholar]
- 2.Banatvala N, Romero Lopez C, Owen R J. Use of the polymerase chain reaction to detect Helicobacter pylori in the dental plaque of healthy and symptomatic individuals. Microb Ecol Health Dis. 1994;7:1–8. [Google Scholar]
- 3.Batchelder M, Fox J G, Hayward A. Natural and experimental Helicobacter mustelae re-infection following successful antimicrobial eradication in ferrets. Helicobacter. 1996;1:34–42. doi: 10.1111/j.1523-5378.1996.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 4.Beauplet G. Study of the distribution of five species of pelagic dolphins in East tropical Pacific ocean. France: École Nat. Vet. Nantes; 1998. p. 107. [Google Scholar]
- 5.Beverley-Burton M. Helminthes of the alimentary tract from a stranded herd of the Atlantic white-sided dolphin, Lagenorhynchus acutus. J Fish Res Board Can. 1978;35:1356–1359. [Google Scholar]
- 6.Coffey D. Dolphins, whales and porpoises and encyclopedia of sea mammals. New York, N.Y: MacMillan Publishing Co. Inc.; 1977. p. 39. [Google Scholar]
- 7.Dewhirst F E, Chien C C, Paster B J, Paster R L, Orcutt R, Schauer D B, Fox J G. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman D. The prevalence of Helicobacter pylori infection in gastric cancer. Aliment Pharmacol Ther. 1995;9:59–69. [PubMed] [Google Scholar]
- 9.Fox J G. The expanding genus of Helicobacter: pathogenic and zoonotic potential. Semin Gastrointest Dis. 1997;8:124–141. [PubMed] [Google Scholar]
- 10.Fox J G. Helicobacter species and in vivo models of gastrointestinal cancer. Aliment Pharmacol Ther. 1998;12:37–60. doi: 10.1111/j.1365-2036.1998.00008.x. [DOI] [PubMed] [Google Scholar]
- 11.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 12.Fox J G, Paster B J, Dewhirst F E, Taylor N S, Yan L, Macuch P J, Chmura L M. Helicobacter mustelae isolation from feces of ferrets: evidence to support fecal-oral transmission of a gastric Helicobacter. Infect Immun. 1992;60:606–611. doi: 10.1128/iai.60.2.606-611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox J G, Blanco M, Yan L, Shames B, Polidoro D, Dewhirst F E, Paster B J. Role of gastric pH in isolation of Helicobacter mustelae from the feces of ferrets. Gastroenterology. 1993;104:86–92. doi: 10.1016/0016-5085(93)90839-5. [DOI] [PubMed] [Google Scholar]
- 14.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox J G, Marini R P. Handbook of animal models of infection. New York, N.Y: Academic Press; 1999. Animal model of Helicobacter (ferrets) pp. 274–284. [Google Scholar]
- 16.Fox J G, Dewhirst F E, Shen Z, Feng Y, Taylor N S, Paster B J, Ericson R L, Lau C N, Correa P, Araya J C, Roa I. Hepatic Helicobacter species identified in bile and gallbladder from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 17.Fox J G, Perkins S, Yan L, Shen Z, Attardo L, Pappo J. Local immune response in Helicobacter pylori-infected cats and identification of H. pylori in saliva, gastric fluid and feces. Immunology. 1996;88:400–406. doi: 10.1046/j.1365-2567.1996.d01-677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo Reynoso J P. Group behavior of common dolphins, Delphinus delphis, during prey capture. An Inst Biol Univ Nac Auton Mex. 1991;62:253–262. [Google Scholar]
- 19.Gaskin D E. Status of the Atlantic white-sided dolphin, Lagenorhynchus acutus, in Canada. Can Field-Nat. 1992;106:64–72. [Google Scholar]
- 20.Geraci J R, Gerstmann K. Relationship of dietary histamine to gastric ulcers in the dolphin. J Am Vet Med Assoc. 1966;149:884–890. [PubMed] [Google Scholar]
- 21.Gobert B, Korwin J D, Conroy M C, Bene M C, Faure G C. Polymerase chain reaction for Helicobacter pylori. In: Pajares J, Pena A, Malfertheiner P, editors. Helicobacter pylori and gastroduodenal pathology. Berlin, Germany: Springer Verlag; 1993. pp. 8–12. [Google Scholar]
- 22.Harrison R J, Johnson F R, Young B A. The esophagus and stomach of dolphins (Tursiops, Delphinus, Stenella) J Zool (Lond) 1970;160:377–390. [Google Scholar]
- 23.Herreras M V, Kaarstad S E, Balbuerna J A, Kinze C C. Helminth parasites of the digestive tract of the harbor porpoise Phocoena phocoena in Danish waters: a comparative geographical analysis. Dis Aquat Org. 1997;28:163–167. [Google Scholar]
- 24.Holcombe C, Omotara B A, Eldridge J, Jones D M. H. pylori, the most common bacterial infection in Africa: a random serological study. Am J Gastroenterol. 1992;87:28–30. [PubMed] [Google Scholar]
- 25.Kuipers E J, Thijs J C, Festen H P M. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment Pharmacol Ther. 1995;9:71–76. [PubMed] [Google Scholar]
- 26.Leatherwood S, Reeves R. The bottlenose dolphin. New York, N.Y: Academic Press; 1990. p. 176. [Google Scholar]
- 27.Megraud F. Transmission of Helicobacter pylori: fecal-oral versus oral-oral route. Aliment Pharmacol Ther. 1995;9:85–91. [PubMed] [Google Scholar]
- 28.Mendes E N, Quieroz D M M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 29.Ohizumi H, Yoshioka M, Mori K, Miyazaki N. Stomach contents of common dolphins (Delphinus delphis) in the pelagic western North Pacific. Mar Mamm Sci. 1998;14:835–844. [Google Scholar]
- 30.Paster B J, Dewhirst F E. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides urealyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 31.Ridgway S H. The bottlenose dolphin in biomedical research. In: Gay W I, editor. Methods of animal experimentation. III. New York, N.Y: Academic Press; 1968. p. 435. [Google Scholar]
- 32.Ridgway S H. Mammals of the sea, biology and medicine. Springfield, Ill: Charles C Thomas, Publisher; 1972. p. 52. [Google Scholar]
- 33.Saunders K E, Shen Z, Dewirst F E, Paster B J, Dangler C A, Fox J G. Novel intestinal Helicobacter species isolated from cotton-top tamarins (Saguinus oedipus) in chronic colitis. J Clin Microbiol. 1999;37:146–151. doi: 10.1128/jcm.37.1.146-151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shomer N H, Dangler C A, Whary M T, Fox J G. Experimental Helicobacter pylori infection induces antral gastritis and gastric mucosa-associated lymphoid tissue in guinea pigs. Infect Immun. 1998;66:2614–2618. doi: 10.1128/iai.66.6.2614-2618.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweeney J C, Ridgway S H. Common diseases of small cetaceans. J Am Vet Med Assoc. 1975;167:533–540. [PubMed] [Google Scholar]
- 36.Sweeney J. Marine mammals in zoo and wild animal medicine. Philadelphia, Pa: The W. B. Saunders Co.; 1978. p. 600. [Google Scholar]
- 37.Versalovic J, Fox J G. Helicobacter. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 727–738. [Google Scholar]