Abstract
Due to the presence of a transmembrane domain, the subcellular mobility plan of membrane-bound or membrane-tethered transcription factors (MB-TFs) differs from that of their cytosolic counterparts. The MB-TFs are mostly locked in (sub)cellular membranes, until they are released by a proteolytic cleavage event or when the transmembrane domain (TMD) is omitted from the transcript due to alternative splicing. Here, we review the current knowledge on the proteolytic activation mechanisms of MB-TFs in plants, with a particular focus on regulated intramembrane proteolysis (RIP), and discuss the analogy with the proteolytic cleavage of MB-TFs in animal systems. We present a comprehensive inventory of all known and predicted MB-TFs in the model plant Arabidopsis thaliana and examine their experimentally determined or anticipated subcellular localizations and membrane topologies. We predict proteolytically activated MB-TFs by the mapping of protease recognition sequences and structural features that facilitate RIP in and around the TMD, based on data from metazoan intramembrane proteases. Finally, the MB-TF functions in plant responses to environmental stresses and in plant development are considered and novel functions for still uncharacterized MB-TFs are forecasted by means of a regulatory network-based approach.
Keywords: membrane-bound transcription factors, proteolytic activation, regulated intramembrane proteolysis, intracellular signaling, stress response, Arabidopsis thaliana
Introduction
Membrane-bound TFs (MB-TFs) are TFs with at least one transmembrane domain (TMD) and they are present in all kingdoms, including viruses (Zupicich et al., 2001). Although TFs anchored to the membrane via lipid modifications (Eisenhaber et al., 2011; Duan et al., 2017) are sometimes classified as MB-TFs as well (Liu et al., 2018), we will use the most ‘stringent’ definition, namely that MB-TFs are “proteins that contain both a (predicted) TMD and a transcription factor family domain (TFFD)” (Kim et al., 2010; Yao et al., 2017). By their attachment to (sub)cellular membranes, MB-TFs are generally assumed to reside outside the nucleus in a dormant state until their release from the membrane in response to an intra- or extra-cellular trigger. Currently two mechanisms are known to reroute MB-TFs to the nucleus, either at the posttranscriptional level, by the generation of an alternative TMD-lacking transcript or by a proteolytic cleavage releasing the TF from the TMD (Kim et al., 2010). During the proteolytic activation, the MB-TF protein is cleaved between the TFFD and the TMD, thereby liberating the active TF from the membrane and enabling relocation to the nucleus because of the occurrence of a nuclear localization signal (NLS) in the remaining TF part. When the TMD is present at either the C- or N-terminus, a single proteolytic event is sufficient for release (Kim et al., 2010). The best-known mechanism is cleavage inside or proximal to the TMD by an intramembrane protease, also referred to as regulated intramembrane proteolysis (RIP) (Liu et al., 2018; Ye, 2020). Currently, one mechanism described thus far for the RIP activation of MB-TFs is conserved in plant, animal and yeast systems and mediated by a set of two intramembrane metalloproteases that release BASIC LEUCINE ZIPPER (bZIP) TFs from the endoplasmic reticulum (ER) during the unfolded protein response (Ye et al., 2000a; Stirling and O’Hare, 2006; Liu et al., 2007; Tajima et al., 2008). In metazoan and yeast systems, alternative release mechanisms have been described including auto-proteolytic activation, in which the proteolytic activity occurs in the MB-TF itself, as is the case for the vertebrate MYELIN REGULATORY FACTOR that is required for ER homeostasis maintenance (Bujalka et al., 2013; Milan et al., 2020). A related, but protease-independent, release mechanism translocates vertebrate epidermal growth factors upon hormone recognition to the nucleus by budding from the Golgi membranes via coat protein complex I-coated vesicles (Sigismund et al., 2008). In another mechanism, referred to as ubiquitin/proteasome-dependent processing (RUP), the ER luminal and transmembrane regions are degraded upon ubiquitination, leading to the release and nuclear translocation of the cytosolic MB-TF segment (Hoppe et al., 2000).
Recently, a comprehensive computational inventory of plant MB-TFs was generated by integrating the Plant Transcription Factor DataBase v4.0 with seven membrane topology predictors, resulting in 64 high-confidence Arabidopsis MB-TFs, further referred to as atMB-TFs (Yao et al., 2017). The atMB-TFs are found in 24 different TF families with the highest representation in the Arabidopsis NO APICAL MERISTEM/ARABIDOPSIS THALIANA ACTIVATING FACTOR/CUP-SHAPED COTYLEDON (NAC) and bZIP families, with 17 and 5 atMB-TFs, respectively. Here, we built on this inventory of atMB-TFs, assessed their subcellular localization and membrane topology, and identified novel potential cleavage events and structural features for proteolytic release based on in silico analyses. In this review, we also provide a thorough summary of the current knowledge on the proteolytic release mechanisms, the cellular and environmental MB-TF-activating triggers, and the MB-TFs involvement in plant responses to stresses and in plant development.
MB-TF Activation Mechanisms
Alternative Transcription
Posttranscriptional activation of MB-TFs involves the generation of a variant transcript isoform that no longer contains the TMD-encoding sequence. In plants, thus far, only two MB-TFs have been proven to be activated by alternative transcription: Arabidopsis bZIP60 and NAC with transmembrane motif 1-like 5 (NTL5) (Nagashima et al., 2011; Li et al., 2014). The bZIP60 transcript is spliced by the INOSITOL REQUIRING 1 (IRE1) endoribonuclease/kinase. Two IRE1 isoforms (IREa and IREb) are localized to the ER. Upon ER stress, provoked, for instance, by heat stress, dithiothreitol or tunicamycin treatment, with an unfolded protein response (UPR) as a consequence, they form homodimers that trigger their autoactivation, and consequently, the binding and splicing of an alternative 23-nucleotide intron that precedes the bZIP60 TMD (Deng et al., 2011; Nagashima et al., 2011). This IRE1-dependent alternative splicing causes a frameshift, resulting in a premature stop codon that excludes the TMD-encoding sequence from the mature transcript. This IRE1-dependent bZIP60 activation mechanism is similar to the previously discovered activation mechanism of the mammalian X-BOX BINDING PROTEIN 1 (XBP1) and the yeast HOMOLOGOUS TO ATF/CREB 1 (HAC1) bZIP TFs, hinting at the conservation of the UPR activation in eukaryotes. (Yoshida et al., 2001; Calfon et al., 2002; Cairrão et al., 2022). For NTL5, intron retention, leading to a premature TMD-preceding stop codon, was based on a single-nucleotide polymorphism (SNP) in the third intron of the NTL5 gene in the Columbia-0 accession of Arabidopsis, leading to a permanent nuclear localization in this accession. In most of the other accessions, NTL5 is stored in the ER in a dormant state, until it is activated by still unidentified mechanisms in response to abscisic acid (ABA) stimulation (Li et al., 2014). This SNP-dependent activation caused by a mutation in the genome instead of an mRNA modification caused by the plant’s splicing machinery is designated differential (instead of alternative) splicing.
MB-TFs potentially activated by alternative splicing can be detected by searching for transcript variants in RNA-sequencing data (RNA-seq). Genome-wide analysis revealed that 18 (∼ 30%) of the 64 atMB-TF genes express annotated TMD-lacking alternative transcripts (Yao et al., 2017; Table 1). Remarkably, in the bZIP and NAC (or NAC with transmembrane motif-like, NTL) families, which represent the largest MB-TFs subfamilies, no transcripts without the predicted TMD were identified, except for ANAC050. This NAC TF is one of the two special cases, in which the TMD overlaps with its TFFD, hence the TMD could only be removed by alternative splicing. However, transcript variants can be missed from RNA-seq data, because particular alternative splicing events are often specific and depend on certain stimuli; for instance, the bZIP60 alternative transcript had only been detected under ER stress conditions (Nagashima et al., 2011). Of the 18 atMB-TF TMD-lacking transcript variants identified in RNA-seq data, 15 lost their TMD due to alternative splicing resulting in exon skipping. An example is the LSD ONE LIKE 1 (LOL1) zinc finger TF involved in the regulation of oxidative stress-induced cell death during the hypersensitive response (Epple et al., 2003). LOL1 has, according to RNA-seq data, seven different transcripts, of which only two contain a predicted N-terminal TMD (Epple et al., 2003). Thus far, LOL1 functional studies had been based only on a coding sequence without the predicted TMD. Thus, the existence of a TMD-containing protein isoform and its activation mechanism remain subjects for further study. For ZINC FINGER NUCLEASE 2 (ZNF2), an alternative transcript leads to a premature stop codon, similarly as for bZIP60, whereas for AINTEGUMENTA-LIKE 6 (AIL6) and LONESOME HIGHWAY LIKE 2 (LHL2), an N-terminal TMD-lacking alternative transcript hints at alternative splicing with a new translation initiation site as a result (Yao et al., 2017). Theoretically, activation of an MB-TF by alternative splicing does not rule out proteolytic cleavage, but activation by these two mechanisms in parallel has not been reported yet.
TABLE 1.
Overview of protein domain organization, membrane topology, subcellular localization, structural features indicative for regulated intramembrane proteolysis (RIP) and alternative transcripts of membrane-bound TFs in Arabidopsis (atMB-TFs).
| Protein name | TMD position relative to the TFFD | TMD score predictiona | Membrane topologyb | Positionc (AA) of |
Subcellular localizationd | Postranslational modificationse | Helix-breaking residues in the TMDf | No. positively charged AA (K,R) in theg |
No. alternative transcripts resulting in TMD loss Yao et al., 2017 | ||
| TFFD | TMD | Predicted| | experimental | TMD | 20-AA TM-flanking region | |||||||
| AIL6 | N | 0.702 | I | 389-440 | 301-321 | mt, nuc| | mt | ph(4) | 0 | K(3) R(1) | 2 | |
| ANAC028 | C | 0.570 | 6-143 | 609-630 | Nuc | nt(1)* | K(2) | K(5) R(1) | |||
| AT2G13960 | N | 0.582 | II | 40-86 | 9-29 | sec, nuc, PM | [GxxN] | 0 | K(4)* | ||
| AT2G29660 | N | 0.477 | I | 126-149 | 9-29 | sec, mt, nuc | ph(1)* | R(1) | K(6) R(4)* | ||
| AT3G04930 | C | 0.486 | II | 137-235 | 372-395 | nuc| | PM | ph(5)* | 0 | K(1) | 1 | |
| AT5G25475 | N | 0.606 | I | 78-155 | 52-75 | Mt | [PxxN] | K(1) | K(1)R(5)* | 2 | |
| AT5G63280 | C | 0.836 | II | 105-128 | 223-246 | sec, nuc | nt(1) ng(1)* ac(1) ro(1) | 0 | K(4) R(1)* | ||
| bHLH035 | C | 0.581 | II | 58-101 | 203-224 | Nuc | ph(1)* | 0 | K(2) | 3 | |
| 0.360 | II | 190-213 | K(1) | K(2) R(1) | |||||||
| 0.871 | I | 214-245 | [PN] | R(1) | |||||||
| bHLH115 | C | 0.363 | 136-182 | 190-211 | Nuc | K(5) | K(1) R(2) | ||||
| bHLH131 | N | 0.463 | 1351-1397 | 1242-1262 | nuc, sec| | pl and mt | ph(1) | 0 | K(2) | 2 | ||
| 0.341 | 1277-1297 | 0 | |||||||||
| bZIP7 | N | 0.662 | 197-254 | 38-59 | Nuc | ph(3)* ng(1) | K(1) | 0 | 3 | ||
| bZIP17 | C | 0.798 | I | 226-287 | 364-387 | nuc| | ER, PM, nuc [1] | ph(1) | [GA] | K(1) | K(7)* | |
| bZIP28 | C | 0.848 | I | 192-237 | 321-344 | nuc, sec, ER| | ER, PM, nuc [2] | K(1) | K(7) R(1)* | |||
| bZIP49 | C | 0.755 | I | 172-219 | 286-309 | Nuc | [GA] | K(1) | K(6) R(1)* | ||
| bZIP60 | C | 0.693 | II | 141-183 | 217-240 | Nuc | 0 | K(2) R(1) | |||
| CAMTA1 | C | 0.380 | I | 81-188 | 192-211 | Nuc | ph(10)* na(1) nt(1) | R(1) | K(1) R(3) | ||
| CAMTA5 | C | 0.683 | 30-146 | 579-600 | nuc| | Golgi(4), PM, nuc | ph(3)* sm(1) | 0 | K(4) | 1 | ||
| FRF3 | C | 0.424 | 25-110 | 115-136 | mt, cyt | R(1) | K(2) R(1) | 1 | |||
| GPL2 | C | 0.786 | 61-155 | 328-348 | Nuc | ph(4)* | [PxxP] | 0 | 0 | ||
| HHO5 | N | 0.567 | 218-272 | 64-85 | Nuc | ph(1) | R(1) | K(3) R(1) | 3 | ||
| LD | C | 0.496 | 66-122 | 222-243 | Nuc | ph(2)* ub(1) | K(1) | K(2) R(1) | |||
| LHL2 | N | 0.537 | 529-566 | 123-144 | nuc| | nuc [3] | 0 | K(1) R(4) | 2 | |||
| LOL1 | N | 0.488 | II | 70-171 | 24-47 | sec, nuc | 0 | K(3) R(1) | |||
| MAMYB | N | 0.770 | I | 159-193 | 35-55 | nuc| | ER(2), Golgi(4), PM(3), nuc [4] | ph(14)* na(1) | 0 | K(1) | ||
| 0.575 | II | 61-84 | [PxxP] | K(1) | |||||||
| NFXL2 | C | 0.462 | II | 247-452 | 840-863 | Nuc | ph(1)* | K(1) | K(2) R(2) | ||
| NGAL2 | C | 0.469 | II | 29-142 | 198-221 | Nuc | 0 | K(2) | 2 | ||
| NLP3 | N | 0.550 | I | 495-546 | 41-64 | mt, nuc | 0 | 0 | |||
| NOK | N | 0.345 | 57-155 | 30-51 | nuc| | nuc [5] | nt(1) | K(1) R(1) | K(1) R(1) | |||
| NTL1 | C | 0.792 | I | 10-135 | 497-520 | nuc| | ER, PM, nuc [6] | [GA] | 0 | K(7) R(1)* | ||
| NTL2 | C | 0.760 | II | 24-151 | 605-625 | nuc| | ER, PM, nuc [6] | 0 | K(5) R(1)* | |||
| NTL3 | C | 0.718 | II | 17-143 | 535-555 | nuc| | ER, PM, nuc [6] | [GA] | K(1) | K(2) R2 | ||
| NTL4 | C | 0.714 | II | 9-136 | 522-545 | nuc, PM| | ER, PM, nuc [6] | K(1) | K(1) | |||
| NTL5 | C | 0.528 | II | 15-140 | 316-334 | nuc| | ER, PM, nuc [6] | R(1) | K(1) R(1) | |||
| NTL6 | C | 0.523 | II | 13-141 | 442-462 | nuc| | ER, PM, nuc [6] | ph(9)* | R(1) | K(3) R(1) | ||
| NTL7 | C | 0.856 | II | 17-143 | 525-548 | nuc, cyt| | ER, PM, nuc [6] | ph(1)* | [GA] | R(1) | K(3) R(2) | |
| NTL8 | C | 0.759 | II | 14-140 | 312-332 | nuc, PM| | ER, PM, nuc [6] | 0 | K(4) R(2) | |||
| NTL9 | C | 0.528 | II | 9-135 | 488-511 | nuc, Golgi| | ER, PM, nuc [6] | K(1) R(1) | K(2) R(5)* | |||
| NTL10 | C | 0.773 | II | 5-138 | 410-428 | nuc| | ER, PM, nuc [6] | ph(3)* | 0 | K(2) | ||
| NTL11 | C | 0.571 | II | 9-136 | 541-564 | nuc| | ER, PM, nuc [6] | ph(2)* | K(1) R(1) | K(1) | ||
| NTL13 | C | 0.480 | II | 22-147 | 319-339 | nuc| | ER, PM, nuc [6] | 0 | K(2) R(1) | |||
| NTM1 | C | 0.432 | II | 6-136 | 445-468 | nuc| | ER, PM, nuc, cytoskeleton [6] | ph(4)* | K(2) R(2) | K(4) R(2) | ||
| OBP3 | N | 0.595 | 118-177 | 42-62 | mt, nuc | ph(2)* | 0 | 0 | 5 | ||
| RLT2 | C | 0.554 | 18-74 | 518-539 | nuc| | Golgi | ph(24)* ac(1)* | R(1) | K(4) | 3 | ||
| SCP | N | 0.550 | II | 36-136 | 14-37 | sec, nuc| | nuc [7] | [GA] | R(1) K(1) | K(1) R(3) | ||
| SPL1 | C | 0.641 | II | 105-182 | 835-858 | nuc| | ER, PM, nuc [8] | ph(3)* | R(1) | K(2) R(2) | ||
| SPL7 | C | 0.627 | II | 137-213 | 762-782 | nuc| | nuc [9] | ph(1)* | 0 | K(2) R(3) | ||
| SPL12 | C | 0.641 | I | 126-203 | 881-904 | nuc| | ER, PM, nuc [8] | ph(10)* | R(1) | K(3) R(1) | ||
| SPL14 | C | 0.614 | II | 119-196 | 995-1018 | nuc| | ER, PM, nuc [8] | ph(3)* | 0 | K(1) R(3) | ||
| SPL16 | C | 0.614 | II | 82-158 | 978-1001 | nuc| | PM | 0 | K(2) R(3) | |||
| SRS8 | C | 0.375 | II | 46-140 | 144-164 | Nuc | ph(1) | 0 | K(1) R(1) | 3 | |
| WIP4 | N | 0.468 | 256-362 | 42-63 | nuc, sec | 0 | K(1) R(1) | ||||
| ZFN2 | C | 0.720 | 45-334 | 444-465 | Nuc | ph(8)* na(1) nt(1) my(1) ub(1) | K(1) | 0 | 2 | ||
cyt, cytosol; pl, plastid; nuc, nucleus; sec, secretory pathway; PM, plasma membrane; mt, mitochondria; ph, phosphorylation; na, N-terminal acetylation; nt, N-terminus proteolysis; ng, N-glycosylation; ac, lysine acetylation; ro, reversible cysteine oxidation; sm, lysine SUMOylation; ub, lysine ubiquitination; my, myristolysation.
aMean hydrophobicity within the TMD obtained from Aramemnon (Schwacke et al., 2003), with mean hydrophobicity value > 0.68 representing a high score and < 0.42 representing a low score.
bMembrane topology prediction obtained from TMHMM (Krogh et al., 2001), with type-I and type-II referring to a membrane-bound protein with the C-terminus and the N-terminus in the cytosol, respectively.
cPosition of the TFFD and TMD obtained from the PlnTFDB v5.0 database (Riaño-Pachón et al., 2007) and TMHMM (Krogh et al., 2001) or from Aramemnon (Schwacke et al., 2003), respectively.
dThe predicted subcellular localization obtained from SeqNLS (Lin et al., 2012), SignalP 6.0 (Teufel et al., 2022), DeepSig (Savojardo et al., 2018), and Aramemnon (Schwacke et al., 2003) is presented in Supplementary Table 2; the experimentally determined subcellular localization based on fluorescent protein fusion (regular text), mass spectrometry analysis of subcellular fractions (bold) and both methods (underlined) was obtained from SUBA4 [2] Liu et al. (2007); [7]Oh et al. (2010); [1] Liu et al. (2008); [4] Slabaugh et al. (2011); [3] Ohashi-Ito et al. (2013); [6] Liang et al. (2015); [8] Chao et al. (2017), Hooper et al. (2017); [9] Ramamurthy et al. (2018); [5] Hong et al. (2021).
eBetween parentheses, the number of modifications; *indicates that at least one of them is present in between the TMD and TFFD, with the amino acid positions indicated in Supplementary Table 3.
fAmino acid position in the TMD are presented in Supplementary Table 4.
gNumber of lysines (K) and number of arginine (R) are indicated between parentheses; *significantly enriched compared to the Arabidopsis proteome or compared to the TMD regions encompassing the 5–, 10–, 15– or 20-amino-acid flanking regions of the Arabidopsis membrane-bound proteome (Bonferroni-corrected hypergeometric P values < 0.05)., with corresponding P values presented in Supplementary Table 7.
Proteolytic Cleavage
The Arabidopsis bZIP17 and bZIP28 are the only known plant MB-TFs, for which the proteolytic release mechanisms are well characterized and the responsible proteases have been discovered (Liu et al., 2007; Tajima et al., 2008). For years, the SITE-1 PROTEASE (S1P) metalloprotease has been assumed to be responsible for cleavage of bZIP17 and bZIP28 at their > 300 amino acid-long tail in the ER lumen, resulting in their translocation to the Golgi whereas S2P to be responsible for the subsequent release of both TFs from the Golgi membranes, likewise to the well-described activation of STEROL REGULATORY ELEMENT-BINDING PROTEIN (SREBP) bZIP TFs in mammalian systems (Ye et al., 2000a; Stirling and O’Hare, 2006). This hypothesis was based on mutation of the S1P canonical sequence in bZIP17 and bZIP28 that resulted in the loss of the bZIP17 and bZIP28 target genes activation as well as in a deficient UPR (Sun et al., 2013, 2015). Cleavage patterns of bZIP17 in an s1p mutant background showed that bZIP17 is cleaved by S1P, but the second cleavage, anticipated to be carried out by S2P, has not been explicitly demonstrated. However, analysis of bZIP28 cleavage patterns in the s1p and s2p mutant backgrounds, confirmed cleavage only by S2P and not by S1P, indicating that the first cleavage event of bZIP28 is done by another, still unknown protease (Iwata et al., 2017). This implies that, in addition to S1P and S2P, in plants, (an)other protease(s) are involved in the proteolytic release of these bZIP TFs (Figure 2).
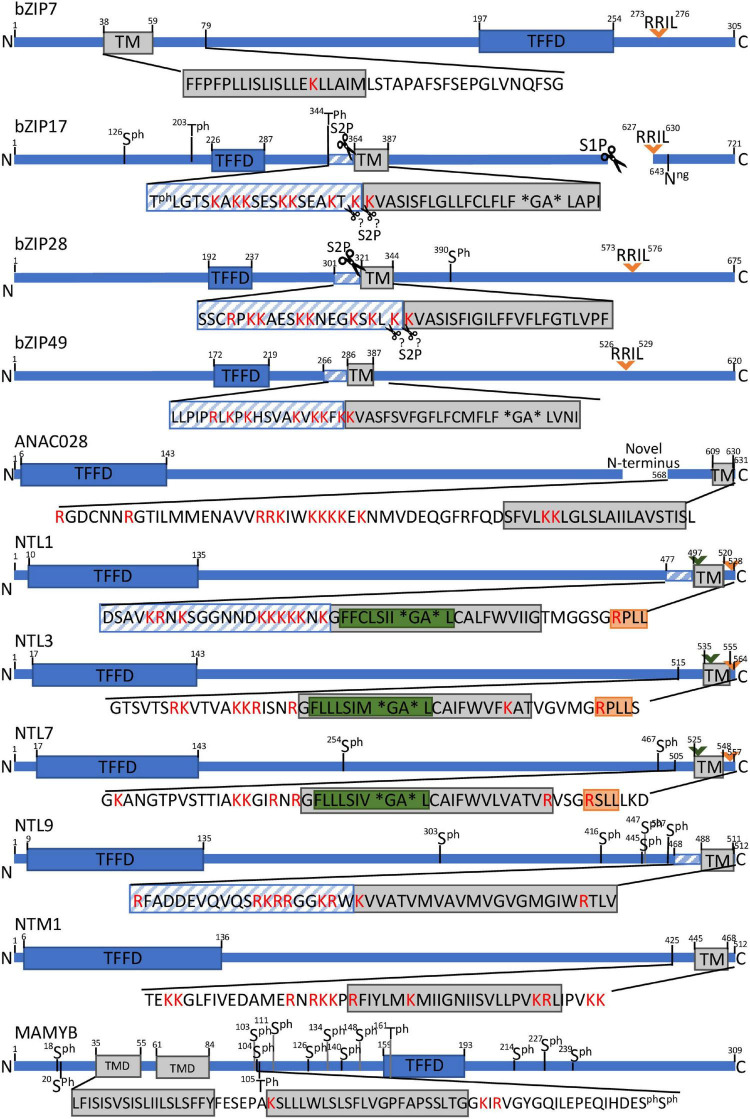
FIGURE 2.
Schematic presentation of protease recognition motifs and structural features indicative of RIP activation for a selection of MB-TFs. Predicted rhomboid recognition sites (LxLSIxGA) are indicated in green and (predicted) SIP recognition sites (Rx[LIT][KL]) in orange. Diagonal blue stripes mark TMD-neighboring regions that are significantly (corrected P value < 0.05) enriched for positive amino acids (R and K. lysine and arginine) and scissors indicate known or predicted cleavage sites. Asterisks specify helix-breaking residues. Ph. phosphorylated amino acid.
Furthermore, members of the NAC family have been studied for proteolytic activation. NAC WITH TRANSMEMBRANE MOTIF1 (NTM1) exchanges an ER to a nuclear localization pattern during cell division induced by kinetin or cytokinin (Kim et al., 2006; Kim and Park, 2007). Accordingly, a shortened protein isoform was detected by protein immunoblot analysis, with a size corresponding to that of a truncated isoform without TMD. Interestingly, pretreatment with the calpain protease inhibitor N-acetyl-leucinyl-leucinyl-norleucinal (ALLN) attenuated the nuclear relocalization and the truncated isoform detection, hinting at cleavage by a cytosolic non-membranous protease (Kim et al., 2006). In mammals, only the Nuclear Respiratory Factor 1 (Nrf1) MB-TF is known to be activated by a cytosolic protease, the cytosolic aspartic protease DNA damage-inducible 1 homolog 2 (DDI2), but this occurs after retrotranslocation of the TFFD from the luminal to the cytosolic side of the ER membrane through ER-associated degradation (ERAD) complex (Chen et al., 2022).
NTL1, NTL3, and NTL7 have been proposed to be activated by a comparable mechanism due to the high sequence similarity in their C-terminal region that harbors the (predicted) TMD (Figure 3). NTL1 and NTL7 are localized to the ER and function under conditions that perturb the mitochondrial reactive oxygen species (ROS)/redox status or induce mitochondrial dysfunction, resulting in their nuclear translocation and activation of genes involved in oxidative stress responses (De Clercq et al., 2013; Ng et al., 2013). How perturbed mitochondria signal to the ER to trigger and release these TFs is still not understood, but clearly hint at a proteolytic event, because the mitochondrial stress-induced expression of the NTL1/3/7 target genes was attenuated by pretreatment with the serine protease inhibitor N-p-Tosyl-L-phenylalanine chloromethyl ketone (TPCK). Moreover, the TMD of NTL7 contains a conserved dual cleavage site of the well-studied Drosophila melanogaster Rhomboid 1 (Rho-1) at the presumed cytosolic side, indicating cleavage by a yet unidentified rhomboid protease (Figure 2).
FIGURE 3.
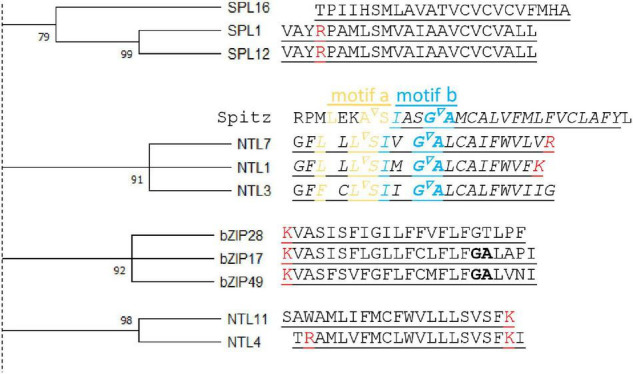
Transmembrane domain similarities of atMB-TFs. Left, A bootstrap (1,000 replicates) consensus tree was constructed from all atMB-TF transmembrane domains (TMDs) (Tamura et al., 2021) (for the complete tree, see Supplementary Figure 1). Here, only clusters with bootstrap values higher than 70 are displayed. Right, Alignment of the TMD amino acid sequences with positive amino acids (lysine and arginine, red) and helix-breaking residues (bold) indicated. For NTL7, the alignment is presented with the Drosophila melanogaster Rhomboid-1 recognition (motifs a and b) and cleavage (inverted triangle) sites from the Spitz substrate TMD region.
Membrane Targeting and Topology of atMB-TFs
The subcellular localization of MB-TFs is predominantly defined by their TMD, but once the TMD is removed, the TF is directed to the nucleus by the presence of a NLS. We assessed the subcellular localization of the atMB-TFs by combining in silico predictions and experimental evidence based on fluorescence labeling and mass spectrometry analysis of purified subcellular compartments. According to the SeqNLS algorithm (Lin et al., 2012), 73% of the atMB-TFs contain a NLS (Table 1), but because SeqNLS was shown to predict only a NLS for 80% of the known nuclear proteins in a yeast training dataset (Lin et al., 2012), other atMB-TFs might have a still undiscovered NLS. In addition, a dual, nuclear and non-nuclear, localization for one and 15 (1.5 and 23.4%) and an exclusive non-nuclear localization for eight and three (12.5 and 4.7%) out of the 64 atMB-TFs were predicted by SUBA4 (Hooper et al., 2017) and Aramemnon (Schwacke et al., 2003), respectively (Table 1). The reason for this low number of dual and extranuclear predictions is that these programs mostly rely on targeting or signaling peptides and not on the TMD-based localization. Presumably, the TMD is sufficient for extranuclear targeting, because the TMD of NTL7 was shown to have a similar localization pattern as the full length protein (Ng et al., 2013). Moreover, proximity-specific ribosome profiling experiments revealed that ribosomes that translate membrane-anchored proteins, including MB-TFs, typically target and bind to the ER-localized translocons just before or after the TMD translation (Jan et al., 2014). However, how these proteins further traverse the secretory pathway to reach their final destination, and whether this is dictated by the TMD and/or by additional signal peptides, is still not clear (Yang et al., 1997).
Eleven atMB-TFs were experimentally identified in one or more isolated subcellular compartments, including the plasma membrane (AGAMOUS-LIKE 69, NTL8, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 [SPL14], SPL16 and a DUF573 family TF [AT3G04930]), the Golgi (RINGLET 2 [RLT2]), the ER, Golgi and plasma membrane (MEMBRANE ANCHORED MYB [MAMYB]), the mitochondria [AIL 6], the plastids and mitochondria (basic HELIX-LOOP-HELIX [bHLH131]), the cytoskeleton [NTL12], and the Golgi, plasma membrane and nucleus (CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 5 [CAMTA5]) (Hooper et al., 2017; Table 1). Surprisingly, with the exception of CAMTA5, none of the atMB-TFs were identified in isolated nuclei (Hooper et al., 2017). Possibly, these TFs might be present only in the nucleus when they are activated under certain conditions and, hence, escape detection in the nucleus under basal conditions. However, based on fluorescent protein tagging, both nuclear and membrane-bound localization patterns could be shown for 18 atMB-TFs (Liu et al., 2007, 2008; Oh et al., 2010; Slabaugh et al., 2011; Ohashi-Ito et al., 2013; Liang et al., 2015; Chao et al., 2017; Ramamurthy et al., 2018; Hong et al., 2021; Table 1).
To enable nuclear translocation after cleavage, the TFFD-containing protein part should principally reside on the cytosolic membrane side. The best-studied mechanism for proteolytic release of such MB-TFs is through a single proteolytic event inside or close to the TMD by an intramembrane protease (Liu et al., 2018; Ye, 2020). However, the TFFD may not always be present in the cytosol under basal conditions. For example, the TFFD of the mammalian NRF1 occurs in the ER lumen, but is retrotranslocation to the cytosolic side through the ERAD complex that ubiquitinates and shuttles ER proteins to the cytosol for their degradation by the proteasome. The cytosolic-oriented NRF1 TFFD is subsequently cleaved between the TMD and the TFFD by the cytosolic DDI2 protease to escape from proteasomal degradation and eventually translocate to the nucleus (Zhang et al., 2014; Chen et al., 2022). To assess the membrane organization of the atMB-TFs, we computationally evaluated the positions relative to the TFFD of the TMDs and their membrane topology. Similar to the study of Yao et al. (2017), the TFFD position within the MB-TF and the TMD position and orientation were retrieved from the PlnTFDB v5.0 (Riaño-Pachón et al., 2007) and the TMHMM predictor (Krogh et al., 2001) and Aramemnon tool that integrates 18 different TMD-predicting algorithms (Schwacke et al., 2003; Schwacke and Flügge, 2018), respectively (Table 1). Based on the TMD position, we distinguished three atMB-TF groups with the TMD (i) on the C-terminal side of the TFFD (60.3%) (Figures 1A,B), (ii) on the N-terminal side of the TFFD (23.8%) (Figures 1C,D), and (iii) overlapping with the TFFD (15.9%) (Figure 1E). Most atMB-TFs, including all NTLs and all bZIP MB-TFs, except bZIP7 belong to group (i), of which 59.5% and 21.5% are predicted to have a type-II and a type-I membrane topology, i.e., the N-terminal TFFD is present in the cytosol (Figure 1A) and the organelle lumen or cell exterior (Figure 1B), respectively, whereas for 19% no membrane topology was predicted. For MB-TFs with the TMD on the N-terminal side (group ii), only 26.6% are predicted to have a type-I orientation that would result in the C-terminal TFFD at the cytosolic side (Figure 1C) and 26.6% to have the opposite topology (Figure 1D), whereas for 46.6% the prediction was lacking. This resulted in 28 (54%) atMB-TFs with the TFFD predicted at the cytosolic side of the membrane, including all NTLs, except NTL1. However, for most MB-bZIPs that have a predicted membrane topology, the TFFD is predicted in the lumen. Nevertheless, the prediction of the membrane topology remains challenging and is based on homology to proteins with experimentally verified topologies (Käll et al., 2007). The TFFDs of bZIP17 and bZIP28 had been predicted to be on the luminal side of the ER and Golgi, respectively, but treatment of permeabilized cells with a non-specific protease revealed that the TFFDs of both TFs were accessible to protease digestion, thus residing on the cytosolic side (Gao et al., 2008). In addition to protease digestion assays, the predicted membrane topology of TMDs is sometimes experimentally validated by a self-assembling split fluorescent protein (FP) system, in which one half is targeted to one side of the TM protein and the other to the cytosol or organelle lumen (Bujalka et al., 2013). When the TF is present at the cytosolic side, the two GFP parts spontaneously assemble and a fluorescent signal is detected.
FIGURE 1.
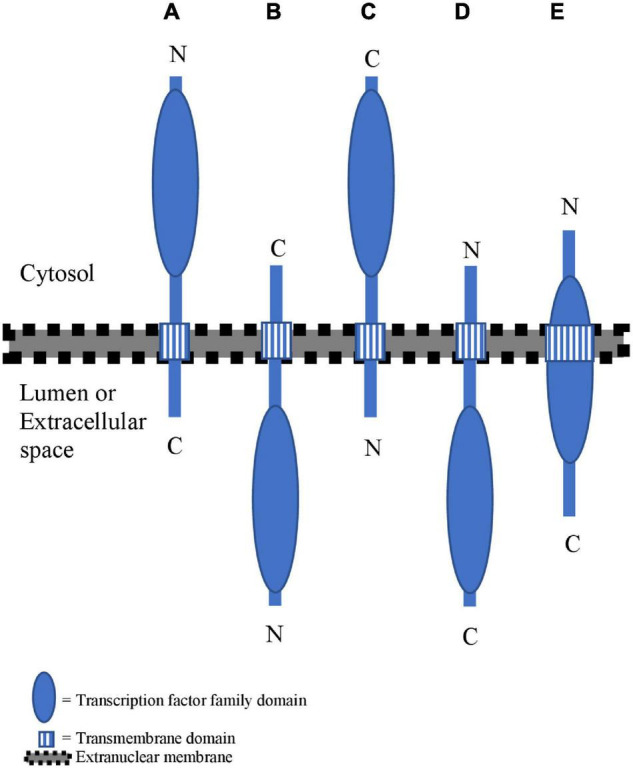
Overview of different membrane topologies for membrane-bound TFs (MB-TFs). (A) MB-TFs with the transmembrane domain (TMD) at the C-terminal side of the transcription factor family domain (TFFD) and a type-II membrane topology. (B) MB-TFs with the TMD at the C-terminal side of TFFD and a type-I membrane topology. (C) MB-TFs with the TMD at the N-terminal side of TFFD and a type-I membrane topology. (D) MB- TFs with the TMD at the N-terminal side of TFFD and a type-II membrane topology. (E) MB-TFs with the TMD overlapping with the TFFD.
The presence of a predicted TMD within the TFFD (group iii) seems illogical and if it were a true TMD, activation through alternative splicing instead of proteolytic cleavage would be the only manner to ‘activate’ these TFs. Two of the 10 atMB-TFs with a TFFD-overlapping ‘hydrophobic peptide’, ANAC050 and S1Fa1, have a transcript isoform lacking it, hinting at activation of these MB-TFs by alternative splicing (Supplementary Table 1; Yao et al., 2017). These hydrophobic peptides in the TFFD might plausibly be important for DNA binding or for proper folding of the TFs instead of for membrane anchoring (Zhou et al., 1995; Ogata et al., 1996). atMT-TFs for which the TMD overlaps with the TFFD were therefore excluded for further in silico analysis of proteolytic activation mechanisms in this review.
In silico Evidence for Proteolytic Activation of atMB-TFs
Intramembrane Protease Recognition Sites in atMB-TFs
Before the identification of specific sequences and structural features indicative of intramembrane proteolysis, we carried out a comparative analysis of the transmembrane domains among the different atMB-TFs. Previously, the TMD of substrates of intramembrane proteases from the same family had been shown to be similar in their TMD amino acid sequence (Beel and Sanders, 2008; Strisovsky et al., 2009). We assessed whether similarities between TMDs could infer proteolytic activation of atMB-TFs. Therefore, we constructed a bootstrap consensus tree for all atMB-TF TMDs and identified five clusters with bootstrap values higher than 90 (Figure 3). The TMDs of bZIP17 and bZIP28, both known to be activated through S2P cleavage, and bZIP49, predicted to be activated by S2P (Tajima et al., 2008; Iwata et al., 2017), but not bZIP60 that is regulated by alternative splicing (Nagashima et al., 2011), cluster together in our analysis, hinting at the competence to differentiate between RIP and alternative splicing regulation by means of TMD sequence similarity. Moreover, the ER membrane-bound NTL7, NTL1, and NTL3 with partially redundant functions in mitochondrial retrograde signaling have highly conserved TMDs as well (De Clercq et al., 2013; Ng et al., 2013). As mentioned, NTL7 is possibly activated by RIP (Ng et al., 2013) and interestingly, we also found the Rho-1 recognition site in NTL1 and NTL3 (see below), indicating that these TFs are probably also activated through RIP and, presumably, by (the same or different) rhomboid proteases (Figure 3). NTL4 and NTL11, which are localized to the ER as well and are, among other functions in abiotic stress responses, also involved in mitochondrial retrograde signaling (Morishita et al., 2009; Yabuta et al., 2011; Lee et al., 2012, 2014; De Clercq et al., 2013; Shih et al., 2014; Gladman et al., 2016; Van Aken et al., 2016), have a TMD dissimilar from that of NTL1/3/7 (Figure 3). Although the proteolytic activation of NTL4 and NTL11 has not been evidenced, NTL11 contains a phosphorylation site that is essential for production of a nuclear isoform, indicating activation at the posttranslational level and through proteolysis (Tang et al., 2016). NTL4 and NTL11 have no Rho-1 recognition site and are, therefore, likely cleaved by other proteases.
Next, we analyzed all atMB-TF protein sequences for previously identified recognition sequences for intramembrane proteases, because, thus far, no MB-TF is known to be released from the membranes by a soluble protease. Naturally, the release by a soluble protease cleaving the MB-TF in its cytosolic part cannot be excluded, as this activation means has been proposed for NTM1. Cleavage by a cytosolic calpain has been put forward, after immunoblot analysis had revealed that the calpain inhibitor ALLN altered the NTM1 cleavage pattern (Kim et al., 2006).
In plants, three intramembrane protease families are distinguished: intramembrane metalloproteases, rhomboid proteases, and intramembrane aspartyl proteases. Currently only one plant intramembrane protease recognition sequence has been identified, namely the S1P recognition site “RRIL”. This sequence was deduced from the consensus “Rx[LIT][KL]” (with x any amino acid) of the mammalian S1P substrates and in Arabidopsis first characterized in the bZIP28 (Denard et al., 2011; Iwata et al., 2017; Ye, 2020). In contrast to that of S1P, the S2P recognition site still awaits identification, both in mammals and in plants but is known to depend on structural elements rather than on sequence motifs in the TMD (see below). The S1P recognition sequence “RRIL” is present in four out of the 57 atMB-TFs, namely the previously reported bZIP17, bZIP28, and bZIP49 (Liu et al., 2007, 2008; Sun et al., 2015), and the additionally found hit in bZIP7 (Supplementary Table 1). Similarly as the mammalian S1P substrates that are cleaved inside the ER lumen, RRIL is present in the luminal tail of bZIP17, bZIP 28, and bZIP49, but its location (cytosolic or luminal) for bZIP7 remains unclear due to the lack of confident orientation prediction (Table 1). To discover whether other atMB-TFs also contain a potential S1P recognition site, we mapped the more degenerate consensus S1P recognition motif Rx[LIT][KL] (Denard et al., 2011). This motif was found in 38 atMB-TFs, corresponding to a 1.50-fold overrepresentation (67 vs. 45%; P value = 0.001, hypergeometric distribution) and a 1.33-fold enrichment (67% vs. 50%; P value = 0.02, hypergeometric distribution) when compared to the background Arabidopsis proteome (obtained via UniProt; Bateman et al., 2021) and all membrane-bound proteins in Arabidopsis (obtained via the Arabidopsis membrane protein library; Ward, 2001), respectively (Supplementary Table 4). As intramembrane proteases are generally expected to cleave in or close to the TMD, we restricted the search window to the TMD and the TMD region encompassing the 20-amino-acid flanking sequences. We identified Rx[LIT][KL] in seven atMB-TFs (NTL1, NTL3, NTL7, FAR1-RELATED SEQUENCES-RELATED FACTOR 3 [FRF3], CAMTA5, NIN-LIKE PROTEIN 3 [NLP3], and AT5G63280) and more specifically, in the 20-amino-acid surrounding, but not in the TMD, region. However, this approach did not result in a clear overrepresentation, based on the comparison to the 20-amino-acid region around the TMD of all membrane-bound proteins in Arabidopsis (12% versus 8%; P value = 0.3, hypergeometric distribution). These enrichment analyses indicate that this sequence is too small and degenerate and that additional sequence and/or structural features need to be taken into account to identify bona fide S1P targets. Another important criterium is the subcellular localization of the S1P cleavage, for several bZIP MB-TFs known to occur in the luminal tail. Based on the membrane topology predictions (Table 1), the Rx[LIT][KL] sequence was predicted, besides for bZIP17, bZIP28, and bZIP49, also on the luminal side for NTL3, NTL7, NPL3, and AT5G63280.
Thus far, for rhomboid proteases, only one potential recognition site has been identified in plants, of which the sequence was inferred from the Drosophila Rho-1 recognition site in the SPITZ substrate (Strisovsky et al., 2009). This sequence motif “LxxASIxxGA” includes two redundant cleavage sites: between alanine (A) and serine (S) and between glycine (G) and alanine (A). When the AS-encoding sequence is mutated, SPITZ is cleaved at the second, but less favored, cleavage site between G and A (Strisovsky et al., 2009). On the other hand, the “GA” sequence is also known as a structural feature for RIP (see below). A similar recognition sequence, “LxLSIxGA” had been identified in the TMD of Arabidopsis NTL7 (Ng et al., 2013). Although this predicted rhomboid recognition site still awaits validation in plants, pharmacological inhibition assays with the rhomboid inhibitor TPCK demonstrated attenuation of the NTL7 target gene promoter activation in response to mitochondrial stress (Ng et al., 2013). The “[LF]xLSIxGA” sequence also occurs in the closely related NTL1 and NTL3 and only in one additional protein in the Arabidopsis proteome, i.e., the tonoplast-located nitrate transporter NPF5.11 (He et al., 2017), corresponding to a 399-fold enrichment among the MB-TFs when compared to the background proteome (P value = 8e-166; hypergeometric distribution) (Supplementary Table 4). Other rhomboid recognition sequences, namely those of the bacterial proteases, AarA and GlpG/YqgP, and Drosophila Rho-1 and Human PARL, identified in the TatA, LacYTM2, Gurken/SPITZ, and PINK1, substrates, respectively, “[ILMF]xx[GAS][AHS][IMLF]” and “[ILMF]x[GAS][AHS][IMLF]”, could not be identified in or around the TMD of the atMB-TFs (Strisovsky et al., 2009; Deas et al., 2011) (Supplementary Table 4). Therefore, we searched the minimal consensus rhomboid recognition sequence conserved for both animal and bacterial rhomboids ([^WP][IMYFWLV][^WPD][^WF][AGCS][^P][FIMVACLTW]) (with ^ corresponding to all, but the amino acids indicated) (Strisovsky et al., 2009) and found it in the TMD and in the TMD region encompassing the 20-amino-acid flanking sequences of most (86 and 93%, respectively) atMB-TFs, demonstrating that this sequence is too degenerate to predict RIP in plants (Supplementary Table 4).
The third group of intramembrane proteases are the aspartyl proteases that encompass signal peptidases, presenilins, and γ-secretase. We did not carry out a sequence analysis, because no recognition sequences are well defined, the sequence conservation among substrates is minimal, and targeted substrate mutations are well tolerated. Thus, secondary and higher-order structures are more important for substrate recognition than sequence motifs (Beel and Sanders, 2008).
Structural Features for Regulated Intramembrane Proteolysis
Substrate specificity for intramembrane proteolysis is, besides a protease recognition sequence, highly dependent on structural features in the TMD. Mutations resulting in conservative amino acid changes, such as replacement of a small hydrophobic amino acid with another small hydrophobic acid, have mostly no effect on the activation, whereas major changes, such as exchange of small hydrophobic amino acids by large ones, even when far from the cleavage site, often reduce or even completely abolish the proteolytic activation (Ye et al., 2000a,b; Strisovsky et al., 2009). Moreover, inversion of the TMD, removal of helix-breaking residues, or mutation of positively charged amino acids close to the TMD also reduced or even completely abolished cleavage, as reported for S1P substrates (Ye et al., 2000a,b; Beel and Sanders, 2008; Ye, 2020). Similarly, for rhomboid and aspartyl intramembrane protease substrates, which are cleaved inside the TMD, secondary protein structures, such as broken α-helixes or the α-helix and random coil interface, have been shown to be important for cleavage by Signal Peptide Peptidases, λ-secretase, and rhomboids (Beel and Sanders, 2008). Such secondary structures are considered essential for protease accessibility and binding to the recognition sequence because they weaken the protein structure within the membrane (Strisovsky et al., 2009).
We searched the atMB-TF TMDs for helix destabilization motifs based on the presence of helix-breaking residues, i.e., “NP” (Ye et al., 2000a), “NxxP” (Ye et al., 2000b), “PxxP” (Denard et al., 2011), and “GxxN” (Ye, 2020), found to be necessary for S1P-dependent RIP events in animal models, and “GA”, required for cleavage by animal and bacterial rhomboids (Hooper and Lendeckel, 2007). As the orientation of the protease relative to the substrate is not always determined in plants, the helix destabilization motifs were examined both in forward and reverse orientation. We identified one helix destabilization motif within the TMDs of 11 out of the 57 atMB-TFs, including GEBP-like protein 2 and MAMYB (“PxxP”), the Myb TF AT2G13960 (“GxxN”), bHLH035 (“PN”), the AP2/B3-like TF AT5G25475 (“PxxN”), NTL1, NTL3, NTL7, bZIP17, bZIP49, and SIDECAR POLLEN (SCP) (“GA”) (Table 1). These motifs are equally represented in TMDs of all Arabidopsis membrane-bound proteins and none of them appears significantly overrepresented in the atMB-TFs when compared to the Arabidopsis membrane-bound proteins (Supplementary Table 5). Interestingly, the “GA” motif is present in five atMB-TFs, NTL1/3/7 and bZIP17/49, known or predicted to be regulated by RIP.
Besides helix-breaking residues, positively charged amino acids in the TMD or in the TMD-flanking region could also destabilize the TMD of RIP substrates, as previously reported for rhomboid and S2P substrates that were enriched for the positively charged amino acids arginine (R) and lysine (K) in the TMD-neighboring region (Fleig et al., 2012; Greenblatt et al., 2012; Liu et al., 2020). Moreover, R and K mutation in the Transitional ER ATPase p97 TMD decreased the cleavage efficiency by the mammalian Rhomboid-like 4 (RHBL4), whereas insertion of a patch of arginine amino acids in the TMD of non-RHBL4 substrates led to cleavage by RHBL4 (Fleig et al., 2012). Positively charged amino acids were overrepresented in the TMD of the atMB-TFs in comparison to the TMD of Arabidopsis membrane proteins (57% versus 30%; P value = 78e-5, hypergeometric distribution; Supplementary Table 5). Next, we assessed the overrepresentation of positively charged amino acids in the TMD-surrounding region of the atMB-TFs. However, as the criteria on the required number of positively charged amino acids and their distance from the TMD were not defined, we searched the atMB-TFs for overrepresentation of arginine and lysine within a 5–, 10–, 15–, and 20-amino-acid window flanking the TMD at the TFFD side, relative to the background Arabidopsis proteome and to the corresponding TMD-surrounding region for all Arabidopsis membrane proteins. Ten atMB-TFs (NTL1, NTL9, NTL2, bZIP28, bZIP17, bZIP49, AT5G63280, AT2G13960, AT2G29660, and AT5G25475) were detected with a significant (adjusted hypergeometric P value < 0.05) overrepresentation of positively charged amino acids in at least one of the tested TMD-flanking regions (Table 1 and Supplementary Table 7). The most significant results, based on the lowest hypergeometric P values, were obtained with the 15-amino-acid TMD-flanking region and included NTL1, bZIP17, bZIP28, and bZIP49, known or predicted to be regulated by RIP. The overrepresentation of positively charged amino acids observed in the TMD and in the TMD-flanking regions, respectively, was mainly due to the presence of lysine (38% versus 12%; P value = 1.79e-6, hypergeometric distribution and Supplementary Table 7, respectively).Moreover, NTL7 and NTL3, predicted to be activated through (a) rhomboid protease(s), contain at least one lysine/arginine in their TMD and bZIP17, bZIP28, and bZIP49, including bZIP17 and bZIP28 known to be cleaved by S2P, all possess one lysine in the TMD and enrichment of lysine/arginine in the TMD-flanking region, stressing the importance for RIP in plants of positively charged amino acids, and specifically, lysine in and close to the TMD.
Identification of Proteolytic N-Termini of atMB-TFs
Another strategy to identify proteolytic events is the profiling of N-terminal peptides resulting from cellular or in vivo proteolysis. We searched the atMB-TFs for in vivo proteolytic N-termini from N-terminomics experiments adopted by the PTM Viewer database (Willems et al., 2019). For more details on N-terminomics assays, we refer the reader to Kaushal and Lee (2021). A proteolytic N-terminus was identified for ANAC028 in a 41-amino-acid proximity of the TMD at the side of the TFFD (Figure 2). The N-terminal arginine of ANAC028 is a N-terminus typically to be expected from RIP, for instance from S1P cleavage. The presence of two lysine residues in its TMD further support ANAC028 as a novel candidate for RIP regulation. However, identification of N-termini that are the consequence of proteolytic MB-TF activation is not straightforward. TFs are generally low-abundant proteins and fragments caused by juxta- or intramembrane proteolysis might be difficult to detect by mass spectrometry, because they are highly hydrophobic and short. Indeed, RIP cleavage regions in and around the TMD are enriched for lysine and arginine, the preferred cleavage sites for trypsin, which is the most widely used peptide generator in mass spectrometry.
Posttranslational Modifications of atMB-TFs
Phosphorylation and glycosylation of the ER luminal part of human bZIPs CREB4 and NRF1 have been shown to be a first trigger for their proteolytic release (Stirling and O’Hare, 2006; Zhang et al., 2014). For NRF1, glycosylation is essential for its topological repartitioning across the ER membrane by the ERAD complex, whereas deglycosylation of the same amino acid in the translocated TFFD part is crucial for its subsequent proteolytic release from the ER-membrane (Zhang et al., 2014). For the plant NTL11 as well, phosphorylation by the phosphatidylinositol 4-kinase 5 is indispensable for its release and relocalization during auxin-regulated cell division (Tang et al., 2016), but the corresponding phosphorylation site and the exact function of this phosphorylation event have not been identified yet. Moreover, NTM1 was found to be stable in the presence of the proteasome inhibitor MG132, indicating it is ubiquitinated and regulated by rapid protein turnover (Kim et al., 2006). By means of publicly available mass spectrometry data from the PTM Viewer (Willems et al., 2019), posttranslational modifications (PTMs), including phosphorylation events, were searched in the atMB-TFs (Table 1). In total, 27 atMB-TFs had at least one phosphorylation site in their protein sequence and it was located in-between the TFFD and TMD for 19 of them. In addition to phosphorylation, other PTMs, such as a K-acetylation (in RLT2) and an N-glycosylation (in AT5G63280) sites were found between the TFFD and the TMD domain. However, for NTL11, none of the mass spectrometry studies revealed a phosphorylation event, indicating that its phosphorylation only occurs under specific conditions that trigger its activation.
Biological and Cellular Functions of atMB-TFs
Due to the presence of the TMD, MB-TFs remain in a dormant state, until they are either activated by specific environmental or cellular (for example, hormonal) stimuli, or their function is required to control specific plant developmental programs. The number of MB-TFs, and TFs in general, is higher in plants than that of human/animal systems. For instance, in the human proteome, only six MB-TFs have been reported in two TF families, namely bZIP and zinc finger-NF-X1 (Zupicich et al., 2001). This expansion of the (MB-)TF repertoire in plants reflects the need for a tight transcriptional control and prompt responses as a consequence of their sessile lifestyle and the lack of an adaptive immune system (Shiu et al., 2005). Functional studies of MB-TFs are often associated with subcellular dynamics analyses in response to specific intra- and extracellular stimuli, combined with reversed genetics (gain- and loss-of-function) examinations. For the 52 atMB-TFs, we carried out a systematic literature search for subcellular relocalization and/or functional studies and found that 18 atMB-TFs had a function in cellular or environmental stress responses, 11 were involved in plant development, and 6 functioned in both stress and development (Table 2). For 12 of the 22 atMB-TFs, of which the subcellular localization had been studied by fluorescence tagging, an altered localization pattern could be observed under specific conditions or upon triggers corresponding with the MB-TF function based on reverse genetics analysis (Table 1). However, four atMB-TFs, Myc106, bHLH155, SCP, and SPL7, were exclusively detected in the nucleus, implying either that only a TMD-truncated isoform had been produced, or that the TF had been posttranslationally activated under the experimental conditions.
TABLE 2.
Overview of known functions of atMB-TFs in plant development and stress responses.
| Protein name | Function of atMB-TF |
References | ||
| Development | Stress | Description | ||
| AIL6 | X | Flower development | Han and Krizek, 2016 | |
| bHLH115 | X | Fe starvation | Kurt et al., 2019 | |
| bZIP17 | X | X | Unfolded protein response, root elongation and heat stress response | Liu et al., 2008; Sun et al., 2015; Iwata et al., 2017; Gao et al., 2022 |
| bZIP28 | X | X | Unfolded protein response and root elongation | Liu et al., 2007; Sun et al., 2013, 2015; Iwata et al., 2017; Kim et al., 2018 |
| bZIP60 | X | Unfolded protein response and heat stress response | Deng et al., 2011; Nagashima et al., 2011; Singh et al., 2021 | |
| CAMTA1 | X | Drought stress response | Pandey et al., 2013 | |
| CAMTA5 | X | X | Calcium- dependent root development and drought stress response | Iqbal et al., 2022. |
| HHO5 | X | Floral meristem development | Moreau et al., 2016 | |
| LHL2 | X | Early seed development | Ohashi-Ito et al., 2013 | |
| LOL1 | X | Programmed cell death during hypersensitive response | Epple et al., 2003 | |
| MAMYB | X | Root hair development | Slabaugh, 2011; Slabaugh et al., 2011 | |
| NFXL2 | X | X | Cuticle biosynthesis and speed of the circadian clock | Johansson et al., 2011; Lisso et al., 2012 |
| NGAL2 | X | Seed size and abiotic stress response | Chen et al., 2019 | |
| NLP3 | X | Nitrogen starvation | Konishi and Yanagisawa, 2013; Tian et al., 2017 | |
| NOK | X | Petal morphogenesis and flowering | Baumann et al., 2007; Hong et al., 2021 | |
| NTL1 | X | X | Mitochondrial retrograde signaling and seed dormancy | De Clercq et al., 2013; Jurdak et al., 2021 |
| NTL3 | X | Mitochondrial retrograde signaling and dark-induced senescence | Broda et al., 2021 | |
| NTL4 | X | Proteasome activity, ROS damage, senescence and ABA-dependent programmed cell death, drought stress and heat stress | Lee et al., 2012, 2014; Shih et al., 2014,Gladman et al., 2016; | |
| NTL5 | X | ABA signaling | Li et al., 2014 | |
| NTL6 | X | Unfolded protein response | Seo et al., 2010; Yang et al., 2014a | |
| NTL7 | X | X | Mitochondrial retrograde signaling, drought stress, flooding and (mitochondrial) unfolded protein response, and (dark-induced) senescence. | Ng et al., 2013; Van Aken et al., 2016; Meng et al., 2019, 2020; Kacprzak et al., 2020; Broda et al., 2021 |
| NTL8 | X | X | Abiotic stresses-induced flowering and trichome development | Kim et al., 2007; Tian et al., 2017 |
| NTL9 | X | Calcium-dependent programmed cell death during biotic stress responses | Yoon et al., 2008; Block et al., 2014; | |
| NTL11 | X | Proteasome activity during abiotic stress | Morishita et al., 2009; Yabuta et al., 2011; Gladman et al., 2016; Tang et al., 2016 | |
| NTL13 | X | ER-stress induced programmed cell death | Yang et al., 2014b | |
| NTM1 | X | Cytokinin-mediated cell division | Kim and Park, 2007 | |
| OBP3 | X | Light signaling from phytochrome and cryptochrome | Ward et al., 2005 | |
| RLT2 | X | Phaseolin (major seed storage protein)production | Sundaram et al., 2013 | |
| SCP | X | Asymmetric cell division of the gametophyte during pollen development | Chen and McCormick, 1996; Oh et al., 2010; Kim et al., 2015 | |
| SPL1 | X | Heat-induced inflorescence | Chao et al., 2017 | |
| SPL7 | X | Cu and Fe starvation response | Bernal et al., 2012; Ramamurthy et al., 2018 | |
| SPL12 | X | Heat-induced inflorescence | Chao et al., 2017 | |
| SRS8 | X | Gynoecium development | Kuusk et al., 2006 | |
| WIP4 | X | Embryogenesis | Listiawan et al., 2015 | |
Six atMB-TFs (NTL6, NTL7, NTL13, bZIP17, bZIP28, and bZIP60) play a role during ER stress by regulating the expression of unfolded protein-responsive genes (Liu et al., 2008; Tajima et al., 2008; Nagashima et al., 2011; Yang et al., 2014a,b; Fuchs et al., 2022; Table 2). Also in rice (Oryza sativa) and maize (Zea mays), orthologs of bZIP17/28/60 were discovered in ER stress regulation (Hayashi et al., 2012; Li et al., 2012; Takahashi et al., 2012; Yang et al., 2013, 2022; Table 3). The involvement of membrane-bound bZIPs in the UPR is well studied in different eukaryotes and the activation mechanisms are largely conserved. Transcripts of the plant bZIP60, mammalian XBP1, and yeast HAC1 are alternatively spliced by IRE1 isoforms that are activated upon ER stress (for instance, tunicamycin treatment) by oligomerization and autophosphorylation. Consequently, the bZIP mRNAs are alternatively spliced, with a premature stop codon to exclude the TMD as a result (Yoshida et al., 2001; Calfon et al., 2002; Nagashima et al., 2011; Jäger et al., 2012; Diwan et al., 2021). Moreover, for the plant bZIP17 and bZIP28 and the mammalian SREBPs/ATF6, the accumulation of unfolded proteins is sensed in the luminal TF part and results in the translocation of the TFs from the ER to the Golgi, where they are released by the S2P metalloprotease (Ye et al., 2000a,b; Stirling and O’Hare, 2006; Liu et al., 2008; Tajima et al., 2008; Iwata et al., 2017). In mammalian systems, bZIP MB-TFs are activated through phosphorylation (SREBPs) or glycosylation (ATF6) of their luminal part upon unfolded protein accumulation (Hong et al., 2004; Stirling and O’Hare, 2006; Bujalka et al., 2013), but PTMs of their plant counterparts remain to be discovered.
TABLE 3.
Overview of MB-TFs functions in crop development and stress responses.
| Crop | Protein name | TF family | A. thaliana ortholog | Function | References |
| Brassica napus | BnaNAC60 | NAC | AtNTL5 | Programmed cell death and age-triggered leaf senescence | Yan et al., 2021a |
| BnaNTL1 | NAC | AtNTL7 | Leaf senescence | Yan et al., 2021b | |
| Glycine max | GmbHLHm1 | bHLH | Nodule development, NH+ transport | Chiasson et al., 2014 | |
| GmNTL1 | NAC | AtNTL1 | H2O2 sensitivity | Li et al., 2016 | |
| GmNTL1 GmNTL4 GmNTL10 | NAC | Al toxicity response | Lin et al., 2021 | ||
| Lactuca sativa | LsNAC069 | NAC | AtNTL1/3/7 | Downy mildew resistance | Meisrimler et al., 2019 |
| Nicotiana benthamiana | NbNAC089 | NAC | AtNTL14 | Virus resistance | Li et al., 2018 |
| NbNTP1 | NAC | AtNTL6 | Phytophthora resistance | McLellan et al., 2013 | |
| NbNTP2 | NAC | AtNTL1/3/7 | Phytophthora resistance | McLellan et al., 2013 | |
| Oryza sativa | OsbZIP39 | bZIP | AtbZIP28 | ER stress response | Takahashi et al., 2012 |
| OsbZIP50 | bZIP | AtbZIP60 | ER stress response | Hayashi et al., 2012 | |
| OsbZIP60 | bZIP | ER stress response and grain chalkiness | Yang et al., 2022 | ||
| OsMADS18 | AGL | Seed germination, tiller development and ABA response | Yin et al., 2019 | ||
| OsNTL3 | NAC | Thermotolerance, ER stress and unfolded protein response | Liu et al., 2020 | ||
| OsNTL5 | NAC | AtNTL4 | Flower development | Guo et al., 2018 | |
| Raphanus raphanistrum | RsNAC013 | NAC | AtNTL1 | Oxidative stress response, programmed cell death and pithiness | Hoang et al., 2022 |
| Solanum lycopersicum | SlNACMTF2 | NAC | Drought and heat stress | Bhattacharjee et al., 2017 | |
| SlNACMTF3 SlNACMTF11 | NAC | Viral infection response | Bhattacharjee et al., 2017 | ||
| SlNACMTF8 | NAC | Drought stress | Bhattacharjee et al., 2017 | ||
| SlSRN1 | NAC | AtNTL1/3/7 | Pathogen resistance | Liu et al., 2014 | |
| Solanum tuberosum | StNTP1 | NAC | AtNTL6 | Phytophthora resistance | McLellan et al., 2013 |
| StNTP2 | NAC | AtNTL1/3/7 | Phytophthora resistance | McLellan et al., 2013 | |
| Triticum aestivum | TaNAC8 | NAC | Abiotic stress response and fungal pathogen (Rust) resistance | Xia et al., 2010 | |
| TaNTL1 | NAC | ZmNTL1 OsNTL3 | Drought resistance and ABA response | Sun et al., 2022 | |
| Zea mays | ZmbZip17 | bZIP | AtbZIP17 | ER quality control and ABA signaling | Yang et al., 2013 |
| ZmbZIP53 | bZIP | Gibberellin-regulated germination and plant growth | Lv et al., 2021 | ||
| ZmbZIP60 | bZIP | AtbZIP60 | ER and heat stress response | Li et al., 2012, 2020 | |
| ZmNTL1 | NAC | H2O2 sensitivity | Wang et al., 2016 | ||
| ZmNTL2 | NAC | AtNTL4/11 | H2O2 sensitivity | Wang et al., 2016 | |
| ZmNTL5 | NAC | H2O2 sensitivity | Wang et al., 2016 |
Besides the conserved function of MB-bZIPs in UPR in eukaryotes, the plant UPR is also regulated by ER membrane-bound NAC TFs. A nuclear isoform of NTL6 was detected during ER stress, after tunicamycin treatment, as well as during different biotic and abiotic stresses and treatment with the abiotic stress hormone ABA, and resulted in the regulation of UPR, pathogenesis-regulated, and cold-responsive genes (Seo and Park, 2010; Yang et al., 2014a). However, further research is still needed on the exact activation mechanisms and it remains to be assessed whether NTL6 is activated by the accumulation of un- or misfolded proteins in the ER, resulting from excessive demands on the protein folding machinery during environmental stresses (Yang et al., 2014a). NTL13 regulates ER stress-induced programmed cell death in response to ER overreduction by the reducing agent dithiothreitol (DTT) and, in contrast to NTL6, is seemingly not directly involved in UPR, but is responsive to lipid composition changes in the plasma membrane caused by ER dysfunction (Yang et al., 2014b). Interestingly, NTL13 also controls programmed cell death in response to DTT overreduction of the chloroplasts by repressing stromal ascorbate peroxidase-encoding genes (Klein et al., 2012). Moreover, NTL7, well-characterized for its function in mitochondrial stress responses, also plays a role during DTT-induced ER stress by boosting mitochondrial respiration to enable oxidization of excess reducing equivalents from the ER (Fuchs et al., 2022). Whether these membrane NAC and bZIP TFs have distinct and/or overlapping functions in UPR and which are the precise mechanisms of ER stress sensing and their consequent activation are still not understood.
Regulation of the proteasome activity is another function that is mediated by MB-TFs. NTL4 and NTL11 control the expression of proteasome stress regulon-encoding genes, a set of genes discovered co-expressed and essential during short and long proteotoxic stresses provoked by the proteasome inhibitors MG132 and bortezomib. Moreover, their function had been shown to be essential during proteotoxic stress and during heat, drought and high light stress that also impair the plant’s ability to recycle polyubiquitinated proteins (Morishita et al., 2009; Yabuta et al., 2011; Lee et al., 2012, 2014; Shih et al., 2014; Gladman et al., 2016). Also human NRF1 functions in maintaining proteostasis by coordinating the expression of all proteasome subunit genes during proteotoxic stress and NRF1 itself is attenuated by the proteasome as its protein levels were stabilized by MG132 treatment (Sha and Goldberg, 2014). Similarly, for Arabidopsis NTL4 and NTL6, rapid protein turnover by the proteasome had been shown (Lee et al., 2012; Gladman et al., 2016).
In total, 11 atMB-TFs have, based on gain- and/or loss-of-function analyses, a proven function in the plant’s responses to environmental stresses, including heat and cold stress (NTL4, NTL6, NTL11, SPL1, SPL12, bZIP17, and bZIP60), drought and salt stress (NTL4, NTL7, NTL8 and NTL9), high light (NTL11), flooding (NTL7) and pathogen attack (LOL1, NTL1, NTL6, and NTL9) (Epple et al., 2003; Kim et al., 2007; Yoon et al., 2008; Morishita et al., 2009; Seo et al., 2010; Deng et al., 2011; Yabuta et al., 2011; Lee et al., 2012, 2014; De Clercq et al., 2013; Ng et al., 2013; Block et al., 2014; Chao et al., 2017; Meng et al., 2020; Singh et al., 2021; Gao et al., 2022; Table 2). Also in non-model species there are multiple examples of stress phenotypes from altered expression of MB-TFs, namely during heat stress (Oryza sativa [Os]NTL3 and Solanum lycopersicum NAC MEMBRANE-BOUND TRANSCRIPTION FACTOR 2 [SlNACMTF2]), drought (SlNACMTF2, SlNACMTF8, Triticum aestivum [Ta]NTL1), Al toxicity (Glycine max [Gm]NTL1/4/10) and biotic stress (Lactuca sativa LaNAC069, Nicotiana benthamiana [Nb]NAC089, NAC TARGETED BY PHYTOPHTHORA 1 [NbNTP1] and NbNTP2, SlNACMTF3, SlNACMTF11 and STRESS-RELATED NAC1 [SlSRN1], Solanum tuberosum [St]NTP1 and StNTP2, TaNAC8) (McLellan et al., 2013; Xia et al., 2010; Liu et al., 2014, 2020; Bhattacharjee et al., 2017; Li et al., 2018; Meisrimler et al., 2019; Lin et al., 2021; Sun et al., 2022; Table 3). Pre-existing dormant TFs provide an efficient way of gene regulation and enable prompt responses to environmental changes that are necessary for stress adaptation and survival. As several MB-TFs have been reported to mediate reactions to multiple stresses, not surprisingly, several MB-TFs (NTL1, NTL3, NTL4, NTL7, and NTL11) mediate responses to increased cellular ROS production, a common factor under various abiotic and biotic stress conditions (Table 2; Morishita et al., 2009; Lee et al., 2012; De Clercq et al., 2013; Ng et al., 2013). ROS are not just toxic molecules, but also act as secondary messengers under various stress conditions, similarly to calcium. NTL9 is regulated by calcium through binding to calmodulin and plays a role in the regulation of calcium-dependent programmed cell death during effector-triggered immunity (ETI) in addition to its role in osmotic stress-induced leaf senescence (Yoon et al., 2008; Block et al., 2014). Its function during the ETI-induced hypersensitive response is impaired by the Pseudomonas syringae pathogenicity-dependent outer protein D1 (HopD1) effector, a strong repressor of ETI. Interaction of HopD1 with NTL9 inhibits the calcium-induced translocation of NTL9 from the ER to the nucleus and the ETI response induction (Block et al., 2014). This finding, i.e., that the NTL9 regulation is affected at the posttranslational level by binding an effector protein, indicates that its activation is most probably regulated posttranslationally through proteolysis. Similarly to the Arabidopsis NTL9, the lettuce LsNAC069, an ortholog of Arabidopsis NTL1, NTL3, and NTL7, had been shown to be targeted by effectors (i.e., from downy mildew Bremia lactucae) that abolished its relocalization to the nucleus (Meisrimler et al., 2019). NTL1, NTL3, and NTL7 have mainly been studied for their function in mitochondrial retrograde signaling, in which stressed or dysfunctional mitochondrial status (for instance, by treatment with the mitochondrial complex III inhibitor antimycin A) is communicated to the nucleus to induce adaptation responses for the maintenance of the mitochondrial as well as the whole cellular homeostasis (De Clercq et al., 2013; Ng et al., 2013; Van Aken et al., 2016; Broda et al., 2021). Since their discovery as regulators of mitochondria-to-nucleus communication pathways, NTL1, NTL3, and NTL7 were studied in the context of various cellular and environmental stress responses, including the mitochondrial and ER unfolded protein responses (Kacprzak et al., 2020; Fuchs et al., 2022), drought (Van Aken et al., 2016), and flooding stress (Meng et al., 2020). NTL7 is constitutively produced and acts as a ‘master switch’ that regulates the expression of NTL1, NTL3 and NTL4 as downstream targets (Van Aken et al., 2016; Broda et al., 2021). Mutant analyses have revealed that the fine-tuned expression of these NTLs is necessary to sustain a normal development, because misregulation of their expression resulted in deficient seed dormancy breaking (NTL3) (Jurdak et al., 2021), accelerated dark-induced senescence (NTL3 and NTL7) (Meng et al., 2019; Broda et al., 2021), and growth retardation and altered leaf development due to decreased cell size and viability (NTL7) (Meng et al., 2019).
Among the MB-TFs with a described function in plant development, the majority plays a role in the control of developmental stages from flower development to seed generation (SCP, bHLH155, WOUND-INDUCED POLYPEPTIDE 4, AINTEGUMENTA-LIKE 6, HRS1 HOMOLOG, NTL4, NTL8, NTL11, SHI-RELATED SEQUENCE 8, RNASE THREE-LIKE PROTEIN 2, and NF-X LIKE 2), and in root hair development or in root elongation (MAMYB, bZIP17, bZIP28, and FRF3) (Chen and McCormick, 1996; Kuusk et al., 2006; Kim et al., 2007; Morishita et al., 2009; Oh et al., 2010; Johansson et al., 2011; Lisso et al., 2012; Sundaram et al., 2013; Shih et al., 2014; Kim et al., 2015; Listiawan et al., 2015; Han and Krizek, 2016; Moreau et al., 2016; Tian et al., 2017; Kurt et al., 2019; Table 2). Also in non-model organisms several MB-TFs are involved in development, including flower development (OsNTL5), seed germination (OsMINICHROMOSOME MAINTENANCE 1/AGAMOUS/DEFICIENS/SERUM RESPONSE FACTOR 18 and ZmbZIP53), aging (BnaNAC60 and BnaNTL1) and nodule formation GmbHLH membrane 1) (Chiasson et al., 2014; Guo et al., 2018; Yin et al., 2019; Lv et al., 2021; Yan et al., 2021a,b; Table 3). The ER-to-nucleus mobilization of MAMYB specifically takes place in root epidermal cells adjacent to the sites of lateral root initiation and loss-of-function mutation results in absence of lateral roots (Slabaugh et al., 2011). Another well-studied MB-TF is SCP that plays a role in the regulation of cell division during pollen development. SCP has been detected in the nucleus, specifically during early and polarized microspore stages, and functions in the control of asymmetric cell division of the gametophyte during pollen development (Chen and McCormick, 1996; Oh et al., 2010; Kim et al., 2015). Although these TFs seem to be activated in specific cell types, in which their activity is required to mediate developmental programming, their specific activation mechanism in a cellular and developmental stage specific manner, has not been elucidated yet. Developmental programming is known to be also controlled by environmental stimuli. Flower development and seed germination are triggered by changes in photoperiod and temperature (Baskin and Baskin, 2004; Song et al., 2013), whereas lateral root growth that is regulated by hormones, such as auxin, is also responsive to environmental stimuli, such as nutrient deficiency and soil water content (Banda et al., 2019). For instance, NTL4 and NTL11 regulate jasmonic acid (JA)- and ABA-dependent and high-light-induced florescence, respectively (Yabuta et al., 2011; Shih et al., 2014; Tang et al., 2016). However, how various intra-, inter- and extracellular signals contribute to and are intersected for the MB-TF activation is not understood.
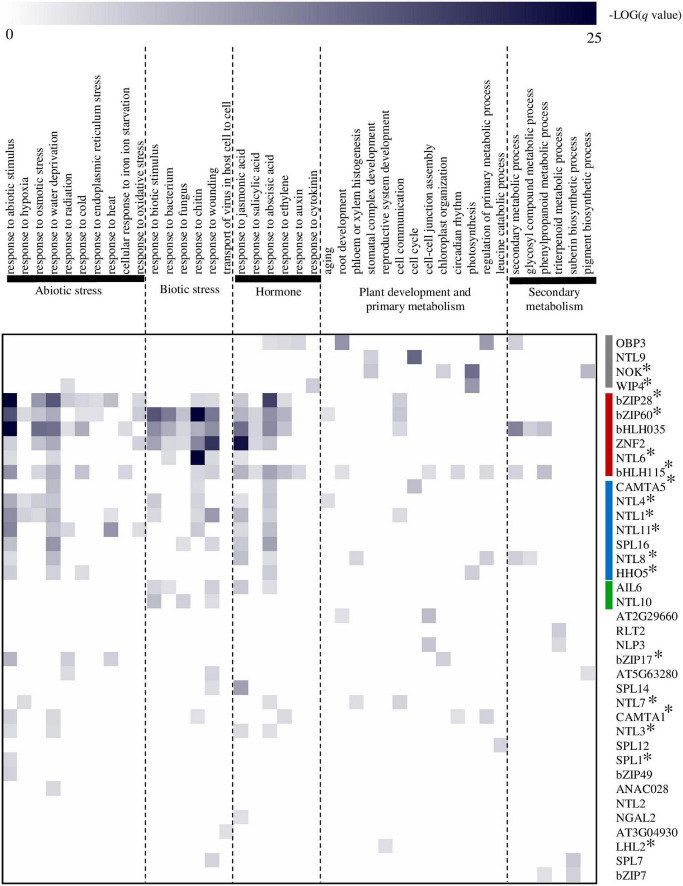
To discover novel functions for yet uncharacterized atMB-TFs, we used a transcriptional regulatory network approach for the identification of TF functions based on their target genes (De Clercq et al., 2021). This method has a high predictive power to correctly infer functions for both functionally characterized and novel TFs involved in various biological processes. For 38 atMB-TFs, at least one enriched gene ontology (GO) biological process (BP) term was obtained, with in total 291 GO-BP terms used for hierarchical clustering of the MB-TFs according to their predicted function(s) (De Clercq et al., 2021; Figure 4 and Supplementary Table 3). We could distinguish atMB-TFs that regulate (i) a broad range of abiotic and biotic stress responses, (ii) mainly responses to water deficit and ABA, (iii) mostly biotic stress responses, and (iv) specific cellular and/or developmental processes (Figure 4). This network-based identification could assign one or more of the known functions for 18 of the 35 functionally characterized atMB-TFs (Table 2), whereas for the remaining part, no enriched GO terms were found (9/35) or novel functions were assigned (9/35). MB-TFs involved in the same or similar biological processes cluster together in our meta-analysis and are, among other functions, enriched for unfolded protein response (i.e., bZIP28 and bZIP60; group i) and water deprivation functions (i.e., NTL4, NTL8, and HHO that control drought stress-triggered flowering; group ii) (Kim et al., 2007; Shih et al., 2014; Moreau et al., 2016). For 12 out of the 17 atMB-TFs without known functions, novel roles could be predicted. These yet uncharacterized atMB-TFs are expected to function in stress responses, namely bHLH035 and SPL16 to be involved in a broad range of biotic and abiotic stresses, bZIP49 and NTL2 in abiotic stress responses, and NTL10 and ZFN2 in biotic stress responses. Furthermore, specific functions were assigned to the other atMB-TFs: water deprivation (ANAC028), cell-to-cell transport of viruses (AT3G04930), JA responses (SPL14), regulation of root development and cell junctions (AT2G29960), regulation of pigment biosynthesis (AT5G63280), and regulation of the phenylpropanoid pathway and suberin biosynthesis (bZIP7). However, further experimental studies are needed to validate these functional predictions.
FIGURE 4.
Prediction of atMB-TF functions by means of a regulatory network-based approach. Functional terms were obtained per TF by Gene Ontology (GO) Biological Process (BP) enrichment analysis of their respective target genes (De Clercq et al., 2021). MB-TFs were hierarchically clustered (average linkage) based on their functional categories using Genesis software version 1.6 (Sturn et al., 2002). atMB-TFs that regulate a broad range of abiotic and biotic stress responses are in indicated in red; those involved mainly in responses to water deficit and ABA, in blue; mostly involved in biotic stress responses, in green; and those in specific cellular and/or developmental processes, in gray. A selection of GO-BP terms are displayed for the heatmap presentation. For the complete list of enriched GO-BP terms per atMB-TF, see Supplementary Table 3. *Indicates atMB-TFs with correctly predicted function(s).
Conclusion and Further Perspectives
MB-TFs play an important role in the regulation of various cellular processes and unraveling their mode of action provides important insights into the molecular mechanisms of how plants sense and coordinate intra- and intercellular and environmental signals into appropriate responses. However, our knowledge on the activation mechanisms of MB-TFs in plants is limited to that of bZIP17 and bZIP28, and bZIP60 that are regulated through RIP and alternative spicing, respectively, during the unfolded protein response (Liu et al., 2008; Tajima et al., 2008; Nagashima et al., 2011). Both activation mechanisms are highly conserved in multicellular life (Sun et al., 2015; Diwan et al., 2021). Experimental indication for proteolytic activation of MB-TFs is often based on N-terminomics or pharmacological methods with certain protease inhibitors. Both approaches have their limitations, because peptides derived from RIP are often difficult to detect by mass spectrometry and pharmacological systems are restricted to currently available bacterial and mammalian small molecule inhibitors, of which action mode in plants is often not well known. Moreover, chemical inhibitor studies rely on previous knowledge of a readout (specific trigger and timing) of the MB-TFs activity. In plants, this approach has been applied only to NTL7 and NTM1, by means of a broad-spectrum serine/rhomboid and calpain protease inhibitor, respectively (Kim et al., 2006; Ng et al., 2013). Furthermore, in silico-based prediction of proteolytic activation is unsatisfactory because of the short and degenerate nature of known protease recognition sequences. Structural features within or adjacent to the TMD, such as helix-breaking motifs and positively charged amino acids, rather than sequence motifs are seemingly more relevant to predict cleavage events (Ye, 2020). Here, we identified 11 and 30 atMB-TFs with at least one helix-breaking motif and positively charged amino acid inside the TMD, respectively, among which MAMYB, SCP, NTL3, NTL7, bZIP17, bZIP49, bHLH035, and AT5G25475 have both features, hinting at activation through RIP.
For the subsequent identification of the responsible protease, a forward genetics approach is not straightforward, because the protease activity readout is often indirect and based on the expression of downstream target genes. In the case of bZIP28, the downstream target gene analysis in the s1p mutant identified S1P as the responsible protease, although its cleavage was not affected in this mutant (Iwata et al., 2017). A genetics approach is also hampered by potential redundancy between proteases that can cleave one MB-TF (Iwata et al., 2017). Therefore, a CRISPR-Cas library screen targeting multiple proteases simultaneously, in random combinations, may be a useful alternative (Callies et al., 2019). A more direct method for the detection of the responsible protease is chemical genomics using a chemical protease inhibitor as a bait. State-of-the-art techniques to discover protein targets of small molecules are Target Identification by Chromatographic Co-Elution (TICC) (Chan et al., 2012), Drug Affinity Responsive Target Stability (DARTS) (Lomenick et al., 2011), and Activity-Based Protein Profiling (ABPP) (Sieber et al., 2004), but require specific research resources and expertise. Traditional affinity purification experiments using the MB-TF substrate as a bait are rather unlikely to pull down the responsible protease, because the protease–substrate interactions are weak and transient and, thus, will probably get lost during the purification steps. On the contrary, proximity-dependent labeling techniques circumvent this problem by tagging all interacting/neighboring proteins before pull-down and purification and they are also ideal to detect protein interactions in a membranous environment (Arora et al., 2020). A combination of N-terminomics, chemical biology, and proximity labeling-based interactomics is promising to elucidate the activation mechanism of plant MB-TFs.
Manipulation of the MB-TFs levels by gene knockout or overexpression in Arabidopsis and non-model species has been shown to impact the plant’s tolerance or resistance to stresses, but as a drawback, often perturbs growth and development, making this approach unfavorable for agricultural applications. In contrast, expression of a constitutively active isoform of the endogenous MB-TF, for example, by CRISPR-Cas-mediated gene editing of the TMD, provides a valuable alternative. This was shown for Arabidopsis NTL7 for which overexpression of the full length protein resulted in growth and developmental retardation in addition to increased oxidative stress tolerance, whereas exclusion of the TMD from the endogenously expressed transcript induced stress tolerance without affecting the growth and development (Ng et al., 2013; Meng et al., 2019; Broda et al., 2021). Moreover, understanding of the cleavage mechanisms offers possibilities to fine tune the proteolytic activation by the native or by alternative proteases. For example, mutation of the cleavage site or addition of positively charged amino acids in or around the TMD was shown to promote cleavage or enable cleavage by alternative proteases in mammals (Beel and Sanders, 2008; Fleig et al., 2012; Greenblatt et al., 2012; Liu et al., 2020; Silber et al., 2020; Spitz et al., 2020; Ye, 2020). Different MB-TFs have already been studied in agricultural important crops showing their involvement in responses to drought stress and infection by viral and fungal pathogens and in various developmental traits such as flowering time and seed germination (Xia et al., 2010; Liu et al., 2014; Bhattacharjee et al., 2017; Li et al., 2018; Guo et al., 2018; Yin et al., 2019; Table 3). A thorough understanding of their activation mechanisms will offer perspectives to fine tune their activity with respect to applications for crop improvement.
Author Contributions
IDC conceived this review manuscript. JDB summarized and analyzed the literature and performed the in silico analyses. IDC and FVB contributed to the discussion and writing of the manuscript. JDB and IDC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Steven Verhelst for discussions about the RIP-predicting in silico analyses and Martine De Cock for help in preparing the manuscript.
Funding
This work was supported by the Research Foundation-Flanders-Fonds de la Recherche Scientifique (Excellence of Science project no. 30829584) to FVB and the European Research Council (ERC) under the Horizon 2020 research and Innovation Program (Grant agreement No. 949808) to IDC. JDB is indebted to the Research Foundation-Flanders for a predoctoral fellowship in Fundamental Research (1126821N).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.927746/full#supplementary-material
References
- Arora D., Abel N. B., Liu C., Van Damme P., Yperman K., Eeckhout D., et al. (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32 3388–3407. 10.1105/tpc.20.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda J., Bellande K., von Wangenheim D., Goh T., Guyomarc’h S., Laplaze L., et al. (2019). Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci. 24 826–839. 10.1016/j.tplants.2019.06.015 [DOI] [PubMed] [Google Scholar]
- Baskin J. M., Baskin C. C. (2004). A classification system for seed dormancy. Seed Sci. Res. 14 1–16. 10.1079/SSR2003150 [DOI] [Google Scholar]
- Bateman A., Martin M. J., Orchard S., Magrane M., Agivetova R., Ahmad S., et al. (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49 D480–D489. 10.1093/NAR/GKAA1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K., Perez-Rodriguez M., Bradley D., Venail J., Bailey P., Jin H., et al. (2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134 1691–1701. 10.1242/DEV.02836 [DOI] [PubMed] [Google Scholar]
- Beel A. J., Sanders C. R. (2008). Substrate specificity of γ-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 65 1311–1334. 10.1007/S00018-008-7462-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M., Casero D., Singh V., Wilson G. T., Grande A., Yang H., et al. (2012). Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24 738–761. 10.1105/TPC.111.090431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee P., Das R., Mandal A., Kundu P. (2017). Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol. Biol. 93 511–532. 10.1007/S11103-016-0579-Z [DOI] [PubMed] [Google Scholar]
- Block A., Toruño T. Y., Elowsky C. G., Zhang C., Steinbrenner J., Beynon J., et al. (2014). The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 201 1358–1370. 10.1111/nph.12626 [DOI] [PubMed] [Google Scholar]
- Broda M., Khan K., O’Leary B., Pružinská A., Lee C. P., Millar A. H., et al. (2021). Increased expression of ANAC017 primes for accelerated senescence. Plant Physiol. 186 2205–2221. 10.1093/PLPHYS/KIAB195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalka H., Koenning M., Jackson S., Perreau V. M., Pope B., Hay C. M., et al. (2013). MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 11:e1001625. 10.1371/JOURNAL.PBIO.1001625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrão F., Santos C. C., Le Thomas A., Marsters S., Ashkenazi A., Domingos P. M. (2022). Pumilio protects Xbp1 mRNA from regulated Ire1-dependent decay. Nat. Commun. 13:1587. 10.1038/s41467-022-29105-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., et al. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415 92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Callies L. K., Tadeo D., Simper J., Bugge T. H., Szabo R. (2019). Iterative, multiplexed CRISPR-mediated gene editing for functional analysis of complex protease gene clusters. J. Biol. Chem. 294 15987–15996. 10.1074/jbc.RA119.009773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. N. Y., Vuckovic D., Sleno L., Olsen J. B., Pogoutse O., Havugimana P., et al. (2012). Target identification by chromatographic co-elution: monitoring of drug-protein interactions without immobilization or chemical derivatization. Mol. Cell. Proteomics 11 M111.016642. 10.1074/mcp.M111.016642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L.-M., Liu Y.-Q., Chen D.-Y., Xue X.-Y., Mao Y.-B., Chen X.-Y. (2017). Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 10 735–748. 10.1016/J.MOLP.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang N., Zhang Q., Zhou G., Tian H., Hussain S., et al. (2019). Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 20:2695. 10.3390/IJMS20112695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Ho M., Briere J., Moscvin M., Czarnecki P. G., Anderson K. C., et al. (2022). Multiple myeloma cells depend on the DDI2/NRF1-mediated proteasome stress response for survival. Blood Adv. 6 429–440. 10.1182/bloodadvances.2020003820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., McCormick S. (1996). sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122 3243–3253. 10.1242/dev.122.10.3243 [DOI] [PubMed] [Google Scholar]
- Chiasson D. M., Loughlin P. C., Mazurkiewicz D., Mohammadidehcheshmeh M., Fedorova E. E., Okamoto M., et al. (2014). Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc.Natl. Acad. Sci. U S A 111 4814–4819. 10.1073/PNAS.1312801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I., Van de Velde J., Luo X., Liu L., Storme V., Van Bel M., et al. (2021). Integrative inference of transcriptional networks in Arabidopsis yields novel ROS signalling regulators. Nat. Plants 7 500–513. 10.1038/s41477-021-00894-1 [DOI] [PubMed] [Google Scholar]
- De Clercq I., Vermeirssen V., Van Aken O., Vandepoele K., Murcha M. W., Law S. R., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25 3472–3490. 10.1105/TPC.113.117168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S. H. Y., et al. (2011). PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20 867–867. 10.1093/HMG/DDQ526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denard B., Seemann J., Chen Q., Gay A., Huang H., Chen Y., et al. (2011). The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host Microbe. 10 65–74. 10.1016/J.CHOM.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U S A 108 7247–7252. 10.1073/PNAS.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan D., Liu X., Andrews C. F., Pajerowska-Mukhtar K. M. (2021). A quantitative Arabidopsis IRE1a ribonuclease-dependent in vitro mRNA cleavage assay for functional studies of substrate splicing and decay activities. Front. Plant Sci. 12:707378. 10.3389/FPLS.2021.707378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M., Zhang R., Zhu F., Zhang Z., Gou L., Wen J., et al. (2017). A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29 1748–1772. 10.1105/tpc.17.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B., Sammer M., Lua W. H., Benetka W., Liew L. L., Yu W., et al. (2011). Nuclear import of a lipid-modified transcription factor: mobilization of NFAT5 isoform a by osmotic stress. Cell Cycle 10 3897–3911. 10.4161/cc.10.22.18043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P., Mack A. A., Morris V. R. F., Dangl J. L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A 100 6831–6836. 10.1073/pnas.1130421100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig L., Bergbold N., Sahasrabudhe P., Geiger B., Kaltak L., Lemberg M. K. (2012). Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol. Cell 47 558–569. 10.1016/j.molcel.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Fuchs P., Bohle F., Lichtenauer S., Ugalde J. M., Araujo E. F., Mansuroglu B., et al. (2022). Reductive stress triggers ANAC017-mediated retrograde signaling to safeguard the endoplasmic reticulum by boosting mitochondrial respiratory capacity. Plant Cell 34 1375–1395. in press, 10.1093/PLCELL/KOAC017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Brandizzi F., Benning C., Larkin R. M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A 105 16398–16403. 10.1073/pnas.0808463105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang M.-J., Wang J.-J., Lu H.-P., Liu J.-X. (2022). bZIP17 regulates heat stress tolerance at reproductive stage in Arabidopsis. aBIOTECH 3 1–11. 10.1007/S42994-021-00062-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N. P., Marshall R. S., Lee K.-H., Vierstra R. D. (2016). The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 28 1279–1296. 10.1105/TPC.15.01022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt E. J., Olzmann J. A., Kopito R. R. (2012). Making the cut: intramembrane cleavage by a rhomboid protease promotes ERAD. Nat. Struct. Mol. Biol. 19 979–981. 10.1038/NSMB.2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Dai S., Singh P. K., Wang H., Wang Y., Tan J. L. H., et al. (2018). A membrane-bound NAC-like transcription factor OsNTL5 represses the flowering in Oryza sativa. Front. Plant Sci. 9:555. 10.3389/FPLS.2018.00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Krizek B. A. (2016). AINTEGUMENTA-LIKE6 can functionally replace AINTEGUMENTA but alters Arabidopsis flower development when misexpressed at high levels. Plant Mol. Biol. 92 597–612. 10.1007/S11103-016-0535-Y [DOI] [PubMed] [Google Scholar]
- Hayashi S., Wakasa Y., Takahashi H., Kawakatsu T., Takaiwa F. (2012). Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69 946–956. 10.1111/J.1365-313X.2011.04844.X [DOI] [PubMed] [Google Scholar]
- He Y.-N., Peng J.-S., Cai Y., Liu D.-F., Guan Y., Yi H.-Y., et al. (2017). Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 7:6417. 10.1038/s41598-017-06744-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang N. V., Park S., Park C., Suh H., Kim S. T., Chae E., et al. (2022). Oxidative stress response and programmed cell death guided by NAC013 modulate pithiness in radish taproots. Plant J. 109 144–163. 10.1111/TPJ.15561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Niu F., Lin Y., Wang S., Chen L., Jiang L. (2021). MYB106 is a negative regulator and a substrate for CRL3BPM E3 ligase in regulating flowering time in Arabidopsis thaliana. J. Integr. Plant Biol. 63 1104–1119. 10.1111/JIPB.13071 [DOI] [PubMed] [Google Scholar]
- Hong M., Luo S., Baumeister P., Huang J.-M., Gogia R. K., Li M., et al. (2004). Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J. Biol. Chem. 279 11354–11363. 10.1074/JBC.M309804200 [DOI] [PubMed] [Google Scholar]
- Hooper C. M., Castleden I. R., Tanz S. K., Aryamanesh N., Millar A. H. (2017). SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45 D1064–D1074. 10.1093/nar/gkw1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper N. M., Lendeckel U. (2007). Intramembrane-Cleaving Proteases (I-CLiPs). Dordrecht: Springer. [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. (2000). Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102 577–586. 10.1016/S0092-8674(00)00080-5 [DOI] [PubMed] [Google Scholar]
- Iqbal S., Wang X., Mubeen I., Kamran M., Kanwal I., Díaz G. A., et al. (2022). Phytohormones trigger drought tolerance in crop plants: outlook and future perspectives. Front. Plant Sci. 12:799318. 10.3389/fpls.2021.799318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Ashida M., Hasegawa C., Tabara K., Mishiba K.-I., Koizumi N. (2017). Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J. 91 408–415. 10.1111/tpj.13572 [DOI] [PubMed] [Google Scholar]
- Jäger R., Bertrand M. J. M., Gorman A. M., Vandenabeele P., Samali A. (2012). The unfolded protein response at the crossroads of cellular life and death during endoplasmic reticulum stress. Biol. Cell 104 259–270. 10.1111/BOC.201100055 [DOI] [PubMed] [Google Scholar]
- Jan C. H., Williams C. C., Weissman J. S. (2014). Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346:1257521. 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., McWatters H. G., Bakó L., Takata N., Gyula P., Hall A., et al. (2011). Partners in time: early bird associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol. 155 2108–2122. 10.1104/PP.110.167155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurdak R., Launay-Avon A., Paysant-Le Roux C., Bailly C. (2021). Retrograde signalling from the mitochondria to the nucleus translates the positive effect of ethylene on dormancy breaking of Arabidopsis thaliana seeds. New Phytol. 229 2192–2205. 10.1111/NPH.16985 [DOI] [PubMed] [Google Scholar]
- Kacprzak S. M., Dahlqvist A., Van Aken O. (2020). The transcription factor ANAC017 is a key regulator of mitochondrial proteotoxic stress responses in plants. Philos. Trans. R. Soc. B. 375:20190411. 10.1098/rstb.2019.0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35 W429–W432. 10.1093/nar/gkm256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal P., Lee C. (2021). N-terminomics – its past and recent advancements. J. Proteomics 233:104089. 10.1016/j.jprot.2020.104089 [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Yamaguchi-Shinozaki K., Shinozaki K. (2018). ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176 2221–2230. 10.1104/PP.17.01414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Kim M., Lee M. R., Park S. K., Kim J. (2015). Lateral organ boundaries domain (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 81 794–809. 10.1111/tpj.12767 [DOI] [PubMed] [Google Scholar]
- Kim S.-G., Kim S.-Y., Park C.-M. (2007). A membrane-associated NAC transcription factor regulates salt-responsive flowering via flowering locus T in Arabidopsis. Planta 226 647–654. 10.1007/s00425-007-0513-3 [DOI] [PubMed] [Google Scholar]
- Kim S.-G., Lee S., Seo P. J., Kim S.-K., Kim J.-K., Park C.-M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95 56–65. 10.1016/J.YGENO.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Kim Y.-S., Kim S.-G., Park J.-E., Park H.-Y., Lim M.-H., Chua N.-H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18 3132–3144. 10.1105/TPC.106.043018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-S., Park C.-M. (2007). Membrane regulation of cytokinin-mediated cell division in Arabidopsis. Plant Signal. Behav. 2 15–16. 10.4161/PSB.2.1.3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Seidel T., Stöcker B., Dietz K.-J. (2012). The membrane-tethered transcription factor ANAC089 serves as redox-dependent suppressor of stromal ascorbate peroxidase gene expression. Front. Plant Sci. 3:247. 10.3389/FPLS.2012.00247/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4:2621. 10.1038/ncomms2621 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305 567–580. 10.1006/JMBI.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kurt F., Filiz E., Kurt B. (2019). A key gene bHLH115 in iron homeostasis: comprehensive bioinformatics analyses in Arabidopsis, tomato, rice, and maize. Biometals 32 641–656. 10.1007/S10534-019-00199-Z [DOI] [PubMed] [Google Scholar]
- Kuusk S., Sohlberg J. J., Eklund D. M., Sundberg E. (2006). Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 47 99–111. 10.1111/J.1365-313X.2006.02774.X [DOI] [PubMed] [Google Scholar]
- Lee S., Lee H.-J., Huh S. U., Paek K.-H., Ha J.-H., Park C.-M. (2014). The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 227 76–83. 10.1016/J.PLANTSCI.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Lee S., Seo P. J., Lee H.-J., Park C.-M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70 831–844. 10.1111/J.1365-313X.2012.04932.X [DOI] [PubMed] [Google Scholar]
- Li F. F., Sun H. J., Jiao Y. B., Wang F. L., Yang J. G., Shen L. L. (2018). Viral infection-induced endoplasmic reticulum stress and a membrane-associated transcription factor NbNAC089 are involved in resistance to virus in Nicotiana benthamiana. Plant Pathol. 67 233–243. 10.1111/PPA.12707 [DOI] [Google Scholar]
- Li P., Zhou H., Shi X., Yu B., Zhou Y., Chen S., et al. (2014). The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 10:e1004213. 10.1371/journal.pgen.1004213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang N., Ji D., Xue Z., Yu Y., Jiang Y., et al. (2016). Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol. 172 1804–1820. 10.1104/PP.16.01132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Humbert S., Howell S. H. (2012). ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes 5:1–11. 10.1186/1756-0500-5-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tang J., Srivastava R., Bassham D. C., Howell S. H. (2020). The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 32 3559–3575. 10.1105/TPC.20.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Li H., Zhou F., Li H., Liu J., Hao Y., et al. (2015). Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16 1062–1074. 10.1111/TRA.12311 [DOI] [PubMed] [Google Scholar]
- Lin J.-R., Mondal A. M., Liu R., Hu J. (2012). Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinformatics 13:157. 10.1186/1471-2105-13-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Liu G., Xue Y., Guo X., Luo J., Pan Y., et al. (2021). Functional characterization of aluminum (Al)-responsive membrane-bound NAC transcription factors in soybean roots. Int. J. Mol. Sci. 22:12854. 10.3390/IJMS222312854/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J., Schröder F., Schippers J. H. M., Müssig C. (2012). NFXL2 modifies cuticle properties in Arabidopsis. Plant Signal. Behav. 7 551–555. 10.4161/PSB.19838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listiawan D. A., Tanoue R., Kobayashi K., Masuda T. (2015). Expression analysis of transcription factors involved in chloroplast differentiation. Procedia Chem. 14 146–151. 10.1016/J.PROCHE.2015.03.021 [DOI] [Google Scholar]
- Liu B., Ouyang Z., Zhang Y., Li X., Hong Y., Huang L., et al. (2014). Tomato NAC transcription factor slsrn1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS One 9:e102067. 10.1371/JOURNAL.PONE.0102067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Beaton S. E., Grieve A. G., Evans R., Rogers M., Strisovsky K., et al. (2020). Bacterial rhomboid proteases mediate quality control of orphan membrane proteins. EMBO J. 39:e102922. 10.15252/EMBJ.2019102922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-X., Srivastava R., Howell S. H. (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31 1735–1743. 10.1111/j.1365-3040.2008.01873.x [DOI] [PubMed] [Google Scholar]
- Liu J.-X., Srivastava R., Che P., Howell S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 4111–4119. 10.1105/TPC.106.050021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. H., Lyu Y. S., Yang W., Yang Z. T., Lu S. J., Liu J. X. (2020). A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 18 1317–1329. 10.1111/PBI.13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li P., Fan L., Wu M. (2018). The nuclear transportation routes of membrane-bound transcription factors. Cell Commun. Signal. 16:12. 10.1186/s12964-018-0224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., Jung G., Wohlschlegel J. A., Huang J. (2011). Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3 163–180. 10.1002/9780470559277.ch110180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Li X., Li H., Hu Y., Liu H., Wen S., et al. (2021). Gibberellin induced transcription factor bZIP53 regulates CesA1 expression in maize kernels. PLoS One 16:e0244591. 10.1371/JOURNAL.PONE.0244591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H., Boevink P. C., Armstrong M. R., Pritchard L., Gomez S., Morales J., et al. (2013). An RxLR effector from phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathogens 9:e1003670. 10.1371/JOURNAL.PPAT.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisrimler C.-N., Pelgrom A. J. E., Oud B., Out S., Van den Ackerveken G. (2019). Multiple downy mildew effectors target the stress-related NAC transcription factor LsNAC069 in lettuce. Plant J. 99 1098–1115. 10.1111/tpj.14383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Li L., De Clercq I., Narsai R., Xu Y., Hartmann A., et al. (2019). ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 180 634–653. 10.1104/PP.18.01603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Li L., Narsai R., De Clercq I., Whelan J., Berkowitz O. (2020). Mitochondrial signalling is critical for acclimation and adaptation to flooding in Arabidopsis thaliana. Plant J. 103 227–247. 10.1111/tpj.14724 [DOI] [PubMed] [Google Scholar]
- Milan M., Balestrieri C., Alfarano G., Polletti S., Prosperini E., Nicoli P., et al. (2020). Pancreatic cancer cells require the transcription factor MYRF to maintain ER homeostasis. Dev. Cell 55 398–412. 10.1016/j.devcel.2020.09.011 [DOI] [PubMed] [Google Scholar]
- Moreau F., Thévenon E., Blanvillain R., Lopez-Vidriero I., Franco-Zorrilla J. M., Dumas R., et al. (2016). The Myb-domain protein ultrapetala1 interacting factor 1 controls floral meristem activities in Arabidopsis. Development 143 1108–1119. 10.1242/DEV.127365 [DOI] [PubMed] [Google Scholar]
- Morishita T., Kojima Y., Maruta T., Nishizawa-Yokoi A., Yabuta Y., Shigeoka S. (2009). Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50 2210–2222. 10.1093/PCP/PCP159 [DOI] [PubMed] [Google Scholar]
- Nagashima Y., Mishiba K.-i, Suzuki E., Shimada Y., Iwata Y., Koizumi N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1:29. 10.1038/srep00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Ivanova A., Duncan O., Law S. R., Van Aken O., De Clercq I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25 3450–3471. 10.1105/tpc.113.113985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K., Kanei-Ishii C., Sasaki M., Hatanaka H., Nagadoi A., Enari M., et al. (1996). The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 3 178–187. 10.1038/nsb0296-178 [DOI] [PubMed] [Google Scholar]
- Oh S. A., Park K. S., Twell D., Park S. K. (2010). The sidecar pollen gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 64 839–850. 10.1111/J.1365-313X.2010.04374.X [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Matsukawa M., Fukuda H. (2013). An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant Cell Physiol. 54 398–405. 10.1093/PCP/PCT013 [DOI] [PubMed] [Google Scholar]
- Pandey N., Ranjan A., Pant P., Tripathi R. K., Ateek F., Pandey H. P., et al. (2013). CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14:216. 10.1186/1471-2164-14-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy R. K., Xiang Q., Hsieh E.-J., Liu K., Zhang C., Waters B. M. (2018). New aspects of iron–copper crosstalk uncovered by transcriptomic characterization of Col-0 and the copper uptake mutant spl7 in Arabidopsis thaliana. Metallomics 10 1824–1840. 10.1039/C8MT00287H [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón D. M., Ruzicic S., Dreyer I., Mueller-Roeber B. (2007). PlnTFDB: an integrative plant transcription factor database. BMC Bioinfo. 8:42. 10.1186/1471-2105-8-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savojardo C., Martelli P. L., Fariselli P., Casadio R. (2018). DeepSig: deep learning improves signal peptide detection in proteins. Bioinformatics 34 1690–1696. 10.1093/BIOINFORMATICS/BTX818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Flügge U.-I. (2018). Identification and characterization of plant membrane proteins using aramemnon. Methods Mol. Biol. 1696 249–259. 10.1007/978-1-4939-7411-5_17 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., et al. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131 16–26. 10.1104/PP.011577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Park J.-Y., Kim S.-Y., Jeon J., Lee Y.-H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61 661–671. 10.1111/J.1365-313X.2009.04091.X [DOI] [PubMed] [Google Scholar]
- Seo P. J., Park C.-M. (2010). A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav. 5 481–483. 10.4161/PSB.11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z., Goldberg A. L. (2014). Proteasome-Mediated Processing of Nrf1 Is Essential for Coordinate Induction of All Proteasome Subunits and p97. Curr. Biol. 24 1573–1583. 10.1016/j.cub.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C.-F., Hsu W.-H., Peng Y.-J., Yang C.-H. (2014). The NAC-like gene anther indehiscence factor acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 65 621–639. 10.1093/jxb/ert412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Shih M.-C., Li W.-H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139 18–26. 10.1104/pp.105.065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S. A., Mondala T. S., Head S. R., Cravatt B. F. (2004). Microarray platform for profiling enzyme activities in complex proteomes. J. Am. Chem. Soc. 126 15640–15641. 10.1021/ja044286+ [DOI] [PubMed] [Google Scholar]
- Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. (2008). Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell 15 209–219. 10.1016/j.devcel.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Silber M., Hitzenberger M., Zacharias M., Muhle-Goll C. (2020). Altered hinge conformations in APP transmembrane helix mutants may affect enzyme-substrate interactions of γ-secretase. ACS Chem. Neurosci. 11 4426–4433. 10.1021/ACSCHEMNEURO.0C00640 [DOI] [PubMed] [Google Scholar]
- Singh M. B., Lohani N., Bhalla P. L. (2021). The role of endoplasmic reticulum stress response in pollen development and heat stress tolerance. Front. Plant Sci. 12:661062. 10.3389/fpls.2021.661062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh E. (2011). Characterization of a Plant-Specific Membrane-Tethered MYB Transcription Factor in Arabidopsis Thaliana. Ph.D. Michigan, MICH: Michigan State University. [Google Scholar]
- Slabaugh E., Held M., Brandizzi F. (2011). Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 67 395–405. 10.1111/J.1365-313X.2011.04602.X [DOI] [PubMed] [Google Scholar]
- Song Y. H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18 575–583. 10.1016/j.tplants.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz C., Schlosser C., Guschtschin-Schmidt N., Stelzer W., Menig S., Götz A., et al. (2020). Non-canonical shedding of TNFα by SPPL2a Is determined by the conformational flexibility of its transmembrane helix. IScience 23:101775. 10.1016/J.ISCI.2020.101775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling J., O’Hare P. (2006). CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol. Biol. Cell 17 413–426. 10.1091/mbc.E05-06-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strisovsky K., Sharpe H. J., Freeman M. (2009). Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36 1048–1059. 10.1016/J.MOLCEL.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., Trajanoski Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18 207–208. 10.1093/bioinformatics/18.1.207 [DOI] [PubMed] [Google Scholar]
- Sun H., Xie Y., Yang W., Lv Q., Chen L., Li J., et al. (2022). Membrane-bound transcription factor TaNTL1 positively regulates drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 182 182–193. 10.1016/J.PLAPHY.2022.04.023 [DOI] [PubMed] [Google Scholar]
- Sun L., Lu S.-J., Zhang S.-S., Zhou S.-F., Sun L., Liu J.-X. (2013). The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Mol. Plant 6 1605–1615. 10.1093/MP/SST059 [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang S.-S., Lu S.-J., Liu J.-X. (2015). Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Sci. China Life Sci. 58 270–275. 10.1007/S11427-015-4807-6 [DOI] [PubMed] [Google Scholar]
- Sundaram S., Kertbundit S., Shakirov E. V., Iyer L. M., Juříček M., Hall T. C. (2013). Gene networks and chromatin and transcriptional regulation of the phaseolin promoter in Arabidopsis. Plant Cell 25 2601–2617. 10.1105/TPC.113.112714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H., Iwata Y., Iwano M., Takayama S., Koizumi N. (2008). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374 242–247. 10.1016/J.BBRC.2008.07.021 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kawakatsu T., Wakasa Y., Hayashi S., Takaiwa F. (2012). A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 53 144–153. 10.1093/PCP/PCR157 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 3022–3027. 10.1093/MOLBEV/MSAB120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Zhao C.-Y., Tan S.-T., Xue H.-W. (2016). Arabidopsis type II phosphatidylinositol 4-kinase PI4Kγ5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PLoS Genet. 12:e1006252. 10.1371/journal.pgen.1006252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel F., Almagro Armenteros J. J., Johansen A. R., Gíslason M. H., Pihl S. I., Tsirigos K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 1–3. 10.1038/s41587-021-01156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Wang X., Guo H., Cheng Y., Hou C., Chen J.-G., et al. (2017). NTL8 regulates trichome formation in Arabidopsis by directly activating R3 MYB genes TRY and TCL1. Plant Physiol. 174 2363–2375. 10.1104/pp.17.00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O., Ford E., Lister R., Huang S., Millar A. H. (2016). Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 88 542–558. 10.1111/tpj.13276 [DOI] [PubMed] [Google Scholar]
- Wang D., Yu Y., Liu Z., Li S., Wang Z., Xiang F. (2016). Membrane-bound NAC transcription factors in maize and their contribution to the oxidative stress response. Plant Sci. 250 30–39. 10.1016/J.PLANTSCI.2016.05.019 [DOI] [PubMed] [Google Scholar]
- Ward J. M. (2001). Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 17 560–563. 10.1093/bioinformatics/17.6.560 [DOI] [PubMed] [Google Scholar]
- Ward J. M., Cufr C. A., Denzel M. A., Neff M. M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17 475–485. 10.1105/TPC.104.027722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P., Horne A., Van Parys T., Goormachtig S., De Smet I., Botzki A., et al. (2019). The plant PTM viewer, a central resource for exploring plant protein modifications. Plant J. 99 752–762. 10.1111/tpj.14345 [DOI] [PubMed] [Google Scholar]
- Xia N., Zhang G., Sun Y. F., Zhu L., Xu L. S., Chen X. M., et al. (2010). TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 74 394–402. 10.1016/J.PMPP.2010.06.005 [DOI] [Google Scholar]
- Yabuta Y., Osada R., Morishita T., Nishizawa-Yokoi A., Tamoi M., Maruta T., et al. (2011). Involvement of Arabidopsis NAC transcription factor in the regulation of 20S and 26S proteasomes. Plant Sci. 181 421–427. 10.1016/j.plantsci.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Yan J., Chen Q., Cui X., Zhao P., Gao S., Yang B., et al. (2021a). Ectopic overexpression of a membrane-tethered transcription factor gene NAC60 from oilseed rape positively modulates programmed cell death and age-triggered leaf senescence. Plant J. 105 600–618. 10.1111/TPJ.15057 [DOI] [PubMed] [Google Scholar]
- Yan J., Li Y., Zhao P., Mu B., Chen Q., Li X., et al. (2021b). Membrane-bound transcriptional activator NTL1 from rapeseed positively modulates leaf senescence through targeting genes involved in reactive oxygen species production and programmed cell death. J. Agric. Food Chem. 69 4968–4980. 10.1021/ACS.JAFC.1C00182 [DOI] [PubMed] [Google Scholar]
- Yang M., Ellenberg J., Bonifacino J. S., Weissman A. M. (1997). The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272 1970–1975. 10.1074/jbc.272.3.1970 [DOI] [PubMed] [Google Scholar]
- Yang W., Xu P., Zhang J., Zhang S., Li Z., Yang K., et al. (2022). OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. J. Genet. Genom. 49 414–426. 10.1016/J.JGG.2022.02.002 [DOI] [PubMed] [Google Scholar]
- Yang Y. G., Lv W. T., Li M. J., Wang B., Sun D. M., Deng X. (2013). Maize membrane-bound transcription factor zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol. 54 2020–2033. 10.1093/PCP/PCT142 [DOI] [PubMed] [Google Scholar]
- Yang Z.-T., Lu S.-J., Wang M.-J., Bi D.-L., Sun L., Zhou S.-F., et al. (2014a). A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 79 1033–1043. 10.1111/tpj.12604 [DOI] [PubMed] [Google Scholar]
- Yang Z.-T., Wang M.-J., Sun L., Lu S.-J., Bi D.-L., Sun L., et al. (2014b). The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 10:e1004243. 10.1371/journal.pgen.1004243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Deng L., Zeng K. (2017). Genome-wide in silico identification of membrane-bound transcription factors in plant species. PeerJ 5:e4051. 10.7717/peerj.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. (2020). Transcription factors activated through RIP (regulated intramembrane proteolysis) and RAT (regulated alternative translocation). J. Biol. Chem. 295 10271–10280. 10.1074/jbc.rev120.012669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Davé U. P., Grishin N. V., Goldstein J. L., Brown M. S. (2000a). Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease. Proc. Natl. Acad. Sci. USA 97 5123–5128. 10.1073/pnas.97.10.5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Rawson R. B., Komuro R., Chen X., Davé U. P., Prywes R., et al. (2000b). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6 1355–1364. 10.1016/S1097-2765(00)00133-7 [DOI] [PubMed] [Google Scholar]
- Yin X., Liu X., Xu B., Lu P., Dong T., Yang D., et al. (2019). OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice. J. Exp. Bot. 70 3895–3909. 10.1093/JXB/ERZ198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.-K., Kim S.-G., Kim S.-H., Park C.-M. (2008). Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 25 438–445. [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107 881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ren Y., Li S., Hayes J. D. (2014). Transcription factor Nrf1 is topologically repartitioned across membranes to enable target gene transactivation through its acidic glucose-responsive domains. PLoS ONE 9:e93458. 10.1371/journal.pone.0093458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.-X., Bisanz-Seyer C., Mache R. (1995). Molecular cloning of a small DNA binding protein with specificity for a tissue-specific negative element within the rps1 promoter. Nucleic Acids Res. 23 1165–1169. 10.1093/nar/23.7.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupicich J., Brenner S. E., Skarnes W. C. (2001). Computational prediction of membrane-tethered transcription factors. Genome Biol. 2:research0050. 10.1186/gb-2001-2-12-research0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.