Abstract
The discovery and development of new potent antimicrobial and antioxidant agents is an essential lever to protect living beings against pathogenic microorganisms and free radicals. In this regard, new functionalized pyrazoles have been synthesized using a simple and accessible approach. The synthesized aminobenzoylpyrazoles 3a-h and pyrazole-sulfonamides 4a-g were obtained in good yields and were evaluated in vitro for their antimicrobial and antioxidant activities. The structures of the synthesized compounds were determined using IR, NMR, and mass spectrometry. The structure of the compound 4b was further confirmed by single crystal X-ray diffraction. The results of the in vitro screening show that the synthesized pyrazoles 3 and 4 exhibit a promising antimicrobial and antioxidant activities. Among the tested compounds, pyrazoles 3a, 3f, 4e, 4f, and 4g have exhibited remarkable antimicrobial activity against some microorganisms. In addition, compounds 3a, 3c, 3e, 4a, 4d, 4f, and 4g have shown a significant antioxidant activity in comparison with the standard butylhydroxytoluene (BHT). Hence, compounds 3a, 4f, and 4g represent interesting dual acting antimicrobial and antioxidant agents. In fact, pyrazole derivatives bearing sulfonamide moiety (4a-g) have displayed an important antimicrobial activity compared to pyrazoles 3a-h, this finding could be attributed to the synergistic effect of the pyrazole and sulfonamide pharmacophores. Furthermore, Molecular docking results revealed a good interaction of the synthesized compounds with the target proteins and provided important information about their interaction modes with the target enzyme. The results of the POM bioinformatics investigations (Petra, Osiris, Molinspiration) show that the studied heterocycles present a very good non toxicity profile, an excellent bioavailability, and pharmacokinetics. Finally, an antiviral pharmacophore (O δ−, O δ−) was evaluated in the POM investigations and deserves all our attention to be tested against Covid-19 and its Omicron and Delta mutants.
Key-words: Pyrazole Linked Sulfonamide Conjugates, Antimicrobial/Antioxidant bioactivity, Docking study, Petra/Osiris/Molinspiration (POM) analyses, Identification of the pharmacophore sites
Abbreviations: POM, Petra/Osiris/Molinspiration; PSH, Pyrazoles-Sulfonamide Hybrids
Graphical Abstract
1. Introduction
Pathogenic microbes remain a constant threat for human health [1]. They are responsible for serval serious infectious diseases that lead to death [2]. In addition, these pathogenic agents have the ability to undergo genetic modification through spontaneous mutations and become therefore resistant to existing antimicrobials drugs [3,4]. Furthermore, the production of free radicals in living organisms is a physiological process, regulated by various chemical or enzymatic detoxification processes. However, when the protective system of the human organism shows some failure and loses its efficiency, the number of free radicals increases significantly and leads to oxidative stress [5]. This oxidative effect leads to the damage of certain biomolecules, including lipids, DNA and proteins, …etc. [6]. This damage causes serious threat to human health, such as cancer, Alzheimer's, and Parkinson's [7,8]. Hence, the development of new antimicrobial and antioxidant agents able to overcome these health problems is urgently needed.
In this context, chemists have devoted great efforts to design new compounds with excellent therapeutic effects. In fact, pyrazole pharmacophore have attracted considerable attention since their discovery as a key building blocks of many drugs [9,10]. Moreover, sulfonamides are among of antibiotics widely used as preventive and curative agents against various infectious diseases [11,12]. Recently, different innovative strategies have been proposed in the investigation of molecules with potential antimicrobial and antioxidant activities. The association of two or more pharmacophores in the one molecular skeleton may lead to a synergistic effect [13,14]. The conception of molecular scaffolds containing pyrazole heterocycle and sulfonamide moiety is part of this strategy [11,15,16]. Hence, molecules containing the two pharmacophores sulfonamide and pyrazole exhibit a wide array of biological activities, such as: antioxidant [13], antimicrobial [13,17,18], anti-inflammatory [17,19], anticancer [20], selective inhibitor of carbonic anhydrase [21], …etc. Some of these compounds are commercially available as antibiotics like sulfaphenazole and anti-inflammatory, in particular, celecoxib [15,16].
Chemo-informatics approaches have become an effective and rapid tool in the design of new molecules with significant biological activity [22,23]. Recently, various structure-based virtual screening techniques are used to design new drug candidates against the novel coronavirus [24], [25], [26]. In this context, several studies have been conducted on many organic molecules [25], [26], [27]. In silico simulations have identified new compounds with potential antiviral activity, which need further experimental studies. Pyrazoles and sulfonamides are known for their remarkable antiviral activities [28,29]. In the best of our knowledge, there is no previous study performed on molecules that incorporate these two motifs towards SARS-CoV-2 proteins. This prompted us to carry out an in-silico study on these compounds with the SARS-CoV-2 main protease, in order to identify new active molecules against this pathogenic agent.
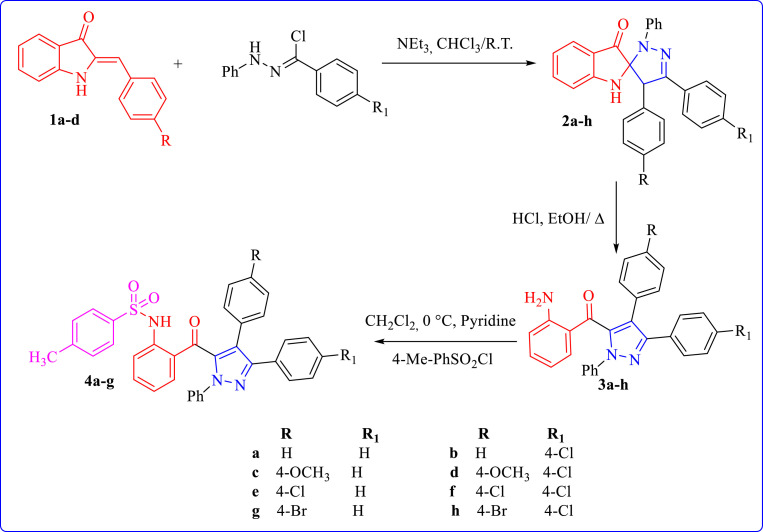
Based on literature data, in particular, those related to pharmacological interest of pyrazole and sulfonamide moieties, and in continuation of our ongoing research focused on the synthesis of new heterocyclic systems [30,31]. We describe herein, the synthesis of a new series of hybrid heterocyclic molecules, which the pyrazole and sulfonamide moieties were linked together via a benzoyl group (Fig. 1 ). The synthesized compounds were evaluated in vitro for their antimicrobial and antioxidant properties. The antimicrobial activity was performed using the conventional broth microdilution method against the different bacterial and fungal strains, and the antioxidant activity through the DPPH radical scavenging assay. The results obtained indicate that some studied compounds display interesting antimicrobial and antioxidant activities. Furthermore, a molecular docking study was carried out to predict the possible binding interactions between the studied compounds and the target enzyme. In addition, a docking study was done against SARS-CoV-2 main protease, to identify inhibitors that could the potentially effective anti-COVID-19 drug candidates. as well as to identify the pharmacophores sites in the synthesized compounds for the antibacterial, antifungal, antiviral activities using Petra/Osiris/Molinspiration (POM) analyses were also carried out.
Fig. 1.
Target compounds design concept based on commercially antibiotic drug containing pyrazole and sulfonamide scaffolds.
2. Results and discussion
2.1. Chemistry
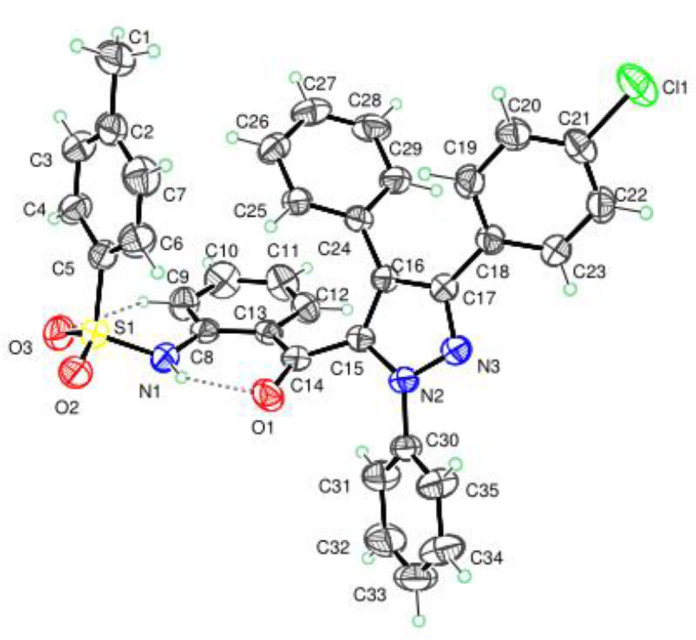
The synthesis of the targeted pyrazole derivatives bearing sulfonamide moiety was carried out using the procedure described in scheme 1 . The intermediates 5-(2-aminobenzoyl)-3,4-diaryl-1-phenylpyrazoles 3a-h were prepared in two steps from aza-aurones 1a-d: 1,3-dipolar cycloaddition reaction followed by ring opening of spiropyrazolines 2a-h [30]. Then, the pyrazoles 3a-h were used as a key synthon to synthesize the target pyrazole-sulfonamide hybrid compounds 4a-g. The chemical structure of the synthesized compounds was established using FT-IR, 1H NMR, 13C NMR and HRMS techniques. In addition, the structure of compound 4b was further confirmed by single crystal X-ray diffraction (Fig. 3).
Scheme 1.
Route of synthesis of compounds 4a-g.
Fig. 3.
Ortep drawing the structure of the title compound (4b) and its numbering scheme. Thermal ellipsoids were drawn at the 50 % probability level at 296 K.
For example, the infrared spectrum of compound 4a shows the presence of two absorption bands at 3303 cm−1 and 1622 cm−1 corresponding to N-H and -C=O stretchings, respectively. In addition, its 1H NMR spectrum reveals the presence of a signal at 2.45 ppm owing to CH3 protons of tosyl group. It showed also a signal at 11.07 ppm exchangeable with D2O attributable to the proton of NH group. Furthermore, 13C NMR spectrum displays tow characteristic signals at 21.65 ppm and 192.25 ppm belonging to methyl carbon (CH3) and ketone carbon (C=O), respectively. The mass spectrum of compound 4a shows a peak for the molecular ion [M+Na]+ at m/z = 592.16635. The spectral data are in perfect agreement with the proposed structures.
2.2. Tautomeric analysis and prediction of bioactivity
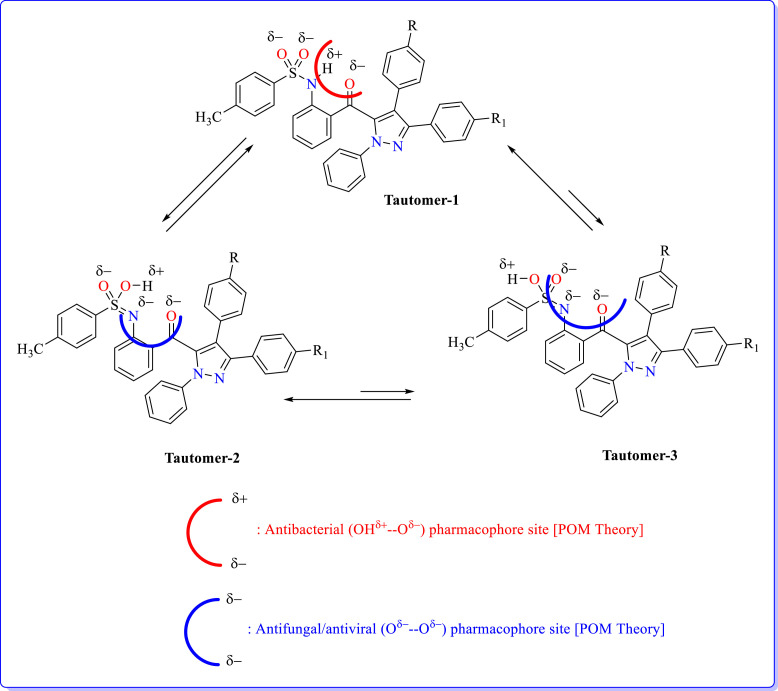
For PSH and certainly for their analogues, depending on the pH and position of the dissociate amidogen hydrogen atom, thre possible PSH tautomerisation can be described for the neutral forms. These relevant structures are sketched in Fig. 2 .
Fig. 2.
Possible tautomers and their corresponding pharmacophore sites [POM Theory].
In past, attention was mainly devoted to the N-H structure. However, from a chemical point of view, all other structures are possible.
For the development of binding approaches for PSH and their analogues in the environment, the identification of the active sulfonamide structures present is important. Neither experimental nor theoretical data is available for the identification of water-solved PSH species. Theoretically, NMR spectroscopy could be useful for identifying chemical structures. Theoretical ab initio studies could supplement these measurements. Additionally, calculations of energetics, atomic charges, minimum energy structures, geometry, and natural bond orbital (NBO) could indicate the electronic density distribution of each atom. Finally, by taking NBO results showing the presence of S–O single bonds in consideration, realistic Lewis structures can be determined. These systematic data, regarding the variation of molecular properties, are important for the chemical structure and could therefore provide first insights into the still poorly understood chemical bonding of PSH complexes to soil.
In brief, the objective of this study is to investigate the potential pharmacophore sites of PSH species using antibacterial and antifungal screenings dependence on pH and comparison with the calculated molecular properties. To verify these structures, further X-ray analysis becomes necessary to fix the predominant tautomer. Then the Petra/Osiris/Molinspiration (POM) analyses were carried out for example calculation of net atomic charges, bond polarity, atomic valence, electron delocalization and lipophicity. Finally, to investigate the combined antibacterial/antifungal/Antiviral bioactivity of the PSH species, tautomeric structure was performed.
Current thinking in the generation of specific drug leads embodies the concept of achieving high molecular diversity within the boundaries of reasonable drug-like properties. Natural and semi-natural products, examples penicillin and, imipenemhave high chemical diversity, biochemical specificity and other molecular properties that make them favourable as lead and standard references (SD) structures for drug discovery, and which serve to differentiate them from libraries of synthetic and combinatorial compounds. Various investigators have used computational methods to understand differences between natural products and other sources of drug leads. Modern drug discovery is based in large part on high throughput screening of small molecules against macromolecular disease targets requiring that molecular screening libraries contain drug-like or lead-like compounds. We have analyzed known standard references (SD) for drug-like and lead-like properties. With this information in hand, we have established a strategy to design specific drug-like or lead-like PSH (4a-g).
To be sure of the real and dominant tautomeric form, it becomes necessary to get some crystallographic data of someone of tested compounds 3a-h and/or 4a-g.
2.3. X-ray diffraction data and crystal structure of compound 4b
The X-ray measurement conditions, crystal data and structure refinement details are summarized in Table S1. The selected interatomic distances and angles listed in Table S2 are close to those observed in the structures of similar compounds [31], [32], [33]. The plot of the molecular structure of the title compound is shown in Fig. 3 . The central pyrazoline ring (N2–N3–C15–C16–C17) is almost planar with the largest deviation from the mean plan of -0.012(2) at C17 atom. It is connected to three benzene rings and to a sulfonamide conjugates as shown in Fig. 3. The dihedral angles between the pyrazole ring and the different substituents namely: chlorobenzene ring (C18—C23) and the two benzene rings (C24—C29) and (C30—C35) are of 37.35(11)°, 49.98(11)° and 34.75(13)°, respectively. The mean plan through the aminobenzene ring (C8—C13) is nearly orthogonal to that of the chlorobenzene ring as indicated by the dihedral angle between them of 87.95(11)°. On the other hand, the dihedral angle formed by the two benzene rings (C2—C7) and (C8—C13) linked to sulfoamine group is 79.30(12) °, which indicates that these two rings are almost orthogonal to each other.
In the crystal, the molecules are interconnected through C4—H4…Cl1 hydrogen bond building a chain running along the b-axis. The chains are linked by π…π interaction between the pyrazoline and one benzene ring (C30—C35), with inter-centroid distance of 3.92 (2) Å as shown in Fig. 4 and Table S3. The resulting three-dimensional framework is represented in Fig. 4. Moreover, the molecular conformation is stabilized by two stark intra-molecular hydrogen bonds completing two S(6) rings (see Fig. 4 and Table S3).
Fig. 4.
The crystal packing for the title compound, showing molecules linked by hydrogen bonds (dashed cyan lines) and π…π interactions (dashed green lines).
3. Biological activities
3.1. Antimicrobial activity
The new series of synthesized compounds (3a-h and 4a-g) were tested in vitro for their antimicrobial activity against Gram-positive bacteria: Staphylococcus aureus (S. A.), Gram-negative bacteria: Escherichia coli (E. C.), Klebsiella pneumoniae (K. P.) and Pseudomonas aeruginosa (P. A.), and yeasts: Candida albicans (C. A.) and Saccharomyces cerevisiae (S.C.). The antibacterial and antifungal activities of the target compounds were determined as minimum inhibitory concentrations (MICs) by the broth microdilution method. The antibiotics Ampicillin and Streptomycin were used as positive controls against bacterial strains, and Fluconazole as a positive control for fungal strains. The results of the antimicrobial activity are given in Table 1 .
Table 1.
Antimicrobial activity (MIC, µg/mL) of the synthesized compounds (3a-h and 4a-g)
| Compd. | R | R1 |
Bacterial strains |
Fungal strains |
||||
|---|---|---|---|---|---|---|---|---|
| E. C. | K. P. | P. A. | S. A. | C. A. | S. C. | |||
| 3a | H | H | 125 | 250 | 250 | 125 | 62.5 | 62.5 |
| 3b | H | Cl | 250 | 125 | 250 | 250 | 125 | 125 |
| 3c | OCH3 | H | 250 | 125 | 250 | 250 | 125 | 125 |
| 3d | OCH3 | Cl | 250 | 125 | 250 | 250 | 125 | 125 |
| 3e | Cl | H | 250 | 250 | 250 | 125 | 250 | 250 |
| 3f | Cl | Cl | 250 | 125 | 250 | 250 | 125 | 62.5 |
| 3g | Br | H | 250 | 250 | 125 | 125 | 125 | 250 |
| 3h | Br | Cl | 250 | 125 | 250 | 250 | 125 | 125 |
| 4a | H | H | 250 | 250 | 250 | 125 | 250 | 125 |
| 4b | H | Cl | 250 | 125 | 250 | 250 | 125 | 125 |
| 4c | OCH3 | H | 250 | 125 | 125 | 250 | 125 | 125 |
| 4d | OCH3 | Cl | 250 | 125 | 125 | 250 | 125 | 125 |
| 4e | Cl | H | 125 | 125 | 125 | 250 | 125 | 62.5 |
| 4f | Cl | Cl | 250 | 125 | 250 | 250 | 62.5 | 31.25 |
| 4g | Br | H | 125 | 125 | 125 | 62.5 | 125 | 125 |
| Streptomycin | — | — | 25 | 3.12 | — | 12.5 | — | — |
| Ampicillin | — | — | 50 | 25 | 1.56 | 25 | — | — |
| Fluconazole | — | — | — | — | — | — | 40 | 20 |
Bacteria strains : E. C.(Escherichia coli (ATB:57) B6N),K. P.(Klebsiella pneumoniae),P. A.(Pseudomonas aeruginosa),S. A.(Staphylococcus aureus); Fungal strains: C. A.(Candida albicans ATCC10231) and S. C.(Saccharomyces cerevisiae ATCC9763).
The results of the preliminary studies show that the two series of pyrazoles (3a-h and 4a-g) exhibit antimicrobial activity at concentrations between 31.25 and 250 µg/mL against the pathogenic microorganisms tested. Some compounds of these two series display significant antibacterial and antifungal activity, in particular, those containing the sulfonamide moiety. Compound 4f shows remarkable activity against Candida albicans (62.5µg/mL) and Saccharomyces cerevisiae (31.25 µg/mL) compared to fluconazole. The antifungal activity obtained for compound 4cb can be attributed to the presence of two chlorine atoms (Cl) in para position on the benzene rings [34,35]. Compounds 3a, 3f, and 4e show significant inhibitory activity against Saccharomyces cerevisiae with a MIC value of 62.5 μg/mL, whereas they manifested moderate activity against Candida albicans (MIC values between 62.5 and 125 μg/mL), compared to the same standard reference. The antifungal activity observed for these compounds is comparable to that described in the literature for analogous compounds [17,35,36]. However, for antibacterial activity compound 4g shows remarkable inhibition against the gram-positive strain Staphylococcus aureus with a MIC value of 62.5 μg/mL and moderate activity against other bacteria with MIC values in the range of 125 μg/mL. The activity observed for the 4g product compared to the other compounds can be attributed to the synergistic effect of the bromine atom and the sulfonamide moiety [17,36].
3.2. Antioxidant activity
To evaluate the antioxidant potential of our prepared heterocyclic compounds, the free radical scavenging capacity was determined by the DPPH assay, using butylhydroxytoluene (BHT) as a standard antioxidant. It is a commonly used assay that provides information on the ability of a compound to donate an electron or proton to the DPPH radical, and allows for the measurement of the free radical scavenging capacity in a in a high speed, simple, and economic way. The percentages of inhibition at different concentrations are expressed as an average and summarized in Table 2 .
Table 2.
The antioxidant activity of the synthesized compounds and the BHT reference.
| Percentage inhibition of antioxidant activity of tested compounds | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Comp | R | R1 | R2 | 3 µg/mL | 15 µg/mL | 31 µg/mL | 62.5 µg/mL | 250 µg/mL | 1000 µg/mL |
| 3a | H | H | - | 41.81±0.67 | 43.03±0.09 | 45.63±0.23 | 46.20±0.19 | 46.51±0.45 | 64.84±0.88 |
| 3b | H | Cl | - | 21.06±0.42 | 36.51±0.35 | 37.87±0.79 | 39.09±0.41 | 42.87±0.45 | 58.93±0.34 |
| 3c | OCH3 | H | - | 23.18±0.56 | 24.84±0.45 | 28.03±0.27 | 28.48±0.45 | 50.15±0.3 | 65.75±0.66 |
| 3d | OCH3 | Cl | - | 2.7±0.63 | 18.93±0.34 | 22.27±0.3 | 23.78±0.28 | 42.72±0.43 | 62.87±0.68 |
| 3e | Cl | H | - | 38.03±0.76 | 45±0.6 | 45.30±0.31 | 46.06±0.51 | 46.36±0.44 | 68.63±0.35 |
| 3f | Cl | Cl | - | 3.78±0.69 | 17.12±0.32 | 20.15±0.96 | 20.60±0.46 | 23.33±0.56 | 64.54±0.65 |
| 3g | Br | H | - | 15±0.09 | 18.33±0.97 | 19.84±0.8 | 20.30±0.63 | 22.12±0.32 | 63.48±0.97 |
| 3h | Br | Cl | - | 13.18±0.82 | 28.48±0.72 | 30.60±0.54 | 46.51±0.25 | 49.84±0.43 | 56.51±0.72 |
| 4a | H | H | tosyl | 25.30±0.67 | 25.75±0.85 | 29.39±0.94 | 35±0.54 | 38.63±0.22 | 62.27±0.37 |
| 4b | H | Cl | tosyl | 3.63±0.64 | 5.75±0.24 | 14.39±0.35 | 32.27±0.63 | 43.78±0.82 | 59.69±0.93 |
| 4c | OCH3 | H | tosyl | 14.84±0.8 | 24.24±0.86 | 44.84±0.24 | 45.60±0.34 | 47.42±0.76 | 64.54±0.17 |
| 4d | OCH3 | Cl | tosyl | 9.24±0.42 | 10.60±0.23 | 15±0.32 | 38.33±0.92 | 51.21±0.35 | 57.27±0.43 |
| 4e | Cl | H | tosyl | 14.01±0.93 | 19.84±0.22 | 26.66±0.43 | 30±0.43 | 33.78±0.06 | 59.24±0.77 |
| 4f | Cl | Cl | tosyl | 14.39±0.39 | 22.87±0.98 | 36.66±0.7 | 43.18±0.23 | 43.93±0.76 | 66.21±0.76 |
| 4g | Br | H | tosyl | 17.12±0.12 | 19.84±0.64 | 40.45±0.24 | 46.81±0.65 | 48.18±0.34 | 67.87±0.26 |
| BHT | — | — | — | 43.93±0.93 | 59.09±0.09 | 63.63±0.63 | 75.75±0.75 | 80.30±0.30 | 87.87±0.87 |
The results of this preliminary study show that some compounds have a remarkable free radical scavenging capacity (DPPH) compared to the BHT standard. The study reveals that compounds 3a (% PI = 41.81) and 3e (% PI = 38.03) show an antioxidant activity comparable to that of the BHT (% PI = 43.93) at a low concentration (3 µg/mL). For compounds 3b, 3c, 4a, 4d, 4f, and 4g we noticed that the antioxidant activity increases with concentration and becomes close to that of the standard at high concentrations. The analysis of the obtained results shows that the substituents OCH3, Cl, and Br have no significant influence on the antiradical activity of the studied compounds, this suggests that this activity is related to the amino group NH and NH2 in both types of compounds [37,38].
4. Molecular Docking
The docking results obtained from Auto-Dock showed that the studied compounds can be accommodated in the binding pocket of DHPS with a comparable orientation to the one observed in the STZ-DHPP covalent adduct in the reported crystal structure [39]. The top-ranked docking poses reproduce the key interactions observed in the STZ-DHPP–DHPS complex (Fig. 5 ).
Fig. 5.
Re-docking of the co-crystallized ligand (PDB ID: 3JQ9)
Analysis of molecular docking results of pyrazoles-sulfonamide hybrids derivatives 4a-4g shows that most of these derivatives exhibit the same polar H-interaction and hydrophobic interaction of STZ ligand (referent) with the site of action DHPs, this interaction similarity is due to the sulfonamide moiety (Fig. 6 , Table 3 and figure S1). The good antibacterial activity presented by the good interaction of the oxazaphosphinanes compounds with the DHPs target can be explained by the presence of hydrophobic interactions identical to those of the co-crystallized ligand and the interesting stability inside the DHPs cavity with a binding energy varying between -5.96 and -7.01 (kcal / mol).
Fig. 6.
Molecular docking analysis of PSH derivatives (4c, 4e, 4f and 4g). Pose view of interaction with receptor DHPs.
Table 3.
Ranking of the PSH derivatives 4a-4g derivatives after docking study.
| Compds | Binding Energy (kcal/mol) | Vdw Energy | Electrostatic Energy | Nature of interactions | Amino acids on active sites with |
|---|---|---|---|---|---|
| 4a | -6.01 | -8.46 | 0.06 | -Hydrophobic interaction- Polar H interactions | -Phe190- Ser222, Arg255 |
| 4b | -6.48 | -8.72 | -0.14 | -Hydrophobic interaction- Polar H interactions | -Lys221, Phe190- Ser222, Arg255 |
| 4c | -6.92 | -9.46 | -0.14 | -Hydrophobic interaction- Polar H interactions | -Phe190-Arg255, Thr62 |
| 4d | -6.41 | -9.01 | -0.08 | -Hydrophobic interaction- Polar H interactions | -Arg255, Thr62 |
| 4e | -6.96 | -9.2 | -0.14 | -Hydrophobic interaction- Polar H interactions | -Arg255, Lys221, Phe190-Arg255, Thr62 |
| 4f | -7.01 | -9.02 | -0.38 | -Hydrophobic interaction- Polar H interactions | -Arg255- Lys221, Thr62 |
| 4g | -6.89 | -9.18 | -0.1 | -Hydrophobic interaction- Polar H interactions | -Phe190, Lys221-Arg255, Thr62 |
| co-crystallized ligand | -9.41 | -10.13 | -1.37 | -Hydrophobic interaction- Polar H interactions | -Phe190, Lys221-Asn115, Ser222, Asp155, Gly189 |
Compounds 4f and 4g present the best interaction with the target with binding energy respectively 7.1 and 6.89.
5. POM Analyses
Now, it becomes easier, by using the POM (Petra/Osiris/Molinspiration) Theory, to identify and to optimize most of the antibacterial [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], antifungal [50], [51], [52], antiviral [53], [54], [55], antiparasital [56,57], and antitumor [58], [59], [60] pharmacophore sites, one by one, on the basis of their different physico-chemical parameters and their different electronic charge repartition of corresponding heteroatoms. This young POM Theory was extended to other various and different biotargets [61,62]. Here we treate the series of compounds 3a-h and 4a-g in the goal to identify their pharmacophore sites.
The Osiris analysis of series of tested compounds shows that most of compounds represent no side effects, except compounds 3c, 3d, 4c and 4d (Table S4). The Molinspiration analysis of series of tested compounds shows clearly that there are more substituents than the necessary because the molecular weight depasses 500 g/mol. This constitutes the first violation of Lipinski 5 rules. A second violation will appear when the cLogP depasses 5 (Table S5).
6. Identification of a combined Antibacterial/Antiviral pharmacophore site
Recently, we discovered the Dithymoquinone as potential anti-Covid-19 with MIC in range of nano-Molar [63]. Interestingly, we compare the pharmacophore site of the Dithymoquinone (dO1-O2 = 4.5 A) and the combined antibacterial/antiviral pharmacophore site of compound 4b (dO1-O2 = 4.6 A). This important similarity encourages us to go ahead without hesitation in anti-Covid screening of series 4a-g. Who knows?! Maybe a great surprise is waiting for our group (Figs. 7 and 8 ).
Fig. 7.
Atomic charge of tested compounds 3a-h and 4a-g.
Fig. 8.
Identification of potential Antiviral (O1, O2)-pharmacophore site.
7. In-silico screening the anti-Covid-19 activity of PSH by Molecular Docking
In order to understand the interactions between protein and ligand, molecular docking study was performed to explore the binding mode of the prepared pyrazole-sulfonamide derivatives to the SARS-CoV-2 main protease, Autodock Tools were employed for identifying the torsion angles in the ligand, by adding the solvent model and assigning the Kollman atomic charges to the protein. The Methyl 4-sulfamoylbenzoate was taken as reference ligand to investigate the binding mode of the studied synthesized derivatives 4a-g.
The RMSD value is 0.84 Ǻ, which allowed us to validate our docking methodology and to obtain a good prediction of the ligand-protein confirmation in absence of water molecules. Fig. 9 shows docked Methyl 4-sulfamoylbenzoate RZG and co-crystallized one in almost same position among the receptor.
Fig. 9.
Superimposition of docked pose and crystal structure pose of ligand RZG in wild type of SARS-CoV-2 main protease (PDB ID: 5R80).
The co-crystallized ligand RZG (PDB ID: 5R80) forms a polar hydrogen interaction with the amino acid Glu166 and several hydrophobic alkyl and Pi-alkyl interactions in the receptor tyrosine kinase transmembrane EGFR, the hydrophobic interactions are shown in the Fig. 10 .
Fig. 10.
Binding site structure of SARS-CoV-2 main protease-RZG.
The docking results of the synthesized compounds and Methyl 4-sulfamoylbenzoate were reported in Table 4 . Compounds 4a-g show interesting stability inside the SARS-CoV-2 main protease cavity (Fig. 11 , figures S2-S8 in Supplementary information) with a binding energy varying between -6.10 and -10.10 (kcal/mol) (Table 4).
Table 4.
Ranking of the pyrazole-sulfonamide derivatives after docking study.
| N° | protein | Compound | RMSD | Free Energy of Binding | Inhibition Constant, Ki | Amino acids involved in interactions | |
|---|---|---|---|---|---|---|---|
| 1 | 5R80 | RZG | 0.00 | -5.27 | 137.38 uM | hydrogen bonding | GLU166 |
| hydrophobic interaction | GLN189, MET165, MET49, HIS41 | ||||||
| 2 | 5R80 | 4a | 0.70 | -8.72 | 403.64 nM | hydrogen bonding | GLN189 |
| hydrophobic interaction | GLN189, MET165, HIS41, PRO168 | ||||||
| 3 | 5R80 | 4b | 0.00 | -6.85 | 9.59 uM | hydrogen bonding | ASN142, GLU166(2), CYS145 |
| hydrophobic interaction | GLN 189, MET165, MET49, HIS41, GLU166 | ||||||
| 4 | 5R80 | 4c | 0.00 | -10.10 | 39.32 nM | hydrogen bonding | GLN189 |
| hydrophobic interaction | GLN 189, HIS41, PRO168, ALA191, GLU166 | ||||||
| 5 | 5R80 | 4d | 0.00 | -7.64 | 1.98 uM | hydrogen bonding | GLU166(2), GLN189 |
| hydrophobic interaction | MET165, MET49, GLU166, GLN 189, PRO168, ALA191 | ||||||
| 6 | 5R80 | 4e | 0.00 | -9.39 | 130.62 nM | hydrogen bonding | GLN189 |
| hydrophobic interaction | GLN 189, HIS41, PRO168, MET165, CYS145, GLU166 | ||||||
| 7 | 5R80 | 4f | 0.00 | -6.10 | 33.57 uM | hydrogen bonding | GLU166 (2), ASN142 |
| hydrophobic interaction | GLN 189, MET165, MET49, CYS145, GLU166, ASN142 | ||||||
| 8 | 5R80 | 4g | 1.64 | -9.74 | 435.66 nM | hydrogen bonding | GLN 189 |
| hydrophobic interaction | GLN 189, PRO168, HIS41, CYS145, GLU166, ARG188, LEU167 | ||||||
Fig. 11.
Binding site structure of SARS-CoV-2 main protease- 4f.
Analysis of the molecular docking results showed that the interactions within the active site of SARS-CoV-2 main protease were attributed to hydrogen bonds, hydrophobic and electrostatic attraction forces. Compounds 4b, 4d and 4f were the most stable compounds, they form a hydrogen bond with the Glu166 residue as the binding of the reference ligand. all compounds form other important hydrogen bonds with the residues Gln192 and Asn142. Compound 4c, which has the least binding energy (-10.10 kcal/mol) is most favorable, with the most interesting interaction inside the pocket. In addition, most of the compounds developed hydrophobic and electrostatic attraction forces and aromatic π–π stacking interactions with GLN 189, MET165 and His 41.
8. Materials and methods
8.1. General Information
All chemicals were purchased from Sigma-Aldrich and were of analytical grade. The used instruments are mentioned in the Supplementary Material file.
8.2. X-ray diffraction data measurement
Single crystals suitable for X-ray diffraction of the compound 4b were grown by slow evaporation of its ethanolic solution at room temperature. Crystal data of compound 4b were deposited in the CCDC Database Centre, with the deposition number 2151661. The crystallographic data and refinement information are mentioned in Supplementary Material Section 2.
8.3. Antimicrobial activity
The in vitro antibacterial and antifungal activities of all synthesized compounds were determined using the microdilution technique, according to procedures described in the literature [64,65], and following the guidelines of the Clinical and Laboratory Standards Institute (CLSI, approved standard M7-A8 and M27-A3) [66,67]. The bacterial and fungal strains used in this study were obtained from the Microbiology Laboratory, Faculty of Medicine and Pharmacy (FMP-Fez, Morocco). The products were tested against Gram-positive bacteria strains; Staphylococcus aureus, Gram-negative bacteria; Escherichia coli (ATB:57) B6N, Pseudomonas aeruginosa, Klebsiella pneumoniae and yeasts; Candida albicans ATCC10231 and Saccharomyces cerevisiae ATCC9763.
8.4. Antioxidant activity
The antioxidant activity of the target compounds was assessed using the DPPH radical scavenging assay following the protocol described in our previous work [31]. The detailed protocol is given in Supplementary Material Section 3.
In silico studies
The X-ray crystal structure of SARS-CoV-2 main protease (PDB ID: 5R80) was obtained from the Protein Data Bank [68]. All the selected molecules were drawn using 2D and 3D option of Chem. Draw Ultra 16.0.and saved in mol2 format. The three-dimensional structures of all compounds were performed using Maestro software, and prepared with Ligprep using Optimized Potentials for Liquid Simulation (OPLS3e) force field with a convergence of heavy atoms of 0.30 Å [69]. molecular docking study was performed to explore the binding mode of the prepared pyrazole-sulfonamide derivatives to the SARS-CoV-2 main protease, we have performed our studies with AUTODOCK 4.2 [70,71] using the improved force field. Docked compounds along with SARS-CoV-2 main protease protein were visualized on Discovery software. The structures of proteins Yersinia pestis Dihydropteroate Synthase DHPs (PDB ID: 3JQ9) was selected as a receptor for docking study that were prepared and energetically minimized using the Protein Preparation wizard protocol of the Schrodinger Suite [39].
9. Conclusion
In conclusion, in the present work, we report the synthesis, crystallographic, biological, and computational studies of the new pyrazolic compounds bearing sulfonamide moiety. The target compounds were obtained via a multi-step reaction sequence with an efficient and reliable strategy from 2-arylidene-indolin-3-ones (aza-aurones). The structures of the newly synthesized compounds were determined based on usual spectroscopic data (IR, 1H-NMR, 13C-NMR) and high-resolution mass spectrometry. In addition, an X-ray diffraction analysis was performed on the single crystals of compound 4b further confirms the structure of the target molecules. The target compounds of the two series 3a-h and 4a-g were evaluated for their antioxidant activity using DPPH assay, and antimicrobial activity in vitro through broth microdilution method. Among the tested products, compounds 3a, 3f, 4e, 4f, and 4g showed promising antimicrobial activity against selected pathogenic bacteria and yeasts. In addition, compounds 3a, 3c, 3e, 4a, 4d, 4f, and 4g exhibited good free radical scavenging ability. In addition, a docking study was performed on targeted compounds to support the experimental results, and showed high docking score and good binding energy with the target enzyme. Analysis of molecular docking results of pyrazole-sulfonamide derivatives 4a-g shows that most of these derivatives exhibit the same hydrophobic interaction of RZG ligand (referent) with the site of action SARS-CoV-2 main protease, this interaction similarity is due to the sulfonamide moiety. The antiviral activity presented by the good interaction of the pyrazole-sulfonamide derivatives with SARS-CoV-2 main protease target can be explained by the presence of hydrophobic interactions identical to those of the co-crystallized ligand and the interesting stability inside the target cavity with a binding energy varying between -6.10 and -10.10 (kcal/mol). Hence, the identified most active compound, by using Docking and POM Theory, may be considered as lead for further study in the search of novel pathogenic viruses’ inhibitory agent. The right formulation will be needed to improve the drug-like properties of these compounds, especially their lipophilicity and solubility.
ORCID identification
-
•
Mohammed CHALKHA: https://orcid.org/0000-0002-9400-2183.
-
•
Asmae NAKKABI: https://orcid.org/0000-0002-9845-243X.
-
•
Taibi BEN HADDA: https://orcid.org/0000-0002-5633-6203.
-
•
Malika BERREDJEM: https://orcid.org/0000-0002-4312-8771.
-
•
Abdelfattah EL MOUSSAOUI: https://orcid.org/0000-0001-5261-570X.
-
•
Mohamed BAKHOUCH: https://orcid.org/0000-0003-0732-6524.
-
•
Lahcen EL AMMARI: https://orcid.org/0000-0001-7980-974X.
-
•
Mohamed SAADI: https://orcid.org/0000-0003-2655-0230.
-
•
Faisal A. ALMALKI: https://orcid.org/0000-0003-4048-1526.
-
•
Violeta JEVTOVIC: https://orcid.org/ 0000-0002-2180-8303.s
-
•
Mohamed EL YAZIDI: https://orcid.org/ 0000-0002-2772-6632.
Credit author statement
Mohammed Chalkha and Asmae Nakkabi: Synthesis of molecules
Malika Berredjem, Mohamed El Yazidi and Taibi ben Hadda: Docking, POM, Conceptualization, Methodology, Writing- Reviewing and Editing Software.
Abdelfattah El Moussaoui and Mohamed Bakhouch: characterization of molecules
Hamid Laaroussi and Violeta Jevtovic: Biological Evaluation
Faisal A. Almalki: English language visualization.
Mohamed Saadi and Lahcen El Ammari: Theoretical study.
Declaration of Competing Interest
The authors declare that there is no conflict of interests.
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: 19-MED-1-01-0001 (KSA). We also thank the staff members of the "Cité de l'Innovation" of the Sidi Mohamed Ben Abdellah University (Morocco), and the “Centre Sciences des Matériaux”, Faculty of Sciences, Mohammed V University, Rabat. We also thank the General Directorate for Scientific Research and Technological Development (DG-RSDT), Algerian Ministry of Scientific Research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.133605.
Appendix. Supplementary materials
References
- 1.Morens D.M., Fauci A.S. Emerging Infectious Diseases: Threats to Human Health and Global Stability. PLoS Pathog. 2013;9:7–10. doi: 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/mmbr.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkey P.M., Jones A.M. The changing epidemiology of resistance. J. Antimicrob. Chemother. 2009;64:i3–i10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 5.Levine R.L., Stadtman E.R. Oxidative modifacation of proteins during aging. Exp. Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti B., Chakravarti D.N. Oxidative Modification of Proteins: Age-Related Changes. Gerontology. 2007;53:128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- 7.Cai H., Harrison D.G. Endothelial dysfunction in cardiovascular diseases: The role of oxidant stress. Circ. Res. 2000;87:840–844. doi: 10.1161/01.res.87.10.840. [DOI] [PubMed] [Google Scholar]
- 8.Valko M., Leibfritz D., Moncol J., Cronin M.T.D., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Boustany-Kari C.M., Jackson V.M., Gibbons C.P., Swick A.G. Leptin potentiates the anti-obesity effects of rimonabant. Eur. J. Pharmacol. 2011;658:270–276. doi: 10.1016/j.ejphar.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Wyde P.R., Gilbert B.E., Ambrose M.W. Comparison of the anti-respiratory syncytial virus activity and toxicity of papaverine hydrochloride and pyrazofurin in vitro and in vivo. Antiviral Res. 1989;11:15–26. doi: 10.1016/0166-3542(89)90017-X. [DOI] [PubMed] [Google Scholar]
- 11.Shingare R.M., Patil Y.S., Sangshetti J.N., Patil R.B., Rajani D.P., Rajani S.D., Madje B.R. Benzene sulfonamide pyrazole thio-oxadiazole hybrid as potential antimicrobial and antitubercular agents. Res. Chem. Intermed. 2018;44:4437–4453. doi: 10.1007/s11164-018-3396-y. [DOI] [Google Scholar]
- 12.Gulçin İ., Taslimi P. Sulfonamide Inhibitors: a Patent Review 2013- Present. Expert Opin. Ther. Pat. 2018;28:541–549. doi: 10.1080/13543776.2018.1487400. [DOI] [PubMed] [Google Scholar]
- 13.Badgujar J.R., More D.H., Meshram J.S. Synthesis, Antimicrobial and Antioxidant Activity of Pyrazole Based Sulfonamide Derivatives. Indian J. Microbiol. 2017;58:93–99. doi: 10.1007/s12088-017-0689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azab M.E., Youssef M.M., El-bordany E.A. Synthesis and Antibacterial Evaluation of Novel Heterocyclic Compounds Containing a Sulfonamido Moiety. Molecules. 2013;18:832–844. doi: 10.3390/molecules18010832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan G.S., Abou-Seri S.M., Kamel G., Ali M.M. Celecoxib analogs bearing benzofuran moiety as cyclooxygenase-2 inhibitors: Design, synthesis and evaluation as potential anti-inflammatory agents. Eur. J. Med. Chem. 2014;76:482–493. doi: 10.1016/j.ejmech.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Giancarlo G.M., Venkatakrishnan K., Granda B.W., Von Moltke L.L., Greenblatt D.J. Relative contributions of CYP2C9 and 2C19 to phenytoin 4-hydroxylation in vitro: Inhibition by sulfaphenazole, omeprazole, and ticlopidine. Eur. J. Clin. Pharmacol. 2001;57:31–36. doi: 10.1007/s002280100268. [DOI] [PubMed] [Google Scholar]
- 17.Keche A.P., Hatnapure G.D., Tale R.H., Rodge A.H., Kamble V.M. Synthesis, anti-inflammatory and antimicrobial evaluation of novel 1-acetyl-3,5-diaryl-4,5-dihydro (1H) pyrazole derivatives bearing urea, thiourea and sulfonamide moieties. Bioorganic Med. Chem. Lett. 2012;22:6611–6615. doi: 10.1016/j.bmcl.2012.08.118. [DOI] [PubMed] [Google Scholar]
- 18.Sharma P.K., Chandak N., Kumar P., Sharma C., Aneja K.R. Synthesis and biological evaluation of some 4-functionalized-pyrazoles as antimicrobial agents. Eur. J. Med. Chem. 2011;46:1425–1432. doi: 10.1016/j.ejmech.2011.01.060. [DOI] [PubMed] [Google Scholar]
- 19.Gedawy E.M., Kassab A.E., El Kerdawy A.M. Design, synthesis and biological evaluation of novel pyrazole sulfonamide derivatives as dual COX-2/5-LOX inhibitors. Eur. J. Med. Chem. 2020;189 doi: 10.1016/j.ejmech.2020.112066. [DOI] [PubMed] [Google Scholar]
- 20.Yamali C., Gul H.I., Ece A., Bua S., Angeli A., Sakagami H., Sahin E., Supuran C.T. Synthesis, biological evaluation and in silico modelling studies of 1,3,5-trisubstituted pyrazoles carrying benzenesulfonamide as potential anticancer agents and selective cancer-associated hCA IX isoenzyme inhibitors. Bioorg. Chem. 2019;92 doi: 10.1016/j.bioorg.2019.103222. [DOI] [PubMed] [Google Scholar]
- 21.Yamali C., Sakagami H., Uesawa Y., Kurosaki K., Satoh K., Masuda Y., Yokose S., Ece A., Bua S., Angeli A., Supuran C.T., Gul H.I. Comprehensive study on potent and selective carbonic anhydrase inhibitors: Synthesis, bioactivities and molecular modelling studies of 4-(3-(2-arylidenehydrazine-1-carbonyl)-5-(thiophen-2-yl)-1H-pyrazole-1-yl) benzenesulfonamides. Eur. J. Med. Chem. 2021;217 doi: 10.1016/j.ejmech.2021.113351. [DOI] [PubMed] [Google Scholar]
- 22.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alagöz M.A. New molecule design with in-silico methods for Covid-19 treatment. Bioorganic Med. Chem. Reports. 2020;2:32–40. doi: 10.25135/acg.bmcr.23.20.08.1773. [DOI] [Google Scholar]
- 24.Kanhed A.M., Patel D.V, Teli D.M., Patel N.R., Chhabria M.T., Ram M. Identification of potential Mpro inhibitors for the treatment of COVID‑19 by using systematic virtual screening approach. Mol. Divers. 2021;25:383–401. doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obakachi V.A., Kushwaha N.D., Kushwaha B., Mahlalela M.C. Design and Synthesis of Pyrazolone-based Compounds as Potent Blockers of SARS-CoV-2 Viral Entry into the Host Cells. J. Mol. Struct. 2021 doi: 10.1016/j.molstruc.2021.130665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negi M., Chawla P.A., Faruk A., Chawla V. Role of heterocyclic compounds in SARS and SARS CoV-2 pandemic. Bioorg. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elewa M., Mohamed M., Nafie M.S., Said M.M., Elgendy A., Aziz Y.M.A. Synthesis and in silico studies of novel pyrazole-based anti-MERS-CoV agents. Rec. Pharm. Biomed. Sci. 2021;5:121–134. doi: 10.21608/rpbs.2021.73630.1100. [DOI] [Google Scholar]
- 28.Dawood K.M., Abdel-Gawad H., Mohamed H.A., Badria F.A. Synthesis , anti-HSV-1, and cytotoxic activities of some new pyrazole- and isoxazole-based heterocycles. Med. Chem. Res. 2011;20:912–919. doi: 10.1007/s00044-010-9420-4. [DOI] [Google Scholar]
- 29.Scozzafava A., Owa T., Mastrolorenzo A., Supuran C.T. Anticancer and Antiviral Sulfonamides. Curr. Med. Chem. 2003;10:925–953. doi: 10.2174/0929867033457647. [DOI] [PubMed] [Google Scholar]
- 30.Chalkha M., Bakhouch M., Akhazzane M., Bourass M., Nicolas Y., Al Houari G., El Yazidi M. Design, synthesis and characterization of functionalized pyrazole derivatives bearing amide and sulfonamide moieties from aza-aurones. J. Chem. Sci. 2020;132:86. doi: 10.1007/s12039-020-01792-3. [DOI] [Google Scholar]
- 31.Chalkha M., El Moussaoui A., Ben Hadda T., Berredjem M., Bouzina A., Almalki F.A., Saghrouchni H., Bakhouch M., Saadi M., El Ammari L., Abdellatiif M.H., El Yazidi M. Crystallographic study, biological evaluation and DFT/POM/Docking analyses of pyrazole linked amide conjugates: Identification of antimicrobial and antitumor pharmacophore sites. J. Mol. Struct. 2022;1252 doi: 10.1016/j.molstruc.2021.131818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benharref A., El Ammari L., Saadi M., Oukhrib A., Berraho M. (1 S ,3 R ,8 R ,9 S ,10 R )-2,2-Dichloro-3,7,7,10-tetramethyltricyclo[6.4.0.0 1,3 ]dodecan-9-yl 4-methylbenzene-1-sulfonate. IUCrData. 2016;1 doi: 10.1107/s2414314616004223. [DOI] [Google Scholar]
- 33.Kouakou A., Rakib E.M., Abbassi N., Saadi M., El Ammari L. Ethyl 3-[7-(N-acetyl-4-methoxybenzenesulfonamido)-3-chloro-2H-indazol-2-yl] -propionate. Acta Crystallogr. Sect. E Struct. Reports Online. 2014;70:o307–o308. doi: 10.1107/S1600536814003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivakumar C., Balachandran V., Narayana B., Salian V.V., Revathi B., Shanmugapriya N., Vanasundari K. Molecular spectroscopic assembly of 3-(4-chlorophenyl)-5-[4-(propane-2-yl) phenyl] 4, 5-dihydro-1H pyrazole-1-carbothioamide, antimicrobial potential and molecular docking analysis. J. Mol. Struct. 2020;1210 doi: 10.1016/j.molstruc.2020.128005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rangaswamy J., Kumar H.V., Harini S.T., Naik N. Functionalized 3-(benzofuran-2-yl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazole scaffolds: A new class of antimicrobials and antioxidants. Arab. J. Chem. 2017;10:S2685–S2696. doi: 10.1016/j.arabjc.2013.10.012. [DOI] [Google Scholar]
- 36.Puthran D., Poojary B., Purushotham N., Harikrishna N., Nayak S.G., Kamat V. Synthesis of novel Schiff bases using 2-Amino-5-(3-fluoro-4-methoxyphenyl)thiophene-3-carbonitrile and 1,3-Disubstituted pyrazole-4-carboxaldehydes derivatives and their antimicrobial activity. Heliyon. 2019;5:e02233. doi: 10.1016/j.heliyon.2019.e02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamada N.M.M., Abdo N.Y.M. Synthesis, characterization, antimicrobial screening and free-radical scavenging activity of some novel substituted pyrazoles. Molecules. 2015;20:10468–10486. doi: 10.3390/molecules200610468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali S.A., Awad S.M., Said A.M., Mahgoub S., Taha H., Ahmed N.M. Design, synthesis, molecular modelling and biological evaluation of novel 3-(2-naphthyl)-1-phenyl-1H-pyrazole derivatives as potent antioxidants and 15-Lipoxygenase inhibitors. J. Enzyme Inhib. Med. Chem. 2020;35:847–863. doi: 10.1080/14756366.2020.1742116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., Shadrick W.R., Wallace M.J., Wu Y., Griffith E.C., Qi J., Yun M., White S.W., Lee R.E. Pterin–sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents. Bioorg. Med. Chem. Lett. 2016;26:3950–3954. doi: 10.1016/j.bmcl.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Faydy M., Dahaieh N., Ounine K., Rastija V., Almalki F., Jamalis J., Zarrouk A., Ben Hadda T., Lakhrissi B. Synthesis and antimicrobial activity evaluation of some new 7-substituted quinolin-8-ol derivatives: POM analyses, docking, and identification of antibacterial pharmacophore sites. Chem. Data Collect. 2021;31 doi: 10.1016/j.cdc.2020.100593. [DOI] [Google Scholar]
- 41.Rbaa M., Hichar A., Dohare P., Anouar E.H., Lakhrissi Y., Lakhrissi B., Berredjem M., Almalki F., Rastija V., Rajabi M., Ben Hadda T., Zarrouk A. Synthesis, Characterization, Biocomputational Modeling and Antibacterial Study of Novel Pyran Based on 8-Hydroxyquinoline, Arab. J. Sci. Eng. 2021;46:5533–5542. doi: 10.1007/s13369-020-05089-y. [DOI] [Google Scholar]
- 42.Bhat A.R., Dongre R.S., Almalki F.A., Berredjem M., Aissaoui M., Touzani R., Ben Hadda T., Akhter M.S. Synthesis, biological activity and POM/DFT/docking analyses of annulated pyrano[2,3-d]pyrimidine derivatives: Identification of antibacterial and antitumor pharmacophore sites. Bioorg. Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104480. [DOI] [PubMed] [Google Scholar]
- 43.Grib I., Berredjem M., Rachedi K.O., Djouad S.E., Bouacida S., Bahadi R., Ouk T.S., Kadri M., Ben Hadda T., Belhani B. Novel N-sulfonylphthalimides: Efficient synthesis, X-ray characterization, spectral investigations, POM analyses, DFT computations and antibacterial activity. J. Mol. Struct. 2020;1217:1–10. doi: 10.1016/j.molstruc.2020.128423. [DOI] [Google Scholar]
- 44.Rbaa M., Jabli S., Lakhrissi Y., Ouhssine M., Almalki F., Ben Hadda T., Messgo-Moumene S., Zarrouk A., Lakhrissi B. Synthesis, antibacterial properties and bioinformatics computational analyses of novel 8-hydroxyquinoline derivatives. Heliyon. 2019;5:e02689. doi: 10.1016/j.heliyon.2019.e02689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Titi A., Messali M., Alqurashy B.A., Touzani R., Shiga T., Oshio H., Fettouhi M., Rajabi M., Almalki F.A., Hadda T.Ben. Synthesis, characterization, X-Ray crystal study and bioctivities of pyrazole derivatives: Identification of antitumor, antifungal and antibacterial pharmacophore sites. J. Mol. Struct. 2020;1205 doi: 10.1016/j.molstruc.2019.127625. [DOI] [Google Scholar]
- 46.Rbaa M., Oubihi A., Anouar E.H., Ouhssine M., Almalki F., Ben Hadda T., Zarrouk A., Lakhrissi B. Synthesis of new heterocyclic systems oxazino derivatives of 8-Hydroxyquinoline: Drug design and POM analyses of substituent effects on their potential antibacterial properties. Chem. Data Collect. 2019;24 doi: 10.1016/j.cdc.2019.100306. [DOI] [Google Scholar]
- 47.Dongre R.S., Meshram J.S., Selokar R.S., Almalki F.A., Hadda T.Ben. Antibacterial activity of synthetic pyrido[2,3-d]pyrimidines armed with nitrile groups: POM analysis and identification of pharmacophore sites of nitriles as important pro-drugs. New J. Chem. 2018;42:15610–15617. doi: 10.1039/C8NJ02081G. [DOI] [Google Scholar]
- 48.Messali M., Aouad M.R., El-Sayed W.S., Ali A.A.S., Ben Hadda T., Hammouti B. New eco-friendly 1-alkyl-3-(4-phenoxybutyl) imidazolium-based ionic liquids derivatives: A green ultrasound-assisted synthesis, characterization, antibacterial activity and POM analyses. Molecules. 2014;19:11741–11759. doi: 10.3390/molecules190811741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben Hadda T., Khardli F.Z., Mimouni M., Daoudi M., Kerbal A., Salgado-Zamora H., Gandhare N., Parvez A., Lahsasni S. Impact of Geometric Parameters, Charge, and Lipophilicity on Bioactivity of Armed Quinoxaline, Benzothiaole, and Benzothiazine : POM Analyses of Antibacterial and Antifungal Activity, Phosphorus , Sulfur. Silicon Relat. Elem. 2014;189:753–761. doi: 10.1080/10426507.2013.855763. [DOI] [Google Scholar]
- 50.Mabkhot Y.N., Arfan M., Zgou H., Genc Z.K., Genc M., Rauf A., Bawazeer S., Hadda T.Ben. How to improve antifungal bioactivity: POM and DFT study of some chiral amides derivatives of diacetyl-L-tartaric acid and amines. Res. Chem. Intermed. 2016;42:8055–8068. doi: 10.1007/s11164-016-2578-8. [DOI] [Google Scholar]
- 51.Hatzade K., Sheikh J., Taile V., Ghatole A., Ingle V., Genc M., Lahsasni S., Hadda T.Ben. Antimicrobial/antioxidant activity and POM analyses of novel 7-o-β-D-glucopyranosyloxy-3-(4,5-disubstituted imidazol-2-yl)-4H-chromen-4-ones. Med. Chem. Res. 2015;24:2679–2693. doi: 10.1007/s00044-015-1326-8. [DOI] [Google Scholar]
- 52.Mabkhot Y.N., Aldawsari F.D., Al-Showiman S.S., Barakat A., Ben Hadda T., Mubarak M.S., Naz S., Ul-Haq Z., Rauf A. Synthesis, bioactivity, molecular docking and POM analyses of novel substituted thieno[2,3-b]thiophenes and related congeners. Molecules. 2015;20:1824–1841. doi: 10.3390/molecules20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rachedi K.O., Ouk T.S., Bahadi R., Bouzina A., Djouad S.E., Bechlem K., Zerrouki R., Ben Hadda T., Almalki F., Berredjem M. Synthesis, DFT and POM analyses of cytotoxicity activity of α-amidophosphonates derivatives: Identification of potential antiviral O,O-pharmacophore site. J. Mol. Struct. 2019;1197:196–203. doi: 10.1016/j.molstruc.2019.07.053. [DOI] [Google Scholar]
- 54.Ben Hadda T., Berredjem M., Almalki F.A., Rastija V., Jamalis J., Bin Emran T., Abu-Izneid T., Esharkawy E., Rodriguez L.C., Alqahtani A.M. How to face COVID-19: proposed treatments based on remdesivir and hydroxychloroquine in the presence of zinc sulfate. Docking/DFT/POM structural analysis. J. Biomol. Struct. Dyn. 2021;0:1–14. doi: 10.1080/07391102.2021.1930161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarrahpour A., Heiran R., Sinou V., Latour C., Bouktab L.D., Brunel J.M., Sheikh J., Hadda T.Ben. Synthesis of new β-lactams bearing the biologically important morpholine ring and POM analyses of their antimicrobial and antimalarial activities. Iran. J. Pharm. Res. 2019;18:34–48. doi: 10.22037/ijpr.2019.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alawadi D.Y., Saadeh H.A., Kaur H., Goyal K., Sehgal R., Ben Hadda T., ElSawy N.A., Mubarak M.S. Metronidazole derivatives as a new class of antiparasitic agents: synthesis, prediction of biological activity, and molecular properties. Med. Chem. Res. 2014;24:1196–1209. doi: 10.1007/s00044-014-1197-4. [DOI] [Google Scholar]
- 57.Ben Hadda T., Kerbal A., Bennani B., Al Houari G., Daoudi M., Leite A.C.L., Masand V.H., Jawarkar R.D., Charrouf Z. Molecular drug design, synthesis and pharmacophore site identification of spiroheterocyclic compounds: Trypanosoma crusi inhibiting studies. Med. Chem. Res. 2013;22:57–69. doi: 10.1007/s00044-012-0010-5. [DOI] [Google Scholar]
- 58.Berredjem M., Bouzina A., Bahadi R., Bouacida S., Rastija V., Djouad S.E., Sothea T.O., Almalki F.A., Ben Hadda T., Aissaoui M. Antitumor activity, X-Ray crystallography, in silico study of some-sulfamido-phosphonates. Identification of pharmacophore sites. J. Mol. Struct. 2022;1250 doi: 10.1016/j.molstruc.2021.131886. [DOI] [Google Scholar]
- 59.Farghaly T.A., Abbas I.M., Hassan W.M.I., Lotfy M.S., Al-Qurashi N.T., Hadda T.Ben. Structure Determination and Quantum Chemical Analysis of 1,3-Dipolar Cycloaddition of Nitrile Imines and New Dipolarophiles and POM Analyses of the Products as Potential Breast Cancer Inhibitors. Russ. J. Org. Chem. 2020;56:1258–1271. doi: 10.1134/S1070428020070210. [DOI] [Google Scholar]
- 60.Elsharkawy E.R., Almalki F., Ben Hadda T., Rastija V., Lafridi H., Zgou H. DFT calculations and POM analyses of cytotoxicity of some flavonoids from aerial parts of Cupressus sempervirens: Docking and identification of pharmacophore sites. Bioorg. Chem. 2020;100 doi: 10.1016/j.bioorg.2020.103850. [DOI] [PubMed] [Google Scholar]
- 61.Hakkou Z., Maciuk A., Leblais V., Bouanani N.E., Mekhfi H., Bnouham M., Aziz M., Ziyyat A., Rauf A., Ben Hadda T., Shaheen U., Patel S., Fischmeister R., Legssyer A. Antihypertensive and vasodilator effects of methanolic extract of Inula viscosa: Biological evaluation and POM analysis of cynarin, chlorogenic acid as potential hypertensive. Biomed. Pharmacother. 2017;93:62–69. doi: 10.1016/j.biopha.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 62.Mabkhot Y.N., Barakat A., Yousuf S., Choudhary M.I., Frey W., Ben Hadda T., Mubarak M.S. Substituted thieno[2,3-b]thiophenes and related congeners: Synthesis, β-glucuronidase inhibition activity, crystal structure, and POM analyses. Bioorganic Med. Chem. 2014;22:6715–6725. doi: 10.1016/j.bmc.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 63.Esharkawy E.R., Almalki F., Hadda T.Ben. In vitro potential antiviral SARS-CoV-19- activity of natural product thymohydroquinone and dithymoquinone from Nigella sativa. Bioorg. Chem. 2022;120 doi: 10.1016/j.bioorg.2021.105587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El Moussaoui A., Jawhari F.Z., Almehdi A.M., Elmsellem H., Fikri Benbrahim K., Bousta D., Bari A. Antibacterial, antifungal and antioxidant activity of total polyphenols of Withania frutescens.L. Bioorg. Chem. 2019;93 doi: 10.1016/j.bioorg.2019.103337. [DOI] [PubMed] [Google Scholar]
- 65.zahra Jawhari F., El Moussaoui A., Bourhia M., Imtara H., Saghrouchni H., Ammor K., Ouassou H., Elamine Y., Ullah R., Ezzeldin E., Mostafa G.A.E., Bari A. Anacyclus pyrethrum var. pyrethrum (L.) and Anacyclus pyrethrum var. depressus (Ball) Maire: Correlation between Total Phenolic and Flavonoid Contents with Antioxidant and Antimicrobial Activities of Chemically Characterized Extracts. Plants. 2021;10:149. doi: 10.3390/plants10010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institue; Wayne, PA: 2008. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard Third Edition. CLSI document M27-A3. [Google Scholar]
- 67.Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, PA: 2012. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Ninth Edition. CLSI document M07-A9. [Google Scholar]
- 68.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-damerell C., Aimon A., Ábrányi-balogh P., Brandão-neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keserü G.M., Klein H.F., Martin M.P., Noble M.E.M., Brien P.O., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., Von Delft F., Walsh M.A. screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Release S. Schrödinger, LLC; New York, NY: 2015. LigPrep. version 3.4.26400175. [Google Scholar]
- 70.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Software News and Updates AutoDock4 and AutoDockTools4 : Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santos-Martins D., Forli S., Ramos M.J., Olson A.J. AutoDock4 Zn An improved AutoDock forcefield for small-molecule docking to zinc metalloproteins. J. Chem. Inf. Model. 2014;54:2371–2379. doi: 10.1021/ci500209e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.