Abstract
In etiolated seedlings, red light (R) activates phytochrome and initiates signals that generate major changes at molecular and physiological levels. These changes include inhibition of hypocotyl growth and promotion of the growth of primary roots, apical hooks, and cotyledons. An earlier report showed that the sharp decrease in hypocotyl growth rapidly induced by R was accompanied by an equally rapid decrease in the transcript and protein levels of two closely related apyrases (APYs; nucleoside triphosphate-diphosphohydrolases) in Arabidopsis (Arabidopsis thaliana), APY1 and APY2, enzymes whose expression alters auxin transport and growth in seedlings. Here, we report that single knockouts of either APY inhibit R-induced promotion of the growth of primary roots, apical hooks, and cotyledons, and RNAi-induced suppression of APY1 expression in the background of apy2 inhibits R-induced apical hook opening. When R-irradiated primary roots and apical hook-cotyledons began to show a gradual increase in their growth relative to dark controls, they concurrently showed increased levels of APY protein, but in hook-cotyledon tissue, this occurred without parallel increases in their transcripts. In wild-type seedlings whose root growth is suppressed by the photosynthesis inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea, the R-induced increased APY expression in roots was also inhibited. In unirradiated plants, the constitutive expression of APY2 promoted both hook opening and changes in the transcript abundance of Small Auxin Upregulated RNA (SAUR), SAUR17 and SAUR50 that help mediate de-etiolation. These results provide evidence that the expression of APY1/APY2 is regulated by R and that APY1/APY2 participate in the signaling pathway by which phytochrome induces differential growth changes in different tissues of etiolated seedlings.
The expression of apyrase genes is needed for light to induce increased growth of primary roots, apical hooks, and cotyledons during de-etiolation of Arabidopsis seedlings.
Introduction
When seedlings emerge into the light after their growth in underground darkness, they undergo major changes in growth and development. Many of these changes are induced by the photoreceptor phytochrome, and they include inhibition of hypocotyl growth and promotion of the growth of primary roots, apical hooks, and cotyledons (Montgomery, 2016). Growth-regulating hormones play a key role in mediating light effects on de-etiolation (Halliday et al., 2009; Abbas et al., 2013). Early studies also implicated enzymes that regulate wall properties, such as peroxidases and polyamine oxidases, as important mediators of light-induced growth changes in seedlings (Laurenzi et al., 1999; Casal et al., 2006; Kim et al., 1989). More recently, apyrase (APY) enzymes, whose expression level strongly impacts both the transcript abundance of wall peroxidases (Lim et al., 2014) and auxin transport in seedlings (Liu et al., 2012a), have been implicated as potential mediators of phytochrome-induced growth inhibition changes in hypocotyls during de-etiolation (Wu et al., 2007). However, the question of whether APY also participates in those light-induced signaling changes that occur in tissues whose growth is promoted during de-etiolation remains unresolved.
APYs have been linked to growth control in Arabidopsis (Arabidopsis thaliana) (Meng et al., 2019) and diverse other plants, including, among others, peas (Pisum sativum) (Sharma et al., 2014), poplar (Populus euphratica) (Deng et al., 2015), potatoes (Solanum tuberosum) (Riewe et al., 2008), and soybeans (Glycine max) (Tanaka et al., 2011). In Arabidopsis, there is evidence to support a potential role for two specific APYs in growth promotion. Among seven APY genes in Arabidopsis, APY1 and APY2 are the ones most highly expressed in actively growing tissues, such as etiolated hypocotyls, pollen tubes, and elongation zones of roots (Wu et al., 2007). The suppression of their expression radically inhibits seedling growth (Wolf et al., 2007; Wu et al., 2007; Lim et al., 2014). These two APYs have 87% structural similarity (Wu et al., 2007), but ˂30% structural similarity to the other five APYs (Clark et al., 2014, 2021), and they can partially complement each other’s functions (Wu et al., 2007). However, the apy1apy2 double knockout is lethal, because pollen grains fail to germinate in homozygous lines (Steinebrunner et al., 2003).
When APY1 and APY2 expression is suppressed by RNAi in the inducible mutant R2-4A, polar auxin transport in hypocotyls and hypocotyl growth is both significantly inhibited (Liu et al., 2012a). In contrast, constitutive expression of APY1 promotes both auxin transport and growth in hypocotyls of Arabidopsis (Liu et al., 2012a). Growth promotion is also induced by the ectopic expression of a pea ectoapyrase in Arabidopsis and soybean, where it expands the root system architecture and enhances the biomass, drought tolerance, and seed yield of these plants (Veerappa et al., 2019).
The fact that both APY1 and APY2 have promoter elements known to be regulated by phytochrome (Wu et al., 2007) would predict that the expression of both genes could be modulated by this photoreceptor. In rapidly growing etiolated hypocotyls, which have high levels of APY1 and APY2 based on promoter-GUS assays (Wu et al., 2007), both the transcript and protein levels of APY1 and APY2 in whole seedings are suppressed within 15 min by the red light (R) activation of phytochrome, coincident with the rapid suppression of growth by R in hypocotyls. The R-induced changes in the transcript levels of APY1 and APY2 were separately assessed by RT-qPCR, and the R-induced changes in the combined protein levels of APY1/APY2 were evaluated by immunoblots, using highly specific polyclonal antibodies that recognized only APY1 and APY2, but no other Arabidopsis APYs (Wu et al., 2007). The R-induced suppression of APY expression and growth in hypocotyls does not occur in phytochromeA (phyA), phyB, or phyA/phyB mutants (Wu et al., 2007). Although the aerial portions of seedlings used for these experiments included apical hooks, cotyledons, and hypocotyls, in the short time frame assayed (˂30 min), only the hypocotyl growth rate changed significantly. An hour or more later, R induces the opening of apical hooks and increased growth of both cotyledons and roots.
These prior results were consistent with the hypothesis that the R-induced decrease in APY expression could, by inhibiting auxin transport, play an important role in mediating the R-induced decrease in hypocotyl growth. Because the constitutive expression of APY in seedlings enhances auxin transport in both hypocotyls and roots of seedlings (Liu et al., 2012a), this raises the question of whether R promotes the expression of APY1/2 in the primary roots, apical hooks, and cotyledons of seedlings when it induces their increased growth during de-etiolation, and whether these changes could play a role in the R-induced growth changes of these tissues.
In darkness, seedling resources are mainly allocated to the development of the hypocotyl, so its growth dominates, and, for seedlings germinating underground, this allocation would accelerate its emergence into light needed for autotrophic growth (van Gelderen et al., 2018). Root development in darkness has less priority, thus roots in etiolated seedlings are shorter compared with those grown in light (Dyachok et al., 2011). However, when the primary roots of etiolated plants are provided with exogenous sugar, they can initially grow faster while remaining in darkness than the roots of R-irradiated plants (Kircher and Schopfer, 2012). Thus, in roots of etiolated seedlings grown on sucrose, R induces an initial, transient, decrease in growth before it promotes a longer-term increase in growth (Correll and Kiss, 2005; Kircher and Schopfer, 2012), so the question of whether R induces kinetically parallel down-and-up changes in APY expression is pertinent. Answers to these questions would reveal whether the temporal association of R-induced changes in APY levels with R-induced changes in growth occurs not only in hypocotyls, whose growth is inhibited by R, but also in seedling tissues whose growth is ultimately stimulated by R, such as apical hooks, cotyledons, and roots. This report provides data to address these questions.
Results
R-induced increases in the growth of cotyledons and primary roots and in hook opening are inhibited in mutants suppressed in APY1 and APY2 expression
The kinetics of R-induced apical hook opening typically begins within 2 h after R treatment (Wang et al., 2009; Abbas et al., 2013). At the beginning of the R treatment (time 0), the apical hook angles of wild-type (WT) and mutant seedlings were not significantly different, but after 4 h of R, the apical hook opening of the estradiol-induced R2-4A seedlings, which was null for APY2, and, when induced by estradiol, showed 70% suppression in APY1 expression, was significantly less than that of Wassilewskija (Ws) seedlings. After 6 h of R, the apical hook opening of these mutants was significantly less than that of Ws seedlings, even if the mutant seedlings were not treated with estradiol, that is, were suppressed only in APY2 expression (Figure 1A).
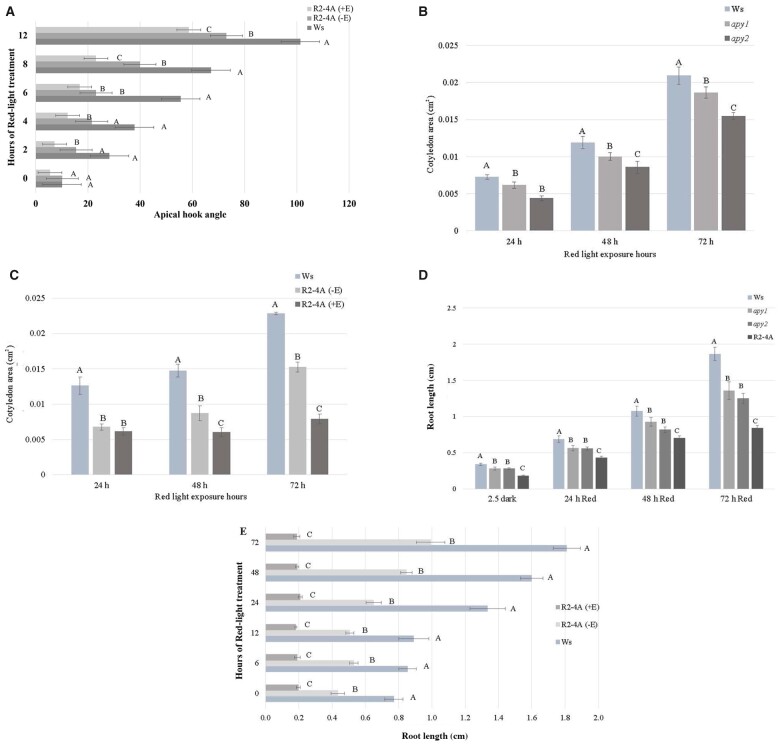
Figure 1.
Suppression of APY expression impairs R-induced hook opening, cotyledon expansion, and root growth. A, Apical hook opening of 3.5-day-old dark-grown seedlings, 2, 4, 6, 8, and 12 h after R treatment of WT Ws, (−E) R2-4A mutants without estradiol, and (+E) R2-4A mutants with estradiol. B, Increase in the growth of cotyledons of 2.5-day-old, etiolated seedlings induced by continuous R (30 mol m−2 s−1) is significantly inhibited in apy1 and apy2. Cotyledon area was measured at 24 h time intervals. C, Cotyledon area measurements with estradiol inducer added to R2-4A mutants at the beginning of the 3.5 days dark period and imaged 1, 2, and 3 days after R, (−E) R2-4A mutants without estradiol, and (+E) R2-4A mutants with estradiol. D, Root length measured at 24-h time intervals. Estradiol inducer was added to R2-4A mutants to suppress APY1 expression by RNAi after the 2.5 days dark period. E, Estradiol inducer is added to R2-4A mutants (+E) at the beginning of the 3.5 days dark period, maximal suppression of APY1 is evident when R treatment begins at the end of the dark period, and R does not induce any increase in primary root growth thereafter. For all panels (A–E), shown is a representative result that was observed in samples independently grown on three separate plates, each of which had an n value of at least 7, and different letters above the bars indicate values that differ significantly from each other (P < 0.05, Student’s t test, error bars are S.E.).
Cotyledon area approximately doubled between 24 and 48 h after R, and continued to increase from 48 to 72 h. At all-time points after R treatment, the cotyledon areas of apy1 and apy2 were significantly less than that of Ws WT seedlings (Figure 1B), and this growth inhibition was even more severe in R2-4A mutants treated with estradiol (i.e. suppressed in both APY1 and APY2 expression) (Figure 1C). After one day of estradiol treatment, the cotyledon area of the R2-4A mutants was not statistically significant from that of those not treated with estradiol, however, the magnitude of the growth suppression continued to increase with estradiol treatment from 1 to 3 days after R, and the difference became statistically significant by 48 h (Figure 1C).
Prior studies revealed that the suppression of APY1 and APY2 in R2-4A mutants severely inhibited the growth of primary roots (Wu et al., 2007; Lim et al., 2014), but Figure 1D shows that root growth can also be inhibited by single knockouts, either apy1 or apy2, though the inhibition is less severe in these mutants than in the estradiol-treated R2-4A mutants. All mutant seedlings grown in dark and R had root lengths that were significantly shorter compared with the Ws WT seedlings at the time point assayed (Figure 1D).
In the R2-4A conditional mutants, the estradiol treatment can induce the RNAi-mediated knock down of APY1 expression up to 70%, but it takes ∼3 days to reach this level of suppression (Lim et al., 2014), so some root growth continued when the inducer was not added until the end of the 2.5 day dark period of seed incubation in the cold to synchronize germination (Figure 1D). However, when the estradiol inducer was added at the start of the 2.5 days dark period, its suppressive effect on root growth of R2-4A mutants was already strong at the start of irradiation, and thereafter R was unable to induce any significant increase in root growth compared to root growth in darkness (Figure 1E).
In primary roots the timing of R-induced changes in root growth approximately coincides with R-induced changes in the level of APY protein
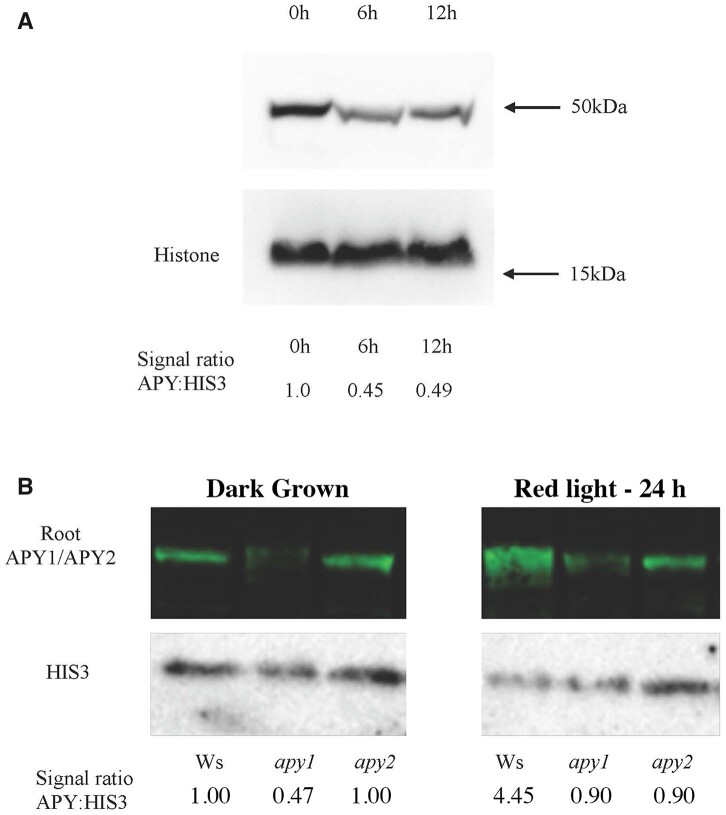
In contrast to the rapid decrease in hypocotyl growth induced by R, which is detectable in 8 min (Parks and Spalding, 1999), the R-induced promotion of the growth of root and hook-cotyledon tissues of seedlings grown on sucrose takes longer to become statistically significant (Correll and Kiss, 2005; Kircher and Schopfer, 2012; Dong et al., 2019). When etiolated seedlings grown on 1% sucrose are irradiated with R, their root growth is initially slower than their growth in darkness for several hours (Correll and Kiss, 2005; Supplemental Figure S1A), but then, as the light treatment is extended, root growth is promoted (Kircher and Schopfer, 2012) and the growth rates of the irradiated roots gradually increase, in parallel with increased sucrose supply from photosynthesizing cotyledons (Kircher and Schopfer, 2012). Consequently, between 12 and 24 h of irradiation, the rate of root growth increases by 2.5-fold, even more than their increased rate in darkness (2.1-fold), so that by 24 h of growth in R, the length of roots becomes equal to that of dark-grown roots (Supplemental Figure S1B). Correspondingly, as estimated by stain quantification of immunoblots, APY protein levels in Ws WT primary roots are lower after 6 h of R (Figure 2A), and higher after 24 h of R (Figure 2B) compared with initial (zero-time) levels.
Figure 2.
R induces changes in the level of APY protein in primary roots. R induces a decrease in APY protein expression in primary roots at early time points (A), and an increase 24 h after R exposure (B). After seedlings were grown for 2.5 days in darkness, they were irradiated with continuous R for 6, and 12 h (A), or maintained in darkness or irradiated with continuous R for 24 h (B). APY1 and APY2 levels were detected by anti-APY antibody, and the same blot was stripped and re-probed with anti-HIS3 antibody for equal loading control. Densitometry measurements of APY and HIS3 levels were measured in cotyledons (A), or in roots (B) by ImageJ.
In the roots of plants that continued to grow in the dark for 24 h, much more of the immunosignal was lost in apy1 than in apy2 (Figure 2B), indicating there is more APY1 than APY2 in WT roots. This finding is consistent with the report of Steinebrunner et al. (2000), who showed that in the primary roots of light-grown seedlings, the mRNA level of APY1 is greater than that of APY2. After 24 h of R, the immunosignal in roots was still evident in both apy1 and apy2 single knock-out mutants, but was barely detectable in apy1, indicating that most of the enhancement of APY expression in WT roots was due to an increase in the level of this APY. The polyclonal antibodies used for these immunoblots were raised against APY peptides, and they recognized APY1 and APY2, which are 87% identical in primary sequence (Steinebrunner et al., 2000).
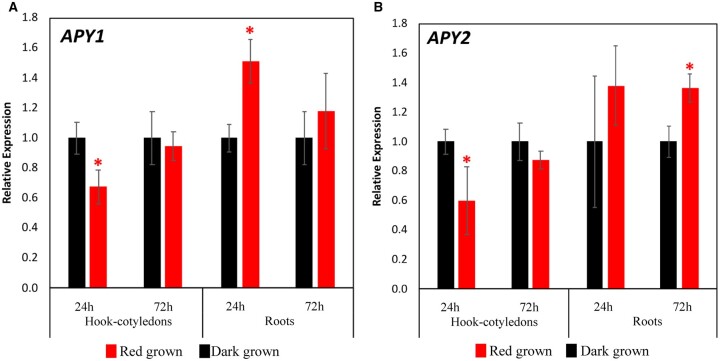
As measured by RT-qPCR, transcriptional changes induced by R in primary roots, like R-induced growth changes, also take many hours to develop. No statistically significant changes in the transcript levels of either APY1 or APY2 were observed after 6 or 12 h of irradiation (Supplemental Figure S2A), but these levels became 60% higher for APY1 (P < 0.05) and 40% higher for APY2 after 24 h of R relative to those of dark-grown roots (Figure 3, A and B).
Figure 3.
R induces changes in the abundance of APY transcripts in roots and hook-cotyledon tissues. R induces changes in the transcript levels of APY1 (A) and APY2 (B) in hook-cotyledon and root tissues of dark-grown Ws WT seedlings after 24 and 72 h of continuous light treatment. Respective dark samples at 24 and 72 h time points are used as calibrators. Experiment performed with three biological repeats, each with three technical replicates. Error bars represent the se. Student’s t test was applied. * Denotes significantly different expression from calibrator (dark) at P < 0.05.
In apical hook-cotyledon tissue, the timing of R-induced promotion of cotyledon expansion is paralleled by R-induced changes in the levels of APY protein, but not paralleled by R-induced changes in APY transcripts
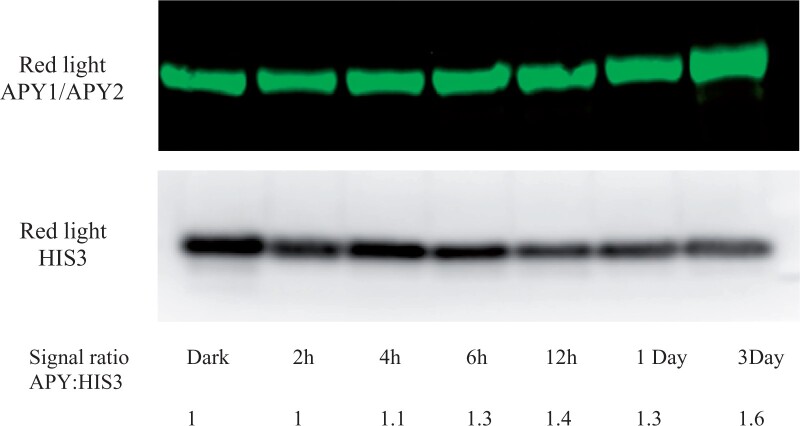
According to Dong et al. (2019) the kinetics of R-induced cotyledon expansion is, overall, slower than that of hook opening, but both growth enhancements become significant by 6 h of irradiation. As estimated by immunoblot, the APY protein level in apical hook-cotyledon tissues of R-treated samples begins to increase over that in dark-grown seedlings by 4–6 h of irradiation, and increases by >60% by 3 days (Figure 4).
Figure 4.
R induces an increase in the level of APY protein in apical hook-cotyledon tissue. The increase in the level of APY protein induced by R in apical hook-cotyledons is gradual, rising to consistently 1.3-fold or higher at and after 6 h of R irradiation. After Ws WT seedlings were grown for 2.5 days in darkness, they were irradiated with continuous R for 2, 4, 6,12, 24, and 72 h. APY1 and APY2 levels were detected by anti-APY antibody, and the same blot was stripped and re-probed with anti-HIS3 antibody for equal loading control. Densitometry measurements of APY and HIS3 levels were measured by ImageJ.
Although the increase in APY protein induced by R within 24 h in primary roots was paralleled by an increase in transcript abundance of APY1 and APY2, these increases did not occur together in hook-cotyledon tissue. After R, the transcript level of APY1 and APY2 remains the same or is lower than the level in the hook-cotyledon tissue of dark-grown seedlings after 6 h (Supplemental Figure S2B) and after 24 and 72 h of irradiation (Figure 3, A and B).
Inhibition of photosynthesis blocks the R-induced increase in APY1 and APY2 protein levels in Ws
To further test the linkage between APY expression and growth, Ws WT seedlings grown without sucrose were treated with DCMU, which blocks the R-induced increase in photosynthesis, thereby reducing the sucrose supply from the photosynthetic cotyledons to the root and blocking the promotion of root growth induced by light (Kircher and Schopfer, 2012). Whereas Ws WT roots grown in darkness for 24 h had lower APY expression compared with R-irradiated roots that were not treated with DCMU, they had a similar expression level to R-irradiated roots that had been treated with DCMU (Figure 5A). Thus, the blockage of R-induced root growth by DCMU in seedlings grown without sucrose also blocked the R-induced increase in APY protein level in roots. In seedlings grown without sucrose, 24 h R significantly promotes root growth in Ws WT seedlings. In apy1 and apy2 mutants, R can still significantly increase the growth of roots relative to their growth in the dark, but the overall growth of roots in both R-treated and dark-grown seedlings is significantly less relative to that observed in Ws WT seedlings (Figure 5B).
Figure 5.
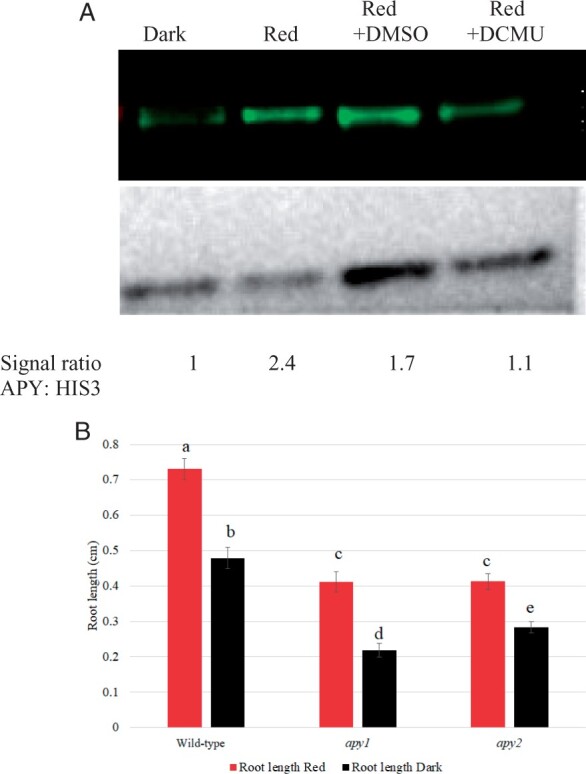
On media without sucrose added, R does not induce a significant increase in APY protein levels if seedling photosynthesis is inhibited by DCMU, and R significantly promotes root growth A, Ws WT seedlings grown in dark, or transferred to continuous R after 2.5 days growth in dark, were grown under R with no treatment, with DMSO (0.001%V/V) or with 50 μM DCMU dissolved in DMSO (0.001% V/V) for 72 h. APYl and APY2 levels were detected by anti-APY antibody, and the same blot was stripped and reprobed with anti-HIS3 antibody for equal loading control. Densitometry measurements of APY and HIS3 levels were measured by ImageJ, and the signal ratio is expressed relative to the dark sample. B, The length of roots in both R-treated and dark-grown apy1 and apy2 seedlings is significantly less than that observed in Ws WT seedlings. Shown is a representative result that was observed in samples independently grown on three separate plates, each of which had an n value of 10. The R treatment in both (A) and (B) was for 24 h. Different letters above the bars indicate values that differ significantly from each other (P < 0.05, Student’s t test; error bars are se).
Constitutive expression of APY1 or APY2 promotes hook opening
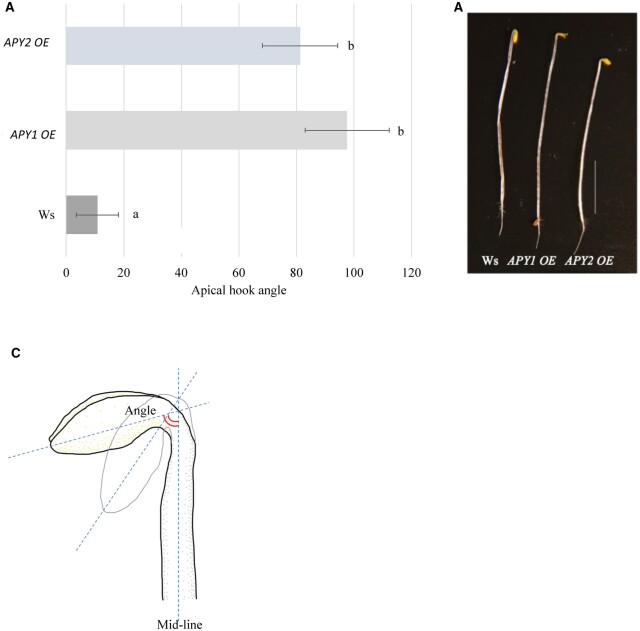
When Apy1 OE and Apy2 OE seedlings were grown in darkness for 2.5 days without sucrose in the medium, apical hooks were more open compared with the WT, as if they had been stimulated by R (Figure 6). Both Apy1 OE and Apy2 OE seedlings had significantly increased apical hook angles compared to the WT seedlings, showing an eight-fold increase in apical hook angle.
Figure 6.
Constitutive expression of APY1 or APY2 promote hook opening in etiolated seedlings. A, 2.5-day-old dark-grown APY1 OE and APY2 OE seedlings without sucrose in the growth medium show significant apical hook opening without the R light stimulus. Shown is a representative result that was observed in samples independently grown on three separate plates, each of which had an n value of 11. The R treatment in both (A) and (B) was for 24 h. Different letters above the bars indicate values that differ significantly from each other (P < 0.05, Student’s t test; error bars are se). B, Representative WT Ws, APY1 OE and APY2 OE seedlings. Bar 5 mm. C, Illustration of an A. thaliana apical hook measurement from the mid-line to the tip of the cotyledon.
In dark-grown seedlings the constitutive expression of APY2 induces changes in the transcript abundance of two SAUR genes that help mediate the de-etiolation of apical hook-cotyledon tissue
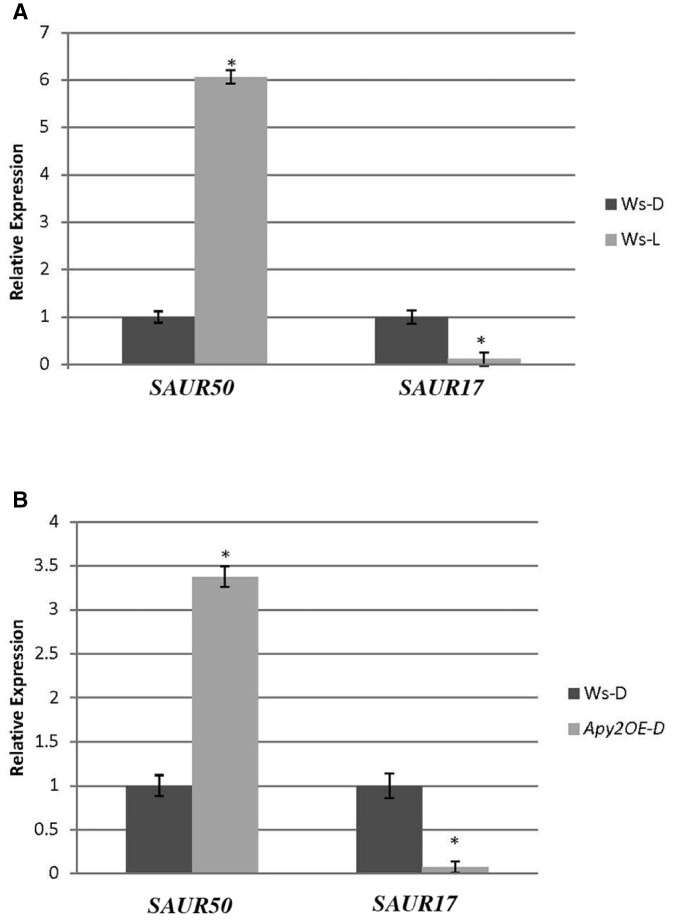
In Col-0, the white-light induced de-etiolation of hook-cotyledon tissue is critically linked to its induction of major changes in the transcript abundance of two Small Auxin Upregulated (SAUR) genes: SAUR17 is downregulated, which promotes the upregulation of SAUR50 (Wang et al., 2020), and the upregulation of SAUR50 is needed for light to promote both hook opening and cotyledon expansion (Sun et al., 2016; Dong et al., 2019). We tested whether light-induced similar changes in the transcript abundance of SAUR17 and SAUR50 in the hook-cotyledon tissue of Ws WT and whether the promotion of hook opening in darkness that was observed in seedlings that constitutively expressed APY2 (Figure 6) was accompanied by the downregulation of SAUR 17 and the upregulation of SAUR50 in hook-cotyledon tissue of APY2 OE mutants. In Ws, 3 h of white light decreased the transcript abundance of SAUR17 by nine-fold, and it increased the abundance of SAUR50 in hook-cotyledon tissue by six-fold (Figure 7A). In the absence of light treatment, the constitutive expression of APY2 in etiolated seedlings downregulated the abundance of SAUR17 by 12-fold and upregulated SAUR50 by 3.4-fold (P < 0.01) (Figure 7B). The suppression of APY in estradiol-treated R2-4A mutants, which inhibits hook opening and cotyledon expansion (Figure 1, A and C), significantly inhibited SAUR50 expression in the hook cotyledon tissue of etiolated seedlings (Supplemental Figure S3).
Figure 7.
Constitutive expression of APY2 promotes light-induced changes in SAUR50 and SAUR17 gene expression in the hook-cotyledon tissue of 4-day-old, etiolated seedlings. Relative expression levels of SAUR50 and SAUR17 (A) WT Ws in dark versus after 3 h of white light. B, WT Ws in dark versus APY2 OE in dark. The data represent average of 4–10 technical replicates of RNA extracted from tissue pooled from five independently grown plates of seedlings. * Denotes significantly different expression from the WT Ws expression level calibrator (P < 0.05, Student’s t test, error bars are se).
Note on normal distribution of growth data
For all the growth data in Figures 1, 5, and 6, tests for normal distribution were performed using the Shapiro–Wilk test. More than 90% of the samples were normally distributed. Variations to the normal distribution observed in some of these samples would be expected. For example, in the apical hook angle measurements (Figure 1A), at the zero-hour time point for the R2-4A (−E/+E) samples, there was a substantial numerical difference between the “closed” and “opened” stages within samples, which would affect normal distribution.
Discussion
The results presented provide data to support the conclusion that APY1 and APY2 play major roles in mediating light-induced promotion of the growth of roots and hook-cotyledon tissue in etiolated seedlings. Key among these data is the results showing that R-induced promotion of root growth is significantly impaired in both apy1 and apy2 mutants, whether the seedlings are grown with or without sucrose, and that suppression of APY expression inhibits both R-induced hook opening and cotyledon expansion. Overall, these results support the conclusion that the expression of both APYs is needed for the full growth promotive effects of R on the root and hook-cotyledon tissues of etiolated seedlings.
In the earlier report of Wu et al. (2007), R-induced suppression of hypocotyl growth was paralleled by the simultaneous suppression of APY1 and APY2 expression, but that report could not address whether APY expression was needed for this growth inhibitory response to R, because hypocotyls of seedlings suppressed in APY expression have impaired growth both in darkness and in light. In contrast, the current report shows that APY expression is needed for the growth promotive effects of R in etiolated seedlings, because in those tissues in which R-induces an increase in growth (roots and hook-cotyledons), this response to R will not occur without substantial expression levels of APY1 and APY2.
To test the need of APY1 and APY2 expression for R-induced changes in seedling development, RNAi-induced knock-downs of APY expression were used instead of apy1apy2 nulls, because pollen null for both these APYs does not germinate (Steinebrunner et al., 2003). Moreover, in conditional mutants, in which transient complementation of apy1apy2 with either APY1 or APY2 only in pollen allows pollen germination and seed formation, by the time the germinated seedlings emerge they have no APY1 or APY2 expression, and their further growth in darkness or in light is totally blocked. These apy1apy2 seedlings die without progressing past this stage (Wolf et al., 2007). The current report, unlike Wu et al. (2007) and Wolf et al. (2007), demonstrates that both apy1 and apy2 single mutants inhibit the R-induced promotion of root and hook-cotyledon growth, although suppressing APY1 expression in the background of an apy2 null mutant inhibits this promotion even more.
Further supporting the important role of APY expression in R-induced de-etiolation responses is the fact that R upregulates APY protein expression in roots and hook-cotyledon tissue at close to the same time it upregulates the growth of these tissues. Here there is an interesting contrast with the results in Wu et al. (2007). In that earlier report, the R-induced inhibition of hypocotyl growth is several-fold and occurs very rapidly (<10 min), whereas, under the same growth conditions, the R-induced promotion of root and hook-cotyledon growth begins to occur only after multi-hour delays and remains less than two-fold for days. Correspondingly, the R-induced changes in APY levels in these more slowly responding tissues are smaller and take longer to become significant.
In roots, the R-induced increase of both transcript and protein abundance of APY is temporally synchronous with its promotion of growth, both occurring after 24 h of irradiation. In hook-cotyledon tissue, however, only the R control of APY protein levels occurs at the same time as when it promotes the expansion of this tissue. Whether this control is due to R regulation of the translation or the turnover rate of APY remains to be determined, although phytochrome control of gene expression via its control primarily of translation is well documented (Paik et al., 2012).
The promoter regions of APY1 and APY2 have phytochrome-regulated elements (Wu et al., 2007), including multiple PIF binding E-box motifs (Leviar and Monte, 2014), and these promoter elements help explain the R-induced changes in the transcription of these APYs in roots. However, as noted above, light affects primarily APY protein levels in hook-cotyledon tissue, and its effects on transcript levels of APY1 and APY2 in this tissue do not parallel its growth effects.
The R-induction of protein changes in etiolated seedlings without corresponding changes in the level of transcripts that encode those proteins has been frequently observed (Cheng et al., 2021). As documented by Liu et al. (2012b) almost 40% of the messages that show a light-induced increase in their translation do not have a corresponding increase in their transcript level. The effects of light on the translation of many proteins during seedling de-etiolation are mainly due to its reversal of the inhibitory effects of cytoplasmic processing bodies on the translation of the transcripts they contain, and this translation de-repression can occur without an increase in the level of these transcripts (Jang et al., 2019).
In Arabidopsis seedlings, light stimuli help control the level and transport of auxin, which, in turn, are needed for light-regulated changes in their photomorphogenic growth (Halliday et al., 2009).
Relatedly, the earlier report of Liu et al. (2012a) documented that the constitutive expression of APY1 and APY2 promotes auxin transport. Thus, the upregulation of these two APYs by light could be a key step in the signaling pathway by which photoactivated phytochrome promotes auxin transport, cotyledon expansion, apical hook opening, and primary root elongation. Correspondingly, the mechanism by which the inhibition of APY1 and APY2 expression by RNAi blocks R-induced promotion of cotyledon expansion, apical hook opening, and enhanced root growth likely involves its inhibition of auxin transport.
Given that auxin transport is critical for apical hook opening (Abbas et al., 2013), our observation that the suppression of APY expression blocks hook opening would be an expected result. However, hook opening requires an asymmetric distribution of auxin, so it is less clear why the promotion of polar auxin transport that is induced by the constitutive expression of APY would, by itself, promote the hook opening observed in dark-grown seedlings (Figure 7), unless that promotion was asymmetric across the two sides of the hook. Additional studies would be needed to resolve this question.
Of course, the effects of R on seedling de-etiolation require more than auxin-mediated growth changes, just as the effects of APY on growth involve more than just its effects on auxin transport. Thus, the impact of APY expression on auxin transport would be only one of many intersecting pathways that help it transduce the effects of R on tissue growth changes in seedlings. As revealed in Supplemental Table S1, another impact of changes in APY expression that could lead to growth changes would be its effect on the expression of peroxidases that help control wall extensibility. In the combined shoot and root tissues of Arabidopsis seedlings grown in the light, the suppression of APY1 and APY2 expression results in significantly higher transcript levels of three type III wall peroxidases that increase wall cross-links (Lim et al., 2014), while the over-expression of APY2 decreases the transcript abundance of these same peroxidases (Supplemental Table S1).
The effects of R on the growth of the primary roots of seedlings are mediated both by phytochrome and by photosynthesis. Kircher and Schopfer (2012) showed that R-induced root growth is driven mainly by sucrose availability transported from photosynthesizing cotyledons to the root, and that media sucrose can maintain strong growth of the root even in darkness. Yet, when seedlings are grown on sucrose, R induces a transient, slight decrease in root growth within 60–90 min, and the irradiated roots continue to be shorter than dark-grown roots for several hours (Correll and Kiss, 2005). They do not reach the same length as those grown in darkness until after ∼18–24 h of irradiation (Supplemental Figure S1B). The fact that both the transient decreased growth of roots and their ultimate increased growth occur close to coincident with parallel changes in R-induced changes in APY levels further establishes the regulatory link between APY protein expression and photomorphogenesis.
Another gene whose transcript abundance is rapidly increased by R is SAUR50, and its enhanced expression has been shown to be a key intermediate step in the molecular changes induced by R that result in de-etiolation, including the opening of apical hooks (Sun et al., 2016; Dong et al., 2019). Conversely, SAUR17 levels are high in etiolated apical hooks and cotyledons of WT seedlings, and its rapid downregulation by light helps promote the light-induced upregulation of SAUR50 expression (Wang et al., 2020). Thus, our finding that the OE of APY2 suppressed SAUR17 expression likely helps explain why it also promoted SAUR50 expression. Although R does not induce an increase in APY transcript abundance in hook-cotyledon tissue, it does induce an increase in APY protein abundance within 6 h. Taken together, these results reveal that the link between APY protein expression and R-induced de-etiolation responses in hook-cotyledon tissue extends also to R-induced gene expression changes in this tissue.
So as to fairly compare the results in this report with those in the earlier study of Wu et al. (2007), most of the assays reported here were carried out on seedlings grown on media containing 1% sucrose. When seedlings are grown without sucrose, they depend on seed food storage and photosynthetically derived sugar for growth, and their growth in light can be arrested by the photosynthesis inhibitor, DCMU (Kurata and Yamamoto, 1997). The fact that this inhibition by DCMU arrests R-stimulated root growth, and this growth change in roots coincides with their lower level of APY, further links root growth changes with changes in their levels of APY1/APY2 proteins (Figure 5A). These results also imply that the R-induced increase in APY expression depends on an R-induced increase in growth. Even without the addition of DCMU, the full promotion of the growth of roots by 24 h of R on media without sugar requires the expression of both APY1 and APY2, because on this media the R-induced increase in root growth is significantly less in apy1 and apy2 mutants.
In R2-4A seedlings, the RNAi-induced suppression of APY1 expression occurs gradually, falling to ∼30% of WT levels in whole seedlings by 3.5 days after estradiol treatment (Lim et al., 2014). Correspondingly, the full growth inhibitory effects of suppressing APY expression in roots did not occur until 3 days after estradiol treatment (Figure 1D). However, if the estradiol inducer was included during the 3-day dark incubation before the R treatment began, then the blockage of R enhancement of root growth was evident even at the first 6-h time point assayed (Figure 1E). Thus, the level of APY1/APY2 suppression in mutants corresponds closely with how much R-induced growth changes are inhibited in these mutants.
As reviewed in Clark and Roux (2018) and discussed in Veerappa et al. (2019), current evidence indicates that APY1 and APY2 could function in the Golgi, in the nucleus, or on the plasma membrane, where it could control the concentration of extracellular ATP. Our report does not address in which subcellular compartment(s) APY1 and APY2 would be functioning to impact seedling growth. Although RNAi-induced suppression of APY1/APY2 expression significantly alters the expression of genes that control growth (Lim et al., 2014), this effect could result from APY functions in either internal organelles or on the plasma membrane (Veerappa et al., 2019).
In this study, the association of APY expression with R-induced growth in early seedling development has been shown in three different ways. Suppression of APY1 and APY2 expression impairs R-induced growth changes in roots, cotyledons, and apical hooks. R-induces an increase in APY1 and APY2 levels in roots and apical hook-cotyledon tissues, and this increase occurs before or near the same time as R-induced growth promotion of these tissues, Constitutive expression of APY1 and APY2 stimulate the R-induced hook opening and R-induced SAUR 50 expression in etiolated tissues even without the R stimulus. The fact that the constitutive expression of neither APY1 nor APY2 could duplicate the effects of R on cotyledon expansion or on primary root growth clearly shows that enhanced expression of these APYs alone does not suffice to mediate the R-induced growth changes of these tissues in etiolated seedings.
Overall, the combined results of this study support the hypothesis that in the signaling pathway between R activation of phytochrome and the increase of root and cotyledon growth during early stages of seedling de-etiolation, the light-induced expression of APY1 and APY2 genes in these tissues is both required and occurs close to when the growth changes occur. They are also consistent with the hypothesis proposed by Wu et al. (2007) that the suppression of APY1 and APY2 expression is a key step in the R-induced suppression of hypocotyl growth during seedling de-etiolation.
Materials and methods
Plant materials and growth conditions
In this study Arabidopsis (A. thaliana) WT Ws was used in all the experiments unless otherwise mentioned. R2-4A, apy1, and apy2 mutants were used in RNA, protein and growth assays, and APY1OE and APY2OE mutants were used in Figures 6 and 7 and Supplemental Table S1. Seeds were sterilized with 40% (V/V) bleach for 8.5 min and washed thoroughly with sterile deionized water for five times. Seeds were stratified at 4°C for 48 h and grown on solid Murashige Skoog (MS) medium with 0.8% (W/V) plant agar and 1% (W/V) sucrose. Growth assays performed with APY1 OE and APY2 OE in Figure 6 were grown in dark on solid MS medium with 0.8% (W/V) plant agar without sucrose. After their stratification at 4°C, the seeded plates were light treated for 2 h induce germination, then grown upright in darkness at 22°C until light treatments began at 2.5 days. After 2.5 days of growth in darkness, R-treated plants were grown under 30 μmol m−2 s−1 R for the time mentioned in the figures. For growth measurements, seedlings were grown on the same plate. Seeds were planted on three different plates, and in in several rows. Seedlings were randomly collected from different rows for all growth measurement assays. Once the seedlings were harvested, plates were placed back in the growth chamber until the next time point. The lengths of their primary roots were measured after different time periods of continuous R, as designated in the Figure legends. The cotyledon areas were measured after 2 h, 4 h, 6 h, 1 day, 2 days, and 3 days after continuous R. The angle of hook opening was measured in R2-4A mutants at 2, 4, and 6 h after continuous R, and was measured in APY1OE and APY2OE seedlings after 2.5 days of dark growth.
The R2-4A mutant has an estradiol-inducible RNAi construct (Wu et al., 2007). It is null for APY2, and 2 days after treatment with estradiol its expression of APY1 is suppressed ∼70% by RNAi. These mutants were grown directly on MS plates with 4 μM estradiol after 48 h stratification and 2 h light treatment or grown 2.5 days on MS medium without estradiol and transfer to MS plates with 4 μM estradiol before the R treatment.
Growth measurements and statistical analysis
For the growth rate assays, plants were immediately photographed in a black background under white light. Seedling measurements were taken using ImageJ software. For the hook angle measurements, a midline was drawn in the hypocotyl and the cotyledon tip, and the hook angle was measured. For all the growth data in Figures 1, 5, and 6, tests for normal distribution were performed using the Shapiro–Wilk test. Student’s t test was applied to compare the significance between groups.
Protein level analysis
Plants were grown vertically on solid MS medium with or without sucrose in dark for 2.5 or 3.5 days and treated with R for different time periods. Changes to the growth medium or the time grown in dark are mentioned specifically in figures. Etiolated seedlings were harvested with a green safe light and R-treated plants were harvested under R. Once the hook-cotyledon tissues were separated they were immediately frozen with dry ice. Tissues were homogenized with a bead beater and immediately dissolved in 150 μL protein isolation buffer containing, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton, 0.1% SDS, 140 mM NaCl, 5 mM β-mercaptoethanol, Sigma Halt protease inhibitor cocktail. Protein quantity was measured with Bradford assay with BioRad reagent and 30 μg protein was loaded in each lane. Proteins were separated by SDS PAGE and transferred to a Nitrocellulose membrane by semi-Dry transfer method. Blots were incubated with 1:200 APY peptide antibody and 1:10,000 Rockland Guinea Pig IgG (H&L) Antibody IR Dye800CW Conjugated Pre-Adsorbed florescent secondary antibody. Bio-Rad Precision Plus Protein Dual color marker was used to estimate the molecular weight of protein bands. Florescent images were generated using default Odyssey infrared imaging system at intensity = 5, sensitivity = auto, channel = 800 nm, resolution = high. All other imaging parameters kept unaltered for the default setting. For equal loading control, blots were stripped with Sigma restore stripping buffer according to manufacturer’s recommendations. Blots were re-probed with Abcam anti His3 1791 primary 1:10,000 and anti-rabbit HRP secondary 1:10,000 and developed with Thermo Fisher Supersignal west Femto substrate according to manufacturer’s recommendations.
RT-qPCR of APY, SAUR17, and SAUR50 expression
Total RNA was extracted from whole seedlings, or from separated seedling roots or apical hook-cotyledon tissue. For APY gene expression studies, three biological replicates were used for each treatment, and ∼100 individuals were pooled to extract RNA from each biological replicate, using Qiagen RNeasy plant mini kit. For the measurements of SAUR50 and SAUR17 gene expression, four to ten technical replicates of RNA was extracted (using Spectrum RNA isolation kit) from apical hook tissue pooled from five independently grown plates of seedlings of Ws (WT) and/or APY2 OE (APY2 overexpressor) and/or estradiol treated R2-4A that had been grown in the dark for 4 days or were exposed to 3 h of light after dark treatment.
Total RNA concentration and purity were measured using Thermo Scientific NanoDrop 1000 spectrophotometer. About 0.1 µg of total RNA treated with Amplification Grade DNase I was reverse transcribed using a High Capacity cDNA Reverse Transcription kit (AB Biosystems/Thermo Fisher, Waltham, MA, USA), following the manufacturer’s protocol.
Gene-specific primers of APY1, APY2, and ACT2 for RT-qPCR were designed using PerlPrimer software (perlprimer.sourceforge.net). Primer standardization was performed to select the primers that had 90%–110% amplification efficiency and to determine optimum cDNA template amount for RT-qPCR reactions. The selected primer sequences were: APY1 Forward: 5′-AAGGGAAATCGAAGTTCCCAC-3′, APY1 Reverse: 5′-TGATGGCTTCAATCCGAATCC-3′; APY2 Forward: 5′-CTGAGGCTGGATTCGTTGAC-3′, APY2 Reverse: 5′-AGGCAAATTCTCTTCCTCCAC-3′. Primers for SAUR50 gene and for SAUR17 gene were as stated in Wang et al. (2020).
RT-qPCR was performed using the Life Technologies/AB Biosystems ViiA 7 Real-Time PCR System. Power SYBR Green PCR Master Mix (Life Technologies/AB Biosystems, Waltham, MA, USA) was used for RT-qPCR reactions. The RT-qPCR conditions used were: hold stage: 50.0°C, 2 min and 95.0°C, 10 min; PCR stage: 40 cycles of 95.0°C, 15 s and 60.0°C, 1 min. Melting curves generated from machine dissociation conditions were used to identify primer dimers and multiple targets.
For each biological replicate, technical triplicates with ±0.5 CT were used for RT-qPCR analysis. The relative gene expression was calculated by ΔΔCt method. ACT2 (At3g18780) was used as the reference gene in all RT-qPCR assays. Statistical significance between treatments was calculated by Student’s t test, using ΔCT for biological replicates.
Root growth and cotyledon area assays
Plants were grown with 1% sucrose in dark for 2.5 days and kept in dark or treated with R for different time periods. Thirty plants for each time point were measured for root and lengths were measured by ImageJ software. For cotyledon area measurements, plants were collected at timepoints specified in the figure legends and excised ∼2 mm from the base of the cotyledons and placed upright on a dark surface. Cotyledons were imaged immediately, and the area was calculated by ImageJ software. Seedlings for root growth and cotyledon area assays with R2-4A mutants were grown for 3.5 days in dark and treated with R for indicated times in the figure.
Treatment with the photosynthesis inhibitor DCMU
Ws seedlings were grown without sucrose for 2.5 days in the dark, and pretreated with DCMU or dimethyl sulfoxide (DMSO) for 4 h in the dark before they were moved to the R. DMSO solvent control (0.001% V/V) and 50 μM DCMU in DMSO (0.001% V/V) were mixed with MS liquid media without sucrose, and 5 mL fresh media was added to the filter paper during the pretreatment and every 24 h during the treatment. Roots were excised and flash frozen and later ground with the bead beater 23s for 2 min. Tissues were dissolved in 150 μL buffer, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton, 0.1% SDS, 140 mM NaCl, 5 mM β-mercaptoethanol, Sigma Halt protease inhibitor cocktail. Protein level was quantified using the Bradford method, and 30 μg was loaded into each lane for separation by SDS-PAGE.
Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane by semi-dry transfer method. Blots were incubated with 1:200 APY peptide antibody and 1:10,000 Rockland Guinea Pig IgG (H&L) Antibody IR Dye800CW Conjugated Pre-Adsorbed florescent secondary antibody. Bio-Rad Precision Plus Protein Dual color marker was used to estimate the molecular weight of the protein bands. Florescent images were generated using default Odyssey infrared imaging system at intensity = 5, sensitivity = auto, channel = 800 nm, resolution = high. All other imaging parameters kept unaltered for the default setting. For equal loading control, blots were stripped with Sigma restore stripping buffer according to manufacturer’s recommendations. Blots were re-probed with Abcam anti His3 1791 primary 1:10,000 and anti-rabbit HRP secondary 1:10,000 and developed with Thermo Fisher Supersignal west Femto substrate according to manufacturer’s recommendations.
Accession numbers
APY1 and APY2 accession numbers are At3g04080 and At5g18280, respectively. SAUR50 and SAUR17 accession numbers are At4g34760 and At4g09530, respectively.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root growth rates and lengths of seedlings grown in different periods of darkness or R.
Supplemental Figure S2. In roots and hook-cotyledons, APY1 and APY2 transcript levels do not significantly change after 6 and 12 h of red-light treatment.
Supplemental Figure S3. SAUR50 transcript level in cotyledon-hooks of etiolated seedlings treated with light is significantly lower in estradiol-induced R2-4A mutants than in WT (Ws) seedlings.
Supplemental Table S1. Transcript abundance of peroxidase genes in etiolated seedlings of WT (Ws WT) compared to that in APY mutants.
Supplementary Material
Acknowledgments
We thank Dr. Enamul Huq and members of his laboratory and Dr. Robert Slocum for help in the planning and design of experiments.
Funding
This work was supported by the National Science Foundation (IOS-1027514) and by Texas Crop Science, LLC (UTA13-000682) to S.J.R. and G.C.
Conflict of interest statement. S.J.R. and G.C. are consultants to Texas Crop Science, which partially funded some of the research presented here. No other authors have conflicts of interest to declare.
Contributor Information
Gayani Weeraratne, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Huan Wang, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Tharindu P Weeraratne, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Tanya Sabharwal, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Han-Wei Jiang, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Araceli Cantero, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Greg Clark, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
Stanley J Roux, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA.
G.W. and S.J.R conceived and designed the study. G.W, T.P.W., H.W., T.S., A.C., and H.W.J. performed one or more of the experiments. G.W., T.P.W., H.W., A.C., and T.S. analyzed the data. S.J.R. and G.C. supervised the experiments. G.W., G.C., and S.J.R. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Stanley J. Roux (sroux@austin.utexas.edu).
References
- Abbas M, Alabadi D, Blazquez MA (2013) Differential growth at the apical hook: all roads lead to auxin. Front Plant Sci 4: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Mella A, Ballare CL, Maldonado S (2006) Phytochrome-mediated effects on extracellular peroxidase activity, lignin content and bending resistance in etiolated Vicia faba epicotyls. Physiol Plant 92:555–562 [Google Scholar]
- Cheng MC, Kathare PK, Paik I, Huq E (2021) Phytochrome signaling networks. Ann Rev Plant Biol 72:217–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Morgan RO, Fernandez MP, Salmi ML, Roux SJ (2014) Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development. Plant Sci 225:107–116 [DOI] [PubMed] [Google Scholar]
- Clark G, Roux SJ (2018) Role of Ca2+ in mediating plant responses to extracellular ATP and ADP. Int J Mol Sci 19: 3590 doi:10.3390/ijms19113590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Brown KA, Tripathy MK, Roux SJ (2021) Recent advances clarifying the structure and function of plant apyrases (nucleoside triphosphate diphosphohydrolases). Int J Mol Sci 22:3283 doi:10.3390/ijms22063283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46:317–323 [DOI] [PubMed] [Google Scholar]
- Deng SR, Sun J, Zhao R, Ding MQ, Zhang YN, Sun YL, Wang W, Tan YQ, Liu DD, Ma XJ, et al. (2015) Populus euphratica APYRASE2 enhances cold tolerance by modulating vesicular trafficking and extracellular ATP in Arabidopsis plants. Plant Physiol 169:530–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Sun N, Yang J, Deng ZG, Lan JQ, Qin GJ, He H, Deng XW, Irish VF, Chen HD, et al. (2019) The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31:1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao FQ, He J, Huq E, Blancaflor EB (2011) SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23:3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Martinez-Garcia JF, Josse EM (2009) Integration of light and auxin signaling. Cold Spring Harb Perspect Biol a001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang GJ, Yang JY, Hsieh HL, Wu SH (2019) Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc Natl Acad Sci USA 116:6451–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Shinkle JR, Roux SJ (1989) Phytochrome induces changes in the immundetectabe level of a wall peroxidase that precede growth changes in maize seedlings. Proc Natl Acad Sci USA 86:9866–9870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P (2012) Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc Natl Acad Sci USA 109:11217–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Yamamoto KT (1997) Light-stimulated root elongation in Arabidopsis thaliana. J Plant Physiol 151:346–351 [Google Scholar]
- Laurenzi M, Rea G, Federico R, Tavladoraki P, Angelini R (1999) De-etiolation causes a phytochrome-mediated increase of polyamine oxidase expression in outer tissues of the maize mesocotyl: a role in the photomodulation of growth and cell wall differentiation. Planta 208:146–154 [Google Scholar]
- Leviar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26:56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MH, Wu J, Yao JC, Gallardo IF, Dugger JW, Webb LJ, Huang J, Salmi ML, Song J, Clark G, et al. (2014) Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol 164:2054–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wu S, Chen H, Wu S (2012b) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8:566–n/a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu J, Clark G, Lundy S, Lim M, Arnold D, Chan J, Tang WQ, Muday GK, Gardner G, et al. (2012a) Role for apyrases in polar auxin transport in Arabidopsis. Plant Physiol 160:1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng R, Zhu LQ, Yang YF, Zhu LC, Hou ZK, Jin L, Wang BC (2019) Apyrases in Arabidopsis thaliana. Biol Plant 63:38–42 [Google Scholar]
- Montgomery B (2016) Spatiotemporal phytochrome signaling during photomorphogenesis: from physiology to molecular mechanisms and back. Front Plant Sci 7:480 doi:10.3389/fpls.2016.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G (2012) Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci USA 109:1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Spalding EP (1999) Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc Natl Acad Sci USA 96:14142–14146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Grossman L, Fernie AR, Wucke C, Geigenberger P (2008) The potato-specific apyrase is apoplastically localized and has influence on gene expression, growth, and development. Plant Physiol 147:1092–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T, Morita EH, Abe S (2014) Expression pattern of PsAPY1 during apical hook development in pea. Biologia 69:293–299. [Google Scholar]
- Steinebrunner I, Jeter C, Song C, Roux SJ (2000) Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem 38:913–922 [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131:1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113:6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nguyen CT, Libault M, Cheng J, Stacey G (2011) Enzymatic activity of the soybean ecto-apyrase GS52 is essential for stimulation of nodulation. Plant Physiol 155:1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang CK, Pierik R (2018) Light signaling, root development, and plasticity. Plant Physiol 176:1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappa R, Slocum RD, Siegenthaler A, Wang J, Clark G, Roux SJ (2019) Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ 42:337–353 [DOI] [PubMed] [Google Scholar]
- Wang L, Uilecan IV, Assadi AH, Kozmik CA, Spalding EP (2009) HYPOTrace: image analysis software for measuring hypocotyl growth and shape demonstrated on Arabidopsis seedlings undergoing photomorphogenesis. Plant Physiol 149:1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun N, Zhang F, Yu R, Chen H, Deng XW, Wei N (2020) SAUR17 and SAUR50 differentially regulate PP2C-D1 during apical hook development and cotyledon opening in Arabidopsis. Plant Cell 32:3792–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C, Hennig M, Romanovicz D, Steinebrunner I (2007) Developmental defects and seedling lethality in apyrase AtAPY1 and AtAPY2 double knockout mutants. Plant Mol Biol 64:657–672 [DOI] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ (2007) Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol 144:961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.