Abstract
The salt overly sensitive (SOS) pathway plays an important role in plant salt stress; however, the transcriptional regulation of the genes in this pathway is unclear. In this study, we found that Linker histone variant HIS1-3 and WRKY1 oppositely regulate the salt stress response in Arabidopsis (Arabidopsis thaliana) through the transcriptional regulation of SOS genes. The expression of HIS1-3 was inhibited by salt stress, and the disruption of HIS1-3 resulted in enhanced salt tolerance. Conversely, the expression of WRKY1 was induced by salt stress, and the loss of WRKY1 function led to increased salt sensitivity. The expression of SOS1, SOS2, and SOS3 was repressed and induced by HIS1-3 and WRKY1, respectively, and HIS1-3 regulated the expression of SOS1 and SOS3 by occupying the WRKY1 binding sites on their promoters. Moreover, WRKY1 and HIS1-3 acted upstream of the SOS pathway. Together, our results indicate that HIS1-3 and WRKY1 oppositely modulate salt tolerance in Arabidopsis through transcriptional regulation of SOS genes.
HIS1-3 and WRKY1 oppositely modulate salt tolerance in Arabidopsis through transcriptional regulation of SOS genes.
Introduction
Soil salinity is one of the most important factors limiting agricultural development and crop production worldwide (Godfray et al., 2010; Mark and Peter, 2010; Agarwal et al., 2013). Salt stress can disrupt a variety of cellular and physiological processes as a result of ion toxicity, osmotic stress, and secondary oxidative stress.
To adapt to or resist salt stress, plants have evolved several strategies including osmotic adjustment (such as osmoprotectant accumulation), ionic balance (especially for Na+/K+ homeostasis), and anti-oxidation (antioxidant enzyme accumulation and activity enhancement to resist oxidative stress; Munns and Tester, 2008; Jaleel et al., 2009; Arbona et al., 2013; Zhang et al., 2013; Tang et al., 2015; Fu et al., 2018). Ion transporters, such as the salt overly sensitive (SOS), Na+/H+ antiporter (NHX), and high-affinity potassium transporter, are important mediators of the ionic balance during salt stress. In Arabidopsis thaliana, the SOS pathway contains three main components, including SOS3, SOS2, and SOS1, which have been identified as being involved in the exclusion of excess Na+ ions out of the cell across the plasma membrane in the roots (Shi et al., 2000; Zhu, 2003; Martinez-Atienza et al., 2007; Munns and Tester, 2008). SOS3 (CBL) encodes a Ca2+-binding protein and senses transient increases in cytosolic Ca2+ concentration, which is elicited by salt stress (Liu and Zhu, 1998; Tuteja, 2007). SOS2 encodes a serine/threonine protein kinase and can be activated by SOS3 in a calcium-dependent manner (Halfter et al., 2000; Yang et al., 2009). SOS1, a plasma membrane Na+/H+ antiporter, can be phosphorylated and activated by the SOS2–SOS3 kinase complex directly, resulting in Na+ efflux (Shi et al., 2000; Qiu et al., 2002). In addition, Yang et al. (2009) found that the overexpression of SOS1/SOS3 led to enhanced salt tolerance in A. thaliana (Yang et al., 2009). These results have extensively indicated that the expression of SOS1, SOS2, and SOS3 plays an important role in plant salt stress; however, the transcriptional regulation of these regulatory proteins is not clear. In eukaryotes, chromatin remodeling is critical for the regulation of stress-responsive gene expression (Kim et al., 2010; Luo et al., 2012; Zhu et al., 2013). Linker histones, the main structural components of chromatin, bind to the linker DNA between nucleosome cores, thereby stabilizing higher-order chromatin structure and limiting the accessibility of DNA to the regulatory factors (Horn et al., 2002; Koop et al., 2003; Kim et al., 2010). The linker histones may control specific processes during development and respond to environmental stress by changing the expression of H1 variants (Jerzmanowski et al., 2000). Some linker histone variants are induced by drought stress, such as HIS1-3 of Arabidopsis (A. thaliana; Ascenzi and Gantt, 1997), H1.S of tomato (Lycopersicon esculentum; Scippa et al., 2000, 2004), H1-D of wild tomato (Lycopersicon pennellii; Wei and O'Connell, 1996), and H1.S of cotton (Gossypium herbaceum; Trivedi et al., 2012). Ascenzi and Gantt (1997) found that the expression of HIS1-3 was induced by drought stress and abscisic acid (ABA) treatment (Ascenzi and Gantt, 1997). They also found that HIS1-3 was primarily expressed in the root meristem and elongation zone in drought-stressed plants (Ascenzi and Gantt, 1999). The expression of HIS1-3 was upregulated in ABA-responsive element-binding protein AREB1▵QT-overexpressing plants but downregulated in a dominant loss-of-function mutant of AREB1 (Fujita et al., 2005). These data suggest that HIS1-3 may be a stress-inducible H1 variant. However, whether HIS1-3 is involved in the regulation of salt stress is unclear.
The WRKY transcription factors, which belong to a larger family of plant-specific regulatory proteins, are another important regulatory component of salt stress tolerance. Members of this family contain a highly conserved WRKY domain at the N-terminus and a zinc finger motif (C2H2 or C2HC) at the C-terminus. Based on their primary structure, the WRKY proteins have been categorized into three groups (I, II, and III). The WRKY proteins specifically recognize the W-box (contains a TGAC core sequence; Eulgem et al., 2000; Rushton et al., 2010). It has been proven that WRKY proteins play important roles in many plant processes, including plant development and responses to biotic and abiotic stress (Eulgem et al., 2000; Johnson et al., 2002; Ulker et al., 2007; Agarwal et al., 2011; Phukan et al., 2016). In Arabidopsis, 18 WRKY genes were induced by 150-mM NaCl (Jiang and Deyholos, 2006). The expression of AtWRKY25 and AtWRKY33 is induced by salt stress, and increased AtWRKY25 or AtWRKY33 expression can enhance salinity tolerance in transgenic plants (Jiang and Deyholos, 2009). AtWRKY8 plays a crucial role in the response to salt stress through directly binding the promoter of Response to Desiccation 29A to modulate salinity tolerance, and the VQ motif-containing protein 9 protein acts as a repressor of AtWRKY8 to maintain an appropriate balance of the AtWRKY8-mediated signaling pathway (Hu et al., 2013). In addition, many WRKY genes in other plants are also involved in the response to salt stress, such as ZmWRKY33, ZmWRKY17, GhWRKY34, and OsWRKY45 (Tao et al., 2011; Li et al., 2013; Zhou et al., 2015; Cai et al., 2017). WRKY1 (also known as zinc-responsive transcriptional activator ZAP1) was the first WRKY gene identified from Arabidopsis and belongs to the group I WRKY family (de Pater et al., 1996). WRKY1 may be involved in the salicylic acid (SA) signaling pathway, the induction of which is partially dependent on nonexpresser of PR gene 1 (Duan et al., 2007), while WRKY1 regulates stomatal movement in the guard cells (Qiao et al., 2016).
In this report, we showed that the expression of HIS1-3 could be inhibited by salt stress, and the loss-of-function of HIS1-3 enhanced salt tolerance in Arabidopsis. The HIS1-3 protein was found to act as a negative regulator in the response to salt stress through the SOS pathway. Conversely, WRKY1 positively regulated plant salt tolerance and maintained Na+/K+ homeostasis via the SOS pathway. Further investigation showed that HIS1-3 and WRKY1 recognized and bound to the same W-boxes in the promoter regions of SOS1 and SOS3. The functional antagonism between HIS1-3 and WRKY1 may be a specific mechanism by which Na+/K+ homeostasis is maintained in order to regulate salt tolerance.
Results
The his1-3 mutant is tolerant to salt stress
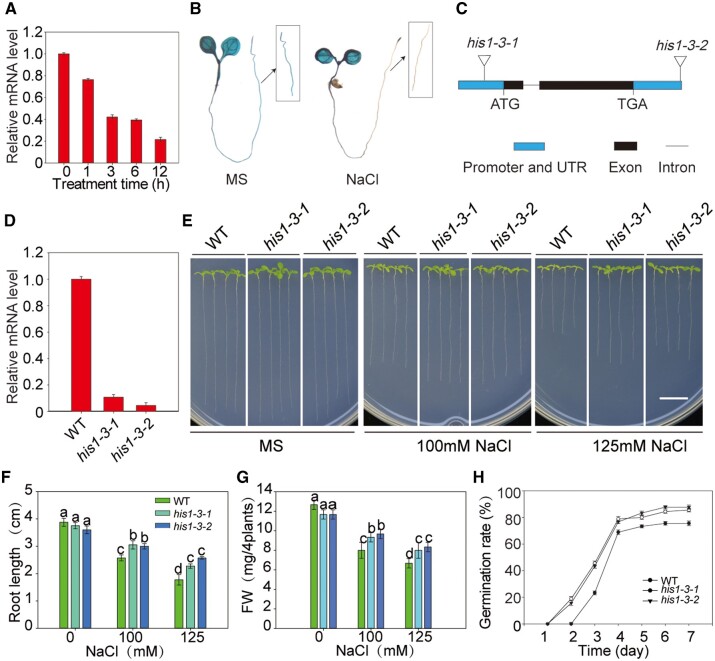
Many studies have confirmed that HIS1-3 plays an important role in many abiotic stresses, such as water deficit and ABA treatment. Thus, we speculated that HIS1-3 might also respond to salt stress. We used reverse transcription-quantitative PCR (RT-qPCR) and ProHIS1-3::β-glucuronidase (GUS) lines to analyze the expression of HIS1-3 under salt stress. The results showed that the expression of the HIS1-3 gene was inhibited by salt stress (Figure 1, A and B). We also tested the expression profiles of HIS1-3 in various tissues using RT-qPCR and histochemical analysis and found that HIS1-3 was expressed in most of the examined tissues, with high expression in the rosette leaf and inflorescence (Supplemental Figure S1).
Figure 1.
Loss of function of HIS1-3 leads to enhanced salt tolerance. A, The effect of salt stress on HIS1-3 transcript levels were analyzed by RT-qPCR, with Actin8 as the internal control. Two-week-old WT seedlings were treated with NaCl for 0, 1, 3, 6, and 12 h. The vertical bars indicate the mean ± se of three biological replicates. B, GUS staining of 7-d-old ProHIS1-3::GUS seedlings treated with NaCl. Seven-day-old seedlings were treated with NaCl (100 mM) for 0 and 6 h and then stained with X-Gluc for 12 h at 37°C. C, The location of the T-DNA insertion in the HIS1-3 gene. Exons are shown as black boxes and introns are shown as lines. The triangle indicates the locations of the T-DNA insertion sites. D, The RT-qPCR analysis of the WT and his1-3 mutants. The Actin8 gene was amplified as an internal control. Similar results were obtained in at least three biological replicates. E, Salt tolerance assay of the WT and his1-3 mutants. The seedlings were grown on MS medium for 3 d and then transferred to MS medium with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. F and G, Root length (F) and fresh weight (G) of the plants described in (E). H, Germination rate of the WT and his1-3 mutants. The seedlings were grown on MS medium with NaCl and then counted at a fixed time every day. Data are presented as means ± se of three replicate experiments. Statistical significance was determined by Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
To further verify this, seeds of two T-DNA mutants (SAIL_799_A07 and SALK_025209C) were screened from the SALK Arabidopsis T-DNA mutant collection, named his1-3-1 and his1-3-2. The data showed dramatically decreased transcript levels in the his1-3 mutants compared with the wild-type (WT) plants (Figure 1, C and D). The root length and fresh weight of the mutants were significantly increased compared with the WT plants (Figure 1, E–G). The seed germination rate of the mutants was higher than that in the WT under salt stress (Figure 1H). Together, these results indicated that the loss-of-function of HIS1-3 resulted in increased salt tolerance.
To test whether the his1-3 mutants responded specifically to sodium, the his1-3 mutants were grown on Murashige and Skoog (MS) media containing KCl, LiCl, and mannitol. We observed that there was no morphological difference between the his1-3 mutants and WT (Supplemental Figure S2). This result suggested that HIS1-3 may perform a critical function in Na+ toxicity.
Overexpression of HIS1-3 leads to increased salt sensitivity
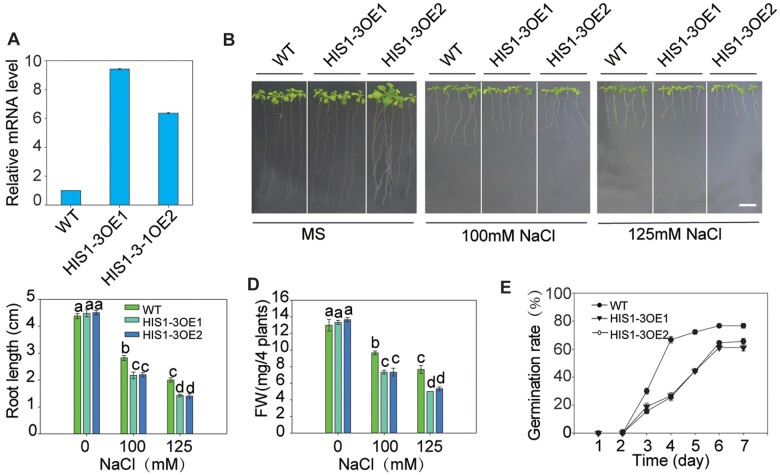
To further confirm the role of HIS1-3 in salt stress, we built the 35S::HIS1-3 construct and then transformed it into WT plants to obtain the overexpression lines. The independent homozygous lines (OE1 and OE2) were chosen to further analyze the salt stress phenotype, and the expression of HIS1-3 was found to be much higher than that in the WT (Figure 2A). Under salt stress, the overexpression seedlings were more sensitive than the WT (Figure 2, B–D). Furthermore, the seed germination rate of the overexpressing lines was lower than that in the WT under salt stress (Figure 2E). Under normal growth conditions, the his1-3 mutants, HIS1-3-overexpressing lines, and WT exhibited similar growth and development in both the vegetative and reproductive phases (Supplemental Figure S3). These results showed that the overexpression of HIS1-3 was responsible for the enhanced salt sensitivity.
Figure 2.
HIS1-3-overexpressing plants show increased salt sensitivity. A, The RT-qPCR analysis of the WT and HIS1-3-overexpressing plants. The Actin8 gene was amplified as an internal control. Similar results were obtained in at least three biological replicates. B, Salt sensitivity assay of the WT and HIS1-3-OE seedlings. Three-day-old WT and HIS1-3-OE seedlings were transferred to MS medium with or without 100 mM or 125 mM of NaCl. Bar = 1 cm. C and D, Root length (C) and fresh weight (D) of the plants described in (B). E, Germination rate of the WT and HIS1-3-OE seedlings. The seedlings were grown on MS medium with NaCl and then counted at a fixed time every day. Data are presented as means ± se of three replicate experiments. Statistical significance was determined by Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
HIS1-3 mediates Na+/K+ homeostasis via the SOS pathway
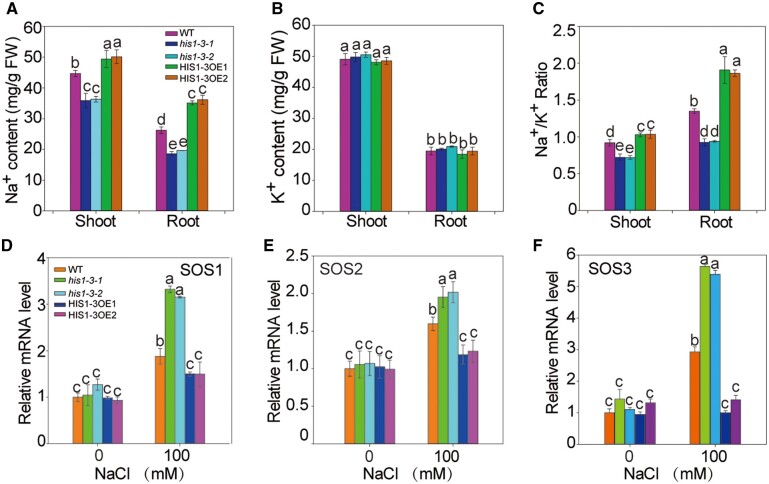
To study whether HIS1-3 responds to salt stress by regulating the Na + balance, we analyzed the Na+ and K+ contents in the WT, his1-3 mutants, and HIS1-3-overexpressing lines subjected to salt treatment. Compared with the WT, the Na+ contents in the roots and the aboveground parts of the mutants were much lower, and so, the ratios of Na+/K+ were reduced (Figure 3, A and C). The contents of K+ did not differ among the WT, his1-3 mutants, and HIS1-3-overexpressing lines (Figure 3B). On the contrary, the Na+ contents and the Na+/K+ ratio in the HIS1-3-overexpressing lines were higher (Figure 3, A and C). Thus, HIS1-3 may respond to salt stress by mediating Na+/K+ homeostasis.
Figure 3.
HIS1-3 mediates Na+/K+ homeostasis via the SOS pathway. A and B, Na+ and K+ content in the shoots and roots of WT, his1-3 mutants, and HIS1-3-OE seedlings. The seedlings were treated with 100 mM of NaCl for 2 weeks, and then, the roots and shoots of these samples were collected. C, The Na+–K+ ratio was calculated from the Na+ and K+ content. D–F, Expression patterns of genes involved in the SOS pathway in the WT, his1-3 mutants, and HIS1-3-OE seedlings under salt stress. Using RT-qPCR, gene expression was compared between 2-week-old seedlings of WT, his1-3 mutants, and HIS1-3-OE seedlings after 0-h and 6-h treatment with 100 mM of NaCl. Actin8 was used as the internal control. All data shown here are presented as the means ± se of three replicate experiments. Statistical significance was determined using Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
The Ca2+-dependent SOS signaling pathway is an Na+-specific signaling pathway that mainly consists of three protein components: SOS1, SOS2, and SOS3. We, therefore, examined the transcription levels of SOS1, SOS2, and SOS3 in the WT, his1-3 mutants, and HIS1-3-overexpressing lines using RT-qPCR. The results showed that the expression of SOS1, SOS2, and SOS3 in the mutants were significantly higher than that in the WT. In contrast, the expression of these genes were significantly decreased in the HIS1-3-overexpressing lines (Figure 3, D–F). All the above results demonstrated that HIS1-3, as a negative transcriptional regulator, mediated Na+/K+ homeostasis through the SOS pathway.
WRKY1 positively regulates salt tolerance
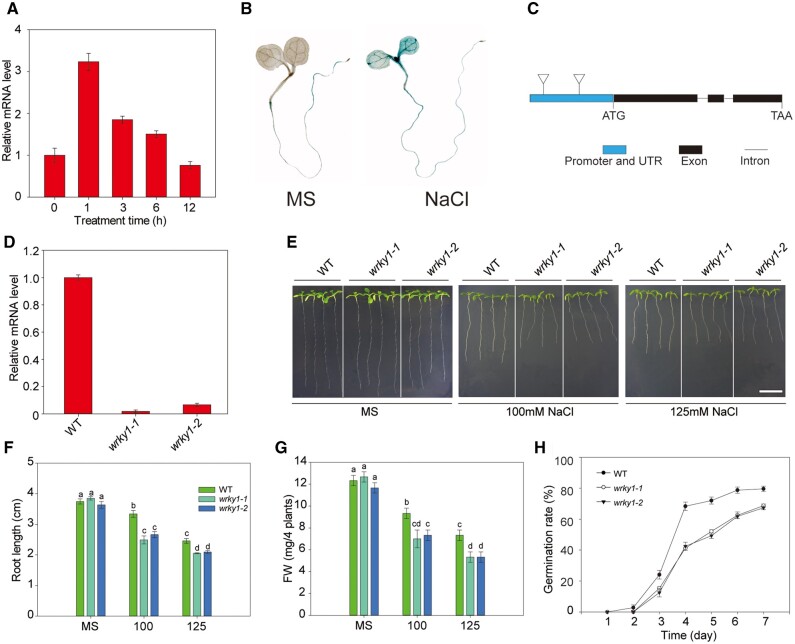
The HIS1-3 protein has been found to interact with the transcription factor WRKY1 in banana (Musa acuminata), which is a unique transcription factor in plants containing one or two highly conserved WRKY domains (Wang et al., 2012). A large number of studies have confirmed that WRKY transcription factors are involved in the regulation of various abiotic stresses such as low Pi, high temperature, and pathogen invasion. Therefore, we speculated that WRKY1 may be involved in the response to salt stress. First, we tested the expression profiles of WRKY1 under salt stress in various tissues. The results of RT-qPCR and histochemical analysis showed that the expression of WRKY1 was induced by salt stress (Figure 4, A and B). The RT-qPCR and histochemical analysis also showed that WRKY1 was expressed in most of the examined tissues, with high expression detected in the cauline leaf and inflorescence (Supplemental Figure S4).
Figure 4.
The wrky1 mutants are sensitive to salt stress. A, Effect of salt stress on WRKY1 transcript levels analyzed by RT-qPCR, with Actin8 as the internal control. Two-week-old WT seedlings were treated with NaCl for 0, 1, 3, 6, and 12 h. The vertical bars indicate the means ± se of three biological replicates. B, The GUS staining of 7-d-old ProWRKY1::GUS seedlings treated with NaCl for 6 h. The 7-d-old seedlings were treated with NaCl (100 mM) for 0 and 6 h and then stained with X-Gluc for 12 h at 37°C. C, The T-DNA insertion sites of the WRKY1 gene. Exons are shown as black boxes, and introns are shown as lines. The triangle indicates the locations of the T-DNA insertion sites. D, Transcription analysis of the WRKY1 gene in the wrky1-1 and wrky1-2 mutants by RT-qPCR. The Actin8 gene was amplified as an internal control. Similar results were obtained in at least three biological replicates. E, The WRKY1 loss-of-function mutation led to sensitivity to salt stress. Seedlings were grown on MS medium for 3 d and then treated with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. F and G, Measurements of the root length (F) and fresh weight (G) described in (E). H, Germination rate of the WT, wrky1-1, and wrky1-2 mutants grown in MS medium with NaCl and then counted at a fixed time every day. Data are presented as means ± se of three replicate experiments; statistical significance was determined by Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
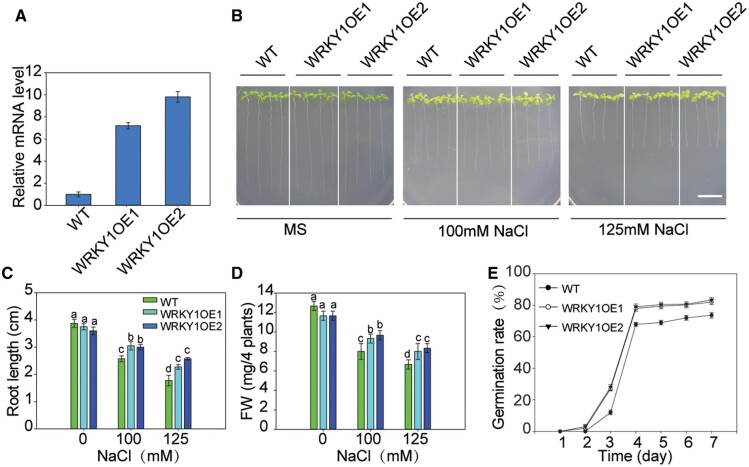
We then used two independent T-DNA insertion mutants (wrky1-1: SALK_070989C and wrky1-2: SALK_136009C) and two WRKY1 overexpression lines (WRKY1-OE1 and WRKY1-OE2) to test the salt tolerance phenotypes. We identified the T-DNA insertion site (Figure 4C), and the homozygous mutants were used for phenotype analysis. The expression of WRKY1 was blocked in the two mutants (Figure 4D). Under salt stress, the root length and fresh weight of the mutants were significantly decreased compared with the WT plants (Figure 4, E–G). The seed germination rate of the mutants was lower than that in the WT under salt stress (Figure 4H). In contrast, WRKY1 was highly accumulated in the WRKY1-OE lines and led to enhanced salt tolerance (Figure 5). We then observed the growth phenotypes of the wrky1 mutants, WRKY1-overexpressing lines, and WT in the soil and found that they exhibited similar growth and development in both the vegetative and reproductive phases (Supplemental Figure S5). Together, these results indicated that WRKY1 played a positive role in salt tolerance.
Figure 5.
WRKY1 positively regulates salt tolerance. A, The expression of WRKY1 in the WT and WRKY1-overexpressing lines using RT-qPCR. Actin8 was used as the internal control. Data are presented as the means ± se of three replicate experiments. B, Growth of WT and WRKY1-overexpressing lines under salt stress. Seedlings were grown on MS medium for 3 d and then transferred to MS medium with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. C and D, Root length (C) and fresh weight (D) of plants described in (B). E, Germination rate of WT and WRKY1-overexpressing lines grown in MS medium with NaCl and then counted at a fixed time every day. These experiments were repeated 3 times with similar results, and the data are presented as the means ± se. Statistical significance was determined by Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
To test whether the wrky1 mutants were also specific to the sodium response, the wrky1 mutants were grown on MS media containing KCl, LiCl, and mannitol. No significant difference was observed between the wrky1 mutants and WT (Supplemental Figure S6), which suggests that WRKY1 may play an important role in Na+ toxicity.
WRKY1-mediated salt tolerance by maintaining Na+/K+ homeostasis
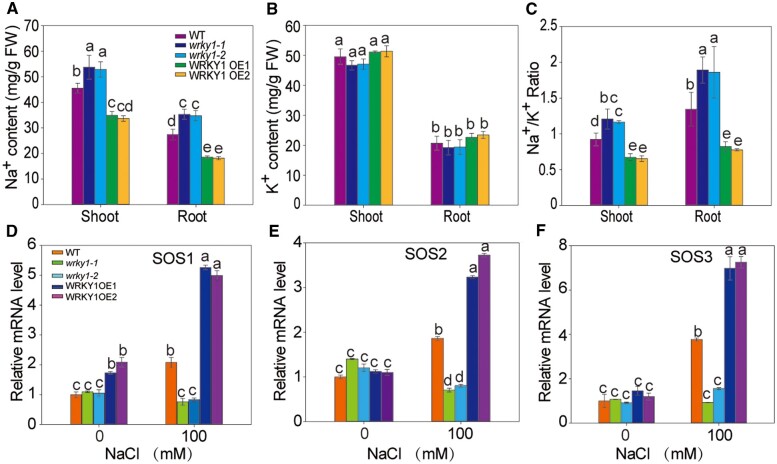
To further examine whether WRKY1 also regulates salt stress through the SOS pathway, we tested the contents of Na+ and K+ in the WT, wrky1 mutants, and WRKY1-overexpressing lines. The results showed that the Na+ content and the Na+/K+ ratio were much higher in the mutants and lower in the WRKY1-overexpressing lines (Figure 6, A–C).
Figure 6.
WRKY1-mediated salt tolerance by mediating Na+/K+ homeostasis. A–B, The Na+ and K+ content in the shoots and roots of WT, wrky1 mutants, and WRKY1-OE seedlings. The seedlings were treated with 100 mM of NaCl for 2 weeks. The roots and shoots of these samples were collected. C, The Na+–K+ ratio of WT, wrky1 mutants, and WRKY1-OE seedlings was calculated from the Na+ and K+ content. D–F, Transcript levels of SOS1, SOS2, and SOS3 in the WT, wrky1 mutants, and WRKY1-OE seedlings under salt stress. Gene expression was compared by RT-qPCR among 2-week-old seedlings of WT, wrky1 mutants, and WRKY1-OE seedlings after 0- and 6-h treatment with 100 mM of NaCl. Actin8 was used as the internal control. All data shown here are presented as the means ± se of three replicate experiments. Statistical significance was determined using Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
The transcript levels of SOS1, SOS2, and SOS3 in the WT, wrky1 mutants, and WRKY1-overexpressing lines were then analyzed by RT-qPCR. The results showed that the expression of SOS1, SOS2, and SOS3 in the mutants was significantly decreased compared to that in the WT but increased in the WRKY1-overexpressing lines (Figure 6, D–F). Overall, WRKY1-mediated Na+/K+ homeostasis positively regulates salt tolerance.
HIS1-3 and WRKY1 bind to the promoters of three SOS genes
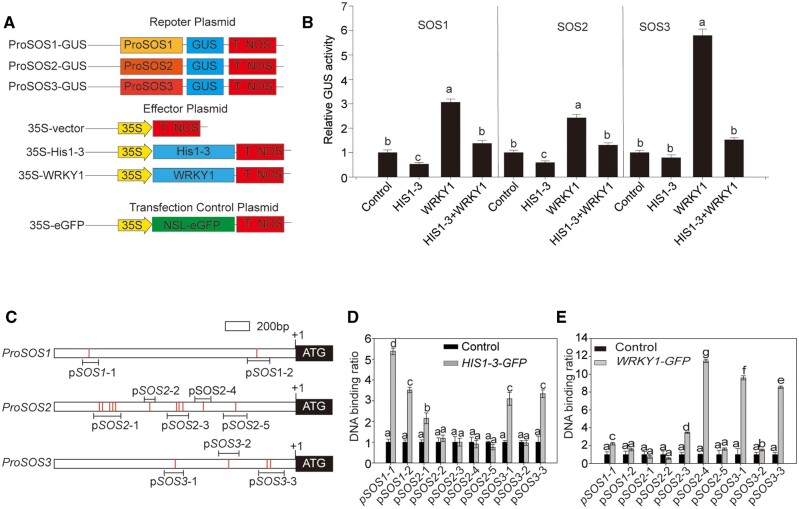
To assess whether HIS1-3 and WRKY1 synergistically regulated the response to salt stress, we first confirmed that both were localized in the nucleus (Supplemental Figure S7, A and B). We then studied whether there was an interactive relationship between them by using yeast two-hybridization and bimolecular fluorescence complementation (BiFC), but the results showed that HIS1-3 did not interact with WRKY1 (Supplemental Figure S7, C and D). Subsequently, we explored whether HIS1-3 and WRKY1 synergistically affect the expression of SOS pathway genes by transient transactivation assay. Consistent with the above results, WRKY1 activated the expression of SOS1, SOS2, and SOS3, whereas HIS1-3 reduced the expression of SOS1, SOS2, and SOS3 (Figure 7, A and B). Under the co-expression of HIS1-3 and WRKY1, the expression of SOS1, SOS2, and SOS3 exhibited no obvious change compared with the WT (Figure 7B). These results showed that HIS1-3 reduced the expression of WRKY1-activated SOS1, SOS2, and SOS3.
Figure 7.
HIS1-3 and WRKY1 regulate three SOS genes by binding to their promoters. A, Schematics of all constructs used for transient expression assays in N. benthamiana leaves. The SOS1, SOS2, or SOS3 promoter was fused to the GUS reporter gene. 35S::HIS1-3 and 35S::WRKY1 acted as an effector. The 35S promoter was fused to the GFP gene as an internal control. B, Relative GUS activity showed the expressions of ProSOS1, ProSOS2, or ProSOS3 after co-expression with HIS1-3 and/or WRKY1. The constructs of ProSOS1:GUS, ProSOS2:GUS, or ProSOS3:GUS were co-transformed into N. benthamiana epidermal cells with 35S::HIS1-3 or 35S::WRKY1, and GUS activity was assessed at 48 h after injection using a MUG assay. The 35S-empty vector was used as an effector plasmid control. C, Schematic diagram of the SOS1, SOS2, and SOS3 promoters showing the W-box present in different regions. Bars indicate the W-box (TGAC); lines beneath the bars represent the sequences for ChIP assays. D and E, The direct binding of HIS1-3 and WRKY1 to the W-box of the SOS1, SOS2, and SOS3 promoters using ChIP-qPCR assay. The DNA binding ratio of the promoter regions of SOS1, SOS2, and SOS3 was confirmed by qPCR using the ChIP products from the WT HIS1-3-GFP or WRKY1-GFP. Input DNAs were used as the internal control. All data shown here are presented as the means ± se of three replicate experiments. Statistical significance was determined using Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
It has been reported that WRKY can regulate target gene expression by binding to the typical cis-element W-box in the promoters of these genes, and so, we analyzed the promoters of the SOS1, SOS2, and SOS3 genes by bioinformatics and found that W-boxes existed on the promoters of these genes (Figure 7C). To determine whether the SOS genes were directly regulated by HIS1-3 and WRKY1, we performed chromatin immunoprecipitation (ChIP) experiments using 35S::HIS1-3::GFP and 35S::WRKY1::GFP transgenic plants. Enrichment of specific promoter fragments in the precipitate was measured using qPCR. These results showed that WRKY1 was enriched in pSOS1-1, pSOS2-3, pSOS2-4, pSOS3-1, pSOS3-2, and pSOS3-3 with the GFP antibody, and HIS1-3 was enriched in pSOS1-1, pSOS1-2, pSOS2-1, pSOS3-1, and pSOS3-3, with the binding sites on SOS1 and SOS3 being consistent with WRKY1 and those on SOS2 being adjacent to WRKY1 (Figure 7, D–E). We also intended to further verify this result by yeast one-hybrid (Y1H) assay, but WRKY1 exhibited self-activation (Supplemental Figure S8). These data supported the hypothesis that HIS1-3 may suppress the expression of SOS1 and SOS3 by occupying the binding sites of WRKY1 on the promoters of SOS1 and SOS3.
WRKY1 and HIS1-3 act upstream of the SOS pathway
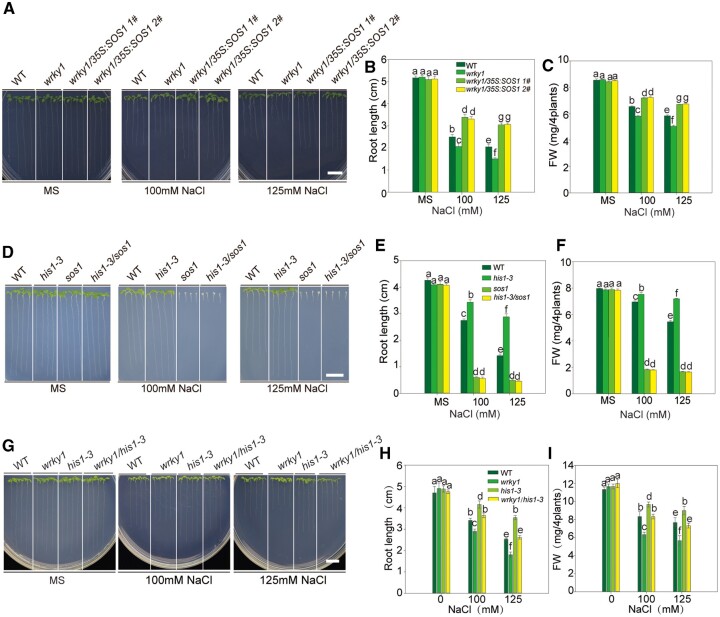
SOS1, a putative plasma membrane Na+/H+ antiporter, is a downstream target of the SOS2–SOS3 complex. The sos1 mutations render plants extremely sensitive to salt stress, and the overexpression of SOS1 improves salt tolerance in A. thaliana (Wu et al., 1996; Shi et al., 2003). In order to genetically study the role of HIS1-3, WRKY1, and SOS1 in regulating salt stress, we generated the double mutants wrky1/35S::SOS1, his1-3/sos1-1, and his1-3/wrky1 (Supplemental Figure S9). Consistent with our results (Figure 4), the wrky1 mutant was hypersensitive to salt stress and the his1-3 mutant was tolerant to salt stress. The salt sensitivity of the wrky1 mutant was rescued by 35S::SOS1 (Figure 8, A–C), and the increased salt tolerance of the his1-3 mutant was eliminated by the loss-of-function of sos1-1 (Figure 8, D–F). Meanwhile, there was no significant difference between the growth of the his1-3wrky1 double mutant and WT under salt stress (Figure 8, G–I). This result suggested that WRKY1 and HIS1-3 acted upstream of the SOS genes and competed with each other.
Figure 8.
Genetic interaction of HIS1-3 and WRKY1 with SOS1. A, Growth of the WT, wrky1, and wrky1/35S:SOS1 lines under salt stress. Seedlings were grown on MS medium for 3 d and then transferred to MS medium with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. B and C, Root length (B) and fresh weight (C) of the plants described in (A). D, Growth of the WT, his1-3, sos1, and his1-3/sos1 double mutants under salt stress. Seedlings were grown on MS medium for 3 d and then transferred to MS medium with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. E and F, Root length (E) and fresh weight (F) of the plants described in (D). G, Growth of WT, wrky1, his1-3, and wrky1/his1-3 lines under salt stress. Seedlings were grown on MS medium for 3 d and then transferred to MS medium with or without 100 or 125 mM of NaCl for 2 weeks. Bar = 1 cm. H and I, Root length (G) and fresh weight (I) of the plants described in (G). These experiments were repeated 3 times with similar results, and data are presented as the means ± se. Statistical significance was determined by Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
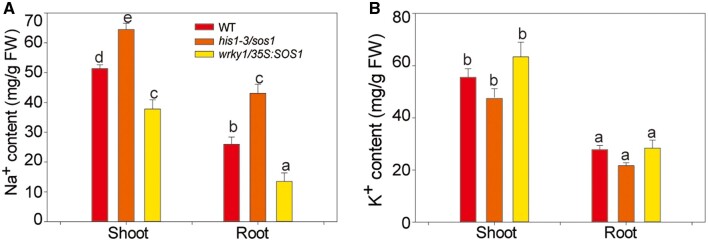
Subsequently, we detected the content of Na+ and K+ in the his1-3/sos1 and wrky1/35S:SOS1 plants. The result showed that the Na+ content in the roots and the aboveground parts of the his1-3/sos1 mutant was much higher than that of the WT (Figure 9A). On the contrary, the Na+ contents in the wrky1/35S:SOS1 plants were lower. The content of K+ did not differ among the WT, his1-3/sos1 mutant, and wrky1/35S:SOS1 plants (Figure 9B). The result suggested that WRKY1 and HIS1-3 may respond to salt stress through SOS pathway-dependent Na+/K+ homeostasis.
Figure 9.
The Na+ and K+ content in the shoots and roots of WT, his1-3/sos1, and wrky1/35S:SOS1 mutants. A, The Na+ content in the shoots and roots of the WT, his1-3/sos1, and wrky1/35S:SOS1 mutants. B, The K+ content in the shoots and roots of the WT, his1-3/sos1, and wrky1/35S:SOS1 mutants. The seedlings were treated with 100 mM of NaCl for 2 weeks. The roots and shoots of these samples were collected. All data shown here are presented as the means ± se of three replicate experiments. Statistical significance was determined using Student’s t tests; significant differences (P < 0.05) are indicated by different lowercase letters.
Discussion
HIS1-3 and WRKY1 play opposing roles in salt stress
The Arabidopsis genome encodes three H1 variants (HIS1-1, HIS1-2, and HIS1-3), and HIS1-3 encodes a linker histone protein that is an abundant component of chromatin fibers. The quantitation of stoichiometry of linker histone in chromatin may play important roles in a variety of physiological processes (Jerzmanowski, 2007). Previous studies found that HIS1-3 plays an important role in drought-responsive gene expression mediated by chromatin remodeling. However, the role of HIS1-3 in salt stress is far less known. In our study, we showed that the expression of HIS1-3 was inhibited by salt stress and the loss-of-function of HIS1-3 led to increased salt tolerance (Figure 1). Moreover, the overexpression of HIS1-3 enhanced salt sensitivity in Arabidopsis (Figure 2). Thus, HIS1-3 is involved in salt stress as a negative factor.
Many studies have proved that WRKY transcription factors regulate the response to multiple biotic or abiotic stresses in plants (Rushton et al., 2010; Agarwal et al., 2011). WRKY1 was the first member of this family to be isolated, and it was found to be inducible by SA and drought (de Pater et al., 1996; Duan et al., 2007; Qiao et al., 2016). In this study, WRKY1 was significantly induced by salt stress, which was similar to that of AtWRKY25 and AtWRKY33 in Arabidopsis (Jiang and Deyholos, 2009), ZmWRKY17/33 in maize (Zea mays L.; Li et al., 2013; Cai et al., 2017), and CmWRKY17 in Chrysanthemum morifolium (Li et al., 2015). Phenotypic analysis showed that the wrky1 mutants exhibited salt sensitivity, and the overexpression lines exhibited salt tolerance (Figures 4 and 5). These data suggested that WRKY1 was able to mediate the salt stress response as a positive factor. Thus, HIS1-3 and WRKY1 have opposing effects on plant tolerance to salt stress.
HIS1-3 and WRKY1 respond to salt stress via the SOS pathway
Intracellular Na+ accumulation leads to ionic toxicity, resulting in plant growth cessation or cell death (Zhu, 2003). Thus, the maintenance of ion homeostasis is a major determinant of salt tolerance, such as by increasing Na+ extrusion. Herein, compared to the WT, the Na+ content was much lower in the his1-3 mutants and higher in the HIS1-3-overexpressing lines (Figure 3). However, compared to the WT, the Na+ content was much higher in the WRKY1-overexpressing lines and lower in the wrky1 mutants (Figure 6). These results indicated that both HIS1-3 and WRKY1 may maintain Na+ balance in cells, thereby enhancing plant salt tolerance.
The Ca2+-dependent SOS pathway is an important regulatory system activated by salt stress. When SOS3 is activated by Ca2+, the concentration of which increased under salt stress, the SOS3 protein binds to SOS2 to release the SOS2 kinase domain for substrate access. SOS2 interacts with SOS3, and then, the SOS3/SOS2 protein kinase complex activates SOS1. SOS1, a PM Na+/H+ antiporter, exports Na+ from cells (Shi et al., 2000; Qiu et al., 2002; Quintero et al., 2011). SOS2 is a key regulator in the SOS pathway and transmits the stress signal downstream through phosphorylation (Halfter et al., 2000; Guo et al., 2001). The SOS pathway is not activated when plants are not subjected to salt stress, but the molecular mechanisms underlying this process remain poorly understood. Kim et al. (2013) reported that the GI protein represses SOS2 activity in the absence of salt stress, and salt stress causes the degradation of the GI protein (Kim et al., 2013). The SOS2 kinase activity is repressed by the 14-3-3 protein, and salt stress reduces the interaction between 14-3-3 and SOS2, leading to the release of SOS2 activity (Zhou et al., 2014). In our study, the loss-of-function of HIS1-3 led to the increased expression of SOS1, SOS2, and SOS3, and the overexpression of HIS1-3 repressed the expression of these genes (Figure 3). Furthermore, the GUS activity assay showed that HIS1-3 repressed the expression of SOS1, SOS2, and SOS3 (Figure 7B). Linker histone H1, unlike core histones (H2A, H2B, H3, and H4), regulates specific gene expression but not global transcription (Shen and Gorovsky, 1996; Jerzmanowski et al., 2000). Thus, our results indicated that HIS1-3 suppressed the expression of SOS1, SOS2, and SOS3 under conditions of no stress in Arabidopsis, and salt stress reduced HIS1-3 gene expression, thereby inducing SOS gene expression.
However, many genes have been found to induce the expression of SOS genes, such as FcWRKY40 and GhWRKY34 (Zhou et al., 2015; Dai et al., 2018). Herein, WRKY1 induced the expression of three SOS genes (Figure 7B). The transcript abundances of three SOS genes were increased in the WRKY1-overexpressing lines but decreased in the wrky1 mutants, indicating that WRKY1-mediated salt tolerance depends on its positive regulation of the SOS genes. Our GUS activity analysis further confirmed that the expression of SOS1, SOS2, and SOS3 was inhibited when HIS1-3 and WRKY1 were co-expressed in Nicotiana benthamiana (Figure 7B). One possible explanation for the different roles of HIS1-3 and WRKY1 in regulating the SOS pathway is that HIS1-3 binds to SOS1, SOS2, and SOS3 more avidly than WRKY1 in the absence of salt stress. In order to prove that specific competition at the promoter regions of the SOS1 and SOS3 genes existed between HIS1-3 and WRKY1, we also performed a reciprocal competitive electrophoretic mobility shift assay (EMSA). The result showed that there was no specific competition at the promoter regions of the SOS1 and SOS3 genes between HIS1-3 and WRKY1. This may indicate that the two genes do not respond to salt stress by competing for the same sites of the SOS1 and SOS3 genes. One possible mechanism is that HIS1-3 binds to the linker DNA between nucleosome cores and occupies the binding site at the promoter regions of the SOS1 and SOS3 genes, thereby limiting the accessibility of WRKY1 to the SOS1 and SOS3 genes (Supplemental Figure S10).
In addition, owing to WRKY transcription factors being able to specifically recognize and bind W-box elements in the promoter regions of the target genes, we analyzed the promoter sequences of SOS1, SOS2, and SOS3 (Figure 7C). The ChIP-qPCR data showed that HIS1-3 and WRKY1 could bind to the same region in the promoters of SOS1 and SOS3 and to the adjacent region in the promoter of SOS2 (Figure 7D). Further genetic analysis indicated that the phenotypes of his1-3/sos1-1 and wrky1/35S::SOS1 were similar to the mutant and SOS1 overexpression lines, and the growth of the double mutant his1-3/wrky1 did not differ significantly from the WT under salt stress (Figure 8). The Na+ contents were consistent with the phenotypes (Figure 9). Taken together, these results suggest that HIS1-3 and WRKY1 regulate salt stress via a Na+ extrusion mechanism by directly activating the expression of SOS genes, and HIS1-3 and WRKY1 act upstream of the SOS genes.
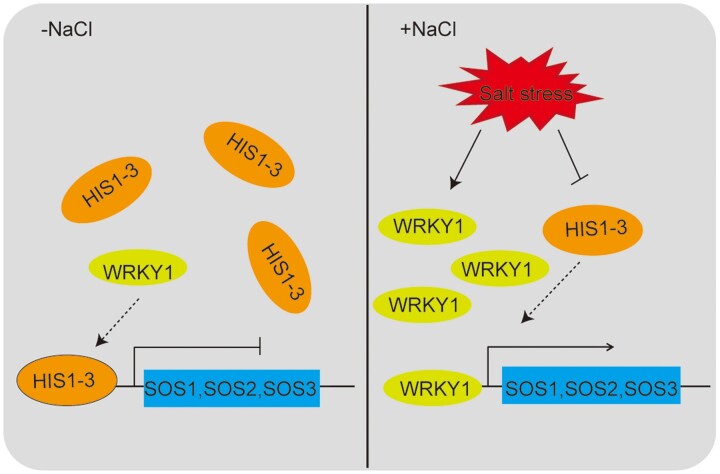
In summary, we propose the possible action mode for HIS1-3 and WRKY1 in regulating salt stress in Arabidopsis (Figure 10). Under normal physiological conditions, the HIS1-3 protein binds to the W-boxes of the SOS1, SOS2, and SOS3 promoter sequences by occupying the binding sites of WRKY1 on their promoters, thereby inhibiting the expression of these genes. Under salt stress, HIS1-3 protein expression decreased, resulting in the W-boxes of the SOS1, SOS2, and SOS3 promoters becoming exposed. Subsequently, the WRKY1 protein was directly combined with the W-boxes to activate gene expression, thereby regulating the Na+/K+ balance in the plant in order to respond to salt stress.
Figure 10.
A working model for the regulation of salt stress in Arabidopsis by HIS1-3 and WRKY1. HIS1-3 is a negative regulator under salt stress. It inhibits the expression of SOS1, SOS2, and SOS3 by binding to the W-boxes of the SOS1, SOS2, and SOS3 promoter sequences under conditions of no stress, which are also the binding sites of WRKY1 on these genes. Once plants are subjected to salt stress, the expression of HIS1-3 is decreased, leading to the exposure of the W-boxes of SOS1, SOS2, and SOS3. Subsequently, the WRKY1 protein activates the expression of these genes and regulates the Na+/K+ balance in the plant in order to respond to salt stress.
Materials and methods
Plant materials, growth conditions, and treatments
The plant materials used in this study included Arabidopsis (A. thaliana) WT (Col-0), his1-3 mutants (his1-3-1, SAIL-799-A07; his1-3-2, SALK-025209), and wrky1 mutants (wrky1-1, SALK-070989C; wrky1-2, SALK-136009C). The seeds of the mutants were obtained from the Arabidopsis Biological Resource Centre. The T-DNA insertion sites were confirmed by PCR amplification using gene-specific primers and a left border T-DNA primer (Supplemental Table S1). The sos1-1 mutant contains a 14-bp deletion that causes a frameshift, and the seeds were provided by Professor Jiankang Zhu (Shi et al., 2000). The Arabidopsis seeds were vernalized in the dark at 4°C for 2 d and then grown in a growth chamber maintained at 22°C with 16 h of daylight.
For the salt tolerance tests, seeds of the WT and the mutants or transgenic lines were germinated on MS medium for 3 d and then transferred to MS medium with or without NaCl (100 mM, 125 mM) for 2 weeks. The plants were then sampled for root length and fresh weight determination. To determine the germination rate, seeds of the WT and the mutants or transgenic lines were germinated on MS medium containing NaCl (100 mM), and the germination of the seeds was counted at a fixed time every day from the second day of sowing until 7 d later.
Vector construction and generation of transgenic plants
To generate HIS1-3-overexpressing and WRKY1-overexpressing plants, the sequences of HIS1-3 and WRKY1 were amplified with the specific primers (the primers are presented in Supplemental Table S1) and then cloned into pXB94 (pART27 with expanded restriction sites, 35S promoter and GFP reporter) or pBI121, respectively (named 35S::HIS1-3, 35S::HIS1-3::GFP, 35S::WRKY1, and 35S::WRKY1::GFP). These constructs were introduced into the Agrobacterium tumefaciens GV3101 strain and then transformed into the WT using the floral-dip method (Clough and Bent, 1998). All transgenic lines used in this study were T3 homozygous plants with a single-copy insertion.
The construct for the overexpression of SOS1 was generated as described by Shi et al. (2003) and then transformed into the wrky1-1 mutant (named wrky1/35S::SOS1). The his1-3/sos1 and his1-3/wrky1 double mutants were obtained by crossing sos1 or wrky1 to his1-3. All homozygous lines of the double mutant were selected for further study.
RNA extraction and RT-qPCR analysis
Six-week-old WT seedlings were used for analyzing the expression of HIS1-3 and WRKY1 in the different tissues. Treated and nontreated 2-week-old seedlings of the mutants and overexpression lines were used for analyzing the expression of HIS1-3, WRKY1, SOS1, SOS2, and SOS3. The total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed with PrimeScript Reverse Transcriptase. The qPCR reaction was performed according to the instructions provided for the Bio-Rad iCycleriQ system (Bio-Rad Laboratories, Hercules, CA, USA) and using platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). The PCR amplifications for each sample were quantified at least in triplicate and normalized using Actin8 as an internal control. The primers used are listed in Supplemental Table S1.
GUS staining
The full-length promoters of HIS1-3 and WRKY1 were amplified by PCR from Arabidopsis genomic DNA and then cloned into the transformation vector pXB93 (pART27 with expanded restriction sites and the GUS reporter) using specific primers (Supplemental Table S1). The plasmids of ProHIS1-3::GUS and ProWRKY1::GUS were induced into the Agrobacterium strain GV3101 and transformed into Arabidopsis. Homozygous transgenic lines were germinated on MS medium for 7 d and then treated with 100-mM NaCl for 6 h. All samples were collected and stained with 5-bromo4-chloro-3-indoyl-β-d-glucuronide (X-Gluc) in the dark at 37°C for 12 h (Fan et al., 2013). The samples were bleached in 75% (v/v) alcohol for 2–6 d to remove chlorophyll and then photographed under a Volume microscope (Zeiss Axio Zoom.V16, Germany).
Analysis of Na+ and K+ content
Seeds of the WT, mutants, and overexpression lines were germinated on MS medium with 100-mM NaCl for 2 weeks, and then, the roots and aboveground parts were collected separately to determine the ion content. The contents of Na+ and K+ in the roots and aboveground parts were detected by atomic absorption spectrometry (Solar 316 M6; Thermo Fisher Scientific, Waltham, MA, USA).
Transient expression assay
The full-length promoters of SOS1, SOS2, and SOS3 were amplified by PCR from Arabidopsis genomic DNA and then cloned into the transformation vector pXB93 (pART27 with expanded restriction sites and GUS reporter) using specific primers (Supplemental Table S1). Transient expression assays were performed as described (Sparkes et al., 2006; Chen et al., 2009). The A. tumefaciens cells were collected by centrifugation and suspended to an optical density (600 nm) of 0.1 using a solution containing 50 mM of 2-(N-morpholino)ethanesulfonic acid, 5 g/L of D-Glc, 2 mM of Na3PO4, and 0.1 mM of acetosyringone. The constructs of ProSOS1:GUS, ProSOS2:GUS, or ProSOS3:GUS were then co-transformed into N. benthamiana epidermal cells with 35S::HIS1-3 or 35S::WRKY1 using a needle-free syringe. The GUS activity was assessed at 48 h after injection and quantified by the fluorometric 4-methylumbelliferyl-β-d-glucuronide (MUG) assay, as described by Xu et al. (2006).
ChIP
The ChIP experiments were performed as described in published protocols (Kaufmann et al., 2010) with an anti-GFP antibody (Abmart, Shanghai, China). In brief, 3 g of 10-d-old 35S:WRKY1::GFP seedlings, 35S:HIS1-3::GFP seedlings, and 35S::GFP seedlings were collected and cross-linked with 35 mL of MC buffer (1 M of Na3PO4, 5 M of NaCl, 0.1 M of sucrose), to which 1 mL of 37% (v/v) formaldehyde was added, and the samples were placed under vacuum for 30 min. A total of 4 mL of 1.25-M Gly was added to stop the cross-linking. After rinsing the seedlings with MC buffer, the tissues were ground with liquid nitrogen and resuspended in 15 mL of extraction buffer I (1 M of Na3PO4, 5 M of NaCl, 1 M of hexylene glycol, 10 mM of β-mercaptoethanol, and 1× protease inhibitor) and then filtered through a nylon mesh. The filtrate was centrifuged at 1,000g at 4°C for 20 min. The pellet was resuspended in 5 mL of extraction buffer II (1 M of Na3PO4, 5 M of NaCl, 1 M of hexylene glycol, 10 mM of β-mercaptoethanol, 20 mM of MgCl2, 0.5% (v/v) Triton X-100, and 1× protease inhibitor) and centrifuged at 1,000g and 4°C for 20 min. The pellet was resuspended in 5 mL of extraction buffer III (1 M of Na3PO4, 5 M of NaCl, 10 mM of β-mercaptoethanol, and 1× protease inhibitor) and loaded on top of an equal amount of clean extraction buffer III, and then centrifuged at 1000g for 10 min. The crude nuclear pellet was resuspended and sonicated with ultrasonic cell disruption. The sonicated chromatin was centrifuged, and the insoluble pellet was discarded. Then, the samples were divided into three parts: one part was used for DNA, and the other two parts were incubated with an anti-GFP antibody. One microliter of anti-GFP-specific monoclonal antibody was added to the chromatin solution and incubated for 1 h at 4°C. The immunocomplexes were extracted by incubating with 40 µL of 50% (v/v) protein A–Sepharose beads for 1 h at 4°C. After several washes, the immunocomplex was eluted twice from the beads with 100 µL of elution buffer (0.1 M of glycine, 5 M of NaCl, 0.05% (v/v) Tween-20) and then reversed cross-linked. All proteins were removed. The relative concentrations of the DNA fragments were analyzed by qPCR with the different primers listed in Supplemental Table S1.
BiFC
The full length CDS of HIS1-3 were amplified and inserted into the plant binary vector pBSPYCE to generate HIS1-3-CYFP; the full-length CDS of WRKY1 were amplified and inserted into the plant binary vector pBSPYNE to generate WRKY1-NYFP. BiFC assay was carried out via the Agrobacterium-mediated transient expression of WRKY1-NYFP with HIS-CYFP in N. benthamiana leaves as described previously (Walter et al., 2004). Two 4-week-old N. benthamiana leaves were injected with Agrobacterium GV3101strains containing individual BiFC construct pairs and a binary plasmid expressing the p19 protein to suppress gene silencing. After 4′,6-diamidino-2-phenylindole staining, the epidermal cell layers were examined using confocal microscope. The acquire YFP signals using the LSM 710, 488 nm was used for excitation and fluorescence was detected at a 410–550 nm range.
Yeast two-hybrid assays
Arabidopsis WRKY 1 was cloned into the pGADT7 vector to generate AD-WRKY1, whereas HIS1-3 cDNA was cloned into the pGBKT7 vector to generate BD-HIS1-3. The corresponding combinations were transformed into yeast (AH109), respectively. The mated yeast cells were selected on SD–Leu–Trp–His plates which contain 20-mM 3-Amino-1,2,4-triazole.
Y1H assays
To generate AD-HIS1-3 and AD-WRKY1, the full-length CDS of HIS1-3 and WRKY1 were inserted into pJG4-5 vector (Clontech, Mountain View, CA, USA). The Y1H assay was performed according to the Yeast Protocols Handbook (Clontech). Briefly, the AD fusion constructs were co-transformed with various LacZ reporter plasmids into yeast strain EGY48. Transformants were grown on SD/Trp-Uradropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for blue color development.
EMSA
The full-length CDS of HIS1-3 and WRKY1 were cloned into the pMAL-c2 vector and MBP-HIS1-3 and MBP-WRKY1 fusion protein were expressed in Escherichia coli Rosseta2 strain. Free probes containing W-box motifs were synthesized with biotin label at the 5′–end by Sangon Biotech (Sangon Biotech, Shanghai, China). EMSA was performed using a LightShift EMSA Optimization and Control Kit (Thermo Fisher Scientific, Waltham, MA, USA). The results were detected using a CCD camera system (Image Quant LAS 4000).
Statistical analyses
The experiment used a completely random design. The above indicators were repeated for 3 times. The data were analyzed by SPSS version l9.0. Tukey’s multiple comparison method was used for difference significance analysis, and the significance level was 0.05.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HIS1-3 (At2g18050), WRKY1 (At2g 04880), SOS1 (At2g 01980), SOS2 (At5g 35410), and SOS3 (At5g 24270).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profile of the HIS1-3 gene.
Supplemental Figure S2. Responses of the his1-3 mutants to KCl, LiCl, and mannitol.
Supplemental Figure S3. Phenotypes of the WT, his1-3 mutants, and HIS1-3-OE lines in soil-filled pots.
Supplemental Figure S4. Tissue-specific expression of WRKY1.
Supplemental Figure S5. Phenotypes of the WT, wrky1 mutants, and WRKY1-OE lines in soil-filled pots.
Supplemental Figure S6. Phenotype test of the WT and wrky1 mutant seedlings in response to KCl, LiCl, and mannitol.
Supplemental Figure S7. The relationship between HIS1-3 and WRKY1.
Supplemental Figure S8. WRKY1 exhibits self-activation by Y1H assay.
Supplemental Figure S9. Identification of transgenic plants.
Supplemental Figure S10. No specific competition at the promoter regions of SOS3 genes between HIS1-3 and WRKY1 by reciprocal competitive EMSA.
Supplemental Table S1. Primers used for cloning, qPCR, and ChIP assay.
Supplementary Material
Acknowledgments
We thank Prof. Jian-Kang Zhu for providing the sos1-1 mutant. We would like to thank Na Li for their technical assistances. We thank Mengxiang Sun and Xiongbo Peng for providing the pXB93 and pXB94 vectors.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 31500213, 31872803, and 31571250).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Xi Wu, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Jiena Xu, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Xingnan Meng, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Xue Fang, School of Horticulture, Anhui Agricultural University, Hefei 230009, China.
Minghui Xia, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Jing Zhang, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Shuqing Cao, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
Tingting Fan, School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, China.
T.F. conceived the original research plans. X.W., J.X., X.M., X.F., M.X., and J.Z. performed the experiments. S.C. and T.F. designed the experiments and analyzed the data. T.F. and S.C. wrote the article with contributions of all the authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Tingting Fan (fantting@hfut.edu.cn).
References
- Agarwal P, Reddy MP, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38: 3883–3896 [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Shukla PS, Gupta K, Jha B (2013) Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol 54: 102–123 [DOI] [PubMed] [Google Scholar]
- Arbona V, Manzi M, de Ollas C, Gomez-Cadenas A (2013) Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int J Mol Sci 14: 4885–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS (1997) A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol 34: 629–641 [DOI] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS (1999) Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol Biol 41: 159–169 [DOI] [PubMed] [Google Scholar]
- Cai RH, Dai W, Zhang CS, Wang Y, Wu M, Zhao Y, Ma Q, Xiang Y, Cheng BJ (2017) The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 246: 1215–1231 [DOI] [PubMed] [Google Scholar]
- Chen Y, Li L, Xu Q, Kong Y, Wang H, Wu W (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3555–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai W, Wang M, Gong X, Liu JH (2018) The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol 219: 972–989 [DOI] [PubMed] [Google Scholar]
- de Pater S, Greco V, Pham K, Memelink J, Kijne J (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res 24: 4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD (2007) DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res 35: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fan TT, Zhai HH, Shi WW, Wang J, Jia HL, Xiang Y, An LZ (2013) Overexpression of profilin 3 affects cell elongation and F-actin organization in Arabidopsis thaliana. Plant Cell Rep 32: 149–160 [DOI] [PubMed] [Google Scholar]
- Fu L, Shen Q, Kuang L, Yu J, Wu D, Zhang G (2018) Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol Biochem 130: 248–257 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C, (2010) Food security: the challenge of feeding 9 billion people. Science 327: 812–818 [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9: 263–267 [DOI] [PubMed] [Google Scholar]
- Hu YR, Chen LG, Wang HP, Zhang LP, Wang F, Yu DQ (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Jaleel CA, Riadh K, Gopi R, Manivannan P, Ines J, Al-Juburi H, Zhao CX, Shao HB, Panneerselvam R (2009) Antioxidant defense responses: physiological plasticity in higher plants under abiotic constraints. Acta Physiol Plant 31: 427–436 [Google Scholar]
- Jerzmanowski A, Przewloka MR, Grasser KD (. 2000) Linker histones and HMG1 proteins of higher plants. Plant Biol 2: 586–597 [Google Scholar]
- Jerzmanowski A (2007) SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta 1769: 330–345 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YQ, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protocol 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kim JM, To TK, Nishioka T, Seki M (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, et al. (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4: 1352. [DOI] [PubMed] [Google Scholar]
- Koop R, Di Croce L, Beato M (2003) Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J 22: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gao Y, Xu H, Dai Y, Deng DQ, Chen JM (2013) ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul 70: 207–216 [Google Scholar]
- Li P, Song A, Gao C, Wang L, Wang Y, Sun J, Jiang J, Chen F, Chen S (2015) Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep 34: 1365–1378 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819: 129–136 [DOI] [PubMed] [Google Scholar]
- Mark T, Peter L (2010) Breeding technologies to increase crop production in a changing world. Science 327: 818–822 [DOI] [PubMed] [Google Scholar]
- Martinez-Atienza J, Jiang XY, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7: 760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Li CL, Zhang W (2016) WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol Biol 91: 53–65 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci U S A 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, et al. (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Nat Acad Sci USA 108: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QXJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Scippa GS, Di Michele M, Onelli E, Patrignani G, Chiatante D, Bray EA (2004) The histone-like protein H1-S and the response of tomato leaves to water deficit. J Exp Bot 55: 99–109 [DOI] [PubMed] [Google Scholar]
- Scippa GS, Griffiths A, Chiatante D, Bray EA (2000) The H1 histone variant of tomato, H1-S, is targeted to the nucleus and accumulates in chromatin in response to water-deficit stress. Planta 211: 173–181 [DOI] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA (. 1996) Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86: 475–483 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee B, Wu S, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protocol 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Tang XL, Mu XM, Shao HB, Wang HY, Brestic M (2015) Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit Rev Biotechnol 35: 425–437 [DOI] [PubMed] [Google Scholar]
- Tao Z, Kou YJ, Liu HB, Li XH, Xiao JH, Wang SP (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62: 4863–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi I, Ranjan A, Sharma YK, Sawant S (2012) The histone H1 variant accumulates in response to water stress in the drought tolerant genotype of Gossypium herbaceum L. Protein J 31: 477–486 [DOI] [PubMed] [Google Scholar]
- Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428: 419–438 [DOI] [PubMed] [Google Scholar]
- Ulker B, Shahid Mukhtar M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang JN, Kuang JF, Shan W, Chen J, Xie H, Lu WJ, Chen JW, Che JY (2012) Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep 31: 1485–1494 [DOI] [PubMed] [Google Scholar]
- Wei T, O’Connell MA (1996) Structure and characterization of a putative drought-inducible H1 histone gene. Plant Mol Biol 30: 255–268 [DOI] [PubMed] [Google Scholar]
- Wu SJ, Lei D, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z (2009) Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2: 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BG, Liu KD, Zheng Y, Wang YX, Wang JX, Liao H (2013) Disruption of AtWNK8 enhances tolerance of Arabidopsis to salt and osmotic stresses via modulating proline Content and Activities of Catalase and Peroxidase. Int J Mol Sci 14: 7032–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26: 1166–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang NN, Gong SY, Lu R, Li Y, Li XB (2015) Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol Biochem 96: 311–320 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Shen WH (2013) Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta 1819: 343–348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.